Abstract
Nef is a multifunctional virulence factor of primate lentiviruses that facilitates viral replication in the infected host. All known functions of Nef require that it be myristoylated at its N terminus. This reaction is catalyzed by N-myristoyltransferases (NMTs), which transfer myristate from myristoyl coenzyme A (myristoyl-CoA) to the N-terminal glycine of substrate proteins. Two NMT isoforms (NMT-1 and NMT-2) are expressed in mammalian cells. To provide a better mechanistic understanding of Nef function, we used biochemical and microsequencing techniques to isolate and identify Nef-associated proteins. Through these studies, NMT-1 was identified as an abundant Nef-associated protein. The Nef-NMT-1 complex is most likely a transient intermediate of the myristoylation reaction of Nef and is modulated by agents which affect the size of the myristoyl-CoA pool in the cell. We also examined two other proteins that bear an N-terminal myristoylation signal, human immunodeficiency virus type 1 Gag and Hck protein tyrosine kinase, and found that Gag bound preferentially the NMT-2 isoform, while Hck bound mostly to NMT-1. Recognition of different NMT isoforms by these viral and cellular substrate proteins suggests nonoverlapping roles for these enzymes in vivo and reveals a potential for the development of inhibitors that target the myristoylation of specific viral substrates more selectively.
Nef proteins are important virulence factors of primate lentiviruses (human immunodeficiency virus type 1 [HIV-1], HIV-2, and simian immunodeficiency virus [SIV]) (21, 22). These multifunctional proteins modify the cellular environment of infected cells to facilitate viral replication and evade detection by cells of the immune system (9, 27, 41). Nef usurps at least two distinct mechanisms as it downregulates the cell surface expression of receptors that are important for normal immune function. Both HIV-1 and SIV Nef proteins induce endocytosis of CD4 and CD28 via the AP-2 clathrin adaptor pathway (1, 4, 15, 24, 33, 44). SIVmac 239 Nef induces T-cell receptor (TCR):CD3 endocytosis through the AP-2 mechanism as well (3, 45). Interestingly, lentiviral Nef also downregulates cell surface class I major histocompatibility complexes, through a mechanism that involves PACS-1 and possibly ARF GTPases (6, 16, 23, 34, 40). Finally, Nef targets signal transduction machinery in T cells and enhances replication and infectivity of viral particles (8, 18, 20, 25, 27, 39).
These diverse functions are executed through independent interactions of Nef with a broad array of downstream effectors in protein sorting and signaling machineries. For example, the accelerated endocytosis of TCR:CD3 by Nef is mediated by formation of higher-order protein complexes in which Nef, the CD3ζ subunit of TCR:CD3, and the AP-2 clathrin adaptor bind cooperatively (18, 45). Specific recruitment of other cell surface receptors to the endocytic machinery is likely mediated by similar mechanisms. Nef also forms stable complexes with several elements in the Rac signaling pathway that control various aspects of cell signaling and the cytoskeleton, including DOCK2-ELMO1 and possibly Vav guanine nucleotide exchange factors, Rac, and p21-activated kinase (PAK2) (2, 10, 20, 31, 36, 37).
All previously described Nef functions require that a functional myristoylation signal be present at the N terminus of Nef (26, 32, 38, 42, 50). N myristoylation is catalyzed by the enzyme myristoyl coenzyme A (myristoyl-CoA):protein N-myristoyltransferase (NMT), which transfers a myristic acid moiety (C14:0) from myristoyl-CoA and attaches it via an amide bond to the N-terminal glycines of both cellular and viral substrate proteins (5, 7). At least two mammalian NMT isoforms (NMT-1 and NMT-2) exist and are encoded by separate genes in both the mouse and human genomes (12, 13).
Molecular mechanisms that mediate a subset of Nef functions such as downregulation of cell surface receptors have been described in considerable detail, whereas interactions with other cellular machineries are less clear. To reveal other events leading to Nef biogenesis and function, we carried out biochemical and protein microsequencing experiments intended to identify host cell proteins with which Nef associates. Previously, we reported that the DOCK2-ELMO1-Rac complex is a major target of Nef for signal transduction in T cells and that HIV-1 Nef activates Rac through binding to this complex (20). Here, using a similar strategy, we found that Nef forms a stable complex specifically with N-myristoyltransferase isoform 1. The association requires an N-terminal region of Nef containing an intact myristoylation signal, is modulated by agents that alter the cellular myristoyl-CoA pool, and, therefore, represents a transient intermediate of the myristoylation reaction of Nef. We further demonstrated that two other NMT substrates, HIV-1 Gag and cellular protein tyrosine kinase Hck, also associate with NMT but show different isoform preferences.
MATERIALS AND METHODS
Mammalian expression plasmids and epitope tagging.
Dual-epitope-tagged nef genes were constructed by fusing a sequence encoding the AU-1 epitope (29) (DTYRYI, A epitope) derived from bovine papillomavirus major capsid protein, followed by the hemagglutinin (HA) epitope (30) (ANATYPYDVPDYAG, H epitope) derived from influenza virus hemagglutinin, to the C-terminal amino acid in HIV-1 NA7 nef (NA7.AH) and in SIVmac 239 nef (239.AH). The wild-type NA7 and 239 nef allele as well as NA7(G2A), 239(P104A,P107A), and 239(Δ23-74) mutants were described previously (19, 26, 35, 43). In 239(G2▿).AH, the glycine at position 2 in SIVmac 239 Nef was replaced with the AH epitope. Epitope-tagged nef genes were subcloned into pCG (16) and BABE(puro) (28) mammalian expression vectors. The AH epitope-tagged green fluorescent protein (GFP) gene was constructed as described above for nef. Genes encoding human NMT-1 and NMT-2 were generously provided by B. Cravatt (Scripps Research Institute) and modified to encode a T7 epitope (MASMTGGQQMG) at the N terminus of NMT. Genes encoding Nef-GFP fusion protein and H-tagged HIV-1 NL4-3 Gag and human Hck protein tyrosine kinase were constructed by using overlap PCR as previously described (15) and subcloned into pCG mammalian expression vectors.
Cell lines and retroviral transduction.
Jurkat T cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 10 mM HEPES (pH 7.4), and antibiotics (100 U of penicillin per ml and 100 μg of streptomycin per ml) (16). Phoenix-ampho packaging cells (kindly provided by G. Nolan, Stanford University) and Cos-7 cells (14) were maintained in Dulbecco modified Eagle medium supplemented as described above. BABE(puro) plasmids were transfected into Phoenix-ampho cells by calcium phosphate coprecipitation method (46). Virus-containing cell culture medium was removed 48 h after transfection, filtered (0.45-μm-pore-size filter; Millipore) and used to infect 1 × 105 to 2 × 105 Jurkat T cells in complete RPMI medium supplemented with Polybrene (4 μg/ml; Sigma). Infected cell populations were selected with puromycin (0.4 μg/ml) for 7 to 10 days.
