Abstract
The smallest capsid proteins (SCPs) of the human herpesviruses differ substantially in size and sequence and are thought to impart some unique aspects of infection to their respective viruses. We used electron cryomicroscopy and antibody labeling to show that the 8-kDa SCP of human cytomegalovirus is attached only to major capsid protein subunits of the hexons, not the pentons. Thus, the SCPs of different herpesviruses illustrate that a protein can evolve significantly in sequence, structure, and function, while preserving its role in the architecture of the virus by binding to a specific partner in a specific oligomeric state.
The herpesviruses are large, double-stranded DNA viruses whose capsids share a high degree of structural similarity. The representative human herpesviruses—herpes simplex virus type 1 (HSV-1, a member of the alphaherpesvirus subfamily), human cytomegalovirus (HCMV, a member of the betaherpesvirus subfamily), and Kaposi's sarcoma-associated herpesvirus (KSHV, a member of the gammaherpesvirus subfamily)—all possess icosahedral capsids built from four structural proteins. The major capsid protein (MCP) is known as VP5 in HSV-1, pORF25 in KSHV, and MCP (pUL86) in HCMV. It can form hexons, which make up the faces of the icosahedral capsid, and pentons, which make up the vertices. The pentons and hexons are connected by trimers formed by the two minor capsid proteins. Finally, the smallest capsid protein (SCP)—known as VP26 in HSV-1, pORF65 in KSHV, and SCP (pUL48/49) in HCMV—decorates the outside of the capsid. These SCPs range in size from 16 kDa in KSHV to 8 kDa in HCMV.
The SCPs are of great interest since they share the least sequence homology among the capsid proteins of herpesviruses (21). The other major and minor capsid proteins of the herpesviruses share relatively high sequence similarity, so the overall capsid structure is very similar among species, but the conformations, locations, and functions of the SCPs may be more diverse. The SCP of HCMV has been shown to be essential for HCMV infection in vivo (3), whereas its counterpart in HSV-1, VP26, is dispensable for HSV-1 infection (6, 10). In KSHV, although the presence of SCP (pORF65) is used as a hallmark of KSHV infection for diagnostic purposes, whether it is essential for KSHV infection remains unknown.
The HCMV SCP has been difficult to study because of its small size and because the inherent difficulties in culturing HCMV have limited the achievable resolution of its three-dimensional (3D) structure. Recent biochemical and immunoprecipitation studies by Lai and Britt confirmed that the SCP interacts specifically with the MCP (13). Based on architectural similarities, it was proposed that the HCMV SCP, like the SCPs of HSV-1 and KSHV (15, 19, 24), binds to MCP hexon subunits only (5, 7). However, these suggestions could not be verified by these structural studies because of the low resolutions (∼35 Å) of the HCMV capsid maps. At such resolutions, protein boundaries cannot be resolved, and it is difficult to recognize structural features of proteins as small as SCP. In fact, even at 8.5-Å resolution, at which α-helices can be resolved, it has proven difficult to clearly delineate VP26 in HSV-1 hexons because of its extensive and interdigitated interactions across a large area of the MCP upper domain (23) and because its secondary structure consists mainly of β-sheets (20). Because there are no reverse-engineering systems to generate an SCP-minus HCMV capsid for comparative structural studies as has been done with HSV-1 (19, 24), biochemical techniques, such as antibody labeling, seem to be the only available practical means by which to directly localize the HCMV SCP.
We have improved our HCMV purification and electron cryomicroscopy (cryoEM) imaging techniques such that we were able to obtain sufficient data to reconstruct the HCMV capsid to a higher resolution (22 Å) than previously achieved. Using anti-SCP antibody labeling, we show that the SCP of HCMV binds only to the tips of MCP hexon subunits.
Culture and imaging of HCMV capsids.
