Abstract
A significant proportion of the human genome consists of stably inherited retroviral sequences. Most human endogenous retroviruses (HERVs) became defective over time. The HERV-K(HML-2) family is exceptional because of its coding capacity and the possible involvement in germ cell tumor (GCT) development. HERV-K(HML-2) transcription is strongly upregulated in GCTs. However, regulation of HERV-K(HML-2) transcription remains poorly understood. We investigated in detail the role of CpG methylation on the transcriptional activity of HERV-K(HML-2) long terminal repeats (LTRs). We find that CpG sites in various HERV-K(HML-2) proviral 5′ LTRs are methylated at different levels in the cell line Tera-1. Methylation levels correlate with previously observed transcriptional activities of these proviruses. CpG-mediated silencing of HERV-K(HML-2) LTRs is further corroborated by transcriptional inactivity of in vitro-methylated 5′ LTR reporter plasmids. However, CpG methylation levels do not solely regulate HERV-K(HML-2) 5′ LTR activity, as evidenced by different LTR activities in the cell line T47D. A significant number of mutated CpG sites in evolutionary old HERV-K(HML-2) 5′ LTRs suggests that CpG methylation had already silenced HERV-K(HML-2) proviruses millions of years ago. Direct silencing of HERV-K(HML-2) expression by CpG methylation enlightens upregulated HERV-K(HML-2) expression in usually hypomethylated GCT tissue.
About 8% of the human genome is composed of sequences with retroviral origins. The human endogenous retroviruses (HERVs) stem from germ line infections of ancient exogenous retroviruses. A large number of proviral elements and in particular the solitary long terminal repeats (LTRs) are present in the human genome. Despite the long-time presence of HERV LTRs in the human genome, several studies demonstrated transcriptional activity of both proviral and solitary HERV LTRs. Some HERV elements are involved in the transcription of cellular genes (for instance, see references 10, 22, 26, and 31).
The HERV-K(HML-2) family displays a number of remarkable features. First, despite their long-time presence in the human genome, a number of proviruses still display open reading frames for retroviral Gag, Prt, Pol, and/or Env proteins (3, 4, 29, 30, 40). Second, transcriptional activity of the HERV-K(HML-2) family is strongly upregulated in human germ cell tumors (GCTs) and GCT-derived cell lines, as opposed to transcriptional silencing in non-GCT cell lines. Third, GCT patients, particularly seminoma patients, display high antibody titers against HERV-K(HML-2) Gag and Env proteins (6, 17, 37, 38). Fourth, HERV-K(HML-2) encodes the so-called Rec protein, an additional splicing product from the proviral env gene that associates with the promyelocytic zinc finger protein and that may be involved in the development of GCT (5, 27, 42). Therefore, HERV-K(HML-2) can be considered a tumor marker for GCT and is potentially involved in GCT development.
Despite the possible involvement of HERV-K(HML-2) in GCT, the regulation of the transcriptional activity of HERV-K(HML-2), including cellular factors influencing that activity, has hitherto been vaguely understood. An early study noted upregulation of HERV-K(HML-2) expression in T47D cells after treatment of cells with estradiol and progesterone (34). A complex of at least three otherwise-unidentified proteins was reported to bind to the 5′ portion of the LTR U3 region (2). Furthermore, the transcription factor YY1 and other unidentified proteins were shown to bind to an enhancer region within the proviral LTR U3 region. However, the ubiquitous transcription factor YY1 is not responsible for different activities in cell lines expressing or not expressing HERV-K(HML-2) (25).
Methylation of cytosines in CpG dinucleotides can have profound impacts on gene expression. About 80% of the CpG dinucleotides in the human genome are methylated, and methylation has been implicated, among other functions, in silencing of repetitive sequences (9, 21, 41). Methylation can repress transcription when directly interfering with binding of sequence-specific transcription factors (20) or by an indirect mechanism involving the methyl-CpG binding domain proteins (16).
Transposable elements in genomic DNA are usually methylated, and cytosine methylation has been suggested to act as a defense mechanism against such “intragenomic parasites” (43). For example, the role of methylation in repressing transcription of mouse intracisternal A-type particles is well documented (18, 19, 41). There is evidence that transcription of HERVs is likewise influenced by methylation. For instance, transcription of HERV sequences in systemic lupus erythematosus may be due to methylation defects (33). Abrink et al. (1) related upregulation of H-plk, a human Kruppel-related zinc finger gene that is probably activated by an upstream ERV3 locus to tissue-specific demethylation of the corresponding gene region in several human cell lines.
Cellular methylation was also reported to influence HERV-K(HML-2) transcription. Treatment of the HERV-K(HML-2)-expressing cell line Tera-1 with 5-azacytidine, a demethylating agent, further increased expression of HERV-K(HML-2) Gag protein (14). Sequence regions flanking human-specific HERV-K(HML-2) LTRs were reported to be differently methylated in cerebellum versus lymph node tissue (24). Hypomethylation was also implicated to influence HERV-K(HML-2) expression in urothelial cancer (11). Thus, methylation levels seem to regulate transcription of HERV-K(HML-2) proviruses. However, an indirect effect of methylation is conceivable where general demethylation actually altered expression of cellular activators or repressors involved in the transcriptional regulation of these HERV elements.
