Abstract
Abnormalities in the circadian clock system are known to affect the body’s metabolic functions, though the molecular mechanisms responsible remain uncertain. In this study, we achieved continuous knockdown of B. mori Period (BmPer) gene expression in the B. mori ovary cell line (BmN), and generated a Per-KD B. mori model with developmental disorders including small individual cells and slow growth. We conducted cell metabolomics assays by gas chromatography/liquid chromatography-mass spectrometry and showed that knockdown of BmPer gene expression resulted in significant inhibition of glycometabolism. Amino acids that used glucose metabolites as a source were also down-regulated, while lipid metabolism and nucleotide metabolism were significantly up-regulated. Metabolite correlation analysis showed that pyruvate and lactate were closely related to glycometabolism, as well as to metabolites such as aspartate, alanine, and xanthine in other pathways. Further validation experiments showed that the activities of the key enzymes of glucose metabolism, hexokinase, phosphofructokinase, and citrate synthase, were significantly decreased and transcription of their encoding genes, as well as that of pyruvate kinase, were also significantly down-regulated. We concluded that inhibition of the circadian clock gene BmPer repressed glycometabolism, and may be associated with changes in cellular amino acid metabolism, and in cell growth and development.
Metabolic regulation is the most important output of circadian clock signals1 across a wide range of species from Chlorococcus2 to humans3, though the specific molecular mechanisms involved remain unclear1,3,4. Studies have shown that important metabolic processes, such as glycometabolism and cholesterol metabolism, are controlled by the circadian clock5. Circadian clock disorders thus lead not only to sleep and mental disorders, but also to severe metabolic diseases6, including obesity7,8, diabetes9, hypertension and other cardiovascular diseases10,11, and may even be associated with accelerated aging12 and tumor development13.
The period protein (PER) is a core component of the circadian clock. It includes a PAS (Per-Art-Ser) domain14, and plays a role in signal conduction in the negative feedback loop involved in circadian clock core oscillations. The mammalian PER protein is able to bind the CLOCK/BMAL protein dimer to regulate the expression of downstream genes15. Recent studies have found that PER not only plays a role in regulating gene expression, but also has an important impact on metabolism; however, the results of studies have been inconsistent or even contradictory. Feillet et al.16 reported no significant changes in liver glycogen content in mPer2 mutant mice, while later studies found that silencing the expression of mPer2 led to changes in blood sugar, liver glycogen, and insulin17,18,19. mPer1 knockout caused increased blood pressure in mice20, while simultaneous knockout of mPer1/mPer2 or of mPer2 alone both led to lower triglyceride levels in mouse liver21,22. Similarly, the results of two studies on corticosterone metabolism in mPer mutant mice produced conflicting results in terms of metabolic damage and enhancement23,24.
Biological metabolism is a complex regulatory process, largely controlled by endocrine hormones25, and subject to interactions between different metabolic pathways26. However, systematic studies of the correlation between the circadian clock and metabolism are lacking, and most studies have only investigated single indicators, such as blood glucose, liver glycogen, and triglycerides, and have not fully assessed changes in all metabolites. An overall metabolomics analysis is therefore required. However, Per genes in mammals, such as mice and humans, include multiple gene subtypes, and the gene expression levels of mouse mPer1, mPer2, and mPer3 differ among different tissues and developmental stages, along with differences in the functions of the protein products27,28,29,30. No studies to date have investigated the metabolic effects of knockdown/out of all mPer genes simultaneously. It may therefore be possible to obtain a clearer understanding of the metabolic effect of the circadian clock by selecting a model animal with only one type of Per gene, thus avoiding interactions among multiple genes. We therefore investigated this relationship in the silkworm Bombyx mori, which only has one type of PER protein (Figure S1). We further knocked down PER protein expression continuously in a cell line in vitro, to eliminate any potential endocrine effects. We performed metabolomic analysis in B. mori using gas chromatography/liquid chromatography-mass spectrometry (GC/LC-MS), to evaluate the effect of the peripheral circadian clock on cellular metabolism, and to investigate the relationship between repression of glycometabolism and other metabolic pathway abnormalities caused by changes in the circadian clock system, by detailed evaluation of changes in glucose metabolic pathways.
Results
Effect of knockdown of BmPer gene expression on circadian clock system and growth of BmN cells
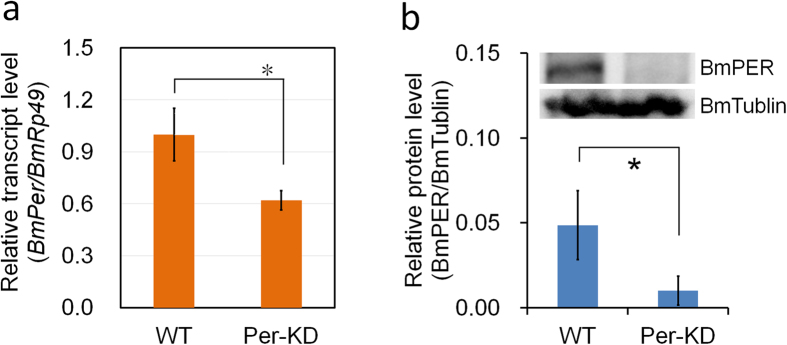
Screening revealed that BmPer gene expression was continuously knocked down in B. mori ovary cells transfected with a recombinant vector expressing the interference sequence of the BmPer gene (Per-KD). BmPer gene transcription in Per-KD cells, determined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR), was downregulated by about 40% compared with wild type (WT) cells (Fig. 1a), while BmPER protein levels were significantly reduced by about 80%, according to western blotting (Fig. 1b). BmPer gene knockdown thus significantly reduced both gene and protein expression of Per. We investigated transcription of the genes encoding proteins involved in circadian clock oscillation, BmCRY1 (Cryptochrome1), BmCRY2 (Cryptochrome2), BmTIM (Timeless), BmCLK (Clock), and BmCYC (Cycle), and showed that rhythms in expression levels of all five genes were decreased or even absent after synchronization with dexamethasone (Dex), with expression levels of BmTim and BmClk genes being significantly down-regulated (Figure S2). These results indicated that continuous down-regulation of BmPer gene transcription and protein translation in Per-KD cells further disrupted the transcription of genes encoding the circadian oscillators, suggesting that circadian signal outputs may be affected in Per-KD cells.
Figure 1. Knockdown efficiency of BmPer gene.
(a) BmPer transcripts were analyzed by qRT-PCR with BmRp49 as an internal control. (b) BmPER protein levels determined by western blot with BmTublin protein as an internal control. WT, wild-type BmN cells; Per-KD, BmPer knockdown BmN cells. *p ≤ 0.05 (repeated three times).
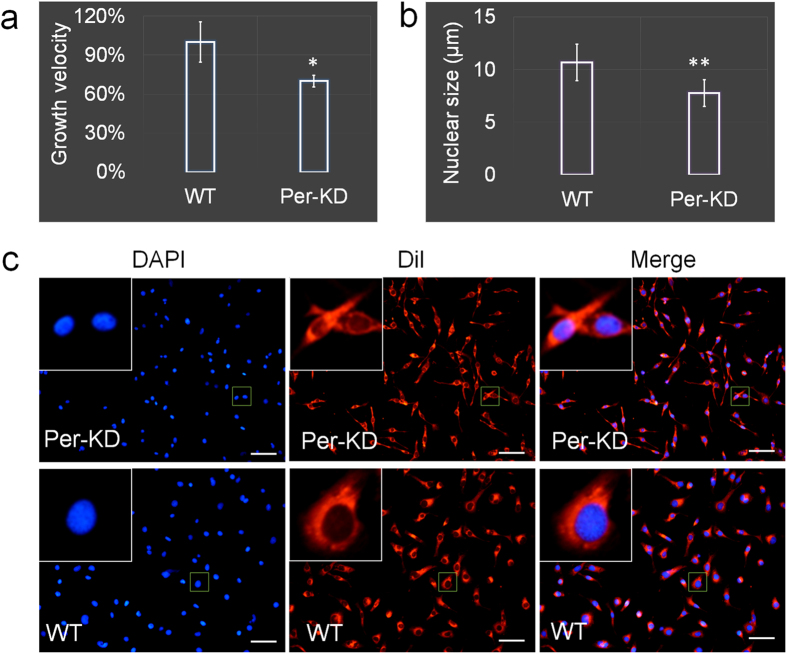
Per-KD cells showed distinctly different phenotypic traits from WT cells. Cell growth rate measured by MTT showed that Per-KD cells grew slower than WT cells; the growth velocity of Per-KD cells was decreased by about 30% within 4 h of measurement (Fig. 2a). Staining of cell membranes and nuclei also revealed that Per-KD cells were smaller than WT cells (Fig. 2b), and the average diameter of WT nuclei stained with DAPI was 10.5 μm, compared with 7 μm in Per-KD nuclei (p ≤ 0.05) (Fig. 2b,c).
Figure 2. Effect of BmPer gene knockdown on BmN cell growth.
