Abstract
Simian immunodeficiency viruses (SIVcpz) infecting chimpanzees (Pan troglodytes) in west central Africa are the closest relatives to all major variants of human immunodeficiency virus type 1 ([HIV-1]; groups M, N and O), and have thus been implicated as the source of the human infections; however, information concerning the prevalence, geographic distribution, and subspecies association of SIVcpz still remains limited. In this study, we tested 71 wild-caught chimpanzees from Cameroon for evidence of SIVcpz infection. Thirty-nine of these were of the central subspecies (Pan troglodytes troglodytes), and 32 were of the Nigerian subspecies (Pan troglodytes vellerosus), as determined by mitochondrial DNA analysis. Serological analysis determined that one P. t. troglodytes ape (CAM13) harbored serum antibodies that cross-reacted strongly with HIV-1 antigens; all other apes were seronegative. To characterize the newly identified virus, 14 partially overlapping viral fragments were amplified from fecal virion RNA and concatenated to yield a complete SIVcpz genome (9,284 bp). Phylogenetic analyses revealed that SIVcpzCAM13 fell well within the radiation of the SIVcpzPtt group of viruses, as part of a clade including all other SIVcpzPtt strains as well as HIV-1 groups M and N. However, SIVcpzCAM13 clustered most closely with SIVcpzGAB1 from Gabon rather than with SIVcpzCAM3 and SIVcpzCAM5 from Cameroon, indicating the existence of divergent SIVcpzPtt lineages within the same geographic region. These data, together with evidence of recombination among ancestral SIVcpzPtt lineages, indicate long-standing endemic infection of central chimpanzees and reaffirm a west central African origin of HIV-1. Whether P. t. vellerosus apes are naturally infected with SIVcpz requires further study.
Chimpanzees (Pan troglodytes) are dispersed across tropical sub-Saharan Africa and are currently classified into four subspecies on the basis of genetic differences in mitochondrial DNA (mtDNA) sequences (3, 8, 10, 16). These include the western Pan troglodytes verus, the Nigerian Pan troglodytes vellerosus, the central Pan troglodytes troglodytes, and the eastern Pan troglodytes schweinfurthii. Thus far, infection with simian immunodeficiency virus of chimpanzees (SIVcpz) has only been documented in members of the P. t. troglodytes and P. t. schweinfurthii subspecies (Table 1 and references therein). The western subspecies does not seem to be naturally infected with SIV since testing of over 1,500 P. t. verus apes failed to uncover evidence of SIVcpz infection (23, 25; W. M. Switzer, B. Prakesh, V. Shanmugam, V. Bhullar, S. Phillips, T. M. Folks, J. Ely, and W. Heneine, Abstr. 9th International Workshop on HIV Dynamics and Evolution, Lake Arrowhead, Calif., abstr. 43, 2002). The infection status of P. t. vellerosus remains unknown, because only very few such apes have been screened for SIVcpz (4). A single SIVcpz-positive P. t. vellerosus (CAM4) has been reported, but this ape is believed to have acquired his infection in captivity from a naturally infected P. t. troglodytes cage mate (4).
TABLE 1.
Captive and wild-living chimpanzees with documented SIVcpz infection
| Ape | History | Yeara | Ageb | Sexc | Countryd | Locale | Subspeciese | Diagnosisf | SIVcpz strain | Seqg | Current status | Remarks | No. of infected animals/no. tested (%)i | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAM13 | Wild-born | 2001 | <2 | M | Cameroon | Littoral province | P.t.troglodytes | Plasma Ab | CAM13 | FL | Deceased, 2002 | Died of massive parasite infection | 1/39 (2.6)j | This study |
| CAM3 | Wild-born | 1992 | 1 | M | Cameroon | Dja Forest Reserve | P.t.troglodytes | Plasma Ab | CAM3 | FL | Deceased, 1998 | Died of pneumonia | 2/29 (6.9) | 4, 17 |
| CAM5 | Wild-born | 1998 | <1 | F | Cameroon | Central province | P.t.troglodytes | Plasma Ab | CAM5 | FL | Deceased, 1998 | Died of severe diarrhea | 2/29 (6.9) | 4, 17 |
| Marilyn | Wild-born | N/A | N/A | F | N/A | N/A | P.t.troglodytes | Plasma Ab | US | FL | Deceased 1985 | Died in childbirth at age of ≈26 | 1/98 (1) | 7 |
| GAB1 | Wild-born | 1987 | 2-3 | F | Gabon | Oyem region | P.t.troglodytes | Plasma Ab | GAB1 | FL | Deceased 1990 | Died of unknown causes | 2/50 (4) | 13, 19 |
| GAB2 | Wild-born | 1988 | 2 | F | Gabon | Macatamagoye village | P.t.troglodytes | Plasma Ab | GAB2 | P | Deceased 1988 | Died of wounds inflicted by hunters | 2/50 (4) | 14, 19 |
| CAM4 | Wild-born | 1993 | 2.5 | M | Cameroon | Southwest province | P.t.vellerosus | Plasma Ab | CAM4 | P | Alive | Presumed recipient of cage transmission | N/A | 4 |
| Ch-No | Wild-born | 1987 | 2-3 | M | DRC | N/A | P.t.schweinfurthii | Plasma Ab | ANT | FL | Alive | Confiscated en route to Belgium | 1/44 (2.2) | 20, 29 |
| DRC1 | Wild-living | 2003 | N/A | N/A | DRC | Parisi Forest, Kisangani | P.t.schweinfurthii | Fecal vRNA | DRC1 | P | N/A | Nonhabituatedh | N/A | 30 |
