Abstract
Medulloblastoma is one of the most prevalent pediatric brain malignancies, accounting for approximately 20% of all primary CNS tumors in children under the age of 19. OTX2 is the member of a highly conserved family of bicoid-like homeodomain transcription factors responsible for the regulation of cerebellar development and of current investigational interest in the tumorigenesis of medulloblastoma. Recent studies have revealed that Group 3 and Group 4 medulloblastomas show marked overexpression of OTX2 with a concurrent amplification of the MYC and MYCN oncogenes, respectively, correlating with anaplasticity and unfavorable patient outcomes. More recent attempts at elucidating the mechanism of OTX2-driven oncogenesis at the cellular level has also revealed that OTX2 may confer stem-cell like properties to tumor cells via epigenetic regulation. The review seeks to define the interaction pathways and binding partners involved in OTX2 function, its usefulness as a molecular marker for risk stratification and prognosis, and the mechanism by which it drives tumor maintenance. Additionally, it will preview unpublished data by our group highlighting the unanticipated involvement of OTX2 in the control of cellular metabolism.
Keywords: OTX2, medulloblastoma, MYC, histone modification, cancer metabolism
Introduction
Medulloblastoma is an embryonal solid tumor of the CNS with just over a 60% 5-year survival rate in children, and an even worse prognosis in infants [1]. The prognosis of the disease is largely predicated on which subtype of the neuroectodermal tumor a patient has. Previously established conventions for molecular subtyping utilized the age of the patient and extent of the disease progression. Genomic studies have elaborated a system of subtyping that segregates the tumors into four groups according to mechanisms of pathogenesis and presence of molecular markers. They are WNT, SHH, Group 3, and Group 4 [2]. Sequencing and genomic studies have helped generate a comprehensive snapshot of the genotypic landscape of the more benign medulloblastoma variants, yielding two molecular signatures that link pathogenesis of the WNT and SHH subtypes to over-activation of their namesake signaling pathways. Group 3 and 4 medulloblastoma have yet to be defined to the same extent as the former two subtypes, and Group 3 tumors recur most frequently and have the highest propensity to metastasize [3,4]. Therefore, it is especially important to characterize the gene amplifications that make up Group 3 and 4 tumors in order to create targeted therapies for these aggressive subtypes. Multiple recent large population genomic studies found marked amplification of the MYC gene in Group 3 tumors compared to other subtypes; similar studies found N-MYC amplification to be associated with Group 4 tumors [2,5,6]. Therefore, it has become increasingly important to find novel therapeutic targets that associate directly or indirectly with MYC and contribute to the aggressive nature of Group 3 and Group 4 medulloblastomas.
OTX2 (orthodenticle homeobox 2) is a transcription factor responsible for mediating embryogenesis in the CNS, regulating formation of the brain parenchyma, pineal gland, cerebellum, eye, and the external granule cell layer in cerebellar development, a site extremely relevant in medulloblastoma development [7,8]. In the past, OTX2 has been implicated as a possible oncogene in a number of malignancies [9,10], but has recently emerged as a potential driver of medulloblastoma tumorigenesis. Indeed, contemporary studies have highlighted OTX2 amplification and overexpression to underly particular subtypes of medulloblastoma tumor pathology [11]. While the predictive value of OTX2 as a definitive molecular marker still remains an area of active research, there is consensus that screening for genomic aberrations in OTX2 is an integral component in the establishment of a robust scheme of molecular classification of the Group 3 and Group 4 subtypes [12]. OTX2 has most recently been implicated in the sustenance of tumor anaplasia, promotion of tumor proliferation, and triggering of cell cycle progression [12]. OTX2 preserves stem-like characteristics of medulloblastoma cells via regulation of transcriptional activation of its binding partners, including both epigenetic regulators and cell cycle enzymes [13], as well as inhibition of myogenic and neuronal differentiation by interactions with specific enhancer sequences [14], which will be the focus of the remainder of this review.
OTX2 in brain and cerebellar parenchymal development and cancer
Before delving into the roles that OTX2 plays in cell cycle evasion and preservation of stemness in medulloblastoma, it is important to describe the role OTX2 plays in normal cerebellar development and in other cancer lines. OTX2 is a crucial driver of brain regionalization and development in vertebrates. In mouse embryos, OTX2 has been shown to direct axis formation and morphological regionalization of the brain. Its transcription can be detected as early as the morula stage of development [15]. As development progresses, OTX2 transcription becomes localized namely to the anterior region of the embryo [15]. This localization contributes to the development of the forebrain and midbrain, but OTX2 also serves a role in creating the anatomic border between the hindbrain and midbrain [15]. In later development, OTX2 expression is elevated in the posterior region of the developing cerebellum, helping to define the major functional boundaries of the cerebellum [16]. Furthermore, during embryonic development, OTX2 expression is observed in both the external and internal granular layers of the cerebellum [16]. Embryonic viability is totally compromised in mouse embryos with complete OTX2 deletion, emphasizing the necessity for OTX2 expression in the developing brain [17]. In humans, the OTX2 protein is identical in amino-acid sequence to the protein produced in mice, and the expression pattern described in mice aligns roughly with the expression patterns observed in human fetal brains aged week 7 to 14 [15,18].
