Abstract
This study aims to elucidate the effects of microRNA-27a (miR-27a) on the proliferation and invasion of gastric cancer (GC) cells by targeting SFRP1 via Wnt/β-catenin signaling pathway. GC and normal adjacent tissues were collected from 273 GC patients. Human gastric cancer cell line (MGC803) and normal human gastric mucosal cell line (GES-1) were cultured. The miR-27a mRNA expression was analyzed using Quantitative real-time polymerase chain reaction (qRT-PCR). Immunohistochemistry (IHC) test was used to detect miR-27a and SFRP1 protein expressions. After transfection, cells were divided into five groups: the negative control (NC) group, the miR-27a inhibitor group, the miR-27a mimics group, the miR-27a inhibitor + SFRP1 siRNA group and the miR-27a mimics + SFRP1 overexpression group. Western blotting was conducted to test SFRP1 and Wnt/β-catenin protein expression. Analysis for the target gene of miR-27a was performed using Luciferase assay. Cell proliferation, migration and invasion were determined by CCK8 and Transwell assay. The dual-luciferase reporter assay system was applied to analyze the effects of miR-27a on Wnt/β-catenin signaling pathway. In GC tissue and cell line, miR-27a protein and mRNA expressions were up-regulated, and SFRP1 protein and mRNA expressions were down-regulated. Luciferase assay indicated that miR-27a might target SFRP1 and regulate its expressions. When miR-27a was down-regulated, SFRP1 was up-regulated, and β-catenin, Wnt, p-β-catenin, and p-Wnt were significantly down-regulated. Compared with the NC group, the proliferation, migration and invasion of GC cells were remarkably increased in the miR-27a group, but these were declined in the miR-27a mimics + SFRP1 overexpression group. The proliferation, migration and invasion of GC cells were elevated in the miR-27a inhibitor + SFRP1 siRNA group compared with the miR-27a inhibitor group. These results showed that miR-27a was highly expressed in GC tissues and cells, and it might promote cell proliferation, migration and invasion by targeting SFRP1 via the activation of Wnt/β-catenin signaling pathway.
Keywords: MicroRNA-27a, gastric cancer, SFRP1, Wnt/β-catenin signaling pathway, MGC803 cells, proliferation, migration, invasion
Introduction
Gastric cancer (GC) is derived from epithelial cells of the gastric mucosa, with its incidence ranked No. 4 and the mortality rate ranked No. 2 all over the world [1]. The prevalence of GC shows obvious regional differences with the highest incidence in the northwest and eastern coastal areas [2]. The occurrence and development of GC are multi-step and multi-factor processes in which both genes and environmental factors may be involved [3-5]. In addition, infection plays a very important role in cancer [6]. In recent years, with the rapid development of molecular biology, especially gene cloning techniques, researchers gradually redirected attention towards potential genes related to GC.
MicroRNA is a class of non-coding small RNA molecules, with the capacity of oncogene or anti-oncogene activity, which can negatively regulate target gene, and play an important role in tumorigenesis [7]. Human microRNA-27a gene is located on chromosome 19, which is implicated in regulating the expression of multiple genes [8,9]. Secreted frizzled-related protein (SFRPs), a tumor suppressor protein, is a regulator of Wnt signaling pathway [10]. SFRP1 can inhibit Wnt signaling pathway and also bind protein Wnt in the extracellular compartment to inhibit ligand-receptor interactions and signal transduction [11]. Wnt/β-catenin signaling pathway can affect gene expression and cell migration [12]. β-catenin is of great importance in the signal transduction, stability and accumulation in cytoplasm [13]. Experimental results show that microRNA-27a (miR-27a) can target at gene Myt-1 to involve in cell cycle regulation [14], so we speculated that in other malignancies such as GC, miR-27a may also target on specific genes and play a similar role in the cell regulation. In this regards, the study aims to investigate the effects of miR-27a on the proliferation and invasion of GC cells by targeting SFRP1 via Wnt/β-catenin signaling pathway.
Materials and methods
Ethical statement
This study was carried out in conformity with medical ethical standards and was approved by Ethics Committee of Jiangxi Cancer Hospital and the First Affiliated Hospital of Nanchang University. Written informed consents were obtained from eligible patients.
Subjects
Human gastric cancer (GC) tissue samples were collected from 273 patients who were pathologically diagnosed with GC and underwent surgery in Jiangxi Cancer Hospital and the First Affiliated Hospital of Nanchang University between January 2013 and January 2016. There were 189 patients under 60 years of age and 84 patients over 60 years of age. And there were 147 males and 126 females with a mean age of 52.46 ± 14.56 years. In all patients, 120 patients were diagnosed with moderate or well differentiated GC, and 153 patients were diagnosed with poorly differentiated or undifferentiated GC. According to tumor-node-metastasis (TNM) staging issued by the American Joint Committee on Cancer (AJCC) in 2010 [15], there were 142 patients with stage I/II GC and 131 patients with stage III/IV GC, and there were 105 patients without lymph node metastasis and 168 patients with lymph node metastasis. All patients had no history of chemotherapy or radiotherapy before surgery. GC tissues and adjacent normal tissues (over 5 cm distant from cancer tissue) were obtained, immersed in liquid nitrogen and stored at -80°C in a freezer. One half of the tissues were used for qRT-PCR to detect microRNA-27a (miR-27a) mRNA expressions, and the other half was used for immunohistochemistry to detect SFRP1 protein expressions. Tissues were fixed with 10% formalin and embedded with paraffin, prior to immunohistochemistry test.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from frozen tissues (100 mg) with miRNeasy Mini Kit (Qiagen Company, Hilden, Germany). RNA concentration and purity were determined with an optical density ratio (OD260/OD280) of 1.7-2.1. The cDNA template was synthesized after reverse transcription of RNAs, followed by which qRT-PCR was performed using ABI7500 real time PCR instrument (ABI Company, Oyster Bay, NY, USA). The reaction system (20 μl in total): 2 × Allinone TM qPCR Mix (10 μl; Biomed Gene Technology Co., Ltd, Beijing, China), 1 μl forward primer (1 μl; 10 μM), reverse primer (1 μl; 10 μM), cDNA (1 μl), and distilled deionized H2O (7 μl). The reaction condition: pre-denaturation for 10 min at 95°C, 35 cycles of denaturation for 10 s at 95°C, annealing for 20 s at 60°C and extension for 30 s at 72°C. The primers for qRT-PCR: forward primer of miR-27a: 5’-GGCTTAGCTGCTTGTGAGCA-3’; reverse primer of miR-27a: 5’-GCGGAACTTAGCCACTGTGA-3’; U6 snRNA: 5’-CTCGCTTCGGCAGCACA-3’. With U6 snRNA (Shanghai Genepharma, Co., Ltd, Shanghai, China) selected as internal reference, the relative expression of the target gene was calculated as 2-ΔΔ Ct. ΔCt = Ct(miR-27a) - Ct(U6), ΔΔCt = ΔCt(GC tissue) - ΔCt(adjacent normal tissue). The threshold cycle (Ct) was obtained when the fluorescence intensity reached certain threshold value. All tests were repeated for three times.