| DRC2 | Wild-living | 2000 | N/A | N/A | DRC | Wanie-Rukula Forest, Kisangani | P.t.schweinfurthii | Urine Ab | N/A | N/A | N/A | Nonhabituatedh | N/A | 30 |
| Ch-06 | Wild-living | 2000 | 23 | M | Tanzania | Kasekela, Gombe | P.t.schweinfurthii | Urine Ab | TAN1 | FL | Alive | Habituated; in good health | 2/51 (5)k | 25, 27 |
| Ch-30 | Wild-living | 2001 | 32 | F | Tanzania | Kasekela, Gombe | P.t.schweinfurthii | Urine Ab | N/A | N/A | Alive | Habituated; presumed dead of unknown causes | 2/51 (5)k | 26 |
| Ch-37 | Wild-living | 2000 | 29 | M | Tanzania | Mitumba, Gombe | P.t.schweinfurthii | Urine Ab | N/A | N/A | Deceased, 2000 | Habituated; died of unknown causes | 2/15 (17)k | 26 |
| Ch-45 | Wild-living | 2002 | 26 | M | Tanzania | Mitumba, Gombe | P.t.schweinfurthii | Fecal vRNA | TAN3 | P | Alive | Habituated; in good health | 2/15 (17)k | 26 |
| Ch-64 | Wild-living | 2001 | N/A | F | Tanzania | Kalande, Gombe | P.t.schweinfurthii | Fecal vRNA | TAN2 | P | N/A | Nonhabituatedh | 3/>10 (30)k | 26 |
| Ch-70 | Wild-living | 2002 | N/A | F | Tanzania | Kalande, Gombe | P.t.schweinfurthii | Fecal vRNA | TAN4 | P | N/A | Nonhabituatedh | 3/>10 (30)k | 26 |
| Ch-71 | Wild-living | 2002 | N/A | F | Tanzania | Kalande, Gombe | P.t.schweinfurthii | Fecal vRNA | TAN5 | P | N/A | Nonhabituatedh | 3/>10 (30)k | 26 |
Year of rescue of captive chimpanzees; year of SIVcpz detection in wild-living chimpanzees. N/A, not available.
Estimated age (in years) of captive chimpanzees at the time of rescue; age of wild-living (habituated) chimpanzees at the first time point of SIVcpz detection.
M, male; F, female; N/A, not available; Ch-64, Ch-70, and Ch-71 were identified as females by genetic typing of fecal DNA (26).
Country of capture or home range of wild communities. DRC, Democratic Republic of Congo.
Determined by mitochondrial (mt)DNA analysis (7).
Test performed to diagnose SIVcpz infection. Ab, antibody; vRNA, virion RNA.
SIVcpz sequence information available; FL, full-length sequence; P, partial sequence(s).
Member of nonhabituated community with individual identification performed by mtDNA analysis (26, 30).
Total number of SIVcpz infected chimpanzees per number of chimpanzees screened in study in which the infected ape was identified.
An additional 32 P.t.vellerosus apes were also tested and found negative.
SIVcpz prevalence estimate for entire community (26).
All known SIVcpz strains cluster in two divergent phylogenetic lineages, termed SIVcpzPtt and SIVcpzPts, according to their subspecies of origin (4, 7, 17, 25, 26, 27, 29, 30). The human AIDS viruses (human immunodeficiency virus type 1 [HIV-1] groups M, N, and O) fall into just one of these lineages, i.e., that from P. t. troglodytes, which has implicated the central subspecies as the source of the human infections (7, 11, 17). However, the number of available, fully characterized SIVcpz strains is still rather limited. Only five SIVcpzPtt strains have been reported, all of which were derived from young orphans tested in captivity (Table 1). In contrast, 10 cases of SIVcpzPts infection have been documented in eastern chimpanzees, 9 of which were identified in wild-living apes by using noninvasive detection methods (Table 1). Phylogenetic analyses comparing the more recently characterized SIVcpzPts sequences to those of SIVcpzPtt and HIV-1 have reaffirmed the conclusion that P. t. schweinfurthii apes are not the source of the human AIDS pandemic (26, 30). However, additional SIVcpz strains, especially from chimpanzee subspecies native to west central Africa, are necessary to gain further insight into the chimpanzee reservoir that gave rise to HIV-1.
Cameroon is home to two different chimpanzee subspecies, the Nigerian P. t. vellerosus in the north and the central P. t. troglodytes in the south, with the Sanaga River believed to form the boundary between their habitats (Fig. 1). In this study, we screened 71 chimpanzees (34 females and 37 males) that were captured as infants when their family members were killed by hunters for bushmeat. All of these apes were rescued by the Ministry of Environment and Forestry and placed in zoos and sanctuaries. Sixty-four were less than 8 years of age at the time of rescue (confiscated between December 1998 and March 2002), while the remaining seven represented older individuals (confiscated prior to 1998). Blood samples were obtained by venipuncture and processed into plasma and peripheral blood mononuclear cells (PBMCs) (these studies were carried out in strict accordance with international guidelines for the ethical scientific use and human care of primates in research). DNA was extracted from uncultured PBMCs by using the QIAamp blood kit (QIAGEN, Valencia, Calif.) and subjected to PCR analysis by using primers designed to amplify a 498-bp mitochondrial (D loop) DNA region (7). Phylogenetic analysis of these mtDNA sequences identified 39 of the captive chimpanzees as members of the P. t. troglodytes and 32 as members of the P. t. vellerosus subspecies (E. Nerrienet, unpublished data).
