Abstract
The mTOR pathway was discovered in the late 1970s after the compound and natural inhibitor of mTOR, rapamycin was isolated from the bacterium Streptomyces hygroscopicus. mTOR is serine/threonine kinase belonging to the phosphoinositide 3-kinase related kinase (PIKK) family. It forms two distinct complexes; mTORC1 and mTORC2. mTORC1 has a key role in regulating protein synthesis and autophagy whilst mTORC2 is involved in regulating kinases of the AGC family. mTOR signaling is often over active in multiple cancer types including breast cancer. This can involve mutations in mTOR itself but more commonly, in breast cancer, this is related to an increase in activity of ErbB family receptors or alterations and mutations of PI3K signaling. Rapamycin and its analogues (rapalogues) bind to the intercellular receptor FKBP12, and then predominantly inhibit mTORC1 signaling via an allosteric mechanism. Research has shown that inhibition of mTOR is a useful strategy in tackling cancers, with it acting to slow tumor growth and limit the spread of a cancer. Rapalogues have now made their way into the clinic with the rapalogue everolimus (RAD-001/Afinitor) approved for use in conjunction with exemestane, in post-menopausal breast cancer patients with advanced disease who are HER-2 negative (normal expression), hormone receptor positive and whose prior treatment with non-steroidal aromatase inhibitors has failed. Testing across multiple trials has proven that everolimus and other rapalogues are a viable way of treating certain types of cancer. However, rapalogues have shown some drawbacks both in research and clinically, with their use often activating feedback pathways that counter their usefulness. As such, new types of inhibitors are being explored that work via different mechanisms, including inhibitors that are ATP competitive with mTOR and which act to perturb signaling from both mTOR complexes.
Keywords: mTOR, rapalogues, breast cancer, cell signaling, everolimus, PI3K
Overview of mTOR signaling
The mTOR pathway was not uncovered until the serendipitous discovery of rapamycin in the late 1970s. This compound isolated from the bacterium Streptomyces hygroscopicus and named from the island on which it was discovered (Easter Island/Rapa Nui), was found to have strong anti-fungal, immune-suppressant and anti-cancer properties. Rapamycin was found to inhibit two yeast proteins named the target of rapamycin (TOR) 1 and 2, with the single mechanistic (previously mammalian) TOR (mTOR) then later uncovered. From this point, the mTOR pathway has been built around this central protein which has been shown to be a critical regulator of many important cellular processes [1-8].
mTOR belongs to the phosphoinositide 3-kinase related kinase (PIKK) family and is expressed in most mammalian cells [2,9], causing an increase in cellular protein mass and growth and inhibiting autophagy, with it generally acting as a cellular sensor to nutrients and growth factors, as well as being an important effecter pathway of PI3K signalling [10].
mTOR and mTOR complexes (mTORCs)
Residues 1-1375 of mTOR are not as well defined as the rest of the protein, but predictive modelling techniques and information from related kinases suggest this N-terminal half of the protein consists mostly of HEAT repeats [11]. The remaining structure of the protein is well defined, by crystal structure, consisting of the FAT, FRB, kinase and FATC domains. ATP binds within the kinase domain (KD), whilst rapamycin-FKBP12 binds in the FRB domain [12,13].
mTOR acts in one of two protein complexes; mTORC1 or mTORC2 with a combination of common and unique components (Figure 1). mLST8 binds to mTOR at the kinase domain C-lobe and data suggest that mLST8 is needed for proper mTOR kinase function as well as helping to stabilize the interaction between mTOR and raptor, in mTORC1 [14]. Extremely important to mTORC1 function is raptor, a 149 kDa protein that is usually found in a complex with mTOR, binding to the mTOR HEAT repeats.
Figure 1.
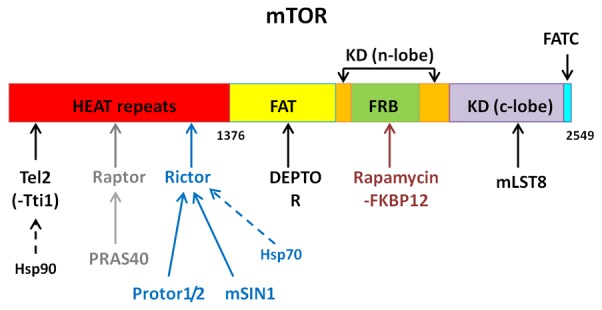
Basic structure of the 2549 residue protein, mTOR. The components of mTORC1 and 2 are marked as to which mTOR domain, or complex protein, they bind to. Components found in both complexes are marked in black, specific mTORC1 components in grey and specific mTORC2 components in blue. Information: [11-13,17,18,23,24].
The sub-complex of Tel2 and Tti1 act as a scaffolding structure to both mTOR complexes and other PIKK proteins; Tel2 also binds to mTOR via the HEAT repeats [15,16], with heat shock protein 90 (Hsp90), acting as a chaperone for the Tel2-Tti1 complex [17,18]. DEPTOR is also an inhibitor of mTOR function, binding to mTOR on its FAT domain via DEPTOR’s PDZ domain [19], with research showing an increase in phosphorylation of mTOR targets after DEPTOR knock down [20]. DEPTOR regulation is via its degradation, with mTOR signaling triggering the phosphorylation of DEPTOR, leading to its ubiquitination by the E3 ligase, SCFβTRCP [21,22].
mTORC1
Raptor acts as a scaffold for mTORC1, not having catalytic activity itself, but is required for full activation of mTORC1 [25-27]. The mTOR complexes also contain sub-units that act as inhibitors of mTOR function. Unique to mTORC1 is the proline-rich Akt substrate of 40 kDa (PRAS40), which binds to the complex via raptor. PRAS40 is believed to have an inhibitory effect on mTORC1 function, with most studies showing increased mTORC1/mTOR activity in the absence of PRAS40, although this may be tissue specific [28,29]; PRAS40’s inhibitory effect is speculated to be due to the inhibition of substrate binding [30].
mTORC2
mTORC2 is less studied than mTORC1, but many years of research have begun to elucidate more components and functions of the second complex. Whilst mTORC2 has a very different set of functions to mTORC1, it does contain many of the same subunits in a similar role; these include mTOR itself, mLST8, DEPTOR and Tel2-Tti1. A defining component of mTORC2 is rictor, which forms the basis of this second complex, also binding to the HEAT repeats of mTOR. Like mLST8, rictor is needed for mTORC2 catalytic activity and also acts as a scaffold for many proteins in the complex [23,31,32]. Research by Martin and colleagues [24] suggested that rictor may act as a point of binding for Hsp70, with this study also implicating Hsp70 as a key regulator of mTORC2 function.
mSIN1 is an mTORC2 scaffold protein, which binds to the complex via rictor. mSIN1 is thought to be required for proper mTORC2 formation, with it stabilizing the mTOR-rictor interaction. mTORC2 targets such as Akt also show markedly decreased phosphorylation without mSIN1, showing mTORC’s role in regulating kinase activity of the complex [33,34]. Protor 1 and 2 are the last major components of mTORC2. Protor-1 and 2 bind to rictor within the complex, but are not needed for stabilisation [28,35,36]. Protor-1 appears to play a role in mTORC2 activity towards one of its substrates, SGK1, with a markedly decreased phosphorylation of this target in protor-1 absence [36,37]. Like protor-1, protor-2 also appears to modulate mTORC2 in a substrate specific manner; with work by Gan and colleagues [38] showing protor-2 may suppress mTORC2 phosphorylation of PKC.
Upstream signaling
mTOR itself is phosphorylated at multiple sites, including a level of auto-phosphorylation at Ser2481 [39], with some of this phosphorylation induced by growth factor signaling. Research suggests many of these phosphorylated sites (such as Ser2448) increase mTOR activity and may be needed for proper mTORC1 function [40-43]. Intriguingly, work by Copp and colleagues [44], suggested that Ser2481 phosphorylation of mTOR could act as a good biomarker for intact mTORC2 complexes as mTORC2 had predominantly Ser2481 phosphorylation, whilst mTORC1 had predominantly Ser2448 phosphorylation.
mTORC1
There are a variety of upstream pathways which control mTORC1 activation, including growth factor signaling, amino acid levels, cellular energy levels and stress (reviewed by Sengupta and colleagues [45]). The tubular sclerosis complex (TSC) is a convergence point for many of these upstream factors and is a key regulator of mTORC1 activity. The complex consists of TSC1 (also known as Hamartin), TSC2 (also known as Tuberin) and TBC1D7 [46], and functions via the Rheb GTPase [47,48]. Lysosomal localization is important for mTORC1 activation with recent research suggesting that the phosphorylation of TSC actually causes TSC to dissociate from the lysosome, away from mTORC1 and Rheb, activating mTORC1 [49].
The PI3K pathway is a key upstream regulator of mTORC1, via TSC. Growth factors such as IGF-1 and insulin activate phosphoinositide 3-kinase (PI3K), which in turn generates PIP3 from membrane-bound PIP2. This recruits downstream effectors such as PDK1 and Akt (also known as protein kinase B/PKB) via their PH domains. Akt can then be activated via phosphorylation by PDK1 on Thr308 and Ser473 [50]. Akt is a critical regulator of TSC, with active Akt phosphorylating TSC2 at multiple sites, to weaken its interaction with TSC1 and destabilize the TSC2 protein. This in turn activates mTORC1, as TSC2 can no longer act as the GTPase activating protein (GAP) for Rheb [51,52]. Akt can also regulate mTORC1 activity by phosphorylating PRAS40, causing it to bind to 14-3-3 proteins, thus relieving its inhibitory effect on the complex [29].
The Ras-Erk MAPK pathway can also lead to downstream activation of mTORC1. Once Erk is activated, it can directly phosphorylate and inactivate TSC2 on Ser664 [53,54] or phosphorylate p90 ribosomal S6 kinase 1 (RSK1), leading to TSC2 inactivation via phosphorylation at Ser1798 [55].
Amino acid levels are critical regulators of mTORC1 function; increased levels of amino acids result in mTORC1 activation, and growth factors are unable to activate mTORC1 without the required level of amino acids [42,56]. The Rag GTPases are central to this regulation, acting as dimers of either RagA or B dimerized with either Rag C or D. In its active state, the complex binds raptor, localizing mTORC1 to the lysosome, and bringing it into contact with Rheb [42,57].
How the cell exactly translates amino acid levels to mTORC1 activation is not well understood, but many proteins are now being revealed to have roles in this amino acid sensing. The molecular pump v-ATPase is required for activation of mTORC1, with it directly interacting with the regulator complex and in turn amino acids directly regulate this interaction [58]. Of interest is work by Pena-Llopis and colleagues [59] which showed that mTORC1 may be involved in positive feedback, with mTORC1 activation increasing v-ATPase expression. It is probable that the full extent of the amino acid sensing ‘machinery’ (in relation to mTORC1) is yet to elucidated, but current candidates include MAP4K3 [60], SLC38A9 [61,62] and PAT1 (SLC36A1) [63].
Cellular energy levels also regulate mTORC1 activity, with low energy generally inhibiting mTORC1, and reducing protein synthesis. This is mainly via cellular levels of AMP decreasing when ATP is low, activating AMPK, and causing raptor phosphorylation and subsequent binding to 14-3-3 proteins, sequestering it away from mTORC1 [64]. Activated AMPK also phosphorylates TSC2 on Thr1227 and Ser1345 to activate (rather than inactivate, as is the case when Akt phosphorylates TSC2 on Ser924 and Thr1518) the TSC to further decrease mTORC1 signalling [51,65]. Since downstream mTORC1 activates protein synthesis it is important the cell only activates mTORC1 signaling when it has the required resources, such as ATP/energy and amino acids. Lower cellular oxygen levels and other cellular stresses also reduce the activity of mTORC1. For example stress such as hypoxia can induce regulated in DNA damage and development 1 (REDD1), which inhibits mTORC1 function [66].
mTORC2
Although knowledge of mTORC2 signaling is less defined than for mTORC1, research is beginning to fill in gaps in our knowledge. It has been known for a while that, like mTORC1, mTORC2 is activated by growth factors such as insulin and IGF-1 [67]; although only mTORC2 complexes containing mSIN1 isoforms 1 and 2 (not 5) are activated by insulin [68]. Recent research has shown that mSIN1 is a critical mediator for growth factors to activate mTORC2, with PI3K signaling linking the two. Membrane bound PIP3 binds mSIN1 via its PH domain, relieving its interactions with mTORC2, thus activating it [69,70]. This is in contrast to earlier findings which show that mSIN1 is needed for mTORC2 activity [33,34]. These seemingly conflicting reports highlight the relatively poor understanding on the precise mechanism of mTORC2 action and activation.