Immunohistochemistry (IHC)
Paraffin-embedded tissues were sliced into 4-μm-thick sections, followed by dewaxing with dimethylbenzene and hydration in graded ethanol. Antigen retrieval was achieved by microwave heating. Endogenous peroxidase was blocked using 3% hydrogen peroxide, and tissues were kept in darkness at room temperature for 10 min. Then primary antibody (sheep anti-human SFRP1, Santa Cruz Biotechnology, Inc, Santa Cruz, CA, USA) was added dropwise into sections, and then they were incubated at 4°C overnight. Horseradish peroxidase-conjugated secondary antibody (Dako North America, Inc. Carpinteria, CA, USA) was added dropwise into sections, and then they were incubated at room temperature for 30 min. After that sections were stained by diaminobenzidine (DAB) and counterstained with hematoxylin, and then they were mounted on slides with neutral balsam. Primary antibody was replaced with PBS as negative control, and human GC biopsies were selected as positive control. Five high-power fields (× 400) were randomly selected for each section, and 100 cells were counted in each visual field. Positive protein expressions were determined by IHC test as the guidelines published by American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) [16]. Positive cells were counted. The ratio of positive GC cells/total GC cells > 10% was recorded as positive (+). The positive cell percentage < 10% was recorded as negative (-) The SFRP1 protein expression was scored by two blinded independent pathologists.
Cell culture
Human gastric cancer cell line (MGC803) and normal human gastric mucosal cell line (GES-1), purchased from American Type Culture Collection (ATCC, VA, USA), were cultured in the RPMI-1640 medium (Invitrogen Inc., Carlsbad, CA, USA) containing 10% fetal calf serum (FCS), and then medium was placed in an incubator (5% CO2, 95% humidity) at 37°C. Grown as monolayers, cells were sub-cultured when they reached 90% confluency. Culture medium was discarded as usual, and then cells were washed twice with PBS and digested with 0.25% trypsin. When the cell gap increased, trypsin were discarded and single-cell suspension was prepared with an addition of new medium.
Cell transfection and grouping
Cells were classified into five groups after transfection: the negative control (NC) group (transfected with negative control plasmid), the miR-27a inhibitor group, the miR-27a mimics group, the miR-27a inhibitor + SFRP1 siRNA group (co-transfected with both miR-27a inhibitor and SFRP1 siRNA plasmids) and the miR-27a mimics + SFRP1 overexpression group (co-transfected with both miR-27a mimics and SFRP1 overexpression plasmids). Both miR-27a inhibitor and miR-27a mimics plasmids were purchased from Shanghai Genepharma, Co., Ltd. (Shanghai, China). The SFRP1 siRNA plasmid was purchased from Guangzhou RiboBio Co., Ltd. (Guangdong, China) and it primer sequence was 5’-AGAAGAAGGACCTGAAGAA-3’. The SFRP1 overexpression plasmid was from Sino Biological Inc. (Beijing, China). Before transfection, cells were cultured in 12-well plates till they reached 70% confluency. MiR-27a inhibitor plasmid (same operation for miR-27a mimics plasmid, SFRP1 siRNA plasmid and SFRP1 overexpression plasmid) was added into Opti-MEM medium (100 μl; Thermo Fisher Scientific Inc., Waltham, MA). And then Lipo3000 reagent (Thermo Fisher Scientific Inc., Waltham, MA) was added, and diluted in Opti-MEM (Thermo Fisher Scientific Inc., Waltham, MA). After 5 min of dilution, the diluted solution was mixed with lipo2000 and maintained at room temperature for 20 min. The serum-free medium (800 μl) was added into each well containing the cells, and then cells were well mixed with the prepared mixture (diluted solution with lipo2000). After cultured for 6 h, cells were transfected for 48 h in the new medium. The result of cell transfection was observed under a fluorescence microscope. Cells were collected for subsequent analysis.
Western blotting
Cultured cells (MGC803 and GES-1) were rinsed with pre-cooled PBS three times. The protein was isolated from cells with the addition of protein lysis buffer (containing cocktail protease inhibitor which was purchased from Hoffmann-La Roche Ltd., Basel, Switzerland). Then cells were allowed to stand on ice for 30 min. Cells were centrifuged at 12000 rpm for 10 min at 4°C. The supernatant was dispensed into centrifuge tubes and stored at -20°C. During electrophoresis, the voltage was changed from 80 V to 120 V once the cells attached to the separation gel. The electrophoresis was continued until bromophenol blue reached the bottom of the gel. Once electrophoresis was over, proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Amersham Pharmacia Biotech Co., Ltd, Tokyo, Japan) using a wet-transferring method (100 V; 90 min; 4°C). Then PVDF membrane was detached, subsequently, samples were sealed with 5% skim milk and incubated with shaking at room temperature for 1-2 h. Primary antibodies: mouse anti-human SFRP1, Wnt and β-catenin, bought from Cell Signaling Technologies (Beverly, MA, USA), were added and kept at 4°C overnight. They were washed with tris buffer solution tween (TBST) (3 times × 10 min). The secondary antibodies were added, and samples were incubated at room temperature for 1 h on a rotating platform, followed by TBST washing (3 times × 10 min). The chemiluminescence was visualized on X-ray film. Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) was used as internal reference.