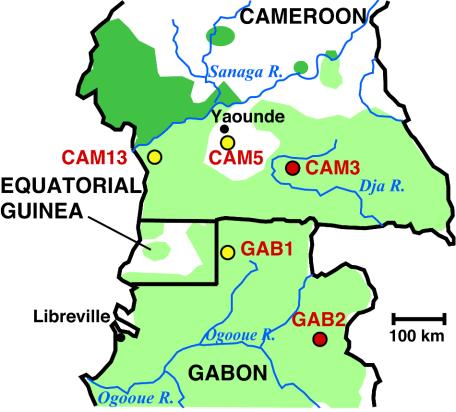
FIG. 1.
Geographic origin of SIVcpz-infected chimpanzees. The rescue location of CAM13 is shown in relation to the geographic origin of four previously reported SIVcpz-infected chimpanzees from southern Cameroon (CAM3 and CAM5) and northern Gabon (GAB1 and GAB2) (4, 19). Red dots indicate known capture locations (CAM3 and GAB2), while yellow dots indicate sites where chimpanzees were first identified in captivity (CAM13, CAM5, and GAB1). The natural ranges of the P. t. troglodytes and P. t. vellerosus subspecies are highlighted in light green (south of the Sanaga River) and dark green (north of the Sanaga River), respectively (major rivers are indicated).
Chimpanzee plasma samples were screened by using two commercially available (HIV-1 antigen-based) enzyme-linked immunosorbent assays (Genscreen Plus version 2 [Bio-Rad, Marnes la Coquette, France] and Wellcozyme, HIV recombinant [Murex Biotech Limited, Rungis, France]). This analysis identified one male infant (CAM13) as harboring antibodies that were strongly cross-reactive with HIV-1 antigens. Blood samples from all other chimpanzees were antibody negative. The seropositive ape was classified as a central chimpanzee (P. t. troglodytes) by mtDNA analysis (Fig. 2), consistent with his capture location south of the Sanaga River (Table 1). Western blot analysis (LAV Blot HIV1/2 Kit; Bio-Rad) confirmed the presence of plasma antibodies that cross-reacted with HIV-1 Env (gp160, gp120, and gp41), polymerase (p66 and p51), and core (p55 and p24) proteins (Fig. 3). A virus isolate was established from CAM13 by coculture of his phytohemagglutinin-stimulated PBMCs with normal human donor lymphocytes, with viral replication monitored by p24 antigen production (Genetic Systems HIV-1 antigen enzyme immunoassay; Bio-Rad) in culture supernatants (data not shown). PCR amplification of subgenomic pol (246 bp) and gp41 (420 bp) fragments confirmed infection with a new SIVcpz strain (data not shown).
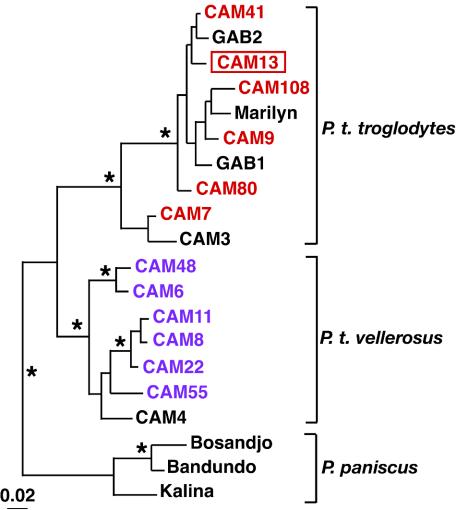
FIG. 2.
Subspecies origin of captive chimpanzees. Chimpanzees were classified as members of the central (P. t. troglodytes) or the Nigerian (P. t. vellerosus) subspecies by mtDNA analysis (7). Representative sequences from newly characterized P. t. troglodytes and P. t. vellerosus apes (GenBank accession no. AY126678 through AY126696) are highlighted in red and magenta, respectively (previously reported mtDNA sequences are shown in black); the SIVcpz-infected ape CAM13 is boxed. A 498-bp (D loop) fragment was amplified from fecal DNA. Nucleotide sequences were aligned by using CLUSTAL W (28); the gap-stripped alignment contained 391 sites. The tree was obtained by the Bayesian method (implemented in MRBAYES) (12) by using the general reversible model of evolution with a gamma distribution of rates across sites. The scale bar indicates 0.02 substitutions per site. Asterisks denote branches with estimated posterior probabilities of 95% or higher. The tree was rooted with human sequences.
FIG. 3.