Aside from typical neurodevelopment, there is a body of emerging literature that implicates OTX2 amplification and overexpression in malignancies. In one such study, Boon & colleagues performed copy number analysis to reveal the presence of OTX2 copy number amplification 8 of 42 primary tumors and in 2 of 5 medulloblastoma cell lines. Intratumor genomic aberrations, localized to two novel amplifications on chromosome 14 via digital karyotyping, were assessed by mRNA transcript quantification, and OTX2 was upregulated at both the DNA and RNA level [11]. Similarly, Adamson and colleagues performed high-density SNP arrays on a larger cohort consisting of 201 primary tumors and 11 medulloblastoma cell lines to pinpoint overall genomic aberrations, which localized the single region of highest focal gains to the OTX2 gene, thus confirming the findings of the Boon study. Subsequent in situ hybridization in three medulloblastoma cell lines was carried out in this study to elucidate the mechanism of OTX2 amplification revealed the predominant form of cellular OTX2 chromosomal gain to be double minutes, which are small circular fragments of extrachromosomal DNA, harboring amplicons of specific oncogenes and genes involved in drug resistance.
Emerging role of OTX2 in Groups 3 & 4 medulloblastoma
With an understanding of the essential role of OTX2 expression in cerebellar development and its general presence in cancer, we can now hone on the reason as to why the gene has emerged as a recent target of medulloblastoma research. After all, neural progenitor cells within the ventricular zone and the external granular layer of the developing cerebellum have been implicated as the source of Group 3 medulloblastoma, (although the origin cells for Group 4 remain unknown) [19]. Focusing on the external granule layer, evidence shows that cell cycle perturbations within this region contribute to medulloblastoma tumorigenesis [20].
Recently, genomic studies have established a significant body of evidence that supports identification of OTX2 as a present oncogene in all four subtypes of medulloblastoma to some extent [11,21,22]. New literature, however, highlights the contrast between the augmented role of OTX2 in Groups 3 and 4 aggressiveness to its relative inaction in SHH/WNT-types [9]. Pietsch & Haberler recommend OTX2 as an essential marker for identifying medulloblastomas as Group 3/4 based on a review of updated WHO classification of CNS tumors [23]. This is due to literature that has shown OTX2 overexpression to be most prominent in Group 3 and Group 4 medulloblastomas [12], and that downregulation of OTX2 has been shown to inhibit growth of medulloblastoma cells that overexpress OTX2 [24]. Additionally, Adamson and colleagues correlated OTX2 overexpression with specificity to tumors, selected from a cohort of 81 samples, that were categorized as either Group C and Group D [12], (the new equivalents of Groups 3 and 4, respectively. These groups are SHH-independent medulloblastomas that carry strong associations with both increased aggressiveness and “classical” histopathological phenotype [25], and are, for all practical purposes, identical to tumors that are considered Group 3 or Group 4 by the current subtyping conventions [2]. The association of OTX2 overexpression with Group 3 and Group 4 tumors has been repeatedly confirmed by subsequently published data [8,12,14]. OTX2 expression was found to be consistently overexpressed in Group 3 tumors in molecular clustering studies, which serves as the current gold standard approach for subtyping medulloblastomas [26]. An additional study that molecularly screened patient samples demonstrated overexpression of OTX2 in the overwhelming majority of Group 3 tumors [2]. Also noteworthy is evidence from Haas and colleagues that OTX2 overexpression contributes to the regulation of Group 3 tumor localization, and that it may actively participate in localization. Medulloblastomas of the classical histopathological designation are usually found to originate in the vermis of the cerebellum, whereas the nodular/desmoplastic variants arise in the cerebellar hemispheres. After analyzing localization data on medulloblastoma tumors at diagnosis, the authors found a strong correlation between OTX2 expression and an origin in the vermis [22]. Further support for OTX2-induced cerebellar localization of Wnt/SHH-independent MB tumor cells can be gleaned from studies involving orthotopic animal models. Wortham and colleagues produced an animal model by generating and introducing a Cre-recombinase-inducible OTX2 construct into the hindbrain of mice and analyzed the subsequent effects of induced OTX2 expression [9]. Compared to wildtype mice, OTX2 overexpression produced marked focal hyperplasia of the cerebellar white matter as well as of the dorsal pontine parenchyma. Immunohistochemical staining for Ki67 showed this so-called “ectopia” to be spheres of neuronal progenitor cells, and additional spatial localization and immunostaining studies characterized these clusters of ectopic hyperplasia as distinct from pre-neoplastic cells induced by SHH/Wnt pathway dysregulation [9].