We recently investigated HERV-K(HML-2) proviruses in the human genome capable of producing Rec protein, which is translated from a splicing product from the HERV-K(HML-2) env gene. We identified four HERV-K(HML-2) proviruses able to produce Rec mRNA, demonstrating transcriptional activity of the respective promoters housed in the proviral 5′ LTRs. Rec transcripts from other proviral loci with potential to produce Rec mRNA were not identified. The latter proviruses therefore may be transcriptionally much less active, or they may be inactive (28). In the present study, we investigated the transcriptional potential of various proviral 5′ LTRs in more detail, and examined the specific influence of CpG methylation on those LTRs in the Tera-1 and the T47D cell lines. We find that CpG methylation immediately affects HERV-K(HML-2) transcriptional activity and that CpG methylation effectively silences HERV-K(HML-2) LTRs. Yet, CpG methylation is not entirely responsible for different transcriptional activities observed between cell lines, e.g., CpG methylation does not silence HERV-K(HML-2) expression in the T47D cell line.
MATERIALS AND METHODS
Bisulfite treatment of DNA.
Genomic DNAs from Tera-1 cells and from T47D cells were isolated by standard methods. To determine CpG methylation levels, we employed the bisulfite sequencing method essentially as described by Fraga and Esteller (12). To demonstrate the efficiency of bisulfite treatment, we added 10 ng of a BamHI-digested pBlueScript plasmid (Stratagene) to the digested genomic DNAs before treatment. Plasmid and genomic DNAs were treated in the same tube. Primers PBSME-1 and PBSME-2 were used to amplify a 134-bp fragment of the vector on bisulfite-treated DNA as described by Goubely et al. (15).
PCR amplification, cloning, and sequencing.
We amplified, in total, five LTR loci from bisulfite-treated DNA by nested PCR. The first round of PCR contained 4 μl of treated DNA (∼100 ng). The Expand High Fidelity PCR system (Roche) was used following the recommended conditions. PCR cycles were as follows: 2 min at 94°C; 10 cycles, each cycle consisting 15 s at 94°C, 30 s at 55°C, and 40 s at 72°C; 25 cycles, each cycle consisting of 15 s at 94°C, 30 s at 55°C, and 40 s at 72°C (plus a 20-s ramp per cycle); and finally 7 min at 72°C. After gel electrophoresis, PCR products were eluted with a gel extraction kit (PeqLab). The second round of PCR contained 1 μl of the DNA eluate. PCR conditions were identical except for primer annealing at 60°C and the use of Taq polymerase (Invitrogen). The various PCR primer sequences are available from the authors on request. PCR products were subsequently cloned into the pGEM-T vector (Promega), and single clones were sequenced with the SequiTherm Excel TM II DNA Sequencing Kit-LC (Biozym) and an automated DNA sequencer (Licor 4000-L).
Plasmid generation.
The reporter vectors pGL3-Basic and pCMVβ were purchased from Promega and Clontech, respectively. In construct pGL3-CMV, luciferase expression is driven by the strong cytomegalovirus (CMV) early promoter. To generate pGL3-CMV, we excised the CMV promoter region from pCMVβ by a EcoRI/SmaI restriction digestion. The blunted fragment was cloned into the SmaI site of pGL3-Basic. Plasmids pLTR-c6, pLTR-c10, and pLTR-c11 harbor HERV-K(HML-2) 5′ LTRs from different proviruses (Table 1) upstream from the luciferase reporter gene. We amplified respective 5′ LTRs from human genomic DNA with a specific PCR primer located in an upstream flanking cellular sequence and a universal reverse primer in the proviral gag gene. Specific PCR primers were located between 32 and 85 bp upstream from the 5′ LTR. Due to a large amount of repetitive elements upstream from some of the LTRs, a nested PCR approach was employed to generate constructs pLTR-c5, pLTR-c7L, and pLTR-7C. Primer sequences included a HindIII site that was employed to clone PCR products into the pGL3-Basic vector previously linearized with HindIII. The Expand High Fidelity PCR system (Roche) was used for all amplifications. PCR primer sequences and PCR conditions are available from the authors on request.
TABLE 1.