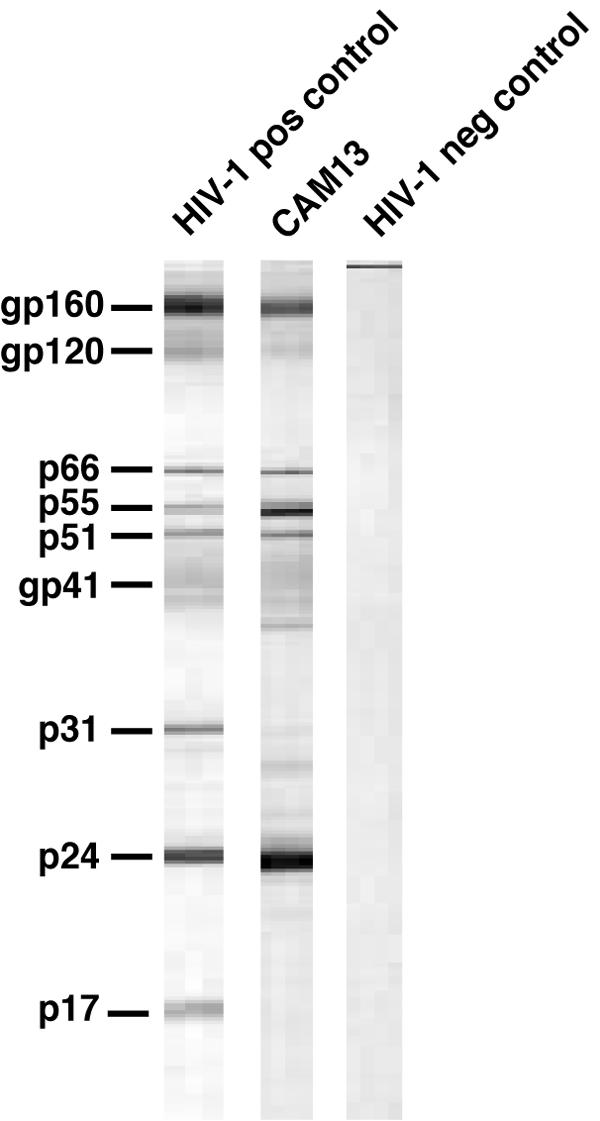
SIVcpz-specific antibodies in chimpanzee plasma. A plasma sample from chimpanzee CAM13 was tested for HIV-1 cross-reactive antibodies by commercial (HIV-1 antigen-based) Western immunoblotting. Molecular weights of HIV-1-specific proteins are indicated. The banding patterns of plasma samples from HIV-1-infected and noninfected humans are shown for positive and negative controls, respectively.
Due to transport restrictions of blood and blood-derived products from endangered primate species (Convention for the International Trade of Endangered Species), fecal samples were collected from CAM13 and sent from Cameroon to the University of Alabama of Birmingham for full-length SIVcpz sequence determination. A similar approach was previously used successfully to derive the full-length sequence of a SIVcpzPts strain infecting a wild chimpanzee in Gombe National Park (27). Briefly, fecal samples were collected on two different occasions (2 March and 5 June 2001), resuspended in an equal volume of RNAlater (Ambion, Austin, Tex.), and shipped at room temperature (RNAlater preserves nucleic acids, allowing storage and shipment at ambient temperature without virion RNA degradation). Total RNA was extracted by using an RNAqueous Midi-Kit (Ambion) and subjected to reverse transcriptase PCR (RT-PCR) amplification by using diagnostic SIVcpz consensus pol and gp41/nef primer pairs (26). Amplification products were sequenced (Fig. 4, fragments 1 and 2), and then used to design SIVcpzCAM13 strain-specific primers. These were used in combination with upstream and downstream consensus primers to amplify the remainder of the SIVcpzCAM13 genome. The order and position of 14 individual amplification products are shown in Fig. 4.
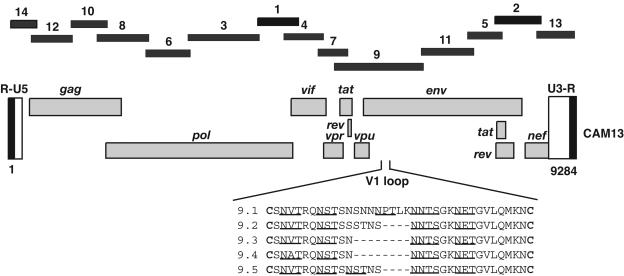
FIG. 4.
Amplification of a complete SIVcpzCAM13 genome from fecal RNA. Partially overlapping subgenomic fragments were amplified from fecal viral RNA by using RT-PCR approaches and a combination of strain-specific and consensus primer pairs. The relative positions of the fragments with respect to the SIVcpzCAM13 genome, along with their order of amplification, are shown. Fragment lengths are drawn to scale; the entire SIVcpzCAM13 sequence (R-U5-gag-pol-env-U3-R) is 9,284 bp in length. Fragment 9 exhibited V1 env sequence length heterogeneity between different quasi-species members (predicted N-linked glycosylation sites are underlined). Five clones (9.1 to 9.5) were sequenced, from which clone 9.1 was chosen for the assembly of the full-length sequence (GenBank accession number AY169968).
All RT-PCR products (ranging in length between 470 and 1,520 bp) were directly sequenced, except for fragments 13 and 14 which were subcloned prior to sequence analysis to obtain sequences in the R region of the long terminal repeat. Fragment 9 was also subcloned since chromatograms from direct sequence analysis could not be resolved. This was due to length variation among quasi-species members in a region corresponding to the first hypervariable loop (V1) of the extracellular envelope (gp120) domain (Fig. 4). Clone 9.1 was arbitrarily selected for the compilation of the full-length SIVcpzCAM13 sequence. In regions of fragment overlap, the 5′ sequence was arbitrarily chosen. The concatenated SIVcpzCAM13 sequence (R-U5-gag-pol-env-U3-R) is 9,284 nucleotides in length.
