Abstract
Isocitrate dehydrogenase 2 (IDH2) is the rate-limiting enzyme in the tricarboxylic acid (TCA) cycle in cellular metabolism. Growing evidence indicates that IDH2 plays a crucial role in the development of cancer. We aimed to investigate the expression level of IDH2 and its prognostic value in esophageal squamous cell cancer (ESCC). We evaluate the IDH2 expression and prognostic value in ESCC by immunohistochemical (IHC) staining, quantitative real-time polymerase chain reaction (qRT-PCR) and Western blotting. The cell counting kit-8 (CCK8), clonogenic and invasion assays were performed to verify the IDH2 function in vitro. The protein expression level of IDH2 was significantly upregulated in ESCC tissues (IHC, Western blotting, all P<0.001) despite no significant difference at mRNA expression level (P>0.05). Kaplan-Meier analysis showed that IDH2 overexpression in ESCC patients was significantly related to worse overall survival (OS) and progression-free survival (PFS), P = 0.003 and 0.002, respectively. The univariate and multivariate analyses revealed that IDH2 overexpression served as an independent prognostic factor for OS and PFS (all P<0.005) in ESCC. The OD450 value, colony formation and invasive cell number were decreased in the shIDH2 groups (all P<0.0001). The upregulation of IDH2 in ESCC cells showed opposite effects (all P<0.05). Additionally, IDH2 knockdown phenotype can be rescued by shRNA-resistant IDH2 (all P<0.05). These results demonstrated that IDH2 was upregulated in ESCC and could be used as a valuable prognostic marker for ESCC patients.
Keywords: ESCC, IDH2, expression, prognosis
Introduction
Esophageal cancer is the eighth most common cancer type and the sixth leading cause of cancer-related mortality globally [1]. According to the study on global cancer statistics of the National Cancer Institute, an estimated 16,910 new esophageal cancer cases and 15,690 death cases will occur in 2016 nationwide [2]. Despite the fact that the incidence of esophageal adenocarcinoma (EAC) is growing faster than other malignancies in the Western countries, squamous cell carcinoma (ESCC) is still the most common histological type of esophageal cancer worldwide [3]. New advances for the diagnosis and treatment of esophageal cancer have not changed the reality that the clinical outcome of ESCC patients remains poor, the 5-year overall survival (OS) rate ranges from 15% to 25% [4]. The most common biological changes during the development of ESCC include activation of oncogenes (such as EGFR and c-MYC), inactivation of tumor suppressor genes (such as p53, TOC and DLC1), and modification of cell cycle control by several mechanisms (such as amplification of Cyclin D1 and homozygous deletion or promoter methylation of MTS1) [5]. However, unlike alpha fetal protein (AFP) in hepatocellular carcinoma and carcinoembryonic antigen (CEA) in colon cancer, no good biomarker for ESCC has been widely applied in clinical practice. Accordingly, identification of novel biomarkers for ESCC should be emphasized.
Besides being involved in maintaining proliferative signaling, evading growth suppressors, immune escape, replicative immortality, tumor-promoting inflammation, invasion and metastasis, angiogenesis, genome instability and mutation and resisting cell death, the reprogramming of energy metabolism has been regarded as an emerging hallmark of cancer [6]. Isocitrate dehydrogenase (IDH) is a reversible enzyme in the tricarboxylic acid (TCA) cycle that can both catalyze the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG) and the reductive carboxylation of the reciprocal process, which depend on NADP+ and NADPH/CO2 respectively [7]. Meanwhile, IDH plays a crucial role in processing reactive oxygen species (ROS) induced by hypoxic tumor microenvironment to support cell growth through the production of nonmitochondrial NADPH [8]. IDH2, which is located in the mitochondria and utilizes NADP+ as a cofactor, is one of the three members of the IDH family [9]. Detoxification mechanisms could be impaired in cancer cells due to the inactivation of IDH2, and further result in DNA damage [10].
