Abstract
Members of the poxvirus family encode multiple immune evasion proteins, including proteins that regulate apoptosis. We recently identified one such protein, F1L, encoded by vaccinia virus, the prototypic member of the poxvirus family. F1L localizes to the mitochondria and inhibits apoptosis by interfering with the release of cytochrome c, the pivotal commitment step in the apoptotic cascade. Sequence analysis of the F1L open reading frame revealed a C-terminal motif composed of a 12-amino-acid transmembrane domain flanked by positively charged lysines, followed by an 8-amino-acid hydrophilic tail. By generating a series of F1L deletion constructs, we show that the C-terminal domain is necessary and sufficient for localization of F1L to the mitochondria. In addition, mutation of lysines 219 and 222 downstream of the C-terminal transmembrane domain resulted in altered localization of F1L to the endoplasmic reticulum. Using F1L protein generated in an in vitro transcription-translation system, we found that F1L was posttranslationally inserted into mitochondria and tightly associated with mitochondrial membranes as demonstrated by resistance to alkaline extraction. Sensitivity to protease digestion showed that the N terminus of F1L was exposed to the cytoplasm. Utilizing various F1L deletion constructs, we found that F1L localization to the mitochondria was necessary to inhibit apoptosis, since constructs that no longer localized to the mitochondria had reduced antiapoptotic ability. Our studies show that F1L is a new member of the tail-anchored protein family that localizes to mitochondria during virus infection and inhibits apoptosis as a means to enhance virus survival.
Apoptosis, or programmed cell death, is a naturally occurring process that specifically eliminates unwanted cells through a tightly controlled pathway (29). The key players are a family of intracellular proteases referred to as caspases (47, 62). Caspases are cysteine proteases that are activated by proteolytic cleavage following an apoptotic stimulus. Activated caspases are responsible for the proteolysis of cellular proteins, resulting in the characteristic morphological and physical changes associated with apoptotic death. Some of these characteristics include chromatin condensation, DNA fragmentation, cell shrinkage, and membrane blebbing (29). Recent evidence indicates that mitochondria function as key organelles and act as the ultimate checkpoint responsible for regulating apoptosis (17, 36). Apoptosis is associated with significant mitochondrial changes, including the loss of the inner mitochondrial membrane potential and the release of cytochrome c (17, 36).
Both pro- and antiapoptotic members of the Bcl-2 family tightly regulate the mitochondrial checkpoint, and the balance between pro- and antiapoptotic members of the family decide the fate of the cell (14, 26). Antiapoptotic family members, such as Bcl-2 and Bcl-xL, localize to the mitochondria, where they function to inhibit the release of cytochrome c (14, 26). However, unlike Bcl-xL, Bcl-2 also localizes to the endoplasmic reticulum (ER), where it has been shown to inhibit apoptosis (35, 60, 61). In contrast, proapoptotic members, such as Bax and Bak, disrupt the inner mitochondrial membrane potential, resulting in the release of cytochrome c and death of the cell (14, 26). Although it is generally accepted that the loss of the inner mitochondrial membrane potential and release of cytochrome c lead to the demise of the cell, the exact mechanism of cytochrome c release is unresolved and controversial.
The detection and elimination of virus-infected cells can occur through the action of cytotoxic T lymphocytes and natural killer cells, resulting in apoptosis and the subsequent destruction of virus-infected cells (6, 55). In response, viruses have developed counterstrategies to inhibit apoptosis. For example, many viruses such as members of the gammaherpesvirus family, Epstein-Barr virus, adenovirus, African swine fever virus, and fowlpox virus encode obvious Bcl-2 homologues that mimic the activity of Bcl-2, thereby maintaining the integrity of the mitochondria and blocking release of cytochrome c (15, 27). Additionally, recent evidence indicates that a subset of viruses which lack obvious Bcl-2 homologues encode unique proteins that directly inhibit mitochondrial events, leading to apoptosis (20, 25, 67). For example, myxoma virus, a member of the poxvirus family, encodes a protein referred to as M11L that localizes to the mitochondria and prevents apoptosis (20, 21). Human cytomegalovirus encodes a novel mitochondria-localized inhibitor of apoptosis denoted vMIA, which inhibits the release of cytochrome c (5, 25, 53). More recently, we have identified an additional mitochondria-localized inhibitor of apoptosis, F1L, encoded by vaccinia virus (VV), the prototypic member of the poxvirus family (67). The F1L open reading frame in VV strain Copenhagen encodes a protein of 226 amino acids that localizes to the mitochondria, where it inhibits the loss of the inner mitochondrial membrane potential and release of cytochrome c (67).
A common trait shared by several members of the Bcl-2 family is that they belong to a growing family of proteins referred to as tail-anchored (TA) proteins and are specifically targeted to intracellular membranes by virtue of a C-terminal transmembrane domain (9, 10, 56, 68). F1L possesses a putative C-terminal transmembrane domain flanked by positively charged amino acids and a short C-terminal hydrophilic tail similar to that of Bcl-2 (34, 56). This observation led us to speculate that F1L might be a new member of the TA family of proteins. TA proteins are characterized as containing a membrane anchor located at their C terminus (9, 10, 68). This membrane anchor is composed of a helical hydrophobic domain of generally 12 to 24 amino acids flanked by positively charged amino acids (68). TA proteins do not possess N-terminal signal sequences and are therefore inserted into membranes posttranslationally, since the C-terminal membrane anchor is only exposed upon completion of translation (38, 68). The C-terminal membrane anchor encodes all the necessary information that allows each unique TA protein to be targeted to its specific destination. TA proteins are found in a wide variety of cellular membranes, including the ER, mitochondria, nuclear membrane, Golgi apparatus, and plasma membrane, where they carry out a wide range of biological functions (9, 10, 38).
Based on the presence of a putative C-terminal membrane anchoring domain in F1L that exhibits similarity to domains in TA proteins, we hypothesized that F1L would be a new member of the TA family and that localization of F1L to the mitochondria would be necessary for efficient apoptosis inhibition. In support of this hypothesis, our studies revealed that the C-terminal domain of F1L is necessary and sufficient for localization to the mitochondria. By constructing a series of F1L mutants containing deletions and point mutations, we further show that the positively charged residues within the C-terminal domain are necessary for mitochondrial localization of F1L. Using in vitro transcription-translation studies, we show that the mitochondrial targeting information is present in the C-terminal domain of F1L, and we provide evidence that F1L demonstrates the characteristics and membrane orientation of a TA protein. Importantly, we now report that localization of F1L to the mitochondria is necessary for efficient inhibition of apoptosis. Our studies indicate that the VV-encoded F1L protein is a TA protein that localizes to the mitochondria during infection, where it retains classical TA orientation in order for it to inhibit apoptosis.
MATERIALS AND METHODS
Cell culture and viruses.
HeLa, CV-1, and TK-H143 cells were obtained from the American Type Culture Collection and maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (GIBCO Invitrogen Corp.), 50 U of penicillin (GIBCO Invitrogen Corp.)/ml, 50 μg of streptomycin (GIBCO Invitrogen Corp.)/ml, and 200 μM glutamine (GIBCO Invitrogen Corp.). Jurkat cells were maintained in RPMI 1640 medium (GIBCO Invitrogen Corp.) supplemented with 10% fetal bovine serum, 100 μM 2-mercaptoethanol, 50 U of penicillin/ml, and 50 μg of streptomycin/ml at 37°C and 5% CO2. Rabbitpox virus strain Utrecht and cowpox virus strain Brighton Red were a kind gift from Richard Moyer, University of Florida, Gainesville. Ectromelia virus strain Moscow containing lacZ was a generous gift from Mark Buller, St. Louis University, St. Louis, Mo. VV811 and VV65 strain Copenhagen were generated as previously described (50). All viruses were propagated in baby green monkey kidney cells and grown in Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum (GIBCO Invitrogen Corp.), 50 U of penicillin/ml, 50 μg of streptomycin/ml, and 200 μM glutamine at 37°C and 5% CO2.
Generation of F1L antisera.
The F1L open reading frame (ORF) was amplified by PCR from pEGFP-F1Lwt using the forward oligonucleotide F1LBamHI (5′-GGATCCATGTTGTCGATGTTTATG-3′) containing a BamHI restriction site and the reverse oligonucleotide F1LNotI (5′-AGCGGCAATAGATGCCATATCATA-3′) to construct pGEX4T-3:F1L (1-120) (Amersham Biosciences) containing amino acids 1 to 120 of F1L appended to glutathione S-transferase (GST). PGEX4T-3:F1L (1-120) was transformed into BL31(DE3) cells, and protein expression was induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (Rose Scientific Ltd.). GST-F1L (1-120) was purified using glutathione-Sepharose 4B according to the manufacturer's instructions (Amersham Biosciences). Rabbits were immunized by injection of 500 μg of purified GST-F1L (1-120) in Freund's complete adjuvant. At biweekly intervals the animals were boosted with 500 μg of GST-F1L (1-120) in Freund's incomplete adjuvant.
Generation of recombinant VV.
The construction of pSC66-FLAG-F1Lwt, which was used to generate VV strain Western Reserve containing a FLAG-tagged copy of F1L, has been described elsewhere (67). VV strain Western Reserve was used to generate VV-WR-FLAG-F1Lwt as previously described (45). Recombinant virus was selected on TK-H143 cells in the presence of 25 μg of 5-bromodeoxyuridine and plaque purified three times using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside to visualize recombinant viruses. The presence of FLAG-F1Lwt in VV-WR-Flag-F1L was confirmed by PCR and by anti-FLAG M2 antibody (Sigma Aldrich).
Plasmid construction.
The F1Lwt ORF was amplified from virus DNA as described elsewhere (67). Plasmids pEGFP-F1LHTR (1-218), pEGFP-F1LTR (1-206), pEGFP-F1Ltail(+) (206-226), pEGFP-F1Ltail(−) (207-226), pEGFP-F1L-K219A, pEGFP-F1L-K222A, pEGFP-F1L-K219/222A, and pEGFP-F1LCyb5 were generated by PCR. The forward primer used for F1LHTR (1-218), F1LTR (1-206), and F1L-K219/222A was EcoRI-(GAATTCTCATGTTGTCGATGTTTATG). The forward primers used for FILtail(+) (206-226) and FILtail(−) (207-226) were EcoRI-(GGATTCTCAAGATTATTGGCATCACA) and EcoRI-(GAATTCTCCTTATTGGCATCACAGCT), respectively. The reverse primers for F1LHTR (1-218) and F1LTR (1-206) were BamHI-(GGATCCTTAATATGTAGCAAACATGATAGC) and BamHI-(GGATCCTTACTTTAGATATTCACGCGTGCT), respectively. The reverse primer for F1L-tail(+) (206-226) and F1Ltail(−) (207-226) was BamHI-(GGATCCTTATCCTATCATGTATTTGAG). The forward primer used for F1L-K222A and F1L-K219A was XhoI-(CTCGAGATGTTTATG) and for F1L-K219/222A it was EcoRI-(GAATTCTCATGTTGTCGATGTTTATG). Lysine-to-alanine mutations in F1L-K219A, F1L-K222A, and F1L-K219/222A were produced using the reverse primers BamHI-(GGATCCTTATCCTATCATGTATTTGAGAGTTGCATATGTAGCAAACAT), BamHI-(GGATCCTTATCCTATCATGTATGCGAGAGTTTTATA), and BamHI-(GGATCCTTATCCTATCATGTATGCGAGAGTTGCATATGTAGCAAACAT).