(a) Cell growth velocity measured by MTT method. (b) Cells were transferred to a coverslip when they reached 70% confluence. After the cells were completely adherent (at least 12 h) the diameters of the nuclei were measured (average of the longest and shortest diameters). (c) Cell nuclei stained with 4′-6-diamidino-2-phenylindole (DAPI) and cell membranes stained with Dil (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate) (showing cell morphology). WT, wild-type BmN cells; Per-KD, BmPer knockdown BmN cells. *p ≤ 0.05 and **p ≤ 0.01 (n = 3 plates, observations and statistical analyses were repeated at least five times). Bar = 50 μm.
Metabolomic changes in Per-KD cells
The phenotypic differences indicated that BmPer knockdown had a negative effect on the growth and development of Per-KD cells, suggesting potential effects on the metabolism responsible for maintaining cell growth and development. We therefore investigated the metabolomic changes by GC-MS and LC-MS. The ion current chromatograms of eight repeated assays are shown in Figures S3 and S4.
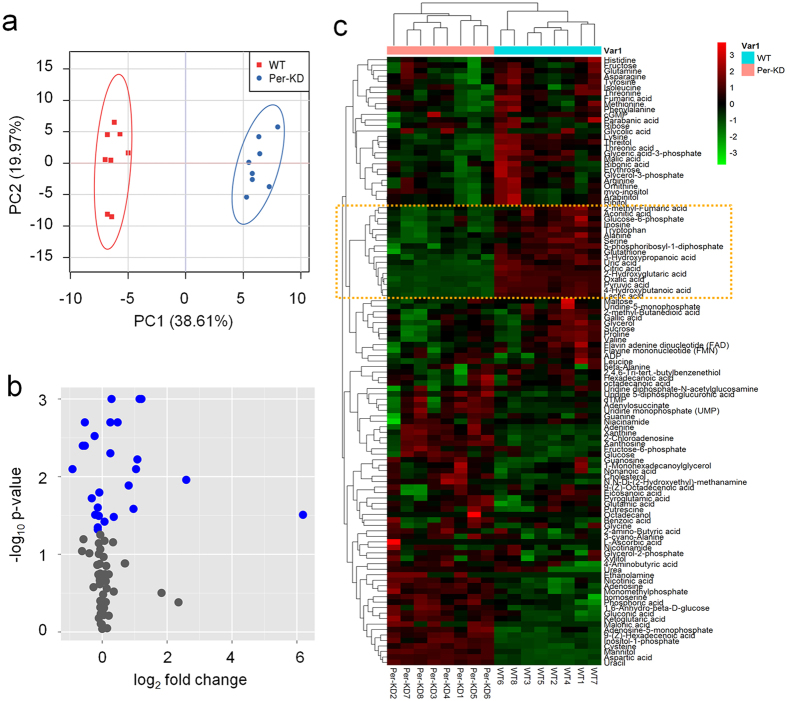
The original data were processed and analyzed using XCMS software (www.bioconductor.org/)31. A total of 112 differential metabolites were detected and identified in Per-KD cells, using WT BmN cells as a control. Prinicipal components analysis (PCA) of differential metabolites was carried out to obtain a PCA score plot (Fig. 3a). PCA simulation identified two principal components that explained 58.58% of the model; PC1 could explain 38.61% of the data model, and PC2 could explain 19.97%. These results indicated that the substances detected by metabolomic method could clearly distinguish between Per-KD BmN and WT cells.
Figure 3. LC/GC-MS assay based metabolomics.
PCA (a), volcano plot (b), and heat map (c). In the volcano plot, differential metabolites (blue) and non-differential substances (gray) were screened out under the conditions of fold change ≥1 and p ≤ 0.05. WT, wild-type BmN cells; Per-KD, BmPer knockdown BmN cells. WT 1–8 and Per-KD 1–8 on the x axis in (c) indicate eight repeated assays.
Partial least squares discriminant analysis (PLS-DA) score plots showed that supervised multivariate analysis revealed more obvious differences between the metabolites in the two kinds of cells, indicating a significant difference in metabolic components between Per-KD and WT cells (Figure S5a). A volcano plot (Fig. 3b) showed the difference in metabolites between the two kinds of cells, calculated from data for all tested substances. Cluster analysis of all the samples further revealed that the repeat results for Per-KD cells and WT cells, respectively, clustered together, indicating a significant difference in metabolism between the two kind of cells (Figure S5b).
All the detected metabolites were analyzed by heat mapping. Some metabolites differed significantly between Per-KD and WT cells, in stable and consistent manners within groups (Fig. 3c). Glucose metabolites such as pyruvate, lactate, and glucose-6-phosphate produced in the glycolytic pathway, and amino acids such as alanine, serine, and tyrosine produced from glycolytic intermediates were abundant in WT cells but low in Per-KD cells (Fig. 3c, orange box), suggesting that knockdown of BmPer gene expression had significant effects on glucose and amino acid metabolism in Per-KD cells.
We further screened for differential metabolites between Per-KD and WT cells using the PLS-DA model first principal component variable importance in projection (VIP) value ≥1 and t-test (p ≤ 0.05) as thresholds for GC-MS assay results. Standard tests were carried out for a certain number of substances, and PLS-DA model first principal component VIP ≥1, p (corr) ≥0.08 of the S-plot and p ≤ 0.05 for t-tests were chosen as thresholds to screen out differential metabolites from LC-MS assay results, while some substances were tested by secondary mass spectrometry. Differential metabolites are summarized in Tables 1 and 2. Differential metabolites between the two types of cells identified by GC-MS were mainly amino acids, glucose metabolites, and organic acids, including fatty acids (Table 1), while differential metabolites identified by LC-MS secondary mass spectrometry were mainly nucleotides and their derivatives (Table 2).
Table 1. Differential metabolites identified by GC-MS assay.
| Metabolite | RI | Q.ion.mz. | RT.s. | vip | p value | q value | fold_change |
|---|---|---|---|---|---|---|---|
| 2-Hydroxyglutaric acid | 1588.9 | 129.0 | 802.6 | 1.57 | 4.79E-13 | 4.50E-11 | −2.63 |
| Pyruvic acid | 1070.7 | 174.0 | 377.4 | 1.57 | 1.25E-12 | 1.16E-10 | −1.51 |
| Citric acid | 1843.5 | 273.1 | 970.8 | 1.50 | 4.92E-08 | 4.18E-06 | −1.43 |
| 4-Hydroxybutanoic acid | 1245.7 | 233.1 | 534.9 | 1.55 | 2.14E-10 | 1.90E-08 | −0.84 |
| Oxalic acid | 1147.6 | 45.0 | 446.4 | 1.57 | 7.88E-12 | 7.17E-10 | −0.80 |
| Aconitic acid | 1760.9 | 229.1 | 918.4 | 1.40 | 7.89E-06 | 6.23E-04 | −0.75 |
| Glyceric acid-3-phosphate* | 1833.5 | 357.1 | 964.4 | 1.15 | 1.57E-03 | 9.54E-02 | −0.60 |
| Uric acid* | 2130.3 | 441.2 | 1137.0 | 1.44 | 1.36E-06 | 1.10E-04 | −0.56 |
| Lactic acid | 1083.1 | 191.0 | 388.3 | 1.57 | 1.55E-12 | 1.43E-10 | −0.54 |
| Glucose-6-phosphate* | 2371.2 | 205.0 | 1260.6 | 1.09 | 3.46E-03 | 1.97E-01 | −0.54 |
| 2-methyl-Fumaric acid | 1361.9 | 184.0 | 631.6 | 1.16 | 1.31E-03 | 8.12E-02 | −0.54 |
| Inosine | 2604.6 | 230.1 | 1371.0 | 1.33 | 6.25E-05 | 4.69E-03 | −0.52 |
| Tryptophan* | 2227.6 | 202.1 | 1188.4 | 1.43 | 2.57E-06 | 2.06E-04 | −0.52 |
| Lysine* | 1865.7 | 174.1 | 984.3 | 1.29 | 1.34E-04 | 9.24E-03 | −0.43 |
| 3-Hydroxypropanoic acid | 1158.4 | 177.0 | 457.7 | 1.46 | 7.21E-07 | 5.99E-05 | −0.31 |
| Threitol* | 1529.4 | 103.0 | 760.0 | 1.36 | 2.59E-05 | 1.99E-03 | −0.28 |
| Threonic acid* | 1565.6 | 292.1 | 786.3 | 1.13 | 2.05E-03 | 1.23E-01 | −0.23 |
| Alanine* | 1122.7 | 116.1 | 424.0 | 1.31 | 8.47E-05 | 6.18E-03 | −0.17 |
| Serine* | 1379.6 | 204.1 | 644.1 | 1.29 | 1.43E-04 | 9.75E-03 | −0.12 |
