Abstract
Ascorbic acid (AsA), known as vitamin C, is an essential nutrient for humans and mainly absorbed from food. Tea plant (Camellia sinensis (L.) O. Kuntze) leaves can be a dietary source of AsA for humans. However, experimental evidence on the biosynthesis, recycling pathway and distribution of AsA during leaf development in tea plants is unclear. To gain insight into the mechanism and distribution of AsA in the tea plant leaf, we identified 18 related genes involved in AsA biosynthesis and recycling pathway based on the transcriptome database of tea plants. Tea plant leaves were used as samples at different developmental stages. AsA contens in tea plant leaves at three developmental stages were measured by reversed-phase high-performance liquid chromatography (RP-HPLC). The correlations between expression levels of these genes and AsA contents during the development of tea plant leaves were discussed. Results indicated that the l-galactose pathway might be the primary pathway of AsA biosynthesis in tea plant leaves. CsMDHAR and CsGGP might play a regulatory role in AsA accumulation in the leaves of three cultivars of tea plants. These findings may provide a further glimpse to improve the AsA accumulation in tea plants and the commercial quality of tea.
The tea plant (Camellia sinensis (L.) O. Kuntze) is an important economic crop in China1. The leaves of tea plants have been classically recognized as a good source for producing tea including oolong tea, black tea, green tea and white tea. The production of tea was estimated at 1,939,457 tons in China in 2013 from the FAOSTAT website (http://faostat3.fao.org). Nowadays, tea is one of the most popular beverages in the world. Tea plants are rich in many nutritious compositions, such as theanine, caffeine, theobromine, theophylline, and ascorbic acid (AsA)2,3. Drinking tea may help reduce the risks of cancer4,5,6.
AsA is an enzyme cofactor in plants and an essential nutrient for humans. Furthermore, AsA possesses a series of observable physiologic functions for reducing the risks of scurvy, lung cancer, and cardiovascular disease7,8,9. l-Gulonolactone oxidase is essential for the synthesis of AsA; however, humans and other primates lack this enzyme10. Therefore, humans must absorb AsA from diet, such as vegetables and fruits which contain a rich concentration of AsA. AsA may improve catechins bioavailability by enhancing intestinal uptake from tea11. Exogenous AsA can increase the flavanol concentration by 20% in green tea12.
Based on previous evidence, four principal biosynthesis pathways of AsA were propounded in plants, namely, l-galactose (l-Gal) pathway, l-gulose pathway, d-galacturonate pathway, and myo-inositol pathway13,14,15,16. l-Gal was important for AsA biosynthesis in higher plants. The l-Gal pathway was predominant in all of the four pathways until now. The other alternative pathways also played a supporting role in the AsA biosynthesis in plants. Wheeler and his colleagues found that mannose and l-Gal were effective precursors for the biosynthesis of AsA, GDP-mannose 3,5-epimerase catalysed the transformation of mannose into l-Gal13. Nine enzymes are involved in the l-Gal pathway17, namely, phosphoglucose isomerase (PGI), phosphomannose isomerase (PMI)18, phosphomannose mutase (PMM)19, GDP-d-mannose-3′,5′-epimerase (GME)20, GDP-d-mannose pyrophosphorylase (GMP)21, GDP-l-galactose phosphorylase (GGP)22, l-galactose-1-P phosphatase (GPP)23; l-galactose dehydrogenase (GalDH)24, and l-galactono-1,4-lactone dehydrogenase (GalLDH)25. In the l-gulose pathway, GDP-d-mannose-3′,5′-epimerase (GME) converts GDP-D-mannose to GDP-l-galactose15. In the d-galacturonate pathway, d-galacturonate reductase (GalUR) catalysed the transformation of d-galacturonic acid (d-GalUA) into l-galactonic acid14. The expression level of the GalUR gene was correlated well with AsA accumulation in strawberry14,26. In the myo-inositol pathway, phosphatase played an important role in the produce of myo-inositol27,28,29. Arabidopsis lines overexpressing miox4, a key gene in the myo-inositol pathway, showed an obvious increase in the AsA content16. In addition, the enzymes involved in the AsA recycling pathway include ascorbate oxidase (AO)30, ascorbate peroxidase (APX)31, dehydroascorbate reductase (DHAR)32, glutathione reductase (GR)33, and monodehydroascorbate reductase (MDHAR)34. Different pivotal enzymes may lead to distinct changes in the AsA concentration in higher plants. Overexpressing a strawberry GalUR gene in Arabidopsis resulted in a two- to three-fold increase in AsA levels14. Both transgenic tobacco and maize plants hosting DHAR gene exhibited higher AsA levels in foliar and kernel35. Overexpression of an acerola GMP gene in tobacco, showed a two-fold increase in the ascorbate content36, whereas overexpression of the MIOX4 gene caused a two- and three-fold increase in the ascorbate content in Arabidopsis leaves16.
Recent studies have demonstrated that the main biosynthesis pathway of AsA was the l-Gal pathway in apple fruits and leaves of different ages37,38. Considerable evidence indicated that the l-Gal pathway was a principal route for AsA biosynthesis in most plants. For instance, the l-Gal pathway was a predominant biosynthetic route of ascorbate in apple leaves38. Similarly, the l-Gal pathway was found to be the primary pathway of AsA accumulation in carrots and radish roots17,39. Meanwhile, l-Gal pathway played a predominant role in AsA biosynthesis in peel and pulp of citrus fruits40.
The tea plant samples of transcriptome sequencing included mid-leaf ‘Yunnanshilixiang’ (Tea_T1) from Yunnan province, small-leaf ‘Chawansanhao’ (Tea_T2) from Jiangsu province, large-leaf ‘Ruchengmaoyecha’ (Tea_T3) from Hunan province, and small-leaf ‘Anjibaicha’ (Tea_T4) from Zhejiang province. These four tea plant samples of transcriptome sequencing were significantly different, including environmental adaptation and leaf size. In the present research, ‘Anjibaicha’ was a kind of small-leaf tea plants. ‘Yingshuang’ was a kind of mid-leaf tea plants. ‘Huangjinya’ was a kind of small-leaf tea plants. The AsA contents were different among the three tea plant cultivars. Based on the different contents of AsA, the three tea plant cultivars (‘Huangjinya’, ‘Anjibaicha’, and ‘Yingshuang’) were used as suitable samples for this research, and were used as samples in gene expression analyses. The related genes that involved in the biosynthesis and recycling pathways of AsA were identified from the tea plant transcriptome database41. Twelve genes involved in AsA biosynthesis and six genes related to the AsA recycling pathways were selected. The AsA content in tea plant leaves at three developmental stages in ‘Yingshuang’, ‘Huangjinya’, and ‘Anjibaicha’ were recorded. Finally, we investigated the expression levels of AsA-related genes in the three tea plant cultivars. This study will provide useful information for exploring of improving the content of AsA in the tea plants.