Inspection of the deduced protein sequences of SIVcpzCAM13 revealed uninterrupted open reading frames for gag, pol, vif, vpr, tat, rev, vpu, env, and nef genes. Regulatory elements in the long terminal repeat, including the predicted binding sites for NF-κB and other transcription factors, the transactivation response element, the TATAA box, and the polyadenylation signal, were also intact. The extracellular envelope domain (gp120 region) of SIVcpzCAM13 contained 18 cysteine residues and 24 potential N-linked glycosylation sites at locations similar to those observed for other SIVcpz isolates (2). As described previously for other SIVcpz strains (11), the V3 loop of the SIVcpzCAM13 gp120 was also highly conserved, differing at only 8 sites from the consensus of the other six known SIVcpz sequences, including both SIVcpzPtt and SIVcpzPts strains (the other six differed from this consensus at 3 to 10 sites). Finally, there were no obvious inactivating mutations in regions of known protein function, such as the YMDD motif in Pol or the PT/SAP domain in Gag p6.
To compare SIVcpzCAM13 to previously characterized SIVcpzPtt and SIVcpzPts strains, we performed diversity plot analyses of concatenated protein sequences (data not shown). Interestingly, SIVcpzCAM13 was most similar to SIVcpzGAB1 from northern Gabon, differing at less than 10% of the residues in the Pol protein. Two other SIVcpzPtt strains from southern Cameroon, SIVcpzCAM3 and SIVcpzCAM5, were considerably more distant, differing from SIVcpzCAM13 at 16 to 17% of the amino acids in Pol. The strains most distant from SIVcpzCAM13 were the two P. t. schweinfurthii viruses SIVcpzANT and SIVcpzTAN1, which differed at up to 30% of the sites in Pol. There was no evidence of recent recombination between SIVcpzCAM13 and other SIVcpz strains.
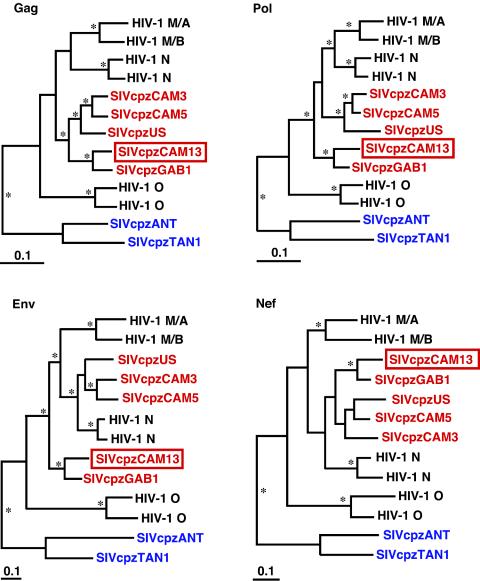
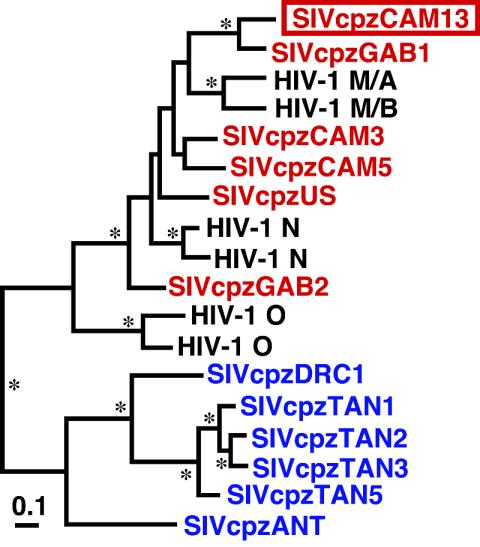
To determine the relationship of SIVcpzCAM13 to other SIVcpz and HIV-1 strains, evolutionary trees were constructed for Gag, Pol, Env, and Nef polypeptides (Fig. 5). Using Bayesian phylogenetic inference, we found that SIVcpzCAM13 was indeed a member of the SIVcpzPtt group of viruses. In all coding regions, SIVcpzCAM13 fell well within the SIVcpzPtt radiation, clustering most closely with SIVcpzGAB1. SIVcpz strains CAM3, CAM5, and US clustered separately, comprising a second lineage within the SIVcpzPtt radiation. Both the CAM13/GAB1 and the CAM3/CAM5/US lineages clustered with HIV-1 groups M and N, forming a monophyletic clade in all four trees, which was supported by high posterior probability values for the two longer regions, Pol and Env (Fig. 5). HIV-1 group O viruses did not fall within this same cluster but formed a closely related sister clade. Analysis of SIVcpzCAM13 thus confirmed and extended the previously described sequence relationships of SIVcpzPtt and HIV-1 (7, 11, 17, 26, 30).
FIG. 5.
Phylogenetic trees depicting the relationship of SIVcpzCAM13 to other HIV-1 and SIVcpz isolates. SIVcpz strains from P. t. troglodytes are shown in red, while those from P. t. schweinfurthii are shown in blue; the new SIVcpzCAM13 is boxed. Deduced amino acid sequences of SIVcpzCAM13 were compared with the sequences of SIVcpzGAB1 (GenBank accession number X52154), SIVcpzUS (AF103818), SIVcpzCAM3 (AF115393), and SIVcpzCAM5 (AJ271369) and of HIV-1 group N strains YBF30 (AJ006022) and YBF106 (AJ271370), group O strains ANT70 (L20587) and MVP5180 (L20571), and group M subtypes A (strain U455; M62320) and B (LAI; K02013). Sequences were aligned by using CLUSTAL W (28), with minor manual adjustment. Trees were estimated by the Bayesian method (implemented in MRBAYES [12]), by using the JTT matrix of amino acid replacement (15) and a gamma distribution of rates across sites and then midpoint rooted. Asterisks denote branches with estimated posterior probabilities of at least 95%. The scale bars indicate 0.1 replacements per site.
