Abstract
Novel metastasis-promoting gene 1 (NVM-1) has a significantly elevated protein level in a variety of tumor tissues and is involved in metastasis. However, its functions in hepatocellular carcinoma (HCC) are not clear. The current study aimed to investigate the functions of NVM-1 in cell proliferation, apoptosis, and epithelial-mesenchymal transition in HCC. NVM-1 protein expression in HCC was assessed by immunohistochemical staining. In vitro, cell proliferation, apoptosis, and aggressiveness were determined by CCK-8, fluorescence-assisted cell sorting, TdT-UTP nick-end labeling, and transwell assays, respectively. For in vivo studies, NVM-1 knockdown HCC cells were transplanted into BALB/c nude mice. NVM-1 was frequently upregulated in HCC tissues and positive NVM-1 expression was linked with poor prognosis. NVM-1 depletion significantly inhibited cell proliferation, migration, and invasion abilities in vitro and in vivo. Apoptosis was induced after NVM-1 knockdown. In conclusion, positive NVM-1 expression confers poor prognosis to HCC patients and the NVM-1 protein level correlates with HCC cell proliferation, apoptosis, and EMT.
Keywords: NVM-1, hepatocellular carcinoma, prognosis, proliferation, metastasis
Introduction
The prevalence of hepatocellular carcinoma (HCC) has progressively increased in recent years; it is currently the forth most common cancer and the second most common cause of cancer mortality in China [1]. The number of new cases diagnosed each year is growing, and it nearly equals the number of deaths from this cancer [2]. HCC usually remains undetected in the early stage, but it deteriorates rapidly once patients develop symptoms. For early HCC patients, radical resection and liver transplantation are still the most effective treatments; however, several recent clinical studies have indicated that the prognosis of these patients remains poor because of high recurrence and early metastasis rates after operation [3-5]. Generally, early diagnosis and treatment result in a better prognosis; therefore, early detection and diagnosis is particularly important in the management of HCC. The rapid development of biotechnology has allowed the identification of HCC prognostic biomarkers [6,7]. However, as the molecular mechanisms involved in HCC progression are complex, the pathogenesis of HCC is still unclear. Thus, it is imperative to identify new biomarkers and explore their functions to provide clues for proper management of HCC.
Novel metastasis-promoting gene 1 (NVM-1; also known as METTL21D, C14orf138, or VCP-KMT), located on chromosome 14 (ORF138), was identified in 2011 as a gene functionally involved in tumor cell motility and metastasis. The protein has a molecular weight of 26-31 kDa and is endogenously expressed in a variety of tissues, with the highest expression observed in the ovaries, duodenum, and pancreas [8-10]. The protein level is significantly elevated in head and neck squamous cell, lung, esophageal, colorectal, thyroid papillary, endometrial, and cervical carcinomas [8]. However, the expression of NVM-1 in HCC and its role in tumorigenesis remain unclear.
In the present study, we investigated NVM-1 function in HCC using in vitro assays for cell proliferation, migration, and invasion as well as in vivo studies using a xenograft mouse model.
Materials and methods
Ethics statement
The Southern Medical University Ethics Committee approved protocols according to the Helsinki Declaration (6th revision, 2008), and informed consent was signed by each patient enrolled in the study. Animal protocols were approved by the Institutional Animal Care and Use Committee of Southern Medical University.
Clinical samples
Ninety-two HCC samples and seven liver tissues with benign lesions were collected from patients who underwent surgical resection in the Department of Hepatobiliary Surgery at Nanfang Hospital of the Southern Medical University between November 2010 and October 2012. The inclusion criteria were as follows: (a) no other treatment before surgery; (b) curative surgical resection was performed; (c) resected specimens were available for pathological examination and examined for metastasis after the operation; (d) the cut edge was confirmed without residual carcinoma; (e) no hepatitis C diagnosis; and (f) a complete medical record. Cancerous tissues were defined as tissues within 1 cm from the tumor edge without necrosis, and noncancerous tissues as tissues exceeding the edge by more than 2 cm. The specimens were subjected to immunohistochemical staining, and the patients were monitored for prognostic analysis via outpatient examinations or telephone follow-up every six months. Demographic features and clinicopathologic data of the patients are shown in Table 1.
Table 1.