Active PI3K signaling promotes mTORC2 activation and binding to ribosomes, possibly as a mechanism to limit its activation only in growing cells with a high enough ribosome content [71]. Remarkably, whilst the TSC inhibits mTORC1 function, it is suggested that, in at least some cell lines (including the breast cancer cell line MCF7), the complex is needed for full mTORC2 activation as well as having a physical interaction with mTORC2, independent of its function with rheb [72].
Considering that DEPTOR was discovered relatively recently, it is possible that there are still mTOR complex components that have not been discovered. If this is the case, it may also explain why there are seemingly conflicting conclusions on the role some of these proteins, as there could be as yet undiscovered interactions. Research by Luo and colleagues [73] found that rapamycin can inhibit mSin1 phosphorylation independently of mTORC1 or 2 (raptor and rictor are not required), but the mechanism of inhibition does involve mTOR and mLST8. This again suggests that there may be further mTOR complexes yet to be discovered, that explain the observed effect.
Downstream signaling
mTORC1
The molecular and cellular effects of mTORC1 activation are well characterized, with a number of processes regulated from this point. Protein synthesis is critically regulated by mTORC1 with mTORC1 phosphorylating both eIF4E-binding proteins (4E-BPs) and S6 kinases including S6K2 and the multiple S6K1 isoforms.
p70-S6K1 is first phosphorylated on multiple sites subsequently allowing phosphorylation of Thr389 by mTORC1, followed by phosphorylation on Thr229 by PDK1 to fully activate the kinase [74]. S6K1/2 then phosphorylates multiple proteins involved in the translation machinery. S6K1 activation is also believed to promote transcription via its interactions with transcription factors such as estrogen receptor α (ERα), as well as regulating ribosomal gene transcription [75,76]. Unsurprisingly negative, feedback loops exist along the mTORC1 axis involving S6K1, with the active protein both repressing the expression of IRS-1 and phosphorylating it on inhibitory serine residues [77]. mTORC1 also serves to feedback to mTORC2, with S6K directly phosphorylating rictor, which may serve to control activation of Akt [78].
mTOR phosphorylation of 4E-BP1 on sites including Thr 37, 46 and 70 and Ser 65 by mTOR, prevents the inhibitory action of the 4E-BPs on eIF4E to allow the latter to initiate cap-dependent translation [79,80].
Autophagy is generally not needed when the cell is healthy with a plentiful nutrient supply activating mTORC1, and inactivating autophagy through phosphorylation of kinases ULK1/2 and ATG13 [81-83]. The ULK complex also cross-talks with the beclin1 (or VSP34) complex. mTORC1 can phosphorylate a member of this complex called AMBRA1, to reduce ubiquitination of ULK1 by the VSP34 complex protein, TRAF6. Unusually, rather than destroy the protein, this ubiquitination actually increases its activity [84]. As AMPK reduces both mTOR signaling, and increases ULK phosphorylation it increases autophagy in cellular stress, in opposition to the mTOR pathway [85].
Aside from these functions, mTORC1 is also partially involved in regulating other important cellular processes related to metabolism, such as glycolysis via hypoxia inducible factor (HIF1α) induction [86-88], lipid metabolism [89], and de novo synthesis of pyrimidines [90].
mTORC2
mTORC2 regulates the activity of several proteins belonging to the AGC kinase family and it can, in one sense, be thought of as ‘upstream’ of mTORC1 as it is one of many regulators of the AGC kinase, Akt. Akt has many downstream effectors of its own, increasing proliferation, cellular growth (e.g. its role in mTORC1 activation via TSC2), cell survival, angiogenesis and metabolic processes [91]. mTORC2 directly phosphorylates Akt on Ser473, which is required for its maximal activation [92]. However mTORC2 is not the only activator of Akt, with Akt substrates such as FoxO1 being impaired by mTORC2 depletion, when others like GSK3β were not affected [33,93].
mTORC2 also phosphorylates the AGC kinase SGK1, thereby contributing to the regulation of proliferation and apoptosis via FoxO3a [94], ion channels such as Na+ [95] and regulating differentiation in cell types such as TH1 and TH2 immune cells [96]. mTORC2 can affect cellular shape, structure and morphology, specifically by altering the actin cytoskeleton, with part of this control, at least, down to mTORC2 regulation of PKC, another member of the AGC kinases [31,32,97].
As well as associating with ribosomes [71], mTORC2 also associates with the endoplasmic reticulum (ER) sub-compartment called the mitochondria-associated ER (MAM). This sub compartment is a key part of calcium and lipid transfer, with mTORC2 deficiency directly leading to a disruption of these functions and MAM integrity [98].
mTOR and breast cancer
Looking at the multitude of cellular events mTOR complexes help regulate, it is of no surprise that the activation of mTOR signaling is associated with cancer and is perceived as being oncogenic. The activation of mTOR complexes will give tumors a vast growth advantage, with an increased amount of protein synthesis, as well increased inhibition of autophagy. Thus whilst growing at an increased rate, these cells are also less likely to die. Research has generally shown that activated mTOR signaling leads to an increase in tumor progression and often a decrease in patient survival [99,100]. mTOR expression correlates for a worse prognosis in breast cancer [101,102] with work by Walsh and Colleagues [103] showing that phospho-mTOR was more common in triple negative breast cancers. Despite the fact that mTORC2 signaling can increase oncogenic signals via Akt and mTOR signaling, research has suggested that rictor expression, which is required for mTORC2 signaling, is actually lower in breast tumors compared to normal breast tissue [102]. This could suggest that mTORC1 signaling is more oncogenic than mTORC2 signaling or that rictor is required in very specific amounts for mTORC2 signaling; with too much or too little ultimately inhibiting the mTORC2 arm.
In terms of how the mTOR pathway is altered in cancer, it is found that the majority of alterations and mutations lie upstream of mTOR itself and lead to an increased activation of mTOR signaling. Common in many cancers, are alterations to PI3Ks, which are key activators of mTOR via Akt and TSC1/2 and have been shown to cause over activation of mTOR signalling [104]. Activating mutations in the PIK3CA gene (which encodes a subunit of PI3K) are common in breast cancer, with the mutations usually centered in kinase and helical domains [105]. Other common mutations upstream of mTOR occur in AKT, with altered or mutated AKT and loss of PTEN detected in breast cancer [106]. Familial mutations in PTEN Cowden Syndrome also increases the risk of developing sporadic cancers of the breast, thyroid and kidneys [107].
Mutations and alterations of core mTOR components (involved in either of the two mTOR complexes) are by and large a lot rarer than upstream mutations, but have still been noted in cancers, within the last few years. With the availability of more powerful sequencing technology combined with large online databases containing sequencing data, many research groups have been able to identify mutations in mTOR itself [108-110]. These pieces of research have shown that mutations have occurred in a variety of cancer types and whilst these alterations occur along the length of mTOR (Figure 2), a high frequency have been found in domains such as the FAT and FATC domains. Since the latter forms part of the kinase domain, it is no surprise that many of the mutations identified in this research resulted in either increased mTORC1 or 2 activity. Some mutations in MTOR also showed decreased binding to the inhibitor DEPTOR, possibly due to mutations in the FAT domain [108].
Figure 2.
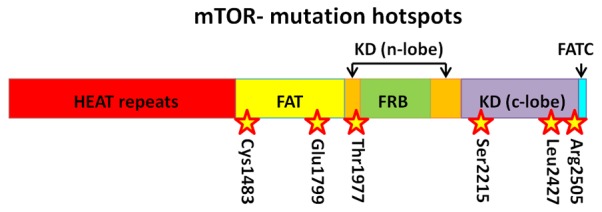
Common mutation hotspots along mTOR. Information: [108].
mTOR and ER
The activation of mTOR signaling in cancer cells is associated with resistance to multiple drug therapies, especially in breast cancer where this affect is well studied. Tamoxifen is a selective estrogen receptor modulator (SERM), binding to the nuclear ERα, to block it’s binding to estrogen and therefore block receptor activation. A majority of breast cancer patients are estrogen receptor positive and so often receive drugs like tamoxifen (if pre-menopausal), but resistance to them is a common issue [111]. Whilst there are multiple mechanisms behind this resistance, mTOR appears to have a major role, with the mTOR pathway phosphorylating ERα at Ser118, making it hyper sensitive to activation and less likely to bind tamoxifen [112]. Research has shown that in the long term, breast cancer cells may use the PI3K/Akt/mTOR axis to escape dependency from ER signaling and thus increase their resistance to tamoxifen [113]. Inhibiting the mTOR pathway has been shown to also help re-sensitize cells to anti-cancerous effects of tamoxifen [114].
mTOR and HER
Also key in breast cancer are the relative expression of ErbB/HER receptors. EGFR appears to be relatively commonly expressed, with 17.1% of a study of 706 invasive ductal breast carcinomas, showing over-expression of EGFR [115]; expression of EGFR appears to correlate well with HER-2 over-expression, suggesting a therapeutic benefit to inhibiting both types of receptor [116].
Since HER family receptors can activate PI3K-mTOR signaling, HER-2 expression is important in the over-activation of mTOR signaling in breast cancer. HER2 is amplified in upwards of 15-20% of all breast cancers, which can result in a nearly 100-fold increase of protein expression. Its status as a key biomarker comes from that fact that HER-2 expression correlates with a much poorer prognosis and a generally more aggressive cancer [117,118]. mTOR signaling has been linked with resistance to HER-2 therapies in breast cancer, such as with the antibody based drug trastuzumab [119], and the dual EGFR (HER-1) and HER-2 inhibitor lapatinib [120]. Activation of mTOR signaling in tumor cells after ErbB inhibition can arise as a result of mutations in the PI3K pathway and the use of other growth factor receptors like IGF-1R (in which HER-2-IGF-1R dimers can form), contributing to drug resistance [121,122]. It is therefore of no surprise that in vivo studies have shown an increased effect when rapamycin is used with trastuzumab [123].
mTOR-targeted therapies
Rapalogues
Since its identification, over four decades ago, rapamycin has been studied as a therapy for a wide variety of diseases. With it being the first mTOR inhibitor to be discovered, work on rapamycin led to a new field devoted to elucidating compounds that inhibited the mTOR pathway. The first, and currently most widely used, set of compounds, are rapamycin and its analogues that are more commonly known as ‘rapalogues’. Rapamycin (structure shown in Figure 3), also known as sirolimus, is a macrocyclic lactone, isolated from the bacterium Streptomyces hygroscopicus initially noted for its strong anti-fungal effect [8]. It was later found to have strong immunosuppressive affects, blocking T-cell activation [3] and in 1999 was originally approved for use as an immunosuppressant drug in the USA [124]; it is used in procedures such as kidney transplantation, to reduce rejection, risk of infections and also to lower the incidence of post-surgery cancer [125].
Figure 3.
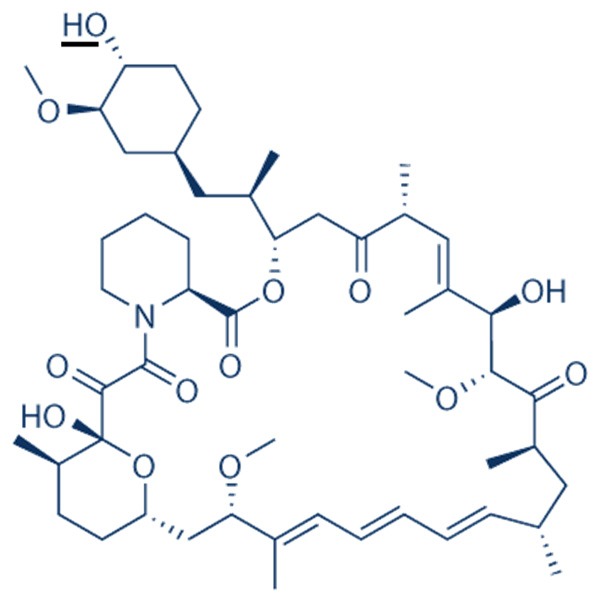
Structure of rapamycin (sirolimus). Rapalogues vary from the parent rapamycin in mostly on one small side group. This occurs as an O-substitution at carbon-40 on rapamycin, underlined on the primary structure [129,130].