Interestingly, OTX2 enrichment in Group 3 and 4 tumors, when mimicked in SHH-type MBs, exert the reverse effect by inducing senescence and inhibiting tumor progression. While increased OTX2 levels is correlated with self-renewal and migration of tumor cells in the Group 3 and 4 subtypes, studies show its levels to be negatively associated with the expression of pluripotency genes in SHH subtypes [9]. OTX2 overexpression in both transformed human embryonic cell models and SHH-positive MB cells by Kaur et al inhibited growth, self-renewal, and migration [8]. The downstream attenuation in tumor growth produced by OTX2 overexpression offers additional evidence that OTX2 exerts its proliferative effects in a subtype-specific manner, limited to group 3 and 4 subtypes. Additionally, Group 3 and 4 subtypes are associated with amplifications of MYC, raising the question of the possible involvement of an OTX2-MYC axis in the oncogenic process, which will be discussed in greater detail later in this review.
Role of OTX2 in the induction and maintenance of stemness
While the specific role of OTX2 in tumor formation in medulloblastomas remains an area of active research, several studies have suggested that OTX2 exerts its primary oncogenic effects through its role as a functional gatekeeper of stemness, regulating transcriptional activity to inhibit cellular differentiation. An investigation by Bai et al revealed that in the context of myogenic and neuronal differentiation, OTX2 functions as a transcription repressor via its homeobox binding domain. OTX2 transcriptionally inhibits myogenic differentiation by binding the core enhancer region of the MyoD DNA binding protein, a key initiator of this process, and siRNA-mediated knockdown of OTX2 in a medulloblastoma cell line led to a remarkable increase in differentiation of primitive neuroectodermal cells to muscle cells [14]. Similarly, an earlier study by Bunt & colleagues illustrated increased neuronal differentiation with decreased OTX2 expression. Doxycycline-induced shRNA knockdown of OTX2 in a medulloblastoma cell line showed a consequent morphological shift of the tumor stem cells from their normal round shape to a more differentiated phenotype with neuritic outgrowths. This phenotypic shift was accompanied by a concurrent upregulation of genes involved in nervous system development and a downregulation of genes mediating cell cycle progression [7]. Ultimately, this induction of neuronal differentiation is a recurrent phenomenon that accompanies OTX2 silencing in medulloblastoma cell lines [7,14]. Also noteworthy are evidence provided by the aforementioned Wortham animal model, which showed mice cerebellar hyperplasia induced by OTX2 overexpression as well as expression of characteristic neuronal progenitor markers. Interestingly, the phenotypic changes mediated by OTX2 overexpression were attenuated following mice maturity, with cells in the periphery of the aforementioned ectopia demonstrating increased differentiation and eventual dispersion, suggesting that the effects of OTX2 on the proliferative duration of these tumor cells are limited [9].
The specific mechanism underlying OTX2-mediated maintenance of pluripotency and self-renewal in tumor cells remains an area of active study, with the most contemporary studies focused on the regulatory role of OTX2 in the epigenome, namely its effects on chromatin remodeling proteins and histone modifiers. A seminal study by Robinson and colleagues studied next generation sequencing data and identified multiple selective mutations in histone modifying genes that were highly enriched in Group 3 and Group 4 tumors, including three genes whose products interact to direct cell differentiation: H3K27 demethylase, KDM6A, and EZH2 [27]. Concurrently, a study by Bunt et al suggested that OTX2 mediates the maintenance of stem-cell like features in MB tumors via the direct transcriptional regulation of these histone modification genes. In particular, OTX2 maintains bivalent state of promoter activation of a number of polycomb genes in Group 3 and Group 4 tumors through maintenance of their H3K27me3 levels [13]. As we will illustrate in the subsequent sections, aberrant expression and dysregulation of these epigenetic modifiers is proposed to be a key driver in the etiology of the corresponding medulloblastoma.