| Malic acid* | 1505.8 | 233.1 | 742.3 | 1.31 | 9.55E-05 | 6.87E-03 | −0.12 |
| Threonine* | 1403.3 | 117.0 | 663.9 | 1.22 | 4.97E-04 | 3.23E-02 | −0.06 |
| Adenine | 1882.3 | 264.1 | 994.6 | 1.37 | 1.62E-05 | 1.27E-03 | 0.24 |
| Guanosine | 2857.1 | 324.1 | 1461.5 | 1.07 | 4.21E-03 | 2.32E-01 | 0.25 |
| Phosphoric acid | 1292.4 | 299.1 | 574.1 | 1.12 | 2.30E-03 | 1.36E-01 | 0.25 |
| Gluconic acid | 2004.4 | 333.1 | 1066.6 | 1.19 | 9.21E-04 | 5.89E-02 | 0.29 |
| Homoserine | 1463.5 | 183.0 | 711.4 | 1.27 | 2.11E-04 | 1.41E-02 | 0.33 |
| Nicotinic acid | 1302.5 | 180.0 | 583.5 | 1.30 | 1.12E-04 | 7.96E-03 | 0.35 |
| Ketoglutaric acid* | 1811.4 | 292.1 | 950.6 | 1.31 | 8.24E-05 | 6.10E-03 | 0.39 |
| 1,6-Anhydro-beta-D-glucose | 1727.1 | 217.1 | 896.1 | 1.27 | 2.13E-04 | 1.41E-02 | 0.43 |
| Cholesterol | 3183.6 | 129.0 | 1633.2 | 1.18 | 1.01E-03 | 6.38E-02 | 0.48 |
| 9-(Z)-Hexadecenoic acid | 2027.9 | 311.2 | 1079.9 | 1.53 | 4.99E-09 | 4.29E-07 | 0.48 |
| Adenosine | 2666.9 | 236.1 | 1398.8 | 1.45 | 9.45E-07 | 7.75E-05 | 0.51 |
| Mannitol* | 1975 | 205.1 | 1048.8 | 1.53 | 2.20E-09 | 1.91E-07 | 0.61 |
| Monomethylphosphate | 1193.0 | 241.0 | 489.3 | 1.30 | 1.21E-04 | 8.47E-03 | 0.71 |
| Adenosine-5-monophosphate | 3112.8 | 230.1 | 1594.3 | 1.34 | 4.72E-05 | 3.59E-03 | 0.76 |
| Uracil | 1350.7 | 241.0 | 622.9 | 1.56 | 1.19E-11 | 1.07E-09 | 0.82 |
| Cysteine* | 1570.3 | 220.1 | 789.5 | 1.54 | 1.29E-09 | 1.14E-07 | 0.99 |
| N,N-Di-(2-Hydroxyethyl)-methanamine | 1433.1 | 160.0 | 690.1 | 1.11 | 2.78E-03 | 1.61E-01 | 1.09 |
| Aspartic acid* | 1535.0 | 232.1 | 765.1 | 1.58 | 8.32E-14 | 7.91E-12 | 1.15 |
| Inositol-1-phosphate | 2468.5 | 318.1 | 1308.1 | 1.46 | 5.76E-07 | 4.83E-05 | 1.45 |
| Malonic acid | 1174.4 | 191.0 | 475.0 | 1.07 | 4.14E-03 | 2.32E-01 | 2.59 |
Note: RI, retention index; Q.ion.mz, quantitative ion; RT.s, retention time/second; vip, PLS-DA first principal component variable importance in projection; pvalue, t-test significance; Qvalue, false-positive correction of t-test significance; fold_change, logarithm of Per-KD/WT fold change (fold change >0 means WT > Per-KD; fold change < 0 means WT < Per-KD). Metabolites with “*” were verified by standards.
Table 2. Differential metabolites identified by LC-MS secondary mass spectrometry.
| Metabolin | Type | Mass | RT.s | vip | p corr | p value | fold_change |
|---|---|---|---|---|---|---|---|
| Glutathione | [M + H]+ | 307.1 | 57.5 | 8.4 | −0.91 | 2.74E-06 | −1.03 |
| cGMP | [M − H]− | 345.0 | 58.0 | 1.4 | −0.89 | 5.71E-04 | −0.86 |
| 5-phosphoribosyl-1-diphosphate | [M − H]− | 390.0 | 35.1 | 2.6 | −0.85 | 2.40E-03 | −0.62 |
| dTMP | [M − H]− | 322.1 | 70.4 | 2.0 | 0.88 | 8.13E-04 | 0.70 |
| Cysteine | [M + H]+ | 121.0 | 59.1 | 2.2 | 0.88 | 1.16E-08 | 0.94 |
| Adenine | [M + H]+ | 135.1 | 195.9 | 3.2 | 0.89 | 1.07E-05 | 1.01 |
| 2-Chloroadenosine | [M + H]+ | 301.1 | 409.6 | 5.5 | 0.81 | 1.58E-06 | 1.07 |
| Xanthine | [M + H]+ | 152.0 | 75.5 | 3.0 | 0.90 | 3.31E-06 | 1.10 |
| Uridine 5-diphosphoglucuronic acid | [M − H]− | 580.0 | 48.5 | 1.6 | 0.92 | 9.11E-04 | 1.19 |
| Xanthosine | [M + H]+ | 284.1 | 171.4 | 1.3 | 0.89 | 5.76E-09 | 1.22 |
| Uridine monophosphate (UMP) | [M − H]− | 324.0 | 46.4 | 10.4 | 0.98 | 2.62E-06 | 1.32 |
| Adenylosuccinate | [M − H]− | 463.1 | 126.0 | 15.3 | 0.96 | 1.07E-04 | 1.35 |
Type, ionization mode; [M + H]+, positive ion mode; [M − H]−, negative ion mode; Mass, exact molecular weight; RT.s, retention time/second; vip, PLS-DA first principal component variable importance in projection; pvalue, t-test significance; Pcorr, p (corr) value of S-plot correlation; fold_change, logarithm of Per-KD/WT fold change (fold change >0 means WT > Per-KD; fold change <0 means WT < Per-KD). All the metabolites listed here did MS/MS.
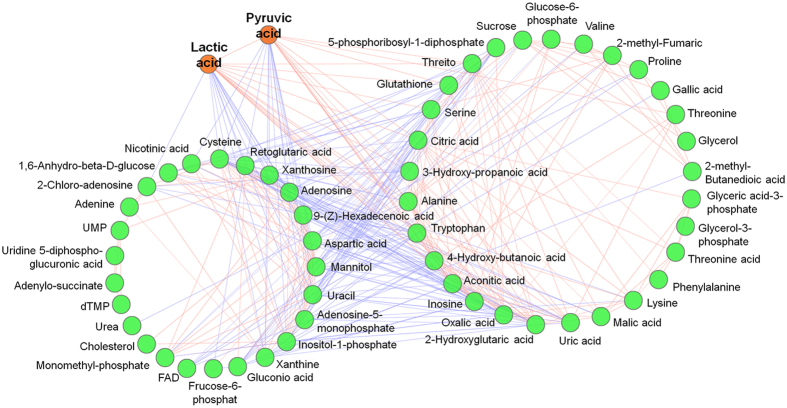
We further analyzed the screened out differential substances using a differential metabolite correlation network. As shown in Fig. 4, 59 differential metabolites constituted numerous edges and formed a complex topological network, indicating that the metabolites in Per-KD cells changed actively. According to the concept of a “clustering factor”32, the more correlations formed by a metabolite, the bigger its clustering factor, indicating that it may act as an important information link in the topological structure. Pyruvic acid and lactic acid constituted up to 25 and 24 edges in the network, respectively, indicating that these two metabolites played important central hub roles in the topological network, and potentially in the regulation of the entire cell metabolism.
Figure 4. Correlation network diagram of differential metabolites.
Metabolite correlation analysis was carried out using Pearson’s correlation coefficient, with the cor() function in the R language package (www.r-project.org). The Pearson’s correlation coefficient threshold was set as 0.8. Red lines represent positive correlations between substances, blue lines represent negative correlations between substances.
We carried out Metabolomics Pathway Analysis (MetPA) analysis of the metabolic pathways of the differential metabolites based on visual analysis33 to clarify the overall statuses of the differential metabolites in the two kinds of cells. There were extensive changes in the activities of pathways including nucleotide, amino acid, carbohydrate, and lipid metabolism, and synthesis of secondary metabolites in Per-KD cells, indicating that the differential metabolites impacted on the overall metabolism in the cells (Fig. 5). The metabolites with most significant impacts were involved in purine metabolism (nucleotide metabolic system), aspartate, alanine, glycine, and glutamate metabolism (amino acid metabolic system), and glycolysis and the tricarboxylic acid (TCA) cycle (glucose metabolism). The effect on lipid metabolism in Per-KD cells was relatively weak, and differential metabolites mainly affected the cholesterol and steroid synthesis pathways. These affected pathways provided clues to guide subsequent study of the effects of BmPER.
Figure 5. MetPA analysis of metabolic pathways.