HCMV has been difficult to culture in sufficient quantities for high-resolution structural studies, since the capsids tend to aggregate within cellular debris. The standard HCMV capsid purification protocol (11) involves growing human foreskin fibroblasts to 90% confluence, infecting them with very-high-multiplicity-of-infection HCMV stocks (∼5 particles/cell), lysing them to release the nuclei, and breaking the nuclei to release the capsids. We improved on this protocol by resuspending the capsids in a 1% solution of NP-40 and sonicating mildly with a Branson probe to dislodge the capsids from the debris before purifying and concentrating them. HCMV-infected cells collected from a total of 20 two-liter roller bottles were used to generate about 40 μl of concentrated HCMV capsids for the experiments in this study.
To generate the antibody-labeled HCMV capsids, we added a fivefold excess of purified monoclonal anti-SCP antibody (11.2.23; 1 mg/ml) (12) to 20 μl of capsid sample and incubated it overnight at 4°C. We used negative-stain electron microscopy with 2% uranyl acetate to evaluate the extent of antibody labeling by observing capsid cross-linkage and aggregation caused by the dual Fab arms of the antibodies. The antibody-labeled capsids were centrifuged in a desktop Eppendorf centrifuge at 13,000 × g for 20 min to remove unbound antibodies and concentrate the capsids. The sample was then resuspended in phosphate-buffered saline and sonicated in a water bath for 30 s to loosen the aggregated capsids.
We used established cryoEM procedures to image the capsids (1). Briefly, 3 μl of purified sample was applied to holey grids and quickly frozen in liquid ethane, such that the capsids were suspended in a thin layer of vitreous ice across the holes of the supporting film. Focal-pair image frames were acquired using a 200-kV JEOL 2010F electron microscope with a field emission gun. This microscope was also equipped with a Gatan US4000 4k- by 4k-pixel charge-coupled device (CCD) camera, which allows us to obtain more image data from a single sample of ice-embedded capsids than is possible using a microscope with a traditional film-loaded camera. Digital cryoEM images were recorded at an effective magnification of 83,100× on the CCD (60,000× on the film plane), corresponding to an effective pixel size of 1.805 Å/pixel at the specimen level, with a specimen dosage of approximately 12 electrons/Å2/micrograph. Focal-pair 4k- by 4k-pixel CCD images were recorded and preprocessed for translation and rotation correction using the JAMES semiautomatic data collection program (1). The electron beam was underfocused, with a 1.5-μm difference between the close-to-focus (∼1 μm under focus) and far-from-focus (∼2.5 μm under focus) images. For data processing, all images were averaged by combining adjacent pixels to yield a final sampling size of 3.61 Å/pixel on the specimen scale.
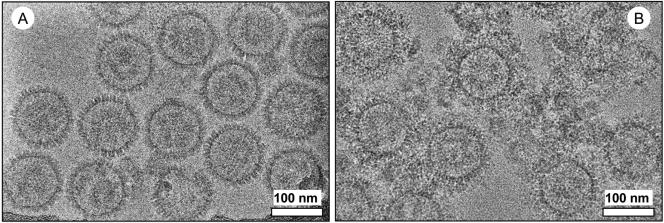
The micrographs of unlabeled HCMV capsids (Fig. 1A) show clean, evenly spaced capsids. In contrast, the antibody-labeled capsids (Fig. 1B) are surrounded by rings of capsid-bound antibodies and interspersed with clumps that may consist of antibodies nonspecifically bound to debris. The antibodies appear to completely surround the capsids, suggesting that most or all of the binding sites on the capsid surface were successfully saturated.
FIG. 1.
CryoEM micrographs of purified HCMV capsids. (A) Unlabeled capsids. (B) Capsids labeled with monoclonal anti-SCP antibodies, visible as clumps of density surrounding the capsids.
Localization of SCP on antibody-labeled capsids.
