Abstract
Increasing evidence indicates that long non-coding RNAs (lncRNAs) can act as crucial regulators of tumor progression. In the present study, UXT-AS1 was found to be significantly upregulated in colorectal cancer (CRC) and high expression levels of UXT-AS1 were significantly associated with poor prognosis in CRC patients. In addition, upregulation of UXT-AS1 resulted in inhibition of cell apoptosis and the promotion of cell proliferation. Moreover, by regulating the alternative splicing of UXT, upregulation of UXT-AS1 decreased the UXT1 transcript which promoted cell apoptosis and increased the UXT2 transcript which promoted cell proliferation. Thus, aberrant high expression of UXT-AS1 can promote CRC progression by changing the alternative splicing of UXT from the UXT1 transcript to the UXT2 transcript. In conclusion, our findings suggest that the regulation of CRC progression is by UXT-AS1-induced alternative splicing of UXT, and the expression level of UXT-AS1 may be a potential prognostic biomarker and therapy target in CRC patients.
Keywords: Colorectal cancer, UXT-AS1, UXT, alternative splicing, tumor progression
Introduction
Colorectal cancer (CRC) is a common disease worldwide, and has one of the highest morbidity and mortality rates of all cancers [1]. As more and more people pursue an unhealthy diet and lifestyle, which are regarded as risk factors, the occurrence of CRC has increased and the morbidity and mortality rates have increased rapidly each year [2]. The primary tumor is unlikely to kill, but tumor progression and metastasis result in mortality [3]. In CRC, over 90% of patienfootts fail treatment and mortality is due to tumor progression and metastatic spread. However, the mechanisms underlying progression of CRC are still poorly understood [4-6].
Recently, it was found that long non-coding RNAs (lncRNAs) play important roles in cancer and help to understand the mechanism of tumor progression, although research on lncRNAs is still in the exploratory stage [7,8]. LncRNA, which is a non-coding RNA of more than 200 nucleotides in length, has been found to play important roles in cell proliferation, cell differentiation and in the regulation of other biological functions, and dysregulation of lncRNAs are associated with many human diseases including cancer [9,10]. According to recent research, lncRNAs can affect the development of cancer as oncogenes or tumor suppressor genes depend on their regulatory functions [11,12]. In cancer, the abnormal structure and expression of lncRNAs, such as mutation, copy number variation, and expression dysregulation can result in disorder of cell function which promotes tumor progression and leads to metastasis [13-15].
In the present study, using RNA microarray chip analysis, we found that the expression of UXT-AS1 was upregulated in CRC tissues compared with paracarcinoma tissues. High expression of UXT-AS1 in CRC tissues was significantly associated with advanced tumor stage. Moreover, in CRC cells, upregulation of UXT-AS1 resulted in inhibition of cell apoptosis and promotion of cell proliferation. In addition, in CRC tissues, the correlation between UXT-AS1 and UXT1 expression was strongly negative but the correlation between UXT-AS1 and UXT2 expression was strongly positive, suggesting that upregulation of UXT-AS1 changed the alternative splicing of UXT from one transcript UXT1 to another transcript UXT2. Consequently, downregulation of UXT1 and upregulation of UXT2, which were induced by aberrant high expression of UXT-AS1, resulted in progression of CRC. These findings demonstrated that UXT-AS1-induced alternative splicing of UXT was involved in the regulation of tumor progression, and UXT-AS1 is a potential biomarker and target for the prognosis and therapy of CRC.
Materials and methods
Cell lines and cell culture
A normal human colonic epithelial cell line, CCD841CoN, and human colonic carcinoma cell lines HT29, HCT116 and SW480 were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum at 37°C in a humidified atmosphere with 5% CO2.
Tissue sample collection
Thirty-seven samples of CRC tissue were obtained during surgery or biopsy from the Cancer Center of Guangzhou Medical University (Guangzhou, China) with informed consent and Institutional Review Board (IRB) permission. The following criteria were met: patients with primary CRC; a histological diagnosis of CRC with at least one measurable lesion; a TNM clinical stage of I to IV. Fresh CRC tissues were immediately snap-frozen and stored in liquid nitrogen until use. All clinical and biological data were available for these samples.
All patients provided written informed consent. This study was approved by the ethics committee of Cancer Center of Guangzhou Medical University (Approval no. (2014) 59).
RNA extraction and RT-PCR
Total RNA was extracted by Trizol (Invitrogen, USA) and used for cDNA synthesis according to the manufacturer’s instructions. cDNA was then used for quantitative real-time RT-PCR (reverse transcription polymerase chain reaction) using the SYBR Green Real-time PCR Master Mix Kit (Toyobo, Japan) on an ABI ViiATM7Dx Real-Time PCR System (Life Technologies, USA). The expression level of mRNA was normalized using GAPDH.
The mRNA RT-PCR primers were designed as follows:
UXT-AS1: Forward 5’-CCAGCAAGAATGGGCCTTTG-3’; Reverse 5’-AGGGCTTAACTTGCTTGGGT-3’. UXT1: Forward 5’-TGAACTGTGTGTGAGGCCAA-3’; Reverse 5’-TAGCTTCCTGGAGTCGCTCA-3’. UXT2: Forward 5’-CGGCCTGAGGAGCCCAT-3’; Reverse 5’-TGTTGCTGAGCTCTGTGAGG-3’. GAPDH: Forward 5’-AGAAGGCTGGGGCTCATTTG-3’; Reverse 5’-AGGGGCCATCCACAGTCTTC-3’.