Expression of NVM-1 in human HCC and its clinical significance
| Characteristics | Cases | NVM-1 (negative) | NVM-1 (positive) | χ2 | P |
|---|---|---|---|---|---|
| Gender | 0.611 | 0.435 | |||
| Male | 80 | 37 (46.3%) | 43 (53.8%) | ||
| Female | 12 | 7 (58.3%) | 5 (41.7%) | ||
| Age (years) | 0.574 | 0.449 | |||
| < 60 | 78 | 36 (46.2%) | 42 (53.8%) | ||
| ≥ 60 | 14 | 8 (57.1%) | 6 (42.9%) | ||
| Hepatitis B | 2.343 | 0.126 | |||
| + | 82 | 42 (51.2%) | 40 (48.4%) | ||
| - | 10 | 2 (20.0%) | 8 (80.8%) | ||
| AFP (μg/L) | 1.093 | 0.296 | |||
| < 400 | 62 | 32 (51.6%) | 30 (48.4%) | ||
| ≥ 400 | 30 | 12 (40.0%) | 18 (60.0%) | ||
| Cirrhosis | 0.650 | 0.420 | |||
| Yes | 61 | 31 (50.8%) | 30 (49.2%) | ||
| No | 31 | 13 (41.9%) | 18 (58.1%) | ||
| Amount | 2.535 | 0.111 | |||
| = 1 | 66 | 35 (53.0%) | 31 (47.0%) | ||
| > 1 | 26 | 9 (34.6%) | 17 (65.4%) | ||
| Tumor size (cm) | 0.031 | 0.860 | |||
| ≤ 3.0 | 14 | 7 (50%) | 7 (50%) | ||
| > 3.0 | 78 | 37 (47.4%) | 41 (52.6%) | ||
| Tumor capsule | 0.565 | 0.452 | |||
| Yes | 58 | 26 (44.8%) | 32 (55.2%) | ||
| No | 34 | 18 (52.9%) | 16 (47.1%) | ||
| Differentiation | 19.723 | 0.000 | |||
| Well | 21 | 13 (61.9%) | 8 (38.1%) | ||
| Moderate | 43 | 27 (62.8%) | 16 (37.2%) | ||
| Poor | 28 | 4 (14.3%) | 24 (85.7%) | ||
| TNM stage | 5.808 | 0.016 | |||
| I+II | 69 | 38 (55.1%) | 31 (44.9%) | ||
| III+IV | 23 | 6 (26.1%) | 17 (73.9%) | ||
| BCLC stage | 8.445 | 0.015 | |||
| A | 58 | 34 (58.6%) | 24 (41.4%) | ||
| B | 12 | 5 (41.7%) | 7 (58.3%) | ||
| C | 22 | 5 (22.7%) | 17 (77.3%) | ||
| Metastasis* | 17.424 | 0.000 | |||
| No | 46 | 32 (69.6%) | 14 (30.40%) | ||
| Yes | 46 | 12 (26.1%) | 34 (73.9%) | ||
| Recurrencea | 11.127 | 0.001 | |||
| Yes | 53 | 18 (34.9%) | 35 (66.0%) | ||
| No | 34 | 24 (70.6%) | 10 (29.4%) | ||
| Recurrence time (months)a | 14.253 | 0.001 | |||
| < 6 | 36 | 9 (25.0%) | 27 (75.0%) | ||
| 6 ≤ X < 12 | 8 | 6 (75.0%) | 2 (25.0%) | ||
| ≥ 12 | 43 | 27 (62.8%) | 16 (37.2%) |
Metastasis was defined as recurrence time < 6 months or cancer embolus (including portal vein tumor thrombus, biliary tract tumor thrombus, and hepatic vein tumor thrombus).
Data are missing for 5 patients.
Immunohistochemical staining
Immunohistochemical staining was performed on paraformaldehyde-fixed paraffin sections. NVM-1 (kindly provided by Professor Wilko Thiele, Center of Biomedical and Medical Technology, Heidelberg University, Germany) (1:200) Ki-67, E-Cadherin, N-Cadherin and Vimentin antibody (Santa Cruz, CA, USA) were used in immunohistochemistry with the streptavidin peroxidase-conjugate method as reported [7]. Cells having brown nuclear granules were defined as positive cells, and positive tumor cells or hepatocytes were classified according to the following staining intensity criteria [11]: 0. no staining; 1. light staining; and 2. deep staining. Additionally, the percentage of cells showing NVM-1 expression was categorized as follows: 1. 1-25% positive cells; 2. 26-50% positive cells; 3. 51-75% positive cells; 4. > 76% positive cells. Finally, expression was evaluated semi-quantitatively based on the product of the color intensity and positive cell percentage scores: < 4, negative expression; ≥ 4, positive expression. Each specimen was evaluated independently by two professional pathologists.
Cell lines
HL-7702, HepG2, SMCC-7721, MHCC-97L, MHCC-97H, and HCCLM3 cells (Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences, China) were cultured in high-glucose DMEM (Gibco, CA, USA) supplemented with 10% fetal bovine serum FBS (Gibco) in a humidified atmosphere containing 5% CO2. Cells in the exponential phase were harvested at approximately 80% confluence. The HL-7702 cell line was used as a control.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA of the tissue samples was extracted with RNAiso Plus reagent (Takara, Dalian, China). cDNA was prepared from total RNA using the PrimeScriptTM First-Strand cDNA Synthesis Kit (TaKaRa, Dalian, China). NVM-1 and GAPDH mRNA expression was measured by PCR using a SYBR green qPCR assay kit (Roche, Shanghai, China). Primer sequences were as follows: NVM-1 sense primer 5’-TGA GCT CCT TCA GCT AGA TTT TGA-3’ and antisense primer 5’-CAG GTT GTT AGA GCC TTG GGT-3’, and GAPDH sense primer 5’-AGC TGA ACG GGA AGC TCA CT-3’ and antisense primer 5’-TGC TTA GCC AAA TTC GTT G-3’. The data were interpreted using the 2-ΔΔCt method with GAPDH serving as a reference gene for normalization.