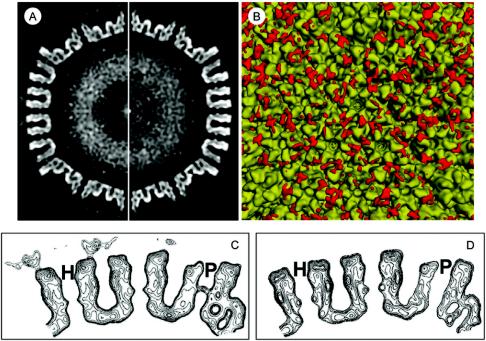
Image processing and 3D reconstruction were performed using the IMIRS software package running on a Dell Dimensional 4500 workstation with Microsoft Windows XP (14, 22). 3D visualization was carried out using Iris Explorer (NAG, Downers Grove, Ill.) with custom-designed modules. We computed a 3D reconstruction of the antibody-labeled HCMV capsid to an effective resolution of 22 Å from 446 particles (Fig. 2A). We also reconstructed the unlabeled capsid from 479 particles and scaled it to the same resolution (Fig. 2B). The effective resolutions of the maps were estimated based on the criterion that the Fourier shell cross-correlation coefficient between two independent reconstructions must be 0.5. A closer view of the antibody-labeled capsid (Fig. 2C) reveals extra densities, not seen in corresponding locations on the unlabeled capsid, that we attributed to antibodies attached to SCP on the capsid surface. These antibodies appear to be bound to the hexons, have a rough Y shape when examined closely (Fig. 2D), and are not seen in the vicinity of the pentons or the triplexes. Although the images of the unlabeled capsids contained enough data to obtain an ∼10-Å structure, the antibody-labeled capsid was reconstructed only to 22 Å because the flexible antibody densities would have been averaged out and would not be as clearly visible at a higher resolution.
FIG. 2.
3D reconstructions of HCMV anti-SCP labeled and unlabeled capsids at 22-Å resolution. For clarity, the structures are colored according to radius (see color bar), so densities at different radial distances from the particle center are colored differently. (A) Antibody-labeled capsid, with antibody-attributed densities colored in purple. (B) Unlabeled capsid. (C) Close-up of boxed area from panel A. Antibody-attributed densities are attached only to MCP subunits of hexons (H), not pentons (arrow). (D) Another close-up clearly showing attachment of antibody to tip of hexon subunit.
We next took 3.61-Å-thick central slices through the density maps, perpendicular to the twofold axes (Fig. 3A). These slices further illustrate the extra antibody-attributed densities attached to the surface of the labeled capsid but not the unlabeled capsid. A closer view of the cross section, illustrated by contour displays, reveals that the antibody densities are bound to the hexons of the labeled capsid (Fig. 3C), whereas the pentons of the labeled and unlabeled (cf. pentons in Fig. 3C and D) capsids are essentially identical.
FIG. 3.
Thin slices and difference mapping of unlabeled and antibody-labeled HCMV capsids. (A) Central slice down the twofold axis of the antibody-labeled (left) and unlabeled (right) HCMV capsid structures from Fig. 2. Extra densities attributed to bound antibodies are visible as a discontinuous ring around the capsid (left). (B) Difference map confirming the binding of anti-SCP antibodies to hexons but not pentons. The 3D structure of the unlabeled capsid (Fig. 2B) was subtracted from that of the labeled capsid (Fig. 2A), and the difference map (red densities) was superimposed onto the structure of the unlabeled capsid. (C and D) Contour maps showing close-up of twofold slice of unlabeled (D) and antibody-labeled (C) capsids. Antibody-attributed densities bound to hexons (H) but not pentons (P) can be seen.
We then obtained a difference map (Fig. 3B) by subtracting the unlabeled capsid structure (Fig. 2B), filtered to 22 Å, from the antibody-labeled structure (Fig. 2A) and superimposing the difference onto the unlabeled map (Fig. 3B). The difference map confirms that the significant extra densities on the antibody-labeled capsid are bound only to hexons, not to pentons or triplexes. There are some small areas of difference that might not be part of the antibody densities. These may be caused by slight surface variations between the labeled and unlabeled capsid maps, since maps of moderate resolution reconstructed from modest numbers of particles are expected to show some statistical fluctuations. In addition, the antibody-attributed densities appear variable and unevenly distributed for several reasons. First, the antibodies are flexible at the Fc hinge, so these regions may not be visible after icosahedral averaging. Second, there may have been uneven antibody binding due to steric hindrance between antibody molecules, such that not all of the possible SCP binding sites were saturated.