HERV-K(HML-2) proviral 5′ LTRs investigated in this studya
| LTR designation | Locus no. | Chromosomal localization | Location in April 2003 Freeze | GenBank accession no. | Or | Transcripts in Tera-1 |
|---|---|---|---|---|---|---|
| LTR-c5 | 2 | 5p13.3 | chr5:30485437-30496686 | AC025757.3 | − | − |
| LTR-c6 | 3 | 6q14.1 | chr6:78375915-78387115 | AL590785.7 | − | + |
| LTR-c7L | 4 | 7p22.1 | chr7:4329521-4354837 | AC072054.10 | − | + |
| LTR-c7C | 5 | 7p22.1 | chr7:4329521-4354837 | AC072054.10 | − | + |
| LTR-c10 | 7 | 10p14 | chr10:7015479-7026716 | AL392086.14 | − | − |
| LTR-c11 | 8 | 11q22.1 | chr11:101598996-101610238 | AP000776.4 | + | + |
| LTR-c12 | 9 | 12q14.1 | chr12:58437090-58448318 | AC025420.26 | + | − |
LTR designation, different construct names; locus numbers, provirus numbers with reference to Mayer et al. (28). The provirus location in the April 2003 Freeze version of the Human Genome Browser (23), the GenBank accession number harboring a particular provirus, and the orientation (Or) within the GenBank entry (+, −) are given. The column “Transcripts in Tera-1” indicates the transcriptional activity of proviral loci as previously determined (28).
Cell lines and cell culture.
Human teratocarcinoma cells (Tera-1) were cultured in McCoy's 5A medium (PAA). The human breast carcinoma cell line T47D was cultured in RPMI 1640 medium (PAA). Cell culture media were supplemented with l-glutamine, 10% fetal calf serum, and penicillin-streptomycin (each at a concentration of 100 μg/ml).
Transfections and reporter gene assays.
Cells were grown in 12-well plates to 60 to 80% confluency. We used FuGENE6 (Roche) for transfections, following the manufacturer's recommendations. Luciferase and β-galactosidase activities were determined using the Luciferase and β-Galactosidase Assay Systems (Promega), following the manufacturer's recommendations. Forty-eight hours after transfection, cells were washed twice with 1× phosphate-buffered saline and were lysed in 1× reporter lysis buffer, followed by two freeze-thaw cycles. Luciferase activities were measured with a Lumat LB 9507 luminometer (Berthold Technologies). All reporter gene assays were performed at least in triplicate.
In vitro methylation.
Two micrograms of reporter plasmid DNA were methylated with SssI methylase (CpG) (New England Biolabs) under recommended conditions. Reaction mixtures contained 160 μM S-adenolsylmethionine (SAM) and were incubated from 30 min to 4 h, followed by heat inactivation for 15 min at 65°C. Negative-control reaction mixtures lacked SAM. To determine methylation levels, a 0.5-μg aliquot from each reaction mixture was treated with the methylation-sensitive restriction enzyme BstUI, and the resulting DNA fragments were separated by gel electrophoresis and were visualized by ethidium bromide staining.
RESULTS AND DISCUSSION
Promoter activity of HERV-K(HML-2) LTRs.
We recently investigated the HERV-K(HML-2) proviruses in the human genome capable of producing the Rec mRNA splicing product, and we investigated transcriptional activity of those proviruses. We determined HERV-K(HML-2) proviral loci on human chromosomes 6, 7, and 11 to be transcriptionally active in the Tera-1 cell line. Transcripts from proviral loci on human chromosomes 5, 10, and 12 were not among cloned and sequenced reverse transcription-PCR (RT-PCR) products (Table 1) (28). Thus, the latter are likely significantly underrepresented in, or missing from, the Tera-1 cellular mRNA pool.
We asked whether different representation of proviral sequences in the cellular mRNA pool was due to different promoter strength of the respective proviral 5′ LTRs. We PCR amplified the complete 5′ LTR sequence, including short stretches of upstream and downstream flanking sequence from seven HERV-K(HML-2) proviral loci (Table 1). We cloned the different LTRs harboring amplicons into a luciferase reporter vector and assayed their transcriptional activity in the Tera-1 cell line. In the following discussion, we designate the proviral 5′ LTR on human chromosome 5 as LTR-c5, for instance. Because the HERV-K(HML-2.HOM) provirus, located on human chromosome 7p22, is organized as a tandem provirus sharing a central LTR between both proviruses (35), we assayed the 5′ LTR as well as the central LTR of the tandem arrangement (designated LTR-c7L and LTR-c7C).
When assaying the different 5′ LTRs in the Tera-1 cell line, we found that all but one 5′ LTRs were transcriptionally active, with activities ranging from 18.34 to 4.25% relative to the activity of a luciferase control vector for which expression was driven by a strong CMV promoter. LTR-c11 displayed the highest activity and LTR-c6 the lowest activity. Only LTR-c10 activity was in the range of an empty reporter vector control. LTR-c10 was therefore concluded to be transcriptionally inactive (Fig. 1). Probably, the latter LTR is inactive because it is present in the genome for a longer time and has therefore acquired more mutations than active LTRs (see below). Taken together, the different representation of proviral transcripts in the Tera-1 cellular mRNA pool is not due to strongly different proviral 5′ LTR promoter activities. Other factors influencing HERV-K(HML-2) transcriptional activity must be considered to explain locus-specific transcription.
FIG. 1.
Transcriptional activities of various HERV-K(HML-2) 5′ LTRs in Tera-1 (left) and T47D (right) cells, as determined by luciferase reporter gene assays. LTR activities are given as percentages of a positive control vector driven by a CMV promoter. Standard deviations are indicated for each LTR. basic, a negative control vector that lacked a promoter sequence. All tested LTRs were inactive in T47D cells, shown on the right, solely to illustrate this finding.