OTX2 works synergistically with MYC in oncogenesis
We now understand that OTX2 works to maintain stemness in Groups 3 and 4 medulloblastoma subtypes. Due to its role as a transcription factor, OTX2 selectively activates and represses genes that ultimately contribute to the formation of the aggressive medulloblastoma subtypes. OTX2 has a high affinity to bind in a complex with many other important oncogenes, one of the most important of which being MYC [28]. OTX2 and MYC were observed as binding in close proximity to one another at transcription start sites of genes contributing to the cancer phenotype, upregulating gene expression in the medulloblastoma condition. Adamson et al further confirmed this association utilizing ChIP analysis and PCR, showing that endogenous OTX2 binds to the promoter region of MYC. Knockdown of OTX2 by OTX2-specific siRNA showed downregulation of MYC [12]. MYC is known to act downstream on the Mad/Max signaling axis [29], which works ultimately to govern cell cycle, apoptotic activity, and cell differentiation. These are traits that define several of the hallmarks of cancer. It has been demonstrated that the acetylation status of histone H4 and the resulting transcription of c-MYC rely on the MYC/Mad/Max signaling axis [30]. When the Mad/Max complex is bound in complex at the promoter of the c-MYC target, the histone H4 is deacetylated, repressing transcription of the c-MYC target gene [31]. Consequently, the cell cycle is arrested, blocking it in the S phase [32]. However, when this histone is acetylated, MYC is bound, activating transcription of genes, including MBII. C-MYC amplification is most commonly observed in Group 3 MB patients and the MYC amplification status of this subgroup is highest of all the subtypes [6]. Therefore, the role of OTX2 in the activation and/or amplification of c-MYC transcription is currently thought to be most important to its contributions of the prognosis of Group 3 and Group 4 MBs. Looking into the role of OTX2 in downstream signaling of the cancer phenotype, Bunt and colleagues worked to demonstrate that the polycomb genes, a family of genes known for silencing HOX genes via chromatin modulation [33], are specifically upregulated in the medulloblastoma condition, and the mechanism by which this is carried out is via histone H3 lysine 27 demethylase (H3K27) [13]. This upregulation is most apparent in Group 3 and Group 4 medulloblastomas [4]. Furthermore, Bunt’s group also correlated OTX2 silencing with a downregulation of Cyclin D1, an oncogene important in the G1-S transition, and upregulation of the G1 inhibitor P21 was noted [7]. Additionally, the phosphorylation status of RB1, a common tumor suppressor, increased with the silencing of OTX2, furthering the notion that OTX2 regulates a number of different transcription start sites to contribute to the aggressive medulloblastoma phenotypes observed in Group 3 and Group 4 (Table 1). In summary, the role of OTX2 in the induction and maintenance of anaplasticity in medulloblastoma is more than likely due to its pleiotropic downstream effects on the oncogenes it activates, most notably MYC. There is, however, an additional area of exploration in the epigenetic modification that is mediated by OTX2 in creating the Group 3 and Group 4 MB phenotype, which will be covered next.
Table 1.
OTX2 interacts with a variety of oncogenic proteins
| Molecule | Connection to OTX2 | Position to OTX2 | Role of OTX2 | Molecule Class |
|---|---|---|---|---|
| Myc/Max | First-degree | Downstream | Stimulatory | Transcription Factor |
| LHX1 | First-degree | Upstream | Stimulatory | Transcription Factor |
| β-catenin | First-degree | Upstream | Stimulatory | Binding Protein |
| FOXP1/2/4 | First-degree | Downstream | Inhibitory | Transcription Factor |
| NuRD complex | First-degree | Upstream | Inhibitory | Kinase |
| Aurora-A | First-degree | Downstream | Stimulatory | Kinase |
| POU3F2 | First-degree | Upstream | Stimulatory | Transcription Factor |
| Transthyretin | First-degree | Downstream | Stimulatory | Binding Protein |
| GLI1 | Second-degree | Downstream | Stimulatory | Transcription Factor |
| HIF1α | Second-degree | Downstream | Stimulatory | Transcription Factor |
| PP2A | Second-degree | Downstream | Stimulatory | Kinase |
| Androgen receptor | Second-degree | Downstream | Both | Transcription Factor |
| DAGK | Second-degree | Downstream | Stimulatory | Kinase |
| HSP60 | Second-degree | Downstream | Stimulatory | Binding Protein |
| HSP61 | Second-degree | Downstream | Stimulatory | Binding Protein |
| HSP62 | Second-degree | Downstream | Stimulatory | Protein |
| HSP63 | Second-degree | Downstream | Stimulatory | Binding Protein |
| HSP64 | Second-degree | Downstream | Stimulatory | Transporter |
| HSP65 | Second-degree | Downstream | Inhibitory | Transcription Factor |
| HSP66 | Second-degree | Downstream | Inhibitory | Binding Protein |
| FADD | Third-degree | Downstream | Inhibitory | Binding Protein |
| HSP70 | Third-degree | Downstream | Inhibitory | Binding Protein |
| INCNEP | Third-degree | Downstream | Stimulatory | Binding Protein |
| DEEPEST | Third-degree | Downstream | Stimulatory | Kinase |
| INPP5E | Third-degree | Downstream | Stimulatory | Kinase |
| HP1 | Third-degree | Downstream | Stimulatory | Binding Protein |
| Tcf (Lef) | Third-degree | Downstream | Stimulatory | Transcription Factor |
| HSP60 | Third-degree | Downstream | Stimulatory | Binding Protein |
The table is a transcription of the interactome demonstrating the upstream and downstream signaling targets of OTX2, how closely the molecules are connected, and which class of molecule the targets are. The Myc/Max complex, FOXP1/2/4, β-catenin, and HIF1α are all present, demonstrating the multifactorial effects of OTX2 as a transcription factor, acting on genes that promote carcinogenesis and stemness in MB (Modified from Metacore, a Thomas-Reuters product.).
Epigenetic modifications in OTX2-mediated oncogenesis
The stemness of Group 3 and Group 4 medulloblastoma tumor cells is largely controlled by epigenetic modifications that are regulated by MYC/Mad/Max signaling pathways. Among these include the aforementioned histone acetylation of targeted downstream genes, mediated by the MYC/Mad/Max complex. However, this system is controlled by a far more complex epigenetic machinery than that of acetylation alone. It is therefore important to fully characterize each of the epigenetic modifications that occur in the pathogenesis of Group 3/4 medulloblastoma in order to understand where future therapeutic efforts may be directed if an attempt is to be made to better treat these resistant subtypes.