Results
Growth analysis of leaves at three developmental stages in three tea plant cultivars
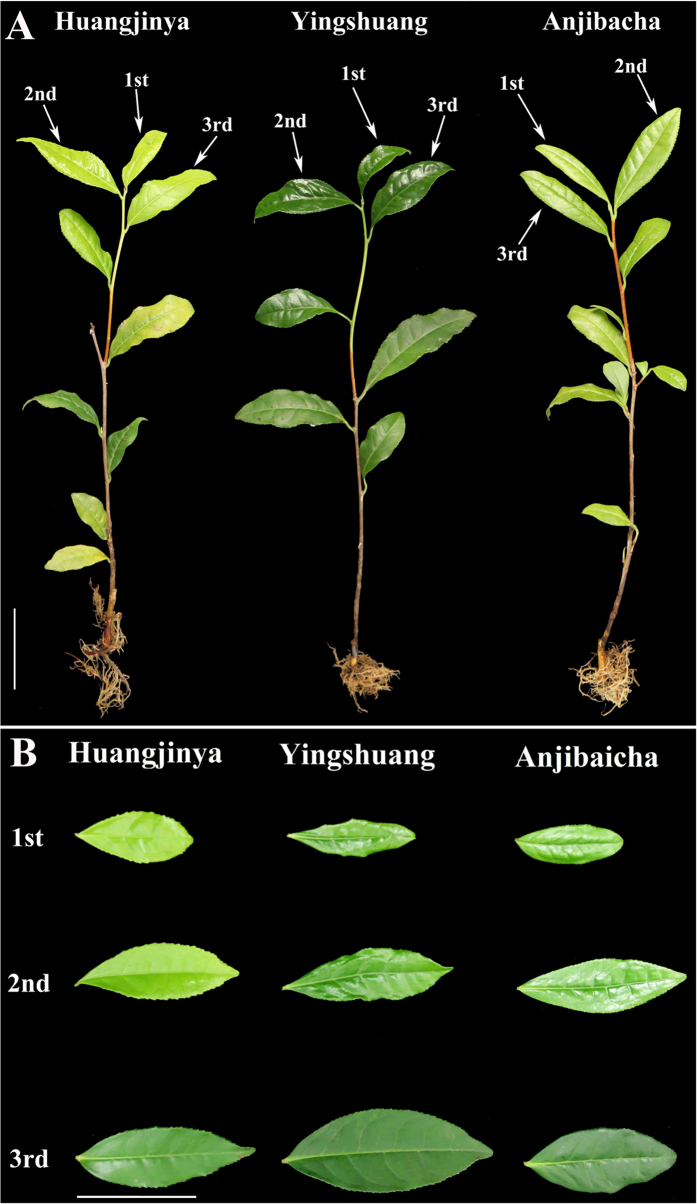
The samples were sorted into three developmental stages, including stage 1 (1st leaf), stage 2 (2nd leaf), and stage 3 (3rd leaf) (Fig. 1). Three tea plants included ‘Anjibaicha’, ‘Yingshuang’, and ‘Huangjinya’.
Figure 1. The three tea plant cultivars.
(A) The plant of ‘Anjibaicha’, ‘Yingshuang’, and ‘Huangjinya’. (B) Tea plant leaves of three developmental stages: Stages 1 (1st leaf), 2 (2nd leaf) and 3 (3rd leaf). The lines beside the leaves represent 5 cm in that pixel.
Changes in AsA content
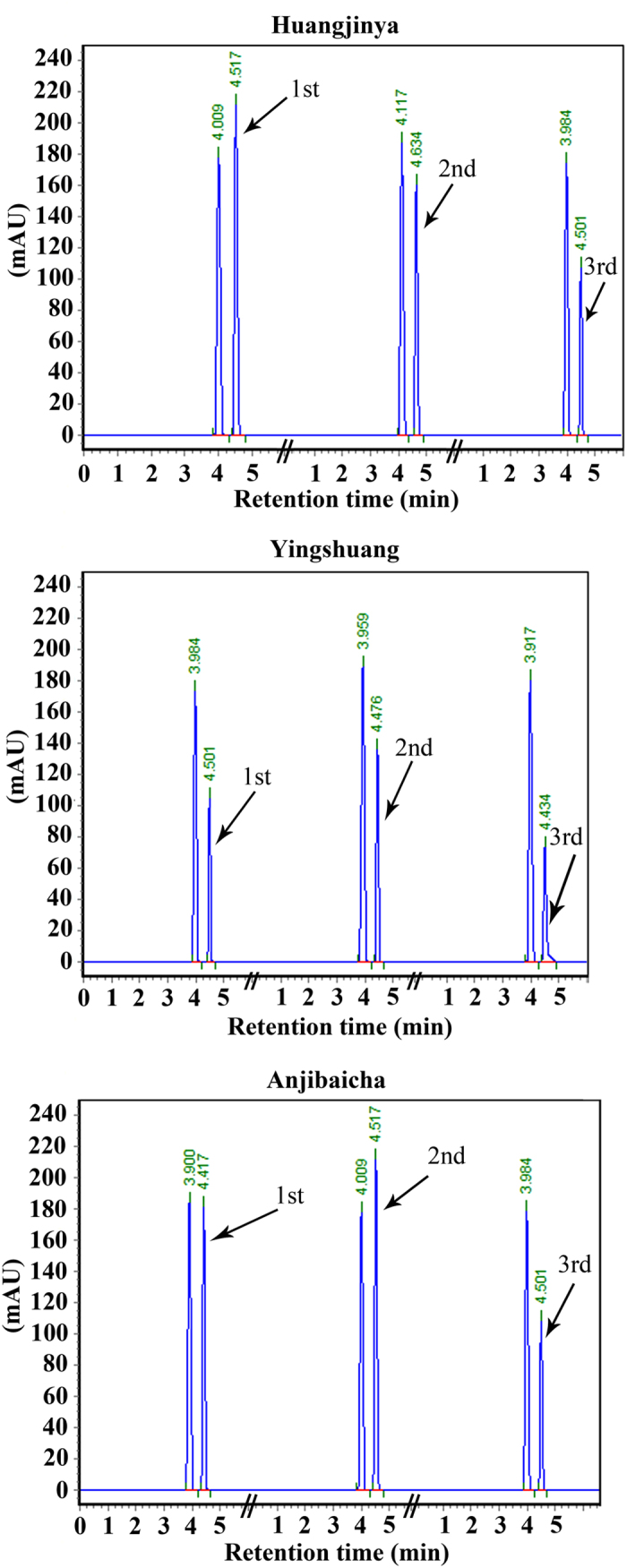
The AsA content was measured at three leaf developmental stages in three tea plant cultivars by RP-HPLC (Figs 2 and 3). The highest concentration of AsA was detected at the first stage in ‘Huangjinya’ (79.81 mg/100 g FW), whereas the lowest content was observed at stage 3 in ‘Yingshuang’ (29.43 mg/100 g FW). The AsA content initially increased and then evidently decreased in ‘Yingshuang’ and ‘Anjibaicha’. A significant reduction of AsA content was observed during leaf development in ‘Huangjinya’.