Although all strains of SIVcpzPtt clustered with HIV-1 groups M and N in all coding regions, inspection of the tree topologies revealed significant differences in the order of divergence of the four main lineages within this group (Fig. 5). For example, the two groups of SIVcpzPtt clustered together in the Gag and Nef trees, but in the Pol and Env trees the GAB1/CAM13 lineage lay as an outgroup to a clade containing the other SIVcpzPtt lineage plus the two HIV-1 groups; this difference was strongly statistically supported in the Gag and Env trees. Furthermore, HIV-1 group N clustered with HIV-1 group M in Gag and Pol trees but with the US/CAM3/CAM5 lineage of SIVcpzPtt in the Env tree, confirming previous results obtained when fewer SIVcpz strains were available (7, 17). Such discordant branching orders are indicative of ancient recombination during the divergence of these four lineages, presumably at a time when the ancestors of the two HIV-1 groups were still infecting chimpanzees. These recombination events would have required the coinfection of individual apes with divergent strains of SIVcpz, implying that such strains were cocirculating in at least some chimpanzee populations.
Partial gp41/Nef sequences are available for one additional SIVcpzPtt strain from Gabon (GAB2), as well as four SIVcpzPts strains from near Kisangani in the Democratic Republic of Congo (DRC1) and from Gombe National Park in Tanzania (TAN2, TAN3, and TAN5) (26, 30; F. Bibollet-Ruche and B. H. Hahn, unpublished results). The phylogenetic tree of these gp41/Nef sequences revealed no correlation between the various SIVcpzPtt strains and their geographic origin in southern Cameroon or northern Gabon, respectively (Fig. 6). By contrast, the four SIVcpzPts strains from Gombe all clustered tightly and were quite distinct from SIVcpzDRC1 from Kisangani. Indeed, the divergences of SIVcpzPtt strains from within Cameroon (CAM13 versus CAM3 or CAM5) or within Gabon (GAB1 versus GAB2), in each case involving viruses obtained from apes captured less than 200 km apart, were as high as the divergences seen between SIVcpzPts strains from Kisangani and Gombe, which are separated by at least 800 km. This lack of intermixing among divergent SIVcpzPts strains from different geographic regions could be the result of limited sampling. Alternatively, since the Gombe viruses are from a small and isolated area (26), their lack of diversity could be the result of a founder effect. However, given the recent finding that SIVcpz originated from recombination between two SIVs infecting prey monkey species in west central Africa (1), it is tempting to speculate that the pattern of diversity and recombination among P. t. troglodytes strains is a reflection of the evolutionary history of SIVcpz; that is, central chimpanzees have likely been infected for a longer period of time, and perhaps at higher prevalence rates, than eastern chimpanzees, who seem to have acquired SIVcpz relatively more recently.
FIG. 6.
Phylogenetic relationships of partial gp41/Nef protein sequences from SIVcpz and HIV-1. See legend of Fig. 5 for details and methods. The additional sequences (with GenBank accession numbers) are SIVcpz strains GAB2 (unpublished), DRC1 (AY542959), TAN2 (AY181991), TAN3 (AY181992), and TAN5 (AY181993).
The new SIVcpzPtt strain reported here came from an infant chimpanzee wild-caught in Cameroon. This chimpanzee is the third seropositive ape from a total of 54 P. t. troglodytes tested in Cameroon since surveys of rescued orphans began in the late 1990s (this study and reference 4 and 17). While the overall prevalence of SIVcpz infection in this group of apes is low (3 of 54) compared to other naturally infected primate species (5, 21), it is similar to that reported previously for wild-caught chimpanzees in Gabon (2 of 50) (Table 1). By contrast, none of the 46 P. t. vellerosus apes screened thus far exhibited evidence of natural SIVcpz infection (this study and reference 4). Although the difference in infection rates between these two subspecies is not statistically significant (P > 0.10; Fisher's exact test), the data suggest that SIVcpz infection of P. t. vellerosus, if it occurs, is rare. Nonetheless, the possibility that Nigerian chimpanzees are naturally infected with SIVcpz cannot be excluded at the present time. The example of CAM4 clearly demonstrates that P. t. vellerosus apes are susceptible to SIVcpzPtt infection (4). Moreover, the Sanaga River, although a significant biogeographical barrier, is not an absolute hindrance to chimpanzee movement. Chimpanzee gene flow across the Sanaga River has been reported (6), indicating that SIVcpz could have similarly spread from P. t. troglodytes to P. t. vellerosus (or from P. t. vellerosus to P. t. troglodytes) in the past. Additional studies of wild chimpanzee communities on both sides of the Sanaga River will be necessary to address this possibility.
