Abstract
T-cell receptor-CD3 complex (TCR) is a versatile signaling machine that can initiate antigen-specific immune responses based on various biochemical changes of CD3 cytoplasmic domains, but the underlying structural basis remains elusive. Here we developed biophysical approaches to study the conformational dynamics of CD3ε cytoplasmic domain (CD3εCD). At the single-molecule level, we found that CD3εCD could have multiple conformational states with different openness of three functional motifs, i.e., ITAM, BRS and PRS. These conformations were generated because different regions of CD3εCD had heterogeneous lipid-binding properties and therefore had heterogeneous dynamics. Live-cell imaging experiments demonstrated that different antigen stimulations could stabilize CD3εCD at different conformations. Lipid-dependent conformational dynamics thus provide structural basis for the versatile signaling property of TCR.
Keywords: T-cell receptor, lipid, conformational dynamics, atomic force microscopy, nuclear magnetic resonance, live-cell imaging
Introduction
A key feature of adaptive immunity is the antigen specificity. Different pathogens can trigger distinct adaptive immune responses, but the underlying molecular mechanism is not fully understood. Adaptive immune response is initiated by the engagement of T-cell receptor-CD3 complex (TCR) with antigenic peptide presented by MHC molecule (pMHC)1,2,3,4,5,6, which can trigger downstream signaling pathways to activate T cells7,8. Recent works demonstrate that TCR is not a binary (active or inactive) but a versatile signaling machinery that can trigger antigen-specific effector function of T cells9,10,11,12,13,14. For example, in a Listeria monocytogenes infection model, antigen with long TCR-pMHC interaction time induces the biased differentiation to T follicular helper cells, while antigen with short TCR-pMHC interaction time induces the biased differentiation to T helper 1 cells11,12. For CD8+ T cells, antigen with high TCR affinity induces asymmetric cell division and differentiation into tissue-infiltrating effector cells, whereas antigen with low TCR affinity induces symmetric cell division and impaired differentiation into effector cells14. These observations demonstrate that diverse T-cell immune responses are determined by versatile TCR signaling.
A TCR contains four subunits, an antigen-binding TCRαβ subunit and three signaling subunits, CD3εδ, CD3εγ and CD3ζζ15. The TCRαβ subunit recognizes antigen on the extracellular side but cannot trigger intracellular signaling because its cytoplasmic domains do not contain signaling motif. All CD3 chains have immunoreceptor tyrosine-based activating motif (ITAM) in their cytoplasmic domains. The phosphorylation of ITAM tyrosines leads to the recruitment of syk kinase member ZAP70, thus triggering downstream activating pathways7,8. Earlier biochemical studies show that different antigens can trigger distinct ITAM phosphorylation programs16. Besides the ITAM, other motifs in CD3 cytoplasmic domains also play important roles in TCR signaling, including the lipid-interacting Basic residue Rich Sequence (BRS) in CD3ε and CD3ζ chains and the Nck-interacting Proline-Rich Sequence (PRS) in CD3ε chain17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32. The physiological importance of BRS and PRS has been demonstrated by in vivo experiments25,28,33. Mutation of CD3ε BRS leads to impaired thymocyte differentiation and positive selection as well as limited peripheral T cell function, due to abnormal TCR surface level and signaling. CD3ε PRS enhances TCR sensitivity to weak ligands20,21. Mutation of PRS inhibits TCR phosphorylation and signaling22. Therefore, the three functional motifs, i.e., ITAM, BRS and PRS, act together to regulate the versatile function of TCR.
A longstanding puzzle of TCR transmembrane signaling is how different interactions between TCR and pMHC at the extracellular side can trigger distinct biochemical changes of the three functional motifs in CD3 cytoplasmic domains. Several lines of evidence strongly suggest that conformational change may play an important role in this process. First, antigen engagement causes the conformational change of the AB loop of TCR extracellular domain, which might be transmitted to the contacting CD3 extracellular domains to trigger signaling34,35. Second, antigen engagement induces the exposure of CD3ε PRS to recruit the adaptor protein Nck23. Intriguingly, only strong antigen but not weak antigen can induce the intracellular PRS exposure24. Third, the conserved cysteine motif in the CD3 stalk regions maintains the conformations of CD3 transmembrane and cytoplasmic domains36. Mutation of a Cys residue in the CD3ε stalk region to prevent the outside-in conformational transition leads to the blockade of αβ T cell development and impaired peripheral T cell function due to abnormal pre-TCR and TCR signaling18,19,37. Fourth, mechanical force has been shown to be critical for TCR signaling and might directly cause CD3 conformational change2,38,39,40,41,42,43,44. These studies highlight the importance of conformational change in TCR triggering. It is reasonable to speculate that different antigens may result in different degrees of conformational change, which leads to distinct TCR signalings.
In resting T cells, no basal phosphorylation of CD3ε is detected and only a small fraction of CD3ζ is partially phosphorylated29,45, which strongly suggests that CD3ε/ζ cytoplasmic domains adopt a closed conformation at quiescent stage. There is emerging evidence showing that phospholipids in the plasma membrane play important roles in regulating CD3 conformations46. The cytoplasmic domains of CD3ε and ζ chains are enriched in basic residues and can ionically interact with the acidic phospholipids in the plasma membrane17,25,26,27,28,29,47. The Nuclear Magnetic Resonance (NMR) structure of the lipid-bound CD3 cytoplasmic domain (CD3εCD) shows that CD3εCD protein backbone is localized at the interface of the lipid hydrophobic acyl-chain region and hydrophilic headgroup region29. The ITAM tyrosine side-chains insert deeply into the membrane hydrophobic core, rendering them inaccessible to Lck in resting T cells. The BRS and PRS regions are also protected by the membrane from being recognized by downstream signaling molecules. This conformation is referred as a “closed” state. Upon antigen stimulation, the lipid-bound CD3ε/ζ cytoplasmic domains need to be dissociated from the membrane and become “open” to allow biochemical modification or binding with downstream molecules. T cell signaling can be divided into three stages: initial triggering by antigen, signaling amplification and signaling sustainment. At the amplification and sustainment stages, the elevation of local Ca2+ concentration can lead to the dissociation of CD3ε/ζ cytoplasmic domains from the membrane and the subsequent ITAM tyrosine phosphorylation, which is caused by the neutralization of negative charges of acidic lipids by Ca2+30. Other factors such as the decrease of local acidic lipid concentration have been shown to contribute to the membrane dissociation of ITAM as well, but the time windows of these effects need to be further clarified27,32. Until now, it is still unknown how CD3 becomes open at the initial antigen triggering stage. Nevertheless, the binary “closed” and “open” CD3 conformations cannot explain the functional versatility of TCR. We hypothesized that there might be multiple intermediate conformations of CD3 cytoplasmic domains with different openness of the three functional motifs. Antigens with different TCR binding kinetics might stabilize different conformations of CD3 cytoplasmic domains to trigger distinct downstream signalings.
In this work, we have studied the functional relevance of this hypothesis in TCR signaling, using CD3ε as an example because it has all three types of functional motifs. We firstly developed an Atomic Force Microscopy (AFM) system to study the closed-to-open conformational transition of CD3ε cytoplasmic domain at the single-molecule level. Our data clearly indicated the presence of multiple conformations of CD3ε cytoplasmic domain with different openness of ITAM, BRS and PRS. NMR spectroscopy experiments, mainly solvent paramagnetic resonance enhancement (sPRE), were performed to probe the conformational dynamics of CD3ε cytoplasmic domain at the atomic resolution. Different regions of CD3ε cytoplasmic domain showed heterogeneous lipid-dependent dynamics, which provided the basis for the multiple conformations. Live-cell imaging experiment confirmed the physiological relevance of heterogeneous CD3ε conformations induced by different antigen stimulations. Our study thus presents a new concept that lipid-dependent CD3 conformational dynamics directly regulates antigen-specific signaling of TCR.
Results
Closed-to-open conformational transition of CD3ε cytoplasmic domain measured at the single-molecule level
We used AFM to study the transition process of CD3εCD from the closed conformation (lipid-bound) to the open conformation (solvent-exposed) at the single-molecule level. An advantage of single-molecule measurement is that weakly populated intermediate states generally hidden in ensemble bulk studies can be readily detected with high accuracy48.
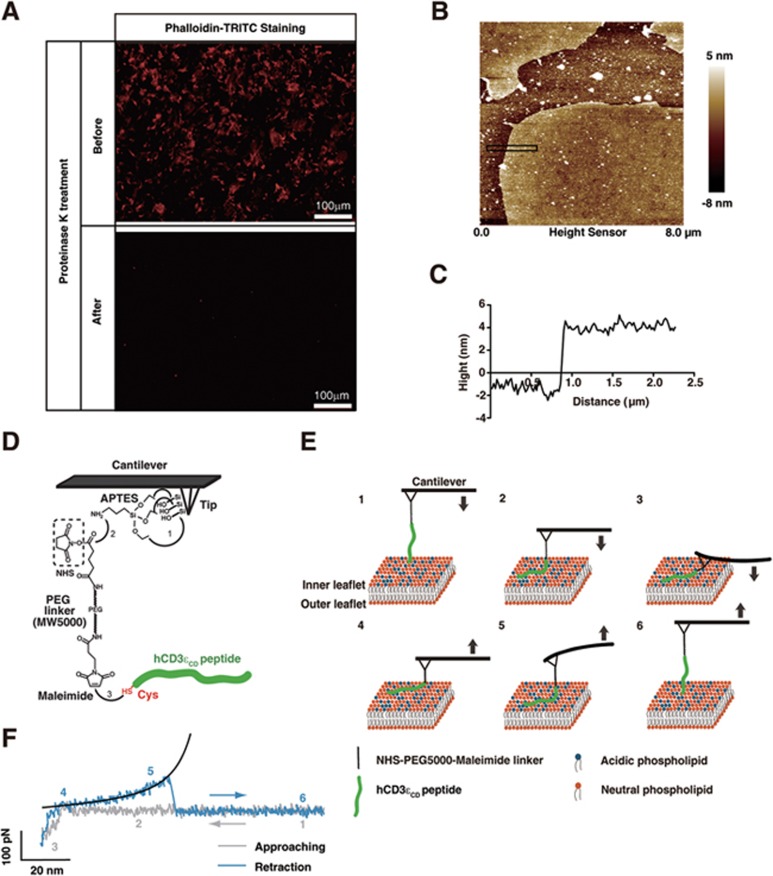
Due to technical difficulties, we cannot perform AFM measurement directly on the inner leaflet of the plasma membrane inside a live cell. Therefore, fresh plasma membrane sheets (PMSs) were prepared instead to provide a physiologically relevant lipid environment49,50,51 for studying CD3ε conformational transition (Supplementary information, Figure S1A). Cells were attached to cover glasses and a swelling-ripping method was used to prepare PMSs with the inner leaflet of the plasma membrane facing upward. Proteinase K was applied to PMSs to digest membrane surface proteins. Fluorescence staining showed that there were nearly no cytoskeleton proteins left on the inner leaflet of the PMSs (Figure 1A). Moreover, AFM scan showed that PMSs had smooth surface and had the physiological height of native lipid bilayer (∼6 nm; Figure 1B and 1C), which agrees well with the previous finding52. These data strongly suggested that the Proteinase K-treated PMSs had fully exposed lipid surface for studying CD3ε cytoplasmic domain binding and dissociation.
Figure 1.