Immunoaffinity purification of Nef and associated proteins from Jurkat T cells.
Immunoaffinity purification of Nef and associated proteins was performed as previously described (20) except for using different epitope tags. Briefly, CD4+ Jurkat T cells were maintained in complete RPMI medium containing puromycin (0.8 μg/ml) in spinner cultures. Every 1 to 2 days, sufficient fresh medium was added to adjust the cell density to 105 cells/ml. Spinner cell cultures were expanded to 6 to 8 liters, and cells were harvested when cell densities reached 5 × 105 to 7 × 105 cells/ml. Detergent extracts were prepared as described previously (20). Monoclonal antibody (MAb) 12CA5, specific for the HA epitope (α-H), was obtained from the Monoclonal Shared Resource at Cold Spring Harbor Laboratory. MAb specific for AU-1 (α-A) was purchased from Babco. For immunoprecipitation experiments, α-H and α-A MAbs were cross-linked to protein G-agarose beads (Roche) (17). Immunoprecipitations were performed for 4 h, beads were washed extensively in lysis buffer, and protein complexes were eluted in lysis buffer containing the appropriate peptide at 0.2 mg/ml. Eluates were concentrated by using Microcon centrifugal filter devices (molecular mass cutoff, 10 kDa; Millipore) according to the manufacturer's instructions. Proteins were resolved on sodium dodecyl sulfate (SDS)-8 to 17% gradient polyacrylamide gels and stained with silver nitrate for analytical experiments (17) or with SimplyBlue Safe Stain (Invitrogen). Liquid chromatography-tandem mass spectroscopy (LC/MS/MS) microsequencing and data analysis were performed at the Cold Spring Harbor Laboratory Cancer Center Protein Chemistry shared resource as described previously (20). Proteomic data were analyzed with Profound software, which is available online from http://prowl.rockefeller.edu. Profound uses “expectation values” to rate matches between experimental spectra and protein sequences in the database. The simple interpretation of an expectation value is the number of matches that would be expected to have a particular score if the matches to the database were completely random. Therefore, the smaller the expectation value, the more likely that a particular match is a true match rather than a random one. The description of the Profound algorithm is available online from http://prowl.rockefeller.edu/.
Transient assays of NMT association with Nef.
Cos7 cells were transfected with plasmid pCG expressing epitope-tagged NMT, Nef, Nef-GFP chimeras, Gag, or Hck by electroporation, collected 36 to 48 h later, and extracted in lysis buffer (16, 19). Extracts were immunoprecipitated with α-H beads. The beads were collected and washed two times with lysis buffer and three times with lysis buffer supplemented with 500 mM LiCl, and samples were analyzed by immunoblotting.
SDS-PAGE and immunoblot analyses.
T7 epitope-specific MAbs conjugated to horseradish peroxidase and to CD3ζ were from Novagen and Santa Cruz Biotechnology, respectively. Serum to recombinant SIVmac 239 Nef was raised in rabbits (α-Nef Ab; Covance) (19). Rabbit polyclonal antibody reactive with both NMT-1 and NMT-2 (α-NMT Ab) was generously provided by Benjamin Cravatt. Polypeptides were separated by SDS-12.5% polyacrylamide gel electrophoresis (PAGE) and electroblotted onto polyvinylidene difluoride membranes (Immobilon-P; Millipore). The membranes were incubated for 1 h in blocking solution containing Tris-buffered saline (150 mM NaCl, 50 mM Tris-HCl [pH 7.5] with 0.1% Tween 20, 1% bovine serum albumin, and 5% nonfat dried milk [Carnation]) and then incubated with appropriate antibodies in blocking solution at the specified dilutions (α-Nef, 1:1,000; α-T7-horseradish peroxidase, 1:5,000; α-H, 1:5,000; α-CD3ζ, 1:1,000; α-NMT, 1:400). Membranes were washed three times with Tris-buffered saline containing 0.1% Tween 20 and, when appropriate, were incubated for 1 h with horseradish peroxidase-conjugated goat α-rabbit immunoglobulin G antibody (Jackson Laboratories) and developed by using an enhanced chemiluminescence (Amersham) detection system as recommended by the manufacturer.
Glycerol gradient sedimentation.
Nef and associated proteins were immunoprecipitated from detergent extracts with α-H beads, eluted with H peptide, and overlaid onto a 5-ml, 10 to 30% glycerol gradient. Gradients were centrifuged at 45,000 rpm in an SW55 Ti rotor for 12 h at 4°C. Fractions (400 μl) were collected from the top of the gradient, and aliquots of each fraction were used for immunoblot analysis with α-NMT Ab, α-CD3ζ, and α-H MAb.
RESULTS
To facilitate the purification of Nef and its associated proteins, we tagged the C-terminal end of SIVmac 239 Nef with AU-1 (A) and HA (H) double-epitope tags (239.AH) and expressed it stably in Jurkat T cells via retroviral transduction and selection with puromycin. The addition of C-terminal AH epitopes had little detectable effect on the ability of Nef to downmodulate CD4, CD28, and CD3 from the cell surface and to associate with p62 phosphoprotein, which was identified previously as p21-associated kinase (PAK) (2, 36), compared to untagged Nef. As controls we also constructed Jurkat cell lines expressing AH epitope-tagged SIVmac 239 Nef proteins with mutations that disrupted the N-terminal myristoylation signal [239(G2▿).AH], the PxxP motif required for association with PAK (37) [239(P104A,P107A).AH], or sequences required for CD4 and CD28 downmodulation and located in the N-terminal region [239(Δ23-74).AH] (45). Finally, we constructed a negative control cell line expressing puromycin N-acetyltransferase selectable marker (PAC) alone.