Western blotting
Total proteins were extracted from the cells and loaded onto 15% SDS-PAGE for analysis. The primary antibody was rabbit polyclonal anti-UXT1 or anti-UXT2 (Cell Signaling USA, 1:1000 dilution). The secondary antibody was goat-anti-rabbit IgG conjugated with HRP (horseradish peroxidase) (1:1000 dilution). The bound antibodies were detected using the ECL Plus Western Blotting Detection system (GE Healthcare). β-actin was used as an internal control.
Construction and transfection of expression plasmids and siRNAs
The expression plasmids of UXT-AS1, UXT1 and UXT2 (pCDNA-UXTAS1, pCDNA-UXT1 and pCDNA-UXT2) were constructed by inserting cDNA into the pCDNA3.1 plasmid. The siRNA of UXT-AS1 (si-UXTAS1) was purchased from company. According to the manufacturer’s protocol, 50 ng plasmids or 100 nM siRNAs were transfected into cells using Lipofectamine Transfection Reagent (Invitrogen).
Cell growth assay
The cell proliferation assay was performed as follows: 48 hours after the transfection of plasmids and siRNAs, the medium was removed and 90 µL of medium plus 10 µL CCK-8 solution were added to each well of the plate. The plates were then incubated for 3-6 hours in an incubator. The absorbance at 450 nm wavelength was measured using an automated reader (Tecan, Switzerland).
Cell apoptosis assay
Cell apoptosis was detected using the Annexin V-FITC apoptosis detection kit (BD, USA). Forty-eight hours after the transfection of plasmids and siRNAs, the cells were collected and fixed in 70% ethanol overnight at 4°C. Subsequently, single-cell suspensions were labeled with Annexin V-FITC and propidium iodide and analyzed using flow cytometry. The cell apoptosis ratio was analyzed using FlowJo software.
Colony formation assay
The colony formation assay was performed as follows: 24 hours after transfection of plasmids and siRNAs, the cells were trypsinized and plated for clonogenic survival (1000 cells per well in six-well plates). After 7 days of colony formation, the cells were stained with Giemsa and counted using ImageJ software.
Statistical analysis
All values are expressed as the mean ± standard deviation (SD) of at least three separate experiments. Student’s unpaired t-test, the chi-square test and Spearman’s correlation were performed using SPSS 21.0 statistical software (IBM). A two-tailed P value test was used in all analyses, and differences were considered statistically significant if the P value was less than 0.05 (P < 0.05).
Results
UXT-AS1 was upregulated in CRC tissues
Using microarray chip assay, we performed RNA expression profiling in five pairs of CRC samples (carcinoma tissues: C, and paracarcinoma tissues: NC). According to the results of chip analysis, 496 lncRNAs showed statistical differences in the expression of UXT-AS1 between C tissues and NC tissues (fold change > 2 or < 0.5; P < 0.001) (Figure 1A). Interestingly, of the significantly changed lncRNAs, the expression of UXT-AS1 was upregulated in C tissues compared with NC tissues (fold change = 2.236; P < 0.001). Using RT-PCR, we measured the expression level of UXT-AS1 in the five pairs of CRC samples, one normal colonic epithelial cell line CCD841CoN, and three human colonic carcinoma cell lines HT29, HCT116 and SW480. Compared with NC tissues and CCD841CoN cells, the expression of UTX-AS1 was upregulated in C tissues and in HT29, HCT116, and SW480 cells (Figure 1B and 1C). These findings suggested that UXT-AS1 is abnormally upregulated in CRC.
Figure 1.
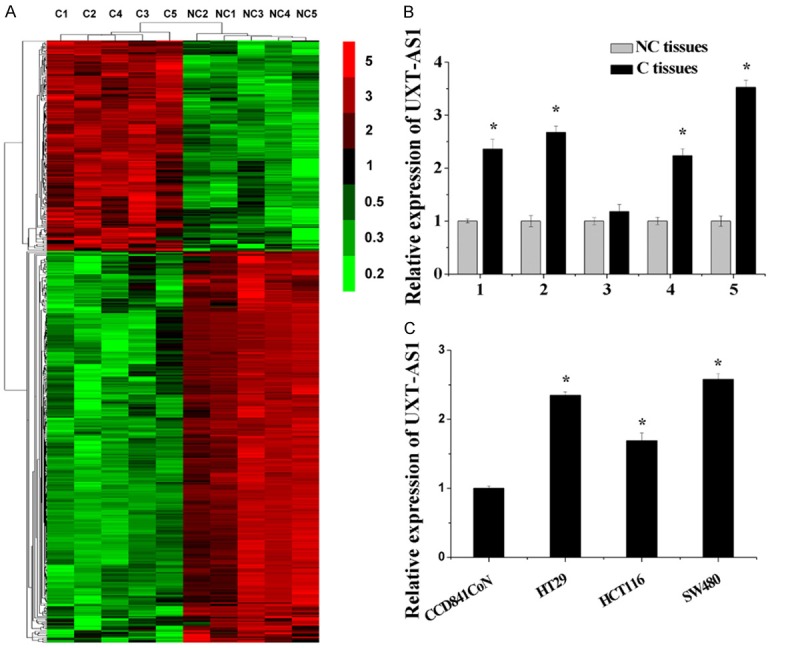
UXT-AS1 was upregulated in CRC. A. Clustering of lncRNAs in five pairs of CRC samples (carcinoma tissues: C and paracarcinoma tissues: NC). The vertical axis corresponds to the expression difference in lncRNAs. The expression levels of lncRNAs are depicted by color variation from red (high expression) to green (low expression) according to the color bar scale. B. Upregulation of UXT-AS1 in microarray experiments was validated by RT-PCR analysis. C. The expression level of UXT-AS1 in one normal colonic epithelial cell line CCD841CoN and three human colonic carcinoma cell lines HT29, HCT116 and SW480. (n = 3, *P < 0.05).
