Abstract
High-density linkage maps can improve the precision of QTL localization. A high-density SNP-based linkage map containing 3207 markers covering 3072.7 cM of the Brassica napus genome was constructed in the KenC-8 × N53-2 (KNDH) population. A total of 67 and 38 QTLs for seed oil and protein content were identified with an average confidence interval of 5.26 and 4.38 cM, which could explain up to 22.24% and 27.48% of the phenotypic variation, respectively. Thirty-eight associated genomic regions from BSA overlapped with and/or narrowed the SOC-QTLs, further confirming the QTL mapping results based on the high-density linkage map. Potential candidates related to acyl-lipid and seed storage underlying SOC and SPC, respectively, were identified and analyzed, among which six were checked and showed expression differences between the two parents during different embryonic developmental periods. A large primary carbohydrate pathway based on potential candidates underlying SOC- and SPC-QTLs, and interaction networks based on potential candidates underlying SOC-QTLs, was constructed to dissect the complex mechanism based on metabolic and gene regulatory features, respectively. Accurate QTL mapping and potential candidates identified based on high-density linkage map and BSA analyses provide new insights into the complex genetic mechanism of oil and protein accumulation in the seeds of rapeseed.
Rapeseed (Brassica napus L., AACC, 2n = 38) is one of the most important oil crops globally. It originated from a spontaneous hybridization between B. rapa (AA, 2n = 20) and B. oleracea (CC, 2n = 18)1. The large amount of unsaturated fatty acid (FA) in rapeseed oil has made it widely accepted as a vegetable oil for human consumption and as a bio-fuel for industry2. The low glucosinolate and high protein content of the seed also make it a valuable feedstuff for animals and a potential protein source for human nutrition3,4. Increasing the seed oil content (SOC) and revealing the complex regulatory mechanism of SOC and the seed protein content (SPC) have become the most important breeding criteria for rapeseed. However, SOC and SPC are two complex quantitative traits that are frequently influenced by the environment5, and the genetic and molecular bases of both traits remain ambiguous.
QTL mapping is an effective strategy to dissect the regulatory loci and genetic mechanism underlying important agronomical traits at the whole genome level. Many studies have focused on SOC-QTL identification in Brassica napus (B. napus). SOC-QTLs were identified in almost all 19 linkage groups based on bi-parental linkage mapping, and varied between 3 and 27 QTLs6,7,8,9,10,11,12,13. Additionally, a genome‑wide association study has revealed SOC variations in B. napus14. Compared with SOC, relatively few studies have addressed SPC-QTLs in B. napus15,16,17,18. In addition, a significant negative relationship between SOC and SPC has been reported, especially in soybean19,20 and Brassica21,22,23. QTLs with pleiotropic effects for SOC and SPC have also been observed in Brassica species15,24. Identifying the underlying candidate genes and recognizing the pleiotropic effect or correlation between these two traits would greatly increase the breeding efficiency.
Nevertheless, in most of these studies, the QTLs spanned large ranges of the genome due to low mapping resolutions, and generally there was a lack of marker sequence information, hindering the integration of these QTLs into the reference physical map and the identification of the underlying candidate genes. In addition, the genomic sequences of the parental cultivars were also unknown in most of the previous studies, which also complicated the identification and analysis of candidate genes. Recent innovations in genome sequencing technology and bioinformatics and single nucleotide polymorphism (SNP) markers, especially the 60 K SNP Infinium Array, have been developed and widely applied for QTL mapping and the genetic dissection of important agronomical traits in B. napus25,26,27,28,29,30,31. Simultaneously, the reference genome of B. napus has been released32, making it feasible to perform a comparative genome analysis between the genetic map and the physical map using SNPs with sequence information. Additionally, bulk segregant analysis (BSA) combined with next-genomic sequencing technologies has been used as a fast track approach to more rapidly identify candidate genomic regions. This approach involves the selection of 20–50 lines with extreme phenotypic values, with pooling at equivalent concentrations followed by sequencing of the pools for subsequent variance analysis33. BSA has been used successfully to map candidate genomic regions for early flowering in cucumber34, 100-seed weight in chickpea35, and fruit weight and locule number in tomato36.
QTL studies have led to the identification of numerous loci that are responsible for variation in SOC and SPC, but the corresponding genomic regions with their complex metabolism and regulatory mechanisms have not been studied with great accuracy. Therefore, candidate genes within QTL regions combined with the relevant metabolic pathway in B. napus have been not reported. There is extensive genome homology and microsynteny between Brassica species and Arabidopsis thaliana (A. thaliana) because of their close evolutionary relationship37. More attention has been focused on genes and regulatory factors involved in acyl-lipid metabolism, and more than 120 enzymatic reactions and at least 600 genes involved in acyl-lipid metabolism in A. thaliana have been well documented by Li-Beisson et al.38. A. thaliana acyl-lipid and storage protein-related orthologous genes mapped in a confidence interval (CI) of QTLs on the B. napus genome might be valuable to identify the candidate genes and dissect the complex metabolism and regulatory mechanism associated with SOC and SPC.
This present report consists of the following three parts: (1) the construction of a high-density genetic linkage map based upon SNP markers combined with non-SNP (SSR and STS) markers that have been used previously to generate a primary linkage map in the KenC-8 × N53-2 (KNDH) population39; (2) precise QTL mapping of SOC and SPC across multiple environments based on a high-density genetic linkage map and rapid detection of associated genomic regions (AGRs) using BSA; (3) identification of candidate genes within QTL regions and provision of primary insights into the complex metabolism and regulatory mechanism of SOC and SPC in B. napus.
Results
Construction of the high-density linkage map
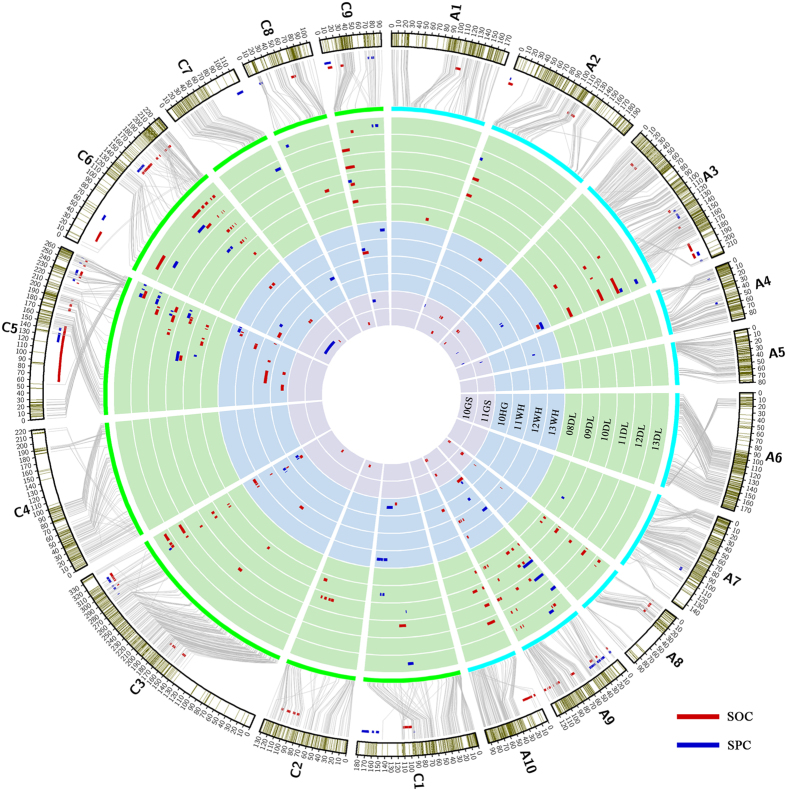
Of the 52157 SNP probes on the Brassica 60 K array, approximately 34.72% were polymorphic between the two parents and displayed segregation within the KNDH population. Finally, 18109 polymorphic SNPs were grouped into 3764 SNP-bins. All 3764 SNP-bins merged with 495 non-SNP markers (SSR and STS) were used for linkage map construction. A set of 3207 markers including 3106 SNP-bins and 101 non-SNPs were successfully assigned to 19 linkage groups (Table 1). The integrated map had a total length of 3072.7 cM with an average distance of 0.96 cM between adjacent markers (covering 1398.6 cM and 1674.1 cM for the A and C genomes, respectively). The number of loci in the 19 linkage groups ranged from 77 (A08) to 324 (A03), and the length ranged from 80.1 (A05) to 337.1 cM (C03), with a mean value of 161.72 cM (Table 1, and Fig. 1). The marker density of the 19 linkage groups varied; the highest marker density was on chromosome A04, with 156 markers distributed over a genetic map distance of 86.8 cM.
Table 1. Characteristics of the KNDH linkage map based on SNP-bins and non-SNP markers.
| Linkage group | SNP-bins (No.) | Non-SNP (No.) | Marker (No.) | Length (cM) | SNP-bins (%) | Average distance between adjacent loci (cM) |
|---|---|---|---|---|---|---|
| A01 | 168 | 10 | 178 | 177.1 | 94.38 | 0.99 |
| A02 | 174 | 12 | 186 | 199 | 93.55 | 1.07 |
| A03 | 321 | 3 | 324 | 218.4 | 99.07 | 0.67 |
| A04 | 156 | 0 | 156 | 86.8 | 100 | 0.56 |
| A05 | 134 | 0 | 134 | 80.1 | 100 | 0.60 |
| A06 | 211 | 13 | 224 | 178.6 | 94.2 | 0.80 |
| A07 | 202 | 0 | 202 | 149.1 | 100 | 0.74 |
| A08 | 77 | 0 | 77 | 92 | 100 | 1.19 |
| A09 | 175 | 0 | 175 | 123.5 | 100 | 0.70 |
| A10 | 130 | 7 | 137 | 94 | 94.89 | 0.69 |
| C01 | 172 | 3 | 175 | 182.2 | 98.28 | 1.04 |
| C02 | 134 | 0 | 134 | 132.4 | 100 | 0.99 |
| C03 | 278 | 13 | 291 | 337.1 | 95.53 | 1.16 |
| C04 | 156 | 4 | 160 | 220.6 | 97.5 | 1.38 |
| C05 | 172 | 7 | 179 | 263.2 | 96.09 | 1.47 |
| C06 | 172 | 12 | 184 | 226.3 | 93.48 | 1.23 |
| C07 | 94 | 6 | 100 | 117 | 94 | 1.17 |
| C08 | 95 | 5 | 100 | 103.9 | 95 | 1.04 |
| C09 | 85 | 6 | 91 | 91.4 | 93.41 | 1.00 |
| A genome | 1748 | 45 | 1793 | 1398.6 | 97.49 | 0.78 |
| C genome | 1358 | 56 | 1414 | 1674.1 | 96.04 | 1.18 |
| Total (A + C) | 3106 | 101 | 3207 | 3072.7 | 96.85 | 0.96 |
Figure 1. KN high-density genetic linkage map, distribution of markers, identified QTLs, consensus QTLs of SOC and SPC on each linkage group, and colinearity of the linkage map and B. napus reference genome.