An AFM system to detect the closed-to-open conformational transition of the CD3ε cytoplasmic domain. (A) Actin staining by phalloidin-TRITC showed that there were nearly no actins left on the membrane surface, strongly suggesting that the prepared PMS had fully exposed lipid surface. Upper panel was a representative image of 14 images before Proteinase K treatment, and lower panel was a representative image of 8 images after Proteinase K treatment in one experiment. (B, C) AFM scan showed that the height of the PMS was ∼6 nm that is the physiological height of native lipid bilayer. It was a representative image of 6 images in one experiment, and this experiment was repeated for three times. (D) AFM tip surface was firstly amino-functionalized by APTES, and then modified by a bi-functionalized PEG linker (molecular weight: 5 000) through an NH2-NHS reaction. The hCD3εCD peptide was then conjugated to the PEG linker through a thiol-maleimide reaction. The additional cysteine added to the terminus of hCD3εCD was labeled in red. (E) A schematic illustration showing the measurement of the closed-to-open conformational transition of hCD3εCD by AFM. First, the cantilever was set at the up position and both the hCD3εCD peptide and the tip were apart from the plasma membrane sheet (PMS). Second, the cantilever was approached toward the PMS to allow the peptide to contact with the PMS. Third, the cantilever was further approached toward the PMS and the tip became in contact with the PMS, which caused a certain degree of cantilever deflection. Fourth, the cantilever was retracted from the PMS, which caused the leave of the tip from the PMS and the disappearance of cantilever deflection. Fifth, the cantilever was further retracted and the interaction between the peptide and the PMS caused opposite-directional deflection of the cantilever. Sixth, the cantilever was further retracted and both the tip and the peptide left the PMS. (F) One typical force-distance curve measured by the AFM setup. Approaching and retraction force curves were labeled in gray and blue colors, respectively. The 6 steps mentioned in E were marked at the corresponding positions in the force-distance curve. The retraction force curve was fitted by the WLC model (black curve).
We then conjugated either the C-terminus or the N-terminus of the CD3εCD peptide to an AFM cantilever tip via a bifunctional PEG5000 linker through thiol-maleimide conjugation and used it for rupture force measurement (Figure 1D). In the conjugation experiment, we have made the density of the peptide attached to the cantilever as low as possible (10-fold less than the regular concentration)53,54 and used 17.5 times molar excess mono-functional PEG to compete with bifunctional PEG to reduce multiple labeling of CD3εCD peptide55,56,57. This protocol has been proven to be effective in single-molecule studies56,57,58. The peptides were chemically-synthesized with more than 95% purity. In the AFM measurement, the CD3εCD-conjugated cantilever was firstly approached to PMSs and remained still at a constant force of 100 pN for 1 s to allow the protein-membrane interaction. Similar low contact forces have been widely used by the AFM community to study physiological systems with membranes to avoid disrupting membrane structures59,60,61. We analyzed the mechanical properties of PMSs by fitting the contact region in the approaching trace using the Hertz model (Supplementary information, Figure S1B). Gauss fitting to the histogram of Young's modulus showed an average of ∼0.7 MPa (Supplementary information, Figure S1C), consistent with previous reports on the supported membrane62, which strongly suggested that the AFM tips actually pressed on the intact PMSs during the detection. The cantilever was then retracted from the membrane at a constant pulling speed to pull CD3εCD off the membrane (Figure 1E and 1F).
The brief contact between CD3εCD peptide and PMS had resulted in low frequency of specific rupture force events (∼3.6% shown in Figure 2A). The specific rupture force events (simplified as force events in the following text) that we selected all can provide a clear force peak above the baseline of thermal noise and the force-extension relationship can be fitted by worm-like chain (WLC) model of polymer elasticity (Supplementary information, Figure S1D). The thermal noise level was ∼10-15 pN, taken as the standard deviation of the baseline of the force-extension curves, which set the force detection limit in our experiments (Supplementary information, Figure S1E). Generally, nonspecific Van der Waals interactions were not detectable in our measurement. Assuming the adhesion bond formation obeyed Poisson statistics, an adhesion frequency of ∼3.6% in the force measurements implies that among the observed force events, the probabilities of forming a single, double, and triple adhesion bonds between CD3εCD peptide and the PMS were 98.2%, 1.7%, and 0.1%, respectively63. Therefore, our experimental condition ensured a > 98% probability that the adhesion event was mediated by a single bond. We observed a substantial amount of rupture events for the CD3εCD peptide pulling from either side of the peptide but not for the PEG linker control (Figure 2A). Previous studies strongly suggest that the juxtamembrane BRS region (KNRKAKAK) is the major lipid-binding site of CD3εCD29. Mutating key basic resides in this site dramatically reduced both the frequency and the force value of rupture force events (Figure 2A and 2B), indicating that the force events observed for CD3εCD wild-type (WT) peptide were specifically caused by the ionic CD3εCD-lipid interaction. This finding was further corroborated by analyzing the mechanical signature of the force peaks. When the C-terminus of CD3εCD peptide was conjugated to the cantilever tip via a PEG linker, the distance (contour length) of the rupture events to the membrane surface should equal the length of the PEG linker (∼30 nm) plus the length of CD3CD peptide before the binding site (in total ∼16.3 nm based on the assumption of 0.34 nm per amino acid residue). As expected, the observed CD3εCD rupture force peak typically occurred at a tip-surface distance of 43.49 ± 1.43 nm (Figure 2C). Normalized force-extension traces to the same contour length showed that all force traces that we measured were superimposable (Supplementary information, Figure S2A and S2B). Fitting the force curves using the WLC model of polymer elasticity yielded a persistence length of 0.372 ± 0.002 nm (Figure 2D), which was the usual value of a single unfolded protein or a PEG linker64. In case of simultaneous rupture of two or more peptides from PMS, the persistence length (p) of WLC fitting will be half or less (pn= p1/n, where pn and p1 denote the persistence lengths for n interactions and a single interaction, respectively). Supplementary information, Figure S2E shows that our force curve was fitted perfectly with p-value of 0.36 nm but not other p-values, therefore supporting the conclusion that our force curves are not from two or more molecules. The membrane-based AFM assay is therefore a reliable new method to study the closed-to-open conformational transition of CD3εCD at the single-molecule level.
Figure 2.
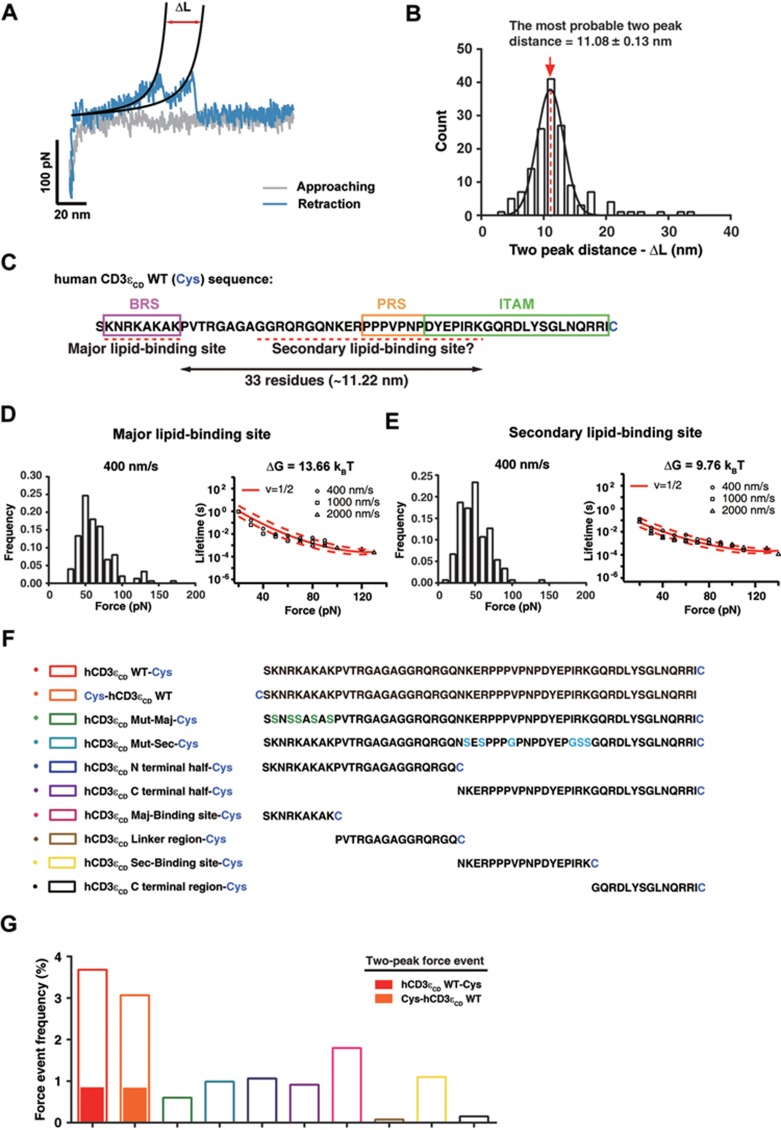
Specific AFM measurements at the single-molecule level. (A) The force event frequencies for hCD3εCD WT-Cys, Cys-hCD3εCD WT, hCD3εCD Mut-Maj-Cys and PEG linker, respectively. The five basic residues in the major lipid-binding site were mutated to Ser in hCD3εCD Mut-Maj to eliminate its ionic interaction with PMS. N.D., no effective data. (B) Scatter dot plot of the force values obtained for hCD3εCD WT-Cys (n = 707), Cys-hCD3εCD WT (n = 209), hCD3εCD Mut-Maj-Cys (n = 191) and PEG linker (n = 0), respectively. Error bars represented mean ± SEM. P-values were determined by the two-tailed Mann-Whitney test. n.s., not significant; ***P < 0.001. N.D., no effective data. (C) The contour length distribution obtained by fitting force curves of hCD3εCD WT with the WLC model (n = 707). Gaussian function analysis showed the most probable contour length was 43.49 ± 1.43 nm, which accounted for the total length of the hCD3εCD WT peptide before the binding site and the PEG linker. (D) The persistence length distribution obtained by fitting force curves of hCD3εCD WT with the WLC model (n = 707). Gaussian function analysis showed that the most probable persistence length was 0.372 ± 0.002 nm.
It is worthy to point out that rupture force of an AFM experiment is measured under non-equilibrium conditions, thus the force value depends significantly on the loading rates65. Therefore we mainly used AFM to measure the dynamics of CD3ε-membrane interaction rather than the absolute force values and binding energies. Our force value cannot be directly compared with the TCR-pMHC interaction forces measured by other systems because of the setup difference2,38,43. Nevertheless, our force value is within the physiological range. For example, the force range of receptor-ligand interaction is within 20-200 pN66.