Luciferase assay
The analysis for target genes of miR-27a was performed on the biological website microRNA.org. The potential target gene of miR-27a was predicted, and finally the sequence of fragments containing the binding site was obtained. And the reporter gene was isolated with cell lysis buffer, and centrifugated for 3-5 min at 10000-15000 g, and then supernatant was collected. Dual Luciferase Reporter Gene Assay Kit (Firefly luciferase and Ranilla luciferase) was added to detect the reporter gene at room temperature. The substrate (100×) of Renilla luciferase was preserved in an ice box for later trials. Certain Renilla luciferase assay kit (100 microliters/sample) was mixed with its substrate of Renilla luciferase (1:100). Fluorometer was operated in accordance with the instruction, and the measurement period was 10 s with the time interval of 2 s. The relative light unit (RLU) value (20-100 microliters of each sample) was detected with an addition of Firefly luciferase assay kit (100 microliters), with the cell lysis buffer of reporter gene as blank control. Then the RLU value was again measured with an addition of prepared Renilla luciferase assay kit (100 microliters), and with Renilla luciferase as the internal reference. Comparisons of the reporter gene activities among different samples were performed with the ratio of the RLU value detected by Firefly luciferase to the RLU value detected by Renilla luciferase.
Cell counting kit-8 (CCK-8) assay
The MGC803 cells were inoculated into 96-well plates at a density of 2 × 103 cells/ml, and 100 μl culture medium was added into each well. At first, the number of cells in each well should be same, and this time point was set as 0 h. After cultured for 24 h, 48 h, 72 h and 96 h, CCK8 reagent (10 μl; CCK8/medium = 1:10) was added in each well (Beyotime Biotechnology Co., Shanghai, China), followed by cell incubation for 1-2 h at 37°C. Microplate reader was used to determine optical density (OP) value at the wavelength of 450 nm/630 nm. Three parallel holes were set for each group. The average value was obtained with three-time repeated tests.
Transwell assay
After 48-h transfection, cells were digested with trypsin. Cell suspension was prepared and cells numbers were counted. Then 1 × 105 cells were seeded into Transwell upper chamber (Corning Inc., Corning, N.Y., USA). The upper chamber was added with serum-free medium, and lower chamber was added with culture medium containing 10% fetal bovine serum (FBS). Cells were cultured in an incubator for 24 h, and then non-migrated cells were removed from the chamber using a cotton swab. Subsequently, migrated cells were fixed with 2% paraformaldehyde for 20 min, stained for 10 min with 1% crystal violet dye, followed by three times PBS washing. Migrated cells were observed under a high power microscope, and photographed. Six fields were selected for each sample. The number of cells across the polycarbonate membrane was counted, and served as the migration ability of cells.
Matrigel (Becton, Dickinson and Company, NJ, USA) was dissolved at 4°C overnight. After diluted (1:3) in serum-free medium, the Matrigel was added into the Transwell upper chamber, and then it was maintained in an incubator for 30 min. After inoculated with 1 × 105 cell suspension into the upper chamber, the upper chamber was added with serum-free medium, and lower chamber was added with culture medium containing 10% FBS. The number of cells in each group across the Matrigel was counted and used as the invasion ability of cells.
Dual-luciferase reporter assay system
Promega Dual-Luciferase Reporter Assay System, TOP/FLASH and FOP/FLASH reporter gene system were also applied to detect the Wnt signaling pathway, where β-catenin migrated into the nucleus to form a complex with the transcription factor TCF/LEF that regulate the transcription of downstream gene expressions. The cultured cells were seeded into 24-well plates for 12-24 h, till the cells reached 80%-90% confluency. The negative control plasmid, miR-27a inhibitor plasmid, miR-27a mimics plasmid, miRNA-27a inhibitor + SFRP1 siRNA plasmid, miR-27a mimics + SFRP1 overexpression plasmid were separately combined with TOP/flash, FOP/flash and Renilla luciferase plasmid (pSV40) and used for cell co-transfection. Dual luciferase reporter gene assay kit was used to test the fluorescence value, and the ratio of TOP/FOP was obtained. The fluorescence values of cells transfected with TOP/flash and Renilla luciferase plasmids were expressed as F(TOP) and R(TOP), respectively. The fluorescence values of cells transfected with FOP-flash and Renilla luciferase plasmids were expressed as F(FOP) and R(FOP), respectively. TOP/FOP = [F(TOP)/R(TOP)]/[F(FOP)/R(FOP)]. All tests were repeated for five times.
Statistical analysis
SPSS 21.0 statistical software (SPSS Inc., Chicago, IL, USA) was applied to for data analysis. Measurement data were expressed as mean ± standard deviation (mean ± SD). Enumeration data were expressed as case number. And t test was used for comparisons of measurement data among groups. The chi-square (χ2) test was applied in comparisons of enumeration data among groups. P < 0.05 was considered statistically significant.
Results
Transfection efficiency among the five groups
After cell transfection, the transfection efficiency observed under the fluorescence microscope was shown as Figure 1. Strong fluorescence signal was observed for all five groups after transfection. When compared with NC group, no significant difference of fluorescence signal was observed in the other four groups (the miR-27a inhibitor group, the miR-27a mimics group, the miR-27a inhibitor + SFRP1 siRNA group and the miR-27a mimics + SFRP1 overexpression group), suggesting a successful transfection.
Figure 1.

Transfection efficiency among the five groups.