Differential CpG methylation of LTR sequences in Tera-1.
Previous data showed that hypomethylation of genomic DNA boosted the overall HERV-K(HML-2) expression level (14). However, the expression levels could have resulted from an indirect effect. General demethylation might simply increase expression of one or more cellular factors that then activate HERV-K(HML-2) expression. We set out to elucidate whether CpG methylation directly influences HERV-K(HML-2) transcription.
In a first set of experiments, we investigated in detail the in vivo methylation status of five out of seven HERV-K(HML-2) proviral 5′ LTRs included in this study in the Tera-1 cell line, employing the bisulfite genomic sequencing method (13). We excluded LTR-c10 from the analysis, as it was shown to be transcriptionally inactive (see above). We further excluded LTR-c7C, because we did not succeed in obtaining specific PCR products for that LTR. The lack of specific PCR product was probably due to limitations of PCR product length in bisulfite genomic sequencing that employs significantly modified DNA as template.
We amplified each 5′ LTR from bisulfite-treated genomic Tera-1 DNA with specific PCR primers in the 5′ LTR upstream flanking region and a reverse primer located downstream from the 5′ LTR. The first PCR was followed by a seminested PCR, employing forward primers in the flanking region and a reverse primer in the 5′ LTR. By doing so, we were able to examine the first 493 bp of the 567-bp U3 region for each 5′ LTR.
We sequenced seven independent clones for each of the five analyzed 5′ LTRs, analyzing between five and nine CpG sites each (Table 2). We found significant methylation levels for three 5′ LTRs. A total of 20 of 35 CpG sites were methylated for LTR-c5, 23 of 63 sites were methylated for LTR-c11, and 35 of 49 CpG sites were methylated for LTR-c12. The overall methylation level of LTR-c11 (36%) appeared lower than that of LTR-c5 (57%) or that of LTR-c12 (71%). Several CpG sites in incompletely methylated LTRs were found to be either methylated or unmethylated. Bisulfite treatment efficiencies of 99%, as determined from the internal control, refute the hypothesis that incomplete methylation seen for these LTRs is due to an experimental artifact. In contrast to the results with the LTRs discussed above, we found that LTR-c7L and LTR-c6 were completely unmethylated in Tera-1 (Fig. 2).
TABLE 2.
Number of C nucleotides and CpG sites analyzed in various HERV-K(HML-2) 5′ LTR sequences employing bisulfite genomic sequencinga
| Provirus | Total C in LTR | Total C analyzed | Total CpG in LTR | Total CpG analyzed |
|---|---|---|---|---|
| LTR-c7L | 240 | 106 | 19 | 9 |
| LTR-c6 | 235 | 101 | 17 | 7 |
| LTR-c11 | 234 | 104 | 17 | 9 |
| LTR-c5 | 230 | 100 | 11 | 5 |
| LTR-c12 | 227 | 95 | 15 | 7 |
Total numbers of C and CpG and respective sites analyzed within the various proviral 5′ LTRs are listed. The proviruses are detailed in Table 1.
FIG. 2.
CpG site methylation status of U3 regions of five HERV-K(HML-2) proviral 5′ LTRs in Tera-1 cells, as determined by bisulfite sequencing. The upper part of the figure depicts the examined LTR U3 region, including PCR primer locations for amplification of LTRs from the human genome. Primers FP2 were specific for flanking cellular sequences, while primer RP2 was specific for the LTR sequence. The different proviral 5′ LTRs examined are indicated on the left. Methylated CpG sites are indicated by solid circles, and unmethylated CpG sites are indicated by open circles. Thus, LTR-c7L and LTR-c6 are unmethylated, while the others are partially methylated. Note that methylation patterns for the latter LTRs are variable. Methylation levels for CpG sites as well as other C nucleotides are summarized as percentages on the right side of the figure. Positive and negative RT-PCR results indicate transcriptional activity of a particular provirus in Tera-1 cells, and fractional amounts of RT-PCR products from respective proviruses, as determined previously (28), are given in parentheses.
Thus, methylation levels of the different examined HERV-K(HML-2) LTRs vary significantly in the Tera-1 cell line. Some LTRs appear significantly methylated, while others are completely unmethylated. When LTR methylation levels are compared to previously observed transcriptional activities (28), there is a good inverse correlation between methylation and transcriptional activity. The previously observed lack of HERV-K(HML-2) transcripts from proviruses on human chromosomes 5 and 12 in Tera-1 cells (28) could be explained by methylation-mediated 5′ LTR promoter silencing, as LTR-c5 and LTR-c12 were found to be methylated. In contrast, the proviruses on chromosomes 6 and 7 could be transcriptionally active in Tera-1 cells because of the observed complete lack of methylation for the corresponding LTR-c6 and LTR-c7. For LTR-c11, only 1 RT-PCR clone from the corresponding proviral locus was isolated, in contrast to 15 clones from the corresponding LTR-c7L provirus (28). Although LTR-c11 is about 60% transcriptionally more active than LTR-c7L (Fig. 1), the Tera-1 RNA pool contains significantly less transcript from LTR-c11 than from LTR-c7L. Incomplete methylation of LTR-c11, and thus partial silencing of LTR-c11, could explain lower representation of corresponding transcript in the Tera-1 cell line.