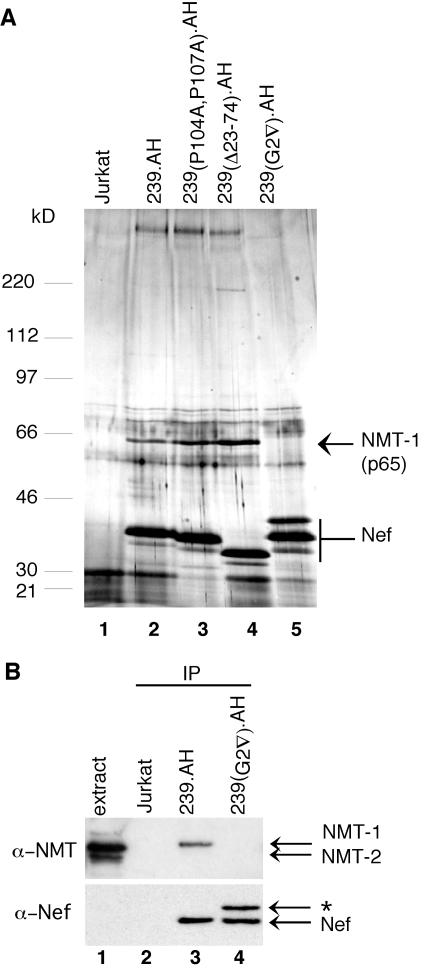
Detergent extracts were prepared from Jurkat T cells expressing wild-type and mutant forms of AH-tagged Nef and from control cells transduced with a control vector expressing PAC alone. Nef and associated proteins were purified by sequential immunoprecipitations with α-H and then α-A epitope antibodies, followed each time by elution with the respective peptide epitope. The immunoprecipitated proteins were separated by SDS-PAGE and stained with silver nitrate. As shown in Fig. 1A, two polypeptides, one with an apparent molecular mass of approximately 65 kDa (p65) and the other with an apparent molecular mass of >220 kDa, copurified with wild-type and mutant SIVmac Nef proteins that contained an intact myristoylation signal (lanes 2 to 4). Notably, these polypeptides were absent in purifications from cells that expressed myristoylation signal-defective Nef [239(G2▿).AH] and from puromycin-resistant cells that did not express 239.AH Nef (lanes 5 and 1, respectively).
FIG. 1.
Nef associates with NMT-1 in a myristoylation signal-dependent manner. (A) p65 specifically copurifies with SIVmac 239 Nef. Nef and associated proteins were isolated by use of a two-step immunoaffinity purification protocol from detergent extracts prepared from Jurkat T cells expressing AH epitope-tagged wild-type (lane 2) and mutant (lanes 3 to 5) forms of SIVmac 239 Nef. Purification from control puromycin-resistant Jurkat cells that did not express Nef is also shown (lane 1). Eluates were separated by SDS-8 to 17% gradient PAGE, stained with silver nitrate, and analyzed by LC/MS/MS. Bands corresponding to NMT-1 and SIV Nef are indicated. The bands migrating just below Nef likely are Nef degradation products. The positions and molecular masses of protein standards are indicated on the left. (B) Immunoblot analysis identifies Nef-associated p65 as N-myristoyltransferase 1. Immunopurifications from Jurkat T cells expressing wild-type (lane 3) and myristoylation signal-mutated (lane 4) SIVmac 239 Nef and from puromycin-resistant Jurkat T cells that did not express Nef (lane 2) were resolved by SDS-PAGE and analyzed by immunoblotting with α-NMT Ab (upper panel) or α-Nef Ab (bottom panel). Extract from Jurkat cells, which express both the NMT-1 and NMT-2 isoforms, provided a positive control for NMT expression (lane 1). The identity of the α-Nef Ab reactive band indicated with an asterisk is not known.
To determine the identity of Nef-associated p65, 239.AH Nef and its associated proteins were immunoprecipitated from approximately 1010 Jurkat T cells. The purified polypeptides were separated on an SDS-8 to 17% gradient polyacrylamide gel and stained with SimplyBlue Safe Stain, and a gel slice containing p65 was excised. Proteins within the gel slice were subjected to in-gel digestion with trypsin, and the resulting peptides were separated and sequenced by LC/MS/MS. LC/MS/MS data for peptides derived from p65 are listed in Table 1. Amino acid sequences of seven peptides matched those for predicted tryptic peptides derived from human NMT-1, while the combined expectation score (the probability that p65 is not NMT-1) was 2.9 × 10−28, thus indicating that Nef-associated p65 is indeed NMT-1. To confirm that Nef-associated p65 is NMT-1, α-Nef immunoprecipitates from Jurkat cells expressing wild-type 239.AH, myristoylation signal-defective 239(G2▿).AH, and control puromycin-resistant Jurkat T cells were analyzed by immunoblotting with a polyclonal antibody that recognizes both the NMT-1 and NMT-2 isoforms of human NMT (α-NMT). Two protein bands migrating at approximately 65 and 60 kDa were detected in Jurkat detergent extracts (Fig. 1B, lane 1). Control experiments using ectopically expressed NMT-1 and NMT-2 revealed that the more intense p65 band corresponded to NMT-1, while the faster-migrating p60 comigrated with the NMT-2 isoform (data not shown; see Fig. 3). Notably, the p65 NMT-1, but not p60, isoform was detected in α-Nef immunoprecipitates from cells expressing wild-type 239.AH, and this association required an intact myristoylation signal in Nef (Fig. 1B, compare lanes 2 and 3). We concluded that SIVmac 239 Nef forms a relatively abundant and stable complex with NMT-1 in vivo.
TABLE 1.
Nef-associated p65 is NMT-1a
| Peptide | Expectation value | Peptide sequence |
|---|---|---|
| 1 | 1.4 × 10−9 | 128LGEVVNTHGPVEPDKDNIR146 |
| 2 | 2.5 × 10−6 | 116SYQFWDTQPVPK127 |
| 3 | 6.3 × 10−6 | 345DIPVVHQLLTR355 |
| 4 | 1.6 × 10−5 | 95AIELFSVGQGPAK107 |
| 5 | 4.1 × 10−4 | 44GGLSPANDTGAK55 |
| 6 | 0.13 | 259VAPVLIR265 |
| 7 | 15.4 | 222LVGFISAIPANIHIYDTEKK241 |
| p65 | 2.9 × 10−28 | N-Myristoyltransferase-1 (Homo sapiens) |
Amino acid sequences and expectation values of peptides derived from SIVmac 239 Nef-associated p65 and analyzed by LC/MS/MS are shown. The relative positions of peptides in the NMT-1 amino acid sequence are indicated. The expectation value is a probability that the actual peptide data do not correspond to the NMT-1 sequence. The combined expectation score for p65 shown at the bottom is the probability that Nef-associated p65 is not NMT-1. The seven peptides cover approximately 19% of the NMT-1 amino acid sequence.
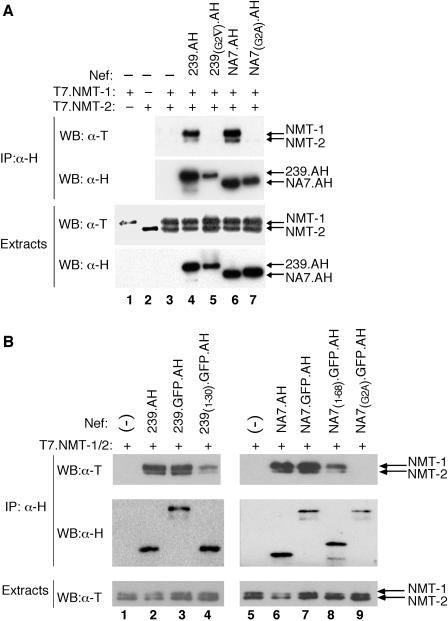
FIG. 3.