MiR-27a and SFRP1 mRNA and protein expressions in GC tissues and cells
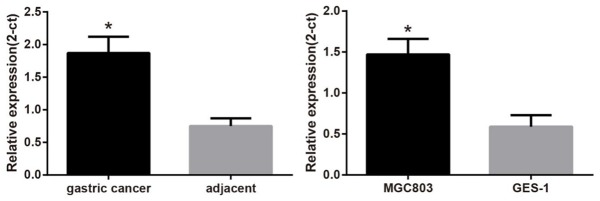
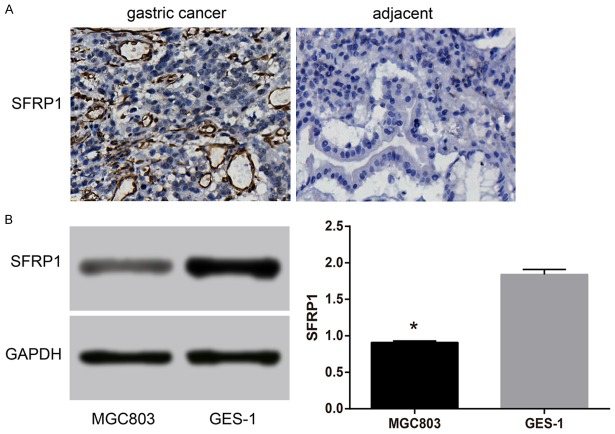
The qRT-PCR results showed that compared with the adjacent normal tissues, miR-27a mRNA expression was significantly increased in GC tissues (P < 0.05) (Figure 2). MiR-27a protein expression was closely associated with lymph node metastasis, degree of tumor differentiation and tumor node metastasis (TNM) staging of GC patients (all P < 0.05), but it was not associated with age and gender of GC patients (both P > 0.05) (Table 1). Meanwhile, human GC cell line (MGC803) also showed a higher mRNA expression of miR-27a in comparison to normal human gastric mucosal cells (GES-1) (Figure 2). The results indicated that SFRP1 protein expression was significantly decreased in GC tissues than that in adjacent normal tissue (P < 0.05) (Figure 3A). The SFRP1 protein expression was closely related to degree of tumor differentiation and gender of patients with GC (both P < 0.05), but was not related to age, lymph node metastasis and TNM staging of patients with GC (all P > 0.05) (Table 1). The SFRP1 protein expression in MGC803 cells was also markedly lower than that in GES-1 cells (P < 0.05) (Figure 3B).
Figure 2.
Comparisons of miR-27a mRNA expression between GC and normal adjacent tissues, between GC and normal gastric mucosa cell line. Note: miR-27a, microRNA-27a; GC, gastric cancer. *, P < 0.05, compared with adjacent normal tissue or normal gastric mucosa cell lines.
Table 1.
Associations of miR-27a and SFRP1 protein expressions with clinicopathologic features of GC patients
| Clinicopathologic feature | Case | MiR-27a | P | SFRP1 | P | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Positive | Negative | Positive rate | |||||
| Age | 0.107 | 0.542 | |||||
| < 60 | 189 | 1.91 ± 0.24 | 135 | 54 | 71.43% | ||
| ≥ 60 | 84 | 1.86 ± 0.22 | 63 | 21 | 75.00% | ||
| Gender | 0.374 | 0.019 | |||||
| Male | 147 | 1.88 ± 0.24 | 98 | 49 | 66.67% | ||
| Female | 126 | 1.91 ± 0.23 | 100 | 26 | 79.37% | ||
| Lymph node metastasis | 0.003 | 0.023 | |||||
| Yes | 168 | 1.93 ± 0.23 | 130 | 38 | 77.38% | ||
| No | 105 | 1.84 ± 0.24 | 68 | 37 | 64.76% | ||
| Tumor differentiation | 0.01 | 0.175 | |||||
| Moderate-high | 120 | 1.85 ± 0.26 | 92 | 28 | 76.67% | ||
| Low-non | 153 | 1.93 ± 0.20 | 106 | 47 | 69.28% | ||
| TNM stage | < 0.01 | 0.174 | |||||
| I-II | 142 | 1.84 ± 0.24 | 108 | 34 | 76.06% | ||
| III-IV | 131 | 1.96 ± 0.21 | 90 | 41 | 68.70% | ||
Note: miR-27a, microRNA-27a, SFRP1, secreted frizzled-related protein 1, GC, gastric cancer; TNM, tumor-node-metastasis.
Figure 3.
Comparions of SFRP1 protein expression between GC and normal adjacent tissues, between GC and normal gastric mucosa cell line. Note: A: SFRP1 protein expression in GC and normal adjacent tissues detected by IHC. Viewed under a microscope (400×), with a proportional scale of 1:50 μm. B: Comparison of SFRP1 protein expression between GC cell line (MGC803) and normal gastric mucosal cell line (GES-1) with gray-scale analysis. GAPDH was used as an internal reference. miR-27a, microRNA-27a; GC, gastric cancer; SFRP1, secreted frizzled-related protein 1; IHC, immunohistochemistry; GAPDH, glyceraldehyde 3-phosphate dehydrogenase. *, P < 0.05, compared with normal gastric mucosal cell line (GES-1).
MiR-27a directly targets SFRP1
The online bioinformatic analysis software predicted the target binding site of SFRP1 and miR-27a in 3’-UTR, and the corresponding sequences were shown in Figure 4A. And the results of the luciferase assay exhibited that the expression of miR-27a was down-regulated in the miR-27a inhibitor group, and there was a higher luciferase activity in the miR-27a inhibitor group when compared with the NC group (Figure 4B). Therefore, miR-27a might directly target SFRP1 and regulate its expression.
Figure 4.