Gene silencing
For transient transfection, MHCC-97L, MHCC-97H, and HCCLM3 cells were plated in 6-well plates and transfected with small interfering RNA (siRNA) for 48 h using Lipofectamine 2000 (Invitrogen, CA, USA) following the manufacturer’s instruction. The sequences were as follows: S1 targeting 5’-GCA TCT TGT CAC TGG TTC TT-3’, S2 targeting 5’-AGA TAT CAG CGG ATT TGA AT-3’), negative control (NC) targeting 5’-TTC TCC GAA CGT GTC ACG TTT-3’. The transfected cells were used in further assays or for RNA/protein extraction. Lentivirus-mediated transfection of short hairpin RNA (shRNA) (GenePharma, Shanghai, China) was performed on HCCLM3 cells according to manufacturer’s protocol. The target and negative control sequences were 5’-GCA TCT TGT CAC TGG TTC TT-3’ and 5’-TTC TCC GAA CGT GTC ACG TTT-3’, respectively.
Western blotting
The following primary antibodies were used in immunoblotting assays: the NVM-1 antibody kindly provided by Professor Wilko Thiele, and apoptosis- and EMT-related antibodies (Bcl-2, Bax, Caspase-3, Caspase-8, p53, β-catenin, slug, MMP2, MMP9) purchased from Santa Cruz (CA, USA). Horseradish peroxidase-conjugated goat anti-mouse/rabbit secondary antibody (Abcam, CA, USA) was used at a 1:4000 dilution and detected using ECL substrate (Millipore, MA, USA) as described [12].
Cell proliferation assay
Cell growth was measured using the CCK-8 assay (KeyGen, Nanjing, China), as described previously [7].
Apoptosis assay
Cell apoptosis was measured by FACS analysis using the Annexin V-FITC/PI Apoptosis Detection Kit (KeyGen). TdT-UTP nick end-labeling (TUNEL) assays were performed with a one-step TUNEL assay kit (Roche, Switzerland), according to the manufacturer’s instructions, as described previously [7].
Transwell migration and invasion assays
Transfected cells were trypsinized and resuspended in serum-free medium. A total of 5 × 105 cells (200 μl) were added to the upper chambers of transwell inserts with 8-μm pores in 24-well plates (Corning, CA, USA). Migration was induced with 20% FBS-supplemented medium added to the lower chambers. After 48 h, non-adherent cells in the chambers were removed and the inserts were fixed in 4% paraformaldehyde and stained with 0.5% crystal violet. Numbers of migrated cells were counted in three different fields under an inverted microscope (Nikon, Tokyo, Japan). For the invasion assay, all conditions were as described for the migration assay, except that cells were added to Matrigel-coated inserts.
In vivo experiments
HCCLM3 cells (5 × 106) stably transfected with a Lentivirus encoding NVM-1 shRNA (sh-NVM-1) or negative control shRNA (sh-NC) were subcutaneously injected into the flanks of BALB/c nude mice as previously described [13]. The tumor volume was determined by measuring two of its dimensions and calculated as tumor volume (V) = length (a) × width (b) × √(a × b) × π/6. Tumor samples were subjected to further detections.
Statistical analysis
Results are displayed as the mean ± SEM. Data were analyzed with the SPSS statistical package for Windows version 13 (SPSS, Chicago, IL, USA) and GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA, USA). Means were compared using the Pearson chi-squared test, multi-variant Cox regression analysis, two-tailed Student’s t-tests, Kaplan-Meier plots, log-rank tests, or ANOVA as appropriate. Differences were considered significant at P < 0.05.
Results
NVM-1 is frequently upregulated in HCC tissues and positive NVM-1 expression confers poor prognosis
To explore NVM-1 expression in HCC tissues, we detected the protein expression in a retrospective cohort of 92 paired HCC samples by immunohistochemical staining. NVM-1 protein expression in HCC cancerous tissues was significantly higher than that in matched noncancerous tissues, while no staining was observed in liver tissues with benign lesions (Figure 1). NVM-1 mRNA expression was higher in various HCC cell lines (HepG2, SMCC-7721, MHCC-97L, MHCC-97H, and HCCLM3) than in the normal hepatocyte cell line, HL-7702 (P = 0.000, Figure 2A). Positive protein expression was detected in 52.2% (48/92) of the HCC specimens, whereas only 28.3% (26/92) of the matched noncancerous tissues showed a positive signal (P< 0.05, Figure 1A). As shown in Table 1, positive expression was significantly associated with the degree of tumor differentiation (P = 0.000), TNM stage (P = 0.016), BCLC stage (P = 0.015), tumor metastasis (P = 0.000), recurrence (P = 0.001), and recurrence time (P = 0.001). These results indicated that elevated expression of NVM-1 is correlated with malignant clinicopathologic parameters of HCC. Kaplan-Meier analysis of clinical survival information from the patients indicated that tumors with NVM-1 positive expression indeed associated with a shorter relapse-free survival time (P = 0.001, Figure 1B and 1C), which suggests that NVM-1 may act as a promising biomarker for predicting prognosis of HCC patients.