Implications of SCPs in viral specificity.
The SCPs of human herpesviruses have several common features: they are small and highly basic, and they decorate the surface of the capsid by binding to MCP subunits of hexons. However, they are highly variable across different herpesviruses (Fig. 4) and appear to have different functions in regard to capsid assembly and infection. In HSV-1, VP26 is not essential for capsid assembly or replication in vitro (6, 8, 10, 17), but it is required for production of infectious virus in vivo (10). In HCMV, the SCP is required for viral infection in vitro (3). The overall capsid structure of the channel catfish virus, a distant relative of the human herpesviruses, is almost identical to that of the other herpesviruses despite the absence of an SCP homolog (2), suggesting that the protein does not contribute to the overall organization of the capsid. The binding of SCP to the tips of the HCMV hexons suggests that it may be involved in interactions with the viral tegument, although previous structural studies have shown that the HCMV tegument proteins bind both hexons and pentons (7).
FIG. 4.
Sequence alignment of HSV-1, HCMV, and KSHV SCPs. CLUSTALW (18) yielded a poor alignment between the HSV-1 VP26 (GenBank accession number P10219), HCMV SCP (GenBank accession number AAR31612), and KSHV pORF65 (GenBank accession number NP_572121) sequences. The average sequence identity among the three SCP homologs was 15.9%, as computed using the program ALIGN (16).
Various methods have been used to show that the SCPs interact with the MCPs. Studies of recombinant SCP-minus HSV-1 capsids revealed that VP26 interacts with VP5, but only in the hexon conformation (19, 24). 3D reconstruction has shown that VP26, which is composed mainly of β-sheets (20, 23), forms a ring of horn-shaped densities around the HSV-1 hexons (19, 24). Similarly, we recently used anti-pORF65 antibody labeling to show that pORF65 binds only to hexons in the KSHV capsid (15). Our reconstruction of the antibody-labeled HCMV capsid shows that SCP, like its homologs in other human herpesviruses, binds the tips of MCP hexon subunits. However, we cannot discern the exact shape of the protein or the precise location of its attachment to the MCP. Because the HCMV SCP is so small, it is not likely to form a ring like VP26 unless it takes on an unusual conformation, such as a ribbon shape. Further experiments designed to remove SCP from the capsids will be required to determine the exact shape of the protein.
It is also not clear why SCP binds only to hexons and not to pentons. This may be related to local differences between MCP subunits in pentons and hexons, such as differences in packing or electrochemical properties. The MCP of HSV-1, VP5, has been shown to have different electrochemical properties in pentons and hexons (4). The key region of VP26 that has been shown to interact with VP5 (9) contains basic residues that may interact with an acidic polyproline loop in the VP5 upper domain (4). A similar mechanism may be at work in HCMV, as the sequence in SCP that has been found to interact with MCP (13) also contains basic residues. An atomic model of the HCMV MCP upper domain and a subnanometer-resolution HCMV capsid map revealing the MCP-SCP interface will be required to identify the corresponding SCP-interacting residues. All these questions await further investigation. Nevertheless, it can be said that the herpesvirus SCPs are unique capsid proteins that represent an unusual example in which a protein evolves extensively in its amino acid sequence, 3D structure, and function while preserving its architectural property of binding to a specific partner in a specific oligomeric state.
Acknowledgments
This research was supported by the National Institutes of Health (NIH grants AI46420 to Z.H.Z. and AI35602 to W.J.B.).
We thank Yong-Hwan Kim for providing an HCMV virion stock and Zhenming Zhao for assistance during the initial cell culture for HCMV capsid isolation. We acknowledge the use of the cryoEM facility at the National Center for Macromolecular Imaging directed by Wah Chiu (supported by NIH grant P41RR02250).