Due to its inhibitory effect on mTOR, and thus cellular growth, rapamycin was explored as an anti-cancer agent. It was shown to inhibit cellular proliferation and/or be effective in several types of cancer including pancreatic [126], colon [4], rhabdomycosarcoma [127] and breast [124]. However, rapamycin has on the whole not been taken forward for cancer therapy due to its poor pharmacokinetic properties, including its low solubility [128].
Rapamycin derivatives/rapalogues have since been developed to tackle these issues, opening up new avenues for treatment for not only cancers but a variety of other conditions. These include everolimus (RAD-001), temsirolimus (CCI-779), ridaforolimus (deforolimus, AB23573) and zotarolimus (ABT-578). Details of these rapalogues can be found in Table 1.
Table 1.
Details, including clinical uses of the rapalogues; everolimus (RAD-001), temsirolimus (CCI-779), ridaforolimus (deforolimus, AB23573) and zotarolimus (ABT-578)
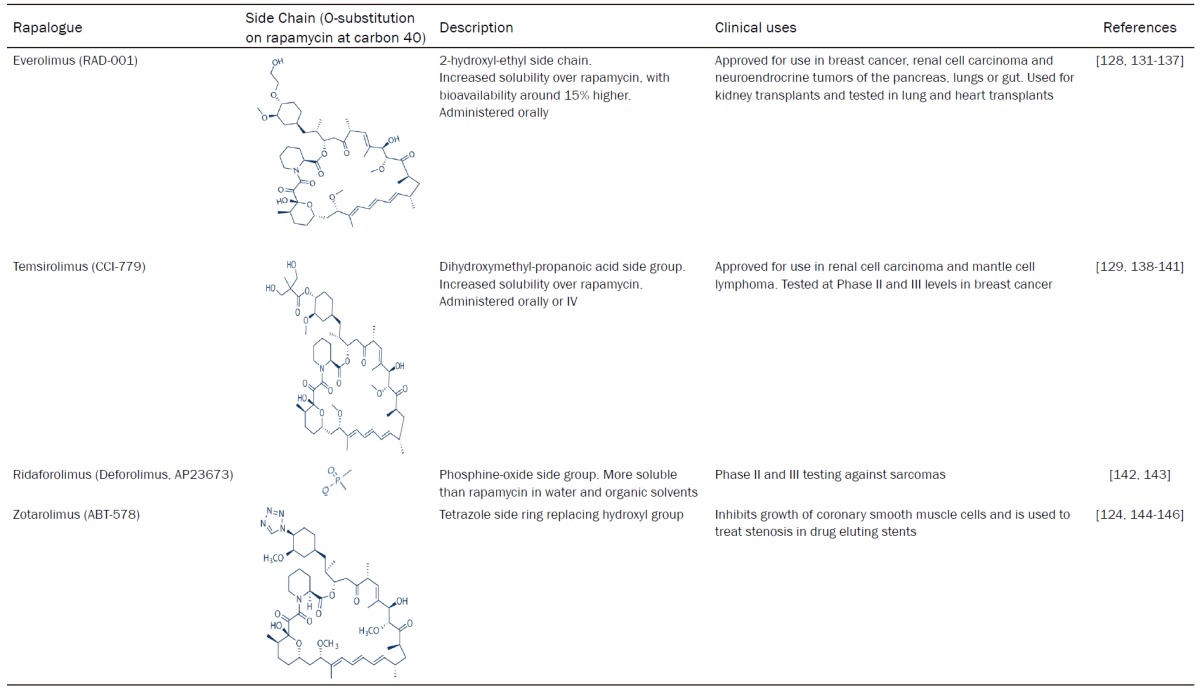
|
Rapalogue mechanism of action
Rapalogues all inhibit mTOR, using the same mechanism of action, which involves the intracellular receptor and immunophilin, FK506 binding protein 12 kDa (FKBP12). FKBP12 binds FK506, and mediates immunosuppressive actions via its alteration of the phosphatase calcineurin, with FKBP12 able to regulate cellular levels of Ca2+ [147,148].
FKBP12 was shown early on to bind rapamycin, and mediate its action through its binding to mTOR, causing an inhibition of cell cycle progression [2]. The FKBP12-rapamycin complex binds to mTOR at the FRB domain, acting through allosteric inhibition and conformational changes in mTOR and mTORC1 [13,149,150], resulting in decreased interaction between mTOR and raptor [151] which could inhibit the phosphorylation and activation of the major mTORC1 downstream targets including S6K and 4E-BP1. However, further, more in depth research about events post-rapalogue treatment has revealed a differentiation in the amount of inhibition actually seen on each mTORC1 substrate, with the levels of inhibition of 4E-BP1 phosphorylation compared to S6K varying greatly over time and between cell types [152,153]. Interestingly, the level of auto-phosphorylation on mTOR in mTORC1 (but not mTORC2) on Ser2481 is also greatly reduced upon rapamycin treatment [39].
Rapalogues were long thought to only inhibit only mTORC1 complexes and their downstream effectors, with evidence at the time supporting this theory [31]. However more in depth study of rapamycin’s effect on mTORC2 has revealed that prolonged treatment does in fact inhibit mTORC2 as well as mTORC1, with rapamycin treatment directly affecting the assembly of mTORC2 components, including rictor. Therefore rather than binding directly to mTORC2, like it does mTORC1, the FKBP12-rapamycin complex binds mTOR and then over time stops the formation of new mTORC2 complexes [154-156].
At a cellular level, rapalogues show many effects useful for the treatment of cancer. Due to the inhibition of protein translation, growth of cells can be severely affected, limiting progression through the cell cycle, usually at the G1 phase, and ultimately inhibiting tumor growth [2,157]. Rapalogues have shown this growth inhibitory effect in a wide variety of cells, with rapamycin inhibiting the growth of cancer cells including prostate [158], small cell lung [159] and rhabdomycosarcoma [127]. Acting through similar mechanisms, everolimus has been shown to inhibit the growth of cancer cells including breast [160], acute lymphoblastic leukemia (ALL) [161] and oral squamous cell carcinoma (OSCC) [162].
Rapalogues are also able to induce autophagy in certain cancer types, including breast cancer [163] and malignant gliomas [164] as well as having an apoptotic effect on human dendritic cells [165]. Whilst this increase in autophagy is not surprising due to mTORC1 control over autophagy initiation and ULK1/2 phosphorylation [81], it is not widely noted in cancer types, where cell cycle arrest and growth inhibition appear to be the primary cellular means by which rapalogues act. Everolimus, like rapamycin, can also cause an increase in apoptosis within breast cancer and rhabdomycosarcoma cell cultures [166-168].
However, inhibiting mTOR signaling in this manner has its drawbacks in terms of the desired molecular effect, highlighting possible issues when applying rapalogues in a clinical setting. Usually, negative feedbacks loops exist to perturb over-active mTOR signaling, with S6K inhibiting IRS-1 to reduce mTOR activation via insulin/IGF-1 signalling [77,169]. Thus, in rapalogue treatment, cells may actually be more sensitive to PI3K-mTOR activation via growth factor signals such as insulin [170]. The inhibition of mTORC1 (on a short term scale) seems to also favor the formation of mTORC2 complexes, shifting mTOR signaling burden from one arm to the other [171]. In line with this, and the fact that mTORC2 leads to increased Akt phosphorylation at Ser473 [32], rapalogue treatment appears to lead to increased Akt activation. This not only further increases upstream signals activating the mTOR pathway but also increases the activation of various survival pathways associated with Akt activation [172]. Everolimus and other rapalogues, have been shown to abolish the negative feedback on IRS-1/insulin signaling, up-regulating and further activating growth factor signaling via PI3K and Akt in both cancer cell cultures and patient samples [173,174]. Patients with metastatic cancer have also shown up-regulation of other signaling pathways including MAPK signaling, when treated with everolimus [175].
Clinical applications of everolimus and rapalogues in breast cancer
Rapamycin
Many rapalogues have now made their way into clinical use, or are being explored for therapeutic in breast cancer patients. Whilst it may be the ‘founding member’ of the rapalogues, rapamycin is not used on a large scale in cancer therapeutics and is unlikely to have future impact as a sole agent. Whilst not yet approved for use in breast cancer, it has shown some small efficacy in the treatment of this disease when used as a combination therapy. Phase II trial data in HER-2 positive patients suggested adding rapamycin may benefit trastuzumab treatment [176] and that the combination of resveratrol with rapamycin may stop Akt feedback activation in breast cancer cells [177].
Temsirolimus
Temsirolimus has been approved for use in renal cell carcinomas since 2007 in the EU [140] and is mainly used as a first line treatment for patients with poor-risk disease, with increased aggressiveness and decreased prognosis. Phase III trial data has shown it improves median survival among this group [178]; however temsirolimus trials in breast cancer have produced inconclusive and ‘mild’ results at best. One phase II study found no objective response in the observed cohort, although the study size was small at only 31 patients [179] and a separate phase II trial, using a larger cohort, showed a very modest response, with only 9.2% patients showing partial response to the drug [138]. Phase III trials of this drug combined with the aromatase inhibitor letrozole, in the HORIZON trials in post-menopausal women, again showed disappointing results and a lack of improved patient survival [141]. Interestingly, Rangwala and colleagues [180] showed that combining a rapalogue like temsirolimus with the autophagy inhibitor hydroxychloroquine (HCQ), was well tolerated and showed anti-tumor activity in melanoma patients, suggesting this may be a valuable area of exploration for breast cancer combination therapy in the future.
Ridaforolimus
Although not currently approved for clinical use in cancer treatment, ridaforolimus has been explored in a number of trials for various cancer types including breast cancer. A phase II trial with ridaforolimus combined with trastuzumab, in HER-2 positive, trastuzumab-refractory metastatic breast cancer patients, showed good anti-tumor activity. The rate of response was similar to that with patients treated with first line trastuzumab, suggesting that a rapalogue like ridaforolimus may help overcome resistance to trastuzumab [181]. Phase II trials of ridaforolimus in endometrial cancer, refractory hematological cancers and soft and bone sarcomas has also shown some promising results in terms of anti-tumor activity, giving cause for possible further investigation [142,182,183].
Everolimus
In breast cancer, everolimus has shown many productive results, across a variety of clinical trials. As such, in 2012 everolimus (marketed as Afinitor) was approved for use in combination with the steroidal aromatase inhibitor exemestane in breast cancer patients with advanced cancer that is hormone receptor positive, HER-2 negative (non-over-expressing), and whose prior treatment with a non-steroidal aromatase inhibitor (such as letrozole or anastrazole) had failed [132,184,185]. Key evidence for the use of everolimus in this subset of cancer patients came from the phase III BOLERO-2 (breast cancer trials of oral everolimus) clinical trial. This trial looked at the effect of combining everolimus with exemestane, in a subset of patients, where the cancer was refractory to non-steroidal aromatase inhibitors (all patients had received prior treatment with either letrozole or anastrazole). The patient set included those who had already been treated with one set of chemotherapy and/or hormonal therapy, and excluded patients who had already been treated with exemestane or other mTOR inhibitors. Patients treated with the combination of everolimus plus exemestane had a statistically significant increase in progression free survival (PFS), compared to exemestane and placebo treated patients; there was a PFS average of 2.8-4.1 months in the placebo arm compared 6.9-10.6 months in the everolimus arm of the trial. In terms of toxicity, the combination treatment was also well tolerated, according to quality of life (QOL) end-points and ECOG status [186-188]. These results are positive compared to the rather flat results of the HORIZON trial; both used a rapalogue in conjunction with an aromatase inhibitor, however it is possible that the use of a steroidal aromatase inhibitor (exemestane) enhanced the effects of the rapalogue in a greater way compared to its non-steroidal counter-part (letrozole).