Figure 1.
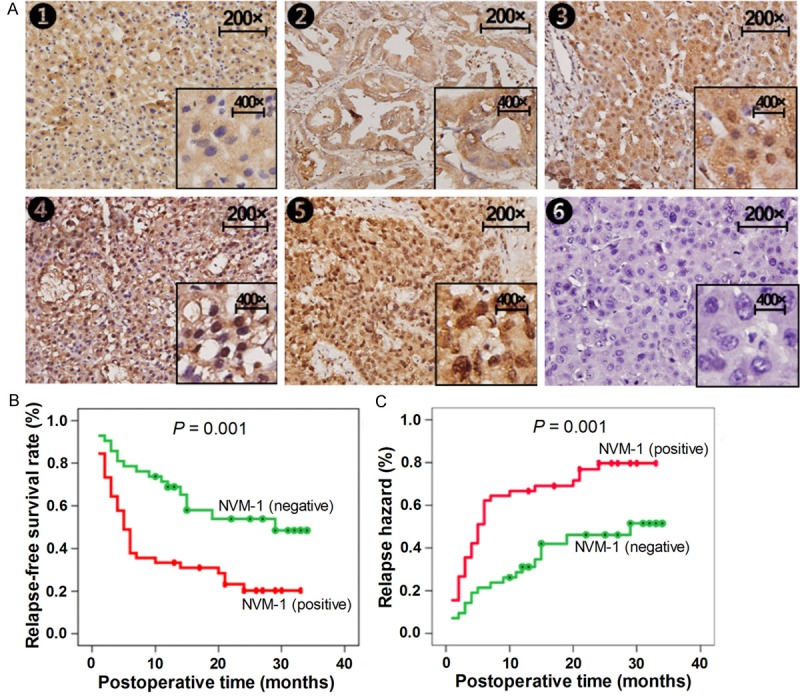
Expression of NVM-1 is upregulated in HCC tissues and negatively correlates with the prognosis for HCC patients. (A) NVM-1 protein expression detected by immunohistochemical staining in liver tissues (200 × and 400 ×): 1, negative expression in normal liver tissues; 2, negative expression in benign liver tissues; 3, negative expression in well-differentiated HCC tissues; 4, positive expression in moderately differentiated HCC tissues; 5, strong positive expression in poorly differentiated HCC tissues; 6, negative control. (B) Relationship between NVM-1 expression in HCC tissues and relapse-free survival rate or relapse hazard: (B) survival curve; (C) relapse hazard.
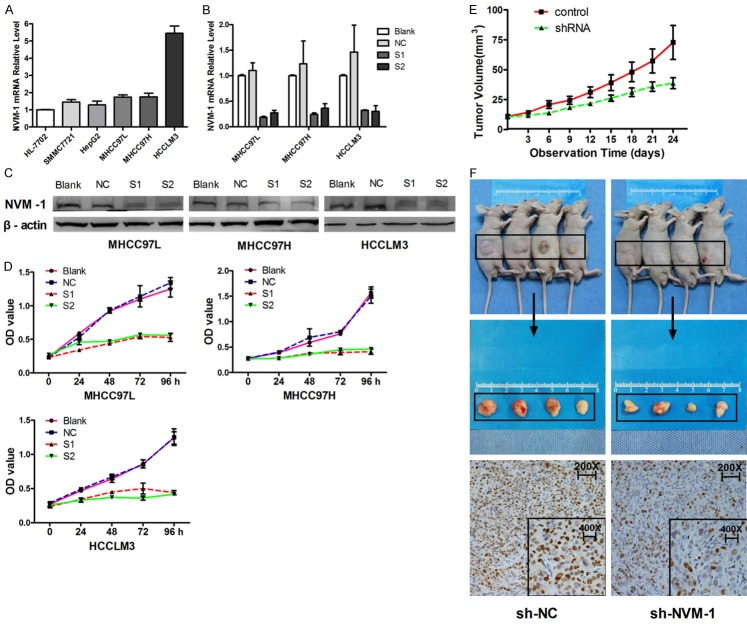
Figure 2.