Detection of a secondary lipid-binding site of CD3εCD
In addition to one-peak events, we also observed a substantial amount of two-peak events showing a secondary peak prior to the major peak when pulling from the C-terminus (Figure 3A). The major rupture force peak occurred at the tip-surface distance that accounted for the N-terminus of the peptide, same as the peak observed in the one-peak events, which indicated that the major peak was contributed by the major lipid-binding site. The secondary rupture force peak occurred at the shorter tip-surface distance that accounted for the middle of the peptide, indicating that there should be a secondary lipid-binding site in CD3εCD. The rupture forces for the secondary lipid-binding site, though smaller, were still comparable with that of the major lipid-binding site, suggesting that it also showed a high dissociation kinetic barrier and could play an important role in CD3εCD conformational dynamics. In AFM measurements, due to the length variation of the PEG linker molecules and the variation of the conjugation position of the PEG linker on the cantilever tip, the contour length alone is not accurate for the prediction of the position of the lipid-binding sites on the peptide sequence, despite that these events are of the same persistence length and are superimposable (Supplementary information, Figure S2C and S2D). However, the relative contour length increment for the rupture of the two lipid-binding sites can be unambiguously measured in the same force-distance trace and used to estimate the relative position of the two lipid-binding sites along the peptide. As shown in Figure 3B, the contour length increment between two rupture events adopted a Gaussian-like distribution with the most probable distance between the major and secondary lipid-binding sites of 11.08 ± 0.13 nm, accounting for ∼33 residues. Although simultaneously picking up two PEG-CD3ε molecules would also result in two-peak events, the frequency would be much less63 and the distribution of the contour length increment would be an exponential decay, rather than a Gaussian-like distribution. Therefore, we are confident that the specific two-peak force events are not from multiple peptide-lipid attachments. Since the major lipid-binding site is located at the N-terminal BRS region, the secondary lipid-binding site should be located in the middle of CD3εCD sequence, which includes the first tyrosine of the ITAM (Figure 3C).
Figure 3.
Detection of multiple lipid-dependent conformations of the CD3ε cytoplasmic domain. (A-E) Analysis of two-peak events when pulling from the C-terminus of the hCD3εCD WT peptide. (A) Typical force-distance curves with two rupture peaks. The retraction force curves (blue) were fitted by the WLC model (black curve). ΔL, the contour length distance between the two peaks. The retraction force curve represented the conformational transition of the CD3ε cytoplasmic domain from the closed state to the open state. (B) Gaussian function analysis of the distribution of distance (ΔL) between two rupture peaks. The most probable distance was 11.08 ± 0.13 nm (red dashed line). n = 150. (C) The major lipid-binding site of CD3εCD has been found to be the polybasic region at the N-terminus. Based on the ΔL value, the secondary lipid-binding site of CD3εCD was estimated to be around the PRS region. The amino acid sequences of BRS, PRS and ITAM were highlighted by magenta, orange and green boxes, respectively. The two lipid-binding sites were highlighted by red dashed lines. (D, E) The rupture-force histograms for the major lipid-binding site (D) and the secondary lipid-binding site (E) at 400 nm/s pulling speed. Data obtained at other speeds were shown in Supplementary information, Figure S6. The lifetime was calculated according to the Dudko-Hummer-Szabo equation and plotted against the force value. Kramers theory was used to fit the dots to a curve for the calculation of free energy (ΔG) required for the disruption of the lipid binding of each site (detailed in Supplementary information, Data S1). n = 150. Dotted curves represent 95% confidence intervals of the best-fit curves obtained by bootstrapping. (F, G) Analysis of force events of WT, truncated and mutant peptides. Cysteine residue was added to the C-terminus or N-terminus for conjugation to AFM tip. (F) The amino acid sequences of hCD3εCD WT, mutant and truncated peptides used in AFM force spectroscopy. Green and cyan labeled residues represent the mutation sites in hCD3εCDMut-Maj-Cys and hCD3εCD Mut-Sec-Cys peptides, respectively. (G) The force event frequencies for peptides shown in F. Solid bars represent the two-peak force event frequencies.
Notably, the finding of a secondary lipid-binding site of CD3εCD should not be an artifact from our data selection criterion. In the data analysis, we tried not to arbitrarily preset a range of persistence length or contour length for data filtering. Only the events that did not show clear mechanical features for the PEG linkers and cannot be fitted with the WLC model were excluded. To ensure that the current selection criterion was valid, we used a more rigorous criterion by selecting only the events with contour length and persistence length falling within one-sigma of the mean value (∼25-61 nm contour length and ∼0.34-0.41 nm persistence length for the one-peak events; ∼30-55 nm contour length and ∼0.33-0.41 nm persistence length for the major peak of the two-peak events). As shown in Supplementary information, Figure S3, the major conclusions under the new criterion were not changed. Especially, the ratio of two-peak events, distance between two peaks, and other conclusions were consistent.
We also observed a similar two-peak event frequency when pulling the peptide from the N-terminus (Figure 3G and Supplementary information, Figure S4). The major rupture force peak with a higher rupture force came before the secondary peak from this pulling direction (Supplementary information, Figure S4A), again confirming that the major peak was contributed by the BRS. The Gaussian fitting analysis showed that the most probable distance between the major and secondary lipid-binding sites was 13.19 ± 0.98 nm (Supplementary information, Figure S4B), which agreed well with the value obtained from the C-terminal pulling experiments (Figure 3B).
To further study whether the major site and the secondary site could bind to membrane independently, we synthesized mutated and truncated peptides that only preserved a single lipid-binding site in CD3εCD. The two-peak events were not observed for any of these mutated or truncated peptides, indicating that the two lipid-binding sites contributed to independent peaks in the two-peak events. The rupture forces can be clearly observed for the mutated or truncated peptides, indicating that the two sites can bind to lipids independently. However, both the force value and event frequency were reduced for the mutated or truncated peptide when compared with the WT peptide, suggesting that the two sites are synergistic in lipid binding (Figure 3F and Supplementary information, Figure S5).
Multiple kinetic intermediates between the closed and open conformations of CD3ε cytoplasmic domain
The presence of two lipid-binding sites leads to the division of CD3εCD into four regions that have different lipid-binding properties: major lipid-binding site, linker, secondary lipid-binding site and C-terminal region (Supplementary information, Figure S6A). Each of the two lipid-binding sites should act as an integrate module to stably interact with the plasma membrane, thus leading to two independent rupture peaks in the force curve. The fact that we did not observe three-peak or four-peak events suggested that the linker and C-terminal region did not act as an integrate module to interact with membrane. Indeed, a peptide that only contains the linker region or C-terminal region showed an extremely low rupture force event frequency compared with the two lipid-binding sites, indicating that the binding of the linker region or C-terminal region to the lipid is extremely dynamic (Figure 3G).
According to the previous NMR structure, the four regions were all inserted into the membrane bilayer when no external force was applied29. During the dissociation of CD3εCD from the membrane, we now observed one stable intermediate state (State II) between the closed state (State I) and the open state (State III) (Supplementary information, Figure S6B). A stable intermediate state (State II) has a conformational state at which the secondary lipid-binding site and the C-terminal region are dissociated from the membrane while the major lipid-binding site and the linker are still associated with the membrane. In between States I and II, there might be a group of transient intermediates, together designated as State Mixture 1 (M1), having a conformational state at which the two lipid-binding sites and the linker are associated with the membrane but the C-terminal region is heterogeneously dissociated from the membrane. In between States II and III, there might be another group of transient intermediates, together designated as State Mixture 2 (M2), having a conformational state at which the C-terminal region and the secondary lipid-binding site are dissociated from the membrane while the major lipid-binding site is still associated with the membrane, along with the heterogeneous dissociation of the linker from the membrane.
We applied Dudko model to further calculate the relative free energies needed for disrupting the membrane binding of the two lipid-binding sites. In addition to the 400 nm/s pulling speed, we also performed experiments at another two pulling speeds, which increased the dynamic force range for the total data set. The data at the three different pulling speeds (i.e., 400, 1 000 and 2 000 nm/s) followed the same trend when being converted to lifetime-force relationships using the method introduced by Dudko and coworkers67 (Supplementary information, Figure S7). The extracted free energy from force spectroscopy data depends on the theoretical model used to describe the shape of the free energy landscape. We have tried three different types of energy potentials for data fitting (see Supplementary information, Data S1 for details). The harmonic well with a cusp-like potential with a scaling factor ν = 1/2 gave rise to the best fit. The other two models (i.e., Bell-Evans-like barrier with ν = 1 or linear and cubic potential with ν = 2/3) could not fully fit to experimental data. The fitting parameters for the three different models were given in Supplementary information, Table S1. Based on the cusp-like potential, free energies of activation (ΔG) are 13.66 kBT for the major site and 9.76 kBT for the secondary site (Figure 3D and 3E). A free energy landscape was then generated to demonstrate the dynamic process of the closed-to-open conformational transition of CD3εCD (Supplementary information, Figure S6C).
In summary, the major conclusion of the AFM study is the presence of the secondary lipid-binding site that leads to a partially open conformation during the dissociation of CD3ε from the membrane. We would like to emphasize that this conclusion is not affected by AFM setups. Next, we used solution NMR to further confirm this conclusion.
A functionally relevant bicelle system for investigating lipid-dependent conformational dynamics of CD3ε cytoplasmic domain by solution NMR
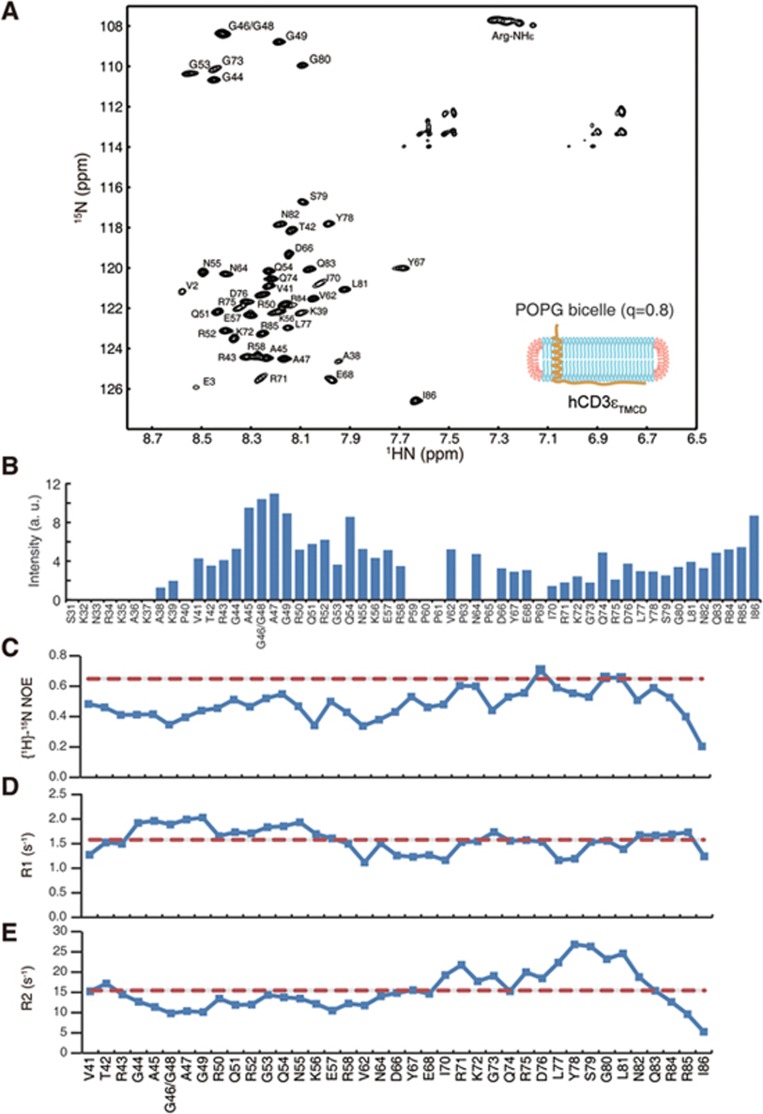
We previously determined the closed conformation of CD3ε cytoplasmic domain based on the NOE measurements, showing that it is embedded into the membrane29. This structure, however, only represents the major conformation but does not contain dynamics information. The CD3ε construct used previously only contains the cytoplasmic domain (CD). Considering that the transmembrane domain (TMD) should have direct impact on the CD dynamics, we designed a new construct CD3εTMCD that is more physiologically relevant. It contains a truncated extracellular domain, the full TMD and the full CD of human CD3ε (Supplementary information, Figure S8A-S8E). Recombinant hCD3εTMCD protein was reconstituted into lipid bicelles by a two-step protocol (Supplementary information, Figure S8F). The far UV Circular Dichroism measurement showed that the reconstituted hCD3εTMCD had correct folding (Supplementary information, Figure S8G). We then performed two-dimensional 1H-15N transverse relaxation optimized spectroscopy (TROSY) of hCD3εTMCD reconstituted in acidic POPG lipid bicelles (q = 0.8) to check the sample quality. All cross-peaks were assigned by a set of NMR triple-resonance experiments (Figure 4A). Every non-proline residue theoretically should have a cross-peak in the spectrum. However, the TMD residues (S8-S31) did not generate detectable signals because the TMD behaved as an integral part of the big lipid bicelle and tumbled too slowly. The juxtamembrane major lipid-binding site in the CD (K32-K39) was also barely detectable, indicating the slow motion of this region. Other parts of the cytoplasmic domain (V41-I86) yielded good signals in the TROSY spectrum, indicating that they had substantial motion relative to the big lipid bicelles (Figure 4B). Similar to the CD3εCD construct, the CD of the CD3εTMCD construct was embedded into the membrane because we observed substantial protein-lipid NOE signals (Figure 6G). Therefore, the reconstituted hCD3εTMCD-bicelle was a physiologically relevant system to probe the conformational dynamics of CD3ε cytoplasmic domain in the context of membrane environment.
