Abstract
Regulation of the Hippo pathway via phosphorylation of Yorkie (Yki), the Drosophila homolog of human Yes-associated protein 1, is conserved from Drosophila to humans. Overexpression of a non-phosphorylatable form of Yki induces severe overgrowth in adult fly eyes. Here, we show that yki mRNA associates with microsomal fractions and forms foci that partially colocalize to processing bodies in the vicinity of endoplasmic reticulum. This localization is dependent on a stem-loop (SL) structure in the 3’ untranslated region of yki. Surprisingly, expression of SL deleted yki in eye imaginal discs also results in severe overgrowth phenotypes. When the structure of the SL is disrupted, Yki protein levels increase without a significant effect on RNA levels. When the SL is completely removed, protein levels drastically increase, but in this case, due to increased RNA stability. In the latter case, we show that the increased RNA accumulation is due to removal of a putative miR-8 seed sequence in the SL. These data demonstrate the function of two novel regulatory mechanisms, both controlled by the yki SL element, that are essential for proper Hippo pathway mediated growth regulation.
Keywords: yki mRNA, mRNA localization, hippo pathway
Introduction
The Hippo pathway is highly conserved signaling pathway from Drosophila to humans. The Hippo pathway regulates tissue size and defect in the pathway leads to tissue overgrowth [1,2]. Yorkie (Yki) and its mammalian homolog Yes-associated protein (YAP) are essential components of the Hippo pathway and can activate the expression of genes related to cell proliferation and anti-apoptosis. The activity of Yki and YAP1 is negatively regulated by phosphorylation through the Hippo signaling pathway. In Drosophila, overexpression of non-phosphorylatable Yki by substitution mutation into phosphorylation site but not wild-type Yki in the eye imaginal discs induces severe overgrowth phenotype in adult compound eyes [3]. While mutation at phosphorylation sites of YAP1 has not been found in human cancer patients, the amplification of genomic region containing the YAP1 gene and overexpression of YAP1 are reported in human and mouse tumors [4-6]. Although it is suggested that perturbation of the regulation by microRNAs leads to up-regulation of YAP1 gene expression in tumors [7], most of the mechanisms underlying YAP1 overexpression in tumors are still unclear.
It is well known that mRNAs encoding secreted or membrane proteins, upon translation, are targeted to the endoplasmic reticulum (ER) in a signal recognition particle (SRP)-dependent manner [8-10]. Recently, however, genome-wide analyses have suggested that many of these mRNAs, as well as many other mRNAs that encode nuclear and cytosolic proteins, are also targeted to the ER in a translationally-independent fashion [11-15]. In several cases, cis-elements and trans-factors that are related to this SRP-independent ER targeting have been identified [16,17]. However, because the biological significance of this ER targeting is still unclear, we attempted to develop a Drosophila model that is able to connect these observations with in vivo biological functions.
In this paper, we identify yki mRNA encoding transcriptional co-activator as one of the mRNAs fractionated into microsome fraction in SRP-independent manner and reveal that yki mRNA localizes to processing bodies (P-bodies), which is dependent on a stem-loop (SL) structure in its 3’ untranslated region (UTR). Furthermore, we indicate that the SL is related to two distinct regulations of yki gene expression at mRNA level and expression of the SL-deleted yki in the eye imaginal discs results in severe overgrowth phenotype in the adult compound eyes.
Materials and methods
DNA constructs
NLS-MCP-GFP was amplified from pCaSpeR-hsp83-NLS-MCP-GFP [18], and inserted into the TA cloning vector pCR2.1 (Invitrogen). The NLS-MCP-GFP was removed from pCR2.1-NLS-MCP-GFP by digesting with BglII and BamHI, and cloned into the BglII/BamHI site of pCaSpeR-hs/act.
ms2 × 12 was isolated from vector pCR-ms2 × 12 using BglII/BamHI, and inserted into the vector pMT/V5-HisB (Invitrogen). Escherichia coli TOP10 was used for constructing all recombinant plasmids containing ms2 sequences. Transformed TOP10 was then spread onto LB plates containing carbenicillin, and incubated at 30°C for 17 h.
To generate an ER maker, the signal sequence (ss) of Calreticulin (Crc), and HDEL coding sequences, were inserted into the NotI/BamHI site of pAc5.1/V5-HisA. DsRed was amplified from pDsRed-Monomer (Clontech) and inserted between the ss and HDEL sequence using the XbaI site of pAc5.1-ss-HDEL. The Vm34Ca, Act5C and yki sequences were amplified from cDNA synthesized from total RNA of S2-DRSC cells (DGRC), and cloned into pMT/V5-HisB-ms × 12 using the XhoI site and the In-fusion HD cloning kit (TaKaRa Clontech).
To make a P-body marker, Me31B was amplified from an EST clone (LD21247), and inserted into the pDsRed-Monomer vector. Me31B-DsRed was amplified from pMe31B-DsRed, and inserted into the XhoI/MluI sites of pAc5.1/V5-HisA.
Deletion constructs or mutants of yki were amplified from pMT-yki-ms2 × 12, and inserted into the XhoI site of pMT/V5-HisB-ms2 × 12.
Plasmids for the expression of miR-8 primary transcripts and miRNA reporters have been described previously [19,20]. The YAP1 3’UTR was amplified from cDNA synthesized from total RNA of HEK293 cells. The yki 3’UTR, yki 3’UTR mutants, and YAP1 3’UTR were then inserted into a styI site that is in the 3’UTR of the firefly luciferase reporter in pGVB using the In-fusion HD cloning kit. The miR-8 expression and reporter plasmids contain an Actin 5C promoter.
To make plasmids for dsRNA of lacZ, Srp54k, AGO1, and HPat-1, each gene was amplified from a pMT/V5-His/lacZ vector, or cDNA synthesized from total RNA of S2-DRSC cells, and cloned into pOT2 using the XhoI and EcoRI sites.
Cell culture and RNAi
S2-DRSC cells were maintained at 25°C in Schneider’s Drosophila medium (Life technologiesTM) supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin (Invitrogen), 50 μg/ml streptomycin (Invitrogen) and 0.1 mg/ml gentamicin (Life technologiesTM).
RNAi was performed as described [21]. dsRNA was synthesized from linearized pOT2 vectors using T7 and SP6 polymerases and RiboMAXTM expression large scale RNA production kit (Promega) according to the manufacturer’s protocol.
S2-DRSC cells were plated into 24-well plates with 2.5 × 105 cells in 450 µl of medium supplemented with 10% heat-inactivated FBS, penicillin, streptomycin and gentamicin per well. After plating, 1 μg of dsRNA was transfected using DharmaFECT4 transfection reagent (GE Dharmacon). After 24 h, knockdown cells were transfected with plasmids for RNA imaging and luciferase assays.