Upregulation of UXT-AS1 indicated poor prognosis of CRC
In order to investigate the relationship between abnormal upregulation of UXT-AS1 and CRC, we analyzed the correlation between the expression levels of UXT-AS1 and clinicopathological factors in CRC patients. The clinical information and demographic characteristics of the 37 CRC patients included in this study are shown in Table 1. Compared with NC tissues, UXT-AS1 was significantly upregulated in C tissues (P = 0.0023) (Figure 2). The clinical correlation between UXT-AS1 and CRC is listed in Table 2. The expression level of UXT-AS1 was significantly associated with the TNM clinical stage and tumor size in CRC patients (P < 0.01). CRC patients with UXT-AS1 upregulation were more likely to have stage III and IV disease (72.0% vs. 16.7%) and large tumor size (84.0% vs. 25.0%) than CRC patients with UXT-AS1 downregulation. Moreover, CRC patients with UXT-AS1 upregulation were more likely to have lymph node metastasis (48.0% vs. 16.7%) and distant metastasis (36.0% vs. 8.3%) than CRC patients with UXT-AS1 downregulation, although the clinical correlation between UXT-AS1 and metastasis was not significant. These data indicated that abnormal upregulation of UXT-AS1 was closely associated with poor prognosis in CRC patients.
Table 1.
Patient characteristics
| Clinicopathological factors | |
|---|---|
| Total | |
| N | 37 |
| Gender, n (%) | |
| Male | 20 (54.1%) |
| Female | 17 (45.9%) |
| Age, years (range) | |
| Mean | 57.9 (34-78) |
| TNM Clinical stage, n (%) | |
| I | 7 (18.9%) |
| II | 10 (27.1%) |
| III | 11 (29.7%) |
| IV | 9 (24.3%) |
Figure 2.
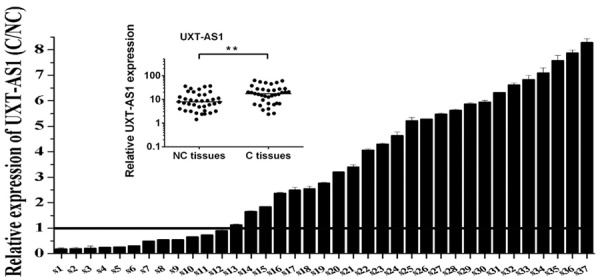
The expression level of UXT-AS1 in 37 CRC tissues. RT-PCR analysis of UXT-AS1 in carcinoma tissues (C) and paracarcinoma tissues (NC). The positions of overexpression are drawn as a solid line. The small picture shows scatter plots of the expression levels of UXT-AS1 in C and NC tissues. (**P < 0.01).
Table 2.
Correlation between the expression of UXT-AS1 and the clinicopathological features of CRC patients
| Clinicopathological factors | Expression of UXT-AS11 | χ2 value | P value | ||
|---|---|---|---|---|---|
|
| |||||
| Downregulation (n = 12) | Upregulation (n = 25) | ||||
| Gender | Male | 6 | 14 | 0.729 | 0.119 |
| Female | 6 | 11 | |||
| Age (years) | < 60 | 8 | 14 | 0.538 | 0.378 |
| ≥ 60 | 4 | 11 | |||
| TNM Clinical stage | I/II | 10 | 7 | 0.001 | 10.012 |
| III/IV | 2 | 18 | |||
| Tumor size (cm) | < 3 | 9 | 4 | < 0.001 | 12.362 |
| ≥ 3 | 3 | 21 | |||
| Invasion depth | T1/T2 | 4 | 4 | 0.229 | 1.448 |
| T3/T4 | 8 | 21 | |||
| Lymph node metastasis | 0 | 10 | 13 | 0.066 | 3.383 |
| 1 | 2 | 12 | |||
| Distant metastasis | 0 | 11 | 16 | 0.076 | 3.139 |
| 1 | 1 | 9 | |||
Effects of UXT-AS1 on cell apoptosis and cell proliferation
Given the abnormal upregulation of UXT-AS1 identified in CRC tissues, several biochemical assays were performed to evaluate the biological effects of UXT-AS1 on CRC cells. Here, we used siRNAs to silence UXT-AS1 and expression plasmids to enhance UXT-AS1 in HCT116 cells (Figure 3A). Cell Counting Kit-8 assays indicated that cell growth was decreased by the downregulation of UXT-AS1 and increased by the upregulation of UXT-AS1 compared with control cells (Figure 3B). Using flow cytometric analysis, cell apoptosis was increased by the downregulation of UXT-AS1 and decreased by the upregulation of UXT-AS1 compared with control cells (Figure 3C). According to colony formation assays, compared with control cells, the colony formation capacity of HCT116 cells was decreased by the downregulation of UXT-AS1 and increased by the upregulation of UXT-AS1 (Figure 3D). Thus, these data indicated that upregulation of UXT-AS1 resulted in inhibition of cell apoptosis and promotion of cell proliferation.