The blocks with green bands at the outermost circle represent the 19 genetic linkage groups and the marker distribution. The third circle (from the outermost one) represents the 19 chromosomes of B. napus. The lines connecting them represent their col-linearity relationship. The 12 inner circles with 3 colors represent 12 microenvironments in winter-type, spring-type and semi winter-type rapeseed growing areas, respectively. The short bars with a red and blue color within the 12 inner circles represent QTLs identified in different environments and linkage groups, and they are located between the outermost two circles representing consensus QTLs.
Phenotypic variation and correlation of SOC and SPC in the parents and the DH population
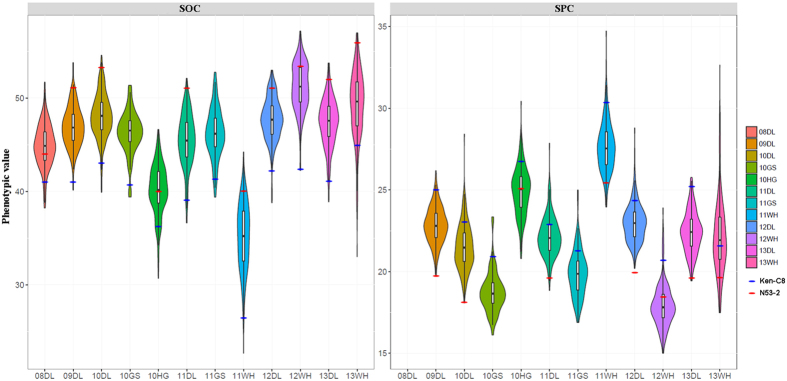
Combined with the data that were previously reported by Wang et al.39, a total of 12 and 11 sets of SOC and SPC trial data were used to analyze the phenotypic variation (PV), respectively (Table S1). The SOC and SPC of the two parents (N53-2 and KenC-8) differed significantly in all investigated environments. The SOC of N53-2 was 10% higher than that of Ken-C8 in all microenvironments except 10HG. While the SPC of Ken-C8 was nearly 5% higher than that of N53-2 in all semi-winter environments, they also showed significant differences, though to a reduced extent for the winter environments. The KN population exhibited a normal or near-normal distribution and transgressive segregation with a small degree of both traits (Fig. 2). These results indicated that quantitative inheritance was suitable for QTL identification, with polygenic effects and favorable alleles mainly inherited from one of the two parents. Further analysis revealed that both parents and populations in different trials all showed significant differences (Table S1), suggesting that both traits were significantly influenced by the environment. In addition, SOC and SPC were significantly negatively correlated with average coefficients of −0.65; the highest correlation up to −0.77 was identified in the 11GS (Table S2), suggesting competition among sinks for assimilates.
Figure 2. Frequency distribution of SOC and SPC in the KNDH population.
In each plot, a marker denotes the median of the data, a box indicates the interquartile range, and spikes extend to the upper and lower adjacent values. The distribution density is overlaid.
Identification, integration and co-localization analysis of QTLs for SOC and SPC
A total of 164 identified QTLs were observed for SOC in 12 trials, with an average CI of 7.43 cM. These QTLs were distributed on 12 chromosomes, excluding A04, A05, A06, A07, C01, C04 and C07, and a single QTL could explain up to 22.24% of the PV. The largest number of QTLs (136) was distributed on A03, A08, A09, C03, C05 and C06, accounting for 82.93% of the total number (Fig. 1 and Table S3). The 164 identified SOC-QTLs were then integrated into 67 consensus QTLs, and the average CI was reduced from 7.36 to 5.26 cM (Tables 2 and S3 and Fig. 1). Fifty-three consensus QTLs were distributed on A03, A08, A09, C03, C05 and C06, accounting for 79.10% of all consensus QTLs, and 35 of them (76.09%) showed environmental stability. There were 59, 30, and 16 QTLs detected in winter type, semi-winter type and spring type environments, respectively (Figure S1a). Only one QTL (cqOC-A9-9) could be detected in all three macroenvironments. Eleven, two and one QTL could be expressed stably at least over 2 years in winter, semi-winter and spring type environments, respectively. Six major QTLs (cqOC-A2-3, cqOC-A9-9, cqOC-A9-10, cqOC-C5-3, cqOC-C5-4, and cqOC-C6-6) were repeatedly detected in at least two trials and displayed a large effect (PV > 10%), among which cqOC-A9-9 and cqOC-C6-6 were detected stably in six microenvironments, while cqOC-C6-6 explained the highest PV (22.24%). Although accounting for less than 10% of the PV value, cqOC-C3-4 could be detected simultaneously in eight microenvironments and was considered a major QTL that merits attention.
Table 2. Consensus QTLs for SOC and SPC identified by a high-density linkage map of the KNDH population.
| Trait | Consensus QTL | Chromosome | Peak | Range | LOD | Additive | PV (%) | Environment |
|---|---|---|---|---|---|---|---|---|
| SOC | cqOC-A1 | A01 | 109.41 | 104.00–112.10 | 4.32047 | 0.7173 | 9.59 | 08DL |
| cqOC-A2–1 | A02 | 2.30 | 0–7.08 | 2.06–2.34 | −0.41– −0.43 | 1.97–2.22 | 10DL/11DL | |
| cqOC-A2-2 | A02 | 107.71 | 105.80–109.20 | 7.69916 | −1.2 | 14.18 | 11GS | |
| cqOC-A2–3 | A02 | 116.26 | 113.42–119.10 | 2.30–7.35 | −0.66– −1.16 | 3.43–13.72 | 11GS/10GS/13WH | |
| cqOC-A3-1 | A03 | 26.01 | 23.58–28.43 | 2.13–3.37 | −0.52– −0.68 | 4.36–6.00 | 10GS/11GS | |
| cqOC-A3-2 | A03 | 32.51 | 31.9–35.8 | 3.38086 | −0.68 | 6.02 | 11GS | |
| cqOC-A3-3 | A03 | 122.37 | 120.45–124.29 | 2.28–3.50 | 0.35–0.50 | 2.12–3.13 | 09DL/11DL/12DL | |
| cqOC-A3-4 | A03 | 151.81 | 150.70–153.90 | 3.65862 | 0.5785 | 3.44 | 11DL | |
| cqOC-A3-5 | A03 | 182.85 | 182.10–183.59 | 2.14–2.45 | 0.37–0.49 | 1.94–2.41 | 11DL/12DL | |
| cqOC-A3-6 | A03 | 191.71 | 183.20–195.70 | 3.71721 | 0.6261 | 4.12 | 11DL | |
| cqOC-A3-7 | A03 | 204.14 | 200.26–208.03 | 2.14–3.59 | 0.38–0.73 | 2.08–3.76 | 11DL/12DL/13WH/12DL | |
| cqOC-A8-1 | A08 | 1.47 | 1.08–1.85 | 2.40–3.48 | 0.40–0.50 | 2.16–3.25 | 12DL/10DL | |
| cqOC-A8-2 | A08 | 8.61 | 7.34–9.88 | 2.331–5.32 | 0.38–0.62 | 2.45–5.32 | 10GS/09DL/13DL/10DL/12DL | |
| cqOC-A8-3 | A08 | 12.32 | 10.85–13.78 | 2.60–3.72 | 0.58–0.72 | 3.50–6.82 | 11GS/11DL/10GS | |
| cqOC-A8-4 | A08 | 18.01 | 17.61–18.41 | 3.45–5.12 | 0.47–0.85 | 3.09–9.36 | 12DL/10DL/11GS/13DL | |
| cqOC-A8-5 | A08 | 20.01 | 19.53–20.49 | 2.84–4.83 | 0.37–0.67 | 2.62–5.89 | 10HG/09DL | |
| cqOC-A8-6 | A08 | 21.91 | 20.9–23.2 | 3.19–5.34 | 0.59–0.86 | 3.90–9.76 | 12WH/11GS | |
| cqOC-A9-1 | A09 | 1.17 | 0.19–2.14 | 2.04–2.69 | 0.36–0.47 | 2.14–2.52 | 10DL/09DL/10HG | |
| cqOC-A9–2 | A09 | 23.27 | 21.81–24.73 | 2.19–4.38 | 0.42–0.57 | 2.54–4.18 | 09DL/10DL/10HG/13DL | |
| cqOC-A9-3 | A09 | 31.21 | 30.4–32.2 | 4.2098 | 0.532 | 3.88 | 13DL | |
| cqOC-A9-4 | A09 | 34.26 | 32.36–36.17 | 2.45–5.35 | 0.41–0.72 | 2.30–5.40 | 11DL/10DL/09DL | |
| cqOC-A9-5 | A09 | 67.61 | 67.1–73.1 | 3.22245 | −0.56 | 3.04 | 11DL | |
| cqOC-A9-6 | A09 | 95.11 | 94.9–96.7 | 4.50709 | 0.5957 | 4.82 | 12DL | |
| cqOC-A9-7 | A09 | 98.41 | 97.23–99.58 | 2.96–4.80 | 0.49–0.68 | 3.06–5.14 | 10DL/12WH | |
| cqOC-A9-8 | A09 | 100.71 | 99.9–101.2 | 9.18561 | 0.8578 | 10.05 | 13DL | |
| cqOC-A9-9 | A09 | 106.28 | 105.79–106.77 | 2.76–14.19 | 0.64–1.05 | 4.88–14.94 | 09DL/11GS/13DL/12DL/12WH/10GS | |
| cqOC-A9-10 | A09 | 112.7 | 111.62–113.79 | 2.87–11.48 | 0.69–0.81 | 3.32–12.41 | 09DL/11DL/12WH/13WH | |
| cqOC-A9-11 | A09 | 116.38 | 116.16–116.59 | 7.33–7.66 | 0.64–0.79 | 7.76–8.46 | 13DL/09DL | |
| cqOC-A9-12 | A09 | 122.01 | 119.88–124.13 | 2.60–7.21 | 0.66–0.89 | 7.25–8.05 | 10GS/11DL | |
| cqOC-A10-1 | A10 | 1.11 | 0–2.59 | 3.64–3.79 | −0.61– −0.64 | 3.39–4.56 | 10DL/09DL | |
| cqOC-A10-2 | A10 | 8.71 | 5.8–16.7 | 2.80147 | −0.68 | 2.73 | 11DL | |
| cqOC-A10-3 | A10 | 18.03 | 14.35–21.71 | 2.12–3.13 | −0.56– −1.02 | 2.00–4.43 | 11WH/10DL/12DL | |
| cqOC-C2-1 | C02 | 84.14 | 82.1–86.18 | 2.60–3.66 | 0.47–0.82 | 2.44–8.68 | 10DL/10GS | |
| cqOC-C2–2 | C02 | 91.10 | 89.17–93.04 | 2.95–3.13 | 0.49–0.76 | 2.76–7.48 | 10DL/10GS | |
| cqOC-C2-3 | C02 | 100.71 | 97.6–103.1 | 3.69768 | 0.5657 | 3.90 | 10DL | |