Cell fractionation
5.0 × 108 S2-DRSC cells were collected by centrifugation (1,000 × g, 5 min), frozen with liquid N2 and resuspended in 5 mL HKM buffer containing 30 mM HEPES [pH 7.3], 50 mM KCl, 2.5 mM MgCl2, 0.88 M sucrose, 1 mM DTT and complete-mini [EDTA-free] protease inhibitors (Roche). The cells were homogenized with 30 strokes in a 21-gauge needle on ice. Nuclei and cell debris were removed by sedimentation at 1,000 × g for 20 min at 4°C, and the supernatant (S1) was further centrifuged at 10,000 × g for 30 min at 4°C to separate into pellet (P10) and supernatant (S10). P10 pellet resuspended in HKM buffer, and the S10 fraction, were loaded onto discontinuous sucrose gradients containing 1.3 M, 2.1 M and 2.7 M sucrose, followed by centrifugation at 190,000 × g for 4 h at 4°C in a RPS40T rotor (HITACHI). After centrifugation, the gradient solutions were withdrawn from the bottom of the tubes through a hole opened by needle to generate 3-drops fractions (~150 µl). Fractions were analysed by UV spectrometry and Western blotting.
Detachment of ribosomes from rough ER
Ribosome removal from rough ER was performed by treatment with puromycin in high-salt buffer [22]. 50 μl of RM fraction was diluted 11-fold with Stripping buffer containing 1 M KCl, 50 mM EDTA, 1 mM DTT, 30 mM HEPES, 1 mM puromycin, and incubated at 37°C for 30 min, then centrifuged at 75,000 × g for 10 min in a RPS40T rotor (HITACHI) at 4°C to produce a naked microsome (NM) pellet and stripped ribosome (SR) supernatant. The membrane pellet was then resuspended in HKM buffer and subjected to Western blot analysis.
RNA FISH and immunofluorescence
RNA in situ hybridization was carried out essentially as described previously [23]. 1 × 105 S2-DRSC cells were plated per well on an 8-well culture Slide (BD FalconTM), and incubated at 25°C for 3 days with 100 µg/ml Gentamycin. 3 μg/ml proteinase K treatment was performed at 4°C for 3 min. For hybridization, DIG-labeled probes were diluted in hybridization buffer at a concentration of 1 μg/ml, and incubated for 16 h at 45°C.
For probe detection, cells were incubated with Biotin-conjugated anti-DIG antibody (Roche: 1:400 dilution) at 25°C for 1.5 h. Streptavidin-HRP secondary antibody (Perkin Elmer life sciences: 1:500 dilution) was incubated at 25°C for 30 min. To amplify signals, FITC-tyramide (1:50 dilution) was incubated at 25°C for 2 h. Following tyramide amplification, cells were fixed with 4% paraformaldehyde in HCMF at 25°C for 10 min, and blocked with 5% normal goat serum (Vector Labs) for 5 min. Cells were incubated for 16 h at 4°C with primary antibody, and then stained with Alexa Fluor 594-conjugated secondary antibody (Molecular Probes: 1:400 dilution) at 25°C for 1 h. Samples were mounted with VECTASHIELD mounting medium with DAPI (Vector Labs). Confocal images were obtained with a Fluoview FV10i confocal microscope, and analysed with MetaMorph software.
RNA imaging
2.5 × 105 cells were plated per well of a 24-well cell culture dish, and grown for 24 h. S2-DRSC cells were transfected with the pCaSpeR-hs-MCP-GFP, the Drosophila expression vector pMT-target RNA-ms2 × 12, pAc5.1-Sp-DsRed-HDEL or pAc5.1-Me31B-DsRed using siLentFectTM lipid reagent (Bio-Rad). After 24 h, cells were heat-shocked at 37°C for 30 min twice with 30 min interval at 25°C to induce MCP-GFP protein. Following heat shock, target RNA was induced by the addition of 500 µM CuSO4 for 24 h. Images were captured using Fluoview FV10i confocal microscope (Olympus) and analysed with MetaMorph software (Molecular devices).
Generation of stable S2-DRSC cell lines
Stable line generation was as previously described [24]. 5 × 105 cells were plated per well in a 24-well cell culture dish, grown for 24 h and transfected with 400 ng pAc5.1-miR-8, together with 10 ng pCoBlast plasmid using siLentFectTM lipid reagent (Bio-Rad). Two days after transfection, cells were collected and incubated in medium containing 1 µg/ml Blasticidin S (Life Technologies), with changes every 4 days over several weeks. Total RNA was isolated for quantitative real-time PCR.
RNA-seq analysis
3 μg Total RNA of RM, cytoplasm, NM, and SR fraction were isolated using TRIzol Reagent (Invitrogen). Isolated RNAs were used for library construction with TruSeq RNA Sample Prep Kit (Illumina) according to the manufacturer’s instructions. All samples were sequenced by illumine Genome Analyzer IIX with 41 bp single-end reads. The deep sequencing data were mapped to the reference D. melanogaster genome sequence (UCSC version dm3) by the TopHat [25] program version 1.3.2 with default parameters. The Cufflinks [26] program version 2.0.0 was then used to assemble transcripts and to calculate fragments per kilobase of transcript per million mapped fragments (FPKM) with option ‘-library-type fr-secondstrand’. All results have been submitted to the Gene Expression Omnibus database (Accession number: GSE85768).
Statistical analysis
All statistical analyses, including Student’s t tests, and ANOVA were performed using GraphPad Prism software.
Quantitative real-time PCR
Total RNA was extracted from intact S2-DRSC cells, cell fractions or the eye imaginal discs using Trizol (Invitrogen) and Dr. GenTLE precipitation carrier (TaKaRa). Total RNA was treated with DNaseI in DNase Buffer containing 40 mM Tris-HCl [pH 8.0], 8 mM MgCl2, 1 mM CaCl2 and 5 mM DTT at 37°C for 30 min. cDNA was synthesized from total RNA using a PrimeScriptTM RT kit (Perfect Real Time, TaKaRa). Real time PCR primers were designed using the IDT Primer Quest Tool. Real-Time PCR was performed using SYBR® Premix Ex Taqtm II (Tli RNaseH Plus, TaKaRa), and analysed using the CFX96 TouchTM Real-Time PCR detection system (Bio-Rad). Srp54k, yki, HPat-1AGO1, RpL32 and spi mRNA expressions were normalised to levels of GAPDH mRNA.
Luciferase assays
2.5 × 105 cells were plated per well of a 24-well cell culture dish and grown for 24 h. Transfection was performed in 24-well dishes using siLentFectTM lipid reagent (Bio-Rad). The following plasmids were cotransfected: firefly luciferase reporter gene construct (0.1 μg), pRL-SV40 Renilla luciferase (0.01 μg, transfection control plasmid), and either pAc5.1-miR-8 or pAc5.1 empty vector (1 μg).