Figure 3.
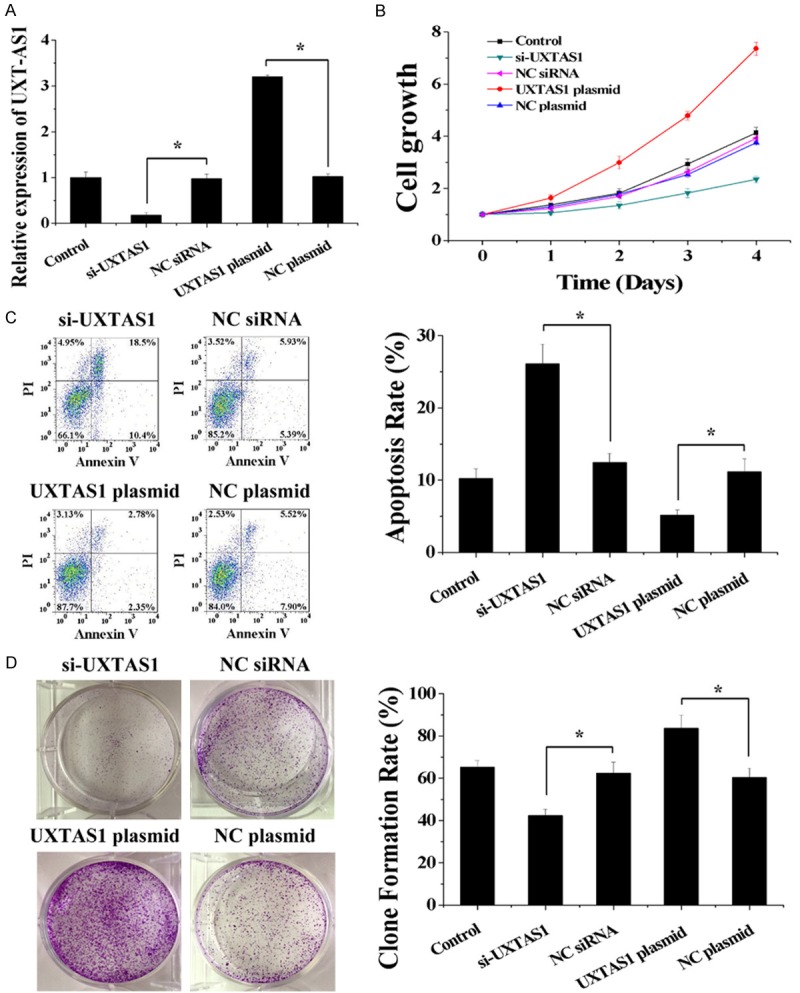
UXT-AS1 regulated cell growth, cell apoptosis and colony formation capacity. A. The expression level of UXT-AS1 was decreased after transfection with si-UXTAS1 and increased after transfection with UXTAS1 plasmid in HCT116 cells. B. Silencing of UXT-AS1 decreased cell growth and overexpression of UXT-AS1 increased cell growth in HCT116 cells. C. Silencing of UXT-AS1 increased cell apoptosis and overexpression of UXT-AS1 decreased cell apoptosis in HCT116 cells. D. Silencing of UXT-AS1 decreased colony formation capacity and overexpression of UXT-AS1 increased colony formation capacity in HCT116 cells. (n = 3, *P < 0.05).
UXT-AS1 regulated the alternative splicing of UXT
UXT-AS1 is a conserved natural antisense transcript corresponding to the 5’ end of UXT which has two alternative splicing isoforms, UXT1 and UXT2. As shown by the RT-PCR and western blotting results, plasmid-induced upregulation of UXT-AS1 decreased the expression of UXT1 and increased the expression of UXT2 (Figure 4A). In contrast, siRNA-induced downregulation of UXT-AS1 increased the expression of UXT1 and decreased the expression of UXT2 (Figure 4A). In order to further confirm the correlation between the expression of UXT-AS1 and the two alternative splicing isoforms of UXT, we also determined the expression of UXT1 and UXT2 in the same 37 pairs of CRC tissues (C tissues) and paracarcinoma tissues (NC tissues). Compared with NC tissues, UXT2 was significantly upregulated in C tissues (P = 0.047), but UXT1 was significantly downregulated in C tissues (P = 0.025) (Figure 4B). Significant negative correlations were observed when the UXT1 expression level was plotted against UXT-AS1 expression level (two-tailed Spearman’s correlation, R = -0.618, P < 0.0001), whereas significant positive correlations were observed between the expression levels of UXT2 and UXT-AS1 (R = 0.532, P = 0.0007) (Figure 4C). Therefore, UXT-AS1 can serve as an “on-and-off switch” to change UXT expression from the UXT1 transcript to the UXT2 transcript, and UXT-AS1-mediated alternative splicing of UXT was associated with CRC development.
Figure 4.