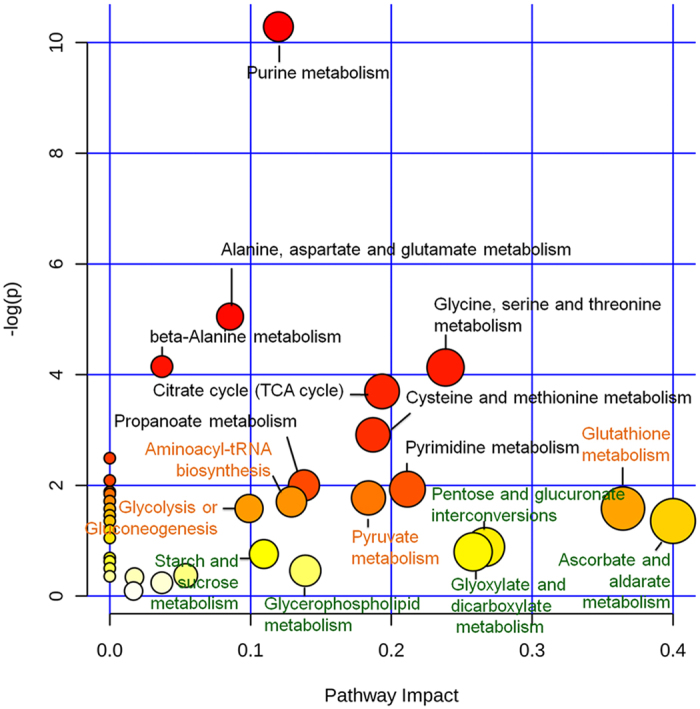
We analyzed the MetPA plot using the online software http://www.metaboanalyst.ca/, with default values as the analysis parameters. The deeper the red color of the metabolic pathway, the greater its -log (p) value, indicating a more significant difference. The darker the yellow color, the greater the impact of the pathway.
Glucose metabolism disorder in Per-KD cells
Per-KD cells grew more slowly and were smaller than WT cells (Fig. 2). The metabolomic results (Fig. 4) further suggested that pyruvate and lactate may be important links among differential metabolites, while the activity of the glycolytic pathway was significantly affected in Per-KD cells (Fig. 5). We therefore speculated that energy metabolism may have been affected by BmPer knockdown in Per-KD cells.
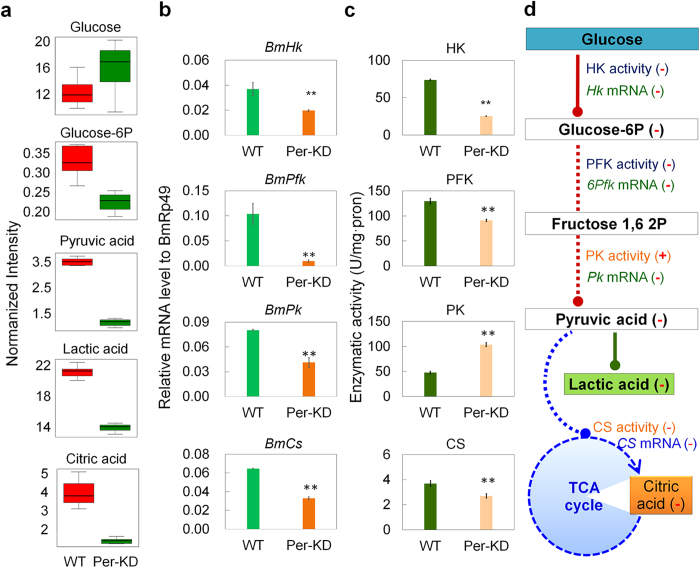
We investigated the effect of Per gene knockdown on glucose metabolism by first selecting important differential metabolites in the glucose metabolic system from the metabolomic assay results (Fig. 6a). Although there was sufficient glucose as a raw material of glycolysis in Per-KD cells, some products in the glycolysis process, especially products after the rate-limiting step, such as glucose-6 phosphate, pyruvate, and lactate, were decreased, indicating repression of the glycolytic pathway.
Figure 6. Effect of BmPer gene knockdown on glucose metabolism in BmN cells.
(a) Changes in contents of main substrates and products, based on metabolomic measurements. (b) Transcription levels of the main rate-limiting enzyme-encoding genes. (c) Activities of the main rate-limiting enzymes. (d) Main affected glucose metabolic pathways. Per-KD, BmPer knockdown BmN cells; WT, wild-type BmN cells; HK, hexokinase; PFK, phosphofructokinase; PK, pyruvate kinase; CS, citrate synthase; +, increased enzymatic activity; −, decreased content or enzyme activity, or down-regulated transcription of coding gene. *p ≤ 0.05 and **p ≤ 0.01.
Furthermore, transcription levels of BmHk(Hexokinase), BmPfk(Phosphofructokinase), and BmPk(Pyruvate kinase), which encode the rate-limiting enzymes HK, PFK, and PK in the glycolytic pathway, were down-regulated by >40% in Per-KD cells compared with WT cells, with the transcript level of BmPfk reduced by 90% (Fig. 6b). The enzymatic activities of BmHK and BmPFK were also significantly reduced in Per-KD cells, consistent with down-regulation of transcript levels of their coding genes (Fig. 6c), thus explaining the mechanisms responsible for repression of the glycolytic pathway in Per-KD cells.
In contrast to the other rate-limiting enzymes, BmPK activity was significantly increased in Per-KD cells, even though transcription of its coding gene BmPk was down-regulated (Fig. 6c). Like pyruvate, the content of its anaerobic metabolite lactate was also reduced in Per-KD cells (Fig. 6a). Furthermore, the TCA cycle starting with pyruvate was also affected. The above metabolomic assays revealed a significant decrease in citrate content in Per-KD cells (Fig. 6a), which was the third largest change among all the metabolites (Table 1). The results for CS (Citrate synthase), the first rate-limiting enzyme in the TCA cycle, indicated that transcription of the gene encoding BmCS (Fig. 6b), as well as its enzymatic activity, were also significantly decreased (Fig. 6c). These results suggested that inhibition of the TCA cycle in Per-KD cells was caused by reduced pyruvate levels, in turn caused by decreased glycolysis and thus lack of feed into the TCA cycle, while the key enzyme of the first step of the TCA cycle was also inhibited.
These results suggest that glucose metabolism and glycometabolism-related pathways, including glycolysis and its further anaerobic and aerobic phases (TCA), were all significantly inhibited by Per gene knockdown.
Discussion
The circadian clock has been reported to regulate glycometabolism in humans34 and other mammals35 containing polymorphic PER proteins. It is possible to estimate the time of the day in mammals based on the expression patterns of various substances36. The major clusters of circadian clock-regulated genes participate in principal functions in the liver, and many comprise rate-limiting steps in their respective pathways37. However, none of the metabolic changes investigated to date have been determined under conditions involving knockdown/out all of mPer genes simultaneously, and without the influence of animal endocrine hormones. We therefore chose to study the silkworm B. mori, which only expresses one PER protein, and to establish a cell line with continuous knockdown of BmPER protein expression. Metabolomic assays revealed that levels of pyruvate and lactate were significantly reduced in cultured Per-KD cells in the absence of any effect of endocrine hormones. Examination of the main rate-limiting steps in glycolysis and the TCA cycle confirmed that glucose metabolic processes were repressed in Per-KD cells (Fig. 6). Knockdown of BmPER levels in BmN cells thus induced consistent changes in the regulation of glucose metabolism, suggesting that lack of expression of this peripheral circadian oscillator core member caused significant changes in metabolic systems, including glucose metabolism.
Circadian control of physiology and behavior in mammals is driven by a master pacemaker located in the suprachiasmatic nucleus in the hypothalamus38,39,40. However, cell-autonomous clocks (peripheral clocks) have been found to be ubiquitous, and most cells in the body contain their own circadian clock41, as verified in cell lines ranging from mammals (mice)42 to lepidoptera species such as Danaus plexippus43. The results of the current study confirmed the existence of a peripheral clock in silkworm BmN cells44. Recent studies have shown that changes in feeding patterns may disrupt the peripheral clock in mice, leading to metabolic syndrome45,46, while a fat-metabolism disorder in Drosophila caused by food restriction was also regulated by the peripheral clock47. However, although individual animal cells have spontaneous rhythms, they are also regulated by the core cranial nerve core. We therefore examined the regulatory effect of the peripheral clock on cell metabolism in BmN cells in vitro, to avoid additional regulatory and interfering effects of the core circadian clock and endocrine hormones on metabolism.
In this study, we constructed the Per-KD subcultured cell line, in which PER protein expression was continuously knocked down, based on the continuous expression of shRNA (short hairpin RNA) by a vector, and observed a stable effect of the circadian clock system on cell growth and development. This result would be difficult to determine by short-term knockdown of the Per gene by treatment with double-stranded RNA or siRNA. Furthermore, 8 biological repeats showed almost the consistent metabolism change, making the evidence for the impact on metabolic pathways more convincing. The results of this study showed that the activities of key enzymes in glycolysis and the TCA cycle, as well as transcription levels of their coding genes, were almost consistently repressed (Fig. 6), suggesting that after knockdown of BmPER, the peripheral clock regulated glucose metabolism by affecting the expression and/or activities of certain key enzymes in glucose metabolism. Comparison of circadian transcription in zebrafish1, flies48, and mammals37 suggests that circadian control of several key processes and pathways have been conserved among organisms and cultured cells in vitro.