Dual Luciferase assays were performed 2 days after transfection according to the manufacturer’s protocol (Promega), and total RNA was isolated for quantitative real-time PCR. F-luc mRNA levels were normalised to R-luc mRNA.
Antibodies
Antibodies used in this study are listed below in the format of name; application; supplier; catalogue; fold dilution. Mouse anti-KDEL; Western, Immunostaining; stressgen; 10C3; 1:2,000 dilution, 1:200 dilution. Mouse anti-α-tubulin; Western; Sigma; T5168; 1:8,000 dilution. Rabbit anti-GM130; Western; abcam; ab30637; 1:1,000 dilution. Mouse anti-HSP60; Western; Enzo; ADI-SPA 806-D; 1:1,000 dilution. Mouse anti-Histone H4; Western; abcam; ab10158; 1:1,000 dilution. Mouse anti-RpS25 antibody was kindly provided by N. Nakashima (Western; 1:1,000 dilution) [27]. Mouse anti-Me31B was kindly provided by A. Nakamura (Western, Immunostaining; 1:4,000 dilution, 1:400 dilution) [28]. Rat anti-Yki was kindly provided by H. McNeill (Western; 1:2,000 dilution) [29].
Fly strains
Fly stocks were raised on medium containing 5% corn flour, 4% dry yeast, 3% rice bran, 10% glucose and 0.65% agar at 25°C except for instances where GMR-GAL4 driven expression was increased by transfer to 28°C.
Transgenic flies and crosses
Transgenic flies were obtained by P element-mediated germline transformation, and selection of w + flies. Several independent lines for each transgene were established. Transgenic flies were crossed to GMR-GAL4 flies to induce expression in eyes, which were subsequently inspected under a scanning electron microscope in high vacuum mode (Keyence VE-7800).
Results
Identification of mRNAs fractionated into microsome fraction in SRP-independent manner
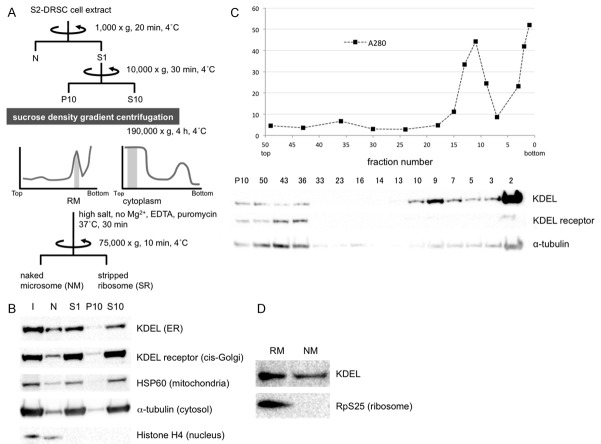
First, to identify mRNAs targeted to rough microsomes (RM) in cultured cells, we performed centrifugal fractionation of cell components (scheme diagrammed in Figure 1A). ER components were enriched within fractions 8-10 of the P10 sucrose gradient and thereby termed RM (Figure 1C), while tubulin-containing fractions from S10 (data not shown) were considered to be cytoplasmic fraction (C). To remove mRNAs that are associated with the RM fractions via ribosomes, a portion of RM was treated with puromycin and EDTA in high-salt buffer to generate naked microsome (NM) and stripped ribosome (SR) fractions. The absence of ribosomes in NM is illustrated by Western blot analysis using anti-RpS25 antibody (Figure 1D).
Figure 1.
Rough microsomal (RM), cytoplasm (C), naked microsome (NM) and stripped microsome (SR) fractions were purified from S2-DRSC cells to identify mRNAs targeted to ER in an SRP-independent manner. A. Scheme for isolation of RM, C, NM and SR fractions. Shaded regions in the graphs indicate fractions that gave rise to the RM and C pools. B. After fractionation, organelle distributions were determined by Western blot analysis with anti-KDEL (ER), KDEL receptor (cis-Golgi), HSP60 (mitochondria), a-tubulin (cytosol) and Histone H4 (nucleus) antibodies, respectively. I = input. C. P10 fraction by sucrose density gradient centrifugation. The graph shows total protein levels detected by UV spectrometry, and the Western blot below illustrates the separation of ER and Golgi. D. Ribosomes on ER-containing fractions were stripped by treatment with EDTA and puromycin in high salt buffer. As expected, the ribosomal marker RpS25 detected by Western blot analysis is no longer detected in the NM fraction.
RNAs fractionated into the RM, C, NM and SR fractions were then identified by deep sequencing. As illustrated in Figure 2A, similar numbers of different transcripts were found in the RM and C fractions, and the NM and SR fractions. Although the numbers of transcripts were similar (Figure 2A), their compositions were quite different (Figure 2B). As expected, Signal peptide prediction using SignalP 4.1 revealed that 81% of the top 200 mRNAs statistics significantly enriched in the soluble C fraction do not harbor canonical signal sequences. Surprisingly though, 78% of the top 200 mRNAs statistics significantly enriched in the NM fraction also do not encode signal peptide- or transmembrane domain-containing proteins. Similar numbers of mRNAs bearing or not bearing signal sequences were found in the RM fraction (Figure 2B). Taken together, these data suggest that SRP-independent microsomal targeting mechanisms will have a major impact on the trafficking and translational control of a large percentage and variety of transcripts.
Figure 2.
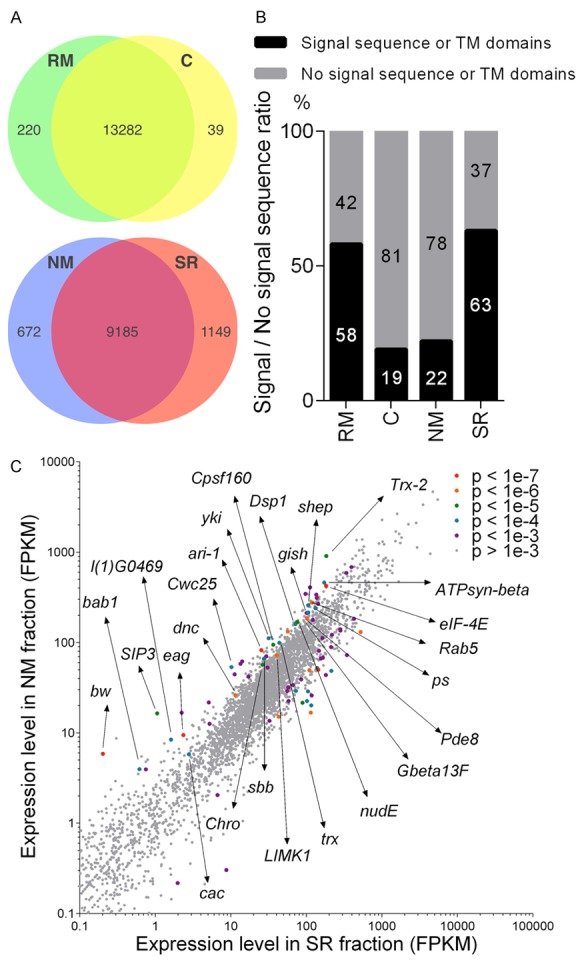
RNA-seq analysis of mRNAs in RM, C, NM or SR fractions. A. Numbers of identified mRNAs in RM, C, NM and SR, respectively. Similar numbers of transcripts were detected in the RM and C fractions and within the NM and SR fractions. B. The percentage of mRNAs bearing signal sequences (black bar) or not (gray bar) in the 200 most highly enriched transcripts in each fraction. C. Scatter plot showing that mRNAs encoding numerous types of proteins are enriched in NM. All analyses were carried out by using the GSE85768 dataset.