Figure 2. AsA contents of leaves at three developmental stages in three tea plant cultivars.

Error bars represent standard deviation among three independent replicates. Data are means of three replicates ± SD.
Figure 3. HPLC chromatogram of AsA in the leaves at three developmental stages from three tea plant cultivars.
Expression levels of the genes involved in AsA biosynthesis in tea plants
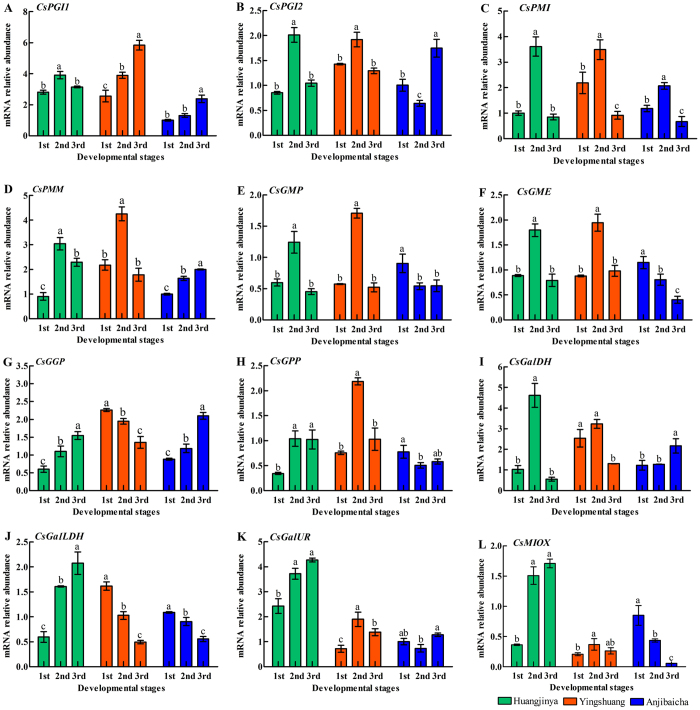
The expression levels of 12 genes involved in AsA biosynthesis were detected in leaves at different developmental stages of three tea plant cultivars by qRT-PCR (Fig. 4). The expression level of CsPGI1 showed an upward trend in both ‘Anjibaicha’ and ‘Yingshuang’ during three developmental stages (Fig. 4A). The expression level of CsPGI2 peaked at stage 2 then declined in ‘Huangjinya’ and ‘Yingshuang’. By contrast, the transcription level of CsPGI2 decreased at the stage 2 and then increased in ‘Anjibaicha’ (Fig. 4B). CsGalLDH displayed a continuous decrease at three developmental stages in both ‘Anjibaicha’ and ‘Yingshuang’ (Fig. 4J). The expression levels of CsGGP and GalLDH experienced a similar upward trend in ‘Huangjinya’ (Fig. 4G,J).
Figure 4. Expression level analyses of genes involved in AsA biosynthesis pathway in tea plant leaves.
Genes involved in AsA biosynthesis pathway (A) Phosphoglucose isomerase (CsPGI1), (B) (CsPGI2), (C) phosphomannose isomerase (CsPMI), (D) phosphomannose mutase (CsPMM), (E) GDP-d-mannose pyrophosphorylase (CsGMP), (F) GDP-d-mannose-3′,5′-epimerase (CsGME), (G) GDP-l-galactose phosphorylase (CsGGP), (H) l-galactose-1-P phosphatase (CsGPP), (I) l-galactose dehydrogenase (CsGalDH), (J) l-galactono-1,4-lactone dehydrogenase (CsGalLDH). Error bars represent standard deviation among three qRT-PCR reaction replicates. Data are means of three replicates ± SD.
Expression levels of the genes involved in AsA recycling in tea plant
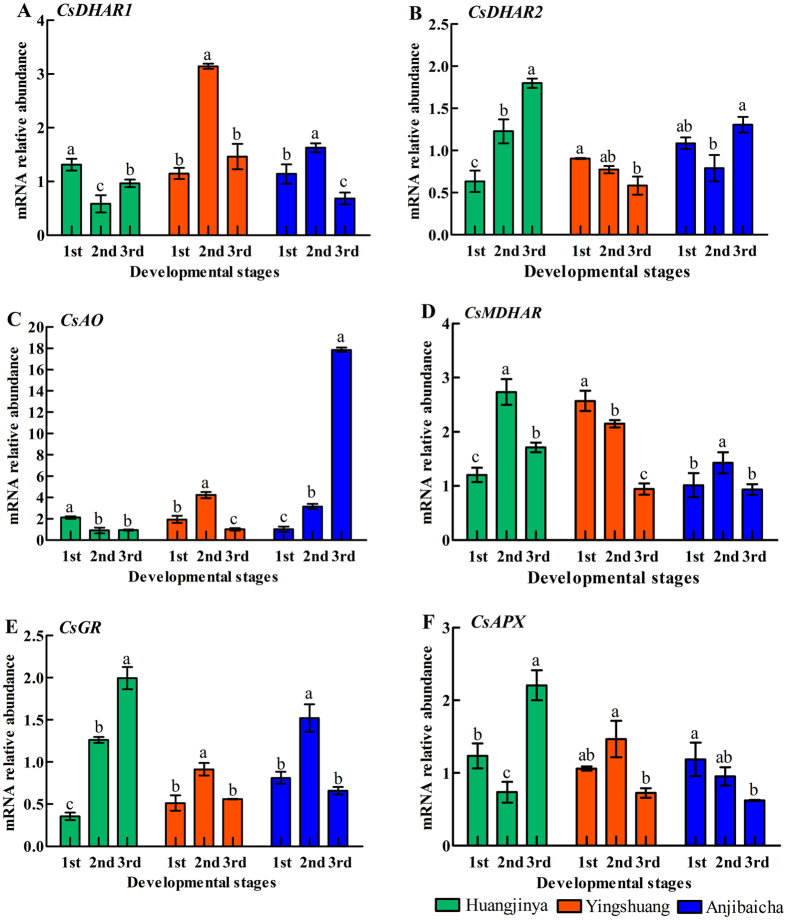
The expression levels of six genes involved in the AsA recycling pathway were also detected in tea plants (Fig. 5). The expression levels of CsDHAR1, CsMDHAR and CsGR initially increased, followed by a decrease at the last stage in ‘Anjibaicha’ (Fig. 5A,D and E). The expression levels of CsDHAR2 and CsMDHAR showed a downward trend during three developmental stages in ‘Yingshuang’ (Fig. 5B,D). Additionally, CsDHAR2 and CsGR exhibited an upward trend of in ‘Huangjinya’ (Fig. 5B,E). CsAO increased prominently at stage 3, which was nearly 16-fold higher than that at stage 1 in ‘Anjibaicha’ (Fig. 5C). The expression level of CsAPX decreased slightly at stage 2 as compared with that at stage 1, and remarkably elevated at stage 3 in ‘Huangjinya’ (Fig. 5F).