NVM-1 knockdown by siRNA inhibits tumor cell proliferation in vitro and in vivo. A: Expression of NVM-1 mRNA in HL-7702 and five HCC cell lines. B: Efficiency of NVM-1 knockdown measured by RT-PCR. C: Efficiency of NVM-1 knockdown measured by western blotting. D: Cell proliferation after NVM-1 knockdown tested by CCK-8. E: Tumor volumes measured on the indicated days to assess the effects of NVM-1 on subcutaneous tumor growth in nude mice. F: Expression of Ki-67 protein in tumor nodules detected by immunohistochemical staining.
Successful NVM-1 knockdown in HCC cell lines
To identify the role of NVM-1 in HCC cells, we chose MHCC-97L, MHCC-97H, and HCCLM3 cells for further functional studies in vitro as they express a high level of NVM-1 mRNA and have high metastatic potential. After transient transfection of NVM-1 siRNA for 48 h, qRT-PCR and western blot analyses showed that both mRNA and protein expression were successfully knocked down in the three HCC cell lines (P < 0.05 in all cases, Figure 2B and 2C).
NVM-1 promotes tumor growth in vitro and in vivo
Limitless replicative potential is a hallmark of cancer cells [14]. To better understand the effects of NVM-1 silencing on cell viability, and more specifically on cell growth, cell viability after transfection was tested at different time points using a CCK-8 assay. A marked reduction in cell viability was observed after NVM-1 depletion as compared with non-transfected control cells in the three HCC cell lines (P < 0.05 in all cases, Figure 2D). Next, we sought to determine whether NVM-1 affects tumor growth in a nude mouse xenograft model. HCCLM3 cells stably transfected with a plasmid harboring sh-NC or sh-NVM-1 were implanted into nude mice through subcutaneous injection. The results showed that subcutaneous tumor volumes in mice of the sh-NVM-1-expressing group were obviously smaller than those in the sh-NC-expressing group (Figure 2F). The tumor growth curves revealed that NVM-1 knockdown slowed down tumor growth significantly (P < 0.05, Figure 2E), which was consistent with the in vitro data. Immunohistochemical staining indicated that the expression of the proliferation marker Ki-67 in subcutaneous tumor was significantly lower in the sh-NVM-1 group than in the sh-NC group (Figure 2F). Thus, we speculated that NVM-1 may act as an oncogene by promoting the proliferative ability of HCC cells.
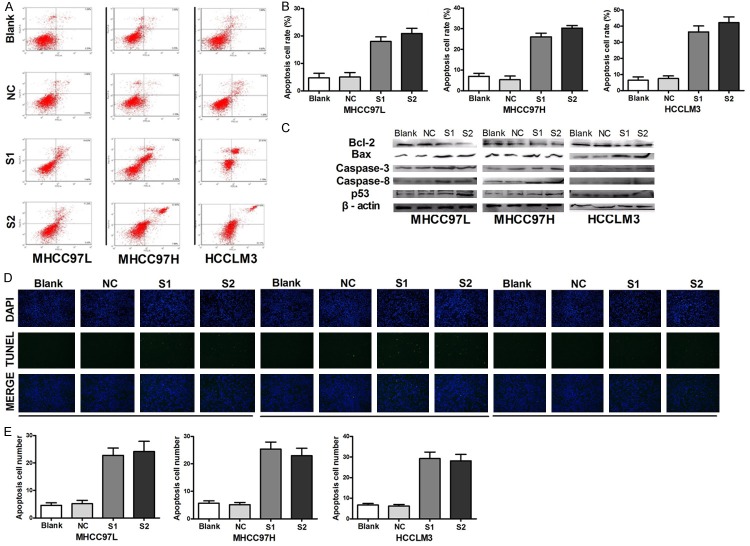
NVM-1 silencing induces apoptosis in HCC cells
Evasion of apoptosis is another hallmark of cancer cells [14]. To explore the potential effect of NVM-1 on apoptosis, flow cytometry was used to detect changes in apoptosis in NVM-1-silenced versus non-silenced HCC cell lines. Remarkable changes were detected in NVM-1 depletion groups (> 17% late apoptotic cells) compared with the control groups (< 8% late apoptotic cells) in the three HCC cell lines (P < 0.05 in all cases, Figure 3A and 3B). Similar results were obtained by TUNEL assay (Figure 3D and 3E). Western blot analysis revealed increased p53, Bax, caspase-3, and caspase-8 expression, and decreased Bcl expression in the three HCC cell lines upon NVM-1 silencing (Figure 3C). Together, these results indicated that NVM-1 plays an important anti-apoptotic role and may be tightly associated with the levels of apoptosis-related proteins.
Figure 3.
NVM-1 silencing by siRNA induces apoptosis in HCC cells. A: Change in apoptotic cell fraction after NVM-1 knockdown analyzed by flow cytometry. B: Proportions of early and late apoptotic cells after NVM-1 silencing. C: Expression of apoptosis-related proteins detected by western blotting after NVM-1 knockdown. D: Apoptosis detected by TUNEL assay after NVM-1 depletion. E: Quantification of apoptotic cells detected by TUNEL after NVM-1 depletion.