Identification of subcellular localization element in yki mRNA
In order to verify whether the mRNAs biochemically fractionated into NM are localized to the ER in intact cells, we used an MS2 coat protein (MCP) and MCP binding element (ms2) to visualize exogenously expressed transcripts in S2-DRSC cells [30,31]. As positive and negative controls, we selected RNAs encoded by Vitelline membrane 34Ca (Vm34Ca), which encodes a secreted protein, and Actin 5C (Act5C), which encodes a cytoplasmic protein. As expected, these transcripts fractionated primarily into the RM and C fractions, respectively (GSE85768 dataset). While Vm34Ca-ms2 RNA formed readily detected foci, Act5C-ms2 RNA did not (Figure 3A and 3B). Deletion of the predicted signal sequence in Vm34Ca-ms2 (Δss; Figure 3E), reduced the number of detected foci to that of the negative control Act5C-ms2 (Act5C; Figure 3E). A similar reduction in Vm34Ca-ms2 foci was obtained by knocking down Signal recognition particle protein 54k (Srp54k), a component of the SRP (Figure 3E). These data indicate that this imaging system faithfully represents the localization of these transcripts in S2-DRSC cells.
Figure 3.
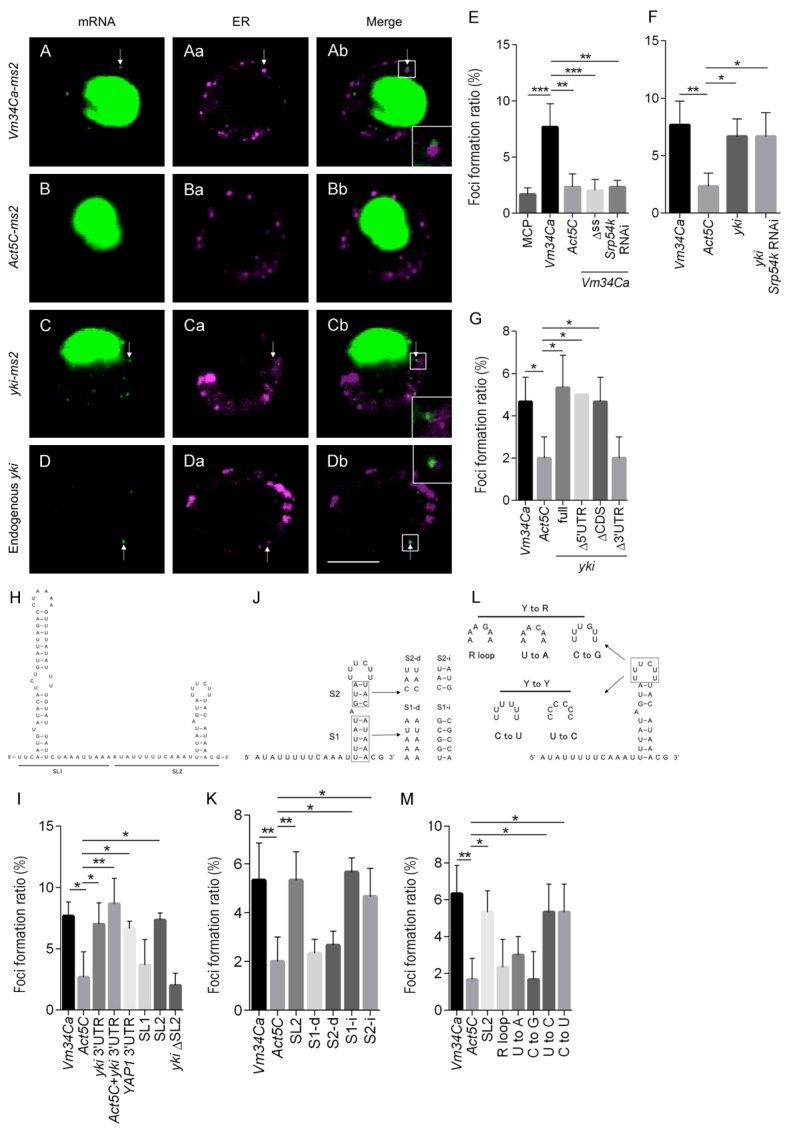
yki mRNA forms foci in S2-DRSC cells in the vicinity of the ER via the SL2 motif in the yki 3’UTR. (A-D) Subcellular RNA localization. (Aa-Da) The ER is marked by ss-DsRed-HDEL. Scale bar, 5 μm. S2-DRSC cells expressing target-ms2 RNA, NLS-MCP-GFP and ss-DsRed-HDEL (A-C) were analyzed by confocal microscopy. While Vm34Ca-ms2 (A-Ab) and yki-ms2 (C-Cb) RNA form foci (arrow), Act5C-ms2 RNA (B-Bb) rarely forms foci. ss-DsRed-HDEL marks ER. NLS = nuclear localization signal; ss = signal sequence; HDEL = ER retention signal. (D-Db) Endogenous yki mRNA and ER are detected by in situ hybridization and anti-KDEL antibody immunostaining, respectively. (Ab-Db) RNA foci are localized adjacent to the ER (white rectangle is magnified in inset, bottom right). (E-G, I, K, M) The foci formation ratio was calculated from the number of cells forming foci in the cytoplasm to the total number of cells expressing GFP in the nucleus. Data are shown as means ± SD (n = 3). (E) Vm34Ca and Act5C-ms2 RNAs were used as positive and negative controls, respectively. Vm34Ca-ms2 RNA produces foci in an SRP-dependent manner. (F, G) The yki-ms2 RNA also forms foci efficiently, but in an SRP-independent, and 3’UTR-dependent manner. (H, I) SL2 in the yki 3’UTR is necessary for foci formation. (H) Predicted secondary structure of the yki 3’UTR: SL1 = stem loop1; SL2 = stem loop2. (J-M) The structure of the SL2 stem regions and pyrimidines in the loop region are critical for foci formation. R = purine; Y = pyrimidine. (J) S1-d and S2-d mutations in SL2 are intended to disrupt secondary structure, while S1-i and S2-i mutations are not. (K) High foci formation ratio is dependent on SL2 secondary structure. (L) Bases in the loop region were exchanged from pyrimidine to purine or from pyrimidine to pyrimidine. (M) The introduction of purines in the loop region decreases foci formation. Data are shown as means ± SD (n = 3). *P<0.05, **P<0.01, ***P<0.001 (one-way ANOVA, Dunnett’s Multiple comparisons test).