Figure 4.
Dynamic feature of the lipid-bound CD3ε cytoplasmic domain. (A) A high-resolution 1H-15N TROSY spectrum of hCD3εTMCD reconstituted in big POPG bicelle (q = 0.8). (B) Signal intensities of backbone amide resonances of hCD3εTMCD construct. The good signals from residues in the cytoplasmic domain indicated that this region had substantial motion. (C-E) The three 15N spin relaxation parameters, including steady-state heteronuclear {1H}-15N NOEs (C), longitudinal spin relaxation rate R1 (D) and transverse spin relaxation rate R2 (E), were plotted as histograms vs the residue number. The {1H}-15N NOE values showed variations in the range of 0.20-0.71 with an average of 0.48. In general, flexible residues give {1H}-15N NOE values less than 0.65 (the red dash line). R1 values showed relatively little variation (1.12-2.03 s−1), with an average value of 1.58 s−1 (the red dash line). Such a large R1 value usually implied larger internal motions. R2 values were generally small, also indicating fast internal motion. R2 values showed a larger variation (5.27-26.84 s−1), giving an average value of 15.46 s−1 (the red dash line). The C-terminal sequence had R2 values larger than average, suggesting the presence of conformational exchange in this region.
Figure 6.
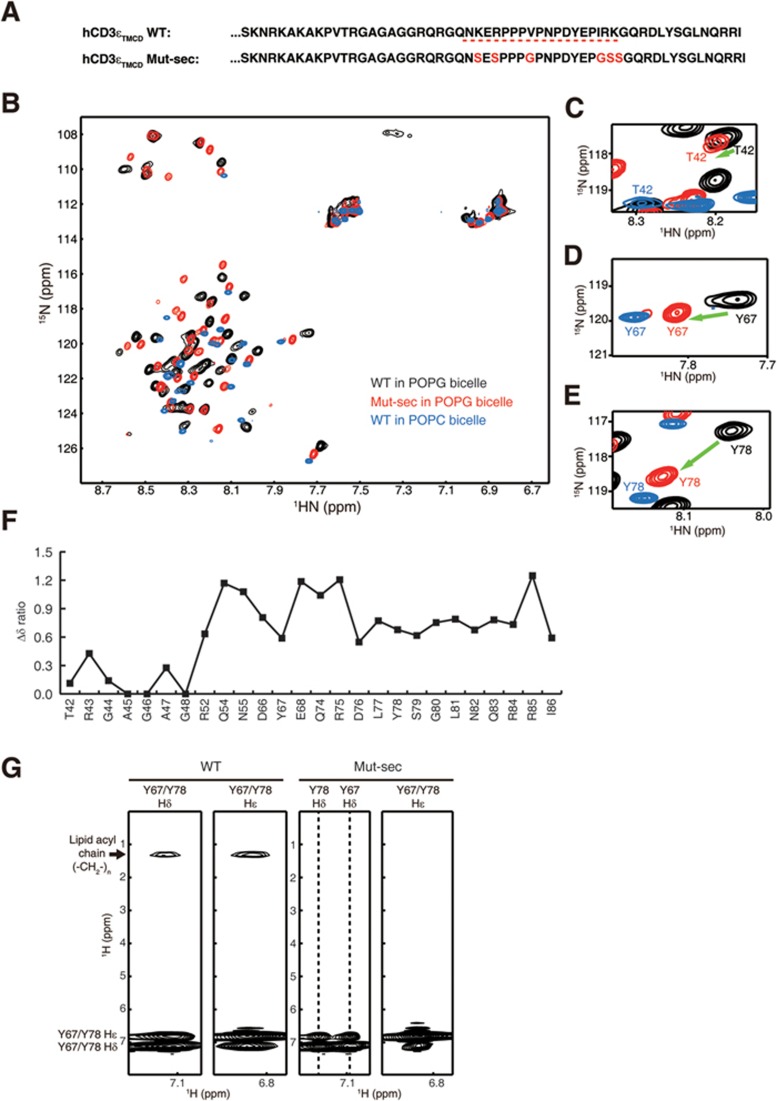
Regulation of the CD3ε cytoplasmic domain conformations by the secondary lipid-binding site. (A) The amino acid sequences of human hCD3εTMCD WT and Mut-sec peptides. The mutation sites were highlighted in red. (B) Superimposed 1H-15N HSQC spectra of hCD3εTMCD WT reconstituted in acidic POPG bicelle (q = 0.8) (black), reconstituted in zwitterionic POPC bicelle (q = 0.8) (blue), and hCD3εTMCD Mut-sec reconstituted in acidic POPG bicelle (q = 0.8) (red). (C-E) The signals of three representative residues, T42 in the N-terminal half of the cytoplasmic domain, Y67 and Y78 in the C-terminal half of the cytoplasmic domain, were enlarged and shown. The signals of T42 of the WT + POPG and Mut-sec + POPG samples appeared at almost the same positions, while the signal positions of Y67 and Y78 of the Mut-sec + POPG sample were close to the corresponding signals of the WT + POPC sample. (F) The ratio of the chemical shift difference (Δδ) induced by mutation (from WT to Mut-sec) against that induced by lipid environment (from POPG to POPC). Larger Δδ ratio value meant that the corresponding residue of the Mut-sec + POPG sample was more solvent-exposed. (G) Strips from aromatic NOESY spectra showing NOEs (distance of < 5 Å) between the aromatic protons Hδ/Hε of Y67 and Y78 and the methylene protons of the lipid acyl chains. Substantial NOE signals were detected for the WT + POPG sample whereas no NOE signals were detected for the Mut-sec + POPG sample, indicating that mutating key residues in the secondary lipid-binding site led to the dissociation of the ITAM from the membrane. These data suggested that mutating the secondary lipid-binding site changed the conformation of CD3ε cytoplasmic domain from State I to State II.
Intrinsic dynamics of CD3ε cytoplasmic domain
NMR spectroscopy provides a wide coverage of timescale of protein dynamics. Motions on the fast timescale (ps-ns) can be probed by the backbone amide 15N spin relaxation experiments. We determined the 15N relaxation parameters of the CD residues, including R1, R2 and the heteronuclear {1H}-15N NOE, with the reconstituted hCD3εTMCD sample (Figure 4C-4E). R1 and R2 are relaxation rates at which the longitudinal and transverse components of magnetization (Mz and Mxy) return to equilibrium after perturbation, respectively, while {1H}-15N NOE provides information and the motion of individual N-H bond vectors. The {1H}-15N NOE values showed variations in the range of 0.20-0.71 with an average of 0.48. In general, flexible residues give {1H}-15N NOE values less than 0.65 (Figure 4C). Therefore, the lipid-bound CD should be very flexible. R1 values of the CD residues were large compared with globular and stably folded proteins and showed relatively little variation (1.12-2.03 s−1), with an average value of 1.58 s−1 (Figure 4D). Large R1 value usually implies large internal motion that is independent on the conformational exchange. R2 values of the CD residues were small, also indicating fast internal motion. In contrast to R1 values, R2 values showed a bigger variation (5.27-26.84 s−1), with an average value of 15.46 s−1 (Figure 4E). The C-terminal part of ITAM had R2 values larger than average (Figure 4E), suggesting the presence of conformational exchange in this region.
These data together showed that the lipid-bound CD was dynamic and the C-terminal part of the ITAM might undergo conformational exchange.
Heterogeneous dynamics of the different regions of CD3ε cytoplasmic domain measured by sPRE
The dynamics of the lipid-bound CD can be at the horizontal direction on the membrane surface or be at the vertical direction to cause the transient dissociation of CD3εCD from the membrane. We developed a sPRE method to specifically probe the vertical motion that is crucial for the closed-to-open conformational transition.
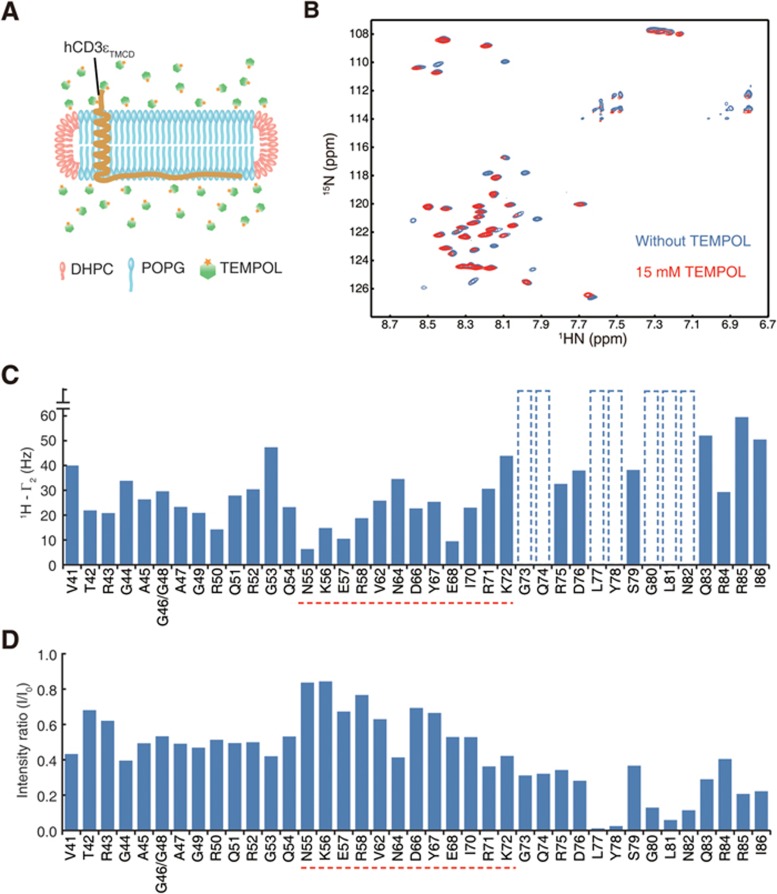
PRE can detect minor conformation of a protein when the exchange kinetics between major and minor conformations is rapid (faster than ∼100 μs) and the nucleus in the minor conformation is much closer to the PRE reagent than that in the major conformation68. We therefore applied a solvent PRE probe, TEMPOL (4-hydroxy-2,2,6,6, tetramethylpiperidin-1-oxyl), into the hCD3εTMCD-POPG bicelle sample (Figure 5A). If the lipid-bound CD would transiently dissociate from lipids and become open, TEMPOL should sense this minor conformation and increase the transverse relaxation rate (R2) and thus reduce the resonance signals of these residues due to paramagnetic effects.