Per gene have important biological significance. It has been reported that RNA interference of the SpPer gene caused a significant delay in the UVD lumen acidification49 and affected the rhythm of sperm release in Spodoptera littoralis moths50. Also, the temporal expression of the yolk protein 2 (yp2) gene at the mRNA and protein level in the testis of Spodoptera littoralis, is regulated by SpPer-based molecular oscillator51. Sandrelli et al.52 found that BmPer silencing produced a small but detectable disruption in the egg-hatching rhythm, as well as a reduction in egg-to-adult developmental time. In the research of Antheraea pernyi, the close anatomical location between PTTH- and PER-expressing cells suggested routes of communication between these two cell populations that may be important for the circadian control of PTTH release53.
The protein of PER binds to nuclear receptors such as PPARα and REV-ERBα to provide another mode of circadian regulation via nuclear receptor function, which regulates numerous life activities including growth, development, endocrine activity, reproduction, and energy metabolism19,22,54. This binding regulation is particularly important for liver glucose metabolism19. Previous studies have also demonstrated that post-translational modifications may play central roles in many biological pathways, including circadian regulation48,55,56. The current results regarding the regulation of pyruvate synthesis showed that PK activity increased in Per-KD cells, despite a significant decrease in gene transcription and pyruvate content.
It is generally known that transcription regulation is a basic to regulate the enzyme activity57,58. In our manuscript, even though the mRNA level of BmPk gene down-regulated, BmPK activity upregulated in Per-KD cells. Meanwhile, content of pyruvic acid, the catalysate of BmPK, was reduced. It was reported that, the phosphorylation level of rate-limiting enzymes such as BmPK is one of the important ways to regulate glycometabolism59. It has been reported that excess alanine which is generated by pyruvate acid, would feedback inhibited the activity of BmPK by phosphorylation60,61. In our result, the content of alanine and pyruvate acid both decreased after BmPer knockdown. Therefore, we speculated that the feedback regulation promoted to dephosphorylation. This kind of post-translational regulation that caused inconsistent of transcription level and enzyme activity has already reported in Bombyx mori eggs62. However, the pyruvate content catalyzed by PK was not restored because of a decrease in metabolic level of the early glycolytic pathway, suggesting that the circadian clock may regulate cell metabolism via complex mechanisms.
The circadian clock and metabolic processes are interactive63. Changes in the alcohol metabolic pathway were observed in a Drosophila Per mutant, resulting in differences in alcohol tolerance and clearance capability64. On the other hand, long-term alcohol intake in mice can lead to changes in the circadian rhythm65, whilst damaging the normal rhythm of liver glycogen metabolism66. Numerous studies have reported such reciprocal regulation of the circadian clock by metabolites, whereby the metabolites regulate expression of core circadian clock genes, thus changing the circadian rhythm. For example, glucose levels can affect the expression of mPer2 in mammalian hypothalamic cells, thus regulating the circadian clock by a feedback mechanism67. This type of regulation is not limited to glucose, and control of other dietary nutrients also changes the circadian rhythm68. In addition, some metabolites, such as cAMP and NAD+ 69,70, can act as input signals to regulate the circadian clock. This regulation can often be seen when the metabolites are also signaling factors. Because of the abnormal transcriptional change of other circadian genes, we speculated that both BmPer gene knockdown and the abnormal metabolism play a role in the change of circadian clock. This interaction between the circadian rhythm and metabolism opens up new areas of research.
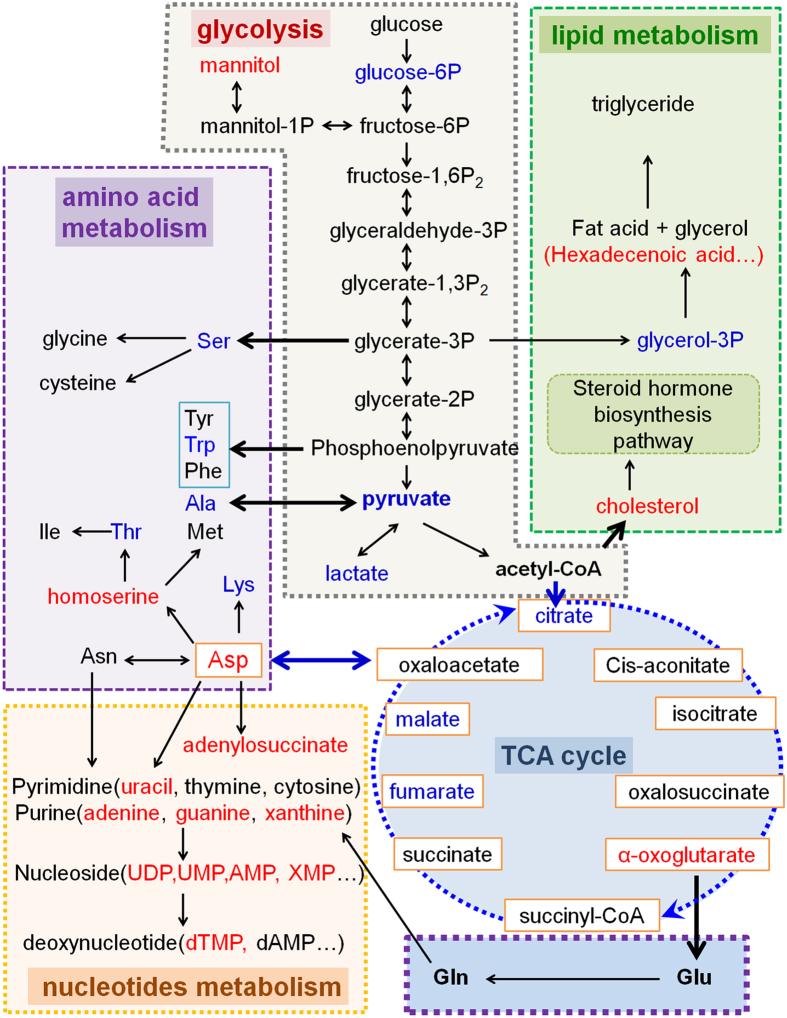
The network of possible effects of BmPer gene knockdown on the metabolism of silkworm BmN cells, based on the results of metabolomic assays, is shown in Fig. 7. The glucose metabolic pathways including glycolysis and TCA cycle were decreased because of the low transcription level and activity of enzyme, and low content of metabolites. Meanwhile, the amino acids which use glucose metabolites as raw materials also decreased. For example, the Ala, who is generated by pyruvic acid, also decreased following pyruvic acid. In contrast, the impact on lipid metabolism was mainly focused on cholesterol and subsequent steroid hormone pathways, while nucleotide metabolism, including purine and pyrimidine metabolism, was increased. These results confirm the complex effect of the circadian clock system on metabolism.
Figure 7. Summary of metabolic pathways affected in Per-KD cells.
The pathways in the figure were simplified according to the KEGG pathways (http://www.kegg.jp/). Metabolites indicated in red were increased after knockdown of BmPer, while metabolites indicated in blue were decreased, as determined by metabolomics analysis. Overall glucose metabolism (including glycolysis and TCA cycle) and amino acid-like substances generated from glucose metabolic intermediates were down-regulated, while overall nucleotide metabolism was upregulated by BmPer knockdown.
Metabolic affect cell size and growth through complicated ways. Some reports showed that a mevalonate pathway regulates cell size homeostasis and proteostasis through autophagy71. The de novo purine synthesis in circadian controlled the cell cycle1. Miettinen et al.72 found that mitochondrial metabolism and lipid synthesis are used to couple cell size and cell proliferation through identification of transcriptional and metabolic programs related to mammalian cell size. Hence, the significant decrease of glycometabolism and relative amino acid metabolism level after BmPer knockdown might be related to cell morphologies such as small size and slow growth.
The circadian clock has been reported to affect lipid metabolism73, as well as having important effects on hormones and the endocrine system, mainly due to the role of the suprachiasmatic nucleus core clock74. Many studies have shown that steroid hormones, such as glucocorticoids75,76 and estrogen77, affect metabolism by regulating the circadian clock through hormone signaling pathways. The results of the current study showed that changes in the circadian clock may also affect cholesterol levels, with subsequent effects on steroid hormone pathways, suggesting that the peripheral clock may impact on hormone signaling pathways.