One of the more interesting transcripts that was highly enriched in the ribosome-free NM fraction, and that contains no signal sequence, was that of the Hippo pathway component yorkie (yki) (Figure 2C). Consistent with this enrichment, expression of yki mRNA fused to ms2 tags in S2-DRSC cells resulted in foci formation at a frequency similar to that of Vm34Ca-ms2 RNA (Figure 3C and 3F). However, in contrast to Vm34Ca-ms2, yki-ms2 foci were not disrupted upon knockdown of Srp54k (Figure 3C and 3F). Endogenous yki transcripts also formed foci in S2-DRSC cells when examined by in situ hybridization (Figure 3D). These data show that yki mRNA accumulates in microsome-associated foci in an SRP-independent fashion.
To identify cis-elements in the yki transcript that mediate foci formation, cells expressing truncated forms of yki-ms2 RNA were generated. While deletion of the 5’UTR (Δ5’UTR) or coding sequence (ΔCDS) had no effect on foci formation, deletion of the 3’UTR (Δ3’UTR) reduced foci formation to levels similar to the negative control (Figure 3G). Furthermore, in cells expressing yki 3’UTR-ms2 or the yki 3’UTR fused to Act5C-ms2 RNA, foci were formed as efficiently as with the positive control (Figure 3I). Taken together, these data indicate that the yki 3’UTR is both necessary and sufficient for this type of localization, and that this activity is SRP-independent. Testing the 3’UTR of YAP1, the human homolog of yki, showed that it was equally efficient at directing the formation of foci (Figure 3I), suggesting that the mechanism of foci formation might be conserved from Drosophila to humans.
In many cases, cis-elements required for RNA localization have defined secondary structures [16,32-37]. Secondary structure prediction performed on the yki 3’UTR using mfold 3.6 revealed two potential SL structures, SL1 and SL2 (Figure 3H). To investigate whether these SLs play a role in yki mRNA subcellular localization, ms2-tagged transcripts deleted for either SL1 or SL2 were expressed and examined in S2-DRSC cells. While the SL1-deleted transcripts showed no significant difference, deletion of SL2 (ΔSL2) reduced the number of foci to that of the Act5C negative control (Figure 3I).
Further analysis of SL2 was then conducted to determine whether the structure and/or sequence is important for foci formation (Figure 3J-M). Base substitutions in either the first (S1-d) or second (S2-d) stem that disrupt base pairing both perturbed foci formation (Figure 3K). On the other hand, mutations that alter the stem sequences, while maintaining structure (S1-i, S2-i) had no effect on foci formation (Figure 3K). Thus, the secondary structure of the two SL2 stems appears to be their primary contributions to foci formation.
Next, we investigated the role of the loop region (Figure 3L). Transversion of pyrimidines to purines extensively reduced foci formation, while transition of pyrimidines to other pyrimidines had no effect (Figure 3M). These results show that the loop sequence appears to be critical for localization, but that there is a significant amount of flexibility so long as residues are pyrimidines. Taken together, it appears that the stem structures and loop sequence act together to promote foci formation.
Degradation of yki mRNA through the SL2
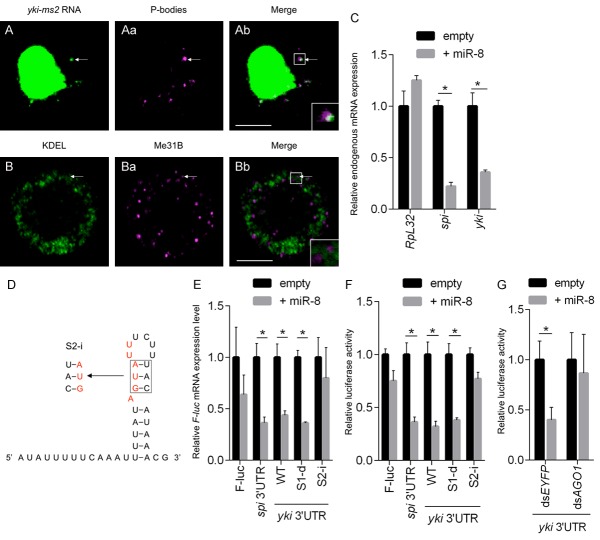
As the foci produced by yki-ms2 RNA are in close proximity to the ER, but did not show a precise overlap (Figure 3C and 3Cb), we examined whether these foci co-localize with other organelles such as Golgi, mitochondria, lysosomes, or cytoplasmic mRNA-protein (mRNP) granules such as processing bodies (P-bodies). The results show that yki-ms2 foci co-localize with a subset of Me31B (a marker for P-bodies)-containing P-bodies (Figure 4A and 4Ab). This co-localization was also confirmed using double labeling of endogenous yki RNA (in situ hybridization) together with endogenous Me31B protein (data not shown). Interestingly, yki may not require P-bodies to form foci, as knockdown of HPat (also known as Protein associated with topo II related-1: Patr-1), a scaffolding component of P-bodies [38,39], did not disrupt foci formation (data not shown).
Figure 4.
yki mRNA is degraded via a miR-8 seed sequence in the 3’UTR. (A-Ab) yki-ms2 RNA is partially localized to P-bodies which are represented by Me31B-DsRed. (B-Bb) Endogenous P-bodies detected by anti-Me31B antibody are localized in the vicinity of anti-KDEL ER marker. (C) Expression of endogenous spi (positive control) and yki mRNAs is decreased in miR-8-overexpressing cells, while the amount of RpL32 mRNA (negative control) is not changed. (D, E) Mutations in the miR-8 seed sequence (S2-i) disrupt degradation of F-luc mRNA bearing the yki 3’UTR, but disruption of SL2 stem structure (S1-d) exerts no effect. (D) Schematic of the miR-8 seed sequence in SL2, and mutation sites of S2-i are shown in red. (E) Relative expression levels of F-luc mRNA harboring several variants of the yki 3’UTR or spi 3’UTR (as positive control) for luciferase based assays. Mutations in the putative miR-8 seed sequence, such as S2-i, perturb the reduction of F-luc mRNA in miR-8 dependent fashion. (F, G) Relative luciferase activity of F-luc harboring several kinds of yki 3’UTR or spi 3’UTR fusions. (G) In AGO1-knockdown cells, yki 3’UTR luciferase activity is not repressed. The region marked with white rectangle is magnified in the inset at bottom right. Scale bars, 5 μm (Ab, Bb). Data shown as are means ± SD (n = 3). *P<0.05 (multiple Student’s t test) (C, E-G).