The MYC/Max complex acts as a recruiting heterodimer that can recruit cofactor and protein complexes that promote histone acetylation (as overviewed above), chromatin remodeling, ubiquitination, and RNA Polymerase II phosphorylation in order to maintain the proliferative and undifferentiated cancer phenotype seen in medulloblastoma [34]. Chromatin remodeling occurs due to MYC-initiated recruitment of the SWI/SNF chromatin remodeling complex via MYC interaction with said chromatin remodeling complex, specifically with the INI1 subunit. SWI/SNF is a multiprotein ATPase complex that activates transcription when recruited by c-MYC transactivation [35,36]. Ubiquitination is promoted by the MYC cofactor SKP2 (S-phase kinase-associated protein 2), an ubiquitin ligase. However, this interaction with SKP2 is also integral indicated in the cell cycle transition from G1 to S phase, which seems to be important in promoting the cell cycle progression seen in the cancer cell phenotype [37,38]. Finally, MYC association with Cyclin Dependent Kinase 9 (CDK9) increases transcriptional elongation due to the direct phosphorylation of RNA Polymerase II by CDK9 [39].
Maintenance of a stem-like state, as seen in all small blue round cell tumors including medulloblastoma, is also achieved by sustaining H3K27 trimethylation (H3K27me3) [13], which leads to the repression of lineage specific genes in stem cells, ultimately inhibiting differentiation [40]. H3K27me3 is regulated by a system of methylases and demethylases; specifically, it is activated by a polycomb repressive complex 2 (PRC2) that includes the methylase EZH2 [41,42]. OTX2 is also known to be a member in its own right of this same PRC2 complex [13]. H3K27me3 is demethylated by enzymes such as KDM6A and a number of other lysine demethylases [43]. It is these factors, then, that are either upregulated (i.e., EZH2) or inactivated (i.e., KDM6A) in group 3/4 medulloblastomas. Indeed, these tumors largely are characterized by marked histone methylation deregulation [13]. Bunt et al also showed the maintenance of H3K27me3 to be a prominent role of OTX2 [13], when compared to its implicated regulation of the expression of downstream genes [7,44]. In this study, OTX2-bound promoters showed increased H3K27me3 activation markers, and silencing OTX2 significantly reduced the levels of H3K27me3. Further, the OTX2 locus is shown to be amplified in group 3/4 medulloblastoma, a finding seen independent of mutations in H3K27 demethylases [12,27], suggesting that both OTX2 and H2K27 demethylases play important and mutually exclusive roles in the high levels of H3K27me3 seen in these tumors.
Interestingly, EZH2, KMD6A, CHD7, and KMYM alteration may also disrupt chromatin marking of genes including OTX2, MYC, and MYCN in medulloblastomas [45,46]. This supports the characterization of the complicated and interrelated nature of genes and factors involved in cell proliferation, and especially the stem-like state we have discussed throughout this review. Interpreting these relationships, and the effects caused by their interactions, presents a role for future studies through which we may glean further understanding of this unforgiving cancer phenotype. The epigenetic and molecular pathways defined for OTX2 in all literature to date focuses on the aforementioned MYC axis and histone modifying roles this gene plays. Our group has recently conducted work to point to a possible additional role of OTX2 in medulloblastoma which is previously uncharacterized: that of metabolic control.
Conclusions: preview of the possible role of OTX2 in metabolic control
To overview, the more aggressive and deadly Group 3/4 medulloblastomas necessitate better prognostic and therapeutic targets in order to bring survivorship to a level of that more comparable with SHH and WNT-types. In this review, we have discussed recent literature related to OTX2 and its role in MB. Typically a mediator of embryogenesis, it is currently implicated in the maintenance of stemness in the more aggressive subtypes of MB as well as the amplification of MYC. First off, OTX2 mediates the maintenance of stemness in tumor cells through the upregulation of epigenetic regulators such as H3K27me3 and the repression of differentiation activators such as MyoD (Figure 1). Second, OTX2 is responsible for increasing tumor cell proliferation via downregulation of tumor suppressor transcriptional regulators and via its interactions with C-MYC, which leads to increased expression of cyclin dependent kinase 9 (CDK9). Through c-MYC expression and via its role in epigenetic modifications that contribute to stemness, OTX2 contributes to the aggressive nature of Group 3 and Group 4 tumors. However, when our group performed mass spectrometry of immunoprecipitated OTX2 in a patient-derived xenograft to validate these conclusions, we received some unexpected results. Interestingly, analysis of mass spec data yielded several key metabolic enzymes that are directly involved in glycolysis, the characteristic energy source of tumor cells due to the Warburg Effect. Because both MYC and OTX2 favor several cell growth and proliferation mechanisms, the MYC-OTX2 regulatory complex must favor the induction of key programs involved in the bioenergetics of cancer cells. The potential role of OTX2 in this regard is unstudied, and may present an opportunity for a novel approach in tumor therapy by targeting the transition to the overtly glycolytic phenotype in tumor cells. Overall, the targeting of OTX2 as a mediator of tumorigenesis, inducer of stemness, and possibly even a controller of metabolic tendencies in the aggressive Group 3 & Group 4 subtypes of medulloblastoma could lead to better outcomes for patients, and will be covered in our future publications.