Few studies have reported on the effects of the circadian clock on amino acid and nucleotide metabolism. The metabolomics results in the current study showed that, when glucose metabolism was repressed, most amino acids produced from the intermediate metabolites of glucose metabolism were also reduced (Fig. 7), suggesting that regulation of amino acid metabolism by the circadian clock may be an indirect effect of its regulation of glucose metabolism. The effect of the circadian clock on nucleotides was an interesting phenomenon, involving up-regulation of pyrimidine and purine metabolism, and the subsequent formation of ribonucleotides and deoxyribonucleosides in Per-KD cells (Fig. 7). However, correlations between these nucleotide substances and other metabolites were not strong (Fig. 3), indicating that nucleotide metabolism may be a relatively independent regulatory process. Purine synthesis has been reported to be controlled by the circadian oscillation system and to affect the regulation of cell cycle by the circadian clock1, presumably linked to the change in growth rate of Per-KD cells in the present experiment. Further studies are needed to clarify the mechanisms whereby the circadian clock regulates nucleotide metabolism.
Conclusion
Knockdown of the core circadian oscillation gene BmPer in the silkworm B. mori demonstrated that the circadian clock system may repress glucose metabolism by regulating the expression of key enzymes of glucose metabolism, leading to abnormalities in other metabolic systems.
Materials and methods
Cell lines
The B. mori ovary cell line (BmN) was preserved in our laboratory and cultured in Grace insect medium at 26 °C, as wild-type (WT) cells.
Pre-microRNA with mir-30 was used to construct transfection plasmid short hairpin RNAs (shRNAs). The BmPer cDNA sequence 270TATATCCATATTCTCAGGTCC290 was chosen for siRNAPer and inserted into shRNA to obtain shRNA-Per, which was then inserted into the pIZT/V5-His/Cat vector position between SacI578 and SpeI651 to generate the shRNA-Per expression vector (Figure S6). BmN cells were cultured in Grace medium containing 10% fetal bovine serum and 1% penicillin–streptomycin solution, and the medium was changed every other day. Lipofectamine LTX reagent (2 μL) and 0.5 mg of DNA plus reagent vector were diluted into 25 μL of Grace medium, incubated for 10 min, and then mixed. The mixture was added to 24-well cell culture plates seeded with BmN cells and shaken gently. At 48 h after transfection, 100 μg/mL bleomycin was added for prescreening. The medium was changed every 48 h to maintain the concentration of bleomycin in the medium. After 12 days of continuous screening, half of the total transfected cells were used for total RNA extraction. The interference efficiency of the BmPer gene was examined by qRT-PCR. BmN mutant cells, with stable interference of the BmPer gene, were identified as BmPer knockdown (Per-KD) cells.
Cell lines were synchronized using Dex (0.1 μM, 2 h) in darkness prior to qRT-PCR and western blotting.
Staining
Cells were seeded into culture dishes for at least 12 h, rinsed with 1 mL phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde at 25 °C for 15 min, and rinsed three times with PBST (PBS + 0.05% Tween-20). Fixed cells were treated with 0.1% Triton X-100 and rinsed three times with PBST. Cells were incubated with Dil (Beyotime, Jiangsu, China) for 1 min followed by three 5-min washes with PBST. Then, cells were incubated with 4′-6-diamidino-2-phenylindole (DAPI) (Beyotime) at room temperature for 5 min. Following three 5-min washes with PBST, the cells were observed under a fluorescence microscope (Olympus SZX16, Tokyo, Japan).
Determination of cell growth rate
Cell growth was determined by the 3-(4,5)-dimethylthiazo (-z-y1) -3,5-diphenytetrazoliumbromide (MTT) method, according to the manufacturer’s instructions (MTT Cell Proliferation Assay Kit C0009; Beyotime). Cells were digested with trypsin and counted, and the cell solution was adjusted to 2 × 104 cells/mL. Cell solution (100 μL) and 10 μL of 5 mg/mL MTT solution were then added to each well of the multi-well plate and incubated at 37 °C for 4 h, followed by the addition of 100 μL formazan solution, and incubation at 37 °C for a further 4 h. The absorbance at 570 nm was then measured using a microplate reader (Eon, Biotek, Winooski, VT, USA). The experiments were repeated three times.
GC/LC-MS assay
Based on the previously described pretreatment method for cell metabolomics78, the cells were counted, 1 × 107 cells were taken, and the cell supernatants were obtained by quenching and centrifugation at 1,000 g for 1 min.
Samples for GC-MS assay were resuspended in 500 μL of methanol pre-cooled at −80 °C, followed by the addition of 60 μL of margaric acid (0.1 mg/mL) and 60 μL of dichlorophenylalanine (0.2 mg/mL), and cryopreservation in liquid nitrogen. The samples were thawed at room temperature, resuspended in 500 μL of pre-cooled methanol, and the supernatant was obtained by centrifugation at 800 g for 1 min. Methanol resuspension and centrifugation were repeated. The cells were resuspended in ultra-pure water and cryopreserved in liquid nitrogen, then centrifuged at 15,000 g for 1 min, and dried at 30 °C in a vacuum to produce the samples to be assayed. Methoxy solution (60 μL) was added and mixed well, and the reaction was carried out at 37 °C for 2 h. Finally, 60 μL of bis-trimethylsilyl-trifluoroacetamide reagent containing 1% trimethylchlorosilane was added and the reaction took place at 37 °C for 90 min. The resulting samples were subjected to GC-MS assay (Agilent 7890A/5975C) using an HP-5MS capillary column (5% phenyl methyl silox: 30 m × 250 μm internal diameter, 0.25 μm; Agilent J & W Scientific, Folsom, CA, USA). Gas chromatographic conditions: split injection with a split ratio of 20:1, injection volume 1 μL, inlet temperature 280 °C, ion source temperature 250 °C, and interface temperature 150 °C. The initial temperature of programmed heating was 80 °C maintained for 2 min, followed by an increase to 300 °C at a rate of 10 °C/min, and then maintained for 5 min. The total operating time was 30 min, the carrier gas was helium, and the carrier gas flow rate was 1 mL/min. Mass spectrometry conditions: electrospray ionization source, full scan mode, electron energy 70 eV; quadrupole scan range 35–780 m/z.
The samples for LC-MS assay were resuspended in 500 μL of pre-cooled methanol and the supernatant was obtained by centrifugation. Methanol resuspension and centrifugation were repeated. The cells were resuspended in ultra-pure water, frozen in liquid nitrogen, and then centrifuged at 15,000 g for 1 min, and dried at 30 °C in a vacuum to produce the samples to be assayed. The samples were then dissolved in 300 μL of methanol solution (v:v = 1:1, 4 °C), filtered through a 0.22-μm membrane, and then subjected to LC-MS assay (Waters UPLC, Waters, Milford, MA, USA) using a C18 column (1.7 μm, 2.1 × 100 mm) (BEH, Waters). The mobile phase A/B was ultrapure water/acetonitrile, consisting of 0.1% (v/v) formic acid, the flow rate was 0.3 mL/min. Linear gradient elution as follows, 0 min–1 min, 2%B; 1 min–1 min, 2–50%B; 11 min–17 min, 50–98%B; 17 min–18 min, 98%B; 18 min–19 min, 98–2%B; 19 min–20 min, 2%. injection volume 2 μL, column temperature 40 °C, autosampler temperature maintained at 4 °C. Leucine enkephalin was used as lock and spray (0.4 ng/L, 0.1% formic acid acetonitrile/H2O 50/50). Mass spectrometry conditions: electrospray ionization source, positive and negative ionization modes. Source temperature 120 °C, desolvation temperature 350 °C, desolvation gas flow 800 L/h, and cone gas flow 50 L/h. The capillary ionization voltages of positive and negative ion mode were 3.0 and 2.8 kV, respectively, sampling cone 30 eV, extraction cone 4 eV, and quadrupole scan range 50–1,000 m/z. After the end of the test, the metabolites were confirmed based on their exact molecular weights and the possible empirical formulae of the metabolites were speculated (molecular weight error <30 ppm). The exact molecular weights and MS/MS fragmentation patterns were then used to identify potential biomarkers by confirmation in the Human Metabolome Database (HMDB) (http://www.hmdb.ca website), Metlin (http://metlin.scripps.edu/website), massbank (http://www.massbank.jp/), and LipidMaps (http://www.lipidmaps.org).
Each sample was extracted 20 μL and mixed as QC sample, which is used to correct the deviation of results and the errors caused by the instrument reason (Figure S7). GC-MS and LC-MS assay were carried out at BioNovoGene Co., Ltd. (Suzhou, China).
qRT-PCR
Total RNA was isolated from WT and Per-KD BmN cells for qRT-PCR analysis using RNAiso™ Plus (TaKaRa, Dalian, China). The concentrations of RNA were determined and the qualities were controlled spectrophotometrically by the A260/A280 ratio (Beckman, Brea, CA, USA). Any DNA in the total RNA samples was digested with RNase-free DNase I (TaKaRa), and 1 μg of the total RNA was used to synthesize first-strand cDNA. qRT-PCR was carried out in a total reaction volume of 20 μL using an ABI StepOnePlus™ PCR system (Ambion, Foster City, CA, USA) and the fluorescent dye SYBR Premix Ex Taq (TaKaRa), according to the manufacturers’ instructions. The reaction conditions were as follows: 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s then 60 °C for 30 s. After PCR, melting curve analysis was used to confirm the amplification of specific products and the data were normalized against endogenous BmRp4962,79. The standard curve method was used to determine the expression levels of the samples. All the experiments were performed in triplicate. The gene-specific primers used in this study are given in Table S1.