N-terminal regions of HIV-1 and SIV Nef proteins associate preferentially with the NMT-1 isoform. (A) Nef associates preferentially with the NMT-1 isoform. Detergent extracts were prepared from Cos7 cells transiently expressing T7 epitope-tagged NMT-1 (T7.NMT-1) (lane 1), or NMT-2 (T7.NMT-2) (lane 2), or both NMT-1 and NMT-2 alone (lane 3) or together with wild-type (239.AH) (lane 4) or myristoylation-defective [239(G2▿).AH] (lane 5) SIV Nef or wild-type (NA7.AH) (lane 6) or myristoylation-defective [NA7(G2A).AH] (lane 7) HIV-1 Nef. Extracts were immunoprecipitated with α-H MAb (IP:α-H), and immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blotting. NMT was detected with a MAb specific for the T7 epitope (WB:α-T), and Nef was detected with H epitope-specific MAb (WB:α-H) in immunoprecipitates (upper panels) and detergent extracts (lower panels). (B) The N-terminal region of Nef preferentially binds the NMT-1 isoform. Detergent extracts prepared from Cos7 cells transiently coexpressing T7.NMT-1 and T7.NMT-2 alone (lanes 1 and 5) or in combination with SIV 239.AH Nef (lane 2), HIV-1 NA7.AH Nef (lane 6), or chimeric molecules consisting of AH epitope-tagged GFP (GFP.AH) fused to the C-terminal end of the full-length SIV Nef (239.GFP.AH) (lane 3), the N-terminal 30 residues of SIV Nef [239(1-30).GFP.AH] (lane 4), full-length NA7 Nef (NA7.GFP.AH) (lane 7), the N-terminal 68 residues of NA7 Nef [NA7(1-68).GFP.AH] (lane 8), or myristoylation-defective NA7 Nef [NA7(G2A).GFP.AH] (lane 9) were immunoprecipitated with α-H MAb (IP:α-H). NMT and Nef were detected in the immunoprecipitates (upper panels), and extracts (lower panel) were resolved by SDS-PAGE.
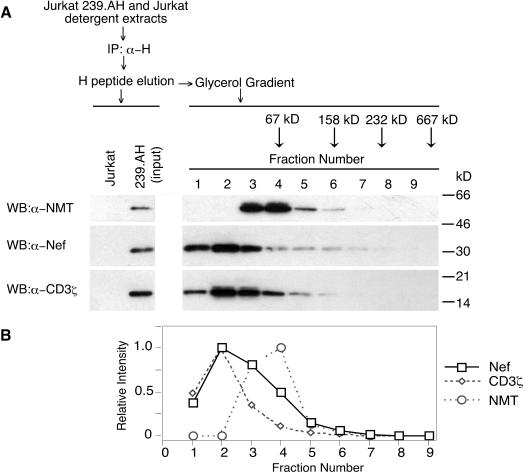
SIV Nef was previously observed to bind cellular proteins such as the CD3ζ subunit of TCR-CD3 (18). To address the relationship between complexes containing Nef and CD3ζ and those containing Nef and NMT-1, we sedimented Nef-containing complexes in a glycerol gradient. Detergent extracts were prepared from Jurkat T cells expressing 239.AH Nef and control puromycin-resistant Jurkat cells. Nef and its associated proteins were immunoprecipitated with α-H MAb and eluted with H peptide. Aliquots of eluates were characterized by immunoblotting. As expected, NMT-1 and CD3ζ were readily detectable in immunoprecipitates from cells expressing 239.AH but not in those from control cells (Fig. 2, left panel).
FIG. 2.
Glycerol gradient sedimentation of complexes containing SIV Nef. Detergent extracts prepared from Jurkat T cells expressing 239.AH Nef were immunoprecipitated with a MAb specific for the H epitope (IP:α-H), and bound proteins were eluted with H peptide. Nef and associated proteins were sedimented through a 10 to 30% glycerol gradient, and fractions were collected from the top of the gradient. The distribution of molecular mass standards sedimented in parallel in an identical gradient is indicated in panel A. (A) Aliquots of glycerol gradient fractions were separated by SDS-PAGE and analyzed by immunoblotting with a polyclonal antibody specific for NMT (WB:α-NMT), a polyclonal antibody specific for SIVmac 239 Nef (WB:α-Nef), or a MAb specific for CD3ζ (WB:α-CD3ζ). (B) The distributions of NMT, 239.AH Nef, and CD3ζ in gradient fractions were quantitated by densitometric scanning of the chemiluminescence images shown in panel A. A representative of two independent experiments is shown.
Next, Nef and associated proteins were sedimented through a glycerol gradient. A set of protein standards of known molecular weights was sedimented in parallel through an identical glycerol gradient to provide an estimate of the sizes of various protein complexes containing Nef. The distributions of 239.AH Nef, NMT-1, and CD3ζ as well as protein standards across the gradient are shown in Fig. 2A. The approximately 35-kDa 239.AH Nef protein sedimented more slowly than the 67-kDa standard and peaked in fractions 2 and 3. This suggested that that the bulk of the immunoprecipitated Nef was either in relatively low-molecular-weight complexes or uncomplexed. Immunoblotting with α-NMT Ab revealed that Nef-associated NMT-1 sedimented more rapidly than the bulk of Nef and peaked in fraction 4. This was consistent with the possibility that the NMT-1 binds Nef directly and that the Nef-NMT-1 complex does not contain other components. Similarly, the CD3ζ (16-kDa) subunit associated with Nef peaked in fraction 2. The small size of the Nef-CD3ζ complex suggested that other TCR:CD3 subunits were absent. This was expected since the association between CD3ζ and the core CD3 complex is known to be unstable under conditions used to prepare detergent extracts (45).
The Nef protein is posttranslationally modified by covalent attachment of the myristoyl moiety to glycine 2, and this reaction is catalyzed by NMT. Therefore, we hypothesized that the Nef-NMT-1 complex could be a transient intermediate of the myristoylation reaction of Nef. To address this possibility, we first developed a transient-expression assay in Cos7 cells in which to study the Nef association with NMT-1.