Figure 5. Expression level analyses of genes involved in AsA recycling pathway in tea plant leaves.
Genes involved in recycling pathway include (A) dehydroascorbate reductase (CsDHAR1), (B) (CsDHAR2), (C) ascorbate oxidase (CsAO), (D) monodehydroascorbate reductase (CsMDHAR), (E) glutathione reductase (CsGR), (F) ascorbate peroxidase (CsAPX). Error bars represent standard deviation among three qRT-PCR reaction replicates. Data are means of three replicates ± SD.
Expression profiles of genes involved in AsA biosynthesis and recycling in four tea plant cultivars
RNA sequencing (RNA-seq) data was extracted from transcriptome database in the four tea plant cultivars (Tea_T1, Tea_T2, Tea_T3, and Tea_T4)41. These tea plants were grown under non-stress conditions. The expression levels of genes involved in AsA biosynthesis and recycling were analyzed in another four other tea plants using RNA-seq data. RPKM values (Reads per kilobase per million mapped reads) were used to analyze the transcript levels of 18 genes, and a heatmap was obtained using HemI software (version1.0; http://hemi.biocuckoo.org/faq.php)42 (Fig. 6). CsAPX was expressed at the highest level (RPKM > 409) in Tea_T3. CsGGP showed a similar expression pattern in Tea_T1, Tea_T2, and Tea_T4. Both CsPMI and CsAO showed relatively low expression levels (RPKM > 2) in Tea_T3. CsMDHAR was highly expressed (RPKM > 291) in Tea_T1. In addition, CsGMP and CsGME, which participate in the AsA biosynthesis pathway were highly expressed in Tea_T1, Tea_T2, and Tea_T4. CsMDHAR and CsAPX were highly expressed in Tea_T1 and Tea_T4, whereas CsGalUR showed relatively low expression levels in the four tea plant cultivars.
Figure 6. Heatmap of the relative expression level of genes involved in AsA biosynthesis and recycling in tea plant.
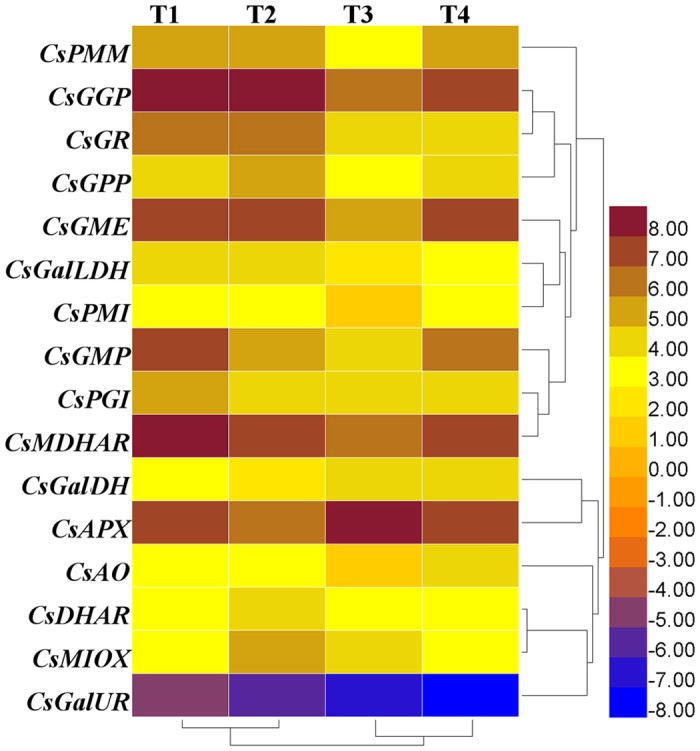
T1 was ‘Yunnanshilixiang’, T2 was ‘Changwansanhao’, T3 was ‘Ruchengmaoyecha’, and T4 was ‘Anjibaicha’.
Discussion
Different enzymes play various roles in AsA accumulation
Various enzymes are involved in ascorbic biosynthesis in higher plants. MIOX is a crucial enzyme in the myo-inositol pathway, but a number of scientific studies observed that the AsA content is insignificantly affected by MIOX. The AsA content did not increase in Miox overexpression lines compared with that in the wild type in Arabidopsis43. Similarly, the levels of AsA content slightly changed in the OsMIOX-overexpressing transgenic rice lines compared with wild type44. GalUR is a pivotal enzyme in the d-galacturonate pathway. The expression level of the VvGalUR gene was correlated with the AsA content level during grape ripening45. By contrast, the AsA content showed no obvious relation to GalUR expression in kiwifruit46. The expression level of CsGalUR was negatively correlated with AsA accumulation from stage 1 to stage 3 in ‘Huangjinya’ and ‘Anjibaicha’. Whereas, the expression level of CsGalUR was positively correlated with AsA accumulation from stage 1 to stage 3 in ‘Yingshuang’. In addition, the expression level of CsMIOX was negatively correlated with AsA accumulation from stage 1 to stage 3 in ‘Huangjinya’. The expression level of CsMIOX was positively correlated with AsA accumulation from stage 1 to stage 3 in ‘Yingshuang’. Therefore, the CsMIOX and CsGalUR might play potential different roles in AsA accumulation in different tea plants.