Rapalogues and drug-resistance
Since mTOR activation can often confer a resistance to trastuzumab [119] it seems a viable option to use a rapalogue to increase patient sensitivity to this therapy once again. Phase II trial data validated this thinking, with results showing that patients on a regime of trastuzumab and paclitaxel (who had progressed whilst on trastuzumab treatment and were HER-2 positive) were showing increased PFS times and response rates to the therapy, with the weekly addition of everolimus [189]. However results from the phase III BOLERO-1 trial in a similar area were not as positive. The trial included patients with HER-2 positive (over-expressing) tumors with advanced disease who had not received chemotherapy (including trastuzumab) within the last 12 months. This time the addition of everolimus to trastuzumab and paclitaxel did not improve outcomes in a significant way although some small advantage to this treatment was noted in women who were hormone/ER receptor negative [190].
The BOLERO-3 phase III trial also studied women with advanced HER-2 positive cancers who were trastuzumab resistant and had previously received taxane treatment. The addition of everolimus to a regime of trastuzumab and vinorelbine increased PFS significantly, albeit by a small amount, compared to the placebo, group from a median of 5.78 months to 7 months. Again the sub-group of patients who were hormone receptor negative showed an increased efficacy of everolimus [191]. The data from the BOLERO-1 and 3 trials suggest that in HER-2 positive patients, hormone/ER receptor status may be key to everolimus efficacy. Since mTOR signaling can directly alter ER signalling [112] and is a direct target of growth factor signaling like that of HER family receptors [42], it is perhaps no surprise that these multiple pathways connect in relation to therapy efficacy.
Considering the importance of selective estrogen receptor modulators (SERMs), the use of everolimus has also been explored in conjunction with tamoxifen. Phase II clinical trial data for this drug combination in post-menopausal advanced breast cancer patients who were HER-2 negative, hormone receptor positive and resistant to aromatase inhibitors, have been positive. Results suggested a significant increase in time to progression and overall clinical benefit [192]. A small phase II study in triple-negative breast cancer patients showed that the combination of carboplatin and everolimus may have clinical benefit in this set of breast cancers [193]; however, the addition of everolimus to a regime of paclitaxel and bevacizumab was not shown to significantly increase efficacy of this combination drug regime [194].
Resistance to rapalogues
Whilst it is clear that the rapalogues have wide potential in the clinic, as in the case of everolimus use in breast cancer, they also are associated with key issues that may ultimately limit their application and range in terms of therapeutic use. Resistance to rapalogues (and a lack of efficacy to treatment) has been noted in many settings and can been caused by a host of factors. The inhibition of mTOR with rapalogues can alter feedback pathways that exist within PI3K-mTOR signaling as well as activate Akt signaling by shifting the burden of signaling towards mTORC2 [32,172]. This reduces the anti-cancer effects that rapalogues have [195], and inhibition of Akt can directly re-sensitize breast and colon cancer cells to rapalogue treatment, partially via increased inhibition of PRAS40 phosphorylation, increasing its inhibitory effect on mTORC1 [196]. This same feedback effect on Akt has been noted in lung cancer cells where PI3K inhibition, again re-sensitized the cells to rapamycin treatment [172]. Since mTOR inhibition can activate apoptosis, a lack of functional apoptotic pathways can reduce their effectiveness as well [195]. Unsurprisingly, breast cancer cells with a higher reliance/activation of mTORC1 signaling, as shown by over-expression of phosphorylated S6K, show increased inhibition by rapalogues [197].
Many other signaling pathways and processes can affect and induce rapalogue resistance. For example MCF-7 breast cancer cells that have developed tamoxifen resistance are intrinsically resistant to everolimus [198]. Research suggests that expression of epithelial-mesenchymal transition (EMT) markers such as snail increase resistance to rapamycin, whilst expression of pre-EMT markers like E-cadherin increase breast cancer cell sensitivity to rapamycin, in vitro [199]. In breast cells (including the MCF-7 cell line) that were induced to be everolimus resistant, MYC was suggested to play a role in the resistance process, as an up-regulation of MYC was seen in the resistant lines and depletion of MYC re-sensitized the cells to everolimus once more [200]. MCF-7 cells treated with rapamycin also showed an up-regulation of transglutaminase 2 (TGM2), seemingly as a compensatory mechanism, with TGM2 inhibition re-sensitizing cells to rapamycin [201]. Work with breast, colorectal and renal cancer cells also implicates Met to be involved a mechanism of rapalogue resistance, with increased Met activation conferring resistance [202]. Mutations could also induce rapalogue resistance breast cell lines; mutations in mTOR’s FRB domain (induced after long term rapamycin treatment) resulted in insensitivity to rapamycin, even over a period of weeks. Cells with this type of mutation are however still sensitive to ATP-competitive inhibitors of mTOR [203].
Alternatives to rapalogues
Inhibiting mTOR via FKBP12 is by no means the only way to achieve the overall effect of blocking mTOR activity. In fact there are now multiple, well explored, ways to block mTOR signaling, many of which circumnavigate the issues that arise with rapalogue use. Whilst these still present with their own issues, such as side effects, they have shown promising efficacy in the field of cancer treatment and early clinical trial stages and it is very possible that they will make their way into the clinical setting [204].
Unlike rapalogues that allosterically inhibit mTOR, ATP-competitive inhibitors block ATP binding and reduce the activity of both mTOR complexes. Due to the related sequence nature of mTOR (and other PIKK family proteins) and PI3K, many of the ATP competitive inhibitors also inhibit PI3K as well as mTOR. These inhibitors therefore reduce signaling across the entire PI3K-Akt-mTOR axis and reduce the problems of feedback activation to PI3K signaling or mTORC2 activation [205]. BEZ235 and PF-04691502 are both dual PI3K-mTOR inhibitor of this class and have been studied for their anti-cancer efficacy in breast cancer. Both shown an anti-proliferative effect on cancer cells (and some tumors) and inhibition of PI3K-mTOR signalling [206,207]. However, since PI3K signaling controls such a broad range of downstream pathways and processes vital for a cell, inhibiting both PI3K and mTOR may have serious side effects that could limit the clinical application of such inhibitors. For example, in a phase II study of BEZ235 in pancreatic neuroendocrine patients who were everolimus resistant, the drug was poorly tolerated, limiting the trial progression [208].
More specific ATP-completive inhibitors, that only target mTOR, and thus block mTORC1 and 2 are becoming more favorable. The drugs MLN0128, CC-223 and ADZ2014 have all shown promising results in terms of their anti-cancerous effects in breast cancer. AZD2014 and MLN0128 both show good anti-proliferative and anti-tumor effect in vitro and in vivo reducing signaling from mTORC1 and mTORC2, with MLN0128 also able to inhibit the growth of rapamycin-resistant breast cancer cells [209-211]. A phase I study of CC-223 has been relatively promising; it is well tolerated, with a partial response noted in a breast cancer patient, and disease stability in multiple types of cancer, as well as good inhibition of mTORC1 and 2 in patients [209].
Despite the issues of inhibiting PI3K, pan-PI3K inhibitors, such as the buparlisib (BKM120) have shown early promise in tackling breast cancers. Buparlisib widely inhibits PI3Ks but not does not directly inhibit mTOR; phase I data with buparlisib in combination with either trastuzumab [212] or fulvestrant [213] shows the drug to be well tolerated in breast cancer patients with some signs of disease management. PI3K inhibition may also be a viable way of avoiding resistance to rapalogues with buparlisib use, in combination with everolimus (or trastuzumab), reducing the occurrence of resistance to these drugs whilst also showing good growth inhibition, in vivo [214].
Inhibiting Akt directly is another alternative therapeutic option to rapalogues that has shown potential at a research stage and early clinical levels. In terms of breast cancer therapeutics, MK-2206, an allosteric inhibitor of Akt is perhaps the most promising of the selective Akt inhibitors. Multiple phase I trials have suggested this may hold key therapeutic benefit and it has been tested in a similar settings to the BOLERO trails. MK-2206 in combination with paclitaxel and trastuzumab, (similar to the BOLERO-1 trial), was well tolerated, with 63% of patients showing a clinical response [215]. Likewise, MK-2206 in combination with anastrazole was also well tolerated and 42% of patients showed clinical benefit. Due to these successes, phase II trials are underway with MK-2206 [216]. Preclinical evidence for the efficacy of the ATP competitive inhibitor, AZD5363, is also positive, with breast cancers cells and xenografts showing some of the best responses to this drug of all malignancies tested [217].
Acknowledgements
SHH is funded by a Brunel University London, Isambard Kingdom Brunel Scholarship for postgraduate research.
Disclosure of conflict of interest
None.