| cqOC-C2-4 | C02 | 111.08 | 109.45–112.72 | 3.72–2.10 | 0.33–0.53 | 1.95–3.50 | 10DL/09DL | |
| cqOC-C3-1 | C03 | 144.94 | 142.23–147.64 | 2.26–2.74 | 0.37–0.4417 | 2.56–2.20 | 12WH/09DL | |
| cqOC-C3–2 | C03 | 166.11 | 163.56–168.65 | 2.35–5.26 | 0.48–0.90 | 2.24–12.72 | 10GS/11DL | |
| cqOC-C3-3 | C03 | 172.61 | 171.6–173.9 | 4.08611 | 0.8218 | 10.25 | 10GS | |
| cqOC-C3–4 | C03 | 300.48 | 299.84–301.12 | 2.69–8.04 | 0.45–1.052 | 2.52–8.12 | 11WH/10HG/12DL/09DL/10DL/11DL/13DL/13WH | |
| cqOC-C3-5 | C03 | 309.01 | 308.17–309.86 | 3.08–8.01 | 0.56–0.78 | 3.84–8.03 | 10HG/09DL/13DL | |
| cqOC-C3-6 | C03 | 312.33 | 311.35–313.31 | 3.44–7.86 | 0.51–1.20 | 3.21–9.32 | 10DL/13WH/12DL/12WH | |
| cqOC-C3-7 | C03 | 318.41 | 314.17–322.64 | 4.59–6.23 | 0.60–1.09 | 4.92–7.86 | 12DL/13DL/13WH | |
| cqOC-C5-1 | C05 | 71.99 | 58.2–85.8 | 5.66153 | 0.9311 | 10.86 | 10HG | |
| cqOC-C5-2 | C05 | 135.39 | 123.4–136 | 3.82137 | −0.5302 | 3.57 | 10DL | |
| cqOC-C5-3 | C05 | 183.19 | 181.38–184.99 | 2.35–10.20 | 0.45–1.00 | 2.86–10.71 | 09DL/11DL/11WH | |
| cqOC-C5-4 | C05 | 191.08 | 190.1–192.06 | 2.17–10.64 | 0.37–1.02 | 2.09–11.11 | 11DL/09DL/12WH | |
| cqOC-C5-5 | C05 | 220.41 | 219.85–220.98 | 3.83–7.55 | 0.47–0.77 | 3.90–7.64 | 09DL/10DL/11DL/12DL/12WH | |
| cqOC-C5-6 | C05 | 230.24 | 229.22–231.26 | 2.42–7.33 | 0.54–0.76 | 2.3–7.43 | 12WH/09DL/10DL/12DL | |
| cqOC-C5-7 | C05 | 234.02 | 232.79–235.24 | 2.42–7.33 | 0.50–0.75 | 2.36–7.43 | 10DL/11DL | |
| cqOC-C5-8 | C05 | 237.39 | 234.66–240.11 | 3.46–4.34 | 0.53–0.89 | 3.87–5.64 | 13DL/13WH | |
| cqOC-C5-9 | C05 | 250.78 | 249.56–252.01 | 4.02–4.37 | 0.53–0.85 | 3.91–5.03 | 13DL/13WH | |
| cqOC-C5-10 | C05 | 258.99 | 257.20–259.40 | 4.20546 | 0.9034 | 5.68 | 11WH | |
| cqOC-C6-1 | C06 | 23.98 | 15.25–32.72 | 2.66–3.26 | 0.47–0.85 | 2.80–5.03 | 11WH/13DL | |
| cqOC-C6-2 | C06 | 149.34 | 147.65–151.02 | 2.29–4.44 | 0.50–0.83 | 2.79–4.32 | 10HG/13DL/13WH | |
| cqOC-C6-3 | C06 | 159.20 | 152.6–162.4 | 2.03–3.72 | 0.73–1.48 | 3.01–3.67 | 13DL/12DL/13WH | |
| cqOC-C6-4 | C06 | 160.61 | 155.2–169.8 | 3.61–4.08 | 0.60–0.68 | 3.73–4.53 | 10HG/13DL | |
| cqOC-C6-5 | C06 | 181.81 | 180.18–183.45 | 2.98–4.35 | 0.52–0.75 | 2.75–4.58 | 13DL/10DL/11DL | |
| cqOC-C6-6 | C06 | 188.71 | 187.58–188.96 | 2.39–13.53 | 0.37–4.85 | 3.78–22.24 | 12DL/10GS/09DL/10DL/11DL/13DL | |
| cqOC-C6-7 | C06 | 195.61 | 195.2–197 | 5.41 | 0.80 | 5.65 | 11DL | |
| cqOC-C6-8 | C06 | 199.11 | 198.88–199.33 | 5.972–6.45 | −2.052–2.12 | 6.17–6.24 | 10DL/12DL | |
| cqOC-C6-9 | C06 | 201.31 | 200.4–202.3 | 3.98846 | 0.5657 | 3.64 | 13DL | |
| cqOC-C6-10 | C06 | 203.8 | 203.56–204.05 | 3.62–4.98 | −1.99–2.06 | 3.42–4.88 | 11DL/12DL | |
| cqOC-C6-11 | C06 | 207.51 | 206.5–211.6 | 3.46766 | 0.5242 | 3.18 | 13DL | |
| cqOC-C8 | C08 | 66.15 | 62.66–69.65 | 2.03–3.75 | 0.40–0.79 | 1.92–9.55 | 10GS/10DL | |
| cqOC-C9-1 | C09 | 7.39 | 4.08–10.7 | 2.01–6.41 | 0.45–0.68 | 1.98–6.49 | 12WH/10DL/11DL/12DL/09DL | |
| cqOC-C9-2 | C09 | 27.11 | 24.6–29.8 | 3.80835 | 0.5388 | 3.70 | 13DL | |
| SPC | cqPC-A2 | A02 | 0.01 | 0–1.13 | 2.91–4.83 | 0.25–0.47 | 3.42–10.09 | 11GS/12DL |
| cqPC-A3-1 | A03 | 127.01 | 124.80–127.70 | 3.46 | −0.34 | 4.23 | 11WH | |
| cqPC-A3-2 | A03 | 131.81 | 131.30–132.30 | 8.7 | −2.12 | 27.48 | 10GS | |
| cqPC-A3-3 | A03 | 135.51 | 133.00–139.90 | 4.64 | −0.4 | 5.68 | 11WH | |
| cqPC-A3-4 | A03 | 203.51 | 200.58–206.43 | 2.11–7.24 | −0.22–0.58 | 2.65–7.75 | 12DL/13DL/13WH | |
| cqPC-A3-5 | A03 | 216.61 | 214.60–216.90 | 6.64 | 1.95 | 20.53 | 10GS | |
| cqPC-A4-1 | A04 | 14.59 | 13.00–15.50 | 3.74 | −0.44 | 7.65 | 11GS | |
| cqPC-A4-2 | A04 | 55.29 | 52.50–56.40 | 4.7 | −0.53 | 6.19 | 12WH | |
| cqPC-A7 | A07 | 97.71 | 95.80–100.90 | 4 | −1.01 | 4.89 | 09DL | |
| cqPC-A9-1 | A09 | 1.17 | 0–5.36 | 2.10–4.42 | −0.27–0.32 | 2.53–5.23 | 10DL/12DL | |
| cqPC-A9-2 | A09 | 16.4 | 13.47–19.32 | 2.93–4.15 | −0.31–0.53 | 3.82–5.39 | 12WH/10HG/11DL | |
| cqPC-A9-3 | A09 | 23.89 | 20.59–27.18 | 2.14–4.20 | −0.26–0.53 | 2.57–5.44 | 10DL/11DL | |
| cqPC-A9-4 | A09 | 32.01 | 31.20–33.40 | 4.42 | −0.38 | 5.34 | 11WH | |
| cqPC-A9-5 | A09 | 37.31 | 33.4–42.21 | 3.04–4.42 | −0.22–0.38 | 3.01–5.34 | 11WH/13DL | |
| cqPC-C1-1 | C01 | 93.79 | 93.05–94.53 | 2.20–4.58 | −0.19–0.39 | 2.18–5.86 | 13DL/10DL | |
| cqPC-C1-2 | C01 | 152.19 | 150.05–154.33 | 4.02–8.22 | 0.38–1.66 | 5.20–10.36 | 10HG/09DL/13WH | |
| cqPC-C1-3 | C01 | 159.36 | 158.0 0−160.72 | 7.80–4.88 | 0.96–1.59 | 6.26–9.86 | 09DL/13WH | |
| cqPC-C1-4 | C01 | 168.01 | 166.00–176.90 | 3.67 | 0.85 | 4.78 | 13WH | |
| cqPC-C3-1 | C03 | 287.81 | 287.50–288.40 | 3.44 | −0.35 | 4.48 | 11WH | |
| cqPC-C3-2 | C03 | 298.77 | 297.38–300.15 | 7.08–8.27 | −0.39–0.49 | 8.74–8.92 | 11WH/13DL | |
| cqPC-C3-3 | C03 | 303.72 | 302.84–304.59 | 3.767–8.02 | −0.35–0.53 | 4.82–9.82 | 10HG/11WH | |
| cqPC-C3-4 | C03 | 312.51 | 310.1–319.9 | 3.47 | −0.34 | 4.47 | 10HG | |
| cqPC-C5-1 | C05 | 133.89 | 121.3–136 | 4.42 | 1.37 | 5.61 | 10DL | |
| cqPC-C5-2 | C05 | 141.96 | 139.89–144.03 | 4.22–5.33 | −0.25–0.32 | 5.42–6.64 | 10DL/09DL | |
| cqPC-C5-3 | C05 | 225.49 | 221.80–225.90 | 5 | −0.38 | 6.55 | 11DL | |
| cqPC-C5-4 | C05 | 231.59 | 231.20–233.90 | 3.63 | −0.41 | 4.78 | 11DL | |
| cqPC-C5-5 | C05 | 237.39 | 234.67–240.1 | 2.48–4.50 | −0.29–0.6 | 5.86–3.34 | 10DL/12DL/13DL | |
| cqPC-C5-6 | C05 | 252.21 | 251.54–252.89 | 2.20–7.91 | −0.24–0.88 | 2.91–8.51 | 10DL/12DL/13WH/13DL | |
| cqPC-C5-7 | C05 | 260.81 | 260.26–261.37 | 2.68–5.72 | 0.38–1.7758 | 3.41–6.25 | 12DL/13DL/13WH | |
| cqPC-C6-1 | C06 | 54.45 | 50.31–58.58 | 2.08–3.17 | −0.27–0.29 | 2.39–4.03 | 12DL/11WH | |
| cqPC-C6-2 | C06 | 149.41 | 145.01–153.8 | 2.53–4.39 | −0.24–0.42 | 2.87–5.59 | 10DL/12DL | |
| cqPC-C6-3 | C06 | 156.6 | 153.85–159.35 | 2.28–3.24 | −0.34–0.37 | 4.31–5.84 | 10GS/10DL | |
| cqPC-C7 | C07 | 109.84 | 105.01–114.66 | 2.21–4.77 | −0.28–0.32 | 2.78–5.68 | 12DL/11WH | |
| cqPC-C8 | C08 | 15.39 | 11.3–16.7 | 4.41 | 0.28 | 4.62 | 13DL | |
| cqPC-C9-1 | C09 | 2 | 0.0–6.7 | 2.23 | −0.28–0.35 | 2.86–4.45 | 12WH/10DL | |
| cqPC-C9-2 | C09 | 8.12 | 6.47–9.78 | 2.055–2.98 | −0.32–0.34 | 3.88–4.18 | 12WH/11GS | |
| cqPC-C9-3 | C09 | 70.54 | 69.35–71.72 | 2.31–3.09 | −0.23–0.603 | 2.92–3.22 | 13DL/13WH | |
| cqPC-C9-4 | C09 | 77.31 | 74.86–79.75 | 2.41–2.60 | −0.2–0.64 | 2.51–3.28 | 13DL/13WH |
A total of 68 identified QTLs for SPC were observed in 11 microenvironments, which were distributed on 12 chromosomes and accounted for 2.19–27.48% of PV with an average CI of 9.31 cM (Fig. 1 and Table S3). The same results were obtained for SPC-QTLs, most of which (64.71%) were distributed on A03, A09, C03 and C05, with the difference that none of the SPC-QTLs were detected on A08. The identified SPC-QTLs were then integrated into 38 consensus QTLs with a narrower average CI of 4.38 cM (Table S3 and Fig. 1). Twenty-three QTLs could be detected in at least two microenvironments, and cqPC-C5-6 could be identified in four microenvironments, while five QTLs (cqPC-A3-4, cqPC-A9-2, cqPC-C1-2, cqPC-C5-5 and cqPC-C5-7) were identified in three microenvironments. None of the consensus QTLs could be detected in all three macroenvironments, and six and one QTL could be expressed stably at least over 2 years in winter and semi-winter type environments, respectively (Fig. S1b). Four QTLs, cqPC-A2 and cqPC-C1-2, which were repeatedly detected in at least two trials with a medial effect (20% > PV > 10%), and cqPC-A3-2 and cqPC-A3-5, which were detected in one trial but had a large effect (PV > 20%), were also treated as major QTLs.