Figure 1.
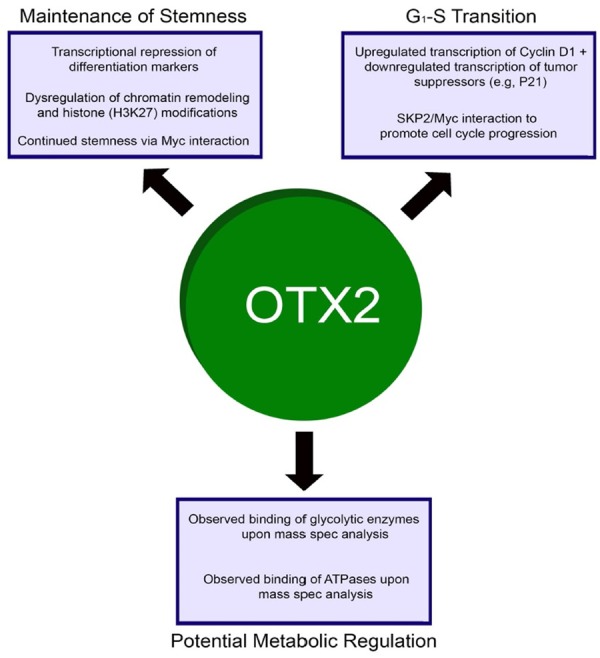
OTX2 has several roles in neurodevelopment and onconeogenesis alike. The 2 roles depicted at the top of the figure, maintenance of stemness and cell cycle progression, are known roles based on current literature. The role depicted at the bottom, metabolic regulation, is a possibility postulated by our lab based on mass spec data and is a target of interest in future studies.
Acknowledgements
This research was supported by foundation grant from William E. McElroy Charitable Foundation, Springfield, Illinois. The authors thank Mark Linder Walk for the Mind, Illinois Neurological Institute and The OSF Foundation, Peoria, IL for their funding support. The authors also thank Christina Constantinidou and Amy Gries for manuscript preparation.
Disclosure of conflict of interest
None.
References
- 1.Smoll NR. Relative survival of childhood and adult medulloblastomas and primitive neuroectodermal tumors (PNETs) Cancer. 2012;118:1313–1322. doi: 10.1002/cncr.26387. [DOI] [PubMed] [Google Scholar]
- 2.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramaswamy V, Remke M, Bouffet E, Faria CC, Perreault S, Cho YJ, Shih DJ, Luu B, Dubuc AM, Northcott PA, Schuller U, Gururangan S, McLendon R, Bigner D, Fouladi M, Ligon KL, Pomeroy SL, Dunn S, Triscott J, Jabado N, Fontebasso A, Jones DT, Kool M, Karajannis MA, Gardner SL, Zagzag D, Nunes S, Pimentel J, Mora J, Lipp E, Walter AW, Ryzhova M, Zheludkova O, Kumirova E, Alshami J, Croul SE, Rutka JT, Hawkins C, Tabori U, Codispoti KE, Packer RJ, Pfister SM, Korshunov A, Taylor MD. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol. 2013;14:1200–1207. doi: 10.1016/S1470-2045(13)70449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, Cho YJ, Koster J, Schouten-van Meeteren A, van Vuurden D, Clifford SC, Pietsch T, von Bueren AO, Rutkowski S, McCabe M, Collins VP, Backlund ML, Haberler C, Bourdeaut F, Delattre O, Doz F, Ellison DW, Gilbertson RJ, Pomeroy SL, Taylor MD, Lichter P, Pfister SM. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roussel MF, Robinson GW. Role of MYC in medulloblastoma. Cold Spring Harb Perspect Med. 2013:3. doi: 10.1101/cshperspect.a014308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stutz AM, Korshunov A, Reimand J, Schumacher SE, Beroukhim R, Ellison DW, Marshall CR, Lionel AC, Mack S, Dubuc A, Yao Y, Ramaswamy V, Luu B, Rolider A, Cavalli FM, Wang X, Remke M, Wu X, Chiu RY, Chu A, Chuah E, Corbett RD, Hoad GR, Jackman SD, Li Y, Lo A, Mungall KL, Nip KM, Qian JQ, Raymond AG, Thiessen NT, Varhol RJ, Birol I, Moore RA, Mungall AJ, Holt R, Kawauchi D, Roussel MF, Kool M, Jones DT, Witt H, Fernandez-L A, Kenney AM, Wechsler-Reya RJ, Dirks P, Aviv T, Grajkowska WA, Perek-Polnik M, Haberler CC, Delattre O, Reynaud SS, Doz FF, Pernet-Fattet SS, Cho BK, Kim SK, Wang KC, Scheurlen W, Eberhart CG, Fevre-Montange M, Jouvet A, Pollack IF, Fan X, Muraszko KM, Gillespie GY, Di Rocco C, Massimi L, Michiels EM, Kloosterhof NK, French PJ, Kros JM, Olson JM, Ellenbogen RG, Zitterbart K, Kren L, Thompson RC, Cooper MK, Lach B, McLendon RE, Bigner DD, Fontebasso A, Albrecht S, Jabado N, Lindsey JC, Bailey S, Gupta N, Weiss WA, Bognar L, Klekner A, Van Meter TE, Kumabe T, Tominaga T, Elbabaa SK, Leonard JR, Rubin JB, Liau LM, Van Meir EG, Fouladi M, Nakamura H, Cinalli G, Garami M, Hauser P, Saad AG, Iolascon A, Jung S, Carlotti CG, Vibhakar R, Ra YS, Robinson S, Zollo M, Faria CC, Chan JA, Levy ML, Sorensen PH, Meyerson M, Pomeroy SL, Cho YJ, Bader GD, Tabori U, Hawkins CE, Bouffet E, Scherer SW, Rutka JT, Malkin D, Clifford SC, Jones SJ, Korbel JO, Pfister SM, Marra MA, Taylor MD. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunt J, Hasselt NE, Zwijnenburg DA, Hamdi M, Koster J, Versteeg R, Kool M. OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J Cancer. 2012;131:E21–32. doi: 10.1002/ijc.26474. [DOI] [PubMed] [Google Scholar]