The pEGFP-F1LCyb5 chimera was generated using a combination of three PCRs. In the first PCR, the primers used in conjunction with pEGFP-F1Lwt template were EcoRI-(GAATTCTCATGTTGTCGATGTTTATG) and (CGATTCAACGGTAGTGATCTTTAGATATTCACGCGT), resulting in a PCR product containing the first 206 amino acids of F1L with an overhang corresponding to amino acids 100 to 105 of cytochrome b5. The C-terminal domain of cytochrome b5 was amplified by PCR from pSPUTK-Bcl2Cyb5 using (ATCACTACCGTTGAATCGAAC) and BamHI-(GGATCCTTAATCTTCAGCCATGTCAG), resulting in a PCR product encoding amino acids 100 to 134 of cytochrome b5. The products from the first two PCRs were combined with primers XhoI-(CTCGAGATGTTTATG) and BamHI-(GGATCCTTAATCTTCAGCCATGTACAG), resulting in a PCR product that contained the N-terminal portion of F1L (amino acids 1 to 206) and C-terminal tail of cytochrome b5 (amino acids 100 to 134). All amplified F1L constructs were verified by DNA sequence analysis and subcloned into pEGFP-C3 (Clontech). pSPUTK-Bcl2Cyb5 and pSPUTK-Bcl2ActA were a generous gift from David Andrews, McMaster University, Hamilton, Ontario, Canada (70).
For construction of pSPUTK-FLAG-F1Lwt, the F1L ORF was amplified using primers BamHI-(GGATCCATGGACTACAAAGACGATGACGACAAGTTGTCGATGTTTATGTGT), which contains an N-terminal FLAG epitope, and XhoI-(CTCGAGTTATCCTATCATGTATTT). The resulting amplified products were subcloned into pSPUTK (22) using the BamHI and SacI restriction sites. Construction of pSPUTK-F1LTR was performed by subcloning F1LTR (1-206) into the BamHI and NcoI restriction sites of pSPUTK.
To determine the subcellular localization of the various enhanced green fluorescent protein (EGFP)-F1L constructs during virus infection, each EGFP-F1L construct was amplified using PWO-Taq DNA polymerase (Roche) and cloned into the Sal1 site of pSC65 to place each construct under the control of a synthetic poxvirus early/late promoter (Roche) (13).
Confocal microscopy.
To determine localization of F1L orthologs in ectromelia virus-, rabbitpox virus-, and cowpox virus-infected cells, HeLa cells were infected with a multiplicity of infection (MOI) of 5 for 8 to 24 h. Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature and permeabilized using 0.02% NP-40 in PBS. For analysis of F1L localization, cells were stained with anti-F1L at a dilution of 1:2,000 followed by the addition of 10 μg of Alexa Fluor 546 goat anti-rabbit antibody (Molecular Probes)/ml. Mitochondrial localization was determined by staining cells with 10 μg of mouse anti-cytochrome c (clone 6H2.B4; Pharmingen)/ml followed by the addition of 10 μg of Alexa Fluor 546 goat anti-mouse (Molecular Probes)/ml. For membrane orientation studies, HeLa cells were infected with VVWR-FLAG-F1Lwt at an MOI of 5 for 8 h and either fixed with 4% paraformaldehyde and permeabilized with 25 μg of digitonin (Sigma Aldrich)/ml for 2 min at room temperature or permeabilized with 1 U of streptolysin O (SLO; Sigma Aldrich)/105 cells for 5 min at 37°C and then fixed (18, 48).
To determine the localization of EGFP-F1L constructs in live cells, HeLa cells were seeded onto 18-mm coverslips (Fisher Scientific) in modified 3.5-cm-diameter cell culture dishes. Cells were transfected with 2 μg of DNA using Lipofectamine 2000 (GIBCO Invitrogen Corp.) according to the manufacturer's specifications. For analysis of mitochondrial localization, the mitochondria of transfected cells were stained with 15 ng of Mitotracker Red CXMRos (Molecular Probes)/ml. To visualize the ER, transfected cells were stained with 1 μM ER-Tracker Blue-White DPX (Molecular Probes). Live cells were examined using a LSM510 laser scanning confocal microscope at 543 nm to assess Mitotracker fluorescence, at 350 nm to assess ER-Tracker, or at 489 nm to assess EGFP fluorescence.
For fixed-cell confocal microscopy, HeLa cells were transfected as described above. Cells were fixed using 4% paraformaldehyde in PBS for 10 min at room temperature and permeabilized using 0.02% NP-40 in PBS. For analysis of mitochondrial localization, cells were stained with 10 μg of mouse anti-cytochrome c (clone 6H2.B4; Pharmingen)/ml followed by the addition of 10 μg of Alexa Fluor 546 goat anti-mouse (Molecular Probes)/ml. For analysis of ER localization, cells were stained with rabbit anticalnexin (residues 575 to 593; Stressgen Biotehnologies) at a dilution of 1:400 followed by the addition of 10 μg of Alexa Fluor 546 goat anti-rabbit (Molecular Probes)/ml. Coverslips were mounted using 50% PBS-50% glycerol containing 4 mg of n-propyl gallate (Sigma Aldrich)/ml. Fixed cells were examined using a LSM510 laser scanning confocal microscope at 543 nm to assess cytochrome c and calnexin staining. Staining intensities were profiled with the use of the Zeiss LSM 510 imaging software.
To determine the localization of EGFP-F1L constructs during virus infection, HeLa cells were infected with VV strain Copenhagen VV65 at an MOI of 3 and simultaneously transfected with 2 μg of DNA as described above. Infection-transfections went for 8 h, after which cells were fixed and stained with anti-cytochrome c or anticalnexin as described above.
Apoptotic killing assays.
For apoptotic killing assays, HeLa cells were transfected as described above and induced to undergo apoptosis by the addition of 10 ng of tumor necrosis factor alpha (TNF-α; Roche Diagnostics)/ml and 5 μg of cycloheximide (ICN Biomedicals Inc.)/ml for 5 to 15 h depending on the experiment. Following treatment with TNF-α, cells were stained with 0.2 μM tetramethylrhodamine ethyl ester (TMRE; Molecular Probes), a dye that preferentially stains healthy respiring mitochondria (19, 23, 41). Loss of mitochondrial membrane potential in EGFP-positive cells was measured as a decrease in TMRE fluorescence by two-color flow cytometry (FACScan; Becton Dickinson). The level of TMRE fluorescence was measured through the FL-2 channel equipped with a 585-nm filter (42-nm band pass). The level of EGFP fluorescence was measured through the FL-1 channel equipped with a 488-nm filter (42-nm band pass). Data were acquired on 10,000 EGFP-positive cells per sample with fluorescent signals at logarithmic gain. Data were analyzed with CellQuest software. Loss of the inner mitochondrial membrane potential in positively transfected cells was calculated using the following equation: (number of EGFP+ TMRE− cells/total number of EGFP+ cells) × 100.
Purification of mitochondria.
Mitochondria used for membrane insertion assays were freshly purified from Jurkat cells as previously described (28, 66). Briefly, 108 Jurkat cells were washed once with buffer A containing 20 mM morpholinepropanesulfonic acid (MOPS), pH 7.4, 100 mM sucrose, and 1.0 mM EGTA. The cells were centrifuged for 10 min at 2,000 × g and resuspended in buffer B, containing 5% Percoll (Sigma Aldrich) and 191 μg of digitonin (Sigma Aldrich)/ml in buffer A at a final concentration of 2 × 107 cells/ml. The cells were incubated on ice for 15 min with occasional inversion. The nuclei were pelleted by centrifugation at 2,500 × g for 10 min at 4°C. The supernatant was collected and further fractionated by centrifugation at 15,000 × g for 15 min at 4°C. The remaining mitochondrial pellet was washed three times with buffer A and resuspended in buffer C, containing 20 mM MOPS, pH 7.4, 300 mM sucrose, and 1 mM EGTA. Protein concentration was determined using a bicinchoninic acid kit (Pierce Chemical Co.).
To determine the membrane orientation of mitochondrial-localized F1L during virus infection, Jurkat cells were infected with VVWR-FLAG-F1Lwt at an MOI of 5 PFU per cell. Five hours after infection mitochondria were purified as described previously (28, 66). Protein concentration was determined using a bicinchoninic acid kit (Pierce Chemical Co.). To determine the membrane orientation of F1L, mitochondria derived from infected cells were treated with either enterokinase, proteinase K, or trypsin as described below.
In vitro transcription-translation.
The TNT SP6-coupled reticulocyte lysate system (Promega Corp.) was used as described by the manufacturer. A 50-μl reaction mixture contained 25 μl of rabbit reticulocyte lysate, 20 μM amino acid mixture (minus methionine), 2.5% TNT reaction buffer, 40 U of RNasin (Promega Corp.), 10 μCi of [35S]methionine (Perkin-Elmer Life Sciences), and 1 μg of either pSPUTK, pSPUTK-FLAG-F1Lwt, pSPUTK-F1LTR (1-206), or pSPUTK-Bcl2ActA. Reaction mixtures were incubated at 30°C for 90 min, and translation was stopped by the addition of 2 μg of cycloheximide (Sigma Aldrich)/ml.
For membrane-insertion assays, 10 μg of purified mitochondria was added to 10 μl of TNT-generated protein and incubated at 30°C for 30 to 60 min to allow for membrane association. The lysate-mitochondria mixture was centrifuged for 20 min at 20,000 × g at 4°C. Supernatants were saved, and pellets were washed with PBS followed by centrifugation at 20,000 × g for 20 min at 4°C. The resulting pellet was resuspended in 50 μl of PBS. The supernatant and pellet samples were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and detection by autoradiography. To determine if TNT-generated proteins were integral membrane proteins, the mitochondrial pellet was washed with 200 μl of 0.1 M Na2CO3 (pH 11.5) to disrupt nonspecific binding to membranes (3, 24). To determine protein orientation in mitochondrial membranes, samples were treated with either 2.5 μg of trypsin (grade III; Sigma Aldrich)/ml or 2.5 μg of proteinase K (Sigma Aldrich)/ml for 1 h at 30°C or 1 to 2 U of enterokinase (Novagen) for 2 h at room temperature.
Immunoblotting.
Purified mitochondria were analyzed on SDS-polyacrylamide gels, and proteins were transferred to nitrocellulose membranes. FLAG-F1Lwt was detected using rabbit anti-F1L (1:5,000) followed by goat anti-rabbit horseradish peroxidase-conjugated (Bio-Rad) antibody at 1:10,000 or anti-FLAG conjugated to horseradish peroxidase (clone M2) at 1:2,500 (Sigma Aldrich). Proteins were visualized with a chemiluminescence detection system according to the manufacturer's protocol (Amersham Biosciences).