AsA accumulation and expression levels of genes involved in the biosynthesis pathway of AsA in tea plant leaves
The expression levels of genes involved in the AsA biosynthesis pathway varied in different tea plant leaves. A correlation was noted between the AsA contents and expression levels of genes involved in AsA biosynthesis in tea plant leaves. The highest expression level of CsGGP was correlated with the lowest content of AsA in each tea plant cultivar (Figs 2 and 4G). This finding indicated that CsGGP played a regulatory role between expression levels and AsA accumulation. Previous reports noted that GGP may function as a regulatory factor47. A positive correlation was found between CsGPP expression level and AsA accumulation from stage 1 to stage 3 in ‘Yingshuang’, whereas a negative correlation was observed from stage 1 to stage 3 ‘Huangjinya’ and ‘Anjibaicha’ (Figs 2 and 4H). In kiwi, Li et al. has also demonstrated that a positive correlation was found between GPP expression and AsA accumulation from 0 to 60 days after anthesis. Meanwhile, a negative correlation was found from 60 to 75 days after anthesis48. In AsA accumulation, l-galactose-1-P phosphatase (GPP) could use myo-inositol-1-phosphate as a substrate49. This finding suggested that CsGPP might be a critical regulatory factor in AsA content levels of leaves of three tea plant cultivars. GPP has been reported as an essential enzyme for AsA accumulation in tomato fruit50. In addition, GME and GGP shared a crucial role in controlling l-ascorbate biosynthesis in tomato, and GME and GGP transcripts were co-regulated20. This finding was consistent with our results in ‘Anjibaicha’ (Fig. 4F and G). Previous studies showed that overexpressed GGP gene from kiwifruit in Arabidopsis resulted in a five-fold increase in ascorbate levels; co-expressing the GME and GGP genes demonstrated a seven-fold increase in ascorbate levels46. The expression profile of CsPMI was positively correlated with CsGR from stage 1 to stage 3 in ‘Yingshuang’ and ‘Anjibaicha’ (Figs 4C and 5E), thereby suggesting a relationship of coordination and cooperation in AsA biosynthesis and recycling.
AsA accumulation and expression levels of genes involved in the AsA recycling pathway
The recycling pathway of AsA is complicated in higher plants. According to theory of AsA-GSH (ascorbate and glutathione) metabolism51. AsA is first oxidized to form mono-dehydroascorbate by APX and AO and then regenerated by mono-dehydroascorbate (MDHA), which is extremely unstable52. Subsequently, DHA regenerates AsA under the action of DHAR and GSH53. Recent research showed that MDHAR negatively regulates ascorbate levels in tomato54. Expression of MDHAR was positively correlated with the AsA content during stage 2 to stage 4 in the radish root flesh, whereas the expression of MDHAR was negatively correlated with the AsA content from stage 1 to stage 2 in the radish root skin55. MDHAR was positively correlated with AsA content from stage 1 to stage 2, whereas it was negatively correlated with AsA content from stage 2 to stage 3 in the development of carrot root17. In the present study, CsMDHAR was negatively correlated with the AsA content from stage 1 to stage 2 in ‘Yingshuang’ and ‘Huangjinya’, whereas CsMDHAR was positively correlated with the AsA content from stage 1 to stage 3 in ‘Anjibaicha’ (Fig. 5D). These combined findings indicated that CsMDHAR might play a regulatory role in AsA content in ‘Anjibaicha’, ‘Huangjinya’, and ‘Yingshuang’. The results might provide more insights into improving the ascorbate levels and investigating molecular mechanisms in plant leaves.
Potential pathway for AsA biosynthesis and recycling in tea plant
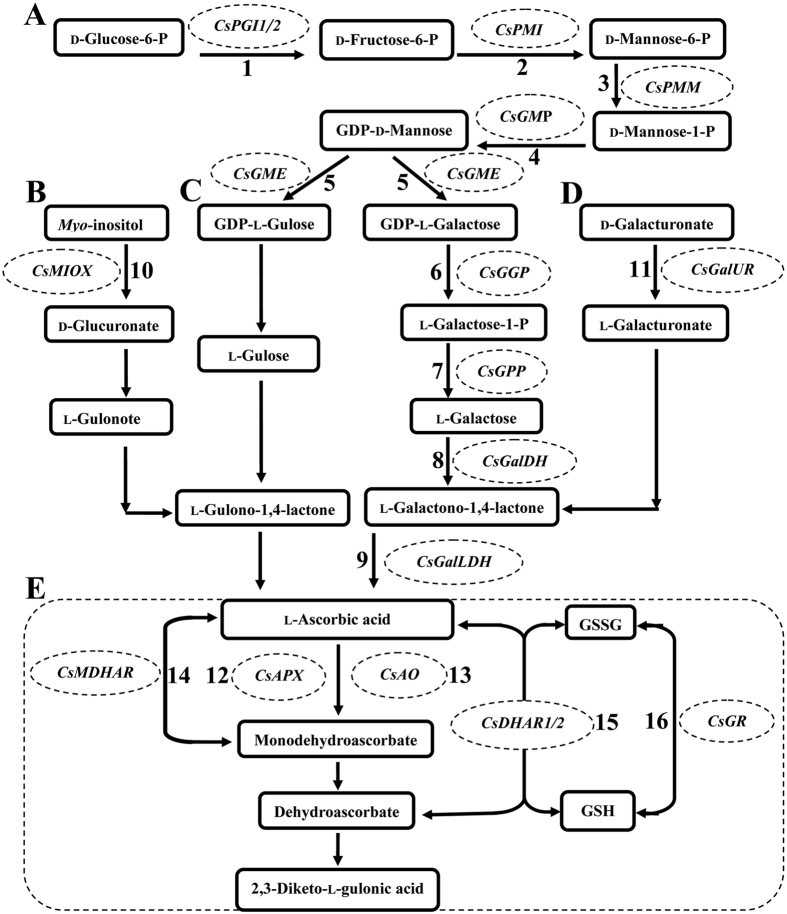
Considerable evidence revealed that the l-Gal pathway was the major AsA biosynthesis pathway in several plant species. However, the network of this pathway is quite complicated. The l-gulose pathway, d-galacturonate pathway, and myo-inositol pathway also play a role in the regulation of AsA biosynthesis in several plants. Based on previous results about the AsA recycling pathways, the potential pathways of AsA biosynthesis and recycling during the development of tea plant leaves were established. Twelve genes involved in the AsA biosynthesis pathway and six genes related to the AsA recycling pathway were identified (Fig. 7).
Figure 7. AsA biosynthetic and AsA recycling routes in tea plant leaves.