- 8.Kaur R, Aiken C, Morrison LC, Rao R, Del Bigio MR, Rampalli S, Werbowetski-Ogilvie T. OTX2 exhibits cell-context-dependent effects on cellular and molecular properties of human embryonic neural precursors and medulloblastoma cells. Dis Model Mech. 2015;8:1295–1309. doi: 10.1242/dmm.020594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wortham M, Jin G, Sun JL, Bigner DD, He Y, Yan H. Aberrant Otx2 expression enhances migration and induces ectopic proliferation of hindbrain neuronal progenitor cells. PLoS One. 2012;7:e36211. doi: 10.1371/journal.pone.0036211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagel S, Ehrentraut S, Meyer C, Kaufmann M, Drexler HG, MacLeod RA. Aberrantly expressed OTX homeobox genes deregulate B-cell differentiation in Hodgkin lymphoma. PLoS One. 2015;10:e0138416. doi: 10.1371/journal.pone.0138416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boon K, Eberhart CG, Riggins GJ. Genomic amplification of orthodenticle homologue 2 in medulloblastomas. Cancer Res. 2005;65:703–707. [PubMed] [Google Scholar]
- 12.Adamson DC, Shi Q, Wortham M, Northcott PA, Di C, Duncan CG, Li J, McLendon RE, Bigner DD, Taylor MD, Yan H. OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Res. 2010;70:181–191. doi: 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bunt J, Hasselt NA, Zwijnenburg DA, Koster J, Versteeg R, Kool M. OTX2 sustains a bivalent-like state of OTX2-bound promoters in medulloblastoma by maintaining their H3K27me3 levels. Acta Neuropathol. 2013;125:385–394. doi: 10.1007/s00401-012-1069-2. [DOI] [PubMed] [Google Scholar]
- 14.Bai RY, Staedtke V, Lidov HG, Eberhart CG, Riggins GJ. OTX2 represses myogenic and neuronal differentiation in medulloblastoma cells. Cancer Res. 2012;72:5988–6001. doi: 10.1158/0008-5472.CAN-12-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beby F, Lamonerie T. The homeobox gene Otx2 in development and disease. Exp Eye Res. 2013;111:9–16. doi: 10.1016/j.exer.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Frantz GD, Weimann JM, Levin ME, McConnell SK. Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 1994;14:5725–5740. doi: 10.1523/JNEUROSCI.14-10-05725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Forebrain and midbrain regions are deleted in Otx2-/- mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- 18.Larsen KB, Lutterodt MC, Mollgard K, Moller M. Expression of the homeobox genes OTX2 and OTX1 in the early developing human brain. J Histochem Cytochem. 2010;58:669–678. doi: 10.1369/jhc.2010.955757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang GH, Xu QF, Cui YH, Li N, Bian XW, Lv SQ. Medulloblastoma stem cells: promising targets in medulloblastoma therapy. Cancer Sci. 2016;107:583–589. doi: 10.1111/cas.12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JY, Nelson AL, Algon SA, Graves O, Sturla LM, Goumnerova LC, Rowitch DH, Segal RA, Pomeroy SL. Medulloblastoma tumorigenesis diverges from cerebellar granule cell differentiation in patched heterozygous mice. Dev Biol. 2003;263:50–66. doi: 10.1016/s0012-1606(03)00434-2. [DOI] [PubMed] [Google Scholar]
- 21.Michiels EM, Oussoren E, Van Groenigen M, Pauws E, Bossuyt PM, Voute PA, Baas F. Genes differentially expressed in medulloblastoma and fetal brain. Physiol Genomics. 1999;1:83–91. doi: 10.1152/physiolgenomics.1999.1.2.83. [DOI] [PubMed] [Google Scholar]
- 22.de Haas T, Oussoren E, Grajkowska W, Perek-Polnik M, Popovic M, Zadravec-Zaletel L, Perera M, Corte G, Wirths O, van Sluis P, Pietsch T, Troost D, Baas F, Versteeg R, Kool M. OTX1 and OTX2 expression correlates with the clinicopathologic classification of medulloblastomas. J Neuropathol Exp Neurol. 2006;65:176–186. doi: 10.1097/01.jnen.0000199576.70923.8a. [DOI] [PubMed] [Google Scholar]