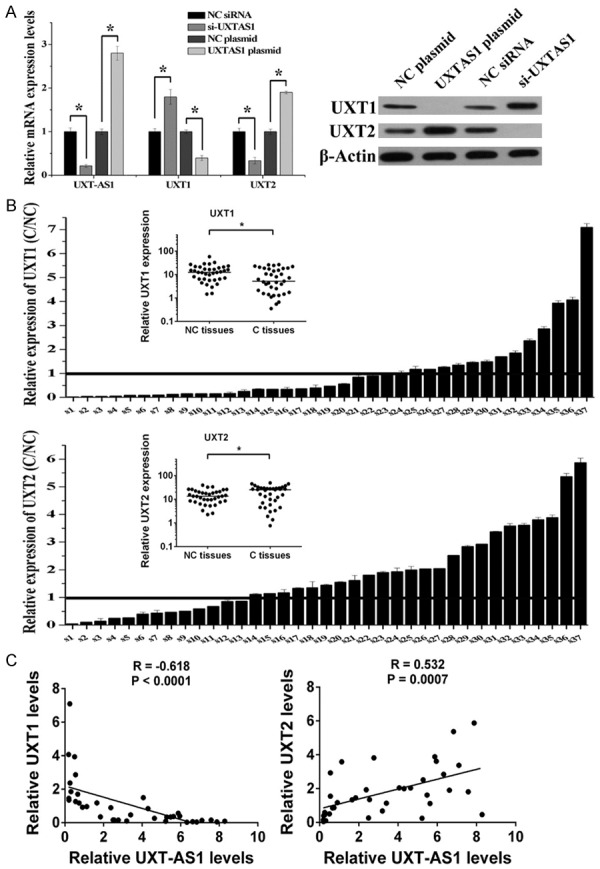
UXT-AS1 induced alternative splicing of UXT. A. Silencing of UXT-AS1 increased UXT1 and decreased UXT2, and overexpression of UXT-AS1 decreased UXT1 and increased UXT2. B. RT-PCR analysis of UXT1 and UXT2 in carcinoma tissues (C) and paracarcinoma tissues (NC). The positions of overexpression are drawn as a solid line. The small picture shows scatter plots of the expression levels of UXT1 and UXT2 in C and NC tissues. C. Correlation between the relative expressions of UXT-AS1 and UXT1, UXT-AS1 and UXT2 in specimens from 37 CRC patients expressed using linear regression (solid line). (n = 3, *P < 0.05).
Finally, given that the alternative splicing of UXT can be regulated by UXT-AS1, several biochemical assays were performed to evaluate the cellular biological effects of UXT1 and UXT2. Here, UXT1 or UXT2 expression plasmids were used to determine the function of UXT1 and UXT2, respectively. In HCT116 cells, overexpression of UXT1 decreased cell growth; however, overexpression of UXT2 increased cell growth (Figure 5A). Moreover, both UXT1 and UXT2 partially reversed the effect of UXT-AS1 on cell growth (Figure 5B). On the one hand, UXT1 but not UXT2 regulated cell apoptosis, as only overexpression of UXT1 resulted in the promotion of cell apoptosis and reversed the effect of plasmid-induced upregulation of UXT-AS1 which inhibited apoptosis (Figure 5C). On the other hand, UXT2 but not UXT1 regulated colony formation capacity, as only overexpression of UXT2 resulted in the promotion of colony formation capacity and reversed the effect of siRNA-induced downregulation of UXT-AS1 which inhibited colony formation capacity (Figure 5D). These findings indicated that downregulation of UXT1 inhibited cell apoptosis and upregulation of UXT2 promoted cell proliferation. Thus, upregulation of UXT-AS1 regulated the progression of CRC by inducing downregulation of UXT1 and upregulation of UXT2.
Figure 5.
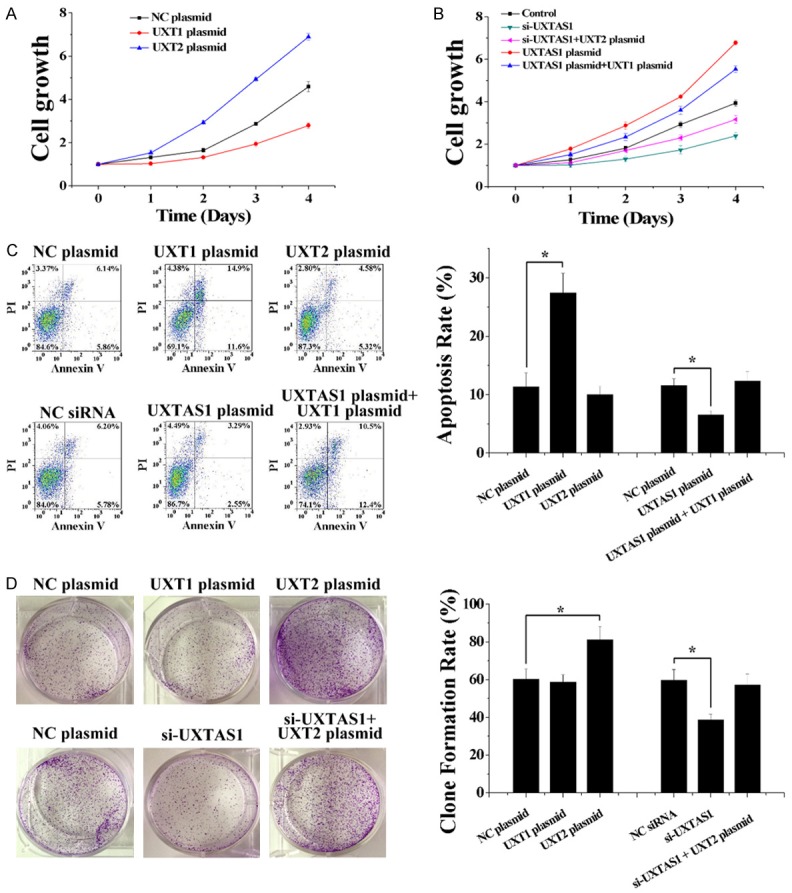
UXT-AS1 regulated cell function by inducing the alternative splicing of UXT. A. Overexpression of UXT1 decreased cell growth and overexpression of UXT2 increased cell growth in HCT116 cells. B. Overexpression of UXT1 and UXT2 reversed the effect of UXT-AS1 on cell growth in HCT116 cells. C. Overexpression of UXT1 increased cell apoptosis and reversed the effect of UXT-AS1 on cell apoptosis in HCT116 cells. D. Overexpression of UXT2 increased colony formation capacity and reversed the effect of UXT-AS1 on colony formation capacity in HCT116 cells. (n = 3, *P < 0.05).