References
- 1.Baker H, Sidorowicz A, Sehgal SN, Vézina C. Rapamycin (AY-22,989), a new antifungal antibiotic. III. In vitro and in vivo evaluation. J Antibiot (Tokyo) 1978;31:539–545. doi: 10.7164/antibiotics.31.539. [DOI] [PubMed] [Google Scholar]
- 2.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 3.Dumont FJ, Staruch MJ, Koprak SL, Melino MR, Sigal NH. Distinct mechanisms of suppression of murine T cell activation by the related macrolides FK-506 and rapamycin. J Immunol. 1990;144:251–258. [PubMed] [Google Scholar]
- 4.Eng C, Sehgal S, Vézina C. Activity of rapamycin (AY-22,989) against transplanted tumors. J Antibiot (Tokyo) 1984;37:1231–1237. doi: 10.7164/antibiotics.37.1231. [DOI] [PubMed] [Google Scholar]
- 5.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 6.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 7.Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 8.Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 9.Lakhlili W, Chevé G, Yasri A, Ibrahimi A. Determination and validation of mTOR kinase-domain 3D structure by homology modeling. Onco Targets ther. 2015;8:1923. doi: 10.2147/OTT.S84200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zarogoulidis P, Lampaki S, Turner JF, Huang H, Kakolyris S, Syrigos K, Zarogoulidis K. mTOR pathway: a current, up-to-date mini-review (review) Oncol lett. 2014;8:2367–2370. doi: 10.3892/ol.2014.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knutson BA. Insights into the domain and repeat architecture of target of rapamycin. J Struct Biol. 2010;170:354–363. doi: 10.1016/j.jsb.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauer E, Imseng S, Maier T, Hall MN. Conserved sequence motifs and the structure of the mTOR kinase domain. Biochem Soc Trans. 2013;41:889–95. doi: 10.1042/BST20130113. [DOI] [PubMed] [Google Scholar]
- 13.Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature. 2013;497:217–223. doi: 10.1038/nature12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 15.Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, Takehana K, Iemura S, Natsume T, Mizushima N. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem. 2010;285:20109–20116. doi: 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takai H, Wang RC, Takai KK, Yang H, de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 17.Izumi N, Yamashita A, Hirano H, Ohno S. Heat shock protein 90 regulates phosphatidylinositol 3-kinase-related protein kinase family proteins together with the RUVBL1/2 and Tel2-containing co-factor complex. Cancer Sci. 2012;103:50–57. doi: 10.1111/j.1349-7006.2011.02112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takai H, Xie Y, de Lange T, Pavletich NP. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. 2010;24:2019–2030. doi: 10.1101/gad.1956410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazi AA, Hong-Brown L, Lang SM, Lang CH. Deptor knockdown enhances mTOR activity and protein synthesis in myocytes and ameliorates disuse muscle atrophy. Mol Med. 2011;17:925–936. doi: 10.2119/molmed.2011.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao D, Inuzuka H, Tan MK, Fukushima H, Locasale JW, Liu P, Wan L, Zhai B, Chin YR, Shaik S, Lyssiotis CA, Gygi SP, Toker A, Cantley LC, Asara JM, Harper JW, Wei W. mTOR drives its own activation via SCF βTrCP-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell. 2011;44:290–303. doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF βTrCP E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011;44:304–316. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin J, Masri J, Bernath A, Nishimura RN, Gera J. Hsp70 associates with rictor and is required for mTORC2 formation and activity. Biochem Biophys Res Commun. 2008;372:578–583. doi: 10.1016/j.bbrc.2008.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 26.Kim D, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 27.Kwak SS, Kang KH, Kim S, Lee S, Lee JH, Kim JW, Byun B, Meadows GG, Joe CO. Amino acid-dependent NPRL2 interaction with raptor determines mTOR complex 1 activation. Cell Signal. 2016;28:32–41. doi: 10.1016/j.cellsig.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Thedieck K, Polak P, Kim ML, Molle KD, Cohen A, Jenö P, Arrieumerlou C, Hall MN. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS One. 2007;2:e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiza C, Nascimento EB, Ouwens DM. Role of PRAS40 in akt and mTOR signaling in health and disease. Am J Physiol Endocrinol Metab. 2012;302:E1453–60. doi: 10.1152/ajpendo.00660.2011. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Harris TE, Roth RA, Lawrence JC Jr. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 31.Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 32.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 33.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 34.Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearce L, Huang X, Boudeau J, Pawłowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. Identification of protor as a novel rictor-binding component of mTOR complex-2. Biochem J. 2007;405:513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woo SY, Kim DH, Jun CB, Kim YM, Haar EV, Lee SI, Hegg JW, Bandhakavi S, Griffin TJ, Kim DH. PRR5, a novel component of mTOR complex 2, regulates platelet-derived growth factor receptor beta expression and signaling. J Biol Chem. 2007;282:25604–25612. doi: 10.1074/jbc.M704343200. [DOI] [PubMed] [Google Scholar]
- 37.Pearce LR, Sommer EM, Sakamoto K, Wullschleger S, Alessi DR. Protor-1 is required for efficient mTORC2-mediated activation of SGK1 in the kidney. Biochem J. 2011;436:169–179. doi: 10.1042/BJ20102103. [DOI] [PubMed] [Google Scholar]
- 38.Gan X, Wang J, Wang C, Sommer E, Kozasa T, Srinivasula S, Alessi D, Offermanns S, Simon MI, Wu D. PRR5L degradation promotes mTORC2-mediated PKC-[delta] phosphorylation and cell migration downstream of G [alpha] 12. Nat Cell Biol. 2012;14:686–696. doi: 10.1038/ncb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soliman GA, Acosta-Jaquez HA, Dunlop EA, Ekim B, Maj NE, Tee AR, Fingar DC. mTOR ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J Biol Chem. 2010;285:7866–7879. doi: 10.1074/jbc.M109.096222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acosta-Jaquez HA, Keller JA, Foster KG, Ekim B, Soliman GA, Feener EP, Ballif BA, Fingar DC. Site-specific mTOR phosphorylation promotes mTORC1-mediated signaling and cell growth. Mol Cell Biol. 2009;29:4308–4324. doi: 10.1128/MCB.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ekim B, Magnuson B, Acosta-Jaquez HA, Keller JA, Feener EP, Fingar DC. mTOR kinase domain phosphorylation promotes mTORC1 signaling, cell growth, and cell cycle progression. Mol Cell Biol. 2011;31:2787–2801. doi: 10.1128/MCB.05437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang K, Fingar DC. Growing knowledge of the mTOR signaling network. Semin Cell Dev Biol. 2014;36:79–90. doi: 10.1016/j.semcdb.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosner M, Siegel N, Valli A, Fuchs C, Hengstschläger M. mTOR phosphorylated at S2448 binds to raptor and rictor. Amino Acids. 2010;38:223–228. doi: 10.1007/s00726-008-0230-7. [DOI] [PubMed] [Google Scholar]
- 44.Copp J, Manning G, Hunter T. TORC-specific phosphorylation of mammalian target of rapamycin (mTOR): phospho-Ser2481 is a marker for intact mTOR signaling complex 2. Cancer Res. 2009;69:1821–1827. doi: 10.1158/0008-5472.CAN-08-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 49.Zheng X, Liang Y, He Q, Yao R, Bao W, Bao L, Wang Y, Wang Z. Current models of mammalian target of rapamycin complex 1 (mTORC1) activation by growth factors and amino acids. Int J Mol Sci. 2014;15:20753–20769. doi: 10.3390/ijms151120753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25:545–555. doi: 10.1016/j.tcb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inoki K, Li Y, Zhu T, Wu J, Guan K. TSC2 is phosphorylated and inhibited by akt and suppresses mTOR signaling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 52.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 53.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by erk: implications for tuberous sclerosisand cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 54.Saini KS, Loi S, de Azambuja E, Metzger-Filho O, Saini ML, Ignatiadis M, Dancey JE, Piccart-Gebhart MJ. Targeting the PI3K/AKT/mTOR and raf/MEK/ERK pathways in the treatment of breast cancer. Cancer Treat Rev. 2013;39:935–946. doi: 10.1016/j.ctrv.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 55.Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci U S A. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hara K, Yonezawa K, Weng Q, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 57.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pena-Llopis S, Vega-Rubin-de-Celis S, Schwartz JC, Wolff NC, Tran TA, Zou L, Xie XJ, Corey DR, Brugarolas J. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30:3242–3258. doi: 10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Findlay G, Yan L, Procter J, Mieulet V, Lamb R. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signaling. Biochem J. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung J, Genau HM, Behrends C. Amino acid dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol Cell Biol. 2015;35:2479–94. doi: 10.1128/MCB.00125-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rebsamen M, Pochini L, Stasyk T, de Araújo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, Scorzoni S, Filipek PA, Huber KV, Bigenzahn JW, Heinz LX, Kraft C, Bennett KL, Indiveri C, Huber LA, Superti-Furga G. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519:477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ogmundsdottir MH, Heublein S, Kazi S, Reynolds B, Visvalingam SM, Shaw MK, Goberdhan DC. Proton-assisted amino acid transporter PAT1 complexes with rag GTPases and activates TORC1 on late endosomal and lysosomal membranes. PLoS One. 2012;7:e36616. doi: 10.1371/journal.pone.0036616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inoki K, Zhu T, Guan K. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 66.Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cybulski N, Hall MN. TOR complex 2: a signaling pathway of its own. Trends Biochem Sci. 2009;34:620–627. doi: 10.1016/j.tibs.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 68.Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM. mSin1 is necessary for akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 69.Liu P, Guo J, Gan W, Wei W. Dual phosphorylation of Sin1 at T86 and T398 negatively regulates mTORC2 complex integrity and activity. Protein Cell. 2014;5:171–177. doi: 10.1007/s13238-014-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan HX, Guan KL. The SIN1-PH domain connects mTORC2 to PI3K. Cancer Discov. 2015;5:1127–1129. doi: 10.1158/2159-8290.CD-15-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 72.Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo Y, Liu L, Wu Y, Singh K, Su B, Zhang N, Liu X, Shen Y, Huang S. Rapamycin inhibits mSin1 phosphorylation independently of mTORC1 and mTORC2. Oncotarget. 2015;6:4286–4298. doi: 10.18632/oncotarget.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 75.Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tavares MR, Pavan IC, Amaral CL, Meneguello L, Luchessi AD, Simabuco FM. The S6K protein family in health and disease. Life Sci. 2015;131:1–10. doi: 10.1016/j.lfs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 77.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signaling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 78.Dibble CC, Asara JM, Manning BD. Characterization of rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol. 2009;29:5657–5670. doi: 10.1128/MCB.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 81.Dunlop EA, Tee AR. mTOR and autophagy: A dynamic relationship governed by nutrients and energy. Semin Cell Dev Biol. 2014;36:121–129. doi: 10.1016/j.semcdb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 82.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim YC, Guan K. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, Cecconi F. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 85.Egan D, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7:643–4. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dodd K, Yang J, Shen M, Sampson J, Tee A. mTORC1 drives HIF-1α and VEGF-A signaling via multiple mechanisms involving 4E-BP1, S6K1 and STAT3. Oncogene. 