The finding of SIVcpzCAM13 in a wild-caught infant chimpanzee also suggests that SIVcpz, like HIV-1 and other primate lentiviruses, is transmitted vertically from infected mothers to their infants. Chimpanzees are not sexually active before the age of 8 years, and biting injuries of infants are extremely rare (9). Since CAM13, CAM3, CAM5, GAB1, and GAB2 were all less than 3 years of age at the time of their capture, mother-to-infant transmission is by far the most likely explanation of their infection. Among HIV-1-infected women in sub-Saharan Africa, only about 20% transmit their virus to their infants in the absence of antiretroviral therapy, and mother-to-infant transmission of HIV-2 has been estimated at 4% (18, 22, 24). Vertical transmission among naturally infected African green monkeys and sooty mangabeys is also rare (5, 21). Thus, the SIVcpz infection rates observed here and in previous studies of captive apes (19, 20) are unlikely to accurately reflect SIVcpz infection rates in wild-living adult chimpanzee populations.
In conclusion, we report here a third SIVcpzPtt strain from a wild-caught P. t. troglodytes ape from Cameroon. Phylogenetic analysis of its sequence revealed the coexistence of multiple diverse SIVcpzPtt lineages in southern Cameroon and northern Gabon. The close phylogenetic relationships of these viruses to HIV-1 groups M, N and O provide further evidence for a west central African origin of the human AIDS viruses. It remains to be determined to what extent wild-living central chimpanzees in Cameroon are infected with SIVcpz. Moreover, SIVcpz strains clustering specifically with HIV-1 group O strains have yet to be identified, and the infection status of P. t. vellerosus requires further investigation. Thus, surveys of wild chimpanzee populations in Cameroon and neighboring countries will need to be conducted. The derivation here of a second full-length SIVcpz sequence from fecal RNA reemphasizes the utility of noninvasive methods for characterizing SIVcpz infections, both in captivity and in the wild.
Nucleotide sequence accession number.
The nucleotide sequence of SIVcpzCAM13 has been deposited in the GenBank database under accession number AY169968. Newly derived mtDNA sequences are available under accession numbers AY126678 through AY126696.
Acknowledgments
We thank the Cameroonian Wildlife Aid Fund, the Limbe Wildlife Foundation, the Pandrillus Organization, and the Ministry of Environment and Forestry of Cameroon for logistical support in obtaining specimens from captive chimpanzees; we also thank Martine Peeters (University of Montpellier, France) for information on the capture location of chimpanzees GAB1 and GAB2, Frederic Bibollet-Ruche and Brandon Keele for helpful discussions, Maria Salazar for technical assistance, and Wendy J. Abbott for manuscript preparation.
This work was supported in part by grants from the National Institutes of Health (R01 AI50529, R01 AI 58715, R01 AI 44596, NO1 AI85338, and P30 AI 27767), the Howard Hughes Medical Institute, and the Centre Pasteur du Cameroun.
REFERENCES
- 1.Bailes, E., F. Gao, F. Bibollet-Ruche, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. 2003. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 2.Bibollet-Ruche, F., E. Bailes, F. Gao, X. Pourrut, K. L. Barlow, J. P. Clewley, J. Mwenda, D. K. Langat, G. K. Chege, H. M. McClure, E. Mpoudi-Ngole, E. Delaporte, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2004. New simian immunodeficiency virus infecting De Brazza's monkeys (Cercopithecus neglectus): evidence for a Cercopithecus monkey virus clade. J. Virol. 78:7748-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butynski, T. M. 2003. The robust chimpanzee Pan troglodytes: taxonomy, distribution, abundance, and conservation status, p. 5-12. In R. Kormos, C. Boesch, M. I. Bakarr, and T. M. Butynski (ed.), West African chimpanzees: status survey and conservation action plan. IUCN, Gland, Switzerland.
- 4.Corbet, S., M. C. Muller-Trutwin, P. Versmisse, S. Delarue, A. Ayouba, J. Lewis, S. Brunak, P. Martin, F. Brun-Vezinet, F. Simon, F. Barre-Sinoussi, and P. Mauclere. 2000. Env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J. Virol. 74:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fultz, P. N., T. P. Gordon, D. C. Anderson, and H. M. McClure. 1990. Prevalence of natural infection with simian immunodeficiency virus and simian T-cell leukemia virus type I in a breeding colony of sooty mangabey monkeys. AIDS 4:619-625. [DOI] [PubMed] [Google Scholar]
- 6.Gagneux, P., M. K. Gonder, T. L. Goldberg, and P. A. Morin. 2001. Gene flow in wild chimpanzee populations: what genetic data tell us about chimpanzee movement over space and time. Philos. Trans. R. Soc. Lond. B 356:889-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 8.Gonder, M. K., J. F. Oates, T. R. Disotell, M. R. J. Forstner, J. C. Morales, and D. J. Melnick. 1997. A new west African chimpanzee subspecies? Nature 388:337. [DOI] [PubMed] [Google Scholar]
- 9.Goodall, J. 1986. The Chimpanzees of Gombe: patterns of behavior. Belknap Press, Cambridge, Mass.
- 10.Groves, C. P. 1993. Order primates, p. 243-277. In D. E. Wilson and D. M. Reader (ed.), Mammalian species of world: a taxonomic and geographic reference, 2nd ed., vol. 2. Smithsonian Institution Press, Washington, D.C.