Discussion
Although the molecular mechanism of CRC has been widely studied, it is still not fully understood what determines tumor progression [16,17]. Recently, abnormal expression of lncRNAs has been found to be directly involved in the occurrence, development and prognosis of human cancers, and lncRNAs can affect tumor progression by different molecular mechanisms, such as protein activity and location, gene expression, and genomic imprinting [18,19]. It has also been found that lncRNAs are associated with the pathogenesis of CRC. HOTAIR, MALAT-1 and H19, which have a close relationship with many types of human malignant tumors, also play important roles in cell proliferation, invasion and metastasis of CRC [20-22]. Moreover, more and more novel lncRNAs are also involved in the initiation and progression of CRC [23-25]. In the present study, abnormal upregulation of UXT-AS1 was found to be significantly associated with poor prognosis of CRC, suggesting that UXT-AS1 may act as a novel lncRNA regulator of tumor progression in CRC. UXT-AS1 is a conserved natural antisense transcript corresponding to the 5’ end of UXT, and it was interesting to find that UXT-AS1 can serve as a switch for the alternative splicing of UXT.
UXT, which is located on the Xp11.23-p11.22 chromosome, was found to be widely expressed in different human tissues [26]. In addition to gene mutation, research has demonstrated that alternative splicing is also disordered in human cancers and the abnormal alternative splicing of genes also results in cancer progression [27-29]. Through alternative splicing, different transcripts are produced from the splicing of a single gene to generate proteomic and functional diversity [30]. UXT has two alternative splicing isoforms: UXT1, the coding transcript which encodes 169 amino acid proteins and is predominantly expressed in the cytoplasm and UXT2, the coding transcript which encodes 157 amino acid proteins and is primarily expressed in the nucleus. According to our results, UXT-AS1 regulated the alternative splicing of UXT and changed the expression of UXT from the UXT1 transcript to the UXT2 transcript. In CRC tissues, the expression of UXT1 was downregulated and the expression of UXT2 was upregulated. It was also found that the correlation between UXT-AS1 and UXT1 was strongly negative and the correlation between UXT-AS1 and UXT2 was strongly positive. Therefore, it is suggested that upregulation of UXT-AS1 changed the expression of these two UXT isoforms from UXT1 to UXT2 by alternative splicing.
It has been reported that UXT1 and UXT2 regulate different cell functions as UXT1 is expressed in the cytoplasm and UXT2 is expressed in the nucleus. On the one hand, UXT1 is associated with TNF-induced apoptosis [31], and on the other hand, UXT2 can interact with transcription factors, such as the AR (androgen receptor) and NF-κB (nuclear factor κB), and regulates the transcription factor-responsive genes as a transcriptional cofactor [32,33]. Moreover, a recent study showed that UXT1 and UXT2 elicited dual opposing regulatory effects on SARM-induced apoptosis [34]. As shown in the present study, UXT1 regulated cell apoptosis and UXT2 regulated cell proliferation in CRC cells, indicating that UXT1 and UXT2 regulated different cell functions and had the opposite effect on the progression of CRC. Thus, it is suggested that abnormal upregulation of UXT-AS1, which resulted in downregulation of UXT1 and upregulation of UXT2, can promote progression of CRC by inhibiting cell apoptosis and promoting cell proliferation.