Taken together, these findings strongly support the idea that CpG methylation of HERV-K(HML-2) LTRs directly regulates transcription of HERV-K(HML-2) proviruses in the Tera-1 cell line.
Influence of methylation on HERV-K(HLM-2) LTRs.
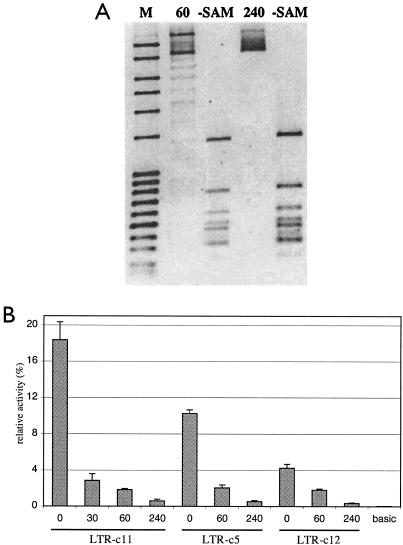
To further substantiate the direct influence of CpG methylation on the transcriptional activity of HERV-K(HML-2) 5′ LTRs, we examined promoter activities of in vitro-methylated LTRs. We methylated CpG sites in luciferase reporter constructs harboring LTR-c5, LTR-c11, and LTR-c12, employing the CpG-specific SssI methylase. Methylation levels of plasmid DNAs were assessed by BstUI restriction enzyme digestions, with BstUI sensitive towards methylated CpG within the recognition sequence. We observed increasing methylation levels for SssI incubation times ranging from 30 min to 4 h. Plasmid DNAs were fully methylated after 4 h of incubation (Fig. 3A).
FIG. 3.
Transcriptional activity of in vitro-methylated HERV-K(HML-2) 5′ LTRs in Tera-1 cells. (A) Different methylation levels of in vitro-methylated reporter constructs. Shown here are in vitro methylation reactions of reporter plasmids that were terminated after 60 and 240 min. Plasmid DNAs were subsequently treated with methylation-sensitive BstUI restriction enzyme, and the fragments were electrophoresed. Note that BstUI partially digests plasmid DNA incubated for 60 min while plasmid incubated for 240 min is completely protected, indicating incomplete and complete methylation levels, respectively.-SAM, control reaction mixtures incubated for similar time periods but lacking SAM substrate. Thus, BstUI is able to completely digest these plasmid DNAs. (B) Effect of different CpG methylation levels on transcriptional activity of HERV-K(HML-2) 5′ LTRs. Three different LTRs with transcriptional activity were examined. Incubation times for in vitro methylation reactions are given in minutes. Transcriptional activities are given as percentages of a CMV-driven positive control vector. basic, a promoter-lacking negative control vector.
We assayed transcriptional activities of the different in vitro-methylated LTR reporter constructs in Tera-1 cells. We observed that transcriptional activity negatively correlated with methylation levels. Increasing methylation levels continuously decreased transcriptional activity of the examined 5′ LTRs. Fully methylated constructs displayed only about 5.5% activity, compared to unmethylated constructs (Fig. 3B). Hence, methylated CpG sites in HERV-K(HML-2) 5′ LTRs strongly inhibit transcriptional activity of HERV-K(HML-2) proviral loci. These findings further corroborate that CpG methylation plays an important and direct role in the transcriptional regulation of the HERV-K(HML-2) family. Furthermore, partial transcriptional silencing of the LTR-c11 due to incomplete methylation levels in the Tera-1 cell line (see above) is corroborated. Partially in vitro-methylated LTRs display transcriptional activity, yet clearly lower activities than unmethylated LTRs. At this point, it remains to be elucidated whether methylated CpG dinucleotides inhibit binding of one or several transcription factors or whether methylated CpGs recruit methyl-CpG binding proteins.
Evidence for CpG methylation in old HERV-K(HML-2) LTRs.
Zsiros et al. (44) noted a significantly lower frequency of CpG dinucleotides in the reverse transcriptase portion of HERV-K(HML-2) proviruses. This finding suggested previous methylation of CpG sites, eventually resulting in fading of CpGs because of higher mutation rates of methylated cytosines (8).