To date, most of the studies about IDH in cancer have focused on its mutation status. Mutant IDH gains a novel enzymatic activity of converting α-KG to the metabolite 2-hydroxyglutarate (2-HG) by a reduction reaction [8]. A tumor-derived IDH mutant inhibits histone and DNA demethylation, which hinder cell differentiation [11,12]. Mutations in IDH1 or 2 (mutant IDH1/2) are detected in several malignancies, including acute myeloid leukemia (AML), glioma, cholangiocarcinoma as well as chondrosarcoma and chondromas [13-16]. Moreover, mutant IDH1/2 is a poor prognostic factor in cytogenetically normal (CN)-AML and a positive prognostic marker in low-grade gliomas [17,18].
However, expression of wild-type IDH2 and its clinical relevance in cancers have been less investigated. IDH2 is downregulated in hepatocellular carcinoma (HCC) and gastric cancer (GC) and patients with low IDH2 had lower 5-year survival [19,20]. In contrast, IDH2 is expressed at high level in endometrial, prostate and testicular cancers [21,22]. In addition, IDH2 expression is downregulated in in situ carcinoma and upregulated in infiltrating carcinoma in colon cancer patients [23]. The expression status and prognostic value of IDH2 has not been examined in patients with ESCC. In this study, the expression of IDH2 was evaluated in cancerous tissue and compared with its expression in paracancerous tissue by quantitative real-time PCR (qRT-PCR), Western blot analysis and immunohistochemistry (IHC). The prognostic potential of IDH2 was also analyzed. Furthermore, we conducted in vitro experiments by interfering IDH2 expression to verify its effect on proliferation and invasion of ESCC cells.
Methods
Patients and specimens
A total of 28 ESCC tissue samples were collected from surgical tissue blocks in the Qilu Hospital of Shandong University from April 2015 to August 2015. Adjacent noncancerous tissue samples were obtained and used as control groups. All the tissues were immediately stored in a freezer at -80°C until used for experiments. A total of 119 formalin-fixed, paraffin-embedded (FFPE) tissue samples from patients who received subtotal esophagectomy and esophagogastric anastomosis plus regional lymph node dissection in 2009 were used for survival analysis. None of the patients enrolled in our cohort had received neoadjuvant therapy (chemotherapy and/or radiotherapy). All the specimens were pathologically confirmed. The AJCC Cancer Staging Manual, 7th edition was used for assessing tumor stages. Our study was approved by the Ethics Boards of Qilu Hospital of Shandong University and informed written consent was obtained from all patients.
IHC
After the ESCC tissue was fixed with 10% formalin and embedded in paraffin, they were cut into 4 μm sections, dried at 80°C for 15 min, dewaxed in xylene, rinsed in ethanol at various concentrations and rehydrated in double-distilled water. Citrate-EDTA buffer (2 mM EDTA, 10 mM citric acid, 0.05% Tween 20, pH 6.2) was used for antigen retrieval. Sections were incubated with hydrogen peroxide for blocking the peroxidase enzyme. Then the anti-IDH2 antibody (1:60; Proteintech, Chicago, IL, USA) was applied to the sections at 4°C overnight. Negative controls were incubated with PBS instead of the primary antibody. The sections were reacted with horseradish peroxidase (HRP)-labeled streptavidin by adding it together with the biotinylated secondary antibody. Then the sections were stained with DAB and counterstained with hematoxylin. We selected five fields (×400 magnification) at random for each sample and invited two pathologists to evaluate and score them independently. The intensity of the dye color and the number of positive cells were both used for assessing the scores. The dye color was classified as: 0 (no staining), 1 (weak), 2 (moderate) and 3 (intense). The number of positive cells was graded as 0 (<5%), 1 (5-25%) and 2 (25-50%), 3 (51-75%) and 4 (>75%). The final score was the multiplication value of these two scores. 0-1 scores (-), 2-4 scores (+), 5-8 scores (++) and 9-12 scores (+++). Samples with (+++) were regarded as overexpression.
qRT-PCR
The TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used for extracting total RNA from fresh tissues following the manufacturer’s instructions. IDH2 expression at the mRNA level was assessed by SYBR Green Real Time PCR Master Mix (TOYOBO, Osaka, Japan) using the Bio-Rad Single Color Real-Time PCR system (Bio-Rad, Hercules, California, USA). The primers (Sangon Biotech, Shanghai, China) were designed and synthesized as follows: IDH2: 5’-CAAAAACATCCCACGCCTAGTC-3’ (forward primer), 5’-CCCGGTCTGCCACAAAGT-3’ (reverse primer); GAPDH: 5’-GAAGGTCGGAGTCAACGGAT-3’ (forward primer), 5’-TGAAACACCGTCTGGCCC-3’ (reverse primer). The 2-∆∆CT method was used for calculating the relative expression of IDH2. We performed all assays in triplicate and present the data as the mean ± SD.