Cos7 cells were transfected to transiently express T7-epitope tagged human NMT isoforms NMT-1 (T7.NMT-1) and NMT-2 (T7.NMT-2) together with wild-type or myristoylation-signal mutated SIVmac 239 Nef, each tagged with the AH epitopes. Nef and associated proteins were immunoprecipitated with α-H MAb from detergent extracts, and the immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting. As shown in Fig. 3A, T7.NMT-1 migrated more slowly than T7.NMT-2 (compare lanes 1 and 2), and the two isoforms could easily be distinguished from each other (lane 3). Notably, wild-type 239.AH Nef precipitated NMT-1 more efficiently than NMT-2 (lane 4), as observed previously with Jurkat cells stably coexpressing SIVmac 239 Nef and endogenous NMT. We then asked whether HIV-1 Nef also forms a complex with NMT. We found that the HIV-1 NA7 Nef isolate also preferentially bound NMT-1 and that this interaction required an intact Nef myristoylation signal (lanes 6 and 7), as observed for SIVmac 239 Nef.
To determine whether elements other than the myristoylation signal in Nef contribute to the interaction of Nef with NMT-1, we constructed chimeric proteins comprised of the amino-terminal regions of SIV and HIV-1 Nef fused to GFP. To facilitate immunoprecipitation experiments, we tagged GFP with the AH epitopes (GFP.AH). We tested the chimeric proteins for their ability to associate with NMT-1 and NMT-2 in Cos7 cells. As shown in Fig. 3B, a short N-terminal fragment of 239 Nef consisting of only the first 30 amino acid residues was sufficient to preferentially bind NMT-1 over NMT-2 (lane 4). Similarly, the (N-terminal) first 68 amino acids of HIV-1 NA7 Nef was also sufficient to preferentially precipitate NMT-1 (lane 8). Notably, both of these Nef fragments precipitated NMT less efficiently than the full-length proteins (compare lanes 3 and 4 and lanes 7 and 8), which suggested that additional elements in Nef contribute to the interaction with NMT.
The catalytic cycle of NMT is thought to involve binding of myristoyl-CoA by NMT, which then opens the NMT substrate binding site and permits substrate binding. This is followed by the transfer of the myristoyl moiety onto glycine 2 in the substrate and the subsequent release of the myristoylated product (7). The release of the product is thought to be the step that limits the reaction rate (11). Therefore, we reasoned that if the Nef-NMT complex is indeed a transient intermediate of Nef myristoylation by NMT-1, it may be possible to modulate its abundance by altering the size of the myristoyl-CoA pool in the cell. To test this possibility, we attempted to alter the intracellular pool of myristoyl-CoA by using pharmacologic approaches, and we then measured the effects on the abundance of Nef-NMT complexes.
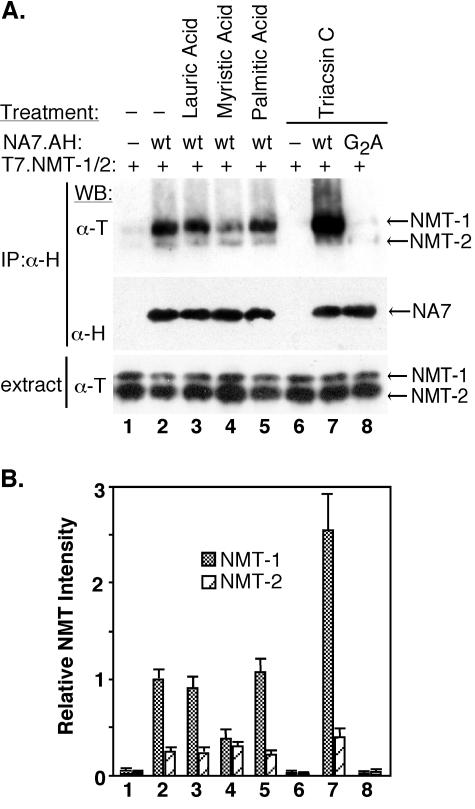
First, we attempted to increase the myristoyl-CoA pool in Cos7 cells transiently coexpressing NMT-1, NMT-2, and NA7 Nef. Since previous studies demonstrated that the availability of fatty acids is limiting to the synthesis of their respective acyl-CoA by fatty acyl-CoA synthetase (48, 49), we cultured cells in the presence of exogenous myristic acid (C14:0). Second, we attempted to decrease the intracellular pool of all fatty acyl-CoA species by treating the cells with an inhibitor of fatty acyl-CoA synthetase, Triacsin C (47). As controls, cells were cultured in the absence of any treatment or in the presence of lauric acid (C12:0) or palmitic acid (C16:0), which do not affect the myristoyl-CoA pool. Following the treatments, Nef was immunoprecipitated from detergent extracts and Nef-associated NMT and Nef were detected by immunoblotting.
Culture of cells in the presence of myristic acid resulted in approximately a two- to threefold decrease in the amount of Nef-bound NMT-1 compared to that in the control cells (Fig. 4A, compare lanes 2 and 4, see also Fig. 4B). In contrast, treatment of cells with Triacsin C resulted in the two- to threefold increase in the amount of Nef-bound NMT-1 (compare lanes 2 and 7). As expected, neither lauric acid nor palmitic acid had a detectable effect on the Nef-NMT association (compare lanes 2, 3, and 5). Notably, none of these treatments affected the overall expression levels of Nef and NMT-1. Thus, the abundance of Nef-NMT-1 complex was inversely correlated to the size of the myristoyl-CoA pool in the cells.
FIG. 4.
Agents that alter the myristoyl-CoA pool modulate the Nef-NMT-1 complex. (A) Cos7 cells coexpressing T7.NMT-1 and T7.NMT-2, either alone (lanes 1 and 6), or together with wild-type (wt) (NA7.AH) (lanes 2 to 5 and 7) or myristoylation-defective [NA7(G2A).H] (lane 8) HIV-1 Nef were cultured in the presence of 50 mM lauric acid (lane 3), 50 mM myristic acid (lane 4), 50 mM palmitic acid (lane 5), or 5 μM Triacsin C (lanes 6 to 8) or left untreated (lane 1). Nef-associated NMT was immunoprecipitated from detergent extracts with α-H MAb (IP:α-H). Immunoprecipitates and aliquots of detergent extracts were resolved by SDS-PAGE, and NMT-1 and NMT-2 were detected by immunoblotting with T7 epitope-specific MAb (WB:α-T), whereas Nef was detected with H epitope-specific MAb (WB:α-H). (B) Nef-associated NMT-1 and NMT-2 were quantitated by densitometry scanning of chemiluminescence images such as those shown in panel A. Mean values and standard deviations from three independent experiments are shown. Data were normalized to the NMT-1 associated with the NA7.AH Nef in untreated cells (lane 2).