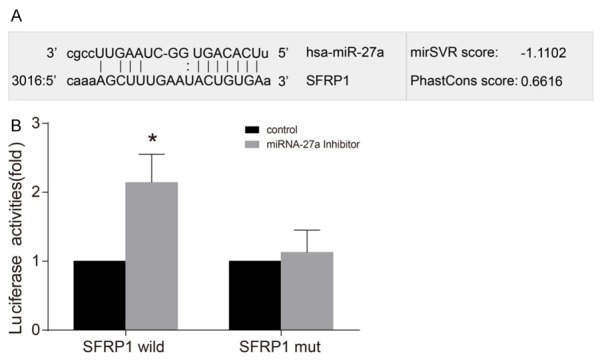
The prediction of miR-27a directly targeting SFRP1 by Luciferase assay. Note: A: The binding site of miR-27a and SFRP1 in the 3’UTR region; B: Effect of miR-27a on the luciferase activity of SFRP1 analyzed by the luciferase assay; *, P < 0.05 compared with the NC group. miR-27a, microRNA-27a; SFRP1, secreted frizzled-related protein 1; 3’UTR, 3’ untranslated region; NC, negative control group.
MiR-27a targets SFRP1 to activate Wnt/β-catenin signaling pathway
Compared with NC group, miR-27a protein and mRNA expressions were up-regulated while SFRP1 protein and mRNA expressions were significantly down-regulated (both P < 0.05), and β-catenin, total protein of Wnt, p-β-catenin, and p-Wnt protein expressions were significantly up-regulated in the miR-27a mimics group (all P < 0.05). When miR-27a protein and mRNA expressions were down-regulated, SFRP1 protein and mRNA expressions were up-regulated, and β-catenin, Wnt, p-β-catenin, and p-Wnt protein expressions were significantly down-regulated (all P < 0.05) (Figure 5). Thus, miR-27a might target SFRP1 to activate Wnt/β-catenin signaling pathway.
Figure 5.
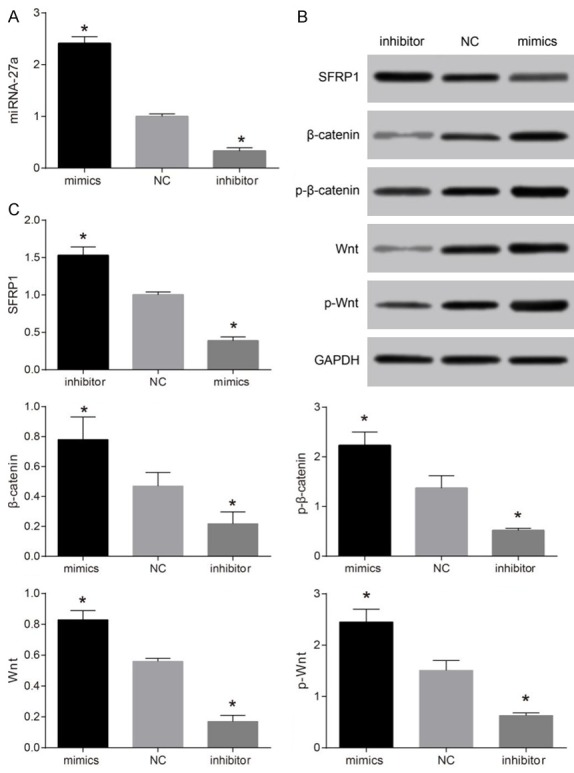
Comparisons of miR-27a mRNA expression, and SFRP1, β-catenin, total protein of Wnt, p-β-catenin, and p-Wnt protein expressions among the NC, miR-27a mimics and miR-27a inhibitor groups. Note: A: Comparion of miR-27a mRNA expressions among the NC, miR-27a mimics and miR-27a inhibitor groups detected by qRT-PCR; B: SFRP1, β-catenin, total protein of Wnt, p-β-catenin, and p-Wnt protein expressions among the NC, miR-27a mimics and miR-27a inhibitor groups detected by Western blotting, GAPDH was used as internal reference; C: Comparions of SFRP1, β-catenin, total protein of Wnt, p-β-catenin, and p-Wnt protein expressions among the NC, miR-27a mimics and miR-27a inhibitor groups. *, P < 0.05, compared with the NC group; miR-27a, microRNA-27a; SFRP1, secreted frizzled-related protein 1; qRT-PCR, quantitative real-time polymerase chain reaction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; NC, negative control group.
The effect of MiR-27a on the proliferation of GC cells
The results showed that the miR-27a mimics group had an elevated proliferation of GC cells than the NC group (P < 0.05). And the proliferation of GC cells was declined in the miR-27a mimics + SFRP1 overexpression group when compared with the miR-27a mimics group (P < 0.05), but there was no evident difference of the proliferation of GC cells between the miR-27a mimics + SFRP1 overexpression group and the miR-27a mimics group. However, when compared with the NC group, cell proliferation was significantly decreased in the miR-27a inhibitor group (P < 0.05). The miR-27a inhibitor + SFRP1 siRNA group exhibited an increased cell proliferation than the miR-27a inhibitor group (P < 0.05), but it was similar to that in the NC group (Figure 6).
Figure 6.
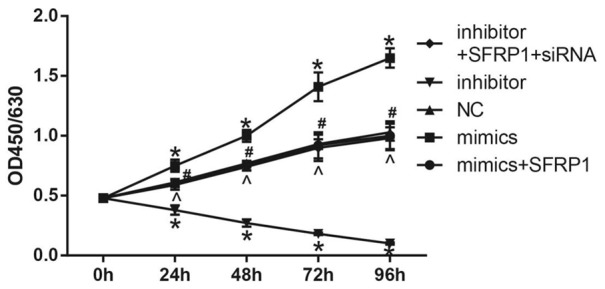
Comparison of the proliferation ability of GC cells among the five groups. Note: *, P < 0.05, compared with the NC group; #, P < 0.05 compared with the miR-27a mimics group; ^, P < 0.05, compared with the miR-27a inhibitor group. GC, gastric cancer; miR-27a, microRNA-27a; NC, negative control group.