(A) L-galactose (L-Gal), (B) myo-inositol, (C)l-gulose, (D) D-galacturonate and recycling pathway (E)). 1, phosphoglucose isomerase (PGI); 2, phosphomannose isomerase (PMI); 3, phosphomannose mutase (PMM); 4, GDP-d-mannose pyrophosphorylase (GMP); 5, GDP-d-mannose-3′,5′-epimerase (GME); 6, GDP-l-galactose phosphorylase (GGP); 7, l-galactose-1-P phosphatase (GPP); 8, l-galactose dehydrogenase (GalDH); 9, l-galactono-1,4-lactone dehydrogenase (GalLDH); 10, myo-inositol oxygenase (MIOX); 11, D-galacturonate reductase (GalUR); 12, ascorbate peroxidase (APX); 13, ascorbate oxidase (AO); 14, monodehydroascorbate reductase (MDHAR); 15, dehydroascorbate reductase (DHAR); and 16, glutathione reductase (GR).
Conclusion
The AsA contents in leaves at three developmental stages of three tea plant cultivars were measured. By analyzing the expression levels of 18 genes, which were involved in the AsA biosynthesis and recycling pathways. The pathways of AsA metabolism were evaluated and predicted. The results indicated that the l-Gal pathway might be the major biosynthetic route for regulating the AsA content of tea plant leaves. Our findings suggested that the AsA biosynthesis and recycling pathways might be controlled by multigene regulation during the development of tea plant leaves. The results also demonstrated that AsA contents were intimately linked to gene expression. Moreover, the AsA biosynthesis and recycling pathways were confirmed. Further studies can explore the possibility to increase the AsA content via metabolic engineering and transgenic engineering in tea plant leaves.
Materials and Methods
Plant materials
The plant materials were two-year-old cutting tea plant seedlings called ‘Anjibaicha’, ‘Yingshuang’, and ‘Huangjinya’. They were planted in a growth chamber at 25 °C at the Tea Science Research Institute, College of Horticulture, Nanjing Agriculture University (Nanjing, China). The tea plants were grown in acidic soil (pH 5.6) with a relative humidity of 70 ± 10% and watered weekly. All of the three tea plant cultivars were collected from Zhejiang Province in China. The tea plant leaves were harvested, quickly frozen in liquid nitrogen, and stored at −80 °C for RNA extraction.
AsA determination by RP-HPLC
AsA content levels were determined according to the method described by Guo and his colleagues56. In brief, 200 mg of fresh samples was ground in a mortar and homogenized 4 mL of 1.0% (w/v) oxalic acid. The mixture was transferred to a 10 mL centrifuge tube and centrifuged at 10,000 rpm for 10 min. Sample analysis by RP-HPLC was performed using the Shimadzu LC-20A series (Shimadzu Co., Kyoto, Japan) with a Hedera ODS-2 C18 analytical column (250 mm × 4.6 mm i.d., 5 μm nominal particle size) at 254 nm. About 20 μL of filtrate was injected in RP-HPLC for AsA determination. Finally, the AsA content was quantified by external calibration and results were recorded as mg/100 g FW.
RNA isolation
Samples from three tea plant cultivars at three developmental stages were separately harvested. Total RNA of the samples was isolated in accordance with the method of a commercial RNA extraction kit (Huayueyang, Beijing, China). The Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE) was used to measure the concentration of isolated RNA.
cDNA synthesis
First-strand cDNA of tea plant leaves at three developmental stages was synthesized with the PrimeScript RT reagent kit (TaKaRa, Dalian, China). The cDNA was diluted 15 times for PCR amplification. To explore the AsA metabolic pathway in tea plants, we selected a total of 18 AsA-related genes and determined their expression levels41.
Gene expression analysis by qRT-PCR
Twelve genes involved in AsA biosynthesis and six genes involved in AsA recycling were identified based on the tea plant transcriptome database. The sequences of these 18 genes were shown on the attached data sheet (Supplementary Table 1). Primer Premier 6.0 software was used to design 18 pairs primer sequences. The primer length was restricted to 20–30 bp for the qRT-PCR (quantitative real-time PCR) (Table 1). The primer sequences with the cDNA template were checked by PCR (Polymerase chain reaction). To ensure the efficiency of optimal polymerization, the amplification length for each gene was restricted to 100–200 bp. The reaction program of qRT-PCR was performed under the following conditions: 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s, and 55 °C for 25 s. The volume reaction was 20 μL, which contained 2 μL of diluted cDNA strand, 7.2 μL of deionized water, 10 μL of SYBR Premix Ex Taq (Tli RNaseH Plus; TaKaRa, Dalian, China), 0.4 μL of forward primer, and 0.4 μL of reverse primer. The mean values and standard deviation were calculated based on three independent biological replicates. CsActin was used as a reference gene to normalize the expression of related genes involved in AsA biosynthesis and recycling of tea plants57.
Table 1. Primers sequences of the related genes and reference gene used for qRT-PCR.
| Name | Forward primer(5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| CsPMM | CCACATTATTAGCTTCCTTCTCGTCAC | CCAACAACACCAACTGTAACAACCTT |
| CsGGP | ATCTTCCTTGTACCACAGTGTTATGCT | TGCCTCCTCGTAGTCCTTCTTCC |
| CsGalUR | GAGCAGCCTCTTGGAGAAGCAAT | ATCACGATGAGCATCAGAACACCAA |
| CsMIOX | GCGTCAATCACATCAACCAAACTTT | GCTCATCTCCACCTTGTCCACTT |
| CsGME | AACTACGGAGCATA CACCTATGAGAAC | CTAGCAATGTGCGAGGCAATGAATC |
| CsGMP | GAACTCGGTTGAGACCATTGACACTT | CCACTTCACTCACTCCAATAGCCTTG |
| CsGPP | GCTGCTGGTGCTGTGGTAGAAT | CTAGAAGTGACTGCTCCACCTTATCG |
| CsGalLDH | GGCGGCATTGTTCAGGTTGGT | GTCCACAGCGAGCAAGATAGAATAGTT |
| CsGalDH | GAGAGTGACTAGGAGCATTGATGAGAG | CCAAGCGGAAGTCCTGTAATACCAA |
| CsPMI | TCTGCGGTCAATATTCACTCAACTCAT | TGTTCCTTATCTGTCAACTGCCTCAC |
| CsPGI 1 | CATTGTGAAGAGTCAGCAACCTGTGTA | CGATTGCCAGAGAAGGTCTTGTGAG |
| CsPGI 2 | CGATGTCGTCAGTGGTAAGATTAAGC | TTATCTTGAGAGGCGGATTATCAGGAG |
| CsAPX | AGCAAGGTCACGAAGCCAACAAT | GCAACAACTCCAGCCAACTGATAGA |
| CsAO | CCAACACCACTCAAGCACTAACAATAC | GAGGATGATACGGCGGTGATGG |
| CsDHAR1 | ATGATGGAACCGAGCAAGCATTACT | GACAAGTCCGCAGCAGATACTCTT |
| CsDHAR2 | ACCCTCCTCTCTGCCATTCTCC | TTCATCCAGTGCCTTCAACTCATCAA |
| CsGR | ACCCTGATGGCTAATAAGAATGCTGAA | TAGTATGTGCCTTGCCGAGTAGAGT |
| CsMDHAR | GGCGGATCAAGTGTTGGAAGGGAG | ACGCTTGGGATTGTATTCGGCATTA |
| CsActin | GATTCCGTTGCCCTGAAGTCCT | CCTTGCTCATACGGTCTGCGATA |
Statistical analysis
Differences in gene expression levels were detected by Duncan’s multiple-range test at a 0.05 probability level.