Double labeling with the P-body marker Me31B, and the ER marker KDEL, shows that the P-bodies are detected in the vicinity of ER, but do not appear to overlap precisely with the marker used (Figure 4B and 4Bb). This was also the case with the use of other ER markers (data not shown). Taken together, it appears that the enrichment of yki transcripts in microsomal fractions does not appear to be due to a direct interaction with ER, at least not in the case of detected foci. Rather, it appears to be largely P-body-related. We note, however, that a significant percentage of yki transcripts may not be localized to foci, as we were unable to detect non-foci-localized transcripts, either before or after disrupting foci formation (e.g; via S2-d mutation).
P-bodies have been shown to regulate both mRNA degradation and translational repression [40,41]. Consistent with this possible role in regulating yki, further analysis of the SL2 sequence revealed the presence of a miR-8 microRNA seed sequence (Figure 4D). To assess the potential regulation of SL2 by miR-8, we first measured the level of endogenous yki mRNA in a miR-8-overexpressing cell line, since S2 cells do not express miR-8 endogenously [42,43]. The data clearly show that endogenous yki mRNA is specifically and significantly reduced in the presence of miR-8 (Figure 4C). In addition, we examined the relative expression levels of luciferase mRNA reporters fused to yki 3’UTRs carrying base substitution mutations in the putative miR-8 seed sequence that do not disrupt the SL2 distal stem-loop structure (e.g; S2-i; Figure 4E, and data not shown). The results show that the effects of miR-8 are clearly dependent on the presence of the miR-8 seed sequence (Figure 4F). This dependence was further validated by showing that knockdown of the RISC complex component Argonaute-1 (AGO1) reverses the effects of miR-8 ectopic expression (Figure 4G).
Biological significance of regulations of yki gene expression at mRNA level
To investigate the biological significance of the SL2 motif on yki gene function in vivo, we generated transgenic fly lines in which full-length (FL), ΔSL2, S1-d, S2-d’ and S2-i yki transcripts can be ectopically expressed using the GAL4/UAS system. While overexpression of the FL construct in eye imaginal discs did not induce any morphological aberrations (Figure 5B), expression of the ΔSL2 variant led to the production of severely enlarged eyes (Figure 5C), similar to those previously seen upon expression of non-phosphorylatable Yki [44-46]. Similarly, ectopic expression of S2-d’, which disrupts both the SL2 distal stem structure and the putative miR-8 seed sequence, and changes some pyrimidine bases to purine bases in the loop region, also showed dramatic overgrowth (Figure 5E). Disruption of just the proximal stem structure (S1-d), or just the miR-8 seed sequence (S2-i), had more modest effects (Figure 5D and 5F). These phenotypes were further enhanced by increasing the number of mutated transgene copies from one to two (Figure 6). The combined effect of disrupting both stem structure and miR-8 seed sequence on eye development reveals that both have important contributions to yki gene expression and eye development. Similar effects on the growth of legs and wings were observed upon ectopic expression of these constructs using a dpp-GAL4 driver (data not shown).
Figure 5.
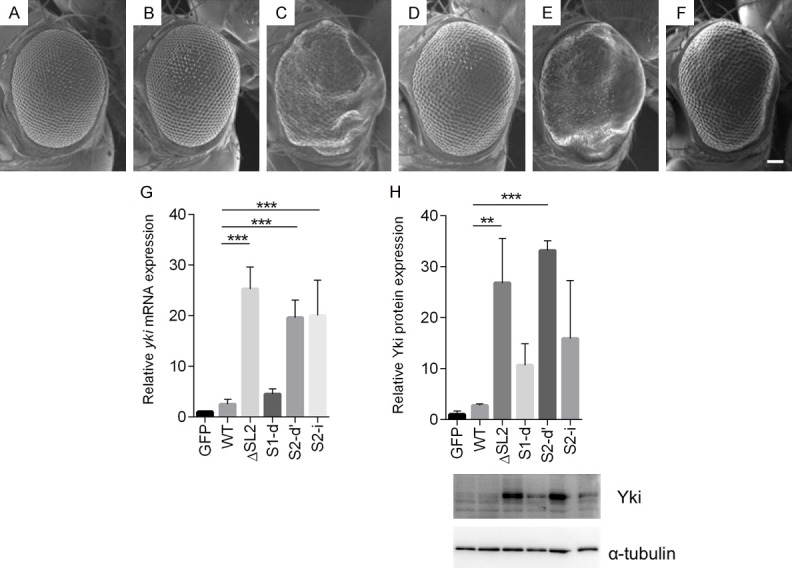
Expression of the ΔSL2 and S2-d yki expression constructs in eye discs induces severe rough eye phenotypes. (A-F) Each panel shows a scanning electron micrograph of an adult compound eye. Scale bar, 66.6 μm. (A) GMR-GAL4/+; UAS-GFP/+; +, (B) GMR-GAL4/+; +; UAS-FL (8-1-1)/+, (C) GMR-GAL4/UAS-ΔSL2 (20-3-4); +; +, (D) GMR-GAL4/+; +; UAS-S1-d (20-3-1)/+, (E) GMR-GAL4/+; UAS-S2-d’ (84-1)/+; +, (F) GMR-GAL4/+; +; UAS-S2-i (13-4)/+, (G) Relative expression levels of yki mRNA in eye imaginal discs, normalised to GAPDH mRNA. Transcripts with a missing or disrupted miR-8 seed sequence over-accumulate (H) Relative Yki protein expression levels normalised to a-tubulin protein expression in eye imaginal discs calculated from Western blots. Level of Yki protein in S2-d or ΔSL2 discs is much higher than controls, while S1-d or S2-i discs yield moderately increases Yki protein expression. Data are shown as means ± SD (n = 3). **P<0.01, ***P<0.001 (one-way ANOVA, Dunnett’s Multiple comparisons test).
Figure 6.