Figure 5.
Heterogeneous dynamics of different regions of CD3ε cytoplasmic domain measured by solvent PRE. (A) A schematic illustration of PRE effect measurement. (B) 1H-15N TROSY spectra of the hCD3εTMCD + POPG bicelle sample before (blue) and after (red) the addition of 15 mM TEMPOL. (C) The transverse PRE rates of the backbone amide protons (1HN-Γ2) of the hCD3εTMCD + POPG bicelle sample upon the addition of TEMPOL. The dotted bars meant that the PRE effect was too significant to be detected in the two time-points PRE measurement. The polyproline region and the first half of the ITAM (N55-K72, highlighted by red dash line), i.e., the secondary lipid-binding site, showed mild PRE effects (less dynamic). The second half of the ITAM (G73-I86), i.e., the C-terminal region, and the linker region showed stronger PRE effects (more dynamic). (D) Intensity ratio (I/I0) of the backbone amide groups in 1H-15N TROSY spectra before (Intensity value I0) and after (Intensity value I) the addition of TEMPOL. The PRE effects of the CD residues showed a similar tendency to C. Again, the secondary lipid-binding site showed the lowest dynamics while the C-terminal region showed the highest dynamics.
The CD residues responded heterogeneously to the TEMPOL treatment (Figure 5B-5D). Figure 5C showed the transverse PRE rates, Γ2, of the backbone amide protons (1HN) measured by a two time-points method69, and Figure 5D showed the PRE effects on peak intensities. The two methods gave similar results on PRE effects. The C-terminal region, starting from the middle of the ITAM toward the C-terminus (G73-I86), showed high PRE effects in response to TEMPOL, strongly suggesting that this region had minor conformation that was solvent-exposed. This agreed well with the 15N spin relaxation data that suggested that the C-terminal part of the ITAM underwent conformational exchange. In contrast, the PRS region and the N-terminal part of ITAM (N55-K72) had low PRE effects, suggesting that this region was stably inserted in the membrane and did not have solvent-exposed minor conformation. To demonstrate that the PRE effects were specifically caused by TEMPOL, we used ascorbic acid (AA) to reduce the free radical of TEMPOL, thereby quenching its PRE effect. We indeed observed the recovery of all resonance signals in the TROSY spectrum (Supplementary information, Figure S9).
The sPRE data showed that N55-K72 was stably inserted in the membrane, which agreed well with the AFM finding showing that the secondary lipid-binding site should be allocated in the middle of CD3εCD and ended at K72. Therefore we defined the N55-K72 as the secondary lipid-binding site that can stably interact with the membrane (Figure 5C and 5D). The linker (P40-Q54) showed a slightly stronger PRE effect than the secondary lipid-binding site but still a significantly less PRE effect than the C-terminal region. The major lipid-binding site (K32-K39) of the CD3εTMCD construct was invisible in NMR spectra due to its slow motion.
Collectively, the PRE data showed that the different regions of the CD had distinct conformational dynamics on the vertical direction, which agreed well with the heterogeneous lipid-binding behaviors of the different regions observed in the AFM experiments. The N55-K72 region is defined as the secondary lipid-binding site that can stably interact with the membrane, which confirms the major conclusion of the AFM experiments. Meanwhile, we observed that the C-terminal region showed significantly higher dynamics than other regions. The heterogeneous lipid-dependent conformational dynamics can provide the basis for the heterogeneous conformations of CD3ε cytoplasmic domain.
Regulation of CD3ε cytoplasmic domain conformations by the secondary lipid-binding site
To further study the role of the secondary lipid-binding site, we mutated all the basic and hydrophobic residues in this site to eliminate its lipid-binding capability (Figure 6A). We compared the HSQC spectrum of hCD3εTMCD Mut-sec reconstituted in acidic POPG bicelles with those of hCD3εTMCD WT reconstituted in acidic POPG bicelles (CD at lipid-bound state, State I) and hCD3εTMCD WT reconstituted in zwitterionic POPC bicelles (CD at solvent-exposed state, State III) (Figure 6B-6F). The residue resonances of the major lipid-binding site and the linker of the mutant overlapped well with those of the hCD3εTMCD WT-POPG sample, indicating that these two regions in the mutant remained lipid-bound. In contrast, the residue resonances of the secondary lipid-binding site and the C-terminal region of the mutant were more similar to those of the hCD3εTMCD WT-POPC sample, strongly suggesting that these two regions should be solvent-exposed. Indeed, the NOE signals between ITAM tyrosine side-chains and lipid acyl chains were undetectable for the hCD3εTMCD Mut-sec-POPG sample, demonstrating the membrane dissociation of the local region (Figure 6G). Mutating the secondary lipid-binding site thus changed the conformation of CD3ε cytoplasmic domain from State I to State II, which further confirmed the importance of lipid binding in regulating CD3ε conformations.
Different antigen stimulations stabilized CD3ε cytoplasmic domain at different open conformations in live T cells
The above biophysical measurements demonstrated that CD3ε cytoplasmic domain had multiple conformations due to the heterogeneous lipid-dependent dynamics of the four regions. We next assessed whether these observations were physiologically relevant in live T cells. It is technically challenging to directly measure CD3 conformational change at the contact site between T cell and antigen presenting cell (APC). We therefore used cover glass-bound peptide-MHC (pMHC) to mimic APC and used TIRF-FRET (Total Internal Reflection Fluorescence-Förster Resonance Energy Transfer) microscopy to specifically measure CD3ε conformational change only at the TCR-pMHC interface. A HA-mCD3ε (YY-FF)-mTFP chimera construct was first transfected into mouse primary OT-I T cells. The mTFP served as a FRET donor and the tyrosines in CD3ε ITAM were mutated to phenylalanines to eliminate the interference of tyrosine phosphorylation on membrane binding. It is known that only the full TCR-CD3 complex but not the subunits can be expressed on the T cell surface70. Only a small fraction of the mTFP signals was on the plasma membrane (Supplementary information, Figure S10A). Surface staining with a HA-tag antibody confirmed the incorporation of HA-mCD3ε (YY-FF)-mTFP into the surface TCR-CD3 complex (Supplementary information, Figure S10B). Moreover, surface staining of CD3ε by 2C11 antibody showed that the surface TCR level was not changed in transfected OT-I T cells (Supplementary information, Figure S10C).
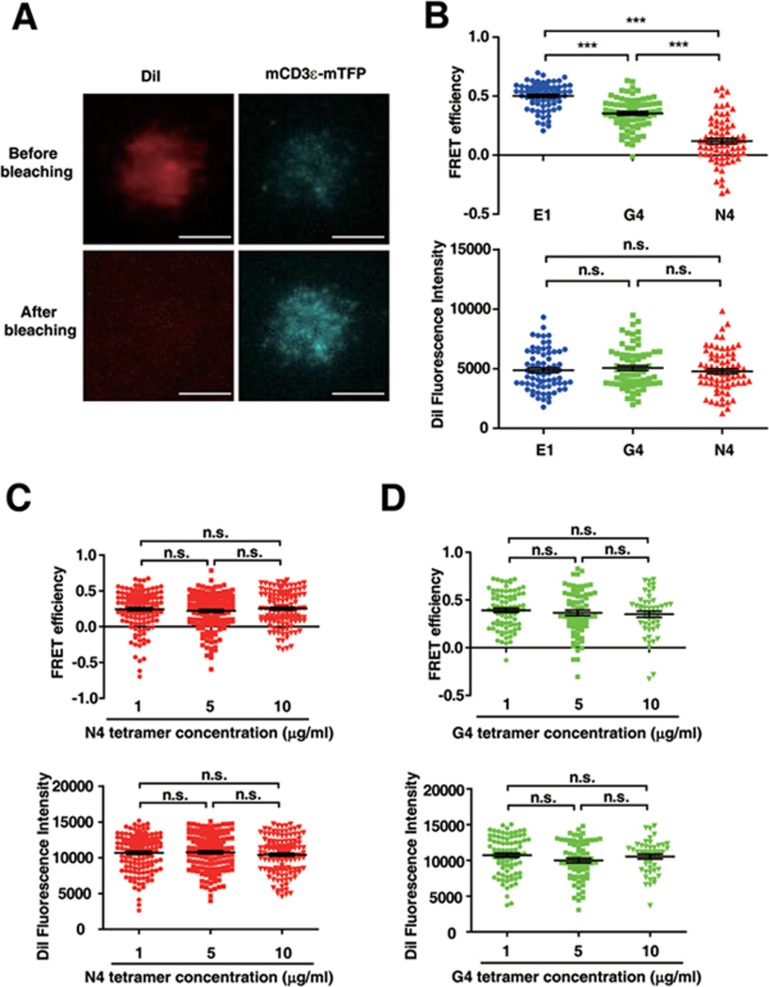
A lipid dye DiI that is a FRET acceptor of mTFP was used to label the plasma membrane of the transfected OT-I cells. The FRET effect between mTFP and DiI, which is sensitive to the distance between CD3ε C-terminus and the plasma membrane, could reflect the opening level of the CD3ε cytoplasmic domain. Three different peptide-MHC tetramers, i.e., H2-Kb N4 (OVA), G4 and E1, were chosen to stimulate TCR with different potencies. The TCR affinity order follows N4 > G4 > E1 and E1 was found to be a weak agonist/antagonist5,71. During the TIRF-FRET measurement, OT-I cells were treated with a Src kinase inhibitor PP2 to suppress downstream TCR signaling for the purpose of explicitly measuring the impact of antigen binding on CD3ε conformation without the influence of feedback regulations of the downstream signaling. The DiI staining was managed at the similar level between groups to avoid any acceptor level difference caused by artifact. A dequenching method was applied to measure the FRET value between CD3εCD and the plasma membrane (Figure 7A). Upon the stimulations of different antigens, the FRET efficiencies displayed significant differences and correlated with antigen potencies. The FRET efficiency of N4-stimulated T cells was at the lowest level, indicating the highest opening level of CD3εCD (Figure 7B). The FRET efficiency of G4-stimulated T cells was in between those of N4-stimulated T cells and E1-stimulated T cells, indicating the intermediate opening level of CD3εCD. The FRET efficiency of E1-stimulated T cells was high, indicating low opening level of CD3εCD. The FRET result agreed well with a previous study showing that the N4 can induce the exposure of the proline-rich sequence of CD3εCD whereas E1 cannot24. The antigen dose (5 μg/ml) that we used in Figure 7B was at the saturating level to trigger all TCR molecules at the cell-plate contact site (Figure 7C and 7D). Under such a setup, the FRET efficiency measured at the population level could reflect the FRET efficiency at the single-molecule level.
Figure 7.