REFERENCES
- 1.Booth, C. R., W. Jiang, M. L. Baker, Z. H. Zhou, S. J. Ludtke, and W. Chiu. Sub-nanometer resolution structure of single particle reconstructed from CCD captured images of a 200 kV electron cryomicroscope. J. Struct. Biol. 147:116-127. [DOI] [PubMed]
- 2.Booy, F. P., B. L. Trus, A. J. Davison, and A. C. Steven. 1996. The capsid architecture of channel catfish virus, an evolutionarily distant herpesvirus, is largely conserved in the absence of discernible sequence homology with herpes simplex virus. Virology 215:134-141. [DOI] [PubMed] [Google Scholar]
- 3.Borst, E. M., S. Mathys, M. Wagner, W. Muranyi, and M. Messerle. 2001. Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J. Virol. 75:1450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman, B. R., M. L. Baker, F. J. Rixon, W. Chiu, and F. A. Quiocho. 2003. Structure of the herpesvirus major capsid protein. EMBO J. 22:757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butcher, S. J., J. Aitken, J. Mitchell, B. Gowen, and D. J. Dargan. 1998. Structure of the human cytomegalovirus B capsid by electron cryomicroscopy and image reconstruction. J. Struct. Biol. 124:70-76. [DOI] [PubMed] [Google Scholar]
- 6.Chen, D. H., J. Jakana, D. McNab, J. Mitchell, Z. H. Zhou, M. Dougherty, W. Chiu, and F. J. Rixon. 2001. The pattern of tegument-capsid interaction in the herpes simplex virus type 1 virion is not influenced by the small hexon-associated protein VP26. J. Virol. 75:11863-11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, D. H., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 8.Chi, J. H., and D. W. Wilson. 2000. ATP-dependent localization of the herpes simplex virus capsid protein VP26 to sites of procapsid maturation. J. Virol. 74:1468-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai, P., J. C. Akpa, and S. Person. 2003. Residues of VP26 of herpes simplex virus type 1 that are required for its interaction with capsids. J. Virol. 77:391-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai, P., N. A. DeLuca, and S. Person. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247:115-124. [DOI] [PubMed] [Google Scholar]
- 11.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39:389-400. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, W., K. S. Clopper, W. J. Britt, and M. K. Baxter. 1996. Human cytomegalovirus (HCMV) smallest capsid protein identified as product of short open reading frame located between HCMV UL48 and UL49. J. Virol. 70:5680-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai, L., and W. J. Britt. 2003. The interaction between the major capsid protein and the smallest capsid protein of human cytomegalovirus is dependent on two linear sequences in the smallest capsid protein. J. Virol. 77:2730-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang, Y., E. Y. Ke, and Z. H. Zhou. 2002. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J. Struct. Biol. 137:292-304. [DOI] [PubMed] [Google Scholar]
- 15.Lo, P., X. Yu, I. Atanasov, B. Chandran, and Z. H. Zhou. 2003. Three-dimensional localization of pORF65 in Kaposi's sarcoma-associated herpesvirus capsid. J. Virol. 77:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myers, E. W., and W. Miller. 1988. Optimal alignments in linear space. Comput. Appl. Biosci. 4:11-17. [DOI] [PubMed] [Google Scholar]
- 17.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trus, B. L., F. L. Homa, F. P. Booy, W. W. Newcomb, D. R. Thomsen, N. Cheng, J. C. Brown, and A. C. Steven. 1995. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J. Virol. 69:7362-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wingfield, P. T., S. J. Stahl, D. R. Thomsen, F. L. Homa, F. P. Booy, B. L. Trus, and A. C. Steven. 1997. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J. Virol. 71:8955-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, L., P. Lo, X. Yu, J. K. Stoops, B. Forghani, and Z. H. Zhou. 2000. Three-dimensional structure of the human herpesvirus 8 capsid. J. Virol. 74:9646-9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou, Z. H., and W. Chiu. 2003. Determination of icosahedral virus structures by electron cryomicroscopy at subnanometer resolution. Adv. Prot. Chem. 64:93-124. [DOI] [PubMed] [Google Scholar]
- 23.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 Å. Science 288:877-880. [DOI] [PubMed] [Google Scholar]
- 24.Zhou, Z. H., J. He, J. Jakana, J. D. Tatman, F. J. Rixon, and W. Chiu. 1995. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 2:1026-1030. [DOI] [PubMed] [Google Scholar]