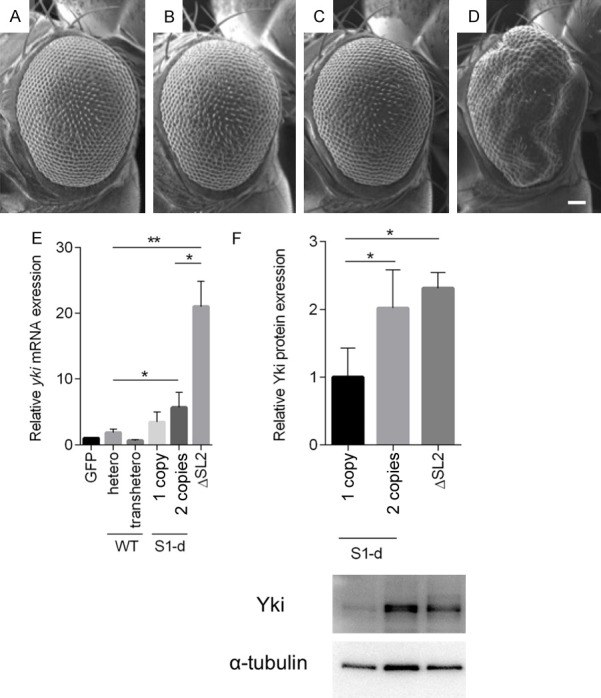
Rough eye phenotypes are enhanced in proportion to the number of copies of UAS-S1-d but not UAS-FL. A-D. In homozygous FL expressing flies, eye morphology is the same as in heterozygotes. In contrast, the rough eye phenotype in S1-d heterozygotes is enhanced in heteroallelic combinations. A-D. Each panel shows a scanning electron micrograph of an adult compound eye. A-D. Scale bar, 66.6 μm. A. GMR-GAL4/+; UAS-GFP/+; UAS-yki FL (8-1-1)/+, B. GMR-GAL4/+; +; UAS-FL (8-1-1), C. GMR-GAL4/+; UAS-GFP/+; UAS-S1-d (18-3-1)/+, D. GMR-GAL4/+; +; UAS-S1-d (18-3-1)/(20-3-1), E. Relative expression levels of yki mRNA normalised to GAPDH mRNA. The relative expression levels of yki mRNA in flies carrying 1 copy and 2 copies of UAS-S1-d are not significantly different. F. Yki protein is significantly increased in fly carrying 2 copies of UAS-S1-d, yielding similar levels of protein as produced by fly carrying 1 copy of UAS-ΔSL2. Data are means SD (n = 3). *P<0.05, **P<0.01 (one-way ANOVA, Dunnetts Multiple comparisons test).
Next we measured the expression levels of yki mRNA and Yki protein in the eye imaginal discs of the lines described above (Figure 5G and 5H). Consistent with the results described above, the levels of yki mRNA and protein generated by the FL construct were not much higher than in control eye imaginal discs. As might be expected from their effects on the miR-8 seed consensus, the S2-d and S2-i mutations caused similar increases in yki RNA stability to that caused by the complete SL2 deletion. However, despite these similarly increased levels of transcript, disruption of just the miR-8 recognition element (S2-i) did not lead to accumulation of the same amounts of protein as disruption of both the sequence and stem (S2-d), indicating that these elements are selective regulators of stability and translation respectively. The resulting levels of proteins correlate well with the phenotypes rendered by the respective constructs (Figures 5 and 6). The relative effects of S2 mutations in vivo also correlate well with those observed in S2-DRSC cells.
Discussion
We have shown that a large fraction of transcripts that do not encode transmembrane or secreted proteins associate with microsomes in a non-ribosome-dependent fashion. This includes yki transcripts, which fractionated into microsomal fractions SRP-independently, and which colocalize with P-bodies in the vicinity of ER. The correlation between foci localization and translational repression suggests that these activities are related. Although the proximity of P-bodies to ER has been reported in several organisms [47-55], the biological meaning or origin of this proximity is still unknown. Taken together, our results showing that the human YAP1 3’UTR also mediates foci formation (Figure 3I), and recent results showing that YAP1 integrates Hippo and unfolded protein responses while controlling liver size and tumorigenesis [56], suggest potentially conserved roles in 3’UTR-mediated post-transcriptional regulatory mechanisms and links to ER stress.
For the P-bodies localization of yki mRNA, we indicated that the pyrimidine bases in the loop is necessary in addition to stem structure in SL2. Consistent with our data, recent study revealed that pyrimidine bases in the loop of stem-loop in the importin β1 3’UTR is important for its axon localization in mammals [57]. Furthermore, Perry R.B. et al. also reported that Nucleolin, mammalian homolog of Drosophila modulo, binds to the pyrimidine bases and regulates the axon localization of importin β1 mRNA. Meanwhile, the other studies have revealed that polypyrimidine tract-binding protein (PTB)/hnRNP I binds to polypyrimidine bases and regulates the subcellular localization or translation of target mRNA in both C. elegance and Drosophila [58,59] and that PTB1 and 2, Arabidopsis thaliana homologs of PTB, are localized to P-bodies [60]. These data suggest that although the factor(s) that regulates foci formation and translational repression of yki mRNA appear to be P-body related, they are yet to be identified.
This study is the first to demonstrate regulation of a Hippo pathway component at the level of RNA post-transcriptional regulation. We have shown that yki gene expression is tightly regulated by a single SL structure in the 3’UTR that has the ability to control transcript localization, RNA stability and translation. While there is a strong correlation between foci formation and translational repression, it seems that RNA degradation occurs independently via miR-8 binding to the corresponding seed sequence in SL2. In the case of miR-8, it is expressed in a complex pattern in numerous stages and tissues including the embryo, and larval brain, wing and leg imaginal discs [61]. Thus, it appears that the fate of yki mRNA depends on multiple trans-acting factors, each recognizing distinct aspects of SL2 sequence and structure.
Acknowledgements
We are very grateful to E.R. Gavis, N. Nakashima, A. Nakamura and H. McNeill for providing reagents, K. Usui for technical assistance and A. Ephrussi for helpful discussions. We also thank the Drosophila Genomics Resource Center for providing the NIH 2P40OD010949-10A cultured cell line. RNA-seq analysis was performed in the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University. This work was supported in part by grants to H.Y. from the Suzuken Memorial Foundation, The Uehara Memorial Foundation, Takeda Science Foundation, Inamori Foundation, Japan Society for the Promotion of Science (JSPS) (Grant Number 26440097) and the JSPS Core-to-Core Program, Asia-Africa Science Platforms, to H.M.K. by the Canadian Institutes of Health Research, and to T.U from The Japan Science Society.
Disclosure of conflict of interest
None.
Authors’ contribution
T.U., H.Y., H.K., M.Y., Y.O., T.S., M.I. and M.S. performed experiments and data analysis, H.Y., H.M.K. and M.Y. designed the research, and T. U., H.Y. and H.M.K. wrote the manuscript.