Additional Information
How to cite this article: Li, H. et al. Transcriptomic analysis of the biosynthesis, recycling, and distribution of ascorbic acid during leaf development in tea plant (Camellia sinensis (L.) O. Kuntze). Sci. Rep. 7, 46212; doi: 10.1038/srep46212 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The research was supported by the National Natural Science Foundation of China (31570691).
Footnotes
The authors declare no competing financial interests.
Author Contributions Conceived and designed the experiments: J.Z., H.L. Performed the experiments: H.L., W.H., G.L.W., W.L.W., X.C., J.Z. Analyzed the data: H.L. Contributed reagents/materials/analysis tools: J.Z. Wrote the paper: H.L. Revised the paper: J.Z., H.L. All authors read and approved the final manuscript.
References
- Chen Y., Yu M., Jie X., Chen X. & Shi J. Differentiation of eight tea (Camellia sinensis) cultivars in China by elemental fingerprint of their leaves. J Sci Food Agr 89, 2350–2355 (2009). [Google Scholar]
- Graham H. N. Green tea composition, consumption, and polyphenol chemistry. Prev Med 21, 334–350 (1992). [DOI] [PubMed] [Google Scholar]
- Aucamp J. P., Hara Y. & Apostolides Z. Simultaneous analysis of tea catechins, caffeine, gallic acid, theanine and ascorbic acid by micellar electrokinetic capillary chromatography. J Chromatogr A 876, 235–242 (2000). [DOI] [PubMed] [Google Scholar]
- Zheng J. et al. Green tea and black tea consumption and prostate cancer risk: an exploratory meta-analysis of observational studies. Nutr Cancer 63, 663–672 (2011). [DOI] [PubMed] [Google Scholar]
- Wang W., Yang Y. E., Zhang W. & Wu W. Association of tea consumption and the risk of oral cancer: A meta-analysis. Oral Oncol 50, 276–281 (2014). [DOI] [PubMed] [Google Scholar]
- Zheng J. S. et al. Effects of green tea, black tea, and coffee consumption on the risk of esophageal cancer: a systematic review and meta-analysis of observational studies. Nutr Cancer 65, 1–16 (2013). [DOI] [PubMed] [Google Scholar]
- Ashor A. W., Lara J., Mathers J. C. & Siervo M. Effect of vitamin C on endothelial function in health and disease: A systematic review and meta-analysis of randomised controlled trials. Atherosclerosis 235, 9–20 (2014). [DOI] [PubMed] [Google Scholar]
- Hodges R. E., Baker E. M., Hood J., Sauberlich H. E. & March S. C. Experimental scurvy in man. Am J Clin Nutr 22, 535–548 (1969). [DOI] [PubMed] [Google Scholar]
- Luo J., Shen L. & Zheng D. Association between vitamin C intake and lung cancer: a dose-response meta-analysis. Sci Rep 4, 6161–6161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster C. L. & Emile V. S. Vitamin C. FEBS J 274, 1–22 (2007). [DOI] [PubMed] [Google Scholar]
- Peters C. M., Green R. J., Janle E. M. & Ferruzzi M. G. Formulation with ascorbic acid and sucrose modulates catechin bioavailability from green tea. Food Res Int 43, 95–102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann B. F. & Gleichenhagen M. The effect of ascorbic acid, citric acid and low pH on the extraction of green tea: How to get most out of it. Food Chem 124, 1543–1548 (2011). [Google Scholar]
- Wheeler G. L., Jones M. A. & Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature 393, 365–369 (1998). [DOI] [PubMed] [Google Scholar]
- Agius F. et al. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat Biotechnol 21, 177–181 (2003). [DOI] [PubMed] [Google Scholar]
- Wolucka B. A. & Marc V. M. GDP-mannose 3′,5′-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J Bio Chem 278, 47483–47490 (2003). [DOI] [PubMed] [Google Scholar]
- Lorence A., Chevone B. I., Mendes P. & Nessler C. L. Myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol 134, 1200–1205 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. L. et al. Regulation of ascorbic acid biosynthesis and recycling during root development in carrot (Daucus carota L.). Plant Physiol Biochem 94, 10–18 (2015). [DOI] [PubMed] [Google Scholar]
- Maruta T. et al. Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid biosynthesis. J Biol Chem 283, 28842–28851 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. et al. Molecular analysis of phosphomannomutase (PMM) genes reveals a unique PMM duplication event in diverse Triticeae species and the main PMM isozymes in bread wheat tissues. BMC Plant Biol 10, 159–169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. et al. GDP-D-mannose 3, 5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J 60, 499–508 (2009). [DOI] [PubMed] [Google Scholar]
- Conklin P. L., Williams E. H. & Last R. L. Environmental stress sensitivity of an ascorbic acid-deficient Arabidopsis mutant. Proc Natl Acad Sci USA 93, 9970–9974 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster C. L. & Clarke S. G. L-Ascorbate biosynthesis in higher plants: the role of VTC2. Trends Plant Sci 13, 567–573 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin P. L. et al. Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J Biol Chem 281, 15662–15670 (2006). [DOI] [PubMed] [Google Scholar]
- Mieda T. et al. Feedback inhibition of spinach L-galactose dehydrogenase by L-ascorbate. Plant Cell Physiol 45, 1271–1279 (2004). [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Dowdle J. & Smirnoff N. Progress in manipulating ascorbic acid biosynthesis and accumulation in plants. Physiol Plant 129, 343–355 (2007). [Google Scholar]
- Cruzrus E., Amaya I., Sánchezsevilla J. F. & Botella M. A. Regulation of L-ascorbic acid content in strawberry fruits. J Exp Bot 62, 4191–4201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumber S. C., Loewus M. W. & Loewus F. A. Further studies on myo-inositol-1-phosphatase from the pollen of Lilium longiflorum Thunb. Plant Physiol 76, 40–44 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus M. W. & Loewus F. A. Myo-Inositol-1-Phosphatase from the Pollen of Lilium longiflorum Thunb. Plant Physiol 70, 765–770 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy G. E., Keddie J. S., Oda K. & Gruissem W. Plant inositol monophosphatase is a lithium-sensitive enzyme encoded by a multigene family. Plant Cell 7, 2175–2185 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cécile G. et al. A diminution in ascorbate oxidase activity affects carbon allocation and improves yield in tomato under water deficit. Plant Cell Environ 36, 159–175 (2013). [DOI] [PubMed] [Google Scholar]