We likewise found evidence for previous CpG methylation in HERV-K(HML-2) 5′ LTR sequences. Notably, the transcriptionally inactive LTR-c10 displays several mutated CpG sites compared to transcriptionally active LTRs (Fig. 4). Obviously, that LTR accumulated more mutations and therefore is probably evolutionarily older than the other LTRs. A higher evolutionary age of LTR-c10 is also indicated from phylogenetic analysis (data not shown). In the light of several mutated CpG sites and higher mutation rates of methylated cytosines, it can be concluded that LTR-c10 used to be methylated. Further evidence comes from evolutionarily older HERV-K(HML-2) proviruses. The so-called HERV-K(OLD) proviruses are ancestral, yet very similar in sequence, to the “modern” HERV-K(HML-2) proviruses. HERV-K(OLD) proviruses were formed in germ cells approximately 28 million years ago (36). We compared 5′ LTR sequences in modern HERV-K(HML-2) proviruses with 5′ LTR sequences in HERV-K(OLD) proviruses that we previously identified in the human genome (unpublished data). Similar to the LTR-c10 sequence, HERV-K(OLD) 5′ LTRs commonly displayed mutated CpG dinucleotides. In particular, (former) CpG sites in the 5′ portion of the LTRs appeared mutated (Fig. 4). We found that 94 of 129 CpG sites (73%) mutated to either TG or CA, which are expected mutations when methylated cytosine is converted to thymine. Thus, there is strong evidence that HERV-K(OLD) proviruses used to be methylated. In combination with above-described silencing of HERV-K(HML-2) LTRs by CpG methylation, we conclude that CpG methylation, at that point in evolution, was employed to silence HERV-K(OLD) proviruses after their integration into the germ line 28 million years ago, just as CpG methylation at present represses the expression of modern HERV-K(HML-2) proviruses. During evolution, CpG methylation appears to have been a universal mechanism to silence HERV-K(HML-2) proviral loci.
FIG. 4.
Occurrence of CpG sites in 5′ LTRs belonging to the HERV-K(HML-2) and the HERV-K(OLD) families (see the text for details). HS consensus is a previously reported consensus sequence for human-specific HERV-K(HML-2) LTRs (7). The entire 5′ LTR sequence is shown for each LTR. Each sequence was analyzed for the presence of CpG sites (triangles), as well as apparently mutated CpG sites (X's). TATA boxes are furthermore indicated for each LTR, when they could be identified. For each LTR, the 5′-most 620 bp were analyzed. Note that HERV-K(OLD) 5′ LTRs frequently display mutated CpG sites, indicating former CpG methylation.
HERV-K(HML-2) inactivity in T47D cells is not due to CpG methylation.
In the course of the study of HERV-K(HML-2) Rec mRNA expression in the Tera-1 cell line (28), Rec mRNA expression in the breast cancer cell line T47D was also examined. However, no RT-PCR products were obtained from T47D cells under experimental conditions identical to the one for the Tera-1 cell line (J. Mayer and M. Sauter, unpublished results). Hence, transcription levels of Rec-producing proviral loci are significantly lower in T47D cells than in Tera-1 cells, or HERV-K(HML-2) proviruses are transcriptionally inactive in T47D. We examined in more detail the observed cell type specificity. We assayed the promoter activity of the seven 5′ LTR luciferase reporter constructs in the T47D cell line. In contrast to the Tera-1 cell line, none of the different reporter constructs displayed promoter activities in T47D cells (Fig. 1). We furthermore determined methylation levels for three HERV-K(HML-2) 5′ LTRs (LTR-c6, LTR-c7L, and LTR-c11) in T47D cells. We observed methylation levels of 71 and 44% for LTR-c6 and LTR-c11, respectively. The overall methylation level of LTR-c11 (44%) appeared lower than that of LTR-c6. On the other hand, no CpG methylation was found for LTR-c7L (Fig. 5). In contrast to the Tera-1 cell line, LTR-c7L is transcriptionally inactive in T47D, although it is likewise unmethylated. Thus, CpG methylation levels are not sufficient to fully explain transcriptional activity of HERV-K(HML-2) proviruses in particular cell types, such as the T47D cells that were examined in this study. In combination with the above-described transcriptional inactivity of LTR reporter constructs in T47D cells, one can conclude that either cellular activators or repressors involved in HERV-K(HML-2) transcription are missing or present, respectively, in T47D cells. It is currently unclear what cellular factors trigger HERV-K(HML-2) activity. Those factors remain to be identified in future studies.
FIG. 5.
CpG site methylation status of U3 regions of three HERV-K(HML-2) proviral 5′ LTRs in T47D cells, as determined by bisulfite sequencing. Please refer to the legend to Fig. 2 for details.
We show in the present study that CpG methylation directly influences transcription of HERV-K(HML-2) sequences. Proviral 5′ LTRs are efficiently silenced by CpG methylation. Our findings have immediate implications for the understanding of upregulated HERV-K(HML-2) expression observed in GCTs. It was previously reported that the genome of seminoma tumor cells is hypomethylated, including CpG dinucleotides (39). Another study showed general hypomethylation in ovarian carcinoma (32). Therefore, changes in the CpG methylation pattern towards CpG hypomethylation, which very likely also concern CpGs located in HERV-K(HML-2) 5′ LTRs, can directly result in transcriptional activation of HERV-K(HML-2) 5′ LTRs. CpG methylation as a repressing factor for HERV-K(HML-2) transcriptional activity is removed from the 5′ LTRs. Presence of thus-far-unidentified cellular (transcription) factors in seminoma cells could then enable expression of HERV-K(HML-2). This may also apply to other tissues. For instance, Khodosevich et al. recently reported a number of human-specific HERV-K(HML-2) LTRs that appeared to be located in differentially methylated genomic regions, when compared among each other and between cerebellum and lymph node tissue (24). So, with appropriate methylation levels and in the presence of required transcription factors, particular HERV-K(HML-2) LTRs may be active in other tissues as well. While our study demonstrates the direct role of CpG methylation in HERV-K(HML-2) transcriptional regulation, the cellular (transcription) factors contributing to high HERV-K(HML-2) expression remain to be elucidated in future studies.