In addition, as observed from the additive effect (Table 2), the N53-2 alleles in the majority of SOC-QTLs (81.25%) increased SOC, but most SPC-QTLs (81.58%) decreased SPC, consistent with the higher SOC and lower SPC of N53-2 compared with Ken-C8 (Table S1).
The close genetic relationship between SOC and SPC could be confirmed by mapping the co-localized loci. All 11 co-localized QTLs affected both traits and showed opposite additive effects (Table 3), and the favorable alleles of the 10 co-localized QTLs that were derived from the high oil content parent N53-2 could increase SOC and decrease SPC, with only uqC5-1 demonstrating the opposite. Close co-localization explained the high genetically negative correlation between SOC and SPC, and suggested a complexity of the competitive mechanism of these two traits.
Table 3. Unique QTLs controlling both SOC and SPC.
| Unique QTL | Peak | Range | Consensus QTL | Peak | Range | LOD | PV (%) | Additive |
|---|---|---|---|---|---|---|---|---|
| uqA3 | 203.73 | 201.4–206.07 | cqOC-A3-7 | 204.14 | 200.26–208.03 | 2.14–3.59 | 2.08–3.76 | 0.38–0.73 |
| cqPC-A3-4 | 203.51 | 200.58–206.43 | 2.11–7.24 | 2.65–7.75 | −0.22–0.58 | |||
| uqA9-1 | 1.16 | 0.25–2.08 | cqOC-A9-1 | 1.17 | 0.19–2.14 | 2.04–2.69 | 2.14–2.52 | 0.36–0.47 |
| cqPC-A9-1 | 1.17 | 0–5.36 | 2.10–4.42 | 2.53–5.23 | −0.27–0.32 | |||
| uqA9-2 | 23.37 | 22.03–24.7 | cqOC-A9-2 | 23.27 | 21.81–24.73 | 2.19–4.38 | 2.54–4.18 | 0.42–0.57 |
| cqPC-A9-3 | 23.89 | 20.59–27.18 | 2.14–4.20 | 2.57–5.44 | −0.26–0.53 | |||
| uqA9-3 | 31.53 | 30.83–32.22 | cqOC-A9-3 | 31.21 | 30.4–32.2 | 4.21 | 3.88 | 0.53 |
| cqPC-A9-4 | 32.01 | 31.20–33.40 | 4.42 | 5.34 | −0.38 | |||
| uqA9-4 | 34.74 | 32.99–36.48 | cqOC-A9-4 | 34.26 | 32.36–36.17 | 2.45–5.35 | 2.30–5.40 | 0.41–0.72 |
| cqPC-A9-5 | 37.31 | 33.4–42.21 | 3.04–4.42 | 3.01–5.34 | −0.22–0.38 | |||
| uqC3 | 312.33 | 311.37–313.29 | cqOC-C3-6 | 312.33 | 311.35–313.31 | 3.44–7.86 | 3.21–9.32 | 0.51–1.20 |
| cqPC-C3-4 | 312.51 | 310.1–319.9 | 3.47 | 4.47 | −0.34 | |||
| uqC5-1 | 134.75 | 129.97–139.53 | cqOC-C5-3 | 135.39 | 123.4–136 | 3.82 | 3.57 | −0.53 |
| cqPC-C5-1 | 133.89 | 121.3–136 | 4.42 | 5.61 | 1.37 | |||
| uqC5-2 | 237.38 | 235.46–239.31 | cqOC-C5-11 | 237.39 | 234.66–240.11 | 3.46–4.34 | 3.87–5.64 | 0.53–0.89 |
| cqPC-C5-5 | 237.39 | 234.67–240.1 | 2.48–4.50 | 3.34–5.86 | −0.29–0.6 | |||
| uqC6-1 | 149.34 | 147.77–150.92 | cqOC-C6-2 | 149.34 | 147.65–151.02 | 2.29–4.44 | 2.79–4.32 | 0.50–0.83 |
| cqPC-C6-2 | 149.41 | 145.01–153.8 | 2.53–4.39 | 2.87–5.59 | −0.24–0.42 | |||
| uqC6-2 | 157.55 | 155.27–159.83 | cqOC-C6-3 | 159.2 | 152.6–162.4 | 2.03–3.72 | 3.01–3.67 | 0.73–1.48 |
| cqPC-C6-3 | 156.6 | 153.85–159.35 | 2.28–3.24 | 4.31–5.84 | −0.34–0.37 | |||
| uqC9 | 7.97 | 6.49–9.45 | cqOC-C9-1 | 7.39 | 4.08–10.7 | 2.01–6.41 | 1.98–6.49 | 0.45–0.68 |
| cqPC-C9-2 | 8.12 | 6.47–9.78 | 2.055–2.98 | 3.88–4.18 | −0.32–0.34 |
Additionally, some QTL clusters associated with both traits distributed on chromosomes A09, C03 and C05 were also detected. For example, one of these clusters falling in interval from 287.50–322.64 cM on C03 influenced SOC and SPC with consistently negative effects (Figure S2). These results indicated that tight linkage was also an important factor in addition to pleiotropy for their competitive relationship. In addition, there were SOC-QTL clusters, but none of the SPC-QTLs were found on A08, which suggested that more allelic variations control SOC compared with SPC on chromosome A08 between two parents.
BSA analysis based on extreme bulks and confirmation of QTL mapping
Twenty-four DH lines with high SOC (47.2–52.7%) and 24 with low SOC (38.0–42.4%) were selected to prepare two extreme bulks. As a significant negative correlation of SOC and SPC, high and low SOC bulks corresponded to low (19.8–23.9%) and high (22.5–27.2%) SPC, respectively.
Illumina high-throughput sequencing was used to generate 274.08 million and 274.85 million reads from the low and the high SOC bulks, with a coverage of 84.30% (35.85-fold) and 90.74% (39.00-fold) of the reference genome of B. napus, respectively. In contrast, 97.47% (17.16-fold) and 96.56% (19.82-fold) genome coverage was obtained for the two parents, KenC-8 (low SOC and high SPC) and N53-2 (high SOC and low SPC), confirming the accuracy of the subsequent analysis (Table S4). The graph of Δ (SNP-index) was generated (Figure S3) using the genome sequence of KenC-8 as a reference.
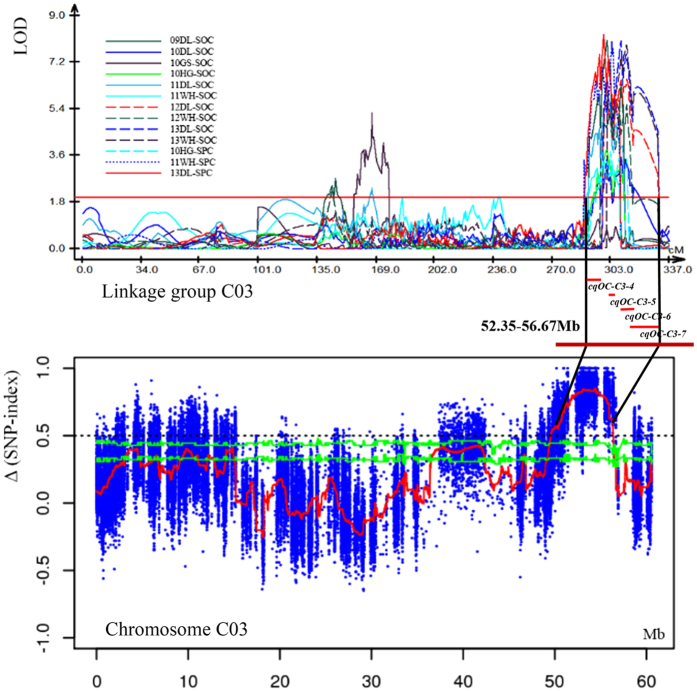
One hundred and thirty-three AGRs underlying SOC were identified on 17 chromosomes (apart from A01 and C07) (Table S5), and 38 AGRs overlapped and/or narrowed the SOC-QTLs from QTL mapping based on the present genetic linkage map (Table S6). The major QTL cqOC-A9-9 was validated by a significant region AGR_A9-8, and the major QTL cqOC-A9-10 was divided into three narrower AGRs (AGR_A9-9, AGR_A9-10 and AGR_A9-11). It is noteworthy that some AGRs with a significantly high Δ (SNP-index) and QTL hotspots were found to co-localize on chromosome A03, A09, C03, and C05 (Table S6). For example, the associated genomic region (AGR_C3-16) was identified to overlap with four SOC-QTLs (cqOC-C3-4, cqOC-C3-5, cqOC-C3-6 and cqOC-C3-7) that could be stably expressed in at least two microenvironments, among which cqOC-C3-4 could be assessed stably in eight microenvironments (Fig. 3).
Figure 3. Overlapping region of AGR and SOC-QTL hotspots on chromosome C03.