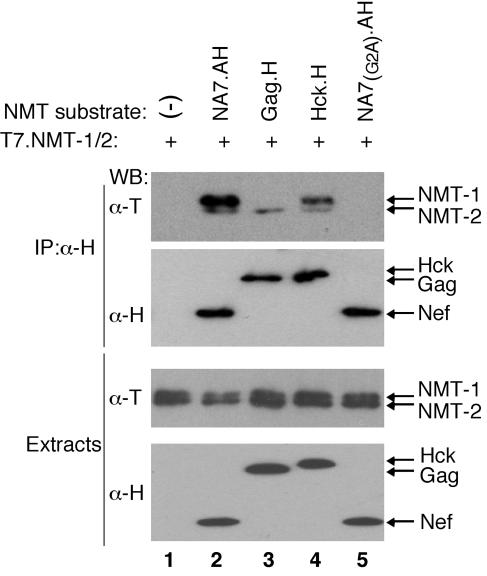
Together, our observations suggested that formation of the NMT-Nef complex is a consequence of the mechanism of the myristoylation reaction. If so, the existence of such enzyme-substrate complexes would not be limited to NMT-1 and Nef but should be seen also with other viral and cellular protein substrates. To address this possibility, two additional NMT substrates, protein tyrosine kinase Hck and HIV-1 Gag, were tested. H epitope-tagged Gag and Hck were coexpressed with NMT isoforms in Cos-7 cells, detergent extracts were immunoprecipitated with α-H, beads and NMT isoforms were detected by immunoblotting. As shown in Fig. 5, NMT was readily detected in both HIV-1 Gag and Hck immunoprecipitates (lanes 3 and 4). Notably, Gag precipitated predominantly NMT-2, while Hck bound more NMT-1 isoform. We concluded that human NMTs form stable intermediate complexes with their protein substrates.
FIG. 5.
HIV-1 Gag and Hck form stable complexes with NMT isoforms. Detergent extracts prepared from Cos7 cells transiently coexpressing T7 epitope-tagged NMT-1 and NMT-2 alone (lane 1) or together with H epitope-tagged HIV-1 Gag (Gag.H) (lane 3), Hck (Hck.H) (lane 4), or, as controls, wild-type NA7 (NA7.AH) (lane 2) or myristoylation-defective [NA7(G2A).AH] (lane 5) HIV-1 Nef were immunoprecipitated with α-H MAb (IP:α-H), and immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blotting as described in the legend to Fig. 3.
DISCUSSION
Nef proteins interact with a large number of downstream effectors. These include components of the signal transduction machinery such as protein tyrosine and protein serine kinases, guanine nucleotide exchange factors, cell surface receptors, and components of the endocytic and protein-sorting machineries, including clathrin adaptor complexes and other proteins that mediate the traffic between various membrane compartments (2, 6, 10, 16, 20, 23, 25, 31, 37, 40). A subset of these proteins, such as CD3ζ and p62 phosphoprotein (data not shown), were readily detectable in the α-Nef immune complexes that we isolated, while others were not. We were unable to detect several of the previously implicated Nef partners despite using numbers of cells that greatly exceeded those used in the previous experiments and employing sensitive immunoblot as well as LC/MS/MS analyses. Specifically, we did not observe Nef associations with subunits of clathrin adaptor complexes, Lck, Fyn, and Vav. The inability to detect these proteins could be due to several factors, including the possibility that these associations were transient or unstable under our experimental conditions or were poorly conserved in the nef alleles that we employed.
The spectrum of proteins that were shown here to copurify with AU-1-HA epitope-tagged SIV Nef (239.AH) differed from that previously reported by us for HA-FLAG epitope-tagged HIV-1 Nef (NA7.hf) (20). Nevertheless, several lines of evidence indicate that HIV-1 NA7 and SIVmac 239 Nef proteins target very similar spectra of cellular proteins and that the use of the hf epitope tag (20) results in a better recovery of Nef-associated protein complexes (A. Janardhan, B. Hill, and J. Skowronski, unpublished data). This is supported by the results of recent experiments in which SIVmac 239 Nef tagged with the hf epitope (239.hf) was expressed in Jurkat T cells and 239.hf Nef-associated polypeptides were purified by using an immunoaffinity strategy described by us previously (20). Microseqencing revealed that 239.hf Nef binds a spectrum of polypeptides that is very similar to that bound by hf-tagged HIV-1 Nef (20), including DOCK2, ELMO1, Rac, and NMT-1 (data not shown). Moreover, microsequencing of HIV-1 NA7.hf Nef-associated proteins (20) resulted in identification of seven NMT-1-derived peptides with a combined expectation score for NMT-1 of approximately 5 × 10−14 (data not shown), thus indicating that NMT-1 binds Nef regardless of the specific epitope tag used. How could the use of different epitope tags at the C terminus of Nef affect the recovery of Nef-associated proteins? The AH-epitope tag is approximately 10 amino acid residues shorter than the hf tag. Thus, the AH tag is more likely to be buried in Nef-bound protein complexes, while the hf tag is more likely to be accessible to antibodies used for immunopurification. Therefore, it is not surprising that in the context of NMT-1, which binds the very N-terminal surface in the Nef molecule, both the AH and hf C-terminal tags were accessible and permitted efficient immunopurification of the Nef-NMT-1 complexes. In contrast, in the context of the DOCK2-ELMO1-Rac complex, which binds a surface located more centrally in the Nef molecule (20), only the longer hf epitope tag was available for immunopurification. Thus, it is important to use an optimal epitope tagging strategy that will not interfere with protein function and that will address potential limitations to protein immunopurification imposed by the topology of protein complexes.
We purified and characterized a novel protein complex containing lentiviral Nef and N-myristoyltransferase-1. Our data showed that the association of Nef with NMT requires an intact myristoylation signal at the N terminus of Nef and that a short N-terminal region of Nef is sufficient for binding. Nevertheless, sequence elements located downstream in Nef likely contribute to the interaction, since the association of the full-length Nef proteins with NMT was more robust than that of chimeras containing only their N-terminal regions. The fact that the intact myristoylation signal in Nef was required for NMT binding suggested that the Nef-NMT-1 complex we purified is an intermediate of Nef myristoylation. This possibility is further supported by the observation that the abundance of the Nef-NMT-1 complex is modulated by altering the myristoyl-CoA pool in the cell and that NMT isoforms also form stable complexes with other substrates. The current model of the NMT catalytic cycle is based on extensive in vitro studies of yeast (Saccharomyces cerevisiae) N-myristoyltransferase (7). This model predicts that an increase in the intracellular myristoyl-CoA pool will facilitate substrate loading and therefore should lead to an increase in the steady-state level of a substrate-NMT complex. Conversely, a decrease in the intracellular myristoyl-CoA pool is predicted to have the opposite effect. Surprisingly, and contrary to these predictions, we observed that the abundance of the Nef-NMT-1 complex is inversely correlated with the size of the myristoyl-CoA pool. One possible explanation for this inconsistency is that the release of the myristoylated product from NMT is coupled to myristoyl-CoA loading onto the product-enzyme complex. This possibility is attractive because dissociation of the product-enzyme complex is thought to be the rate-limiting step of the myristoylation reaction (11). Alternatively, our observations may reflect differences between mechanisms of catalysis by the mammalian and yeast enzymes or levels of regulation in vivo that were not appreciated from in vitro experimentation.