- 11.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-617. [DOI] [PubMed] [Google Scholar]
- 12.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic tress. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 13.Huet, T., R. Cheynier, A. Meyerhans, G. Roelants, and S. Wain-Hobson. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature (London) 345:356-359. [DOI] [PubMed] [Google Scholar]
- 14.Janssens, W., K. Fransen, M. Peeters, L. Heyndrickx, J. Motte, L. Bedjabaga, E. Delaporte, P. Piot, and G. van der Groen. 1994. Phylogenetic analysis of a new chimpanzee lentivirus SIVcpzGAB2 from a wild-captured chimpanzee from Gabon. AIDS Res. Hum. Retrovir. 10:1191-1192. [DOI] [PubMed] [Google Scholar]
- 15.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 16.Morin, P. A., J. J. Moore, R. Chakraborty, L. Jin, J. Goodall, and D. S. Woodruff. 1994. Kin selection, social structure, gene flow, and the evolution of chimpanzees. Science 265:1193-1201. [DOI] [PubMed] [Google Scholar]
- 17.Müller-Trutwin, M. C., S. Corbet, S. Souquière, P. Roques, P. Versmisse, A. Ayouba, S. Delarue, E. Nerrienet, J. Lewis, P. Martin, F. Simon, F. Barré-Sinoussi, P. Mauclère. 2000. SIVcpz from a naturally infected Cameroonian chimpanzee: biological and genetic comparison with HIV-1 N. J. Med. Primatol. 29:166-172. [DOI] [PubMed] [Google Scholar]
- 18.O'Donovan, D., K. Ariyoshi, P. Milligan, M. Ota, L. Yamuah, R. Sarge-Njie, H. Whittle, et al. 2000. Maternal plasma viral RNA levels determine marked differences in mother-to-child transmission rates of HIV-1 and HIV-2 in The Gambia. AIDS 14:441-448. [DOI] [PubMed] [Google Scholar]
- 19.Peeters, M., C. Honore, T. Huet, L. Bedjabaga, S. Ossari, P. Bussi, R. W. Cooper, and E. Delaporte. 1989. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS 3:625-630. [DOI] [PubMed] [Google Scholar]
- 20.Peeters, M., K. Fransen, E. Delaporte, M. Van den Haesevelde, G.-M. Gershy-Damet, L. Kestens, G. van der Groen, and P. Piot. 1992. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS 6:447-451. [DOI] [PubMed] [Google Scholar]
- 21.Phillips-Conroy, J. E., C. J. Jolly, B. Petros, J. S. Allan, and R. C. Desrosiers. 1994. Sexual transmission of SIVagm in wild grivet monkeys. J. Med. Primtol. 23:1-7. [DOI] [PubMed] [Google Scholar]
- 22.Pitt, J., J. Goldfarb, M. Schluchter, A. Kovacs, E. Cooper, D. Hodes, K. McIntosh, H. Peavy, and W. Shearer for the Pediatric Pulmonary and Cardiovascular Complications of Vertically Transmitted HIV Infection Study Group. 1997. HIV vertical transmission rate determinations are subject to differing definitions and therefore different rates. J. Clin. Epidemiol. 51:159-164. [DOI] [PubMed] [Google Scholar]
- 23.Prince, A. M., B. Brotman, D. H. Lee, L. Andrus, J. Valinsky, and P. Marx. 2002. Lack of evidence for HIV type 1-related SIVcpz infection in captive and wild chimpanzees (Pan troglodytes verus) in West Africa. AIDS Res. Hum. Retrovir. 18:657-660. [DOI] [PubMed] [Google Scholar]
- 24.Ryder, R. W., W. Nsa, S. E. Hassig, F. Behets, M. Rayfield, B. Ekungola, A. M. Nelson, U. Mulenda, H. Francis, and K. Mwandagalirwa. 1989. Perinatal transmission of the human immunodeficiency virus type 1 to infants of seropositive women in Zaire. N. Engl. J. Med. 320:1637-1642. [DOI] [PubMed] [Google Scholar]
- 25.Santiago, M. L., C. M. Rodenburg, S. Kamenya, F. Bibollet-Ruche, F. Gao, E. Bailes, S. Meleth, S. J. Soong, J. M. Kilby, Z. Moldoveanu, B. Fahey, M. N. Muller, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, A. E. Pusey, D. A. Collins, C. Boesch, R. W. Wrangham, J. Goodall, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2002. SIVcpz in wild chimpanzees. Science 295:465. [DOI] [PubMed] [Google Scholar]
- 26.Santiago, M. L., M. Lukasik, S. Kamenya, Y. Li, F. Bibollet-Ruche, E. Bailes, M. N. Muller, M. Emery, D. A. Goldenberg, J. S. Lwanga, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, D. P. Watts, A. E. Pusey, D. A. Collins, R. W. Wrangham, J. Goodall, J. F. Brookfield, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2003. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J. Virol. 77:7545-7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santiago, M. L., F. Bibollet-Ruche, E. Bailes, S. Kamenya, M. N. Muller, M. Lukasik, A. E. Pusey, D. A. Collins, R. W. Wrangham, J. Goodall, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2003. Amplification of a complete simian immunodeficiency virus genome from fecal RNA of a wild chimpanzee. J. Virol. 77:2233-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanden Haesevelde, M. M., M. Peeters, G. Jannes, W. Janssens, G. van der Groen, P. M. Sharp, and E. Saman. 1996. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology 221:346-350. [DOI] [PubMed] [Google Scholar]
- 30.Worobey, M., M. L. Santiago, B. F. Keele, J. B. Ndjango, J. B. Joy, B. L. Labama, A. B. Dhed, A. Rambaut, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2004. Origin of AIDS: contaminated polio vaccine theory refuted. Nature 428:820. [DOI] [PubMed] [Google Scholar]