Original QTL identification in the different experiments shown by curves above the line of the linkage group (top). The Δ(SNP-index) plot with statistically significantly associated regions (two green significant threshold lines, P < 0.01 and P < 0.05) is drawn at the bottom. The X-axis represents the position of chromosome C03, and the Y-axis represents Δ(SNP-index). An overlapping region of 52.35–56.67 Mb is drawn in the middle, and the small red bars under the linkage group line within QTL hotspots represent consensus QTLs that overlap with AGR.
Identification and analysis of potential candidates related to acyl-lipid metabolism and seed storage underlying SOC and SPC, respectively
Accurate QTLs for SOC were identified using a high-density linkage map and confirmed by BSA. Through comparative mapping between the linkage map and the physical maps of B. napus based on SNP sequence information, a satisfactory colinearity relationship between the genetic linkage map and the reference genome of B. napus was validated (Fig. 1), which could help us to accurately identify genomic regions and the corresponding candidate genes underlying these QTLs. For example, SOC-QTL cqOC-A9-9 could be stably identified in six microenvironments, explaining up to 14.94% of the PV with 0.98 cM of CI, which was mapped within a 0.07 Mb region in the reference genome (Table S7). Finally, 59 genomic regions of SOC-QTLs (88.06%) and 29 genomic regions of SPC-QTLs (76.32%) could be determined accurately (Table S7).
A total of 448 genes involved in 21 of 22 acyl-lipid pathways were identified within 41 SOC-QTL regions (Figures S4 and S5, and Table S8). Additionally, a whole genome search was implemented to determine the distribution of orthologues related to acyl-lipid metabolism, and a total of 3058 orthologues of A. thaliana acyl-related genes were identified in the whole genome (Figures S4 and S5). Minor acyl-lipid related gene enrichment / prevalence in the CI of SOC-QTLs was found compared to the whole genome (Table S9). For example, six orthologues of AtLACS9 that encoded long-chain Acyl-CoA synthetase (plastidial) were distributed on the chromosome of the B. napus genome, four of which displayed SNP/InDel variation in the SOC-QTL regions. Moreover, four of eight orthologues of AtFAD6 that encoded oleate desaturase fell within SOC-QTL regions and exhibited SNP/InDel variation. According to the re-sequencing results of the two parents from BSA, 355 of 449 acyl-lipid related orthologues had SNP/InDel variation between the two parents (Table S10). These genes were involved in 21 of 22 acyl-lipid metabolism pathways. The 25, 13, 24, 20 and 9 genes among them were involved in FA synthesis in plastids, FA elongation, the synthesis of triacylglycerol (TAG), TAG degradation and beta-oxidation, respectively, accounting for 24.20% of the total potential candidates associated with lipid metabolism within SOC-QTL regions (Figure S6 and Table S10). Additionally, a large proportion of genes involved in lipid signaling and cuticular wax syntheses were mapped, accounting for 11.0% and 19.0% of the total acyl-lipid orthologous genes. This finding supported the complexity of the oil accumulation, which was also likely regulated by some genes related to lipid signaling, and the cuticular wax biosynthesis pathway probably also played a potential role in the accumulation of oil during embryonic development.
Two and eleven potential candidates related to oil body structural protein and seed storage protein were identified in 12 SPC-QTL regions (Table S11). BnaA09g02110D, annotated as an orthologue of oleosin 4, was located in an overlapping region of cqPC-A9-2 and cqOC-A9-2. In addition, seven seed storage protein-related orthologues (BnaA09g13220D, BnaA09g11520D, BnaC03g65080D, BnaC05g02160D, BnaC05g44560D, BnaC05g44510D and BnaC06g18820D) co-localized within the SOC and SPC-QTL interval.
It is also noteworthy that some important potential candidates related to acyl-lipid metabolism were identified in overlapping regions of SOC-QTLs and AGRs of BSA (Table S10). BnaA09g48250D, a homologue of AtCAC2 involved in the key steps of de novo FA synthesis, was identified in AGR_A9-12 and the major QTL cqOC-A9-10, and it contained a nonsynonymous SNP in an exon. BnaA08g12780D, corresponding to AtFAD6 encoding oleate desaturase, was located in AGR_A8-4 and cqOC-A8-3. Twenty-two potential candidates related to acyl-lipid metabolism were identified within three major QTLs overlapping with AGRs: cqOC-A9-9 (1), cqOC-A9-10 (14) and cqOC-C5-4(7) (Table S10). Three genes (BnaA09g48250D, BnaC05g21140D and BnaC05g24510D) involved in plastidial FA synthesis, and two genes (BnaA09g51530D and BnaC05g21070D) involved in FA elongation were identified in the these regions.
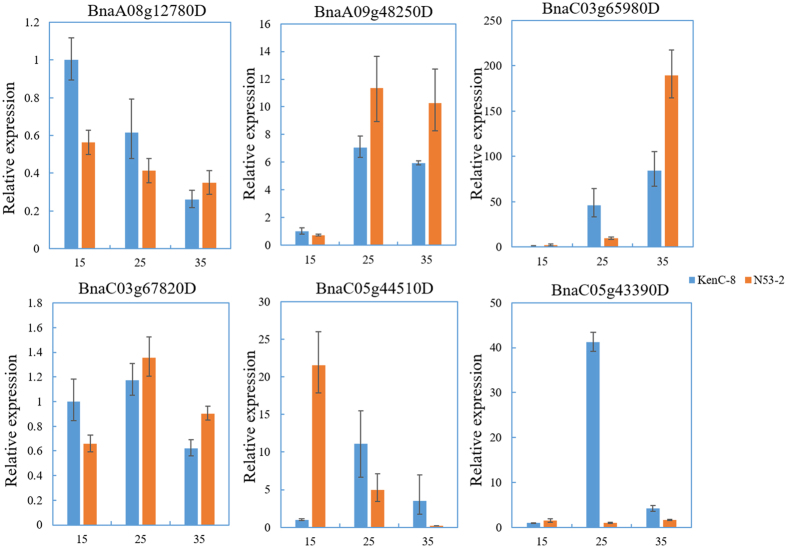
Six potential candidates (BnaA08g12780D, BnaA09g48250D, BnaC03g65980D, BnaC03g67820D, BnaC05g43390D and BnaC05g44510D) that were reported to be involved in acyl-lipid metabolism in B. napus or A. thaliana were selected to measure the expression levels in the developing seed at 15, 25, and 35 days after flowering from both parents. All six genes showed significant expression difference between parents during different developmental periods (Fig. 4). The relative expression level of BnaA09g48250D, which was homologous to the AtCAC2 gene encoding BC (subunit of ACCase), from the overlapping region of AGR_A9-12 and the major QTL cqOC-A9-10 increased significantly in both parents and were clearly higher in N53-2 during the middle and late embryonic development periods. BnaA08g12780D and BnaC03g67820D, which are homologous to AtFAD6 (oleata desaturase), were determined from cqOC-A8-3 and cqOC-C3-7, their relative expression level was reduced but higher in N53-2 during the late developmental period. BnaC03g65980D encoded FAE1 and was involved in FA elongation, showing a rapid increase in expression throughout the process of embryonic development and a significantly higher level in N53-2 than in KenC-8. Of interest, the relative expression level of BnaC05g44510D located in cqOC-C5-7 and involved in cuticular wax synthesis was clearly higher in N53-2 than in KenC-8 but decreased rapidly and was lower in N53-2 during the late embryonic development period. This phenomenon might be associated with the usage of more precursors for the synthesis of TAG in N53-2, especially during late embryonic development.
Figure 4. qRT-PCR results for six potential candidates.
Blue bars represent KenC-8, and orange bars represent N53-2. The X-axis indicates 15, 25, and 35 days after flowering, whereas the Y-axis indicates relative expression levels.
Genetic dissection based on the metabolic pathway and interaction network using potential candidates
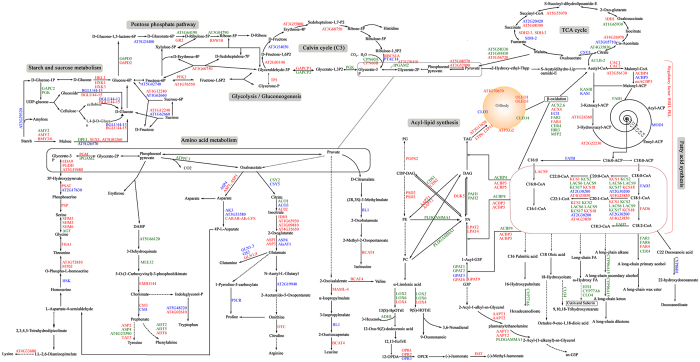
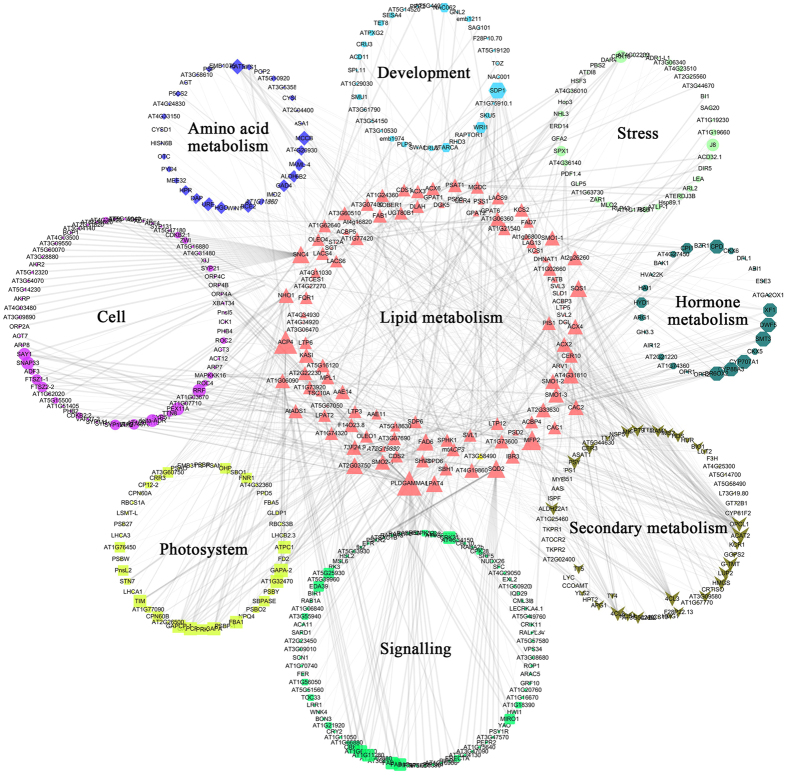
To investigate the genetic basis of oil and protein accumulation in the seeds of B. napus, a magnificent metabolic pathway was constructed with pyruvate as the centric junction based on potential candidates underlying SOC and SPC-QTLs (Fig. 5). The pathway was involved in almost all primary carbohydrate metabolism, including starch and sucrose metabolism, the pentose phosphate pathway, glycolysis/gluconeogenesis, the Calvin cycle, the TCA cycle, FA synthesis, acyl-lipid metabolism, cutin, suberin and wax biosynthesis and amino acid metabolism. There were 167 and 98 orthologues identified in the SOC- and SPC-QTL regions, respectively, which were involved in this enormous pathway, 58 of which could be identified in both traits. IPGAM2, which encodes the 2,3-bisphosphoglycerate-independent phosphoglycerate mutase that catalyzes the formation of glycerate-2P40, was identified in overlapping CIs of qOC-C5-11 for SOC and the QTL qPC-C5-5 for SPC. During glycolysis/gluconeogenesis, some orthologues of key rate-limiting enzymes, such as HKL, PFK, GAPCP and PGK, which are involved in a series of catalyzing the conversion of glucose into pyruvate, were candidates in the co-localized CIs or major QTLs. The Calvin cycle carries out carbon fixation and is an important process for photosynthesis. The TCA cycle is of central importance to many biochemical pathways, and it provides the precursors of certain amino acids and the NADH used in numerous other biochemical reactions through the oxidation of acetyl-CoA derived from carbohydrates, oil and proteins. More than half of the orthologues involved in these two key metabolic pathways could be detected in SOC- and SPC-QTL regions. These results suggested that primary carbohydrate metabolism indeed plays a basic role of providing the initial material for protein synthesis and oil accumulation during embryonic development. To further dissect the mechanism of oil accumulation in seeds, the interaction of potential candidates from SOC-QTL regions was analyzed based on A. thaliana orthologues. The primary results revealed that the whole network incorporated 5817 nodes and 38887 edges (Figure S7). Annotation and clustering elicited an interaction network associated with lipid metabolism (Fig. 6). Some biologic processes, such as protein, secondary metabolism, photosystems, amino acid metabolism, cell size or cycle, and signaling pathways, participated in the regulation of lipid metabolism through some pivotal genes, such as CAC2, SNC4, ACX2 and PLDGAMMA1, which could be expressed during the embryo stage of the plant41.