Detection of protein expression levels for the various EGFP-F1L constructs was determined by transfecting HeLa cells. Cell lysates were analyzed on SDS-polyacrylamide gels, and proteins were transferred to nitrocellulose membranes. The EGFP-F1L constructs were detected using rabbit anti-EGFP (provided by L. Berthiaume, University of Alberta, Edmonton, Alberta, Canada) at 1:20,000 followed by goat anti-rabbit horseradish peroxidase-conjugated antibody at 1:10,000.
RESULTS
F1L orthologs localize to mitochondria.
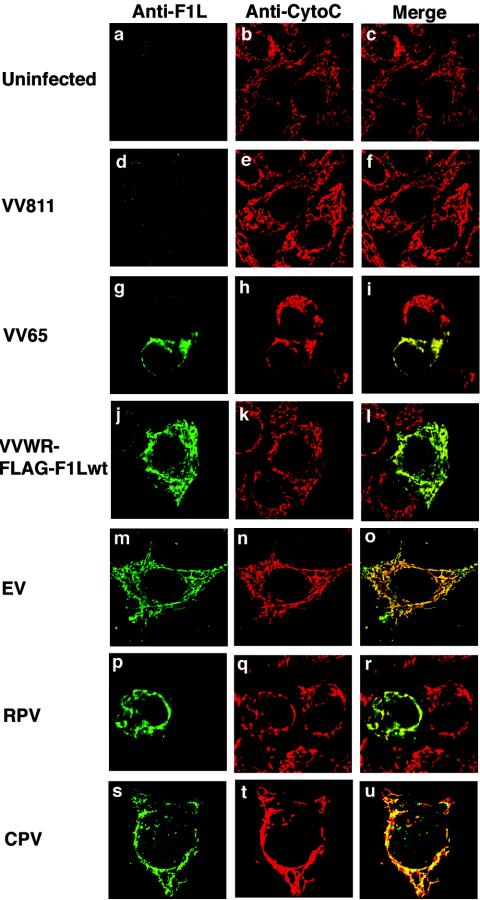
Recently, we identified a VV-encoded protein, F1L, that localizes to mitochondria and inhibits apoptosis (67). Sequence analysis of F1L failed to reveal any obvious similarity to cellular proteins, including members of the Bcl-2 family, which regulate apoptosis at the mitochondria. The poxvirus family, of which VV is the prototypical member, consists of a large family of viruses that infect both vertebrates and invertebrates (44). VV-related F1L ORFs are only found in members of the Orthopoxvirus genus and are highly conserved, with greater than 95% sequence identity over the last 220 amino acids. The greatest sequence diversity among the various F1L orthologs is located within the N-terminal regions, with strains of variola virus, camelpox virus, and ectromelia virus displaying a series of unique repeats, the function of which is currently unknown. To determine if F1L orthologs in other members of the Orthopoxvirus genus also localize to mitochondria, we infected HeLa cells with either the deletion virus VV811, VV strain Copenhagen, a recombinant VV expressing a FLAG-tagged version of F1L (VVWR-FLAG-F1Lwt), ectromelia virus, rabbitpox virus, or cowpox virus. F1L was visualized by confocal microscopy using an anti-F1L antibody. When cells were infected with the deletion virus VV811, which is devoid of the F1L ORF, no F1L was detected within the cells (Fig. 1d to f). Upon infection of cells with the F1L-containing VV65, F1L demonstrated a similar localization pattern to that of cytochrome c, indicating F1L localizes to mitochondria during virus infection (Fig. 1g to i). A similar pattern was detected for cells infected with the recombinant VV expressing a FLAG-tagged version of F1L (Fig. 1j to l). HeLa cells infected with either ectromelia virus (Fig. 1m to o), rabbitpox virus (Fig. 1p to r), or cowpox virus (Fig. 1s to u) and stained with anti-F1L all showed a localization pattern that colocalized with cytochrome c, indicating that F1L orthologs in other Orthopoxvirus genus members also localize to mitochondria during infection.
FIG. 1.
F1L orthologs localize to the mitochondria during infection. HeLa cells were infected with various orthopoxviruses for 8 to 24 h at an MOI of 5. F1L was detected using anti-F1L antibody (green), mitochondria were detected using anti-cytochocrome c (red), and localization was visualized by confocal microscopy. (a to f) F1L expression was not detected in uninfected cells or in cells infected with the deletion VV811. (g to u) F1L localized to mitochondria in cells infected with VV strain Copenhagen VV65, VV-FLAG-F1Lwt, ectromelia virus (EV), rabbitpox virus (RPV), and cowpox virus (CPV).
The C-terminal tail of F1L is critical for mitochondria localization.
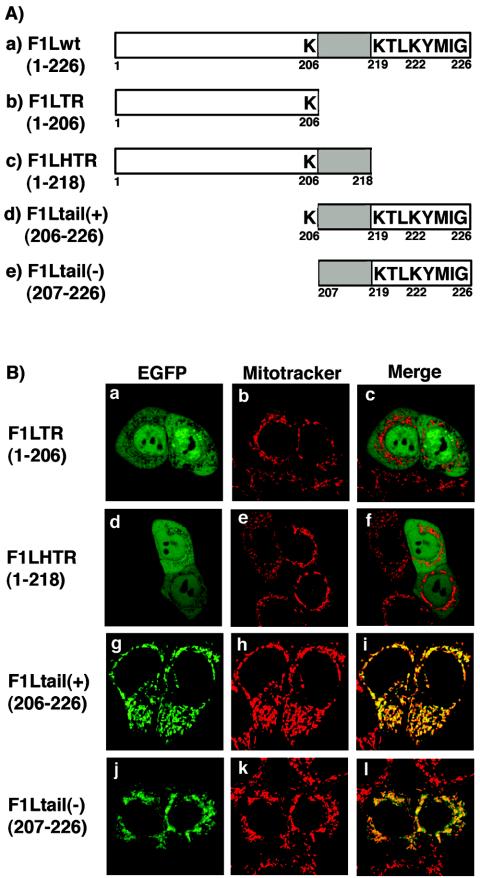
Sequence analysis of all the F1L orthologs revealed the presence of a conserved C-terminal transmembrane domain flanked by positively charged amino acids and a short hydrophilic tail C-terminal to the transmembrane domain (Fig. 2A, construct a). TA proteins, including several members of the Bcl-2 family, localize to membranes through a C-terminal domain containing a transmembrane region (9, 10, 56). In support of this, data from our previous publication showed the last 27 amino acids of F1L were necessary and sufficient for mitochondrial localization (67). To further elucidate the minimal region within the last 27 amino acids that serves as a mitochondrial localization signal for F1L, four additional EGFP-F1L deletion constructs were created (Fig. 2A). When HeLa cells were transfected with either pEGFP-F1LTR (1-206), which no longer contains the C-terminal domain, or pEGFP-F1LHTR (1-218), which is missing the last eight amino acids, a diffuse signal was found throughout the cells which did not colocalize with Mitotracker, indicating that these amino acids were necessary for localization to mitochondria (Fig. 2B, panels a to f). To determine if the last 20 amino acids of F1L were sufficient for mitochondrial localization, we fused the last 20 amino acids of F1L to EGFP and visualized localization by confocal microscopy. Uncertain as to whether the positively charged lysine (206) upstream of the hydrophobic domain was important for localization, two F1L-tail constructs were created: F1L-tail(+) (206-226), which contains lysine 206, and F1L-tail(-) (207-226), which is missing lysine 206. When HeLa cells were transfected with either pEGFP-F1Ltail(+) (206-226) or pEGFP-F1Ltail(-) (207-226), both constructs showed a similar staining pattern to Mitotracker, and when the signals were superimposed a uniform yellow image was produced, indicating that the C-terminal tail of F1L is necessary and sufficient to target F1L to the mitochondria (Fig. 2B, panels g to l).
FIG. 2.
The C-terminal tail of F1L is necessary and sufficient for localization to mitochondria. (A) Schematic representation of F1Lwt deletion constructs fused to EGFP. Wild-type F1L consists of 226 amino acids and contains a C-terminal transmembrane domain (shaded region) flanked by positively charged lysines and an 8-amino-acid hydrophilic tail. F1LTR (1-206) is missing the entire C-terminal domain, while F1LHTR (1-218) is only missing the last eight amino acids. F1Ltail(+) (206-226) and F1Ltail(-) (207-226) contain only the C-terminal region of F1L, with and without lysine 206, respectively. (B) The C-terminal tail is necessary and sufficient for localization. HeLa cells were transiently transfected with either pEGFP-F1LTR (1-206) (a) and pEGFP-F1LHTR (1-218) (d) or with pEGFP-F1Ltail(+) (206-226) (g) or pEGFP-F1Ltail(-) (207-226) (j) containing only the C-terminal tail of F1L. Mitochondria were visualized with Mitotracker Red (b, e, h, and k). Merging of EGFP-F1LTR (1-206) and EGFP-F1LHTR (1-218) with Mitotracker Red (c and f) indicated the two truncated versions of F1L did not localize to the mitochondria. Merging of EGFP-F1Ltail(+) (206-226) or EGFP-F1Ltail(-) (207-226) with Mitotracker Red (i and l) resulted in a uniform yellow image, indicating the C-terminal tail of F1L is sufficient for mitochondrial localization.
To determine the localization of the various F1L constructs during the context of virus infection, each EGFP-F1L construct was subcloned into pSC65 and placed under the control of an early/late poxviral promoter. HeLa cells were infected with VV65 and transfected with pSC65-F1LTR (1-206), pSC65-F1LHTR (1-218), pSC65-F1Ltail(+) (206-226), and pSC65-F1Ltail(-) (207-226), and localization was visualized by confocal microscopy. Similar to the transient transfection of HeLa cells, HeLa cells transfected and infected with VV65 demonstrated similar localization patterns (data not shown).
F1L inserts into mitochondria in vitro as a tail-anchored protein.