Acknowledgments
This study was supported by grants from the Deutsche Forschungsgemeinschaft to J.M. (Ma2298/2-1) and E.M. (Me917/16-1).
REFERENCES
- 1.Abrink, M., E. Larsson, and L. Hellman. 1998. Demethylation of ERV3, an endogenous retrovirus regulating the Kruppel-related zinc finger gene H-plk, in several human cell lines arrested during early monocyte development. DNA Cell Biol. 17:27-37. [DOI] [PubMed] [Google Scholar]
- 2.Akopov, S. B., L. G. Nikolaev, P. P. Khil, Y. B. Lebedev, and E. D. Sverdlov. 1998. Long terminal repeats of human endogenous retrovirus K family (HERV-K) specifically bind host cell nuclear proteins. FEBS Lett. 421:229-233. [DOI] [PubMed] [Google Scholar]
- 3.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout, B., M. Jebbink, and J. Zsiros. 1999. Identification of an active reverse transcriptase enzyme encoded by a human endogenous HERV-K retrovirus. J. Virol. 73:2365-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boese, A., M. Sauter, U. Galli, B. Best, H. Herbst, J. Mayer, E. Kremmer, K. Roemer, and N. Mueller-Lantzsch. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 19:4328-4336. [DOI] [PubMed] [Google Scholar]
- 6.Boller, K., O. Janssen, H. Schuldes, R. R. Tonjes, and R. Kurth. 1997. Characterization of the antibody response specific for the human endogenous retrovirus HTDV/HERV-K. J. Virol. 71:4581-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buzdin, A., S. Ustyugova, K. Khodosevich, I. Mamedov, Y. Lebedev, G. Hunsmann, and E. Sverdlov. 2003. Human-specific subfamilies of HERV-K (HML-2) long terminal repeats: three master genes were active simultaneously during branching of hominoid lineages. Genomics 81:149-156. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, D. N., and M. Krawczak. 1989. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum. Genet. 83:181-188. [DOI] [PubMed] [Google Scholar]
- 9.Costello, J. F., and C. Plass. 2001. Methylation matters. J. Med. Genet. 38:285-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn, C. A., P. Medstrand, and D. L. Mager. 2003. An endogenous retroviral long terminal repeat is the dominant promoter for human β1,3-galactosyltransferase 5 in the colon. Proc. Natl. Acad. Sci. USA 100:12841-12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Florl, A. R., R. Lower, B. J. Schmitz-Drager, and W. A. Schulz. 1999. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br. J. Cancer 80:1312-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraga, M. F., and M. Esteller. 2002. DNA methylation: a profile of methods and applications. BioTechniques 33:632. [DOI] [PubMed] [Google Scholar]
- 13.Frommer, M., L. E. McDonald, D. S. Millar, C. M. Collis, F. Watt, G. W. Grigg, P. L. Molloy, and C. L. Paul. 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 89:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gotzinger, N., M. Sauter, K. Roemer, and N. Mueller-Lantzsch. 1996. Regulation of human endogenous retrovirus-K Gag expression in teratocarcinoma cell lines and human tumours. J. Gen. Virol. 77:2983-2990. [DOI] [PubMed] [Google Scholar]
- 15.Goubely, C., P. Arnaud, C. Tatout, J. S. Heslop-Harrison, and J. M. Deragon. 1999. S1 SINE retroposons are methylated at symmetrical and non-symmetrical positions in Brassica napus: identification of a preferred target site for asymmetrical methylation. Plant Mol. Biol. 39:243-255. [DOI] [PubMed] [Google Scholar]
- 16.Hendrich, B., and A. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herbst, H., M. Sauter, C. Kuhler-Obbarius, T. Loning, and N. Mueller-Lantzsch. 1998. Human endogenous retrovirus (HERV)-K transcripts in germ cell and trophoblastic tumours. APMIS 106:216-220. [DOI] [PubMed] [Google Scholar]
- 18.Hojman-Montes de Oca, F., J. Lasneret, L. Dianoux, M. Canivet, R. Ravicovitch-Ravier, and J. Peries. 1984. Regulation of intracisternal A particles in mouse teratocarcinoma cells: involvement of DNA methylation in transcriptional control. Biol. Cell 52:199-204. [DOI] [PubMed] [Google Scholar]
- 19.Hsiao, W. L., S. Gattoni-Celli, and I. B. Weinstein. 1986. Effects of 5-azacytidine on expression of endogenous retrovirus-related sequences in C3H 10T1/2 cells. J. Virol. 57:1119-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iguchi-Ariga, S. M., and W. Schaffner. 1989. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 3:612-619. [DOI] [PubMed] [Google Scholar]
- 21.Jaenisch, R., and A. Bird. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33(Suppl.):245-254. [DOI] [PubMed] [Google Scholar]