Different antigen stimulations stabilize CD3ε cytoplasmic domain at different conformations. (A) The typical images of dequenching TIRF-FRET measurement. Scale bar, 5 μm. It was a representative image of 70 images in one experiment, and this experiment was repeated for seven times. (B) The TIRF-FRET efficiency (upper) and DiI fluorescence intensity (lower) of cells stimulated by different antigens. The concentrations of antigens were 5 μg/ml. Each dot represented an individual cell. n = 71, 73, 72 for E1, G4, N4, respectively. Error bars represented mean ± SEM. P values were determined by the unpaired two-tailed Student's t-test (P value between G4 and N4 in the upper panel, all P values in the lower panel). The two-tailed Mann-Whitney test was used to determine the P values between E1 and G4 and between E1 and N4 in the upper panel. ***P < 0.001. n.s., not significant. Data are representative of seven independent experiments. (C) The TIRF-FRET efficiency (upper) and DiI fluorescence intensity (lower) of cells stimulated by different concentrations of N4 tetramers. Each dot represented an individual cell. n = 136, 146, 127 for 1, 5, 10 μg/ml, respectively. Error bars represented mean ± SEM. P values were determined by the two-tailed Mann-Whitney test. n.s., not significant. Data are representative of five independent experiments. (D) The TIRF-FRET efficiency (upper) and DiI fluorescence intensity (lower) of cells stimulated by different concentrations of G4 tetramers. Each dot represented an individual cell. n = 82, 70, 48 for 1, 5, 10 μg/ml, respectively. Error bars represented mean ± SEM. P values were determined by the unpaired two-tailed Student's t-test (P-value between 1 and 5 μg/ml in the upper panel, all P-values in the lower panel). The two-tailed Mann-Whitney test was used to determine the P-values between 1 and 10 μg/ml and between 5 and 10 μg/ml in the upper panels. n.s., not significant. Data are representative of six independent experiments.
Taken together, the TIRF-FRET experiments showed that CD3ε could adopt distinct open conformations under different antigen stimulations, which therefore demonstrates the physiological relevance of multiple open CD3ε conformations observed by the above biophysical approaches.
Discussion
Our biophysical experiments present direct evidence of multiple CD3ε conformations that are directly regulated by the heterogeneous lipid-binding properties of the different regions in the CD3ε cytoplasmic domain. Live-cell imaging experiments show that different antigens can indeed stabilize CD3ε cytoplasmic domain at distinct conformations. Lipid-dependent CD3 conformational dynamics thus provides structural basis for the functional versatility of TCR. This mechanism may be relevant to many other membrane proteins whose activities are regulated by lipids.
The different regions of CD3ε cytoplasmic domain (Supplementary information, Figure S6A and S6B) have different lipid-binding properties and dynamics. The juxtamembrane BRS region is a basic residue cluster so that it can stably interact with acidic lipids in the plasma membrane by ionic force. It contributed to the major rupture force peak in the AFM measurement and was designated as the major lipid-binding site. The PRS region and the proximal half of the ITAM contributed to the secondary rupture force peak and they together were designated as the secondary lipid-binding site. This site contains rigid proline cluster in the middle and basic and hydrophobic residues on both sides, thus behaving as an integrative module to interact with acidic lipids. To be noted, the major and the secondary sites had independent binding to the PMS. The linker region (region between the two lipid-binding sites) and the C-terminal region (distal half of the ITAM) only contributed to the force accumulation but did not lead to rupture force peaks. Moreover, they showed higher dynamics than the two lipid-binding sites in the NMR measurements. In particular, the C-terminal region had the highest dynamics among the four regions. These data indicated that the lipid binding of the C-terminal region or the linker region was rather weak and unstable. Most likely, the dissociation of CD3εCD from the membrane in activated T cells would start from the most dynamic C-terminal region and complete at the most stable N-terminal BRS. Here we observed one stable half-open state (State II) in between the closed state (State I) and the open state (State III) in our AFM experiments (Supplementary information, Figure S6). At State II, both the C-terminal region and the secondary site are dissociated from the membrane but the major site and the linker are still in the membrane. In between State I and II, there might be multiple intermediates (together designated as State M1) to transit from the closed conformation to half-open conformation. Similarly, multiple intermediates (together designated as State M2) might also exist between States II and III. These intermediates are too dynamic to be stably measured by the current AFM setup. New technologies will be needed to further study their structural features. Nevertheless, the multiple-conformation concept is supported by our live-cell TIRF-FRET experiments. Antigens with different TCR affinities indeed can stabilize CD3ε cytoplasmic domain at different open conformations.
As mentioned above, the CD3ε cytoplasmic domain has three motifs, i.e., ITAM, BRS and PRS, which have distinct signalings or regulatory roles. The major function of ITAM is to recruit ZAP70 to trigger downstream events such as Ca2+ influx8. ITAM needs to be phosphorylated by Lck first to exert its function. Intriguingly, the phosphorylation of the CD3ε ITAM by Lck is dependent on CD3ε BRS31. We find that the recognition and phosphorylation of CD3ε by Lck is mediated by the interaction between CD3ε BRS and Lck (unpublished data). Therefore, the opening level of BRS can directly affect ITAM phosphorylation. This is supported by in vivo experiments showing that mutating BRS can significantly impair TCR phosphorylation and proximal signaling in peripheral T cells25,28. Other functions of BRS include the regulation of TCR surface expression level and the migration of TCR to immunological synapse25,28,46. Intriguingly, mutating BRS causes augmented preTCR signaling but attenuated TCR signaling, demonstrating the complicated roles of BRS in different T cells. The major function of PRS is to recruit Nck via an atypical PxxDY motif that includes the first tyrosine of the ITAM23,72,73. CD3ε-Nck binding inhibits the phosphorylation of both ITAM tyrosines, and conversely the tyrosine phosphorylation inhibits CD3ε-Nck interaction. These data suggest that PRS and ITAM exert their functions in a mutually exclusive manner. In vivo experiments reveal that CD3ε-Nck interaction not only enhances TCR sensitivity to weak ligands but also contributes to the downregulation of TCR expression on the cell surface20,21. PRS thus may play positive or negative roles in TCR signaling in different scenarios23,24. These studies together show that the functional roles of ITAM, PRS and BRS are complicated and inter-regulated. Their functions should depend on whether they are open and accessible for phosphorylation or interaction with partner proteins. Therefore, different opening levels of the CD3ε cytoplasmic domain will generate functional species with different accessibilities of the three motifs to trigger distinct TCR signalings.
It remains unclear how antigen engagement can directly induce the conformational change of the CD3ε cytoplasmic domain. Two possible models can be considered: allosteric regulation and force-induced movement. The allosteric regulation has been found in many receptors such as G-protein-coupled receptors74. The relevance of allosteric regulation in TCR triggering is supported by the findings that antigen-induced conformational changes have been observed in both extracellular and cytoplasmic domains23,24,34,75. Moreover, mutating CD3ε stalk region to prevent outside-in conformational transition leads to impaired TCR signaling18,19,37. On the other hand, mechanical force generated after T cell-APC conjugation might also affect the binding kinetics between CD3 cytoplasmic domains and the membrane42,44. Either allosteric regulation or force-induced movement should be dependent on the binding kinetics between TCR and pMHC and can cause antigen-specific conformational change to initiate TCR signaling.
In summary, our study shows that TCR can adopt different conformations upon different antigen stimulations. Notably, in addition to the conformational dynamics model shown here, other models such as kinetic segregation and kinase regulation have been shown to be crucial in TCR activation7,8,76,77. Our finding does not contradict other models but rather support the notion that different factors act together to generate a multi-layered regulation of TCR activation. We propose that TCR activation occurs in two steps (Supplementary information, Figure S11A)46. In resting T cells, the cytoplasmic domains of CD3ε/ζ chains are sequestered in the membrane, therefore adopting a “closed” conformation. The cytoplasmic domains of CD3δ/γ are exposed in the cytosol but they are not able to independently recruit Lck to initiate TCR phosphorylation and signaling (unpublished data). In activated T cells, multiple factors can contribute to the dissociation of the sequestered CD3ε/ζ cytoplasmic domains from the membrane to adopt “open” or “partially open” conformation (Step 1)46. Under different antigen stimulations, TCR can adopt distinct open states with different opening levels of CD3 functional motifs (Supplementary information, Figure S11B). Following the opening of CD3 cytoplasmic domains, the exposed functional motifs are either phosphorylated or interact with downstream signaling molecules to become “activated” (Step 2). The kinase-phosphatase equilibrium should play a key role in Step 2 in regulating ITAM phosphorylation. Kinetic segregation of phosphatases and the local enrichment of active Lck are both important to initiate TCR phosphorylation7,8,76,77. Altogether, the initial TCR triggering is regulated by a complex mechanism and modulation of either lipid binding-dissociation equilibrium or kinase-phosphatase equilibrium can affect the final outcome of TCR activation.
Materials and Methods
Reagents
Lipids and detergent (POPG, POPC, DHPC (dihexanoyl-phosphatidylcholine)) were from Avanti polar lipids. Isotopes for NMR experiments were from Cambridge Isotope Laboratories. α-mCD3ε (2C11) was from Biolegend. Mouse H-2Kb tetramers were from Medical & Biological Laboratories (MBL). RPMI 1640, Protein G sepharose beads, Trypsin and Octadecyl Rhodamine B Chloride (R18), DiI (1,1′-Dioctadecyl-3,3,3,3-Tetramethylindocarbocyanine Perchlorate) were from Life Technologies. PP2 was from Selleck Chemicals. IL-2 was from Peprotech. Proteinase K was from Tiangen Biotech (Beijing). Polybrene, F-actin staining reagent Phalloidin-TRITC, complete protease inhibitor cocktail, Iodoacetamide, Trichloroacetic acid, Formic acid, Poly-L-lysine, Aminopropyltriethoxysilane (APTES), diisopropylethylamine (DIPEA), triethylamine, TEMPOL and Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) were from Sigma-Aldrich. Maleimide-PEG-NHS (MW: 5 000) and Methyl-PEG-NHS (MW: 2 000) were from NANOCS (New York, USA). Nickel-affinity column was from QIAGEN. Ascorbic acid was from Shanghai Bio Science & Technology. Co., LTD. 293FT cells were obtained from Cell Bank of Chinese Academy of Sciences, and they have no mycoplasma contamination.
Reconstitution of hCD3εTMCD into lipid bicelles
The hCD3εTMCD peptide powder was first dissolved by 400 mM DHPC solution, and POPG or POPC powder was subsequently added into this solution. After several rounds of freezing and thawing, the POPG or POPC bicelles with reconstituted hCD3εTMCD can be successfully made.
The secondary structure of hCD3εTMCD in bicelles was examined by far UV Circular Dichroism. Spectrum was recorded from 190 to 250 nm at 20 °C in a 0.1 cm path length quartz cuvette using a J-715 spectrometer from JASCO Corporation. The experimental samples contained 10 μM hCD3εTMCD, 4 mM POPG bicelles (q = 0.8) and 10 mM Bis-Tris-HCl buffer, pH 6.7. The control sample contained all the components except hCD3εTMCD.
NMR experiments
Most NMR experiments were acquired at 298K using cryoprobe-equipped Bruker AVANCE 600 MHz and 900 MHz spectrometers with a pulsed field gradient triple-resonance probe. All spectra were processed using the program NMRPipe78 and analyzed by NMRView79 and KUJIRA80. Peak intensities were measured by NMRView.
Chemical shift assignments of the backbone
We performed the two-dimensional 1H-15N TROSY experiment for the reconstituted hCD3εTMCD and assigned all the peaks by a set of NMR experiments including 3D HNCO, HNCACO, HNCA, HNCACB, and CBCACONH. The sample condition was: 0.15 mM 13C, 15N-labeled hCD3εTMCD, 60 mM POPG bicelles, 20 mM Bis-Tris buffer, pH 6.7, 10% D2O.