- 23.Pietsch T, Haberler C. Update on the integrated histopathological and genetic classification of medulloblastoma-a practical diagnostic guideline. Clin Neuropathol. 2016;35:344–352. doi: 10.5414/NP300999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panwalkar P, Moiyadi A, Goel A, Shetty P, Goel N, Sridhar E, Shirsat N. MiR-206, a cerebellum enriched miRNA is downregulated in all medulloblastoma subgroups and its overexpression is necessary for growth inhibition of medulloblastoma cells. J Mol Neurosci. 2015;56:673–680. doi: 10.1007/s12031-015-0548-z. [DOI] [PubMed] [Google Scholar]
- 25.Kaur K, Kakkar A, Kumar A, Purkait S, Mallick S, Suri V, Sharma MC, Julka PK, Gupta D, Suri A, Sarkar C. Clinicopathological characteristics, molecular subgrouping, and expression of miR-379/miR-656 cluster (C14MC) in adult medulloblastomas. J Neurooncol. 2016;130:423–430. doi: 10.1007/s11060-016-2250-6. [DOI] [PubMed] [Google Scholar]
- 26.Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, Pomeroy SL, Korshunov A, Lichter P, Taylor MD, Pfister SM. Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012;12:818–834. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, Chalhoub N, Baker SJ, Huether R, Kriwacki R, Curley N, Thiruvenkatam R, Wang J, Wu G, Rusch M, Hong X, Becksfort J, Gupta P, Ma J, Easton J, Vadodaria B, Onar-Thomas A, Lin T, Li S, Pounds S, Paugh S, Zhao D, Kawauchi D, Roussel MF, Finkelstein D, Ellison DW, Lau CC, Bouffet E, Hassall T, Gururangan S, Cohn R, Fulton RS, Fulton LL, Dooling DJ, Ochoa K, Gajjar A, Mardis ER, Wilson RK, Downing JR, Zhang J, Gilbertson RJ. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bunt J, Hasselt NE, Zwijnenburg DA, Koster J, Versteeg R, Kool M. Joint binding of OTX2 and MYC in promotor regions is associated with high gene expression in medulloblastoma. PLoS One. 2011;6:e26058. doi: 10.1371/journal.pone.0026058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava VK, Nalbantoglu J. The cellular and developmental biology of medulloblastoma: current perspectives on experimental therapeutics. Cancer Biol Ther. 2010;9:843–852. doi: 10.4161/cbt.9.11.11785. [DOI] [PubMed] [Google Scholar]
- 30.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 31.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 32.Sommer A, Hilfenhaus S, Menkel A, Kremmer E, Seiser C, Loidl P, Luscher B. Cell growth inhibition by the Mad/Max complex through recruitment of histone deacetylase activity. Curr Biol. 1997;7:357–365. doi: 10.1016/s0960-9822(06)00183-7. [DOI] [PubMed] [Google Scholar]
- 33.Aranda S, Mas G, Di Croce L. Regulation of gene transcription by Polycomb proteins. Sci Adv. 2015;1:e1500737. doi: 10.1126/sciadv.1500737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 35.Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 36.Kingston RE, Bunker CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 37.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 38.von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, Soderberg O, Kerppola TK, Larsson LG. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 39.Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 40.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 42.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 43.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 44.Bunt J, de Haas TG, Hasselt NE, Zwijnenburg DA, Koster J, Versteeg R, Kool M. Regulation of cell cycle genes and induction of senescence by overexpression of OTX2 in medulloblastoma cell lines. Mol Cancer Res. 2010;8:1344–1357. doi: 10.1158/1541-7786.MCR-09-0546. [DOI] [PubMed] [Google Scholar]
- 45.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-repressive Complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan A, Shover W, Goodliffe JM. Su(z)2 antagonizes auto-repression of Myc in drosophila, increasing Myc levels and subsequent trans-activation. PLoS One. 2009;4:e5076. doi: 10.1371/journal.pone.0005076. [DOI] [PMC free article] [PubMed] [Google Scholar]