Amino acid sequences of NMT-1 and NMT-2 share approximately 80% amino acid sequence identity. Notably, most divergent are the N-terminal regions, and the differences are strictly conserved among mammals. This suggests unique yet presently unknown roles for each enzyme in vivo, even though previous studies using a spectrum of peptide substrates revealed that NMT-1 and NMT-2 show similar substrate preferences in vitro (12). Therefore, it is intriguing that HIV-1 Gag detectably bound only NMT-2, while Nef displayed high preference towards NMT-1. The specific recognition of different NMT isoforms by Gag and Nef suggests that each of these two viral substrates may be myristoylated preferentially by a distinct NMT isoform in vivo. Further studies of the interactions of NMT-1 and NMT-2 isoforms with Nef and other viral and cellular protein substrates will likely reveal previously unappreciated levels of regulation of protein myristoylation. The differential recognition of NMT isoforms by viral and cellular substrates reported here suggests that development of selective NMT inhibitors that target myristoylation of specific viral substrates may be possible.
Acknowledgments
We thank Linda Van Aelst, James Bliska, Nouria Hernandez, and Roger Johnson for helpful discussions. We thank Ajit Janardhan for sharing reagents, many helpful discussions, and critical reading of the manuscript. We thank Benjamin Cravatt for NMT-1 and NMT-2 cDNAs as well as NMT-specific polyclonal antibody. We also thank Mike Myers for protein sequencing.
This work was supported by Public Health Service grant AI-42561.
REFERENCES
- 1.Aiken, C., J. Konner, N. R. Landau, M. E. Lenburg, and D. Trono. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76:853-864. [DOI] [PubMed] [Google Scholar]
- 2.Arora, V. K., R. P. Molina, J. L. Foster, J. L. Blakemore, J. Chernoff, B. L. Fredericksen, and J. V. Garcia. 2000. Lentivirus Nef specifically activates Pak2. J. Virol. 74:11081-11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, I., C. Ashman, J. Maughan, E. Hooker, F. Cook, and T. A. Reinhart. 1998. Association of simian immunodeficiency virus Nef with the T-cell receptor (TCR) zeta chain leads to TCR down-modulation. J. Gen. Virol. 79:2717-2727. [DOI] [PubMed] [Google Scholar]
- 4.Bell, I., T. M. Schaefer, R. P. Trible, A. Amedee, and T. A. Reinhart. 2001. Down-modulation of the costimulatory molecule, CD28, is a conserved activity of multiple SIV Nefs and is dependent on histidine 196 of Nef. Virology 283:148-158. [DOI] [PubMed] [Google Scholar]
- 5.Bhatnagar, R. S., K. Futterer, G. Waksman, and J. I. Gordon. 1999. The structure of myristoyl-CoA:protein N-myristoyltransferase. Biochim. Biophys. Acta 1441:162-172. [DOI] [PubMed] [Google Scholar]
- 6.Blagoveshchenskaya, A. D., L. Thomas, S. F. Feliciangeli, C. H. Hung, and G. Thomas. 2002. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 111:853-866. [DOI] [PubMed] [Google Scholar]
- 7.Boutin, J. A. 1997. Myristoylation. Cell Signal. 9:15-35. [DOI] [PubMed] [Google Scholar]
- 8.Chowers, M. Y., C. A. Spina, T. J. Kwoh, N. J. Fitch, D. D. Richman, and J. C. Guatelli. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J. Virol. 68:2906-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, K. L., B. K. Chen, S. A. Kalams, B. D. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 10.Fackler, O. T., W. Luo, M. Geyer, A. S. Alberts, and B. M. Peterlin. 1999. Activation of Vav by Nef induces cytoskeletal rearrangements and downstream effector functions. Mol. Cell 3:729-739. [DOI] [PubMed] [Google Scholar]
- 11.Farazi, T. A., J. K. Manchester, and J. I. Gordon. 2000. Transient-state kinetic analysis of Saccharomyces cerevisiae myristoylCoA:protein N-myristoyltransferase reveals that a step after chemical transformation is rate limiting. Biochemistry 39:15807-15816. [DOI] [PubMed] [Google Scholar]
- 12.Giang, D. K., and B. F. Cravatt. 1998. A second mammalian N-myristoyltransferase. J. Biol. Chem. 273:6595-6598. [DOI] [PubMed] [Google Scholar]
- 13.Glover, C. J., and R. L. Felsted. 1995. Identification and characterization of multiple forms of bovine brain N-myristoyltransferase. J. Biol. Chem. 270:23226-23233. [DOI] [PubMed] [Google Scholar]
- 14.Gluzman, Y. 1981. SV40-transformed simian cells support the replication of early SV40 mutants. Cell 23:175-182. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg, M. E., S. Bronson, M. Lock, M. Neumann, G. N. Pavlakis, and J. Skowronski. 1997. Co-localization of HIV-1 Nef with the AP-2 adaptor protein complex correlates with Nef-induced CD4 down-regulation. EMBO J. 16:6964-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg, M. E., A. J. Iafrate, and J. Skowronski. 1998. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 17:2777-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Howe, A. Y., J. U. Jung, and R. C. Desrosiers. 1998. Zeta chain of the T-cell receptor interacts with Nef of simian immunodeficiency virus and human immunodeficiency virus type 2. J. Virol. 72:9827-9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iafrate, A. J., S. Bronson, and J. Skowronski. 1997. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 16:673-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janardhan, A., T. Swigut, B. Hill, M. P. Myers, and J. Skowronski. 2004. HIV-1 Nef binds the DOCK2-ELMO1 complex to activate Rac and inhibit lymphocyte chemotaxis. PloS Biol. 2:65-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kestler, H. W., III, D. J. Ringler, K. Mori, D. L. Panicali, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651-662. [DOI] [PubMed] [Google Scholar]
- 22.Learmont, J., B. Tindall, L. Evans, A. Cunningham, P. Cunningham, J. Wells, R. Penny, J. Kaldor, and D. A Cooper. 1992. Long-term symptomless HIV-1 infection in recipients of blood products from a single donor. Lancet 340:863-867. [DOI] [PubMed] [Google Scholar]