Determination of enzyme activity
Soluble protein was isolated from WT and Per-KD cells for enzyme assays. Protein concentrations were measured with a BCA kit (Beyotime), and enzyme activities were determined with an Enzyme Activity Assay Kit (Jiancheng, Nanjing, China), according to the manufacturer’s instructions. The activities of hexokinase (HK), phosphofructokinase (PFK), and pyruvate kinase (PK) were determined by measuring changes in NADPH using glucose, fructose-6-phosphate, and phosphoenolpyruvate as substrates. The activity of citrate synthase (CS) was determined by measuring citrate production with acetyl-CoA and oxaloacetate as substrates, using the dithio-bis-nitrobenzoic acid colorimetric method. One unit of HK, PFK, and PK, respectively, was defined as the consumption of 1 nM NADH/min/mg of cell protein (U/mg protein). One unit of CS was defined as the production of 1 μM citrate/min/mg of cell protein (U/mg protein).
Western blotting
Proteins were extracted from WT and Per-KD cells and subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to a polyvinylidene difluoride membrane. The membrane was blocked with a blocking solution, followed by incubation with the specific antibody, and washed and incubated with horseradish peroxidase (HRP)-labelled anti-rabbit immunoglobulin G (Bioworld Technology, Minneapolis, MN, USA). Immunoblots were visualized using Clarity Western ECL Substrate and ChemiDoc Touch Imaging System (Bio-rad, Hercules, CA, USA). Protein levels were measured by placing a rectangle of the same size over each Per-KD and WT.
The amino acid sequence of the BmPER protein was acquired from NCBI (GenBank accession number NP_001036975). After peptide sequence design, synthesis and purification, the peptide sequence of the PER protein, NH2-ESGESKKRDTRNHCCONH2, was used as an antigen to immunize New Zealand rabbits. Antibody titers were detected by enzyme-linked immunosorbent assay. The antibodies were purified by affinity purification. All these procedures were completed by Abgent Biotechnology Co., Ltd (Suzhou, China). Antibody specificity was analyzed by western blotting.
Statistical analysis
The original GC-MS and LC-MS data were processed and analyzed using XCMS software (www.bioconductor.org/). GC-MS and LC-MS assay results were used to identify differential metabolites between the WT and Per-KD cells using partial least squares-discriminant analysis (PLS-DA) first principal component variable importance in projection (VIP) value (VIP ≥1) and Student’s t-tests (p < 0.05). Statistical tests were performed using R 3.0.3 software (www.r-project.org). PCA was used for unsupervised data analysis, PLS-DA was used for supervised data analysis, and S-plot analysis was performed with Simca-P 13.0 (Umetrics AB, Umea, Sweden)80. The Pheatmap function of the R language package was used for Heatmap analysis (www.r-project.org).
Metabolite correlation analysis was assessed using Pearson’s correlation coefficients. The computing method was the cor() function in the R language package (www.r-project.org), and metabolite correlation analysis and statistical significance tests were performed at the same time. Statistical tests were carried out using the cor.test() function in the R language package. A false positive check was performed on the p value, and the correlation was considered to be significant at a false discovery rate p value ≤ 0.05. The relationships among metabolites for constructing the metabolic pathways were based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database81 (http://www.genome.jp/kegg/). Univariate analysis of variance (ANOVA) was used to determine the significance of differences in relative contents between different groups. Metabolic pathways of differential metabolites were subjected to MetPA analysis33 using the analysis website http://www.metaboanalyst.ca/, and the analysis parameters were the default values.
Enzyme activity and gene expression levels were analyzed by univariate ANOVA.
Additional Information
How to cite this article: Tao, H. et al. Inhibition of expression of the circadian clock gene Period causes metabolic abnormalities including repression of glycometabolism in Bombyx mori cells. Sci. Rep. 7, 46258; doi: 10.1038/srep46258 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31672492), Provincial Key R & D Program of Jiangsu (Grant No. BE2015317), China Agriculture Research System (CARS) (Grant No. CARS-22-ZJ0104), and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The manuscript has got the assistance from native English speaker of International Science Editing scientific services.
Footnotes
The authors declare no competing financial interests.
Author Contributions S.-Q.X., H.T. and Y.-H.S. conceived this project. H.T., X.L., J.-F.Q., and W.-Z.C. performed the research. H.T., X.L. and S.-Q.X. analyzed the data and wrote the paper. And all authors participated in the revision of this paper by providing comments and editing.
References
- Li Y. et al. Integrative analysis of circadian transcriptome and metabolic network reveals the role of de novo purine synthesis in circadian control of cell cycle. PLoS Comput Biol 11, e1004086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond S., Jun D., Rubin B. E. & Golden S. S. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc Natl Acad Sci USA 112, E1916–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C. & Schibler U. Circadian timing of metabolism in animal models and humans. J Intern Med 277, 513–27 (2015). [DOI] [PubMed] [Google Scholar]
- Bass J. & Takahashi J. S. Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa M. Circadian rhythms, metabolism, and insulin sensitivity: transcriptional networks in animal models. Curr Diab Rep 13, 223–8 (2013). [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K. & Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol 16, 462–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summa K. C. & Turek F. W. Chronobiology and obesity: Interactions between circadian rhythms and energy regulation. Adv Nutr 5, 312S–9S (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHill A. W. et al. Impact of circadian misalignment on energy metabolism during simulated nightshift work. Proc Natl Acad Sci USA 111, 17302–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Yeh B., Rakshit K., Colwell C. S. & Matveyenko A. V. Circadian Disruption and Diet-Induced Obesity Synergize to Promote Development of β-Cell Failure and Diabetes in Male Rats. Endocrinology 156, 4426–36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejníková L., Polidarová L., Paušlyová L., Sládek M. & Sumová A. Diverse development and higher sensitivity of the circadian clocks to changes in maternal-feeding regime in a rat model of cardio-metabolic disease. Chronobiol Int 32, 531–47 (2015). [DOI] [PubMed] [Google Scholar]
- Ribas-Latre A. & Eckel-Mahan K. Interdependence of nutrient metabolism and the circadian clock system: Importance for metabolic health. Mol Metab 5, 133–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevy M. F., Giebultowicz J., Pincus Z., Mazzoccoli G. & Vinciguerra M. Aging signaling pathways and circadian clock-dependent metabolic derangements. Trends Endocrinol Metab 24, 229–37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas C. et al. Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell Cycle 13, 3282–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Militi S. et al. Early doors (Edo) mutant mouse reveals the importance of period 2 (PER2) PAS domain structure for circadian pacemaking. Proc Natl Acad Sci USA 113, 2756–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J. C. Molecular bases for circadian clocks. Cell 96, 271–90 (1999). [DOI] [PubMed] [Google Scholar]
- Feillet C. A. et al. Lack of food anticipation in Per2 mutant mice. Curr Biol 16, 2016–22 (2006). [DOI] [PubMed] [Google Scholar]
- Zani F. et al. PER2 promotes glucose storage to liver glycogen during feeding and acute fasting by inducing Gys2 PTG and G L expression. Mol Metab 2, 292–305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. Loss of mPer2 increases plasma insulin levels by enhanced glucose-stimulated insulin secretion and impaired insulin clearance in mice. FEBS Lett 586, 1306–11 (2012). [DOI] [PubMed] [Google Scholar]
- Schmutz I., Ripperger J. A., Baeriswyl-Aebischer S. & Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev 24, 345–57 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solocinski K. et al. Transcriptional regulation of NHE3 and SGLT1 by the circadian clock protein Per1 in proximal tubule cells. Am J Physiol Renal Physiol 309, F933–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamovich Y. et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab 19, 319–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi B. et al. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab 12, 509–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. et al. The role of mPer2 clock gene in glucocorticoid and feeding rhythms. Endocrinology 150, 2153–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann R., Touma C., Palme R., Albrecht U. & Steinlechner S. Impaired daily glucocorticoid rhythm in Per1 (Brd) mice. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 192, 769–75 (2006). [DOI] [PubMed] [Google Scholar]
- Hasenour C. M., Berglund E. D. & Wasserman D. H. Emerging role of AMP-activated protein kinase in endocrine control of metabolism in the liver. Mol Cell Endocrinol 366, 152–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam S. et al. Data-driven integration of genome-scale regulatory and metabolic network models. Front Microbiol 6, 409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K. et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30, 525 (2001). [DOI] [PubMed] [Google Scholar]
- Cermakian N., Monaco L., Pando M. P., Dierich A. & Sassone-Corsi P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the period1 gene. Embo J 20, 3967–3974 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B. et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105, 683–694 (2001). [DOI] [PubMed] [Google Scholar]