References
- 1.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 5.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, Mu D, Lucito R, Powers S, Lowe SW. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen LL, Holm A, Rantala J, Kallioniemi O, Rasmussen MH, Ostenfeld MS, Dagnaes-Hansen F, Øster B, Schepeler T, Tobiasen H. Functional screening identifies miRNAs influencing apoptosis and proliferation in colorectal cancer. PLoS One. 2014;9:e96767. doi: 10.1371/journal.pone.0096767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blobel G, Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91:551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 11.Diehn M, Bhattacharya R, Botstein D, Brown PO. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet. 2006;2:e11. doi: 10.1371/journal.pgen.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- 13.Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pyhtila B, Zheng T, Lager PJ, Keene JD, Reedy MC, Nicchitta CV. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14:445–453. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287:5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: cis sequences act up! Trends Biochem Sci. 2010;35:459–469. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Cui XA, Zhang H, Palazzo AF. p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol. 2012;10:e1001336. doi: 10.1371/journal.pbio.1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrest KM, Gavis ER. Live Imaging of Endogenous RNA Reveals a Diffusion and Entrapment Mechanism for nanos mRNA Localization in Drosophila . Curr Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 19.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 20.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lande MA, Adesnik M, Sumida M, Tashiro Y, Sabatini DD. Direct association of messenger RNA with microsomal membranes in human diploid fibroblasts. J Cell Biol. 1975;65:513–528. doi: 10.1083/jcb.65.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- 24.Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Küchler K, Stoecklin G, Duncan KE, Teleman AA. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014;512:208–212. doi: 10.1038/nature13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinforma Oxf Engl. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 2007;35:1514–1521. doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- 29.Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, McNeill H. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–16420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 31.Querido E, Chartrand P. Using fluorescent proteins to study mRNA trafficking in living cells. Methods Cell Biol. 2008;85:273–292. doi: 10.1016/S0091-679X(08)85012-1. [DOI] [PubMed] [Google Scholar]
- 32.Serano TL, Cohen RS. A small predicted stem-loop structure mediates oocyte localization of Drosophila K10 mRNA. Development. 1995;121:3809–3818. doi: 10.1242/dev.121.11.3809. [DOI] [PubMed] [Google Scholar]
- 33.Bullock SL, Zicha D, Ish-Horowicz D. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 2003;22:2484–2494. doi: 10.1093/emboj/cdg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aragón T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.dos Santos G, Simmonds AJ, Krause HM. A stem-loop structure in the wingless transcript defines a consensus motif for apical RNA transport. Development. 2008;135:133–143. doi: 10.1242/dev.014068. [DOI] [PubMed] [Google Scholar]
- 36.Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol. 2005;25:4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen RS, Zhang S, Dollar GL. The positional, structural, and sequence requirements of the Drosophila TLS RNA localization element. RNA. 2005;11:1017–1029. doi: 10.1261/rna.7218905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pradhan SJ, Nesler KR, Rosen SF, Kato Y, Nakamura A, Ramaswami M, Barbee SA. The conserved P body component HPat/Pat1 negatively regulates synaptic terminal growth at the larval Drosophila neuromuscular junction. J Cell Sci. 2012;125:6105–6116. doi: 10.1242/jcs.113043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 43.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren F, Zhang L, Jiang J. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev Biol. 2010;337:303–312. doi: 10.1016/j.ydbio.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson KK, Lemmon SK. Suppressors of clathrin deficiency: overexpression of ubiquitin rescues lethal strains of clathrin-deficient Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:521–532. doi: 10.1128/mcb.13.1.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilhelm JE, Buszczak M, Sayles S. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila . Dev Cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 49.Decker CJ, Parker R. CAR-1 and trailer hitch: driving mRNP granule function at the ER? J Cell Biol. 2006;173:159–163. doi: 10.1083/jcb.200601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beckham CJ, Light HR, Nissan TA, Ahlquist P, Parker R, Noueiry A. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J Virol. 2007;81:9759–9768. doi: 10.1128/JVI.00844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Squirrell JM, Eggers ZT, Luedke N, Saari B, Grimson A, Lyons GE, Anderson P, White JG. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol Biol Cell. 2006;17:336–344. doi: 10.1091/mbc.E05-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomson T, Liu N, Arkov A, Lehmann R, Lasko P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev. 2008;125:865–873. doi: 10.1016/j.mod.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kilchert C, Weidner J, Prescianotto-Baschong C, Spang A. Defects in the secretory pathway and high Ca2+ induce multiple P-bodies. Mol Biol Cell. 2010;21:2624–2638. doi: 10.1091/mbc.E10-02-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weil TT, Parton RM, Herpers B, Soetaert J, Veenendaal T, Xanthakis D, Dobbie IM, Halstead JM, Hayashi R, Rabouille C, Davis I. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol. 2012;14:1305–1313. doi: 10.1038/ncb2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huch S, Gommlich J, Muppavarapu M, Beckham C, Nissan T. Membrane-association of mRNA decapping factors is independent of stress in budding yeast. Sci Rep. 2016;6:25477. doi: 10.1038/srep25477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu H, Wei L, Fan F, Ji S, Zhang S, Geng J, Hong L, Fan X, Chen Q, Tian J, Jiang M, Sun X, Jin C, Yin ZY, Liu Q, Zhang J, Qin F, Lin KH, Yu JS, Deng X, Wang HR, Zhao B, Johnson RL, Chen L, Zhou D. Integration of Hippo signalling and the unfolded protein response to restrain liver overgrowth and tumorigenesis. Nat Commun. 2015;6:6239. doi: 10.1038/ncomms7239. [DOI] [PubMed] [Google Scholar]
- 57.Perry RB, Rishal I, Doron-Mandel E, Kalinski AL, Medzihradszky KF, Terenzio M, Alber S, Koley S, Lin A, Rozenbaum M, Yudin D, Sahoo PK, Gomes C, Shinder V, Geraisy W, Huebner EA, Woolf CJ, Yaron A, Burlingame AL, Twiss JL, Fainzilber M. Nucleolin-Mediated RNA Localization Regulates Neuron Growth and Cycling Cell Size. Cell Rep. 2016;16:1664–1676. doi: 10.1016/j.celrep.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Besse F, López de Quinto S, Marchand V, Trucco A, Ephrussi A. Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev. 2009;23:195–207. doi: 10.1101/gad.505709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cote CA, Gautreau D, Denegre JM, Kress TL, Terry NA, Mowry KL. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- 60.Stauffer E, Westermann A, Wagner G, Wachter A. Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. Plant J Cell Mol Biol. 2010;64:243–255. doi: 10.1111/j.1365-313X.2010.04321.x. [DOI] [PubMed] [Google Scholar]
- 61.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila . Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]