NVM-1 promotes migration, invasion, and EMT progression in vitro and in vivo
In the process of tumor formation, cancer cells tend to have stronger migration and invasion abilities, and epithelial-mesenchymal transition (EMT) progression is considered to be related to these phenomena [15]. To determine whether NVM-1 affects the HCC cell migration and invasion abilities, transwell migration and invasion assays were conducted after NVM-1 silencing. NVM-1 knockdown led to significantly lower migration and invasion (P < 0.05 for both, Figure 4). In addition, decreased N-cadherin, Slug, MMP2, MMP9, and β-catenin protein expression, and increased E-cadherin protein level were observed in NVM-1-depleted cells (Figure 5A). To further elucidate the effects of NVM-1 on HCC cell migration and invasion, HCCLM3 cells stably transfected with sh-NC or sh-NVM-1 were injected into the caudal vein in 4 pairs of nude mice. Livers and lungs were harvested after 2 months. Pathologic examination revealed no liver or lung metastases in the sh-NVM-1 group, while in the sh-NC group, all mice had liver metastases and 2 out of 4 mice had lung metastases in varying degrees (Figure 5B). Immunohistochemical staining indicated that N-cadherin and Vimentin expression in subcutaneous tumor was significantly lower in the sh-NVM-1 group than in the sh-NC group, while E-cadherin was upregulated (Figure 5C).
Figure 4.
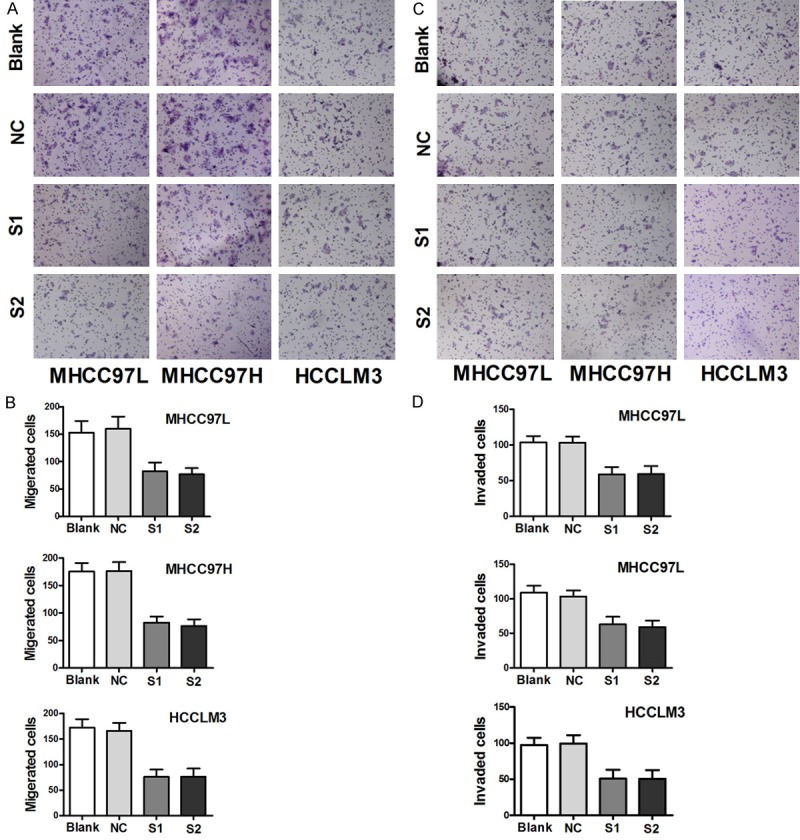
NVM-1 promotes migration and invasion in vitro. A, B: Change in cell migration ability measured by transwell assay after NVM-1 silencing. C, D: Change in cell invasion ability measured by transwell after NVM-1 silencing assay.
Figure 5.
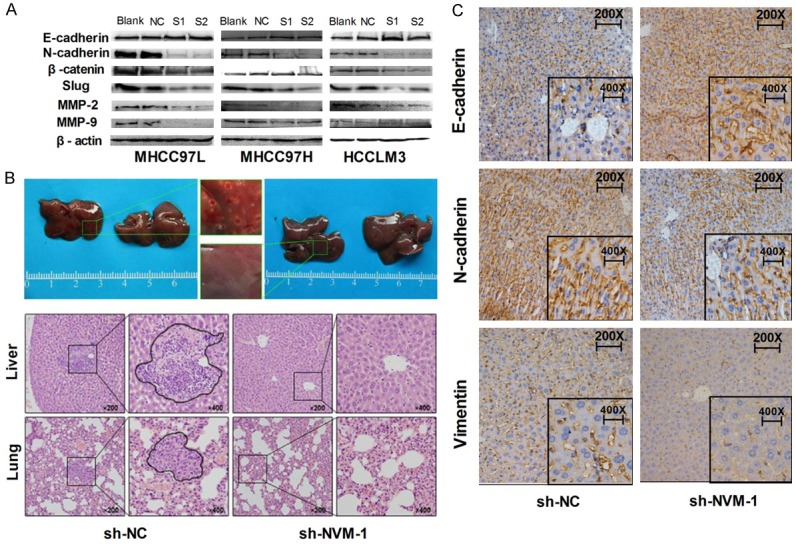
NVM-1 depletion inhibits tumor cell proliferation, metastasis and EMT in vitro and in vivo. A: Expression of EMT-related proteins detected by western blotting in HCC cells after NVM-1 knockdown. B: HE staining of liver and lung metastatic tissues. C: Expression of EMT-related proteins in tumor nodules detected by immunohistochemical staining.