- 22.Kapitonov, V. V., and J. Jurka. 1999. The long terminal repeat of an endogenous retrovirus induces alternative splicing and encodes an additional carboxy-terminal sequence in the human leptin receptor. J. Mol. Evol. 48:248-251. [DOI] [PubMed] [Google Scholar]
- 23.Kent, W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The human genome browser at UCSC. Genome Res. 12:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khodosevich, K., Y. Lebedev, and E. D. Sverdlov. 2004. Large-scale determination of the methylation status of retrotransposons in different tissues using a methylation tags approach. Nucleic Acids Res. 32:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knossl, M., R. Lower, and J. Lower. 1999. Expression of the human endogenous retrovirus HTDV/HERV-K is enhanced by cellular transcription factor YY1. J. Virol. 73:1254-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landry, J. R., A. Rouhi, P. Medstrand, and D. L. Mager. 2002. The opitz syndrome gene mid1 is transcribed from a human endogenous retroviral promoter. Mol. Biol. Evol. 19:1934-1942. [DOI] [PubMed] [Google Scholar]
- 27.Magin, C., R. Lower, and J. Lower. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 73:9496-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer, J., S. Ehlhardt, M. Seifert, M. Sauter, N. Muller-Lantzsch, Y. Mehraein, K. D. Zang, and E. Meese. 2004. Human endogenous retrovirus HERV-K(HML-2) proviruses with Rec protein coding capacity and transcriptional activity. Virology 322:190-198. [DOI] [PubMed] [Google Scholar]
- 29.Mayer, J., E. Meese, and N. Mueller-Lantzsch. 1997. Chromosomal assignment of human endogenous retrovirus K (HERV-K) env open reading frames. Cytogenet. Cell Genet. 79:157-161. [DOI] [PubMed] [Google Scholar]
- 30.Mayer, J., E. Meese, and N. Mueller-Lantzsch. 1997. Multiple human endogenous retrovirus (HERV-K) loci with gag open reading frames in the human genome. Cytogenet. Cell Genet. 78:1-5. [DOI] [PubMed] [Google Scholar]
- 31.Medstrand, P., J. R. Landry, and D. L. Mager. 2001. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J. Biol. Chem. 276:1896-1903. [DOI] [PubMed] [Google Scholar]
- 32.Menendez, L., B. B. Benigno, and J. F. McDonald. 2004. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol. Cancer 3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada, M., H. Ogasawara, H. Kaneko, T. Hishikawa, I. Sekigawa, H. Hashimoto, N. Maruyama, Y. Kaneko, and N. Yamamoto. 2002. Role of DNA methylation in transcription of human endogenous retrovirus in the pathogenesis of systemic lupus erythematosus. J. Rheumatol. 29:1678-1682. [PubMed] [Google Scholar]
- 34.Ono, M., M. Kawakami, and H. Ushikubo. 1987. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J. Virol. 61:2059-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reus, K., J. Mayer, M. Sauter, D. Scherer, N. Muller-Lantzsch, and E. Meese. 2001. Genomic organization of the human endogenous retrovirus HERV-K(HML-2.HOM) (ERVK6) on chromosome 7. Genomics 72:314-320. [DOI] [PubMed] [Google Scholar]
- 36.Reus, K., J. Mayer, M. Sauter, H. Zischler, N. Muller-Lantzsch, and E. Meese. 2001. HERV-K(OLD): ancestor sequences of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 75:8917-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauter, M., K. Roemer, B. Best, M. Afting, S. Schommer, G. Seitz, M. Hartmann, and N. Mueller-Lantzsch. 1996. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res. 56:4362-4365. [PubMed] [Google Scholar]
- 38.Sauter, M., S. Schommer, E. Kremmer, K. Remberger, G. Dolken, I. Lemm, M. Buck, B. Best, D. Neumann-Haefelin, and N. Mueller-Lantzsch. 1995. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 69:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smiraglia, D. J., J. Szymanska, S. M. Kraggerud, R. A. Lothe, P. Peltomaki, and C. Plass. 2002. Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene 21:3909-3916. [DOI] [PubMed] [Google Scholar]
- 40.Tonjes, R. R., F. Czauderna, and R. Kurth. 1999. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J. Virol. 73:9187-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]
- 42.Yang, J., H. P. Bogerd, S. Peng, H. Wiegand, R. Truant, and B. R. Cullen. 1999. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc. Natl. Acad. Sci. USA 96:13404-13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]
- 44.Zsiros, J., M. F. Jebbink, V. V. Lukashov, P. A. Voute, and B. Berkhout. 1999. Biased nucleotide composition of the genome of HERV-K related endogenous retroviruses and its evolutionary implications. J. Mol. Evol. 48:102-111. [DOI] [PubMed] [Google Scholar]