15N spin relaxation
The 15N longitudinal spin-relaxation rates (R1) were measured with relaxation delays of 10, 50, 100, 200, 400, 600, 900, and 1 200 ms. The 15N transverse relaxation rates (R2) were measured with relaxation delays of 5, 15, 30, 60, 90, 120, 150, and 200 ms. {1H}-15N NOEs were recorded with and without proton saturation in an interleaved fashion. The R1, R2, and {1H}-15N NOE values were obtained using the rate calculation module in NMRView. The sample condition was the same as that of the backbone assignment experiments.
sPRE
TEMPOL was added into the hCD3εTMCD-POPG bicelle sample containing 0.15 mM 15N-labeled hCD3εTMCD protein, 30 mM POPG bicelles (q = 0.8) and 20 mM Bis-Tris, pH 6.7. 1H-15N TROSY spectra were measured at the TEMPOL concentration of 0 mM and 15 mM, respectively. To quench the PRE effect of TEMPOL, 11.25 mM AA (molar ratio of TEMPOL:AA = 1:0.75; higher concentration of AA would result in the sample precipitation) was added into the NMR samples. The transverse relaxation rates of the amide protons (1HN-Γ2) were measured using two time-points method69. Peak intensity changes induced by TEMPOL were also measured for the two 1H-15N TROSY spectra at the TEMPOL concentration of 0 mM and 15 mM, respectively.
PMS preparation
293FT cells were cultured on poly-L-lysine coated cover glass for 24-36 h, after that, the cells were swelled by deionized water for 1 min. Then the cell-attached cover glass was covered by another poly-L-lysine coated cover glass, forming a cover glass-cell-cover glass sandwich. After that, the upper cover glass was removed carefully, which ripped off the apical part of the cell membrane but left the basal cell membrane on the bottom cover glass. The inner leaflet of the basal membrane faced upward. The basal PMSs were further treated by Proteinase K to remove nearly all proteins on the membrane and thus made the lipid surface fully exposed. After being washed with PBS, the PMS was treated with 10 mM EGTA for 10 min to remove Ca2+ ions. Then PMS was stained with a membrane dye — Octadecyl Rhodamine B Chloride — R18 (3.75 μM in PBS). After being washed with PBS, each cover glass was stuck on the bottom of a 60 mm dish (Nunc) by double sticky tape for the following AFM experiment. To check the efficiency of Proteinase K treatment, the PMS sample was stained by Phalloidin-TRITC at room temperature for 30 min before EGTA treatment for subsequent fluorescent microscopy imaging (detailed in Supplementary information, Data S1).
Rupture force measurement and data analysis
A NanoWizard II AFM machine (JPK) equipped with an inverted fluorescence microscope (Olympus) was used to measure rupture forces between the hCD3εCD peptide and freshly prepared PMSs. The surface delay time was set to 1 s and the constant force set point was set to 100 pN. The cantilevers (MLCT-D, Bruker, with advertised resonance frequency around 15 kHz in air and spring constant around 30 pN nm−1 in air) were calibrated to get the accurate deflection sensitivities and spring constants. Force-distance curves were recorded by JPK analysis software and analyzed by IGOR Pro 6.04 software (Wave Metrics). Specific force curves were fit with WLC model (IGOR program) to obtain the contour length, persistence length and force amplitude values, which were further analyzed by GraphPad Prism 5 (GraphPad Software) to fit their distribution histograms with Gaussian function to obtain the most probable values (detailed in Supplementary information, Data S1).
Mouse primary OT-I T cell transfection
Splenocytes were isolated from OT-I TCR transgenic mice and cultured in RPMI-1640 medium containing 10% FBS, 1% penicillin/streptomycin, 100 ng/ml IL-2 and 10 nM OVA257-264 peptide for 3 days. After that, most cells were OT-I cytotoxic T lymphocytes (CTLs) in the medium. CTLs were then transfected with the PMXs-HA-mCD3ε (YY-FF)-mTFP1 chimera construct by retrovirus. In this construct, the two ITAM tyrosines were mutated to phenylalanines to avoid the interference of tyrosine phosphorylation on CD3 conformation. 7.5 × 105 CTLs (in 1.5 ml RPMI-1640 medium) and 1 ml of retrovirus were mixed in the well of 12-well culture plate. 100 ng/ml IL-2 was added to the system. The plate was spun at 1 200× g, 30 °C for 2 h. After spin infection, the cells were cultured for 10 h. Then we did the spin infection again with new viruses. After 10 h, the cells were cultured with fresh RPMI-1640 medium for 1 day and the transfection efficiency was examined by FACS.
Dequenching TIRF-FRET
Nunc Lab-Tek II Chamber Cover Glass (1.5 Borosilicate glass, 8 wells; Thermo Fisher Scientific, USA) was coated with H-2Kb OVA tetramer N4, G4, E1, respectively. One million OT-I CTL cells transfected with HA-mCD3ε (YY-FF)-mTFP1 construct were stained with 3 μM DiI lipid dye (Thermo Fisher Scientific, USA) in 37 °C water bath for 15 min. After that, the cells were treated with 50 μM PP2 (a Src kinase inhibitor) in 37 °C water bath for 10 min to block TCR signaling. The cell suspension was then loaded into one well for the subsequent TIRF-FRET measurement (detailed in Supplementary information, Data S1).
Statistical analysis
The normal distribution of the data sets was first analyzed by the D'Agostino and Pearson normality test. Then, the variation within each data set was analyzed to ensure that only data sets with similar variance were statistically compared. For data with normal distribution, the statistic significance was determined by the unpaired two-tailed Student's t-test. For data without normal distribution, the statistic significance was determined by the two-tailed Mann-Whitney test. Data were shown as mean ± SEM, n.s., not significant (P > 0.05), *P < 0.05, **P < 0.01, ***P < 0.001. Data analysis was performed by GraphPad Prism 5 software.
Author Contributions
CX conceived the project. YC supervised the AFM experiments. XG and WH performed the AFM experiments and analyzed the data. HL and CY performed the NMR experiments and analyzed the data. XG and CY performed the TIRF-FRET experiment and analyzed the data. CY, YW, WP, LL, YB and CZ purified the protein samples for NMR and AFM experiments. XS, HM, WW and CW provided technical help for sample preparation and analysis. BO, CT and ZL provided technical help for NMR experiments. YC, HW, XZ, WH, MC and XW provided technical help for AFM experiments. WL provided extensive scientific discussion. CX wrote the manuscript and other authors revised it.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
CX is funded by the Chinese Academy of Sciences (Strategic Priority Research Program XDB08020100), and the National Natural Science Foundation of China (31370860, 31425009, 31530022 and 31621003). HL is funded by the Ministry of Science and Technoloy of China (2014CB541903) and the National Natural Science Foundation of China (31470734). HW is funded by the Ministry of Science and Technoloy of China (2011CB933600) and the National Natural Science Foundation of China (21373200). XG is funded by China Postdoctoral Science Foundation (2015M580357). NMR experiments, part of AFM and imaging experiments were performed at the National Center for Protein Science Shanghai. Part of imaging experiments was performed at the core facility for cell biology of Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Plasma membrane sheet (PMS) preparation and rupture force measurements.
The mechanical signature of the PEG linker and the polypeptide.
AFM data analysis with different data filtering criteria.
Analysis of the two-peak events when pulling from the N-terminus of the hCD3εCD WT peptide.
Histograms of the rupture force values obtained for peptides shown in Figure 3F, respectively.
Multiple kinetic intermediates between the closed and open conformations of the CD3ε cytoplasmic domain.
The estimation of the free energy barrier with different theoretical models.
The expression, purification of hCD3εTMCD proteins and reconstitution of hCD3εTMCD into lipid bicelle.
Quenching of the PRE effect of TEMPOL by Ascorbic Acid.
Expression and localization of HA-mCD3ε (YY-FF)-mTFP in mouse OT-I T cells.
T-cell receptor activation model.
Materials and Methods
The fitting parameters for the three types of potential shapes with different scaling factors.
References
- Birnbaum ME, Mendoza JL, Sethi DK, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell 2014; 157:1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 2014; 157:357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue GP, Pielak RM, Smoligovets AA, Lin JJ, Groves JT. Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. Elife 2013; 2:e00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brameshuber M, Zeng X, et al. A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4(+) T cells. Immunity 2013; 39:846–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zarnitsyna VI, Liu B, et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 2010; 464:932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa JB, Axmann M, Mortelmaier MA, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 2010; 463:963–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen B, Bongrand P. Early T cell activation: integrating biochemical, structural, and biophysical cues. Annu Rev Immunol 2015; 33:539–561. [DOI] [PubMed] [Google Scholar]
- Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling. Nat Immunol 2014; 15:798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazilleau N, McHeyzer-Williams LJ, Rosen H, McHeyzer-Williams MG. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat Immunol 2009; 10:375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Wilson T, Fischer KF, Williams MA. Sustained interactions between T cell receptors and antigens promote the differentiation of CD4(+) memory T cells. Immunity 2013; 39:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubo NJ, Pagan AJ, Taylor JJ, et al. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 2013; 153:785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstead CJ, Weaver CT. Dwelling on T cell fate decisions. Cell 2013; 153:739–741. [DOI] [PubMed] [Google Scholar]
- Yamane H, Paul WE. Early signaling events that underlie fate decisions of naive CD4(+) T cells toward distinct T-helper cell subsets. Immunol Rev 2013; 252:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CG, Koehli S, Hausmann B, Schmaler M, Zehn D, Palmer E. T cell affinity regulates asymmetric division, effector cell differentiation, and tissue pathology. Immunity 2012; 37:709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucherpfennig KW, Gagnon E, Call MJ, Huseby ES, Call ME. Structural biology of the T-cell receptor: insights into receptor assembly, ligand recognition, and initiation of signaling. Cold Spring Harb Perspect Biol 2010; 2:a005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP, Allen PM. How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat Immunol 2012; 13:121–128. [DOI] [PubMed] [Google Scholar]
- Aivazian D, Stern LJ. Phosphorylation of T cell receptor ζ is regulated by a lipid dependent folding transition. Nat Struct Biol 2000; 7:1023–1026. [DOI] [PubMed] [Google Scholar]
- Martinez-Martin N, Risueno RM, Morreale A, et al. Cooperativity between T cell receptor complexes revealed by conformational mutants of CD3ε. Sci Signal 2009; 2:ra43. [DOI] [PubMed] [Google Scholar]
- Blanco R, Borroto A, Schamel W, Pereira P, Alarcon B. Conformational changes in the T cell receptor differentially determine T cell subset development in mice. Sci Signal 2014; 7:ra115. [DOI] [PubMed] [Google Scholar]
- Mingueneau M, Sansoni A, Gregoire C, et al. The proline-rich sequence of CD3ε controls T cell antigen receptor expression on and signaling potency in preselection CD4+CD8+ thymocytes. Nat Immunol 2008; 9:522–532. [DOI] [PubMed] [Google Scholar]
- Tailor P, Tsai S, Shameli A, et al. The proline-rich sequence of CD3ε as an amplifier of low-avidity TCR signaling. J Immunol 2008; 181:243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroto A, Arellano I, Blanco R, et al. Relevance of Nck-CD3ε interaction for T cell activation in vivo. J Immunol 2014; 192:2042–2053. [DOI] [PubMed] [Google Scholar]
- Gil D, Schamel WW, Montoya M, Sanchez-Madrid F, Alarcon B. Recruitment of Nck by CD3ε reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell 2002; 109:901–912. [DOI] [PubMed] [Google Scholar]