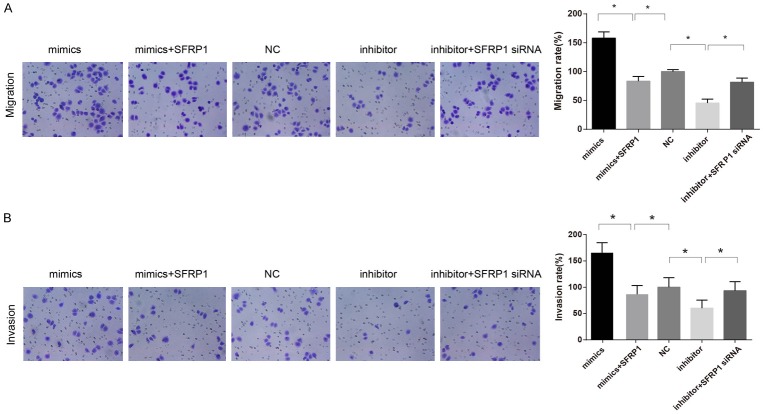
The effect of MiR-27a on the migration and invasion of GC cells
In contrast to the NC group, migration and invasion of GC cells were evidently increased in the miR-27a mimics group (both P < 0.05), but those in the miR-27a mimics + SFRP1 overexpression group were remarkably lower than the miR-27a mimics group (both P < 0.05). Cell migration and invasion were significantly decreased in the miR-27a inhibitor group in comparison to the NC group (both P < 0.05), while those were elevated in the miR-27a inhibitor + SFRP1 siRNA group than the miR-27a inhibitor group (P < 0.05) (Figure 7).
Figure 7.
Comparisons of the migration and invasion abilities of GC cells among the five groups. Note: A: The migration ability of GC cells among the five groups detected by Transwell assay, and comparison of the number of cells across the polycarbonate membrane among the five groups; B: The invasion ability of GC cells among the five groups detected by Transwell assay, and comparison of the number of cells in each group across the Matrigel among the five groups. *, P < 0.05, compared with the NC group. GC, gastric cancer; NC, negative control group.
The effect of MiR-27a on Wnt/β-catenin signaling pathway
The TOP/FOP value in the miR-27a mimics group was higher than that in the NC group, while the TOP/FOP value in the miR-27a mimic + overexpression group was lower than that in the miR-27a mimics group (both P < 0.05). However, the miR-27a inhibitor group presented a markedly down-regulated TOP/FOP value than the NC group (P < 0.05), while the miR-27a inhibitor + SFRP1 siRNA group had an up-regulated TOP/FOP value when compared with the miR-27a inhibitor group (P < 0.05) (Figure 8). Thus, the Wnt/β-catenin signaling pathway might be activated by miR-27a, while it can be inhibited by the SFRP1 overexpression, suggesting that miR-27a might activate Wnt/β-catenin signaling pathway via targeting SFRP1.
Figure 8.
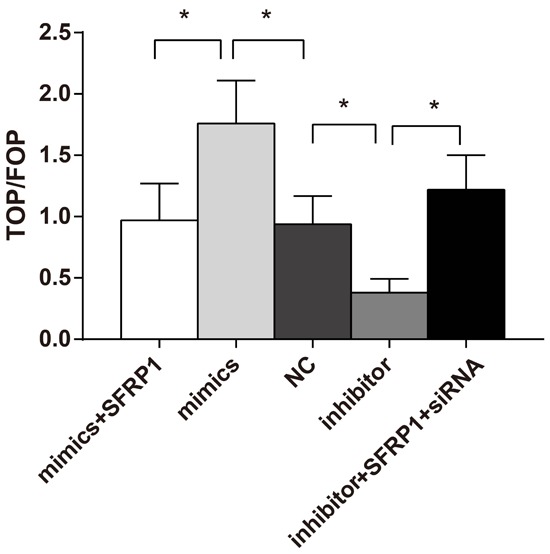
Comparisons of TOP/FOP value among the five groups with the dual-luciferase reporter assay system. Note: *. P < 0.05, compared with the NC group. NC, negative control group.
Discussion
It had been reported that, by base pairing of mRNA with target genes in 3’UTR, mRNA is inhibited for translation, resulting in post-transcriptional gene silencing [17]. Many studies on GC find among the miRNAs, miR-27a is highly expressed in GC, suggesting that miR-27a can be used as a diagnostic marker of GC [18,19]. Song et al. demonstrates that 16 kinds of miRNAs (including miR-27a) are up-regulated in plasma of GC patients [20]. In addition, IHC shows that SFRP1 in GC tissue is significantly reduced compared to adjacent normal tissues, which conforms to the low or no expression of SFRP1 in breast cancer and colorectal cancer [21,22]. SFRP1 gene promoter is hyper-methylated in GC, which may result in the primary mechanism for gene silencing, with SFRP1 expression missing or down-regulated. As a tumor gene suppressor, SFRP1 exerts its suppression role through the promotion of splenic tumor cell apoptosis, and researchers find that miR-27a has potential binding sites on SFRP1 mRNA in the region of 3’UTR [23]. In a variety of physiological and pathological processes, miRNA plays an important role in targeting specific genes via mediating target genes negatively [24].
The results showed that miR-27a was highly expressed in GC cells, SFRP1 was significantly down-regulated, β-catenin, total protein of Wnt, p-β-catenin, and p-Wnt were significantly up-regulated. Thus Wnt/β-catenin pathway might be activated with the upregulation of miR-27a. Meanwhile, with simultaneous SFRP1 overexpression, the phenotype is reversed and Wnt pathway is inhibited. Wnt signaling pathway regulates embryonic development, cell differentiation and cell proliferation, and it is abnormally activated in a variety of tumors [25-27]. SFRP family, by directly binding to Wnt molecule in CaaBD region, can lead to abnormal Wnt signaling pathway downstream molecule [11]. SFRP1 could bind with receptor and compete with Wnt, or direct combine with Wnt protein, thereby blocking the Wnt signaling pathway [28]. In recent years, more and more scholars tend to believe that epigenetic changes of SFRP1 and down-regulation of SFRP1 could lead to the abnormal activation of the Wnt pathway even cause cancer [29]. Abnormal expression of multiple genes mediated Wnt/β-catenin signal activation in GC and ectopic expression of these genes can often inhibit the Wnt/β-catenin signaling and suppress tumor growth [30]. This study also found that miR-27a was highly expressed in the GC tissues and cells, which further targeted SFRP1 to activate Wnt/β-catenin pathway.