Western blot
Tissue blocks were grinded into homogenate. The proteins were extracted from 100 mg of each tissue sample using 1 ml RIPA lysis buffer (50 mM Tris, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, sodium orthovanadate, sodium fluoride, EDTA, leupeptin) and 10 μl PMSF (Beyotime, Shanghai, China). Supernatant was divided and stored at -20°C for later use. Samples containing an identical amount of protein were electrophoresed on 10% polyacrylamide gels and transferred to nitrocellulose membranes. Defatted dry milk (5%) was used to diminish nonspecific binding on the membranes. After incubation with the primary IDH2 antibody and β-actin antibody (1:100, Proteintech, Chicago, IL, USA), the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies. The bands were detected by the chemiluminescence detection system (EMD Millipore, Billerica, MA, USA).
Cell culture
The human ESCC cell lines Eca109 (CCTCC) and Eca9706 (ATCC, Manassas, VA, USA) were cultured in RPMI 1640 media (Gibco, Life Technologies Inc., Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco, Life Technologies Inc.). Cells were incubated in a 5% CO2 atmosphere at 37°C.
Transfection
Plasmid containing a short hairpin RNA (shRNA) targeting IDH2 (Cat. No. HSH009353-LVRU6GP; GeneCopoeia, Inc., Rockville, MD, USA) and a nonsense shRNA (Cat. No. CSHCTR001-LVRU6GP; GeneCopoeia, Inc.) were used for RNA interference experiments. Cells were seeded in 24-well plates for 24 h. Lipofectamine solution was prepared by mixing 2 μl of Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA) and 50 μl of Opti-MEMI medium (Invitrogen) for 5 min. The plasmid solution was prepared by adding 1 μg of recombinant plasmid into 50 μl Opti-MEMI medium, followed by incubation for 5 min. Then, the plasmid and the lipofectamine solutions were combined to obtain the transfection solution. A 100 μl volume of the transfection solution was mixed with pen-strep-free RPMI 1640 and 0.5 ml of this mixture was added per well. After 24 h, the transfection medium was replaced with pen-strep-free normal medium. Puromycin (1.5 mg/ml) was used for selecting stably transfected cells.
Lentivirus transduction
We purchased lentiviruses (GeneCopoeia, Inc.) expressing open reading frame (ORF) of wild-type IDH2 (IDH2W) and a shRNA-resistant form of IDH2 (IDH2R) that harbors eight silent mutations within the sequence targeted by sh-IDH2-1. The targeting sequence by sh-IDH2-1 in the IDH2 was mutated from GACCGACTTCGACAAGAATAA to GACCTACCTCCACCAGGATCA by site-directed mutagenesis. Cells were plated in 24-well plate 24 hours prior to viral infection. 2 μl of lentivirus was diluted in 0.5 ml complete medium with polybrene at a final concentration of 6 μg/ml. The plates were placed for 2 hours at 4-8°C and transferred to a 37°C incubator with 5% CO2 for overnight incubation. The cells were trypsinized and re-seeded onto 6-well plates and incubated for 48 hours. Puromycin selection (1.5 mg/ml) was applied to select stably transduced cells.
Cell viability assays
A cell counting kit-8 assay (CCK8; Dojindo, Japan) was used to evaluate the proliferation of Eca109 and Eca9706 cells. Cells were seeded into 96-well plates at a density of 2×103 per well. After culturing for 0, 24, 48, 72, 96 h, 120, and 144 h at 37°C, the medium was replaced with 100 μl fresh medium and 10 μl CCK-8 solution and incubated for an additional 1 h. The absorbance at 450 nm was measured using the Thermo Scientific Varioskan Flash spectrophotometer (Thermo Scientific, Finland). The mean value of 4 wells was used as the result of each sample. The data are expressed as the mean values ± SD of three independent experiments.