- Asada K. Ascorbate peroxidase-a hydrogen peroxide-scavenging enzyme in plants. Physiol Plant 85, 235–241 (1992). [Google Scholar]
- Yabuta Y. et al. Thylakoid membrane-bound ascorbate peroxidase is a limiting factor of antioxidative systems under photo-oxidative stress. Plant J 32, 915–925 (2002). [DOI] [PubMed] [Google Scholar]
- Potters G. et al. Dehydroascorbate influences the plant cell cycle through a glutathione-independent reduction mechanism. Plant Physiol 134, 1479–1487 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. S., Lee H., Lee I. A., Kim K. Y. & Jo J. Molecular cloning of the monodehydroascorbate reductase gene from Brassica campestris and analysis of its mRNA level in response to oxidative stress. Biochim Biophys Acta 1658, 181–186 (2004). [DOI] [PubMed] [Google Scholar]
- Chen Z., Young T. E., Ling J., Chang S. C. & Gallie D. R. Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc Natl Acad Sci USA 100, 3525–3530 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badejo A., Tanaka N. & Esaka M. Analysis of GDP-D-mannose pyrophosphorylase gene promoter from acerola (Malpighia glabra) and increase in ascorbate content of transgenic tobacco expressing the acerola gene. Plant Cell Physiol 49, 126–132 (2008). [DOI] [PubMed] [Google Scholar]
- Li M. J., Ma F. W., Zhang M. & Pu F. Distribution and metabolism of ascorbic acid in apple fruits (Malus domestica Borkh cv. Gala). Plant Sci 174, 606–612 (2008). [Google Scholar]
- Li M., Ma F., Guo C. & Liu J. Ascorbic acid formation and profiling of genes expressed in its synthesis and recycling in apple leaves of different ages. Plant Physiol Biochem 48, 216–224 (2010). [DOI] [PubMed] [Google Scholar]
- Yao X., Zhu X., Chen Y., Gong Y. & Liu L. Expression profiling of genes involved in ascorbate biosynthesis andrecycling during fleshy root development in radish. Plant Physiol Biochem 70, 269–277 (2013). [DOI] [PubMed] [Google Scholar]
- Alós E., Rodrigo M. J. & Zacarías L. Differential transcriptional regulation of L-ascorbic acid content in peel and pulp of citrus fruits during development and maturation. Planta 239, 1113–1128 (2014). [DOI] [PubMed] [Google Scholar]
- Wu Z. J., Li X. H., Liu Z. W., Xu Z. S. & Zhuang J. De novo assembly and transcriptome characterization: novel insights into catechins biosynthesis in Camellia sinensis. BMC Plant Biol 14, 1–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Wang Y., Liu Z., Cheng H. & Xue Y. HemI: a toolkit for illustrating heatmaps. PloS One 9, e111988–e111988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres S. & Tenhaken R. Myoinositol oxygenase controls the level of myoinositol in Arabidopsis, but does not increase ascorbic acid. Plant Physiol 149, 1042–1049 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J. et al. OsMIOX, a myo-inositol oxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Plant Sci 196, 143–151 (2012). [DOI] [PubMed] [Google Scholar]
- Cruz-Rus E., Botella M. A., Valpuesta V. & Gomez-Jimenez M. C. Analysis of genes involved in L-ascorbic acid biosynthesis during growth and ripening of grape berries. J Plant Physiol 167, 739–748 (2010). [DOI] [PubMed] [Google Scholar]
- Bulley S. M. et al. Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-L-galactose guanyltransferase is a major control point of vitamin C biosynthesis. J Exp Bot 60, 765–778(714) (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Moulé P. An expression analysis of the ascorbate biosynthesis enzyme VTC2. Plant Mol Biol 68, 31–41 (2008). [DOI] [PubMed] [Google Scholar]
- Li M., Ma F., Liang D., Li J. & Wang Y. Ascorbate biosynthesis during early fruit development is the main reason for its accumulation in kiwi. Plos One 5, e14281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing W. A. et al. A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis. Proc Natl Acad Sci USA 101, 16976–16981 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidi E. et al. Expression profiling of ascorbic acid-related genes during tomato fruit development and ripening and in response to stress conditions. J Exp Bot 60, 663–678 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G. & Foyer C. H. Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Biol 49, 249–279 (1998). [DOI] [PubMed] [Google Scholar]
- Smirnoff N. & Wheeler G. L. Ascorbic acid in plants: biosynthesis and function. Crit Rev Biochem Mol Biol 35, 291–314 (2000). [DOI] [PubMed] [Google Scholar]
- Noctor G., Veljovic-Jovanovic S. & Foyer C. H. Peroxide processing in photosynthesis: antioxidant coupling and redox signalling. Philos T R Soc B 355, 1465–1475 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gest N., Garchery C., Gautier H., Jiménez A. & Stevens R. Light-dependent regulation of ascorbate in tomato by a monodehydroascorbate reductase localized in peroxisomes and the cytosol. Plant Biotechnol J 11, 344–354 (2013). [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhu X., Chen Y., Gong Y. & Liu L. Expression profiling of genes involved in ascorbate biosynthesis and recycling during fleshy root development in radish. Plant Physiology & Biochemistry 70, 269–277 (2013). [DOI] [PubMed] [Google Scholar]
- Guo L., Yang R., Wang Z., Guo Q. & Gu Z. Effect of NaCl stress on health-promoting compounds and antioxidant activity in the sprouts of three broccoli cultivars. Int J Food Sci Nutr 65, 476–481 (2014). [DOI] [PubMed] [Google Scholar]
- Li H., Huang W., Wang G. L., Wu Z. J. & Zhuang J. Expression profile analysis of ascorbic acid-related genes in response to temperature stress in the tea plant, Camellia sinensis (L.) O. Kuntze. Genet Mol Res 15, 1–10 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.