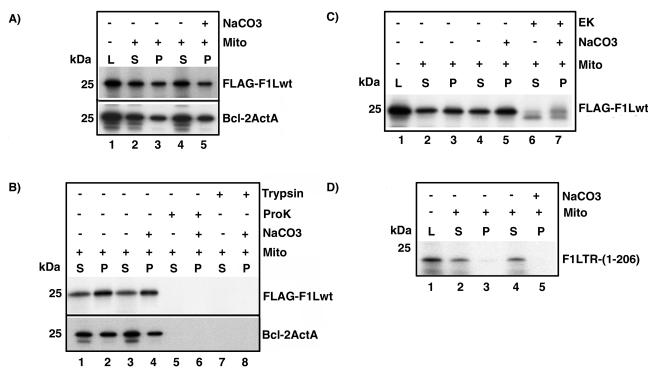
Since our evidence demonstrated that the C-terminal tail of F1L was necessary and sufficient for mitochondrial localization, this suggested that F1L might be a member of the TA family of proteins. Due to the presence of the targeting sequence at the C terminus, TA proteins are inserted posttranslationally into cellular membranes (9, 10). To investigate whether F1L was a representative TA protein and therefore posttranslationally inserted into mitochondrial membranes, we utilized an in vitro transcription-translation (TNT) assay. FLAG-tagged F1Lwt was cloned into the vector pSPUTK to allow for efficient transcription and translation of F1L in the presence of [35S]methionine. Following translation, cycloheximide was added to the 35S-labeled FLAG-F1Lwt to inhibit further translation. The generated [35S]FLAG-F1Lwt protein was added to purified mitochondria, and translated protein was visualized by autoradiography. Results showed that approximately half of the translated FLAG-F1Lwt was recovered in the mitochondrial pellet (Fig. 3A, compare lanes 1 to 3). This observation suggested that F1L was capable of posttranslational insertion into mitochondria, similar to other members of the TA family (Fig. 3A, compare lanes 1 to 3). We routinely found that after the addition of mitochondria approximately half of the FLAG-F1Lwt protein remained in the supernatant (Fig. 3A, lane 2); however, the addition of increasing amounts of mitochondria supported an increased insertion into the mitochondrial pellet (data not shown). As a control, we used a TNT 35S-labeled Bcl-2ActA chimera containing the mitochondrial-targeting polypeptide from Listeria monocytogenes ActA protein, which has been shown to localize Bcl-2 exclusively to the mitochondria (52, 70). Similar to FLAG-F1Lwt, Bcl-2ActA was also recovered in the mitochondrial pellet, with approximately half of the protein remaining in the supernatant (Fig. 3A, lanes 2 and 3).
FIG. 3.
F1L inserts into mitochondria in vitro and demonstrates classical tail-anchored orientation. (A) F1Lwt is anchored into mitochondrial membranes. TNT-generated 35S-labeled FLAG-F1Lwt lysate (L) and Bcl-2ActA (L) were added to 10 μg of purified mitochondria. Mitochondrial pellets (P) and supernatants (S) were separated by centrifugation. Mitochondrial pellet fractions were washed with either PBS or Na2CO3 (pH 11.5). (B) F1Lwt demonstrates tail-anchored membrane orientation with the N terminus exposed to the cytoplasm. FLAG-F1Lwt samples were treated with proteinase K (ProK) or trypsin and incubated for 1 h at 30°C. (C) FLAG-F1Lwt is sensitive to enterokinase (EK). Following insertion into mitochondria, FLAG-F1Lwt samples were treated with EK for 2 h at room temperature. (D) The C-terminal tail of F1L is necessary for insertion into mitochondria. TNT-generated 35S-labeled F1LTR (1-206) (L) was added to 10 μg of purified mitochondria, and pellet (P) and supernatant (S) fractions were separated by centrifugation. Pellets were washed with either PBS or 0.1 M Na2CO3 (pH 11.5).
To determine if F1L was in fact inserted into the mitochondrial membrane and not just loosely associated with the mitochondria, mitochondria were washed with an alkaline solution of Na2CO3 (pH 11.5) following posttranslational insertion of FLAG-F1Lwt. The addition of Na2CO3 (pH 11.5) removes loosely associated proteins but not integral membrane proteins (3, 24). No reduction of FLAG-F1Lwt or Bcl-2ActA in mitochondria was seen between the untreated and treated mitochondrial pellet fractions following Na2CO3 (pH 11.5) washing (Fig. 3A, compare lanes 3 and 5), suggesting that like Bcl-2ActA, F1L is anchored into mitochondrial membranes and is not just loosely associated.
Membrane orientation studies on TA proteins have revealed that they are anchored into the phospholipid bilayer by the C terminus, with the N terminus exposed to the cytosol (9, 10). To determine the orientation of F1L in mitochondria we utilized proteinase K, trypsin, and enterokinase, which will readily degrade exposed portions of F1L. Following the addition of proteases to the mitochondria, we found that the membrane-associated FLAG-F1Lwt was sensitive to both proteinase K and trypsin, as indicated by the complete loss of radiolabeled protein, suggesting that the majority of F1L was in fact oriented towards the cytoplasm (Fig. 3B, lanes 5 to 8). Similarly, the previously characterized TA protein Bcl-2ActA was also sensitive to proteinase K and trypsin (32, 70) (Fig. 3B, lanes 5 to 8). Unfortunately, we were unable to detect the presence of the protected C-terminal fragment of F1L in these assays. This may be due to the presence of only one methionine in the short C-terminal tail of F1L and the small size of the fragment (∼3 kDa). Therefore, to further confirm the membrane orientation of F1L we used enterokinase, which specifically recognizes and cleaves the N-terminal FLAG sequence (Asp-Asp-Asp-Asp-Lys↓) from F1L. Enterokinase was added to either the supernatant fraction containing soluble FLAG-F1Lwt (Fig. 3C, lane 6) or FLAG-F1Lwt associated with mitochondria (Fig. 3C, lane 7) followed by analysis by autoradiography. Using this approach, a noticeable shift in F1L mobility resulted due to the loss of the FLAG tag from the N terminus of F1L, indicating that FLAG-F1Lwt was inserted into the mitochondria with the N terminus exposed (Fig. 3C, lanes 6 and 7). These cumulative data suggest that FLAG-F1Lwt displays a typical TA orientation, with the N terminus facing the cytoplasm.
Since our confocal results demonstrated that expression of EGFP-F1LTR (1-206), which was missing the last 20 amino acids, was unable to localize to the mitochondria (Fig. 2B), we assessed the role of the F1L C terminus in membrane anchoring in the in vitro TNT assay. TNT reactions were performed using the DNA template pSPUTK-F1LTR (1-206), which transcribes and translates a truncated 35S-labeled F1L missing the C-terminal transmembrane domain and the short hydrophilic tail (Fig. 2A). Results showed only a minimal association of F1LTR (1-206) with mitochondria, while most of the protein was found in the supernatant (Fig. 3D, lanes 1 to 3). To determine if the small amount of F1LTR found in the mitochondrial pellet was anchored into the membrane, the mitochondrial fraction was washed with Na2CO3 (pH 11.5). Alkaline extraction resulted in a complete loss of radiolabeled FILTR (1-206), thereby confirming that the C-terminal domain of F1L was necessary for membrane insertion (Fig. 3D, lanes 4 and 5).
F1L inserts into mitochondria during virus infection.
To determine the membrane orientation of mitochondria-localized F1L during virus infection, we performed confocal microscopy utilizing selective permeabilization procedures and antibodies directed specifically to the N terminus of F1L. HeLa cells were infected with VVWR-FLAG-F1Lwt, and F1L localization was visualized using an anti-FLAG or anti-F1L antibody. When HeLa cells were treated with NP-40, which permeabilizes both the plasma membrane and mitochondrial membrane, anti-FLAG and anti-F1L antibodies demonstrated colocalization of F1L with cytochrome c (Fig. 4A, panels a to c, and B, panels a to c). In contrast, when HeLa cells were permeabilized with either digitionin or SLO, which selectively permeabilize the plasma membrane but not the mitochondrial membrane, anti-FLAG antibody and an anti-F1L antibody generated to the first 120 amino acids of F1L still gained access to F1L, indicating that the N terminus of F1L was facing the cytoplasm (Fig. 4A, panels d to i, and B, panels d to i) (18, 48). Notably, however, anti-cytochocrome c was unable to gain access to the inner mitochondrial membrane space, indicating that the digitonin and SLO treatment selectively permeabilized the plasma membrane but not the mitochondrial membrane, as indicated by a loss of cytochrome c staining (Fig. 4A, panels d to i, and B, panels d to i).
FIG. 4.
F1L displays classical tail-anchored orientation in infected cells. (A and B) F1L membrane orientation determined by confocal microscopy. HeLa cells were infected with VVWR-FLAG-F1Lwt and permeabilized with either NP-40, digitonin, or SLO. F1L was detected using anti-F1L (A) or anti-FLAG (B) antibodies, and mitochondria were detected using anti-cytochrome c antibody. (C) F1L inserts into mitochondria during virus infection. Jurkat cells were infected with the recombinant VV VVWR-FLAG-F1Lwt for 6 h at an MOI of 5. Mitochondrial pellet fractions (P) were purified from supernatants (S) by centrifugation. The presence of F1L in mitochondria was detected using either anti-F1L or anti-Flag antibody. (D) F1L is an integral membrane protein. Mitochondria pellet fractions (P) were purified and separated from supernatants (S) by centrifugation. Pellet fractions were washed with either PBS or 0.1 M Na2CO3 (pH 11.5). (E) F1L displays tail-anchored orientation in infected cells. Following purification of mitochondria from virally infected cells, mitochondria were treated with either proteinase K (ProK), trypsin, or enterokinase (EK). Supernatants (S) and mitochondrial pellet (P) fractions were separated by centrifugation and washed with Na2CO3 (pH 11.5), and F1L was detected by immunoblotting using anti-FLAG or anti-F1L antibody.
To further confirm the membrane orientation of F1L during virus infection, Jurkat cells were infected with the recombinant virus VVWR-FLAG-F1Lwt, mitochondria were purified, and the presence of F1L in mitochondria was determined by immunoblotting using both anti-F1L and anti-FLAG antibodies. Mitochondria purified from mock-infected cells showed the absence of F1L in both the supernatant and pellet fractions as expected (Fig. 4C). In contrast, Jurkat cells infected with VVWR-FLAG-F1Lwt revealed the presence of F1L only in the mitochondrial pellet fraction, with undetectable levels in the supernatant (Fig. 4C, lanes 3 and 4). To determine if F1L was an integral membrane protein, mitochondria were washed with Na2CO3 (pH 11.5), and F1L was detected using an anti-FLAG antibody. Similar to our in vitro results, F1L was only detected in the mitochondrial pellet fraction and Na2CO3 washing did not result in a reduction of F1L levels (Fig. 4D, compare lanes 4 and 6). The membrane orientation of F1Lwt during virus infection was determined by isolating mitochondria from virus-infected cells followed by treatment with proteinase K and trypsin. Under these conditions no F1L was detected using either anti-FLAG or anti-F1L, both of which are directed to the N-terminal portion of FLAG-F1L, suggesting that the N terminus of F1L was digested by the proteases (Fig. 4E, lanes 1 to 4). When purified mitochondria were treated with enterokinase and blotted with anti-FLAG antibody, an overall reduction in F1L was observed, indicating that the majority of the FLAG epitope was no longer accessible to the antibody due to cleavage by enterokinase (Fig. 4E, lane 5, and D, lane 6). When the same samples were blotted with the anti-F1L antibody which recognizes amino acids 1 to 120, a doublet was produced, confirming that FLAG-F1L was cleaved by enterokinase and further demonstrating that the N terminus of F1L is facing the cytoplasm (Fig. 3E, lane 5). Together these data indicate that F1L is inserted into mitochondria during virus infection, with the N terminus of the protein in the cytoplasm.
Basic amino acids in the C-terminal tail of F1L are critical for mitochondria targeting.