Clonal efficiency assay
Transfected cells were trypsinized into single cell suspension and diluted in proportion. A total of 200 cells per dish were cultured for 2 weeks for knockdown experiments. 1000 cells per dish were used for overexpression and rescue experiments. Cells transfected with nonsense shRNA were used as controls. Colonies were fixed with ethanol and stained with crystal violet. Clone formation were defined as those containing at least 50 cells.
Transwell invasion assay
Matrigel (BD Bioscience, San Jose, CA, USA) was used for coating the membrane of the transwell chambers (Corning Costar, Cambridge, MA, USA). Eca109 and Eca9706 cells were added into the upper chamber at a density of 2-5×105/ml in serum-free medium, while the lower chamber contained normal medium with 10% FBS. The cells remaining on the upper chamber after 24 h were wiped off with a cotton swab. The cells attached to the lower membrane were fixed with methanol and stained with crystal violet. The number of invasive cells in 5 different fields under the inverted microscope was regarded as the mean value. Each samples were tested in triplicate and each assay was repeated 3 times.
Statistical analysis
The difference between the cancerous and paracancerous tissue groups was assessed by a paired Student’s t-test. The difference between each pair combination among sh-NC, sh-IDH2-1, and sh-IDH2-2 was evaluated using ANOVA with the post-hoc Tukey’s test. The correlation of IDH2 and clinicopathological factors was determined with the Bilateral χ 2 test. Kaplan-Meier method and the log-rank test were used to obtain survival curves and compare the difference between subgroups. Univariate and multivariate Cox survival analyses were used for evaluating the hazard ratios (HRs). Receiver operating characteristic (ROC) curve analysis and the area under the curve (AUC) were used to determine the concordance level between the observed value and the actual value. Statistical analyses were performed with the SPSS software (Statistical Package for Social Sciences, Chicago, IL, USA). P<0.05 was considered statistically significant.
Results
The expression level of IDH2
The expression status of IDH2 in cancerous and paracancerous tissues was evaluated by qRT-PCR, Western blot and IHC analyses. The qRT-PCR analysis results indicated that the expression of IDH2 had no difference between two groups (Figure 1A, 1.02 ± 0.07 vs. 0.91 ± 0.20, P>0.05). The Western blot analysis, which was performed on 8 pairs of cancerous and paracancerous tissues (Figure 1B), revealed that IDH2 was upregulated in cancerous tissues at the protein level (Figure 1C, IDH2/β-actin: 0.57 ± 0.21 vs. 0.18 ± 0.11, P<0.001). To test IDH2 expression further, FFPE tissue samples were prepared and the expression level of IDH2 protein was identified by IHC analysis. IDH2 was mainly distributed in the cytoplasm of ESCC cells (Figure 1D). The scores of the cancerous tissue samples were: 15 (+++), 8 (++), 4 (+) and 1 (-), while those of the paracancerous tissues were: 24 (-) and 4 (+). Overall, the results revealed that IDH2 was overexpressed in 15 cancerous and 0 paracancerous tissue samples, and their difference was statistically significant (Figure 1E, P<0.0001). These results suggest that IDH2 expression was higher in cancerous tissues than in paracancerous tissues.
Figure 1.