Discussion
Tumor metastasis and recurrence are thought to account for nearly 90% of cancer deaths [16]. Understanding the Underlining molecular regulatory mechanism of metastasis formation and recurrence as a prerequisite for the development of novel therapies that improve survival. A previous study indicated a critical role for NVM-1 in tumor invasion and metastasis and suggested NVM-1 as a potential, novel target for anti-metastatic cancer therapy [8]. To our knowledge, the role of NVM-1 in metastasis or recurrence in HCC has not been thoroughly investigated to date.
To explore the role of NVM-1 in HCC, we initially detected NVM-1 protein expression in clinical tissue specimens. NVM-1 protein was observed in both the nucleus and the cytoplasm but was more abundant in the nucleus, and the NVM-1-positive rate was higher in cancerous tissues than in noncancerous tissues and benign hepatic lesions. Additionally, NVM-1 mRNA expression was higher in HCC cell lines than in normal hepatocytes, which is in line with a previous report [8]. We found that NVM-1 expression correlated with tumor stage, tumor recurrence, and metastasis, and NVM-1 was enhanced in poorly differentiated tumors, especially in patients displaying early recurrence (within 6 months). Furthermore, patients with high expression of NVM-1 had a significantly shorter relapse-free survival time. Altogether, these results suggest that NVM-1 expression is critical for prognosis determination in HCC patients, and the protein may have a role in tumor metastasis and recurrence.
In most HCC cases, hepatocarcinogenesis arises in the setting of chronic liver damage and inflammation, which leads to fibrosis and, ultimately, to cirrhosis [17]. In China, the leading risk factor for HCC is hepatitis B or C virus infection [18]. In our study, nearly 90% of HCC patients were diagnosed with hepatitis B; however, we found no competent evidence to confirm the correlation between expression of NVM-1 and hepatitis B infection. Moreover, we found no significant correlation between NVM-1 expression and alpha-1-fetoprotein (AFP) expression, tumor size, lymph node metastasis, etc. However, higher rates of NVM-1 positive expression were found in patients with high AFP level, larger tumors (> 3 cm), multiple tumors, or lymph node metastasis. The fact that we did not find significant correlations may be caused by an insufficient number of cases; further studies are required to explore the relationship between NVM-1 and HCC.
Tumorigenesis is a complex process involving a variety of factors, including uncontrolled cell proliferation [19], cell cycle disorders [20,21], and apoptosis evasion [22]. Using siRNA, NVM-1 expression was successfully knocked down in the three HCC cell lines, MHCC-97L, MHCC-97H, and HCCLM3. The proliferation ability of the NVM-1-silenced cells was significantly weakened, apoptosis was induced, and apoptosis-related proteins changed accordingly. Thus, we speculate that the inhibition of cell proliferation induced by NVM-1 depletion may be achieved through promoting apoptosis.
In recent years, EMT has been considered to be related to tumor metastasis [23]. In the process of EMT, epithelial cells break-up their intercellular connections and cell-matrix junctions, reorganize their cytoskeleton, change shape, and reprogram gene expression and signaling pathways to increase their motility and acquire an invasive phenotype [24,25]. In our study, cell migration and invasion were weakened after NVM-1 silencing. To verify the role of NVM-1 in HCC in vivo, we established a xenograft mouse model to assay tumor growth and metastasis. Silencing of NVM-1 significantly slowed down tumor growth and lowered the incidence of liver and lung metastases. Thus, NVM-1 may play an important role in tumor aggressiveness, which was corroborated by corresponding changes in EMT-related proteins in vitro and in vivo. This may be due to the function of NVM-1 protein as a methyltransferase; on the one hand, NVM-1 can change the methylation status of various proteins, promoting tumor cell proliferation and metastasis potential, and on the other hand, NVM-1 protein can appear in the nucleus and in the cytoplasm, but it functions in the nucleus to ultimately promote tumorigenesis [9]. In the light of these results, we believe that NVM-1 might exert a metastasis-promoting effect by activating EMT in HCC.