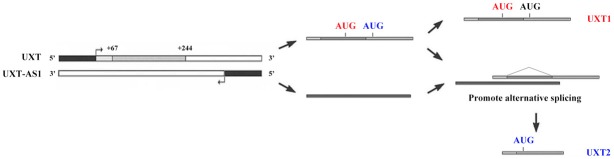
Alternative pre-mRNA splicing, which creates different combinations of exons, is a fundamental regulatory process that results in multiple transcript variants from a single gene locus [35,36]. It is known that numerous human genes are predicted to generate alternatively spliced transcripts by several basic patterns of alternative splicing: exon skipping (cassette alternative exon), intron retention, mutually exclusive, alternative 5’SS, alternative 3’SS, alternative initiation sites, and alternative polyadenylation sites [37]. Normally, pre-mRNA splicing is performed by the spliceosome, which is composed of four small nuclear ribonucleoproteins (snRNPs; U1, U2, U4/U6, and U5) and a number of non-snRNP auxiliary proteins [38]. Recently, it was also found that lncRNAs can participate in the regulation of alternative pre-mRNA splicing and determine the splicing outcome during development and in response to environmental cues [39]. According to the structures of UXT1 and UXT2, the UXT1 isoform is translated from the first AUG; however, the UXT1 isoform is translated using the second AUG as the UXT2 transcript lost a part of the 5’ end through alternative splicing compared with the UXT1 transcript. Moreover, UXT-AS1 is a conserved natural antisense transcript and depends on the activation of a different promoter located at the 5’ end of UXT. Accordingly, it is hypothesized that UXT-AS1 could promote splicing of the UXT 5’ end by binding with UXT pre-mRNA resulting in translation of the UXT2 isoform from the second AUG (Figure 6). Therefore, during the development of CRC, abnormal upregulation of UXT-AS1 promoted splicing of the UXT 5’ end and changed the expression of UXT from the UXT1 transcript to the UXT2 transcript, which finally resulted in the progression of CRC.
Figure 6.
A model of the regulation of UXT-AS1-induced alternative splicing of UXT. UXT-AS1 is a natural antisense transcript of UXT and depends on the activation of a different promoter located at the 5’ end of UXT. UXT-AS1 can promote alternative splicing at the 5’ end of UXT, which results in a change from the UXT1 transcript to the UXT2 transcript.
In summary, this study found that lncRNA UXT-AS1-induced alternative splicing was a post-transcriptional regulatory mechanism to fine-tune homeostatic expression of UXT. We provided clinical evidence to show that UXT-AS1-induced alternative splicing of UXT was closely associated with progression of CRC. Moreover, we showed that abnormal upregulation of UXT-AS1, which mediated downregulation of UXT1 and upregulation of UXT2, was a novel post-transcriptional regulatory mechanism to promote CRC development by inhibiting cell apoptosis and promoting cell proliferation. Thus, this newly identified UXT-AS1-induced alternative splicing of UXT provided novel insight into the molecular mechanisms which regulate the progression of CRC and a promising strategy for future therapy of CRC.
Acknowledgements
This study was supported by grants from the Scientific Research Project of Guangzhou Municipal University (1201410198, 1201630087) and National Natural Science Foundation of China (81501969).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–17. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Robsahm TE, Aagnes B, Hjartåker A, Langseth H, Bray FI, Larsen IK. Body mass index, physical activity, and colorectal cancer by anatomical subsites: a systematic review and meta-analysis of cohort studies. Eur J Cancer Prev. 2013;22:492–505. doi: 10.1097/CEJ.0b013e328360f434. [DOI] [PubMed] [Google Scholar]
- 3.Chakradhar S. Colorectal cancer: 5 big questions. Nature. 2015;521:S16. doi: 10.1038/521S16a. [DOI] [PubMed] [Google Scholar]
- 4.Sridharan M, Hubbard JM, Grothey A. Colorectal cancer: how emerging molecular understanding affects treatment decisions. Oncology. 2014;28:110–8. [PubMed] [Google Scholar]
- 5.Schumacher FR, Schmit SL, Jiao S, Edlund CK, Wang H, Zhang B, Hsu L, Huang SC, Fischer CP, Harju JF, Idos GE, Lejbkowicz F, Manion FJ, McDonnell K, McNeil CE, Melas M, Rennert HS, Shi W, Thomas DC, Van Den Berg DJ, Hutter CM, Aragaki AK, Butterbach K, Caan BJ, Carlson CS, Chanock SJ, Curtis KR, Fuchs CS, Gala M, Giovannucc EL, Gogarten SM, Hayes RB, Henderson B, Hunter DJ, Jackson RD, Kolonel LN, Kooperberg C, Küry S, LaCroix A, Laurie CC, Laurie CA, Lemire M, Levine D, Ma J, Makar KW, Qu C, Taverna D, Ulrich CM, Wu K, Kono S, West DW, Berndt SI, Bezieau S, Brenner H, Campbell PT, Chan AT, Chang-Claude J, Coetzee GA, Conti DV, Duggan D, Figueiredo JC, Fortini BK, Gallinger SJ, Gauderman WJ, Giles G, Green R, Haile R, Harrison TA, Hoffmeister M, Hopper JL, Hudson TJ, Jacobs E, Iwasaki M, Jee SH, Jenkins M, Jia WH, Joshi A, Li L, Lindor NM, Matsuo K, Moreno V, Mukherjee B, Newcomb PA, Potter JD, Raskin L, Rennert G, Rosse S, Severi G, Schoen RE, Seminara D, Shu XO, Slattery ML, Tsugane S, White E, Xiang YB, Zanke BW, Zheng W, Le Marchand L, Casey G, Gruber SB, Peters U. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat Commun. 2015;6:7138. doi: 10.1038/ncomms8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–7. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 9.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 10.Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol. 2016;12:360–73. doi: 10.1038/nrneph.2016.51. [DOI] [PubMed] [Google Scholar]