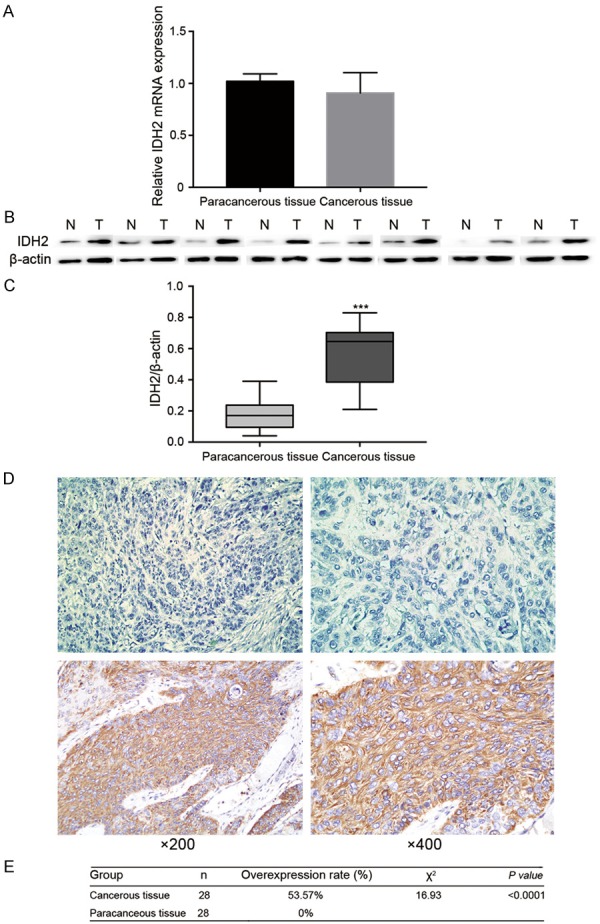
IDH2 expression in ESCC cancerous and paracancerous tissues from 28 patients. A: IDH2 expression at the mRNA level. B: Western blot bands of 8 pairs of cancerous and paracancerous tissues from ESCC patients. C: IDH2/β-actin values of 8 pairs of ESCC tissues. D: Representative immunostaining of IDH2 in ESCC tissues: the upper ESCC tissue did not overexpressed IDH2, while the lower ESCC tissue overexpressed IDH2. E: Comparison of the results of the IHC analysis of cancerous and paracancerous tissues.
The prognostic value of IDH2
During the follow-up period, 43 patients (36.13%) were alive and (63.87%) died. The median survival time was 40 months (11-83 months). The IDH2 expression was assessed in FFPE tissue samples from 119 patients. According to the evaluation criteria, 65 (+++), 32 (++), 17 (+) and 5 (-) samples were observed and the overall overexpression rate was 54.62%. The correlations of IDH2 expression with the clinical features of the patients, as determined by the bilateral χ 2 test, are shown in Table 1. T stage (P=0.011) was significantly related to the IDH2 expression. Age, gender, smoking, drinking, differentiation, N stage and TNM stage did not show any significant correlation with IDH2 expression. The Kaplan-Meier curve (Figure 2A, 2B) demonstrated that ESCC patients without IDH2 upregulation showed longer OS and progression-free survival (PFS) compared to patients with IDH2 upregulation (OS: P=0.003, PFS: P=0002). The univariate survival analysis (Table 2) revealed that IDH2 expression had a negative correlation with OS and PFS (P=0.003 and P=0.002, respectively). Multivariate analysis (Table 2) further verified that IDH2 may be an independent prognostic marker for OS and PFS in ESCC patients (both P<0.001). T stage and N stage, as well as adjuvant therapy were also found to be independent predictive markers for ESCC patients (OS: P=0.016, P<0.001 and P=0.002, respectively; PFS: P=0.008, P<0.001 and P<0.001, respectively). The ROC curves of IDH2 in the OS and PFS models are shown in Figure 2C, 2D, respectively.
Table 1.
The correlations of clinicopathologic variables of ESCC with IDH2 expression in FFPE cancerous tissues
| Clinicopathological features | IDH2 overexpression | P a value | |
|---|---|---|---|
|
| |||
| No (n = 54) | Yes (n = 65) | ||
| Age | 0.852 | ||
| <65 | 30 | 35 | |
| ≥65 | 24 | 30 | |
| Gender | 0.602 | ||
| Female | 25 | 29 | |
| Male | 27 | 38 | |
| Smoking | 0.933 | ||
| No | 27 | 33 | |
| Yes | 27 | 32 | |
| Drinking | 0.235 | ||
| No | 24 | 36 | |
| Yes | 30 | 29 | |
| Differentiation | 0.214 | ||
| Well | 26 | 22 | |
| Moderate | 13 | 24 | |
| Poor | 15 | 19 | |
| T stage | 0.011* | ||
| T1 | 10 | 5 | |
| T2 | 16 | 26 | |
| T3 | 24 | 18 | |
| T4 | 4 | 16 | |
| N stage | 0.381 | ||
| N0 | 25 | 24 | |
| N1 | 11 | 18 | |
| N2 | 8 | 15 | |
| N3 | 10 | 8 | |
| TNM stage | 0.287 | ||
| I | 19 | 15 | |
| II | 11 | 19 | |
| III | 24 | 31 | |
P: Chi-square test.
Abbreviation: FFPE, formalin-fixed paraffin-embedded.