- Shearman L. P., Jin X., Lee C., Reppert S. M. & Weaver D. R. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol cell biol 20, 6269–6275 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Want E. J., O’Maille G., Abagyan R. & Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78, 779–87 (2006). [DOI] [PubMed] [Google Scholar]
- Jourdan F., Breitling R., Barrett M. P. & Gilbert D. MetaNetter: inference and visualization of high-resolution metabolomic networks. Bioinformatics 24, 143–145 (2008). [DOI] [PubMed] [Google Scholar]
- Xia J. & Wishart D. S. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 26, 2342–2344 (2010). [DOI] [PubMed] [Google Scholar]
- Kumar Jha P., Challet E. & Kalsbeek A. Circadian rhythms in glucose and lipid metabolism in nocturnal and diurnal mammals. Mol Cell Endocrinol 418, 74–88 (2015). [DOI] [PubMed] [Google Scholar]
- Morris C. J. et al. Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans. Proc Natl Acad Sci USA 112, E2225–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y. et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA 106, 9890–5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S. et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–20 (2002). [DOI] [PubMed] [Google Scholar]
- Stephan F. K. & Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci USA 69, 1583–6 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. D. et al. Dissociation of circadian and light inhibition of melatonin release through forced desynchronization in the rat. Proc Natl Acad Sci USA 106, 17540–5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper C. B., Scammell T. E. & Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature 437, 1257–63 (2005). [DOI] [PubMed] [Google Scholar]
- Mohawk J. A., Green C. B. & Takahashi J. S. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35, 445–62 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieler J. et al. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol Syst Biol 10, 739 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. et al. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol 6, e4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F. et al. Impact of cyclical changes in temperature on circadian clock genes expression in Bombyx BmN cells. Arch Insect Biochem Physiol 91, 175–86 (2016). [DOI] [PubMed] [Google Scholar]
- Mukherji A. et al. Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc Natl Acad Sci USA 112, E6691–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji A., Kobiita A. & Chambon P. Shifting the feeding of mice to the rest phase creates metabolic alterations, which, on their own, shift the peripheral circadian clocks by 12 hours. Proc Natl Acad Sci USA 112, E6683–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa S. D. et al. Peripheral Circadian Clocks Mediate Dietary Restriction-Dependent Changes in Lifespan and Fat Metabolism in Drosophila. Cell Metab 23, 143–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasik K. et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab 17, 291–302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwica-Rolinska J., Gvakharia B. O., Kedzierska U., Giebultowicz J. M. & Bebas P. Effects of period RNAi on V-ATPase expression and rhythmic pH changes in the vas deferens of Spodoptera littoralis (Lepidoptera: Noctuidae). Insect Biochem Mol Biol 43, 522–32 (2013). [DOI] [PubMed] [Google Scholar]
- Kotwica J., Bebas P., Gvakharia B. O. & Giebultowicz J. M. RNA interference of the period gene affects the rhythm of sperm release in moths. J Biol Rhythms 24, 25–34 (2009). [DOI] [PubMed] [Google Scholar]
- Kotwica J. et al. Diurnal rhythm in expression and release of yolk protein in the testis of Spodoptera littoralis (Lepidoptera: Noctuidae). Insect Biochem Mol Biol 41, 264–72 (2011). [DOI] [PubMed] [Google Scholar]
- Sandrelli F. et al. Phenotypic effects induced by knock-down of the period clock gene in Bombyx mori. Genet Res 89, 73–84 (2007). [DOI] [PubMed] [Google Scholar]
- Sauman I. & Reppert S. M. Molecular characterization of prothoracicotropic hormone (PTTH) from the giant silkmoth Antheraea pernyi: developmental appearance of PTTH-expressing cells and relationship to circadian clock cells in central brain. Dev Biol 178, 418–29 (1996). [DOI] [PubMed] [Google Scholar]
- Costa M. J. et al. Circadian rhythm gene period 3 is an inhibitor of the adipocyte cell fate. J Biol Chem 286, 9063–70 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles M. S., Cox J. & Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet 10, e10040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S. et al. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci USA 110, 3339–44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asanuma N., Kanada K. & Hino T. Molecular properties and transcriptional control of the phosphofructokinase and pyruvate kinase genes in a ruminal bacterium, Streptococcus bovis. Anaerobe 14, 237–41 (2008). [DOI] [PubMed] [Google Scholar]
- Le Moullac G. et al. Transcriptional regulation of pyruvate kinase and phosphoenolpyruvate carboxykinase in the adductor muscle of the oyster Crassostrea gigas during prolonged hypoxia. J Exp Zool A Ecol Genet Physiol 307, 371–82 (2007). [DOI] [PubMed] [Google Scholar]
- Cowan K. J. & Storey K. B. Reversible phosphorylation control of skeletal muscle pyruvate kinase and phosphofructokinase during estivation in the spadefoot toad, Scaphiopus couchii. Mol Cell Biochem 195, 173–81 (1999). [DOI] [PubMed] [Google Scholar]
- Prasannan C. B., Villar M. T., Artigues A. & Fenton A. W. Identification of regions of rabbit muscle pyruvate kinase important for allosteric regulation by phenylalanine, detected by H/D exchange mass spectrometry. Biochemistry 52, 1998–2006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton A. W. & Hutchinson M. The pH dependence of the allosteric response of human liver pyruvate kinase to fructose-1,6-bisphosphate, ATP, and alanine. Arch Biochem Biophys 484, 16–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H. et al. Parental environmental exposure leads to glycometabolic disturbances that affect fertilization of eggs in the silkworm Bombyx mori: the parental transcript legacy. J Comp Physiol B 185, 47–55 (2015). [DOI] [PubMed] [Google Scholar]
- Delezie J. & Challet E. Interactions between metabolism and circadian clocks: reciprocal disturbances. Ann N Y Acad Sci 1243, 30–46 (2011). [DOI] [PubMed] [Google Scholar]
- Liao J., Seggio J. A. & Ahmad S. T. Mutations in the circadian gene period alter behavioral and biochemical responses to ethanol in Drosophila. Behav Brain Res 302, 213–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P. et al. Dissociation between diurnal cycles in locomotor activity, feeding behavior and hepatic PERIOD2 expression in chronic alcohol-fed mice. Alcohol 49, 399–408 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udoh U. S. et al. Chronic ethanol consumption disrupts diurnal rhythms of hepatic glycogen metabolism in mice. Am J Physiol Gastrointest Liver Physiol 308, G964–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterman J. E. & Belsham D. D. Glucose Alters Per2 Rhythmicity Independent of AMPK, Whereas AMPK Inhibitor Compound C Causes Profound Repression of Clock Genes and AgRP in mHypoE-37 Hypothalamic Neurons. PLoS One 11, e0146969 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan K. L. et al. Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–78 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey K. M. et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J. S., Maywood E. S., Chesham J. E., Takahashi J. S. & Hastings M. H. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320, 949–53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen T. P. & Björklund M. Mevalonate Pathway Regulates Cell Size Homeostasis and Proteostasis through Autophagy. Cell Rep 13, 2610–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen T. P. et al. Identification of transcriptional and metabolic programs related to mammalian cell size. Curr Biol 24, 598–608 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnocchi D., Pedrelli M., Hurt-Camejo E & Parini P. Lipids around the Clock: Focus on Circadian Rhythms and Lipid Metabolism. Biology (Basel) 4, 104–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. W., Jeong J. H. & Hong S. C. The impact of sleep and circadian disturbance on hormones and metabolism. Int J Endocrinol 2015, 591729 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusunose N. et al. Mitomycin C modulates the circadian oscillation of clock gene period 2 expression through attenuating the glucocorticoid signaling in mouse fibroblasts. Biochem Biophys Res Commun 467, 157–63 (2015). [DOI] [PubMed] [Google Scholar]
- Honma A. et al. Glucocorticoids Reset the Nasal Circadian Clock in Mice. Endocrinology 156, 4302–11 (2015). [DOI] [PubMed] [Google Scholar]
- Zhu L. et al. Estrogens prevent metabolic dysfunctions induced by circadian disruptions in female mice. Endocrinology 156, 2114–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick C. A., Hansen R., Stephens G. M., Goodacre R. & Dickson A. J. Metabolite extraction from suspension-cultured mammalian cells for global metabolite profiling. Nat Protoc 6, 1241–9 (2011). [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhou Y., Chen K. & Ju X. Suppression of Bm-Caspase-1 Expression in BmN Cells Enhances Recombinant Protein Production in a Baculovirus Expression Vector System. Mol Biotechnol 58, 319–27 (2016). [DOI] [PubMed] [Google Scholar]
- Eriksson L. et al. Multi- and Megavariate Data Analysis. Part I Basic Principles and Applications. Second revised and enlarged edition. Metabonomics in Toxicity Assessment 362–362 (2006). [Google Scholar]
- Kanehisa M. & Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.