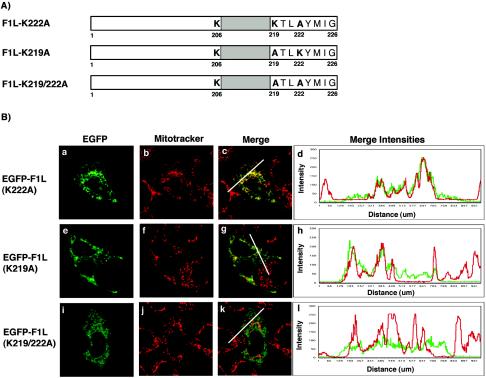
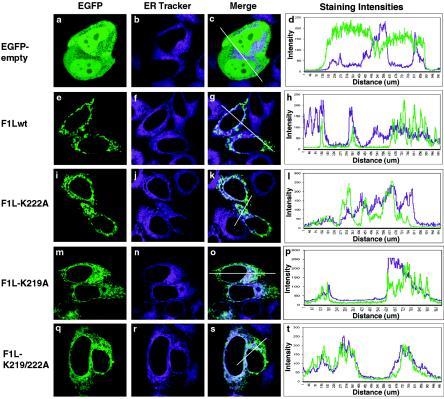
F1L shares characteristics with other TA proteins, including a transmembrane domain flanked by positively charged lysines (Fig. 2A) (30, 34). Mutagenesis of the positively charged amino acids surrounding the transmembrane domain in other TA proteins has revealed a significant role for these amino acids in directing TA proteins to the mitochondria (30, 34). We found that deletion of the last eight amino acids, including lysines 219 and 222, resulted in the inability of F1L to localize to the mitochondria, suggesting that these eight residues are critical for mitochondrial localization (Fig. 2B, panels d and e). Therefore, to investigate if the positively charged amino acids in the C-terminal tail of F1L play a role in localization of F1L to the mitochondria, we generated three F1L mutants by PCR-directed mutagenesis. EGFP-F1L-K219A, EGFP-F1L-K222A, and EGFP-F1L-K219/222A were created by mutating either lysine 219, lysine 222, or both lysines 219 and 222 to neutral alanine residues (Fig. 5A). The localization of the three mutant constructs was determined by transfecting HeLa cells and visualizing protein localization by confocal microscopy. Expression of EGFP-F1L-K222A demonstrated a characteristic mitochondrial staining pattern that colocalized with Mitotracker, indicating that mutagenesis of lysine 222 did not overtly influence mitochondrial localization (Fig. 5B, panels a to c). As an indicator of colocalization, we plotted the merge intensities for both EGFP-F1L-K222A with Mitotracker using a profile option within the Zeiss LSM 510 image software program that accurately compares the intensities of various fluorescent signals along a cellular distance (represented by white lines in the confocal images). This approach nicely confirmed colocalization of EGFP-F1L-K222A with Mitotracker as shown by mirroring of the EGFP and Mitotracker merge intensities (Fig. 5B, panel d). When HeLa cells were transfected with pEGFP-F1L-K219A, we routinely observed only partial localization with Mitotracker (Fig. 5B, panels e to g). The remainder of EGFP-F1L-K219A localized to an unidentified membrane-like structure throughout the cytoplasm. The merge intensity profile of EGFP-F1L-K219A demonstrated some regions of colocalization with Mitotracker and other regions where colocalization was not evident (Fig. 5B, panel h). When HeLa cells were transfected with the double lysine-to-alanine mutant, pEGFP-F1L-K219/222A, the mitochondrial targeting of F1L was reduced and the F1L signal was found localized to a membrane-like structure throughout the cytoplasm (Fig. 5B, panels i to k). This observation was supported by a decrease in colocalization when the merge intensities for EGFP-F1L-K219/222A and Mitotracker were plotted (Fig. 5B, panel l). These data clearly indicated that mutation of the positively charged lysines in the tail of F1L affected localization. Similar localization patterns for EGFP-F1L-K219A, EGFP-F1L-K222A, and EGFP-F1L-K219/222A were also observed during virus infection of HeLa cells (data not shown).
FIG. 5.
Basic amino acids in the C-terminal tail of F1L are critical for mitochondrial targeting. (A) Schematic representation of the various F1L point mutants fused to EGFP. F1L-K222A has lysine 222 mutated to alanine. F1L-K219A has lysine 219 mutated to alanine. F1L-K219/222A has both lysines 219 and 222 mutated to alanines. (B) HeLa cells were transiently transfected with either pEGFP-F1L-K222A (a), pEGFP-F1L-K219A (e), or pEGFP-F1L-K219/222A (i) and stained with Mitotracker Red (b, f, and j). The merge image of EGFP-F1LK222A and Mitotracker Red (c) shows a uniform yellow image, indicating mitochondrial localization. This was further confirmed by the merge intensity plot of EGFP-F1LK222A (green) and Mitotracker (red) over a cellular distance (white line) (d). The merge image of EGFP-F1L-K219A (g) demonstrates mitochondrial localization but, in addition, demonstrates a web-like staining distribution throughout the cell. The merge intensity plot for EGFP-F1L-K219A and Mitotracker Red (h) shows some regions of alignment, indicating mitochondrial localization, but also demonstrates areas lacking colocalization. The merge image of EGFP-F1L-K219/222A (k) demonstrates a staining pattern characteristic of ER and displays minimal colocalization with Mitotracker Red. The merge intensity plot for EGFP-F1L-K219/222A and Mitotracker Red further demonstrates the loss of colocalization between Mitotracker Red and EGFP-F1L-K219/222A (l).
To determine if EGFP-F1L-K219A and EGFP-F1L-K219/222A were rerouted to the ER, a common default membrane when positive charges are eliminated from mitochondria-localized TA proteins, HeLa cells were transfected with the various F1L mutant constructs and stained with ER-Tracker (9, 10, 30, 34). Cells transfected with either pEGFP, pEGFP-F1Lwt, or pEGFP-F1L-K222A showed minimal colocalization with ER-Tracker, and this observation was supported by plotting the merge intensities of EGFP and ER-Tracker (Fig. 6a to l). In contrast, cells transfected with pEGFP-F1L-K219A again showed both a mitochondrial and membrane-like staining pattern throughout the cell (Fig. 6m to p). When the fluorescent signals of pEGFP-F1L-K219A and ER-Tracker were superimposed colocalization was evident, demonstrating that in addition to localizing to the mitochondria F1L-K219A was also localized to the ER (Fig. 6m to o). The merge intensity profile demonstrated some regions of colocalization as well as regions where the merge intensities were very different (Fig. 6p). When cells were transfected with pEGFP-F1L-K219/222A, obvious ER localization was detected, demonstrating that mutating lysines 219 and 222 shifted the localization of F1L from the mitochondria to the ER, and the merge intensity profile supported this observation (Fig. 6q to t). The ER localization of EGFP-F1L-K219A and EGFP-F1L-K219/222A was also confirmed by staining cells with an antibody that recognizes calnexin, an ER-localized protein (Fig. 7), and similar localization patterns were detected during virus infection (data not shown). These observations demonstrated that lysines 219 and 222 played a critical role in localization of F1L to the mitochondria.
FIG. 6.
The loss of basic amino acids in the C-terminal tail of F1L results in ER localization. HeLa cells were transfected with either pEGFP-empty vector (a), pEGFP-F1Lwt (e), pEGFP-F1L-K222A (i), pEGFP-F1L-K219A (m), or pEGFP-F1L-K219/222A (q) and stained with ER-Tracker (b, f, j, n, and r). Merged images and intensity plots (c, d, g, h, k, and l) indicate that EGFP-F1Lwt and EGFP-F1L-K222A do not localize to the ER. The merge image of EGPF-F1L-K219A and ER Tracker (o) indicates partial ER localization, which is further supported by the merge intensity plot (p). The merge image of EGFP-F1L-K219/222A and ER-Tracker (s) indicates EGFP-F1L-K219/222A localizes to the ER, which is further supported by the merge intensity plot (t).
FIG. 7.
EGFP-F1L-K219A and EGFP-F1L-K219/222A colocalize with calnexin. HeLa cells were transfected with either pEGFP-empty vector (a), pEGFP-F1Lwt (e), pEGFP-F1L-K222A (i), pEGFP-F1L-K219A (m), or pEGFP-F1L-K219/222A (q) and stained with anticalnexin, an ER resident protein, to detect the ER (b, f, j, n, and r). Merged images and intensity plots (c, d, g, h, k, and l) indicate that the majority of EGFP, EGFP-F1Lwt, and EGFP-F1L-K222A do not colocalize with calnexin. The merge image of EGPF-F1L-K219A and calnexin (o) indicates partial ER localization, which is further supported by the merge intensity plot (p). The merge image of EGFP-F1L-K219/222A and calnexin (s) indicates EGFP-F1L-K219/222A colocalizes with calnexin, and this is further supported by alignment in the staining intensity plot (t).
Mitochondrial localization of F1L is required for apoptosis inhibition.
Originally we found that VV strain Copenhagen, which is naturally devoid of the caspase 8 inhibitor CrmA/Spi2, inhibits apoptosis by maintaining mitochondrial integrity (66). Our recent findings show that this inhibition is due to expression of F1L, which localizes to the mitochondria and prevents the release of cytochrome c and loss of the inner mitochondrial membrane potential (67). Data from our previous paper showed that F1L was able to inhibit staurosporine-induced apoptosis, similar to the antiapoptotic protein Bcl-2 (67). To determine whether F1L could inhibit an alternative apoptotic stimulus, HeLa cells were transfected with pEGFP or pEGFP-F1Lwt and treated with TNF-α for 5, 8, 10, or 15 h, and cell death was determined by loss of the inner mitochondrial membrane potential, a hallmark feature of apoptosis, in the EGFP-positive population (40). Loss of the inner mitochondrial membrane potential was monitored by staining mitochondria with TMRE, a mitochondria-specific fluorescent dye which is incorporated into healthy respiring mitochondria, and apoptosis was analyzed by two-color flow cytometry (19, 23, 41). HeLa cells transfected with pEGFP and pEGFP-F1Lwt and stained with TMRE showed a population of cells that were both positive for EGFP and TMRE, representing cells that were transfected and containing nonapoptotic mitochondria that retained their inner mitochondrial membrane potential (data not shown). After treatment with TNF-α for 5, 8, 10, and 15 h, 21, 36, 56, and 59% of the cells transfected with pEGFP showed a loss of TMRE fluorescence, indicating that the expression of EGFP alone was not sufficient to inhibit apoptosis (Fig. 8A). In contrast, HeLa cells transfected with pEGFP-F1Lwt and treated with TNF-α exhibited only a 3 to 10% reduction in TMRE fluorescence over the various time points, indicating that expression of F1L protected cells from loss of the inner mitochondrial membrane potential following an apoptotic trigger (Fig. 8A).
FIG. 8.