P<0.05.
Figure 2.
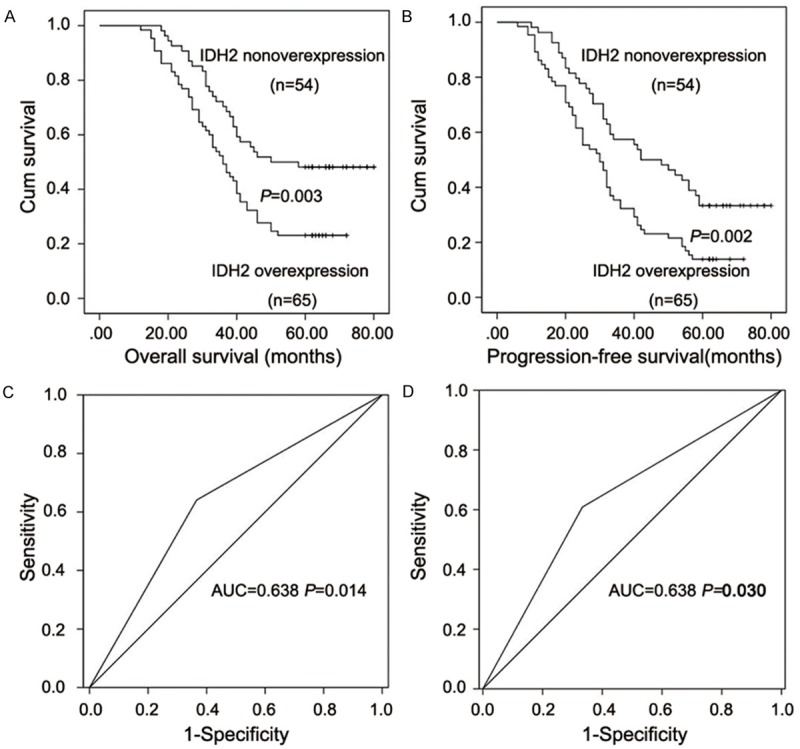
Kaplan-Meier curves and ROC curves analyses of IDH2 in ESCC tissues. A: Kaplan-Meier curve for OS. B: Kaplan-Meier curve for PFS. C: Receiver operating curve for OS. D: Receiver operating curve for PFS.
Table 2.
Univariate and multivariate analyses of prognostic variables
| OS | OS | PFS | PFS | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||
|
| ||||||||
| Variable | P value | P value | HR | 95% CI | P value | P value | HR | 95% CI |
| Gender (Female VS. Male) | 0.792 | 0.751 | 1.086 | 0.652-1.811 | 0.971 | 0.901 | 1.030 | 0.647-1.638 |
| Age (<65 vs. ≥65) | 0.760 | 0.483 | 0.828 | 0.489-1.403 | 0.908 | 0.638 | 0.892 | 0.554-1.436 |
| Smoking (Yes vs. No) | 0.765 | 0.138 | 1.624 | 0.855-3.084 | 0.835 | 0.213 | 1.457 | 0.806-2.635 |
| Drinking (Yes vs. No) | 0.671 | 0.200 | 1.555 | 0.792-3.051 | 0.460 | 0.083 | 1.735 | 0.931-3.236 |
| T stage | 0.001* | 0.016* | <0.001* | 0.008* | ||||
| T1 | 1.000 | Ref. | 1.000 | Ref. | ||||
| T2 | 0.026* | 3.207 | 1.148-8.958 | 0.028* | 2.798 | 1.118-7.003 | ||
| T3 | 0.002* | 5.075 | 1.845-13.96 | 0.001* | 4.701 | 1.971-11.53 | ||
| T4 | 0.062 | 2.638 | 0.951-7.314 | 0.045* | 2.533 | 1.022-6.278 | ||
| N stage | <0.001* | <0.001* | <0.001* | <0.001* | ||||
| N0 | 1.000 | Ref. | 1.000 | Ref. | ||||
| N1 | 0.002* | 3.108 | 1.531-6.312 | <0.001* | 3.542 | 1.847-6.793 | ||
| N2 | 0.004* | 2.804 | 1.383-5.686 | 0.009* | 2.392 | 1.241-4.610 | ||
| N3 | <0.001* | 6.988 | 3.004-16.26 | <0.001* | 6.771 | 3.069-14.94 | ||
| Differentiation | 0.020* | 0.272 | 0.140 | 0.447 | ||||
| Well | 1.000 | Ref. | 1.000 | Ref. | ||||
| Moderate | 0.107 | 1.713 | 0.890-3.296 | 0.275 | 1.393 | 0.768-2.527 | ||
| Poor | 0.475 | 1.253 | 0.675-2.325 | 0.907 | 0.967 | 0.548-1.704 | ||
| Adjuvant therapy (Yes vs. No) | 0.034* | 0.002 | 2.301 | 1.368-3.870 | 0.012* | <0.001* | 2.577 | 1.584-4.192 |
| IDH2 | 0.003* | <0.001* | 3.107 | 1.735-5.565 | 0.002* | <0.001* | 3.473 | 2.040-5.912 |
| (Overexpression VS. nonoverexpression) | ||||||||
Abbreviations: OS, overall survival; PFS, progression-free survival; CI: confidence interval.