- 11.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 12.Haemmerle M, Gutschner T. Long non-coding RNAs in cancer and development: where do we go from here? Int J Mol Sci. 2015;16:1395–405. doi: 10.3390/ijms16011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, Gotoh K, Ariizumi S, Wardell CP, Hayami S, Nakamura T, Aikata H, Arihiro K, Boroevich KA, Abe T, Nakano K, Maejima K, Sasaki-Oku A, Ohsawa A, Shibuya T, Nakamura H, Hama N, Hosoda F, Arai Y, Ohashi S, Urushidate T, Nagae G, Yamamoto S, Ueda H, Tatsuno K, Ojima H, Hiraoka N, Okusaka T, Kubo M, Marubashi S, Yamada T, Hirano S, Yamamoto M, Ohdan H, Shimada K, Ishikawa O, Yamaue H, Chayama K, Miyano S, Aburatani H, Shibata T, Nakagawa H. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500–9. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 14.Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW, Liu XS. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun. 2016;7:10982. doi: 10.1038/ncomms10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie X, Tang B, Xiao YF, Xie R, Li BS, Dong H, Zhou JY, Yang SM. Long non-coding RNAs in colorectal cancer. Oncotarget. 2016;7:5226–39. doi: 10.18632/oncotarget.6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasry A, Zinger A, Ben-Neriah Y. Inflammatory networks underlying colorectal cancer. Nat Immunol. 2016;17:230–40. doi: 10.1038/ni.3384. [DOI] [PubMed] [Google Scholar]
- 17.West NR, McCuaig S, Franchini F, Powrie F. Emerging cytokine networks in colorectal cancer. Nat Rev Immunol. 2015;15:615–29. doi: 10.1038/nri3896. [DOI] [PubMed] [Google Scholar]
- 18.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–7. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 19.Hajjari M, Khoshnevisan A, Shin YK. Molecular function and regulation of long non-coding RNAs: paradigms with potential roles in cancer. Tumour Biol. 2014;35:10645–63. doi: 10.1007/s13277-014-2636-z. [DOI] [PubMed] [Google Scholar]
- 20.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 21.Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3’ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–75. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 22.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–8. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 23.Ma C, Shi X, Zhu Q, Li Q, Liu Y, Yao Y, Song Y. The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 2016;37:1437–44. doi: 10.1007/s13277-015-4521-9. [DOI] [PubMed] [Google Scholar]
- 24.Shi D, Zheng H, Zhuo C, Peng J, Li D, Xu Y, Li X, Cai G, Cai S. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Med Oncol. 2014;31:31. doi: 10.1007/s12032-014-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H, Ueda M, Sakimura S, Takano Y, Kurashige J, Shinden Y, Eguchi H, Sugimachi K, Maehara Y, Mimori K. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35:1385–8. [PubMed] [Google Scholar]
- 26.Schroer A, Schneider S, Ropers H, Nothwang H. Cloning and characterization of UXT, a novel gene in human Xp11, which is widely and abundantly expressed in tumor tissue. Genomics. 1999;56:340–3. doi: 10.1006/geno.1998.5712. [DOI] [PubMed] [Google Scholar]
- 27.Shkreta L, Bell B, Revil T, Venables JP, Prinos P, Elela SA, Chabot B. Cancer-associated perturbations in alternative pre-messenger RNA splicing. Cancer Treat Res. 2013;158:41–94. doi: 10.1007/978-3-642-31659-3_3. [DOI] [PubMed] [Google Scholar]
- 28.Biamonti G, Catillo M, Pignataro D, Montecucco A, Ghigna C. The alternative splicing side of cancer. Semin Cell Dev Biol. 2014;32:30–6. doi: 10.1016/j.semcdb.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Salton M, Misteli T. Small molecule modulators of pre-mRNA splicing in cancer therapy. Trends Mol Med. 2016;22:28–37. doi: 10.1016/j.molmed.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu XD, Ares M Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y, Chen L, Zhou Y, Liu H, Yang J, Liu Z, Wang C. UXT-V1 protects cells against TNF-induced apoptosis through modulating complex II formation. Mol Biol Cell. 2011;22:1389–97. doi: 10.1091/mbc.E10-10-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun S, Tang Y, Lou X, Zhu L, Yang K, Zhang B, Shi H, Wang C. UXT is a novel and essential cofactor in the NF-kappaB transcriptional enhanceosome. J Cell Biol. 2007;178:231–44. doi: 10.1083/jcb.200611081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S, Chen K, Zhang Q, Cheng H, Zhou R. Regulation of the transcriptional activation of the androgen receptor by the UXT-binding protein VHL. Biochem J. 2013;456:55–66. doi: 10.1042/BJ20121711. [DOI] [PubMed] [Google Scholar]
- 34.Sethurathinam S, Singh LP, Panneerselvam P, Byrne B, Ding JL. UXT plays dual opposing roles on SARM-induced apoptosis. FEBS Lett. 2013;587:3296–302. doi: 10.1016/j.febslet.2013.08.033. [DOI] [PubMed] [Google Scholar]
- 35.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345–55. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 36.Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14:153–65. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 37.Han J, Xiong J, Wang D, Fu XD. Pre-mRNA splicing: where and when in the nucleus. Trends Cell Biol. 2011;21:336–43. doi: 10.1016/j.tcb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papasaikas P, Valcárcel J. The spliceosome: the ultimate RNA chaperone and sculptor. Trends Biochem Sci. 2016;41:33–45. doi: 10.1016/j.tibs.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez I, Munita R, Agirre E, Dittmer TA, Gysling K, Misteli T, Luco RF. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol. 2015;22:370–6. doi: 10.1038/nsmb.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]