Localization of F1L to the mitochondria is necessary for apoptosis inhibition. (A) HeLa cells were transfected with either pEGFP or pEGFP-F1Lwt for 16 h and treated with 10 ng of TNF-α/ml for 5, 8, 10, or 15 h. Loss of the inner mitochondrial membrane potential was assessed by TMRE fluorescence in EGFP-positive cells by two-color flow cytometry. Standard deviations were calculated from three ormore independent experiments. (B) HeLa cells were transfected with either pEGFP, pEGFP-F1Lwt, pEGFP-Bcl-2, pEGFP-F1LHTR (1-218), pEGFP-F1LTR (1-206), pEGFP-F1Ltail(+) (206-226), pEGFPtail(-) (207-226), or pEGFP-F1LCyb5 for 16 h and treated with 10 ng of TNF-α/ml for 8 h. Loss of the inner mitochondrial membrane potential was assessed by TMRE fluorescence in EGFP-positive cells by two-color flow cytometry. Standard deviations were calculated from three or more independent experiments. (C) F1LCyb5 localizes to the ER. HeLa cells were transfected with PEGFP-F1LCyb5 (a and d) and stained with either Mitotracker Red (b) or ER-Tracker (e). Merge images indicate that EGFP-F1LCyb5 localized to the ER (f) but not mitochondria (c). (D) Expression levels of various F1L constructs. HeLa cells were transfected, and protein levels were determined by Western blotting with anti-EGFP.
Our studies indicate that F1L localizes predominantly to the mitochondria, suggesting that localization to the mitochondria during virus infection might be critical for apoptosis inhibition (67). To determine if mitochondrial localization of F1L is essential for the antiapoptotic function of F1L, we used the panel of EGFP-F1L deletion constructs shown in Fig. 2. HeLa cells transfected with either pEGFP, pEGFP-F1Lwt, pEGFP-F1LHTR (1-218), pEGFP-F1LTR (1-206), pEGFP-F1Ltail(+) (206-226), or pEGFP-F1Ltail(-) (207-226) were treated with TNF-α for 8 h, a time point at which efficient TMRE loss was detected, and cell death was determined by loss of the inner mitochondrial membrane potential (40). After treatment with TNF-α, 36% of the cells transfected with pEGFP showed a loss of TMRE fluorescence, indicating that expression of EGFP alone was not sufficient to inhibit apoptosis (Fig. 8B). In contrast, HeLa cells transfected with pEGFP-F1Lwt or with the antiapoptotic protein pEGFP-Bcl-2 and treated with TNF-α exhibited only a 3 or 1% reduction in TMRE fluorescence, respectively, indicating that expression of F1L, similar to Bcl-2, protected cells from loss of the inner mitochondrial membrane potential (Fig. 8B). HeLa cells transfected with either pEGFP-F1LHTR (1-218) or pEGFP-F1LTR (1-206) showed no protection from apoptosis following treatment with TNF-α, as demonstrated by losses of TMRE fluorescence of 29 and 32%, respectively (Fig. 8B). This result suggested that expression of the N-terminal region of F1L, which is no longer capable of localizing to mitochondria, is unable to inhibit apoptosis. Similar results were found upon expression of a cytoplasmic form of Bcl-2 and M11L (20, 21, 46). Additionally, we found that expression of the two C-terminal tail constructs EGFP-F1Ltail(+) (206-226) and EGFP-F1Ltail(-) (207-226), both of which localize to the mitochondria, were also unable to provide protection from TNF-α-induced apoptosis (Fig. 8B). These observations indicated that expression of the F1L tail alone was not sufficient to inhibit apoptosis. The results indicate that expression of either the N terminus alone or the mitochondria-localized tail of F1L is not sufficient to inhibit apoptosis.
To determine if localization of F1L to another cellular membrane resulted in apoptosis inhibition, we generated an F1L chimera that would exclusively target F1L to the ER, a location at which Bcl-2 has been reported to function (4, 8, 51, 60, 61, 70). To create this chimera, we swapped the C-terminal tail of F1L (amino acids 207 to 226) with the C-terminal tail of the ER isoform of cytochrome b5, a TA protein that has been shown to localize exclusively to the ER (37, 42, 48, 70). To confirm the localization of F1LCyb5, we transfected HeLa cells with pEGFP-F1LCyb5, stained the cells with Mitotracker and ER-Tracker, and visualized localization by confocal microscopy. The fluorescent signal of EGFP-F1LCyb5 demonstrated an ER-like staining pattern throughout the cell that colocalized with ER-Tracker, indicating that EGPF-F1LCyb5 localized to the ER (Fig. 8C). To determine if EGFP-F1LCyb5 expression could inhibit apoptosis, HeLa cells were transfected with pEGFP-F1LCyb5 and treated with TNF-α. Our data demonstrated that 36% of cells underwent apoptosis, which was comparable to cells expressing EGFP, indicating that EGFP-F1LCyb5 was unable to inhibit apoptosis when targeted to the ER (Fig. 8B).
To ensure that differences in protein expression levels were not influencing the level of inhibition, the expression levels of the various EGFP-F1L constructs were analyzed via Western blotting. HeLa cells transfected with EGFP-F1Ltail(+) (206-226) and EGFP-F1Ltail(-) (207-226) showed higher expression levels compared to wild-type F1L (Fig. 8D). Even at higher expression levels, both of these constructs were clearly unable to inhibit apoptosis. The expression levels of EGFP-F1Lwt and EGFP-F1LCyb5 were virtually equivalent, indicating that expression of EGFP-F1LCyb5, which localizes F1L to the ER, is unable to inhibit apoptosis. Western blot analysis demonstrated that transfection of EGFP-F1LHTR (1-218) had a slightly lower expression level than EGFP-F1Lwt and, unfortunately, the expression level of EGFP-F1LTR (1-206) was significantly lower than that of EGFP-F1Lwt (Fig. 8D). All of our attempts to increase the expression level of this truncated version of F1L were unsuccessful (data not shown). In addition, the transfection efficiencies as measured by EGFP fluorescence were 66, 45, 26, 23, 63, 49, and 50% for EGFP, EGFP-F1Lwt, EGFP-F1LHTR (1-218), EGFP-F1LTR (1-206), EGFP-F1Ltail(+), EGFP-F1Ltail(-) (207-226), and EGFP-F1LCyb5, respectively, which mirrored expression levels determined by Western blotting. Concerned that a lack of inhibition by F1LTR (1-206) was due to lower levels of protein following transfection, we reanalyzed our flow cytometry data for all of the transfected constructs by specifically gating on the population of lower EGFP-expressing cells at 102 and determined the percentage of TMRE loss following the addition of TNF-α. Using this approach to reanalyze only the lower EGFP-positive population, the data again showed the same trend in that expression of the truncated and tail-only versions of F1L were unable to inhibit apoptosis (data not shown). These results indicated that truncated F1L found in the cytoplasm is unable to inhibit apoptosis, while the C-terminal tail of F1L is sufficient to localize F1L to the mitochondria but on its own is not sufficient to inhibit apoptosis.
DISCUSSION
Viruses are under constant pressure from the host immune system. To avoid detection and elimination, many viruses adopt strategies to counteract various facets of the immune response, including strategies that interfere with apoptosis (15, 27, 54). Poxviruses, of which vaccinia virus is a member, express numerous proteins which mediate immune evasion, including proteins aimed at interference with apoptosis (7, 33, 58). Historically, the best-characterized apoptosis modulator encoded by members of the poxvirus family is CrmA/Spi-2, which inhibits apoptosis by preventing the activity of caspase 8 (59, 69). Until recently, it was assumed that CrmA/Spi-2 was the major intracellular antiapoptotic protein encoded by VV. We have recently demonstrated, however, that VV encodes an additional antiapoptotic protein, F1L. F1L localizes to mitochondria and inhibits both the loss of the inner mitochondrial membrane potential, a hallmark feature of apoptotic cells, and prevents the release of cytochrome c, the pivotal commitment step (67).
Currently, within the poxvirus family only members of the Orthopoxvirus genus, which includes VV, variola virus, cowpox virus, monkeypox virus, and ectromelia virus, encode F1L orthologs, suggesting an important role for F1L during Orthopoxvirus infection. All known orthologs share >95% sequence identify over the last 220 amino acids. The greatest sequence diversity among the various F1L orthologs is located within the N-terminal regions, with strains of variola virus, camelpox virus, and ectromelia virus displaying a series of unique repeats, the function of which is currently unknown. Although it is currently unknown if all F1L orthologs are functional, we have shown in this study that F1L orthologs present in ectromelia, rabbitpox, and cowpox viruses localized to mitochondria (Fig. 1).
One obvious feature of F1L is a C-terminal transmembrane domain flanked by positively charged lysine residues followed by a short eight-amino-acid hydrophilic tail (Fig. 2A). Similar domains are important for membrane localization, with both the length of the hydrophobic domain and the presence of flanking positive charges playing a role (9, 10, 68). Therefore, to investigate the sequence requirements necessary for F1L mitochondrial localization, we generated two C-terminal tail F1L constructs: F1Ltail(+) (206-226), which includes amino acids 206 to 226, and F1Ltail(-) (207-226), which includes amino acids 207 to 226 minus lysine 206 (Fig. 2A). Although we were unsure of the role of lysine 206 in mitochondrial localization, both EGFP-F1L tail constructs localized to the mitochondria, indicating that amino acids 207 to 226 were sufficient for mitochondrial localization and that the presence of lysine 206 was not required. Data from our previous publication revealed that the last 27 amino acids of F1L (F1L 199-226) were necessary and sufficient for localization to the mitochondria (67). This construct contained seven additional amino acids upstream of the transmembrane domain, and our present data reveal that these upstream amino acids are dispensable for mitochondrial localization. Additional analysis further revealed that transfection of pEGFP-F1LHTR (1-218), which contains the hydrophobic transmembrane domain but is missing the last eight amino acids of the hydrophilic tail, no longer localized to the mitochondria and was in fact cytosolic, suggesting that the short hydrophilic tail of F1L was playing a role in mitochondrial localization (Fig. 2B). Initially we predicted that F1LHTR (1-218) would be rerouted to the ER, which is a common default membrane for TA proteins (10, 31, 34, 37). However, due to the presence of a short 12-amino-acid hydrophobic transmembrane domain in F1L, it is unlikely that this domain would be of sufficient length to span the ER membrane, thereby resulting in cytoplasmic distribution (9, 49). The tail-anchored proteins Tom5 and outer mitochondrial membrane isoform of cytochrome b5 (OMb) both normally localize to the mitochondria but also demonstrate cytoplasmic staining when the C-terminal hydrophilic tail is deleted (30, 37). These observations indicate that the membrane-targeting signal in F1L is not solely dependent upon the presence and length of a transmembrane domain but, rather, additional amino acids downstream of the transmembrane domain are also required.
The presence of a C-terminal mitochondria-targeting motif in F1L suggests that F1L is a member of a growing group of proteins referred to as TA proteins. This family of proteins has received considerable attention due to their unique, but still-poorly understood, membrane-targeting mechanisms (9, 10, 68). TA proteins constitute a family of proteins that lack N-terminal signal sequences and insert into membranes using a C-terminal transmembrane domain. TA proteins have been discovered in a vast array of organisms, performing a diverse array of functions, and are therefore found in numerous membranes, including the ER, Golgi apparatus, and mitochondria (9, 10, 68). Due to the ordered steps involved in the secretory pathway leading from the ER to Golgi to plasma membrane, TA proteins need only discriminate between the ER and mitochondria (9, 38). The mitochondrial outer membrane contains numerous TA proteins, including Tom proteins involved in protein translocation and members of the Bcl-2 family such as Bcl-2, Bcl-xL, and Bak, which function to regulate the release of mitochondrial proapoptotic proteins (9, 10, 68). TA proteins have also been identified in viruses, such as the VV H3L protein, herpes simplex type II protein UL34, adenovirus E3-6.7K protein, and myxoma virus-encoded antiapoptotic protein M11L (16, 20, 43, 57). Although M11L and F1L display no sequence similarity, they share similar functional features, including localization to mitochondria and the ability to inhibit the loss of the inner mitochondrial membrane potential and release of cytochrome c (20, 21, 65).