Relationship between miRNAs and cancer reveals that its important role in tumor occurrence and development, mainly through regulation of expression of cellular oncogenes, tumor suppressor genes or metastasis-associated genes. It is not only involved in the regulation of tumor cell proliferation and apoptosis, but also in regulation of tumor invasion and metastasis [31,32]. The experimental results showed that compared with the NC group, the proliferation, migration and invasion of GC cells were remarkably increased in the miR-27a group, but these were declined in the miR-27a mimics + SFRP1 overexpression group. The proliferation, migration and invasion of GC cells were down-regulated by inhibiting miR-27a, and were up-regulated by silencing SFRP1. These results suggested that miR-27a might promote proliferation, migration and invasion of GC cells through targeting SFRP1. The mechanism of miR-27a promoting the proliferation of laryngeal carcinoma cells is that the combination of miR-27a and it target gene affects 3’ untranslated coding region of the prohibition gene, therefore, miR-27a owns the function of oncogenes [33]. Guo D et al. report that gene expression of SFRP1 could be knocked down by miR-27a, and tumor cell proliferation could be inhibited, thereby leading to induction of apoptosis and differentiation [34]. In this study, Transwell assay is used to detect cell migration and invasion. The results indicated that, the overexpression of miR-27a led to the improved cell migration and invasion. And the specific mechanism might be that miR-27a target SFRP1 to activate Wnt/β-catenin pathway, thereby regulate cell activities of GC cells.
In summary, our findings demonstrated that miR-27a was highly expressed in GC tissues and cells, and it might promote cell proliferation, migration and invasion by targeting SFRP1 via the activation of Wnt/β-catenin signaling pathway. However, the specific mechanism of miR-27a targets SFRP1 to activate the Wnt/β-catenin signaling pathway in GC remains a future topic which needs to be further elucidated.
Acknowledgements
This study was supported by Natural Science Foundation of Jiangxi province of China (No. 20151BAB205043). We would like to show our gratitude for helpful comments on this paper received from our reviewers.
Disclosure of conflict of interest
None.
References
- 1.Nagini S. Carcinoma of the stomach: A review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J Gastrointest Oncol. 2012;4:156–169. doi: 10.4251/wjgo.v4.i7.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan X, Wang K, Lu W, Qin W, Cui D, He J. CdSe/ZnS Quantum Dot-Labeled Lateral Flow Strips for Rapid and Quantitative Detection of Gastric Cancer Carbohydrate Antigen 72-4. Nanoscale Res Lett. 2016;11:138. doi: 10.1186/s11671-016-1355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Figueiredo C, Garcia-Gonzalez MA, Machado JC. Molecular pathogenesis of gastric cancer. Helicobacter. 2013;18(Suppl 1):28–33. doi: 10.1111/hel.12083. [DOI] [PubMed] [Google Scholar]
- 4.Matsuda K, Tanikawa C, Nakamura Y. [Possible role of genetic factors on reduced risk for gastric cancer among duodenal ulcer patients] . Nihon Rinsho. 2013;71:1491–1496. [PubMed] [Google Scholar]
- 5.Hou IC, Amarnani S, Chong MT, Bishayee A. Green tea and the risk of gastric cancer: epidemiological evidence. World J Gastroenterol. 2013;19:3713–3722. doi: 10.3748/wjg.v19.i24.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 7.Gu YP, Yuan QY, Zhang H, Wang CJ, Zhou F. Association between five common polymorphisms in microRNA genes and the risk of gastric cancer: a meta-analysis. Genet Mol Res. 2015;14:8375–8387. doi: 10.4238/2015.July.28.4. [DOI] [PubMed] [Google Scholar]
- 8.Scheibner KA, Teaboldt B, Hauer MC, Chen X, Cherukuri S, Guo Y, Kelley SM, Liu Z, Baer MR, Heimfeld S, Civin CI. MiR-27a functions as a tumor suppressor in acute leukemia by regulating 14-3-3theta. PLoS One. 2012;7:e50895. doi: 10.1371/journal.pone.0050895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao Y, Chen Z, Guo Y, Feng Y, Li Z, Han W, Wang J, Zhao W, Jiao Y, Li K, Wang Q, Wang J, Zhang H, Wang L, Yang W. Tumor suppressor microRNA-27a in colorectal carcinogenesis and progression by targeting SGPP1 and Smad2. PLoS One. 2014;9:e105991. doi: 10.1371/journal.pone.0105991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito T, Mitomi H, Imamhasan A, Hayashi T, Mitani K, Takahashi M, Kajiyama Y, Yao T. Downregulation of sFRP-2 by epigenetic silencing activates the beta-catenin/Wnt signaling pathway in esophageal basaloid squamous cell carcinoma. Virchows Arch. 2014;464:135–143. doi: 10.1007/s00428-014-1538-1. [DOI] [PubMed] [Google Scholar]
- 11.Kang P, Wan M, Huang P, Li C, Wang Z, Zhong X, Hu Z, Tai S, Cui Y. The Wnt antagonist sFRP1 as a favorable prognosticator in human biliary tract carcinoma. PLoS One. 2014;9:e90308. doi: 10.1371/journal.pone.0090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwai S, Yonekawa A, Harada C, Hamada M, Katagiri W, Nakazawa M, Yura Y. Involvement of the Wnt-beta-catenin pathway in invasion and migration of oral squamous carcinoma cells. Int J Oncol. 2010;37:1095–1103. doi: 10.3892/ijo_00000761. [DOI] [PubMed] [Google Scholar]