Figure 5. Metabolic pathway based on potential candidates underlying SOC- and SPC-QTLs.
The metabolic pathway that was constructed based on potential candidates within QTL regions of SOC and SPC. Red letters indicate orthologues underlying the SOC-QTL regions, blue letters indicate orthologues underlying the SPC-QTL regions, and green letters indicate that these orthologues could be identified in the genomic regions of both SOC- and SPC-QTLs. The broken arrows indicate that there are many steps between the two compounds.
Figure 6. Interaction network associated with lipid metabolism of potential candidates underlying SOC-QTLs.
Genes associated with special types of metabolisms and pathways are shown using special shapes with special colors.
Discussion
Most studies investigating important agronomic traits in B. napus were conducted with lower-density genetic maps6,7,8,9,10,11 and some with high-density linkage maps25,26,29, but QTL analyses of SOC and SPC simultaneously based on high-density linkage maps have not been reported. In this study, we constructed a high-density integrated genetic linkage map with 3106 SNP and 101 non-SNP (SSR and STS) markers covering 3072.7 cM of the KNDH population. The release of the B. napus genome and the application of SNP markers with known sequence information facilitated alignment of the genetic linkage and physical maps of B. napus. Thus, a high-density SNP map was very helpful for the characterization and comparison of QTLs that control important agronomic traits, and the identification of candidate genes and genomic research. The present study allowed us to obtain a high-density KN linkage map that enabled us to localize QTLs more effectively and accurately. Wang et al.39 mapped 24 consensus QTLs associated with SOC integrated from 63 identified QTLs in eight microenvironments using the KNDH population from a previous study39. Here, 67 SOC-QTLs integrated from 164 identified QTLs were mapped, and ten new SOC-QTLs were contributed due to 4 new microenvironments (Table 2). The main cause of the increased QTL number detected in the present map was the higher map density rather than the addition of new environmental data (the number of consensus QTLs increased from 24 to 67, only 10 of which were contributed by new environmental data).
Molecular-assisted breeding is the primary objective for detecting genetic variation in important agronomic traits. This will help us to identify stable QTLs for molecular-assisted selection (MAS) to compare previously published QTLs for oil content in other segregating populations (with different genetic backgrounds) in our current study. This method has been reported in many studies42,43. To validate the consistency and availability of the QTLs detected using the present high-density map for MAS, we compared SOC-QTLs with the published QTL map for oil content in other segregating populations with different genetic backgrounds. Twenty-six SOC-QTLs were found to overlap with loci checked by other populations (Table S12). The major QTL, cqOC-A9-9, was also detected and overlapped with qOC-A9-4-TN in the TN population, which was used to identify the seed-oil content and erucic acid content QTLs44, and the QTLs for many other important agronomic traits45,46,47. Additionally, qOC-A3-4, which could be stably expressed in 8 microenvironments, could also be identified to the co-localized locus in the TN population. These QTLs are very stable and useful for further MAS analyses. It is interesting that some QTLs detected in other populations were identified and divided into multiple QTLs. For example, qOC-A8-1-TN and qOC-A8-RNSL from TN and RNSL, respectively, were overlapping, and both of them were found to cover 5 QTLs in the present study. This result also suggested that the present genetic linkage map had a better resolution than the maps used in previous reports.
The recently proposed NGS-based BSA approach is a cost-effective and rapid method of trait mapping supported by the identification of the target genomic region in rice33. A large number of AGRs were identified rapidly, and significant AGRs with a high Δ (SNP-index) were consistent with the SOC-QTL hotspots, confirming the accuracy of the QTL mapping based on the present map. In addition, we found that highly significant AGRs were subdivided into multiple QTLs (Table S6) rather than being subdivided or narrowed QTLs as previously reported34,35,48,49. These results suggested that the present high-density linkage map had sufficiently high resolution to dissect genetic variation in primary mapping populations.
The application of a high-density linkage map and satisfying linear relationship between the genetic and physical map in our study could be helpful to more accurately identify candidate genes underlying SOC and SPC. A large number of potential candidates involved in acyl-lipid pathways and seed storage were identified within SOC and SPC-QTL regions, respectively (Tables S10 and S11). Additionally, for the most important candidates of SOC, a whole genome search was conducted, followed by a search for enrichment/prevalence in the QTL confidence intervals compared to the whole genome. Minor acyl-lipid related gene enrichment/prevalence was found in the CIs of SOC-QTLs, which indicated that the important genes clustered around QTLs and were consistent with the frequently identified QTL hotspots in the present study. Six potential candidates related to acyl-lipid metabolism were selected as representatives to validate their expression levels between the two parents. The relative differences in the expression of five genes showed a relationship with the SOC phenotype of the two parents in developing seed. The results provided clues for exploring the casual genes underlying SOC. Single gene mutations may affect the expression of some genes in the common metabolic network. Consequently, further identification of the network is necessary, and simultaneously the rapid capture of causal genes requires the use of transcription and fine mapping methods50.
In the present study, the simultaneous identification of QTLs for SPC together with SOC using the high-density linkage map revealed that some QTLs affected both SOC and SPC in an opposite manner (Table 3). SOC and SPC, which have a negative correlation, are controlled by many genes with additive and epistatic effects19,23,51,52. Oil and protein are synthesized in the endoplasmic reticulum (ER) during the same period of seed development53, which suggests that these pathways interfere and/or compete with one another for ER resources. TAG synthesis competes with seed storage protein synthesis due to sucrose utilization, and the reduction of one product would result in higher levels of the other. Pyruvate, which can be converted into carbohydrates via gluconeogenesis, to FAs or energy through acetyl-CoA and amino acids, is a key intersection in the network of metabolic pathways and unites several key metabolic processes. Here, we constructed a magnificent primary carbohydrate pathway in the center of pyruvate using potential candidates underlying SOC- and SPC-QTLs (Fig. 5). A. thaliana orthologues involved in this pathway provide more information to identify the interference and/or competition mechanism from a metabolic perspective. In addition, some genes associated with cuticular waxes, cutin and suberin have also been observed, such as CER4, CYP86, ACC1 and MAH1, and they probably affect seed storage triacylglycerides through competition with the common fatty acid pool54,55. Interestingly, most orthologous genes involved in long chain fatty acid synthesis, especially C20:X and C22:X, fell within SOC-QTL regions. These results were in agreement with the findings of Gu et al.56, in which long chain FAs had a significant and positive relationship with oil content.
Oil and protein biosynthesis during embryo development is a complex and hierarchical biological process that is regulated by some transcription factors and complex gene interaction networks57,58. Therefore, it is not inadequate to dissect the mechanism responsible for oil and protein accumulation in seeds only through primary carbohydrate metabolic pathways. In A. thaliana, metabolic pathways associated with the biosynthesis and degradation of acyl-lipids have been summarized, and most of the genes have been described38. The construction of an interaction network provided new insights into the interaction among related genes (Fig. 6). It has been reported that the yellow seeded Brassica genotypes contained greater oil, higher protein and lower fiber contents than the seeds of the black/brown seeded genotypes59. In plants, flavonoids are the major secondary metabolites involved in the Brassica seed coat pigmentation process60. The orthologues TT4 and TT5, which were classified into secondary metabolism pathways, were involved in flavonoid biosynthetic processes and regulated lipid metabolic pathways by interacting with CAC2 (Fig. 6). In addition, the result verified by the cross-talk between the lipids and potential flavonoid metabolism were driven by the finding that acetyl-CoA acts as a substrate for both pathways61. In addition, 4CL family proteins associated with lignin biosynthesis are linked to lipid metabolism through many nodes, which could help to explain the association with negative additive effects of QTLs for seed oil and lignin content (unpublished data). These results not only enhance our understanding of the complex regulatory mechanisms of oil accumulation but also serve as a basis for follow-up to identify causal loci together with transcriptome and genome-wide association analyses62,63,64.
Materials and Methods
Plant material and trait measurement
The segregating double haploid (DH) population, named KN, was used in this experiment. The KNDH population was derived from a cross between the parental lines KenC-8 with low oil content and N53–2 with high oil content, which was first constructed by Wang et al.39. A total of 300 lines of the KNDH population were implemented in the genotype and phenotype analyses in this study.
The field experiments were conducted in 3 macroenvironments as described by Wang et al.39: Dali of Shaanxi Province (DL), Wuhan and Huanggang of Hubei Province (WH and HG) and Sunan of Gansu Province (GS), which were winter, semi winter and spring-type rapeseed planting areas. Year–location combinations were treated as microenvironments. The materials were planted in Dali, Wuhan, Sunan and Huanggang for, successively, six, three, two and one year, respectively. Four types of microenvironmental data were added in this experiment apart based on a report by Wang et al.39. In total, there were 12 microenvironments. The SOC data from 2012 and 2013 in DL and WH, excluding the report by Wang et al.39, were collected. The SPC data were collected in all microenvironments except 08DL. The trial locations of WH and HG were the experiment bases of Huazhong University of Science and Technology (Wuhan), and DL and GS were the experiment bases of the Hybrid Rapeseed Research Center of Shaanxi Province. No specific permissions were required for the field trials. The field experiment was the same as reported by Wang et al.39.