TA proteins demonstrate a specific membrane orientation with the C terminus, spanning and anchoring the protein into the membrane with the N terminus facing the cytoplasm (38). To determine the membrane orientation of F1L, we performed in vitro TNT assays. Using this in vitro assay we confirmed that the last 20 amino acids of F1L were necessary for insertion of F1L into mitochondria. We also demonstrated using proteinase digestion that the N terminus of F1L was facing the cytoplasm (Fig. 3). This membrane orientation for F1L was also present during virus infection, as determined with purified mitochondria from VVWR-FLAG-F1Lwt-infected Jurkat cells and by confocal microscopy coupled with selective permeabilization procedures (Fig. 4). This specific membrane orientation for TA proteins has led to a common theme where the C-terminal transmembrane domain serves as a membrane anchor while the cytosol-facing N terminus carries out specific functions (9, 10). At present the precise antiapoptotic mechanism of action of F1L is undefined, but the specific membrane orientation of F1L suggests that the N-terminal cytosolic domain may be interacting with cellular proteins to inhibit apoptosis. We are currently pursuing experiments to identify potential F1L-interacting proteins that may elucidate the mechanism of action of this newly identified antiapoptotic protein.
A common trend seen among mitochondrial TA proteins is that, in addition to the transmembrane domain, flanking positively charged amino acids also play a role in localization (30, 31, 34, 37). F1L contains two positively charged lysines downstream of the transmembrane domain: lysines 219 and 222 (Fig. 2A). Mutation of lysine 222 to generate F1L-K222A did not overtly affect the localization of F1L. In contrast, when lysine 219 was mutated to alanine to generate F1L-K219A F1L localized to both the mitochondria and ER, and when both lysines were mutated to generate the construct F1L-K219/222A the majority of the protein localized to the ER. These data clearly indicate a role of lysines 219 and 222 in mitochondrial localization of F1L. Mutation of lysines 219 and 222 to alanine in F1L essentially extended the length of the hydrophobic domain from 12 to 20 amino acids, resulting in a transmembrane domain predicted to be sufficient for ER localization (10, 49). In a similar fashion, mutation of lysine residues downstream of the hydrophobic domain of Tom5 also results in altered localization to the ER (30). In contrast, a single mutation of lysine 219 in F1L to generate F1L-K219A lengthened the hydrophobic domain to 15 amino acids, resulting in obvious localization of F1L to both the mitochondria and ER.
Our data demonstrate that F1L localizes predominantly to the mitochondria, suggesting that the presence of F1L at the mitochondria is important during VV infection (67). HeLa cells transfected with the two F1L truncated constructs, F1LHTR (1-218) and F1LTR (1-206), both of which demonstrate cytoplasmic distribution, were unable to inhibit apoptosis. Similar observations were made with truncated versions of Bcl-2 and M11L, which no longer localized to the mitochondria and were also unable to inhibit apoptosis (20, 46). Although the localization of Bcl-2 at the ER has been associated with inhibition of apoptosis, the altered localization of F1L to the ER by swapping the C-terminal mitochondrial localization domain of F1L with the ER localization domain of cytochrome b5 was unable to inhibit apoptosis (4, 60, 61, 70). Additionally, expression of F1Ltail(+) (206-226) and F1Ltail(-) (207-226) was unable to inhibit apoptosis induced by TNF-α. Collectively, our data indicate that the localization of F1L to the mitochondria is necessary for apoptosis inhibition.
Our studies have now shown that the newly identified antiapoptotic protein F1L is a member of the TA family of proteins that localizes to the mitochondria, where it functions to inhibit apoptosis. Our data support the idea that F1L localization at the mitochondria is important during VV infection, since truncated and chimeric versions of F1L are no longer able to effectively inhibit apoptosis. A VV Copenhagen deletion virus, VV811, missing F1L in addition to other ORFs, is no longer able to inhibit apoptosis, and the addition of F1L restores apoptosis inhibition (67). Although F1L-related ORFs are currently only present in members of the Orthopoxvirus genus, various members of the Leporipoxvirus, Capripoxvirus, Yatapoxvirus, and Suipoxvirus genera encode M11L-like proteins (2, 11, 12, 39, 64). Strikingly, members of the Avipoxvirus genus are the only poxviruses to encode obvious Bcl-2 homologs (1, 63). Recent evidence demonstrates that the M11L protein from myxoma virus functions to inhibit apoptosis by interacting with the peripheral benzodiazepine receptor, a component of the mitochondrial permeability transition pore that has been linked to apoptosis (21). In addition, M11L interacts with Bak, and under some conditions Bax, both of which are proapoptotic members of the Bcl-2 family (65). It is currently unknown if the Avipoxvirus-encoded Bcl-2 proteins or the Orthopoxvirus-encoded F1L proteins function through similar interactions, but the presence of the various poxvirus-encoded antiapoptotic proteins that function to inhibit the mitochondrial apoptotic pathway clearly indicates the importance of interfering with this pathway for members of the poxvirus family.
Acknowledgments
This work was supported by a grant from the Canadian Institutes for Health Research (to M.B.). M.B. is the recipient of a New Investigator Award from the Canadian Institutes for Heath Research and an Alberta Heritage Foundation for Medical Research Scholar award. T.L.S. is the recipient of a studentship from the Natural Sciences and Engineering Research Council of Canada, and S.T.W. is the recipient of a studentship from the Alberta Heritage Foundation for Medical Research.
We thank Xuejun Sun and Gerry Barron at the Cross Cancer Research Institute for technical expertise in confocal microscopy and David Bond for excellent technical assistance.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2000. The genome of fowlpox virus. J. Virol. 74:3815-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, F. A. Osorio, C. Balinsky, G. F. Kutish, and D. L. Rock. 2002. The genome of swinepox virus. J. Virol. 76:783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. J., K. E. Mostov, and G. Blobel. 1983. Mechanisms of integration of de novo-synthesized polypeptides into membranes: signal-recognition particle is required for integration into microsomal membranes of calcium ATPase and of lens MP26 but not of cytochrome b5. Proc. Natl. Acad. Sci. USA 80:7249-7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annis, M. G., N. Zamzami, W. Zhu, L. Z. Penn, G. Kroemer, B. Leber, and D. W. Andrews. 2001. Endoplasmic reticulum localized Bcl-2 prevents apoptosis when redistribution of cytochrome c is a late event. Oncogene 20:1939-1952. [DOI] [PubMed] [Google Scholar]
- 5.Arnoult, D., L. M. Bartle, A. Skaletskaya, D. Poncet, N. Zamzami, P. U. Park, J. Sharpe, R. J. Youle, and V. S. Goldmacher. 2004. Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. USA 101:7988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barry, M., and R. C. Bleackley. 2002. Cytotoxic T lymphocytes: all roads lead to death. Nat. Rev. Immunol. 2:401-409. [DOI] [PubMed] [Google Scholar]
- 7.Barry, M., S. T. Wasilenko, T. L. Stewart, and J. M. Taylor. 2004. Apoptosis regulator genes encoded by poxviruses, p. 19. In C. Alonso (ed.), Viruses and apoptosis. Springer-Verlag, New York, N.Y. [DOI] [PubMed]
- 8.Bassik, M. C., L. Scorrano, S. A. Oakes, T. Pozzan, and S. J. Korsmeyer. 2004. Phosphorylation of BCL-2 regulates ER Ca2+ homeostasis and apoptosis. EMBO J. 23:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borgese, N., S. Brambillasca, P. Soffientini, M. Yabal, and M. Makarow. 2003. Biogenesis of tail-anchored proteins. Biochem. Soc. Trans. 31:1238-1242. [DOI] [PubMed] [Google Scholar]
- 10.Borgese, N., S. Colombo, and E. Pedrazzini. 2003. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J. Cell Biol. 161:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunetti, C. R., H. Amano, Y. Ueda, J. Qin, T. Miyamura, T. Suzuki, X. Li, J. W. Barrett, and G. McFadden. 2003. Complete genomic sequence and comparative analysis of the tumorigenic poxvirus Yaba monkey tumor virus. J. Virol. 77:13335-13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron, C., S. Hota-Mitchell, L. Chen, J. Barrett, J. X. Cao, C. Macaulay, D. Willer, D. Evans, and G. McFadden. 1999. The complete DNA sequence of myxoma virus. Virology 264:298-318. [DOI] [PubMed] [Google Scholar]
- 13.Chakrabarti, S., J. R. Sisler, and B. Moss. 1997. Compact, synthetic, vaccinia virus early/late promoter for protein expression. BioTechniques 23:1094-1097. [DOI] [PubMed] [Google Scholar]
- 14.Cory, S., D. C. Huang, and J. M. Adams. 2003. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene 22:8590-8607. [DOI] [PubMed] [Google Scholar]
- 15.Cuconati, A., and E. White. 2002. Viral homologs of BCL-2: role of apoptosis in the regulation of virus infection. Genes Dev. 16:2465-2478. [DOI] [PubMed] [Google Scholar]
- 16.da Fonseca, F. G., E. J. Wolffe, A. Weisberg, and B. Moss. 2000. Characterization of the vaccinia virus H3L envelope protein: topology and posttranslational membrane insertion via the C-terminal hydrophobic tail. J. Virol. 74:7508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desagher, S., and J. C. Martinou. 2000. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 10:369-377. [DOI] [PubMed] [Google Scholar]
- 18.Duan, S., P. Hajek, C. Lin, S. K. Shin, G. Attardi, and A. Chomyn. 2003. Mitochondrial outer membrane permeability change and hypersensitivity to digitonin early in staurosporine-induced apoptosis. J. Biol. Chem. 278:1346-1353. [DOI] [PubMed] [Google Scholar]