- 23.Le Gall, S., F. Buseyne, A. Trocha, B. D. Walker, J. M. Heard, and O. Schwartz. 1999. Distinct trafficking pathways mediate Nef-induced and clathrin-dependent major histocompatibility complex class I down-regulation. J. Virol. 74:9256-9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lock, M., M. E. Greenberg, A. J. Iafrate, T. Swigut, J. Muench, N. Shohdy, and J. Skowronski. 1999. Two elements target SIV Nef to the AP-2 clathrin adaptor complex, but only one is required for the induction of CD4 endocytosis. EMBO J. 18:2722-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manninen, A., and K. Saksela. 2002. HIV-1 Nef interacts with inositol trisphosphate receptor to activate calcium signaling in T cells. J. Exp. Med. 195:1023-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariani, R., and J. Skowronski. 1993. CD4 down-regulation by nef alleles isolated from human immunodeficiency virus type 1-infected individuals. Proc. Natl. Acad. Sci. USA 90:5549-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, M. D., M. T. Warmerdam, I. Gaston, W. C. Greene, and M. B. Feinberg. 1994. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J. Exp. Med. 179:101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakai, Y., W. D. Lancaster, L. Y. Lim, and A. B. Jenson. 1986. Monoclonal antibodies to genus- and type-specific papillomavirus structural antigens. Intervirology 25:30-37. [DOI] [PubMed] [Google Scholar]
- 30.Niman, H. L., R. A. Houghten, L. E. Walker, R. A. Reisfeld, I. A. Wilson, J. M. Hogle, and R. A. Lerner. 1983. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implication for the structural basis of immune recognition. Proc. Natl. Acad. Sci. USA 59:4949-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nunn, M. F., and J. W. Marsh. 1996. Human immunodeficiency virus type 1 Nef associates with a member of the p21-activated kinase family. J. Virol. 70:6157-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandori, M. W., N. J. Fitch, H. M. Craig, D. D. Richman, C. A. Spina, and J. C. Guatelli. 1996. Producer-cell modification of human immunodeficiency virus type 1: Nef is a virion protein. J. Virol. 70:4283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piguet, V., O. Schwartz, S. Le Gall, and D. Trono. 1999. The downregulation of CD4 and MHC-I by primate lentiviruses: a paradigm for the modulation of cell surface receptors. Immunol. Rev. 168:51-63. [DOI] [PubMed] [Google Scholar]
- 34.Piguet, V., L. Wan, C. Borel, A. Mangasarian, N. Demaurex, G. Thomas, and D. Trono. 2000. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat. Cell Biol. 2:163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Regier, D. A., and R. C. Desrosiers. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retroviruses 6:1221-1231. [DOI] [PubMed] [Google Scholar]
- 36.Renkema, G. H., A. Manninen, D. A. Mann, M. Harris, and K. Saksela. 1999. Identification of the Nef-associated kinas as p21-activated kinase 2. Curr. Biol. 9:1407-1410. [DOI] [PubMed] [Google Scholar]
- 37.Sawai, E. T., I. H. Khan, P. M. Montbriand, B. M. Peterlin, C. Cheng-Mayer, and P. A. Luciw. 1996. Activation of PAK by HIV and SIV Nef: importance for AIDS in rhesus macaques. Curr. Biol. 6:1519-1527. [DOI] [PubMed] [Google Scholar]
- 38.Schaefer, T. M., I. Bell, M. E. Pfeifer, M. Ghosh, R. P. Trible, C. L. Fuller, C. Ashman, and T. A. Reinhart. 2002. The conserved process of TCR/CD3 complex down-modulation by SIV Nef is mediated by the central core, not endocytic motifs. Virology 301:106-122. [DOI] [PubMed] [Google Scholar]
- 39.Schrager, J. A., and J. W. Marsh. 1999. HIV-1 Nef increases T cell activation in a stimulus-dependent manner. Proc. Natl. Acad. Sci. USA 96:8167-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz, O., V. Marechal, S. Le Gall, F. Lemonnier, and J. M. Heard. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2:338-342. [DOI] [PubMed] [Google Scholar]
- 41.Spina, C. A., T. J. Kwoh, M. Y. Chowers, J. C. Guatelli, and D. D. Richman. 1994. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J. Exp. Med. 179:115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stumptner-Cuvelette, P., S. Morchoisne, M. Dugast, S. Le Gall, G. Raposo, O. Schwartz, and P. Benaroch. 2001. HIV-1 Nef impairs MHC class II antigen presentation and surface expression. Proc. Natl. Acad. Sci. USA 98:12144-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swigut, T., A. J. Iafrate, J. Muench, F. Kirchhoff, and J. Skowronski. 2000. Simian and human immunodeficiency virus Nef proteins use different surfaces to downregulate class I major histocompatibility complex antigen expression. J. Virol. 74:5691-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swigut, T., N. Shohdy, and J. Skowronski. 2001. Mechanism for down-regulation of CD28 by Nef. EMBO J. 20:1593-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swigut, T., M. E. Greenberg, and J. Skowronski. 2003. Cooperative interactions of simian immunodeficiency virus Nef, AP-2, and CD3-ζ mediate the selective induction of T-cell receptor-CD3 endocytosis. J. Virol. 77:8116-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka, M., U. Grossniklaus, W. Herr, and N. Hernandez. 1988. Activation of the U2 snRNA promoter by the octamer motif defines a new class of RNA polymerase II enhancer elements. Genes Dev. 12:1764-1778. [DOI] [PubMed] [Google Scholar]
- 47.Tomoda, H., K. Igarashi, and S. Omura. 1987. Inhibition of acyl-CoA synthetase by triacsins. Biochim. Biophys. Acta 921:595-598. [PubMed] [Google Scholar]
- 48.van der Vusse, G. J., M. van Bilsen, J. F. Glatz, D. M. Hasselbaink, and J. J. Luiken. 2002. Critical steps in cellular fatty acid uptake and utilization. Mol. Cell. Biochem. 239:9-15. [DOI] [PubMed] [Google Scholar]
- 49.Watkins, P. A. 1997. Fatty acid activation. Prog. Lipid Res. 36:55-83. [DOI] [PubMed] [Google Scholar]
- 50.Welker, R., M. Harris, B. Cardel, and H. G. Krausslich. 1998. Virion incorporation of human immunodeficiency virus type 1 Nef is mediated by a bipartite membrane-targeting signal: analysis of its role in enhancement of viral infectivity. J. Virol. 72:8833-88340. [DOI] [PMC free article] [PubMed] [Google Scholar]