Open-pollinated seeds were collected for the SOC and SPC measurements. The SOC data in the first eight trials (08DL, 09DL, 10DL, 10HG, 10GS, 11DL, 11GS, 11WH) were measured by nuclear magnetic resonance (NMR) according to Burns et al. (2003) with modifications6. The SOC data for the other four trials and all SPC data were measured by near-infrared reflectance spectroscopy as described by Gan et al.65.
SNP marker analysis
Whole-genomic DNA was extracted from young seedlings using the CTAB method66. A total of 300 DH lines with two parental lines were successfully genotyped using the Brassica 60 K Infinium HD Assay SNP arrays according to the manufacturer’s protocol provided by Illumina Inc. (http://www.illumina.com/). Imaging of the arrays after BeadChip washing and coating was performed using an Illumina HiSCAN scanner. Allele calling data for each probe was exported using Genome Studio genotyping software v2011 (Illumina Inc.). SNP data filtering was accomplished according to Zhang et al.26. Furthermore, SNPs with identical genotypes across the KNDH population were classified into a bin using Perl language based on a close linkage according to Zhang et al.26, and the first SNP of each bin was selected as its name.
Linkage map construction, QTL analysis and integration
The construction of genetic linkage groups, QTL analysis and integration were performed as described by Wang et al.39. Non-SNP (SSR, STS, and SRAP) markers were used to construct the primary linkage map used by Wang et al.39 and also applied to construction of the map, together with the SNP-bin markers. Carthagene software was used to construct the high-density linkage map67. Windows QTL Cartographer 2.5 software was used to detect putative QTLs with a CIM68. For the CIM model, the scan walking speed was 2 cM, and the window size was 10 cM with five background cofactors. The LOD threshold (3.2–3.5) for the detection of significant QTLs was calculated by a 1,000-permutation test based on a 5% experiment-wise error rate. To avoid missing QTLs with small genetic effects, QTLs that appeared repeatedly in at least two microenvironments, below the LOD threshold (3.2–3.5) but above 2.0, were considered micro-real QTLs according to the definition of Long et al.46. All these QTLs, including significant and micro-real ones, were called identified QTLs69. The identified QTLs were named by combining the microenvironment and the chromosome number, e.g., qOC-10DL13-1. The identified QTLs detected in different trials with overlapping CIs were then integrated into consensus QTLs using the BioMercator V4.2 program70. If an identified QTL had no overlapping CIs with others, then it was also regarded as a consensus QTL. Consensus QTLs were designated by the initial letters “cq”, such as cqOC-A1-1. The consensus QTLs with overlapping CIs for SOC and SPC were integrated into unique QTLs, and unique QTLs were named by the initial letter “uq”, for example, uq-A9-1. The method used for QTL nomenclature and integration were as described by McCouch et al.71 and Wang et al.72.
Comparative analysis of the genetic map and physical map, and the identification of genomic regions corresponding to CIs of QTLs and potential candidates underlying QTLs
The method used to identify the homologous locus of SNP probes in B. napus reference genomes was described according to Cai et al.73. The sequences of the 50-mer probes provided by Illumina Inc. were used as queries to search for homologous loci using the NCBI-Blastn local program against the B. napus “Darmor-bzh” reference genomes (http://www.genoscope.cns.fr/brassicanapus/data), as described by Altschul et al.74. The E-value threshold was set to 1e-10. All SNPs assigned to the linkage map were used to validate the alignment relationship of the genetic map and physical map. If an SNP was mapped to multiple loci in the B. napus reference genome, only the location that corresponded to the particular linkage group of the locus was selected30. Additionally, if a SNP had multiple homologous loci on the same chromosome, an accurate position for a particular locus was determined manually by referring to the physical positions of its upstream and downstream SNPs. Genome regions corresponding to the CIs of consensus QTLs, herein denoted QTL regions, were defined through both closely linked SNPs within QTL CIs and co-linearity between the linkage map and the reference genome. Genes that fell within QTL regions were regarded as candidates. According to the BSA re-sequencing, alleles that were within QTL regions and had SNP or InDel variations in introns, exons or within 1 kb up- and downstream between two parents were considered potential candidates, and otherwise were excluded.
The genomic regions of QTLs from other populations, DY (‘Darmor-bzh’ × ‘Yudal’, RNSL (‘Rapid’ × ‘NSL96/25’)75, Z5 (‘zy036’ × ‘51,070’)76, SG (‘Sollux’ × ‘Gaoyou’)77, TN (‘Tapidor’ × ‘Ningyou7’)42, SO (‘Sansibar’ × ‘Oase’)78, PT (‘Polo’ × ‘Topas’)79, and M201 × M20280, were obtained via electronic PCR (e-PCR)81, which was performed with the primer sequences of the molecular markers flanking the QTL confidence intervals using the genomic sequences of ‘Darmor-bzh’ as templates.
BSA sequencing and AGR analysis
DNA was extracted from individuals in the KNDH population with extremely high and low SOC (24 DNA samples for each extreme trait) and bulked equally to generate high and low extreme bulks. Four libraries, including two parents (KenC-8 and N53-2), were constructed and then sequenced using the Illumina HiSeq2500 platform at the Novogene Bioinformatics Institute, and 125-bp paired-end reads were generated with an insert size of approximately 350 bp. Short reads obtained from both parents and two DNA bulks were aligned against the B. napus “Darmor-bzh” reference genomes to obtain the consensus sequence using BWA software82. Reads of high and low SOC bulks were separately aligned to the KenC-8 (low SOC parent) consensus sequence reads to call the SNPs. Variant calling was performed for all samples using the Unified Genotyper function in the GATK software83. SNPs were used in the Variant Filtration parameter in GATK. Homozygous SNPs between two parents were extracted, and the read depth information for homozygous SNPs in the above bulks was acquired to calculate the SNP Index33. The Δ (SNP-index) was calculated based on subtraction of the low SOC bulk from the high SOC bulk SNP-index. The average SNP-index of SNPs in a certain genomic interval was calculated using a sliding window analysis with a 1-Mb window size and 10-kb step size, and the SNP-index graphs and corresponding Δ (SNP-index) graphs were plotted. The genomic regions with a significant Δ (SNP-index) (P < 0.05) were identified as AGRs.
Quantitative PCR (qPCR) analysis of 6 acyl-lipid-related potential candidates
To investigate the potential candidates, we used qRT-PCR for six potential candidates related to acyl-lipids from different associated loci. RNAs were extracted and quantified from developing B. napus seeds at 15, 25 and 35 days after flowering (DAF), following the user manual for the RNA prep Pure Plant Kit (TOYOBO, DP441). cDNA was synthesized from 2 mg of total RNA using a cDNA Synthesis Kit. Expression analysis was performed with the SYBR premix EX TaqTM kit (TaKaRa, Japan) using an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Framingham, USA). For each reaction, three technical replicates were evaluated. The primers of the six target genes and reference gene Actin are listed in Table S13.
The metabolic network and interaction analysis of potential candidates
The orthologues of potential candidates in QTL regions and their annotation were obtained by BLASTn based on the A. thaliana database (http://www.arabidopsis.org/). The same method was used for the whole genome search of acyl-lipid-related orthologues in B. napus. Potential candidates were clustered and analyzed using Mapman software84 according to their orthologues and annotation. The orthologues involved in acyl-lipid metabolism were acquired from the Acyl-Lipid Metabolism chapter from The Arabidopsis Book38 and the ARALIP website (http://aralip.plantbiology.msu.edu/). The interaction was analyzed and constructed using the online software String (http://string-db.org/), clustered again using Mapman software and finally visualized using Cytoscape_V3.2.185. All the interaction and cluster and analyses were based on A. thaliana orthologues.
Additional Information
How to cite this article: Chao, H. et al. Genetic dissection of seed oil and protein content and identification of networks associated with oil content in Brassica napus. Sci. Rep. 7, 46295; doi: 10.1038/srep46295 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors are very grateful to Xiangwei Wu, Beijing igene Code Biotech Co., Ltd. and Qingyong Yang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University for his support with the partial data analysis. The work was supported by the National Key Research and Development Program (2016YFD0100200), National Natural Science Foundation of China (31171582, 31071447) and the New Century Talents Support Program of the Ministry of Education of China (NCET110172).
Footnotes
The authors declare no competing financial interests.
Author Contributions H.B. Chao performed the SNP genotyping, B.S.A.-related experiment and data analysis for the overall study, and wrote the manuscript. H. Wang, W.G. Zhao and B.J. Li implemented part of the field trials and provided most of the phenotypic data. X.D. Wang provided some guidelines for the Q.T.L. analysis and revised the manuscript. L.X. Guo and J.W. Gu performed the data processing, detection and revised the manuscript. N. Raboanatahiry provided the comparative analysis with other populations and helped to modify the language used in the manuscript. D.Y. Chen helped to modify some figures. M.T. Li designed, led and coordinated the overall study. All authors read and approved the final manuscript.