- 19.Ehrenberg, B., V. Montana, M. D. Wei, J. P. Wuskell, and L. M. Loew. 1988. Membrane potential can be determined in individual cells from the Nernstian distribution of cationic dyes. Biophys. J. 53:785-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett, H., M. Barry, S. F. Lee, X. Sun, K. Graham, J. Stone, R. C. Bleackley, and G. McFadden. 2000. M11L: a novel mitochondria-localized protein of myxoma virus that blocks apoptosis of infected leukocytes. J. Exp. Med. 191:1487-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett, H., M. Barry, X. Sun, S. F. Lee, C. Frantz, L. G. Berthiaume, G. McFadden, and R. C. Bleackley. 2002. The myxoma poxvirus protein, M11L, prevents apoptosis by direct interaction with the mitochondrial permeability transition pore. J. Exp. Med. 196:1127-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falcone, D., and D. W. Andrews. 1991. Both the 5′ untranslated region and the sequences surrounding the start site contribute to efficient initiation of translation in vitro. Mol. Cell. Biol. 11:2656-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farkas, D. L., M. D. Wei, P. Febbroriello, J. H. Carson, and L. M. Loew. 1989. Simultaneous imaging of cell and mitochondrial membrane potentials. Biophys. J. 56:1053-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldmacher, V. S., L. M. Bartle, A. Skaletskaya, C. A. Dionne, N. L. Kedersha, C. A. Vater, J. W. Han, R. J. Lutz, S. Watanabe, E. D. Cahir McFarland, E. D. Kieff, E. S. Mocarski, and T. Chittenden. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. USA 96:12536-12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross, A., J. M. McDonnell, and S. J. Korsmeyer. 1999. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 13:1899-1911. [DOI] [PubMed] [Google Scholar]
- 27.Hardwick, J. M., and D. S. Bellows. 2003. Viral versus cellular BCL-2 proteins. Cell Death Differ. 10(Suppl. 1):S68-S76. [DOI] [PubMed] [Google Scholar]
- 28.Heibein, J. A., I. S. Goping, M. Barry, M. J. Pinkoski, G. C. Shore, D. R. Green, and R. C. Bleackley. 2000. Granzyme B-mediated cytochrome c release is regulated by the Bcl-2 family members Bid and Bax. J. Exp. Med. 192:1391-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hengartner, M. O. 2000. The biochemistry of apoptosis. Nature 407:770-776. [DOI] [PubMed] [Google Scholar]
- 30.Horie, C., H. Suzuki, M. Sakaguchi, and K. Mihara. 2002. Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol. Biol. Cell 13:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isenmann, S., Y. Khew-Goodall, J. Gamble, M. Vadas, and B. W. Wattenberg. 1998. A splice-isoform of vesicle-associated membrane protein-1 (VAMP-1) contains a mitochondrial targeting signal. Mol. Biol. Cell 9:1649-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janiak, F., B. Leber, and D. W. Andrews. 1994. Assembly of Bcl-2 into microsomal and outer mitochondrial membranes. J. Biol. Chem. 269:9842-9849. [PubMed] [Google Scholar]
- 33.Johnston, J. B., and G. McFadden. 2003. Poxvirus immunomodulatory strategies: current perspectives. J. Virol. 77:6093-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaufmann, T., S. Schlipf, J. Sanz, K. Neubert, R. Stein, and C. Borner. 2003. Characterization of the signal that directs Bcl-xL, but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 160:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krajewski, S., S. Tanaka, S. Takayama, M. J. Schibler, W. Fenton, and J. C. Reed. 1993. Investigation of the subcellular distribution of the bcl-2 oncoprotein: residence in the nuclear envelope, endoplasmic reticulum, and outer mitochondrial membranes. Cancer Res. 53:4701-4714. [PubMed] [Google Scholar]
- 36.Kroemer, G., and J. C. Reed. 2000. Mitochondrial control of cell death. Nat. Med. 6:513-519. [DOI] [PubMed] [Google Scholar]
- 37.Kuroda, R., T. Ikenoue, M. Honsho, S. Tsujimoto, J. Y. Mitoma, and A. Ito. 1998. Charged amino acids at the carboxyl-terminal portions determine the intracellular locations of two isoforms of cytochrome b5. J. Biol. Chem. 273:31097-31102. [DOI] [PubMed] [Google Scholar]
- 38.Kutay, U., E. Hartmann, and T. A. Rapoport. 1993. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 3:72-75. [DOI] [PubMed] [Google Scholar]
- 39.Lee, H. J., K. Essani, and G. L. Smith. 2001. The genome sequence of Yaba-like disease virus, a yatapoxvirus. Virology 281:170-192. [DOI] [PubMed] [Google Scholar]
- 40.Marchetti, P., M. Castedo, S. A. Susin, N. Zamzami, T. Hirsch, A. Macho, A. Haeffner, F. Hirsch, M. Geuskens, and G. Kroemer. 1996. Mitochondrial permeability transition is a central coordinating event of apoptosis. J. Exp. Med. 184:1155-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metivier, D., B. Dallaporta, N. Zamzami, N. Larochette, S. A. Susin, I. Marzo, and G. Kroemer. 1998. Cytofluorometric detection of mitochondrial alterations in early CD95/Fas/APO-1-triggered apoptosis of Jurkat T lymphoma cells. Comparison of seven mitochondrion-specific fluorochromes. Immunol. Lett. 61:157-163. [DOI] [PubMed] [Google Scholar]
- 42.Mitoma, J., and A. Ito. 1992. The carboxy-terminal 10 amino acid residues of cytochrome b5 are necessary for its targeting to the endoplasmic reticulum. EMBO J. 11:4197-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moise, A. R., J. R. Grant, R. Lippe, R. Gabathuler, and W. A. Jefferies. 2004. The adenovirus E3-6.7K protein adopts diverse membrane topologies following posttranslational translocation. J. Virol. 78:454-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moss, B. 1996. Poxviridae: the viruses and their replication, p. 2637-2671. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 45.Moss, B., and P. L. Earl. 1991. Expression of proteins in mammalian cells using vaccinia viral vectors, p. 16.15.1. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 46.Nguyen, M., D. G. Millar, V. W. Yong, S. J. Korsmeyer, and G. C. Shore. 1993. Targeting of Bcl-2 to the mitochondrial outer membrane by a COOH-terminal signal anchor sequence. J. Biol. Chem. 268:25265-25268. [PubMed] [Google Scholar]
- 47.Nicholson, D. W., and N. A. Thornberry. 1997. Caspases: killer proteases. Trends Biochem. Sci. 22:299-306. [DOI] [PubMed] [Google Scholar]
- 48.Pedrazzini, E., A. Villa, R. Longhi, A. Bulbarelli, and N. Borgese. 2000. Mechanism of residence of cytochrome b5, a tail-anchored protein, in the endoplasmic reticulum. J. Cell Biol. 148:899-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pelttari, A., and H. J. Helminen. 1979. The relative thickness of intracellular membranes in epithelial cells of the ventral lobe of the rat prostate. Histochem. J. 11:613-624. [DOI] [PubMed] [Google Scholar]
- 50.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, E. K. Norton, and E. Paoletti. 1991. Deletion of 55 open reading frames from the termini of vaccinia virus. Virology 180:406-410. [DOI] [PubMed] [Google Scholar]
- 51.Pinton, P., D. Ferrari, E. Rapizzi, F. Di Virgilio, T. Pozzan, and R. Rizzuto. 2001. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. EMBO J. 20:2690-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pistor, S., T. Chakraborty, K. Niebuhr, E. Domann, and J. Wehland. 1994. The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J. 13:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poncet, D., N. Larochette, A. L. Pauleau, P. Boya, A. A. Jalil, P. F. Cartron, F. Vallette, C. Schnebelen, L. M. Bartle, A. Skaletskaya, D. Boutolleau, J. C. Martinou, V. S. Goldmacher, G. Kroemer, and N. Zamzami. 2004. An anti-apoptotic viral protein that recruits Bax to mitochondria. J. Biol. Chem. 279:22605-22614. [DOI] [PubMed] [Google Scholar]
- 54.Roulston, A., R. C. Marcellus, and P. E. Branton. 1999. Viruses and apoptosis. Annu. Rev. Microbiol. 53:577-628. [DOI] [PubMed] [Google Scholar]
- 55.Rudin, C. M., and C. B. Thompson. 1997. Apoptosis and disease: regulation and clinical relevance of programmed cell death. Annu. Rev. Med. 48:267-281. [DOI] [PubMed] [Google Scholar]
- 56.Schinzel, A., T. Kaufmann, and C. Borner. 2004. Bcl-2 family members: intracellular targeting, membrane-insertion, and changes in subcellular localization. Biochim. Biophys. Acta 1644:95-105. [DOI] [PubMed] [Google Scholar]
- 57.Shiba, C., T. Daikoku, F. Goshima, H. Takakuwa, Y. Yamauchi, O. Koiwai, and Y. Nishiyama. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J. Gen. Virol. 81:2397-2405. [DOI] [PubMed] [Google Scholar]
- 58.Smith, G. L. 2000. Secreted poxvirus proteins that interact with the immune system, p. 491-507. In M. W. Cunningham and R. S. Fujinami (ed.), Effects of microbes on the immune system. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 59.Tewari, M., W. G. Telford, R. A. Miller, and V. M. Dixit. 1995. CrmA, a poxvirus-encoded serpin, inhibits cytotoxic T-lymphocyte- mediated apoptosis. J. Biol. Chem. 270:22705-22708. [DOI] [PubMed] [Google Scholar]
- 60.Thomenius, M. J., and C. W. Distelhorst. 2003. Bcl-2 on the endoplasmic reticulum: protecting the mitochondria from a distance. J. Cell Sci. 116:4493-4499. [DOI] [PubMed] [Google Scholar]
- 61.Thomenius, M. J., N. S. Wang, E. Z. Reineks, Z. Wang, and C. W. Distelhorst. 2003. Bcl-2 on the endoplasmic reticulum regulates Bax activity by binding to BH3-only proteins. J. Biol. Chem. 278:6243-6250. [DOI] [PubMed] [Google Scholar]
- 62.Thornberry, N. A., and Y. Lazebnik. 1998. Caspases: enemies within. Science 281:1312-1316. [DOI] [PubMed] [Google Scholar]
- 63.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2004. The genome of canarypox virus. J. Virol. 78:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, J. H. Sur, N. T. Sandybaev, U. Z. Kerembekova, V. L. Zaitsev, G. F. Kutish, and D. L. Rock. 2002. The genomes of sheeppox and goatpox viruses. J. Virol. 76:6054-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, G., J. W. Barrett, S. H. Nazarian, H. Everett, X. Gao, C. Bleackley, K. Colwill, M. F. Moran, and G. McFadden. 2004. Myxoma virus M11L prevents apoptosis through constitutive interaction with Bak. J. Virol. 78:7097-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wasilenko, S. T., A. F. Meyers, K. Vander Helm, and M. Barry. 2001. Vaccinia virus infection disarms the mitochondrion-mediated pathway of the apoptotic cascade by modulating the permeability transition pore. J. Virol. 75:11437-11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wasilenko, S. T., T. L. Stewart, A. F. Meyers, and M. Barry. 2003. Vaccinia virus encodes a previously uncharacterized mitochondrial-associated inhibitor of apoptosis. Proc. Natl. Acad. Sci. USA 100:14345-14350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wattenberg, B., and T. Lithgow. 2001. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2:66-71. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, Q., S. Snipas, K. Orth, M. Muzio, V. M. Dixit, and G. S. Salvesen. 1997. Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J. Biol. Chem. 272:7797-7800. [DOI] [PubMed] [Google Scholar]
- 70.Zhu, W., A. Cowie, G. W. Wasfy, L. Z. Penn, B. Leber, and D. W. Andrews. 1996. Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. EMBO J. 15:4130-4141. [PMC free article] [PubMed] [Google Scholar]