2015;34:2239–2250. doi: 10.1038/onc.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ricoult SJ, Manning BD. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 2013;14:242–251. doi: 10.1038/embor.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 91.Manning BD, Cantley LC. AKT/PKB signaling: Navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 93.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to akt-FOXO and PKCα, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 94.Mori S, Nada S, Kimura H, Tajima S, Takahashi Y, Kitamura A, Oneyama C, Okada M. The mTOR pathway controls cell proliferation by regulating the FoxO3a transcription factor via SGK1 kinase. PLoS One. 2014;9:e88891. doi: 10.1371/journal.pone.0088891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lang F, Pearce D. Regulation of the epithelial na+ channel by the mTORC2/SGK1 pathway. Nephrol Dial Transplant. 2016;31:200–205. doi: 10.1093/ndt/gfv270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heikamp EB, Patel CH, Collins S, Waickman A, Oh MH, Sun IH, Illei P, Sharma A, Naray-Fejes-Toth A, Fejes-Toth G, Misra-Sen J, Horton MR, Powell JD. The AGC kinase SGK1 regulates TH1 and TH2 differentiation downstream of the mTORC2 complex. Nat Immunol. 2014;15:457–464. doi: 10.1038/ni.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Angliker N, Rüegg MA. In vivo evidence for mTORC2-mediated actin cytoskeleton rearrangement in neurons. Bioarchitecture. 2013;3:113–118. doi: 10.4161/bioa.26497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Betz C, Stracka D, Prescianotto-Baschong C, Frieden M, Demaurex N, Hall MN. Feature article: mTOR complex 2-akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci U S A. 2013;110:12526–12534. doi: 10.1073/pnas.1302455110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chiang GG, Abraham RT. Targeting the mTOR signaling network in cancer. Trends Mol Med. 2007;13:433–442. doi: 10.1016/j.molmed.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 100.Xu K, Liu P, Wei W. mTOR signaling in tumorigenesis. Biochim Biophys Acta. 2014;1846:638–654. doi: 10.1016/j.bbcan.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lo SS, Hsueh C. Phosphorylated mTOR expression correlates with poor outcome in early-stage triple negative breast carcinomas. Int J Clin Exp Pathol. 2012;5:806–813. [PMC free article] [PubMed] [Google Scholar]
- 102.Wazir U, Newbold R, Jiang WG, Sharma A, Mokbel K. Prognostic and therapeutic implications of mTORC1 and rictor expression in human breast cancer. Oncol Rep. 2013;29:1969–1974. doi: 10.3892/or.2013.2346. [DOI] [PubMed] [Google Scholar]
- 103.Walsh S, Flanagan L, Quinn C, Evoy D, McDermott EW, Pierce A, Duffy MJ. mTOR in breast cancer: differential expression in triple-negative and non-triple-negative tumors. Breast. 2012;21:178–182. doi: 10.1016/j.breast.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 104.Moschetta M, Reale A, Marasco C, Vacca A, Carratù M. Therapeutic targeting of the mTOR-signaling pathway in cancer: benefits and limitations. Br J Pharmacol. 2014;171:3801–3813. doi: 10.1111/bph.12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Montalto G, Cervello M, Nicoletti F, Fagone P, Malaponte G, Mazzarino MC, Candido S, Libra M, Bäsecke J, Mijatovic S, Maksimovic-Ivanic D, Milella M, Tafuri A, Cocco L, Evangelisti C, Chiarini F, Martelli AM. Mutations and deregulation of ras/raf/MEK/ERK and PI3K/PTEN/akt/mTOR cascades which alter therapy response. Oncotarget. 2012;3:954–987. doi: 10.18632/oncotarget.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Strimpakos AS, Karapanagiotou EM, Saif MW, Syrigos KN. The role of mTOR in the management of solid tumors: an overview. Cancer Treat Rev. 2009;35:148–159. doi: 10.1016/j.ctrv.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 107.Chiarini F, Evangelisti C, McCubrey JA, Martelli AM. Current treatment strategies for inhibiting mTOR in cancer. Trends Pharmacol Sci. 2015;36:124–135. doi: 10.1016/j.tips.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 108.Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, Jordan A, Beck AH, Sabatini DM. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hardt M, Chantaravisoot N, Tamanoi F. Activating mutations of TOR (target of rapamycin) Genes Cells. 2011;16:141–151. doi: 10.1111/j.1365-2443.2010.01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sato T, Nakashima A, Guo L, Coffman K, Tamanoi F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010;29:2746–2752. doi: 10.1038/onc.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Droog M, Beelen K, Linn S, Zwart W. Tamoxifen resistance: from bench to bedside. Eur J Pharmacol. 2013;717:47–57. doi: 10.1016/j.ejphar.2012.11.071. [DOI] [PubMed] [Google Scholar]
- 112.Viedma-Rodríguez R, Baiza-Gutman L, Salamanca-Gómez F, Diaz-Zaragoza M, Martínez-Hernández G, Ruiz Esparza-Garrido R, Velázquez-Flores MA, Arenas-Aranda D. Mechanisms associated with resistance to tamoxifen in estrogen receptor-positive breast cancer (review) Oncol Rep. 2014;32:3–15. doi: 10.3892/or.2014.3190. [DOI] [PubMed] [Google Scholar]
- 113.Miller TW, Hennessy BT, Gonzalez-Angulo AM, Fox EM, Mills GB, Chen H, Higham C, García-Echeverría C, Shyr Y, Arteaga CL. Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J Clin Invest. 2010;120:2406–2413. doi: 10.1172/JCI41680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.deGraffenried LA, Friedrichs WE, Russell DH, Donzis EJ, Middleton AK, Silva JM, Roth RA, Hidalgo M. Inhibition of mTOR activity restores tamoxifen response in breast cancer cells with aberrant akt activity. Clin Cancer Res. 2004;10:8059–8067. doi: 10.1158/1078-0432.CCR-04-0035. [DOI] [PubMed] [Google Scholar]
- 115.Hwangbo W, Lee JH, Ahn S, Kim S, Park KH, Kim CH, Kim I. EGFR gene amplification and protein expression in invasive ductal carcinoma of the breast. Korean J Pathol. 2013;47:107–115. doi: 10.4132/KoreanJPathol.2013.47.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Koletsa T, Kotoula V, Karayannopoulou G, Nenopoulou E, Karkavelas G, Papadimitriou CS, Kostopoulos I. EGFR expression and activation are common in HER2 positive and triple-negative breast tumors. Histol Histopathol. 2010;25:1171–9. doi: 10.14670/HH-25.1171. [DOI] [PubMed] [Google Scholar]
- 117.Iqbal N, Iqbal N. Human epidermal growth factor receptor 2 (HER2) in cancers: overexpression and therapeutic implications. Mol Biol Int. 2014;2014:852748. doi: 10.1155/2014/852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu VS, Kanaya N, Lo C, Mortimer J, Chen S. From bench to bedside: what do we know about hormone receptor-positive and human epidermal growth factor receptor 2-positive breast cancer? J Steroid Biochem Mol Biol. 2015;153:45–53. doi: 10.1016/j.jsbmb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Margariti N, Fox SB, Bottini A, Generali D. “Overcoming breast cancer drug resistance with mTOR inhibitors”. Could it be a myth or a real possibility in the short-term future? Breast Cancer Res Treat. 2011;128:599–606. doi: 10.1007/s10549-010-0986-9. [DOI] [PubMed] [Google Scholar]
- 120.Brady SW, Zhang J, Tsai M, Yu D. PI3K-independent mTOR activation promotes lapatinib resistance and IAP expression that can be effectively reversed by mTOR and Hsp90 inhibition. Cancer Biol Ther. 2015;16:402–411. doi: 10.1080/15384047.2014.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gagliato DM, Jardim DL, Marchesi MS, Hortobagyi GN. Mechanisms of resistance and sensitivity to anti-HER2 therapies in HER2+ breast cancer. Oncotarget. 2016;7:64431–64446. doi: 10.18632/oncotarget.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Thery J, Spano J, Azria D, Raymond E, Llorca FP. Resistance to human epidermal growth factor receptor type 2-targeted therapies. Eur J Cancer. 2014;50:892–901. doi: 10.1016/j.ejca.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 123.Miller TW, Forbes JT, Shah C, Wyatt SK, Manning HC, Olivares MG, Sanchez V, Dugger TC, de Matos Granja N, Narasanna A, Cook RS, Kennedy JP, Lindsley CW, Arteaga CL. Inhibition of mammalian target of rapamycin is required for optimal antitumor effect of HER2 inhibitors against HER2-overexpressing cancer cells. Clin Cancer Res. 2009;15:7266–7276. doi: 10.1158/1078-0432.CCR-09-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou H, Luo Y, Huang S. Updates of mTOR inhibitors. Anticancer Agents Med Chem. 2010;10:571–581. doi: 10.2174/187152010793498663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yanik EL, Siddiqui K, Engels EA. Sirolimus effects on cancer incidence after kidney transplantation: a meta-analysis. Cancer Med. 2015;4:1448–1459. doi: 10.1002/cam4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu M, Bu LM, Wu K, Lu LG, Wang XP. Rapamycin inhibits the proliferation of SW1990 pancreatic cancer cell. Eur Rev Med Pharmacol Sci. 2015;19:3072–3079. [PubMed] [Google Scholar]
- 127.Dilling MB, Dias P, Shapiro DN, Germain GS, Johnson RK, Houghton PJ. Rapamycin selectively inhibits the growth of childhood rhabdomyosarcoma cells through inhibition of signaling via the type I insulin-like growth factor receptor. Cancer Res. 1994;54:903–907. [PubMed] [Google Scholar]
- 128.Bjornsti MA, Houghton PJ. The TOR pathway: a target for cancer therapy. Nat Rev Cancer. 2004;4:335–348. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 129.Ballou LM, Lin RZ. Rapamycin and mTOR kinase inhibitors. J Chem Biol. 2008;1:27–36. doi: 10.1007/s12154-008-0003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Selleckchem. Selleckchem. http://www.selleckchem.com/. Updated 2016. Accessed 08/24, 2016.
- 131.Carmellini M, Garcia V, Wang Z, Vergara M, Russ G. Efficacy of everolimus with reduced-exposure cyclosporine in de novo kidney transplant patients at increased risk for efficacy events: analysis of a randomized trial. J Nephrol. 2015;28:633–639. doi: 10.1007/s40620-015-0180-6. [DOI] [PubMed] [Google Scholar]
- 132.European Medicines Agency. Afinitor everolimus. http://www.ema.europa.eu/ema/index.jsp?curl=pages/medicines/human/medicines/001038/human_med_000633.jsp&mid=WC0b01ac058001d124. Updated 2016a. Accessed 08/10, 2016.
- 133.Granata S, Dalla Gassa A, Carraro A, Brunelli M, Stallone G, Lupo A, Zaza G. Sirolimus and everolimus pathway: reviewing candidate genes influencing their intracellular effects. Int J Mol Sci. 2016;17:735. doi: 10.3390/ijms17050735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kirchner GI, Meier-Wiedenbach I, Manns MP. Clinical pharmacokinetics of everolimus. Clin Pharmacokinet. 2004;43:83–95. doi: 10.2165/00003088-200443020-00002. [DOI] [PubMed] [Google Scholar]
- 135.Kobashigawa JA, Pauly DF, Starling RC, Eisen H, Ross H, Wang SS, Cantin B, Hill JA, Lopez P, Dong G, Nicholls SJ A2310 IVUS Substudy Investigators. Cardiac allograft vasculopathy by intravascular ultrasound in heart transplant patients: substudy from the everolimus versus mycophenolate mofetil randomized, multicenter trial. JACC Heart Fail. 2013;1:389–399. doi: 10.1016/j.jchf.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 136.Moes DJ, Guchelaar H, de Fijter JW. Sirolimus and everolimus in kidney transplantation. Drug Discov Today. 2015;20:1243–1249. doi: 10.1016/j.drudis.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 137.Parada M, Alba A, Sepúlveda C, Melo J. Long-term use of everolimus in lung transplant patients. Transplant Proc. 2011;43:2313–2315. doi: 10.1016/j.transproceed.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 138.Chan S, Scheulen ME, Johnston S, Mross K, Cardoso F, Dittrich C, Eiermann W, Hess D, Morant R, Semiglazov V, Borner M, Salzberg M, Ostapenko V, Illiger HJ, Behringer D, Bardy-Bouxin N, Boni J, Kong S, Cincotta M, Moore L. Phase II study of temsirolimus (CCI-779), a novel inhibitor of mTOR, in heavily pretreated patients with locally advanced or metastatic breast cancer. J. Clin. Oncol. 2005;23:5314–5322. doi: 10.1200/JCO.2005.66.130. [DOI] [PubMed] [Google Scholar]
- 139.Dudkin L, Dilling MB, Cheshire PJ, Harwood FC, Hollingshead M, Arbuck SG, Travis R, Sausville EA, Houghton PJ. Biochemical correlates of mTOR inhibition by the rapamycin ester CCI-779 and tumor growth inhibition. Clin Cancer Res. 2001;7:1758–1764. [PubMed] [Google Scholar]
- 140.European Medicines Agency. Torisel temsirolimus. http://www.ema.europa.eu/ema/index.jsp?curl=pages/medicines/human/medicines/000799/human_med_001098.jsp&mid=WC0b01ac058001d124. Updated 2016b. Accessed 08/10, 2016.