To our knowledge, this study for the first time showed that NVM-1 is upregulated in HCC tissues and negatively correlates with the prognosis of HCC. We found that NVM-1 knockdown significantly inhibits tumor cell proliferation, migration, invasion, and EMT progression. Thus, we expect NVM-1 to become a new, effective biomarker that will open up treatment options for HCC.
Acknowledgements
This project was supported by Guangdong Provincial Science and Technology Projects of China (2013B02200069). The authors thank Professor Wilko Thiele, Center of Biomedical and Medical Technology, Heidelberg university, Germany for providing NVM-1 antibody for our study.
Disclosure of conflict of interest
None.
References
- 1.Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27:2–12. doi: 10.3978/j.issn.1000-9604.2015.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42(Suppl 3):S206–214. doi: 10.1016/S1590-8658(10)60507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tu K, Li C, Zheng X, Yang W, Yao Y, Liu Q. Prognostic significance of miR-218 in human hepatocellular carcinoma and its role in cell growth. Oncol Rep. 2014;32:1571–1577. doi: 10.3892/or.2014.3386. [DOI] [PubMed] [Google Scholar]
- 5.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 6.Cha C, Fong Y, Jarnagin WR, Blumgart LH, DeMatteo RP. Predictors and patterns of recurrence after resection of hepatocellular carcinoma. J Am Coll Surg. 2003;197:753–758. doi: 10.1016/j.jamcollsurg.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Liang B, Jia C, Huang Y, He H, Li J, Liao H, Liu X, Liu X, Bai X, Yang D. TPX2 level correlates with hepatocellular carcinoma cell proliferation, apoptosis, and EMT. Dig Dis Sci. 2015;60:2360–2372. doi: 10.1007/s10620-015-3730-9. [DOI] [PubMed] [Google Scholar]
- 8.Thiele W, Novac N, Mink S, Schreiber C, Plaumann D, Fritzmann J, Cremers N, Rothley M, Schwager C, Regiert T, Huber PE, Stein U, Schlag P, Moll J, Abdollahi A, Sleeman JP. Discovery of a novel tumour metastasis-promoting gene, NVM-1. J Pathol. 2011;225:96–105. doi: 10.1002/path.2924. [DOI] [PubMed] [Google Scholar]
- 9.Kernstock S, Davydova E, Jakobsson M, Moen A, Pettersen S, Maelandsmo GM, Egge-Jacobsen W, Falnes PO. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat Commun. 2012;3:1038. doi: 10.1038/ncomms2041. [DOI] [PubMed] [Google Scholar]
- 10.Cloutier P, Lavallee-Adam M, Faubert D, Blanchette M, Coulombe B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 2013;9:e1003210. doi: 10.1371/journal.pgen.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Huang Y, Yang D, Li X, Liang J, Lin L, Zhang M, Zhong K, Liang B, Li J. Overexpression of TRIM24 is associated with the onset and progress of human hepatocellular carcinoma. PLoS One. 2014;9:e85462. doi: 10.1371/journal.pone.0085462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao H, Huang Y, Guo B, Liang B, Liu X, Ou H, Jiang C, Li X, Yang D. Dramatic antitumor effects of the dual mTORC1 and mTORC2 inhibitor AZD2014 in hepatocellular carcinoma. Am J Cancer Res. 2015;5:125–139. [PMC free article] [PubMed] [Google Scholar]
- 13.Tu K, Zheng X, Zhou Z, Li C, Zhang J, Gao J, Yao Y, Liu Q. Recombinant human adenovirus-p53 injection induced apoptosis in hepatocellular carcinoma cell lines mediated by p53-Fbxw7 pathway, which controls c-Myc and cyclin E. PLoS One. 2013;8:e68574. doi: 10.1371/journal.pone.0068574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 16.Sporn MB. The war on cancer. Lancet. 1996;347:1377–1381. doi: 10.1016/s0140-6736(96)91015-6. [DOI] [PubMed] [Google Scholar]
- 17.Bartosch B. Hepatitis B and C viruses and hepatocellular carcinoma. Viruses. 2010;2:1504–1509. doi: 10.3390/v2081504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. e1261. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claesson-Welsh L. Platelet-derived growth factor receptor signals. J Biol Chem. 1994;269:32023–32026. [PubMed] [Google Scholar]
- 20.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 21.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 22.Daniel PT. Dissecting the pathways to death. Leukemia. 2000;14:2035–2044. doi: 10.1038/sj.leu.2401940. [DOI] [PubMed] [Google Scholar]
- 23.Puchinskaia MV. [Epithelial-mesenchymal transition in health and disease] . Arkh Patol. 2015;77:75–83. doi: 10.17116/patol201577175-. [DOI] [PubMed] [Google Scholar]
- 24.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 25.Medici D, Munoz-Canoves P, Yang PC, Brunelli S. Mesenchymal Transitions in Development and Disease. Stem Cells Int. 2016;2016:5107517. doi: 10.1155/2016/5107517. [DOI] [PMC free article] [PubMed] [Google Scholar]