- Risueno RM, Gil D, Fernandez E, Sanchez-Madrid F, Alarcon B. Ligand-induced conformational change in the T-cell receptor associated with productive immune synapses. Blood 2005; 106:601–608. [DOI] [PubMed] [Google Scholar]
- Deford-Watts LM, Tassin TC, Becker AM, et al. The cytoplasmic tail of the T cell receptor CD3ε subunit contains a phospholipid-binding motif that regulates T cell functions. J Immunol 2009; 183:1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFord-Watts LM, Dougall DS, Belkaya S, et al. The CD3 ζ subunit contains a phosphoinositide-binding motif that is required for the stable accumulation of TCR-CD3 complex at the immunological synapse. J Immunol 2011; 186:6839–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cordoba SP, Dushek O, van der Merwe PA. Basic residues in the T-cell receptor ζ cytoplasmic domain mediate membrane association and modulate signaling. Proc Natl Acad Sci USA 2011; 108:19323–19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettini ML, Guy C, Dash P, et al. Membrane association of the CD3ε signaling domain is required for optimal T cell development and function. J Immunol 2014; 193:258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Gagnon E, Call ME, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3ε cytoplasmic tyrosine-based motif. Cell 2008; 135:702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Bi Y, Yang W, et al. Ca2+ regulates T-cell receptor activation by modulating the charge property of lipids. Nature 2013; 493:111–115. [DOI] [PubMed] [Google Scholar]
- Gagnon E, Xu C, Yang W, et al. Response multilayered control of T cell receptor phosphorylation. Cell 2010; 142:669–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon E, Schubert DA, Gordo S, Chu HH, Wucherpfennig KW. Local changes in lipid environment of TCR microclusters regulate membrane binding by the CD3ε cytoplasmic domain. J Exp Med 2012; 209:2423–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur JF, Li S, da Silva Martins M, Larose L, Dave VP. Critical and multiple roles for the CD3ε intracytoplasmic tail in double negative to double positive thymocyte differentiation. J Immunol 2009; 182:4844–4853. [DOI] [PubMed] [Google Scholar]
- Beddoe T, Chen Z, Clements CS, et al. Antigen ligation triggers a conformational change within the constant domain of the αβ T cell receptor. Immunity 2009; 30:777–788. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Clements CS, Purcell AW, et al. A structural basis for the selection of dominant αβ T cell receptors in antiviral immunity. Immunity 2003; 18:53–64. [DOI] [PubMed] [Google Scholar]
- Brazin KN, Mallis RJ, Li C, et al. Constitutively oxidized CXXC motifs within the CD3 heterodimeric ectodomains of the T cell receptor complex enforce the conformation of juxtaposed segments. J Biol Chem 2014; 289:18880–18892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Becker D, Vass T, White J, Marrack P, Kappler JW. A conserved CXXC motif in CD3ε is critical for T cell development and TCR signaling. PLoS Biol 2009; 7:e1000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das DK, Feng Y, Mallis RJ, et al. Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci USA 2015; 112:1517–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC. Mechanosensing in T lymphocyte activation. Biophys J 2012; 102:L5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Chen BM, Wu PC, et al. Cutting Edge: mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J Immunol 2010; 184:5959–5963. [DOI] [PubMed] [Google Scholar]
- Kim ST, Takeuchi K, Sun ZY, et al. The β T cell receptor is an anisotropic mechanosensor. J Biol Chem 2009; 284:31028–31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Shin Y, Brazin K, et al. TCR mechanobiology: Torques and tunable structures linked to early T cell signaling. Front Immunol 2012; 3:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Blanchfield L, Ma VP, et al. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc Natl Acad Sci USA 2016; 113:5610–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Discher DE, Finkel TH. Mechanical force in T cell receptor signal initiation. Front Immunol 2012; 3:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oers NS, Tao W, Watts JD, Johnson P, Aebersold R, Teh HS. Constitutive tyrosine phosphorylation of the T-cell receptor (TCR) ζ subunit: regulation of TCR-associated protein tyrosine kinase activity by TCR ζ. Mol Cell Biol 1993; 13:5771–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Yan C, Shi X, Li L, Liu W, Xu C. Lipid in T-cell receptor transmembrane signaling. Prog Biophys Mol Biol 2015; 118:130–138. [DOI] [PubMed] [Google Scholar]
- Sigalov AB, Aivazian DA, Uversky VN, Stern LJ. Lipid-binding activity of intrinsically unstructured cytoplasmic domains of multichain immune recognition receptor signaling subunits. Biochemistry 2006; 45:15731–15739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Li H. Single molecule force spectroscopy reveals a weakly populated microstate of the FnIII domains of tenascin. J Mol Biol 2006; 361:372–381. [DOI] [PubMed] [Google Scholar]
- Natkanski E, Lee WY, Mistry B, Casal A, Molloy JE, Tolar P. B cells use mechanical energy to discriminate antigen affinities. Science 2013; 340:1587–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogaart G, Meyenberg K, Risselada HJ, et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature 2011; 479:552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigmann A, van den Bogaart G, Iraheta E, et al. Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat Struct Mol Biol 2013; 20:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi M, Iida K, Kumano S, Wada H. Immune atomic force microscopy of prestin-transfected CHO cells using quantum dots. Pflugers Arch 2009; 457:885–898. [DOI] [PubMed] [Google Scholar]
- Ebner A, Wildling L, Kamruzzahan AS, et al. A new, simple method for linking of antibodies to atomic force microscopy tips. Bioconjug Chem 2007; 18:1176–1184. [DOI] [PubMed] [Google Scholar]
- Wildling L, Unterauer B, Zhu R, et al. Linking of sensor molecules with amino groups to amino-functionalized AFM tips. Bioconjug Chem 2011; 22:1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barattin R, Voyer N. Chemical modifications of AFM tips for the study of molecular recognition events. Chem Commun 2008:1513–1532. [DOI] [PubMed]
- Berquand A, Xia N, Castner DG, et al. Antigen binding forces of single antilysozyme Fv fragments explored by atomic force microscopy. Langmuir 2005; 21:5517–5523. [DOI] [PubMed] [Google Scholar]
- Schmitt L, Ludwig M, Gaub HE, Tampé R. A metal-chelating microscopy tip as a new toolbox for single-molecule experiments by atomic force microscopy. Biophys J 2000; 78:3275–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann JL, Nicolaus T, Neuert G, Blank K. Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nat Protoc 2010; 5:975–985. [DOI] [PubMed] [Google Scholar]
- Puntheeranurak T, Neundlinger I, Kinne RKH, Hinterdorfer P. Single-molecule recognition force spectroscopy of transmembrane transporters on living cells. Nat Protoc 2011; 6:1443–1452. [DOI] [PubMed] [Google Scholar]
- Serdiuk T, Balasubramaniam D, Sugihara J, Mari SA, Kaback HR, Muller DJ. YidC assists the stepwise and stochastic folding of membrane proteins. Nat Chem Biol 2016; 12:911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfreundschuh M, Alsteens D, Wieneke R, et al. Identifying and quantifying two ligand-binding sites while imaging native human membrane receptors by AFM. Nat Commun 2015; 6:8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picas L, Rico F, Scheuring S. Direct measurement of the mechanical properties of lipid phases in supported bilayers. Biophys J 2012; 102:L01–L03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesla SE, Selvaraj P, Zhu C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys J 1998; 75:1553–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt F, Rief M, Gaub HE. Single molecule force spectroscopy by AFM indicates helical structure of poly(ethylene-glycol) in water. N J Phys 1999; 1:6. [Google Scholar]
- Evans EA, Calderwood DA. Forces and bond dynamics in cell adhesion. Science 2007; 316:1148–1153. [DOI] [PubMed] [Google Scholar]
- Müller DJ, Helenius J, Alsteens D, Dufrêne YF. Force probing surfaces of living cells to molecular resolution. Nat Chem Biol 2009; 5:383–390. [DOI] [PubMed] [Google Scholar]
- Dudko OK, Hummer G, Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc Natl Acad Sci USA 2008; 105:15755–15760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev 2009; 109:4108–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara J, Tang C, Marius Clore G. Practical aspects of (1)H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson 2007; 184:185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner RD, Lippincott-Schwartz J, Bonifacino JS. The T cell antigen receptor: insights into organelle biology. Annu Rev Cell Biol 1990; 6:403–431. [DOI] [PubMed] [Google Scholar]
- Daniels MA, Teixeiro E, Gill J, et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 2006; 444:724–729. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Yang H, Ng E, et al. Structural and functional evidence that Nck interaction with CD3ε regulates T-cell receptor activity. J Mol Biol 2008; 380:704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T, Ruppelt A, Wang JH, et al. Reciprocal regulation of SH3 and SH2 domain binding via tyrosine phosphorylation of a common site in CD3ε. J Immunol 2007; 179:878–885. [DOI] [PubMed] [Google Scholar]
- May LT, Leach K, Sexton PM, Christopoulos A. Allosteric modulation of G protein-coupled receptors. Annu Rev Pharmacol Toxicol 2007; 47:1–51. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Dunstone MA, Kostenko L, et al. Crystal structure of the human T cell receptor CD3εγ heterodimer complexed to the therapeutic mAb OKT3. Proc Natl Acad Sci USA 2004; 101:7675–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature 2005; 436:578–582. [DOI] [PubMed] [Google Scholar]
- James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature 2012; 487:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR 1994; 4:603–614. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Iwahara J, Koshiba S, et al. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J Biomol NMR 2007; 39:31–52. [DOI] [PubMed] [Google Scholar]
- Dudko OK, Hummer G, Szabo A. Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys Rev Lett 2006; 96:108101. [DOI] [PubMed] [Google Scholar]
- Friddle RW, Noy A, De Yoreo JJ. Interpreting the widespread nonlinear force spectra of intermolecular bonds. Proc Natl Acad Sci USA 2012; 109:13573–13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GI. Models for the specific adhesion of cells to cells. Science 1978; 200:618–627. [DOI] [PubMed] [Google Scholar]
- Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J 1997; 72:1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. The molecular elasticity of the extracellular matrix protein tenascin. Nature 1998; 393:181–185. [DOI] [PubMed] [Google Scholar]
- Rief M, Fernandez JM, Gaub HE. Elastically coupled two-level systems as a model for biopolymer extensibility. Phys Rev Lett 1998; 81:4764–4767. [Google Scholar]
- Roszik J, Szöllősi J, Vereb G. AccPbFRET: An ImageJ plugin for semi-automatic, fully corrected analysis of acceptor photobleaching FRET images. BMC Bioinformatics 2008; 9:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plasma membrane sheet (PMS) preparation and rupture force measurements.
The mechanical signature of the PEG linker and the polypeptide.
AFM data analysis with different data filtering criteria.
Analysis of the two-peak events when pulling from the N-terminus of the hCD3εCD WT peptide.
Histograms of the rupture force values obtained for peptides shown in Figure 3F, respectively.
Multiple kinetic intermediates between the closed and open conformations of the CD3ε cytoplasmic domain.
The estimation of the free energy barrier with different theoretical models.
The expression, purification of hCD3εTMCD proteins and reconstitution of hCD3εTMCD into lipid bicelle.
Quenching of the PRE effect of TEMPOL by Ascorbic Acid.
Expression and localization of HA-mCD3ε (YY-FF)-mTFP in mouse OT-I T cells.
T-cell receptor activation model.
Materials and Methods
The fitting parameters for the three types of potential shapes with different scaling factors.