- 13.Kim SS, Cho HJ, Lee HY, Park JH, Noh CK, Shin SJ, Lee KM, Yoo BM, Lee KJ, Cho SW, Cheong JY. Genetic polymorphisms in the Wnt/beta-catenin pathway genes as predictors of tumor development and survival in patients with hepatitis B virus-associated hepatocellular carcinoma. Clin Biochem. 2016;49:792–801. doi: 10.1016/j.clinbiochem.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Mertens-Talcott SU, Zhang S, Kim K, Ball J, Safe S. MicroRNA-27a Indirectly Regulates Estrogen Receptor {alpha} Expression and Hormone Responsiveness in MCF-7 Breast Cancer Cells. Endocrinology. 2010;151:2462–2473. doi: 10.1210/en.2009-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 16.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, Spears PA, Vance GH, Viale G, Hayes DF American Society of Clinical Oncology; College of American Pathologists. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 17.Chou J, Wang B, Zheng T, Li X, Zheng L, Hu J, Zhang Y, Xing Y, Xi T. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun. 2016;472:262–269. doi: 10.1016/j.bbrc.2016.02.102. [DOI] [PubMed] [Google Scholar]
- 18.Liu D, Sun Q, Liang S, Xu L, Luo X, Zhao X, Wang X, Yang L. MicroRNA-27a inhibitors alone or in combination with perifosine suppress the growth of gastric cancer cells. Mol Med Rep. 2013;7:642–648. doi: 10.3892/mmr.2012.1191. [DOI] [PubMed] [Google Scholar]
- 19.Park JL, Kim M, Song KS, Kim SY, Kim YS. Cell-Free miR-27a, a Potential Diagnostic and Prognostic Biomarker for Gastric Cancer. Genomics Inform. 2015;13:70–75. doi: 10.5808/GI.2015.13.3.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li JY, Yuasa Y, Kang D, Kim YS, You WC. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One. 2012;7:e33608. doi: 10.1371/journal.pone.0033608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernemann C, Hülsewig C, Ruckert C, Schäfer S, Blümel L, Hempel G, Götte M, Greve B, Barth PJ, Kiesel L, Liedtke C. Influence of secreted frizzled receptor protein 1 (SFRP1) on neoadjuvant chemotherapy in triple negative breast cancer does not rely on WNT signaling. Mol Cancer. 2014;13:174. doi: 10.1186/1476-4598-13-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Xing Y, Liang C, Hu L, Xu F, Chen Y. Crucial microRNAs and genes of human primary breast cancer explored by microRNA-mRNA integrated analysis. Tumour Biol. 2015;36:5571–5579. doi: 10.1007/s13277-015-3227-3. [DOI] [PubMed] [Google Scholar]
- 23.Yang S, Wang K, Qian C, Song Z, Pu P, Zhang A, Wang W, Niu H, Li X, Qi X, Zhu Y, Wang Y. A predicted miR-27a-mediated network identifies a signature of glioma. Oncol Rep. 2012;28:1249–1256. doi: 10.3892/or.2012.1955. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release. 2013;172:962–974. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Chen J, He J, Li J, Shi J, Cho WC, Liu X. Wnt signaling as potential therapeutic target in lung cancer. Expert Opin Ther Targets. 2016;20:999–1015. doi: 10.1517/14728222.2016.1154945. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Shi H, Fan Q, Sun X. Cyclin Y regulates the proliferation, migration, and invasion of ovarian cancer cells via Wnt signaling pathway. Tumour Biol. 2016;37:10161–75. doi: 10.1007/s13277-016-4818-3. [DOI] [PubMed] [Google Scholar]
- 28.Ou Y, Jing G, Liu J, Gao S, Cheng Z, Dong X. [T Cell Factor 4, beta-catenin and SFRP1 Expression of Wnt Signaling Pathway in Colorectal Carcinoma and the Prognosis] . Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2015;32:854–861. [PubMed] [Google Scholar]
- 29.Eskander RN, Ali S, Dellinger T, Lankes HA, Randall LM, Ramirez NC, Monk BJ, Walker JL, Eisenhauer E, Hoang BH. Expression Patterns of the Wnt Pathway Inhibitors Dickkopf3 and Secreted Frizzled-Related Proteins 1 and 4 in Endometrial Endometrioid Adenocarcinoma: An NRG Oncology/Gynecologic Oncology Group Study. Int J Gynecol Cancer. 2016;26:125–132. doi: 10.1097/IGC.0000000000000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiurillo MA. Role of the Wnt/beta-catenin pathway in gastric cancer: An in-depth literature review. World J Exp Med. 2015;5:84–102. doi: 10.5493/wjem.v5.i2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanokura M, Banno K, Iida M, Irie H, Umene K, Masuda K, Kobayashi Y, Tominaga E, Aoki D. MicroRNAS in endometrial cancer: recent advances and potential clinical applications. EXCLI J. 2015;14:190–198. doi: 10.17179/excli2014-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou R, Zhou X, Yin Z, Guo J, Hu T, Jiang S, Liu L, Dong X, Zhang S, Wu G. Tumor invasion and metastasis regulated by microRNA-184 and microRNA-574-5p in small-cell lung cancer. Oncotarget. 2015;6:44609–44622. doi: 10.18632/oncotarget.6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian Y, Fu S, Qiu GB, Xu ZM, Liu N, Zhang XW, Chen S, Wang Y, Sun KL, Fu WN. MicroRNA-27a promotes proliferation and suppresses apoptosis by targeting PLK2 in laryngeal carcinoma. BMC Cancer. 2014;14:678. doi: 10.1186/1471-2407-14-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo D, Li Q, Lv Q, Wei Q, Cao S, Gu J. MiR-27a targets sFRP1 in hFOB cells to regulate proliferation, apoptosis and differentiation. PLoS One. 2014;9:e91354. doi: 10.1371/journal.pone.0091354. [DOI] [PMC free article] [PubMed] [Google Scholar]