- 141.Wolff AC, Lazar AA, Bondarenko I, Garin AM, Brincat S, Chow L, Sun Y, Neskovic-Konstantinovic Z, Guimaraes RC, Fumoleau P, Chan A, Hachemi S, Strahs A, Cincotta M, Berkenblit A, Krygowski M, Kang LL, Moore L, Hayes DF. Randomized phase III placebo-controlled trial of letrozole plus oral temsirolimus as first-line endocrine therapy in postmenopausal women with locally advanced or metastatic breast cancer. J. Clin. Oncol. 2013;31:195–202. doi: 10.1200/JCO.2011.38.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mita M, Sankhala K, Abdel-Karim I, Mita A, Giles F. Deforolimus (AP23573) a novel mTOR inhibitor in clinical development. Expert Opin Investig Drugs. 2008;17:1947–1954. doi: 10.1517/13543780802556485. [DOI] [PubMed] [Google Scholar]
- 143.Vignot S, Faivre S, Aguirre D, Raymond E. mTOR-targeted therapy of cancer with rapamycin derivatives. Ann Oncol. 2005;16:525–537. doi: 10.1093/annonc/mdi113. [DOI] [PubMed] [Google Scholar]
- 144.Burke SE, Kuntz RE, Schwartz LB. Zotarolimus (ABT-578) eluting stents. Adv Drug Deliv Rev. 2006;58:437–446. doi: 10.1016/j.addr.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 145.Chen YW, Smith ML, Sheets M, Ballaron S, Trevillyan JM, Burke SE, Rosenberg T, Henry C, Wagner R, Bauch J, Marsh K, Fey TA, Hsieh G, Gauvin D, Mollison KW, Carter GW, Djuric SW. Zotarolimus, a novel sirolimus analogue with potent anti-proliferative activity on coronary smooth muscle cells and reduced potential for systemic immunosuppression. J Cardiovasc Pharmacol. 2007;49:228–235. doi: 10.1097/FJC.0b013e3180325b0a. [DOI] [PubMed] [Google Scholar]
- 146.Raungaard B, Jensen LO, Tilsted HH, Christiansen EH, Maeng M, Terkelsen CJ, Krusell LR, Kaltoft A, Kristensen SD, Bøtker HE, Thuesen L, Aarøe J, Jensen SE, Villadsen AB, Thayssen P, Veien KT, Hansen KN, Junker A, Madsen M, Ravkilde J, Lassen JF Scandinavian Organization for Randomized Trials with Clinical Outcome (SORT OUT) Zotarolimus-eluting durable-polymer-coated stent versus a biolimus-eluting biodegradable-polymer-coated stent in unselected patients undergoing percutaneous coronary intervention (SORT OUT VI): a randomised non-inferiority trial. Lancet. 2015;385:1527–1535. doi: 10.1016/S0140-6736(14)61794-3. [DOI] [PubMed] [Google Scholar]
- 147.Cameron AM, Steiner JP, Roskams AJ, Ali SM, Ronnettt GV, Snyder SH. Calcineurin associated with the inositol 1, 4, 5-trisphosphate receptor-FKBP12 complex modulates ca 2 flux. Cell. 1995;83:463–472. doi: 10.1016/0092-8674(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 148.Lee CS, Georgiou DK, Dagnino-Acosta A, Xu J, Ismailov II, Knoblauch M, Monroe TO, Ji R, Hanna AD, Joshi AD, Long C, Oakes J, Tran T, Corona BT, Lorca S, Ingalls CP, Narkar VA, Lanner JT, Bayle JH, Durham WJ, Hamilton SL. Ligands for FKBP12 increase Ca2+ influx and protein synthesis to improve skeletal muscle function. J Biol Chem. 2014;289:25556–25570. doi: 10.1074/jbc.M114.586289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci U S A. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 151.Oshiro N, Yoshino K, Hidayat S, Tokunaga C, Hara K, Eguchi S, Avruch J, Yonezawa K. Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells. 2004;9:359–366. doi: 10.1111/j.1356-9597.2004.00727.x. [DOI] [PubMed] [Google Scholar]
- 152.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Choo AY, Blenis J. Not all substrates are treated equally: implications for mTOR, rapamycin-resistance, and cancer therapy. Cell Cycle. 2009;8:567–572. doi: 10.4161/cc.8.4.7659. [DOI] [PubMed] [Google Scholar]
- 154.Rosner M, Hengstschlager M. Cytoplasmic and nuclear distribution of the protein complexes mTORC1 and mTORC2: rapamycin triggers dephosphorylation and delocalization of the mTORC2 components rictor and sin1. Hum Mol Genet. 2008;17:2934–2948. doi: 10.1093/hmg/ddn192. [DOI] [PubMed] [Google Scholar]
- 155.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 156.Schreiber KH, Ortiz D, Academia EC, Anies AC, Liao C, Kennedy BK. Rapamycin-mediated mTORC2 inhibition is determined by the relative expression of FK506-binding proteins. Aging Cell. 2015;14:265–273. doi: 10.1111/acel.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Easton J, Houghton P. mTOR and cancer therapy. Oncogene. 2006;25:6436–6446. doi: 10.1038/sj.onc.1209886. [DOI] [PubMed] [Google Scholar]
- 158.Van Der Poel H, Hanrahan C, Zhong H, Simons J. Rapamycin induces smad activity in prostate cancer cell lines. Urol Res. 2003;30:380–386. doi: 10.1007/s00240-002-0282-1. [DOI] [PubMed] [Google Scholar]
- 159.Seufferlein T, Rozengurt E. Rapamycin inhibits constitutive p70s6k phosphorylation, cell proliferation, and colony formation in small cell lung cancer cells. Cancer Res. 1996;56:3895–3897. [PubMed] [Google Scholar]
- 160.Martin L, Pancholi S, Farmer I, Guest S, Ribas R, Weigel MT, Thornhill AM, Ghazoui Z, A’Hern R, Evans DB, Lane HA, Johnston SR, Dowsett M. Effectiveness and molecular interactions of the clinically active mTORC1 inhibitor everolimus in combination with tamoxifen or letrozole in vitro and in vivo. Breast Cancer Res. 2012;14:R132. doi: 10.1186/bcr3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Saunders P, Weiss J, Welschinger R, Baraz R, Bradstock K, Bendall L. RAD001 (everolimus) induces dose-dependent changes to cell cycle regulation and modifies the cell cycle response to vincristine. Oncogene. 2013;32:4789–4797. doi: 10.1038/onc.2012.498. [DOI] [PubMed] [Google Scholar]
- 162.Naruse T, Yanamoto S, Yamada S, Rokutanda S, Kawakita A, Kawasaki G, Umeda M. Anti-tumor effect of the mammalian target of rapamycin inhibitor everolimus in oral squamous cell carcinoma. Pathol Oncol Res. 2015;21:765–773. doi: 10.1007/s12253-014-9888-1. [DOI] [PubMed] [Google Scholar]
- 163.Lui A, New J, Ogony J, Thomas S, Lewis-Wambi J. Everolimus downregulates estrogen receptor and induces autophagy in aromatase inhibitor-resistant breast cancer cells. BMC Cancer. 2016;16:487. doi: 10.1186/s12885-016-2490-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, Kondo S. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 165.Woltman AM, van der Kooij SW, Coffer PJ, Offringa R, Daha MR, van Kooten C. Rapamycin specifically interferes with GM-CSF signaling in human dendritic cells, leading to apoptosis via increased p27KIP1 expression. Blood. 2003;101:1439–1445. doi: 10.1182/blood-2002-06-1688. [DOI] [PubMed] [Google Scholar]
- 166.Hosoi H, Dilling MB, Shikata T, Liu LN, Shu L, Ashmun RA, Germain GS, Abraham RT, Houghton PJ. Rapamycin causes poorly reversible inhibition of mTOR and induces p53-independent apoptosis in human rhabdomyosarcoma cells. Cancer Res. 1999;59:886–894. [PubMed] [Google Scholar]
- 167.Hurvitz SA, Kalous O, Conklin D, Desai AJ, Dering J, Anderson L, O’Brien NA, Kolarova T, Finn RS, Linnartz R, Chen D, Slamon DJ. In vitro activity of the mTOR inhibitor everolimus, in a large panel of breast cancer cell lines and analysis for predictors of response. Breast Cancer Res Treat. 2015;149:669–680. doi: 10.1007/s10549-015-3282-x. [DOI] [PubMed] [Google Scholar]
- 168.Khairi M, Shamsuddin S, Jaafar H. Effects of rapamycin on cell apoptosis in mcf-7 human breast cancer cells. Asian Pac J Cancer Prev. 2014;15:10659–10663. doi: 10.7314/apjcp.2014.15.24.10659. [DOI] [PubMed] [Google Scholar]
- 169.Saran U, Foti M, Dufour J. Cellular and molecular effects of the mTOR inhibitor everolimus. Clin Sci. 2015;129:895–914. doi: 10.1042/CS20150149. [DOI] [PubMed] [Google Scholar]
- 170.Huang J, Manning BD. A complex interplay between akt, TSC2 and the two mTOR complexes. Biochem Soc Trans. 2009;37:217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.De P, Miskimins K, Dey N, Leyland-Jones B. Promise of rapalogues versus mTOR kinase inhibitors in subset specific breast cancer: old targets new hope. Cancer Treat Rev. 2013;39:403–412. doi: 10.1016/j.ctrv.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 172.Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, Khuri FR. Activation of akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 173.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, Baselga J, Rosen N. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shi Y, Yan H, Frost P, Gera J, Lichtenstein A. Mammalian target of rapamycin inhibitors activate the AKT kinase in multiple myeloma cells by up-regulating the insulin-like growth factor receptor/insulin receptor substrate-1/phosphatidylinositol 3-kinase cascade. Mol Cancer Ther. 2005;4:1533–1540. doi: 10.1158/1535-7163.MCT-05-0068. [DOI] [PubMed] [Google Scholar]
- 175.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, Papa A, Nardella C, Cantley LC, Baselga J, Pandolfi PP. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Acevedo-Gadea C, Hatzis C, Chung G, Fishbach N, Lezon-Geyda K, Zelterman D, DiGiovanna MP, Harris L, Abu-Khalaf MM. Sirolimus and trastuzumab combination therapy for HER2-positive metastatic breast cancer after progression on prior trastuzumab therapy. Breast Cancer Res Treat. 2015;150:157–167. doi: 10.1007/s10549-015-3292-8. [DOI] [PubMed] [Google Scholar]
- 177.Alayev A, Berger SM, Kramer MY, Schwartz NS, Holz MK. The combination of rapamycin and resveratrol blocks autophagy and induces apoptosis in breast cancer cells. J Cell Biochem. 2015;116:450–457. doi: 10.1002/jcb.24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zanardi E, Verzoni E, Grassi P, Necchi A, Giannatempo P, Raggi D, De Braud F, Procopio G. Clinical experience with temsirolimus in the treatment of advanced renal cell carcinoma. Ther Adv Urol. 2015;7:152–161. doi: 10.1177/1756287215574457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fleming GF, Ma CX, Huo D, Sattar H, Tretiakova M, Lin L, Hahn OM, Olopade FO, Nanda R, Hoffman PC, Naughton MJ, Pluard T, Conzen SD, Ellis MJ. Phase II trial of temsirolimus in patients with metastatic breast cancer. Breast Cancer Res Treat. 2012;136:355–363. doi: 10.1007/s10549-011-1910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, Schuchter LM, Torigian DA, Panosian JT, Troxel AB, Tan KS, Heitjan DF, DeMichele AM, Vaughn DJ, Redlinger M, Alavi A, Kaiser J, Pontiggia L, Davis LE, O’Dwyer PJ, Amaravadi RK. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Seiler M, Ray-Coquard I, Melichar B, Yardley DA, Wang RX, Dodion PF, Lee MA. Oral ridaforolimus plus trastuzumab for patients with HER2 trastuzumab-refractory metastatic breast cancer. Clin Breast Cancer. 2015;15:60–65. doi: 10.1016/j.clbc.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 182.Oza AM, Pignata S, Poveda A, McCormack M, Clamp A, Schwartz B, Cheng J, Li X, Campbell K, Dodion P, Haluska FG. Randomized phase II trial of ridaforolimus in advanced endometrial carcinoma. J. Clin. Oncol. 2015;33:3576–3582. doi: 10.1200/JCO.2014.58.8871. [DOI] [PubMed] [Google Scholar]
- 183.Rizzieri DA, Feldman E, Dipersio JF, Gabrail N, Stock W, Strair R, Rivera VM, Albitar M, Bedrosian CL, Giles FJ. A phase 2 clinical trial of deforolimus (AP23573, MK-8669), a novel mammalian target of rapamycin inhibitor, in patients with relapsed or refractory hematologic malignancies. Clin Cancer Res. 2008;14:2756–2762. doi: 10.1158/1078-0432.CCR-07-1372. [DOI] [PubMed] [Google Scholar]
- 184.Geisler J. Differences between the non-steroidal aromatase inhibitors anastrozole and letrozole-of clinical importance & quest. Br J Cancer. 2011;104:1059–1066. doi: 10.1038/bjc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wazir U, Wazir A, Khanzada ZS, Jiang WG, Sharma AK, Mokbel K. Current state of mTOR targeting in human breast cancer. Cancer Genomics Proteomics. 2014;11:167–174. [PubMed] [Google Scholar]