References
- Nagaharu U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jap J Bot 7, 389 (1935). [Google Scholar]
- Meyer M. Rapeseed oil fuel-the crisis-proof home-made eco-fuel. Agrarforschung 16, 262 (2009). [Google Scholar]
- Leckband G., Frauen M. & Friedt W. NAPUS 2000. Rapeseed (Brassica napus) breeding for improved human nutrition. Food Res Int 35, 273 (2002). [Google Scholar]
- Wanasundara J. P. Proteins of Brassicaceae oilseeds and their potential as a plant protein source. Crit Rev Food Sci 51, 635 (2011). [DOI] [PubMed] [Google Scholar]
- Si P., Mailer R. J., Galwey N. & Turner D. W. Influence of genotype and environment on oil and protein concentrations of canola (Brassica napus L.) grown across southern Australia. Crop Pasture Sci 54, 397 (2003). [Google Scholar]
- Burns M. J. et al. QTL analysis of an intervarietal set of substitution lines in Brassica napus: (i) Seed oil content and fatty acid composition. Heredity 90, 39 (2003). [DOI] [PubMed] [Google Scholar]
- Delourme R. et al. Genetic control of oil content in oilseed rape (Brassica napus L.). Theor Appl Genet 113, 1331 (2006). [DOI] [PubMed] [Google Scholar]
- Qiu D. et al. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet 114, 67 (2006). [DOI] [PubMed] [Google Scholar]
- Yan X. Y. et al. Co-location of seed oil content, seed hull content and seed coat color QTL in three different environments in Brassica napus L. Euphytica 170, 355 (2009). [Google Scholar]
- Chen G. et al. Identification of QTL for oil content, seed yield, and flowering time in oilseed rape (Brassica napus). Euphytica 175, 161 (2010). [Google Scholar]
- Sun M. et al. Design of new genome- and gene-sourced primers and identification of QTL for seed oil content in a specially high-oil Brassica napus cultivar. Plos One 7, e47037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. et al. Molecular mapping of Arabidopsis thaliana lipid-related orthologous genes in Brassica napus. Theor Appl Genet 124, 407 (2012). [DOI] [PubMed] [Google Scholar]
- Jiang C. et al. Quantitative trait loci that control the oil content variation of rapeseed (Brassica napus L.). Theor Appl Genet 127, 957 (2014). [DOI] [PubMed] [Google Scholar]
- Liu S. et al. A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor Appl Genet 129, 1203 (2016). [DOI] [PubMed] [Google Scholar]
- Zhao J., Becker H. C., Zhang D., Zhang Y. & Ecke W. Conditional QTL mapping of oil content in rapeseed with respect to protein content and traits related to plant development and grain yield. Theor Appl Genet 113, 33 (2006). [DOI] [PubMed] [Google Scholar]
- Wurschum T., Liu W., Maurer H. P., Abel S. & Reif J. C. Dissecting the genetic architecture of agronomic traits in multiple segregating populations in rapeseed (Brassica napus L.). Theor Appl Genet 124, 153 (2012). [DOI] [PubMed] [Google Scholar]
- Schatzki J., Ecke W., Becker H. C. & Mollers C. Mapping of QTL for the seed storage proteins cruciferin and napin in a winter oilseed rape doubled haploid population and their inheritance in relation to other seed traits. Theor Appl Genet 127, 1213 (2014). [DOI] [PubMed] [Google Scholar]
- Teh L. & Mollers C. Genetic variation and inheritance of phytosterol and oil content in a doubled haploid population derived from the winter oilseed rape Sansibar × Oase cross. Theor Appl Genet 129, 181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskandari M., Cober E. R. & Rajcan I. Genetic control of soybean seed oil: II. QTL and genes that increase oil concentration without decreasing protein or with increased seed yield. Theor Appl Genet 126, 1677 (2013). [DOI] [PubMed] [Google Scholar]
- Mao T. et al. Identification of quantitative trait loci underlying seed protein and oil contents of soybean across multi-genetic backgrounds and environments. Plant Breeding 132, 630 (2013). [Google Scholar]
- Stefansson B. R. & Kondra Z. P. Tower summer rape. Can J Plant Sci 55, 343 (1975). [Google Scholar]
- Grami B., Stefansson B. R. & BAKER R. J. Genetics of protein and oil content in summer rape: heritability, number of effective factors, and correlations. Can J Plant Sci 57, 937 (1977). [Google Scholar]
- Röbbelen G. & Thies W. Biosynthesis of seed oil and breeding for improved meal quality. Brassica crops and wild allies: biology and breeding. Japan Scientific Societies Press, Tokyo 285 (1980). [Google Scholar]
- Mahmood T., Rahman M. H., Stringam G. R. et al. Identification of quantitative trait loci (QTL) for oil and protein contents and their relationships with other seed quality traits in Brassica juncea. Theor Appl Genet 113, 7 (2006). [DOI] [PubMed] [Google Scholar]
- Liu L. et al. A high-density SNP map for accurate mapping of seed fibre QTL in Brassica napus L. Plos One 8, e83052 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. et al. A high-density genetic map identifies a novel major QTL for boron efficiency in oilseed rape (Brassica napus L.). Plos One 9, e112089 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. et al. Genome wide analysis of flowering time trait in multiple environments via high-throughput genotyping technique in Brassica napus L. Plos One 10, e119425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzig S. V. et al. Genome-wide association mapping unravels the genetic control of seed germination and vigor in Brassica napus. Front Plant Sci 6, 221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. High-density SNP map construction and QTL identification for the apetalous character in Brassica napus L. Front Plant Sci 6, 1164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. et al. Linkage and regional association analysis reveal two new tightly-linked major-QTLs for pod number and seed number per pod in rapeseed (Brassica napus L.). Sci Rep 5, 14481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick M., Long Y., Meng J. & Bancroft I. Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnol J 7, 334 (2009). [DOI] [PubMed] [Google Scholar]
- Chalhoub B. et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950 (2014). [DOI] [PubMed] [Google Scholar]
- Takagi H. et al. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. The Plant J 74, 174 (2013). [DOI] [PubMed] [Google Scholar]
- Lu H. et al. QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor Appl Genet 127, 1491 (2014). [DOI] [PubMed] [Google Scholar]
- Das S. et al. Genome-wide insertion-deletion (InDel) marker discovery and genotyping for genomics-assisted breeding applications in chickpea. DNA Resv20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illa-Berenguer E., Van Houten J., Huang Z. & van der Knaap E. Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq. Theor Appl Genet 128, 1329 (2015). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. Integration of linkage maps for the Amphidiploid Brassica napus and comparative mapping with Arabidopsis and Brassica rapa. BMC Genomics 12, 101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y. et al. Acyl-lipid metabolism. Arabidopsis Book 11, e161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. Identification of QTLs associated with oil content in a high-oil Brassica napus cultivar and construction of a high-density consensus map for QTLs comparison in B. napus. Plos One 8, e80569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. & Assmann S. M. The glycolytic enzyme, phosphoglycerate mutase, has critical roles in stomatal movement, vegetative growth, and pollen production in Arabidopsis thaliana. J Exp Bot 62, 5179 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M. et al. A gene expression map of Arabidopsis thaliana development. Nat Genet 37, 501 (2005). [DOI] [PubMed] [Google Scholar]
- Jiang Congcong et al. Quantitative trait loci that control the oil content variation of rapeseed (Brassica napus L.). Theor Appl Genet 127, 57 (2014). [DOI] [PubMed] [Google Scholar]
- Liu Sheng et al. A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor Appl Genet 129, 1203 (2016). [DOI] [PubMed] [Google Scholar]
- Qiu D. et al. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet 114, 67 (2006). [DOI] [PubMed] [Google Scholar]
- Feng Ji et al. Characterization of metabolite quantitative trait loci and metabolic networks that control glucosinolate concentration in the seeds and leaves of Brassica napus. New Phytologist 193, 96 (2012). [DOI] [PubMed] [Google Scholar]
- Long Y. et al. Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics 177, 2433 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Jiaqin et al. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182, 851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Vikas K. et al. QTL‐seq for rapid identification of candidate genes for 100‐seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnol J(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W., Flowers J. M., Sahraie D. J. et al. Extreme QTL mapping of germination speed in Arabidopsis thaliana. Mol Ecol 25, 4177 (2016). [DOI] [PubMed] [Google Scholar]
- Hua Yingpeng et al. Transcriptomics-assisted quantitative trait locus fine mapping for the rapid identification of a nodulin 26-like intrinsic protein gene regulating boron efficiency in allotetraploid rapeseed. Plant, cell & environment 39, 1601 (2016). [DOI] [PubMed] [Google Scholar]
- Gül M. QTL-Kartierung und Analyse von QTL-x-Stickstoff-Interaktionen beim Winterraps (Brassica napus L.). Cuvillier(2002). [Google Scholar]
- Zhao J. QTLs for oil content and their relationships to other agronomic traits in an European × Chinese oilseed rape population. Diss. Grorg-Agust University of Goettingen, Germany(2002). [Google Scholar]
- Basnet R. K. et al. Genome-wide analysis of coordinated transcript abundance during seed development in different Brassica rapa morphotypes. BMC Genomics 14, 840 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L. & Samuels L. Plant cuticles shine: advances in wax biosynthesis and export. Curr opin Plant Biol 12, 721 (2009). [DOI] [PubMed] [Google Scholar]
- Lü S. et al. The glossyhead1 allele of ACC1 reveals a principal role for multidomain acetyl-coenzyme a carboxylase in the biosynthesis of cuticular waxes by Arabidopsis. Plant Physiol 157, 1079 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J. et al. Identification of the relationship between oilbody morphology and oil content by microstructure comparison combining with QTL analysis in Brassica napus. Front Plant Sci 7, 1989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. & McCarty D. R. Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol 11, 548 (2008). [DOI] [PubMed] [Google Scholar]
- Santos Mendoza M. et al. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54, 608 (2008). [DOI] [PubMed] [Google Scholar]
- Lovell J. T. et al. Exploiting Differential gene expression and epistasis to discover candidate genes for drought-associated QTLs in Arabidopsis thaliana. Plant Cell 27, 969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley B. W. Flavonoids in seeds and grains: physiological function, agronomic importance and the genetics of biosynthesis. Seed Sci Res 8, 415 (1998). [Google Scholar]
- Okazaki Y. et al. A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21, 892 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B. et al. Expression quantitative trait locus mapping across water availability environments reveals contrasting associations with genomic features in Arabidopsis. Plant Cell 25, 3266 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. et al. Loci and candidate gene identification for resistance to Sclerotinia sclerotiorum in soybean (Glycine max L. Merr.) via association and linkage maps. Plant J 82, 245 (2015). [DOI] [PubMed] [Google Scholar]
- Wen W. et al. Combining quantitative genetics approaches with regulatory network analysis to dissect the complex metabolism of the maize kernel. Plant Physiol 170, 136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L. et al. Establishment of math models of NIRS analysis for oil and protein contents in seed of Brassica napus (in Chinese with an English abstract). Scientia Agricultura Sinica 36, 1609 (2002). [Google Scholar]
- Porebski S., Bailey L. G. & Baum B. R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15, 8 (1997). [Google Scholar]
- De Givry S., Bouchez M., Chabrier P., Milan D. & Schiex T. Carh ta Gene: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21, 1703 (2005). [DOI] [PubMed] [Google Scholar]
- Zeng Z. Precision mapping of quantitative trait loci. Genetics 136, 1457 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. et al. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182, 851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcade A. et al. BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20, 2324 (2004). [DOI] [PubMed] [Google Scholar]
- McCouch S. R. et al. Report on QTL nomenclature. Rice Genet Newsl 14, 11 (1997). [Google Scholar]
- Wang X. et al. Quantitative trait loci analysis and genome-wide comparison for silique related traits in Brassica napus. BMC Plant Biol 16, 71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G. et al. A complex recombination pattern in the genome of allotetraploid Brassica napus as revealed by a high-density genetic map. Plos One 9, e109910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delourme R. et al. Genetic control of oil content in oilseed rape (Brassica napus L.). Theor Appl Genet 113, 1331 (2006). [DOI] [PubMed] [Google Scholar]
- Sun M. et al. Design of new genome-and gene-sourced primers and identification of QTL for seed oil content in a specially high-oil Brassica napus cultivar. Plos One 7, e47037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Qi L., Zhang X. et al. Characterization of the quantitative trait locus OilA1 for oil content in Brassica napus. Theor Appl Genet 126, 2499 (2013). [DOI] [PubMed] [Google Scholar]
- Teh L. & Möllers C. Genetic variation and inheritance of phytosterol and oil content in a doubled haploid population derived from the winter oilseed rape Sansibar × Oase cross. Theor Appl Genet 129, 181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed N. et al. Identification of QTL influencing seed oil content, fatty acid profile and days to flowering in Brassica napus L. Euphytica 207, 191 (2016). [Google Scholar]
- Cheng X. et al. Identification of quantitative trait loci associated with oil content and development of near isogenic lines for stable qOC-A10 in Brasscia napus L. Can J Plant Sci(2016). [Google Scholar]
- Schuler Gregory D. Sequence mapping by electronic PCR. Genome research 7, 541 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O. et al. Mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37, 914 (2004). [DOI] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.