- 186.Baselga J, Campone M, Piccart M, Burris HA 3rd, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, Beck JT, Ito Y, Yardley D, Deleu I, Perez A, Bachelot T, Vittori L, Xu Z, Mukhopadhyay P, Lebwohl D, Hortobagyi GN. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Beaver JA, Park BH. The BOLERO-2 trial: the addition of everolimus to exemestane in the treatment of postmenopausal hormone receptor-positive advanced breast cancer. Future Oncol. 2012;8:651–657. doi: 10.2217/fon.12.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dorris JR 3rd, Jones S. Everolimus in breast cancer: The role of the pharmacist. Ann Pharmacother. 2014;48:1194–1201. doi: 10.1177/1060028014542415. [DOI] [PubMed] [Google Scholar]
- 189.Hurvitz SA, Dalenc F, Campone M, O’Regan RM, Tjan-Heijnen VC, Gligorov J, Llombart A, Jhangiani H, Mirshahidi HR, Tan-Chiu E, Miao S, El-Hashimy M, Lincy J, Taran T, Soria JC, Sahmoud T, André F. A phase 2 study of everolimus combined with trastuzumab and paclitaxel in patients with HER2-overexpressing advanced breast cancer that progressed during prior trastuzumab and taxane therapy. Breast Cancer Res Treat. 2013;141:437–446. doi: 10.1007/s10549-013-2689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hurvitz SA, Andre F, Jiang Z, Shao Z, Mano MS, Neciosup SP, Tseng LM, Zhang Q, Shen K, Liu D, Dreosti LM, Burris HA, Toi M, Buyse ME, Cabaribere D, Lindsay MA, Rao S, Pacaud LB, Taran T, Slamon D. Combination of everolimus with trastuzumab plus paclitaxel as first-line treatment for patients with HER2-positive advanced breast cancer (BOLERO-1): a phase 3, randomised, double-blind, multicentre trial. Lancet Oncol. 2015;16:816–829. doi: 10.1016/S1470-2045(15)00051-0. [DOI] [PubMed] [Google Scholar]
- 191.André F, O’Regan R, Ozguroglu M, Toi M, Xu B, Jerusalem G, Masuda N, Wilks S, Arena F, Isaacs C, Yap YS, Papai Z, Lang I, Armstrong A, Lerzo G, White M, Shen K, Litton J, Chen D, Zhang Y, Ali S, Taran T, Gianni L. Everolimus for women with trastuzumab-resistant, HER2-positive, advanced breast cancer (BOLERO-3): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol. 2014;15:580–591. doi: 10.1016/S1470-2045(14)70138-X. [DOI] [PubMed] [Google Scholar]
- 192.Bachelot T, Bourgier C, Cropet C, Ray-Coquard I, Ferrero JM, Freyer G, Abadie-Lacourtoisie S, Eymard JC, Debled M, Spaëth D, Legouffe E, Allouache D, El Kouri C, Pujade-Lauraine E. Randomized phase II trial of everolimus in combination with tamoxifen in patients with hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer with prior exposure to aromatase inhibitors: a GINECO study. J. Clin. Oncol. 2012;30:2718–2724. doi: 10.1200/JCO.2011.39.0708. [DOI] [PubMed] [Google Scholar]
- 193.Singh JC, Novik Y, Stein S, Volm M, Meyers M, Smith J, Omene C, Speyer J, Schneider R, Jhaveri K, Formenti S, Kyriakou V, Joseph B, Goldberg JD, Li X, Adams S, Tiersten A. Phase 2 trial of everolimus and carboplatin combination in patients with triple negative metastatic breast cancer. Breast Cancer Res. 2014;16:R32. doi: 10.1186/bcr3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yardley DA, Bosserman LD, O’Shaughnessy JA, Harwin WN, Morgan SK, Priego VM, Peacock NW, Bass JD, Burris HA 3rd, Hainsworth JD. Paclitaxel, bevacizumab, and everolimus/placebo as first-line treatment for patients with metastatic HER2-negative breast cancer: A randomized placebo-controlled phase II trial of the sarah cannon research institute. Breast Cancer Res Treat. 2015;154:89–97. doi: 10.1007/s10549-015-3599-5. [DOI] [PubMed] [Google Scholar]
- 195.Carew JS, Kelly KR, Nawrocki ST. Mechanisms of mTOR inhibitor resistance in cancer therapy. Target Oncol. 2011;6:17–27. doi: 10.1007/s11523-011-0167-8. [DOI] [PubMed] [Google Scholar]
- 196.Mi W, Ye Q, Liu S, She QB. AKT inhibition overcomes rapamycin resistance by enhancing the repressive function of PRAS40 on mTORC1/4E-BP1 axis. Oncotarget. 2015;6:13962–13977. doi: 10.18632/oncotarget.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Noh WC, Mondesire WH, Peng J, Jian W, Zhang H, Dong J, Mills GB, Hung MC, Meric-Bernstam F. Determinants of rapamycin sensitivity in breast cancer cells. Clin Cancer Res. 2004;10:1013–1023. doi: 10.1158/1078-0432.ccr-03-0043. [DOI] [PubMed] [Google Scholar]
- 198.Jordan NJ, Dutkowski CM, Barrow D, Mottram HJ, Hutcheson IR, Nicholson RI, Guichard SM, Gee JM. Impact of dual mTORC1/2 mTOR kinase inhibitor AZD8055 on acquired endocrine resistance in breast cancer in vitro. Breast Cancer Res. 2014;16:R12. doi: 10.1186/bcr3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Holder AM, Akcakanat A, Adkins F, Evans K, Chen H, Wei C, Milton DR, Li Y, Do KA, Janku F, Meric-Bernstam F. Epithelial to mesenchymal transition is associated with rapamycin resistance. Oncotarget. 2015;6:19500–19513. doi: 10.18632/oncotarget.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bihani T, Ezell SA, Ladd B, Grosskurth SE, Mazzola AM, Pietras M, Reimer C, Zinda M, Fawell S, D’Cruz CM. Resistance to everolimus driven by epigenetic regulation of MYC in ER+ breast cancers. Oncotarget. 2015;6:2407–2420. doi: 10.18632/oncotarget.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cao J, Huang W. Compensatory increase of transglutaminase 2 is responsible for resistance to mTOR inhibitor treatment. PLoS One. 2016;11:e0149388. doi: 10.1371/journal.pone.0149388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Raimondo L, D’Amato V, Servetto A, Rosa R, Marciano R, Formisano L, Di Mauro C, Orsini RC, Cascetta P, Ciciola P, De Maio AP, Di Renzo MF, Cosconati S, Bruno A, Randazzo A, Napolitano F, Montuori N, Veneziani BM, De Placido S, Bianco R. Everolimus induces met inactivation by disrupting the FKBP12/met complex. Oncotarget. 2016;7:40073–40084. doi: 10.18632/oncotarget.9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hassan B, Akcakanat A, Sangai T, Evans KW, Adkins F, Eterovic AK, Zhao H, Chen K, Chen H, Do KA, Xie SM, Holder AM, Naing A, Mills GB, Meric-Bernstam F. Catalytic mTOR inhibitors can overcome intrinsic and acquired resistance to allosteric mTOR inhibitors. Oncotarget. 2014;5:8544. doi: 10.18632/oncotarget.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gil EMC. Targeting the PI3K/AKT/mTOR pathway in estrogen receptor-positive breast cancer. Cancer Treat Rev. 2014;40:862–871. doi: 10.1016/j.ctrv.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 205.García-Echeverría C. Allosteric and ATP-competitive kinase inhibitors of mTOR for cancer treatment. Bioorg Med Chem Lett. 2010;20:4308–4312. doi: 10.1016/j.bmcl.2010.05.099. [DOI] [PubMed] [Google Scholar]
- 206.Britten CD, Adjei AA, Millham R, Houk BE, Borzillo G, Pierce K, Wainberg ZA, LoRusso PM. Phase I study of PF-04691502, a small-molecule, oral, dual inhibitor of PI3K and mTOR, in patients with advanced cancer. Invest New Drugs. 2014;32:510–517. doi: 10.1007/s10637-013-0062-5. [DOI] [PubMed] [Google Scholar]
- 207.Dey N, Sun Y, Carlson JH, Wu H, Lin X, Leyland-Jones B, De P. Anti-tumor efficacy of BEZ235 is complemented by its anti-angiogenic effects via downregulation of PI3K-mTOR-HIF1alpha signaling in HER2-defined breast cancers. Am J Cancer Res. 2016;6:714. [PMC free article] [PubMed] [Google Scholar]
- 208.Fazio N, Buzzoni R, Baudin E, Antonuzzo L, Hubner RA, Lahner H, DE Herder WW, Raderer M, Teulé A, Capdevila J, Libutti SK, Kulke MH, Shah M, Dey D, Turri S, Aimone P, Massacesi C, Verslype C. A phase II study of BEZ235 in patients with everolimus-resistant, advanced pancreatic neuroendocrine tumors. Anticancer Res. 2016;36:713–719. [PMC free article] [PubMed] [Google Scholar]
- 209.Bendell JC, Kelley RK, Shih KC, Grabowsky JA, Bergsland E, Jones S, Martin T, Infante JR, Mischel PS, Matsutani T, Xu S, Wong L, Liu Y, Wu X, Mortensen DS, Chopra R, Hege K, Munster PN. A phase I dose-escalation study to assess safety, tolerability, pharmacokinetics, and preliminary efficacy of the dual mTORC1/mTORC2 kinase inhibitor CC-223 in patients with advanced solid tumors or multiple myeloma. Cancer. 2015;121:3481–3490. doi: 10.1002/cncr.29422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Guichard SM, Curwen J, Bihani T, D’Cruz CM, Yates JW, Grondine M, Howard Z, Davies BR, Bigley G, Klinowska T, Pike KG, Pass M, Chresta CM, Polanska UM, McEwen R, Delpuech O, Green S, Cosulich SC. AZD2014, an inhibitor of mTORC1 and mTORC2, is highly effective in ER+ breast cancer when administered using intermittent or continuous schedules. Mol Cancer Ther. 2015;14:2508–2518. doi: 10.1158/1535-7163.MCT-15-0365. [DOI] [PubMed] [Google Scholar]
- 211.Wilson-Edell KA, Yevtushenko MA, Rothschild DE, Rogers AN, Benz CC. mTORC1/C2 and pan-HDAC inhibitors synergistically impair breast cancer growth by convergent AKT and polysome inhibiting mechanisms. Breast Cancer Res Treat. 2014;144:287–298. doi: 10.1007/s10549-014-2877-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Saura C, Bendell J, Jerusalem G, Su S, Ru Q, De Buck S, Mills D, Ruquet S, Bosch A, Urruticoechea A, Beck JT, Di Tomaso E, Sternberg DW, Massacesi C, Hirawat S, Dirix L, Baselga J. Phase ib study of buparlisib plus trastuzumab in patients with HER2-positive advanced or metastatic breast cancer that has progressed on trastuzumab-based therapy. Clin Cancer Res. 2014;20:1935–1945. doi: 10.1158/1078-0432.CCR-13-1070. [DOI] [PubMed] [Google Scholar]
- 213.Ma CX, Luo J, Naughton M, Ademuyiwa F, Suresh R, Griffith M, Griffith OL, Skidmore ZL, Spies NC, Ramu A, Trani L, Pluard T, Nagaraj G, Thomas S, Guo Z, Hoog J, Han J, Mardis E, Lockhart C, Ellis MJ. A phase I trial of BKM120 (buparlisib) in combination with fulvestrant in postmenopausal women with estrogen receptor-positive metastatic breast cancer. Clin Cancer Res. 2016;22:1583–1591. doi: 10.1158/1078-0432.CCR-15-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yu F, Zhao J, Hu Y, Zhou Y, Guo R, Bai J, Zhang S, Zhang H, Zhang J. The combination of NVP-BKM120 with trastuzumab or RAD001 synergistically inhibits the growth of breast cancer stem cells in vivo. Oncol Rep. 2016;36:356–64. doi: 10.3892/or.2016.4799. [DOI] [PubMed] [Google Scholar]
- 215.Chien AJ, Cockerill A, Fancourt C, Schmidt E, Moasser MM, Rugo HS, Melisko ME, Ko AH, Kelley RK, Korn WM, Esserman LJ, van’t Veer L, Yau C, Wolf DM, Munster PN. A phase 1b study of the Akt-inhibitor MK-2206 in combination with weekly paclitaxel and trastuzumab in patients with advanced HER2-amplified solid tumor malignancies. Breast Cancer Res Treat. 2016;155:521–530. doi: 10.1007/s10549-016-3701-7. [DOI] [PubMed] [Google Scholar]
- 216.Ma CX, Sanchez C, Gao F, Crowder R, Naughton M, Pluard T, Creekmore A, Guo Z, Hoog J, Lockhart AC, Doyle A, Erlichman C, Ellis MJ. A phase I study of the AKT inhibitor MK-2206 in combination with hormonal therapy in postmenopausal women with estrogen receptor-positive metastatic breast cancer. Clin Cancer Res. 2016;22:2650–2658. doi: 10.1158/1078-0432.CCR-15-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Davies BR, Greenwood H, Dudley P, Crafter C, Yu DH, Zhang J, Li J, Gao B, Ji Q, Maynard J, Ricketts SA, Cross D, Cosulich S, Chresta CC, Page K, Yates J, Lane C, Watson R, Luke R, Ogilvie D, Pass M. Preclinical pharmacology of AZD5363, an inhibitor of AKT: pharmacodynamics, antitumor activity, and correlation of monotherapy activity with genetic background. Mol Cancer Ther. 2012;11:873–887. doi: 10.1158/1535-7163.MCT-11-0824-T. [DOI] [PubMed] [Google Scholar]