P<0.05.
Downregulation of IDH2 expression in ESCC cell lines
In order to determine whether the sh-IDH2 plasmids were successfully transfected or not, qRT-PCR analysis (Figure 3A) and Western blot analysis (Figure 3B, 3C) were used to detect IDH2 expression at mRNA and protein levels, respectively, in Eca109 and Eca9706 cells. The result indicated that IDH2 mRNA was successfully downregulated by sh-IDH2-1 and sh-IDH2-2 compared to sh-NC (Eca109: 1.02 ± 0.13 vs. 0.42 ± 0.09 and 0.47 ± 0.08, P<0.001, respectively; Eca9706: 1.01 ± 0.15 vs. 0.45 ± 0.05 and 0.40 ± 0.05, P<0.001, respectively). In addition, the result also indicated that IDH2 protein expression was effectively downregulated by sh-IDH2-1 and sh-IDH2-2 compared to sh-NC (Eca109: 0.87 ± 0.09 vs. 0.40 ± 0.08 and 0.37 ± 0.11, P<0.01 and P<0.001 respectively; Eca9706: 0.84 ± 0.07 vs. 0.44 ± 0.04 and 0.44 ± 0.10, P<0.001 respectively).
Figure 3.
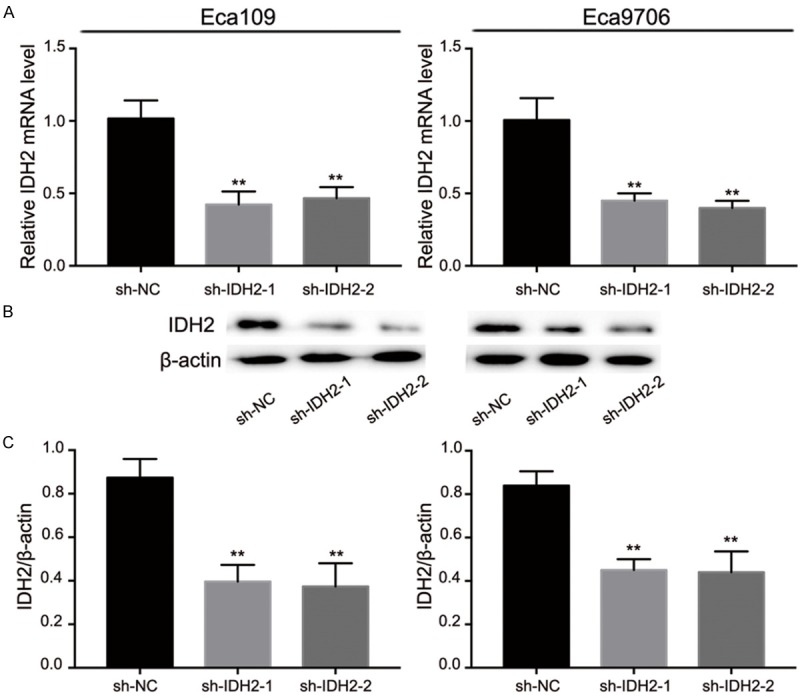
The downregulation effect of sh-IDH2-1 and sh-IDH2-2. A: sh-IDH2-1 and sh-IDH2-2 decreased mRNA expression of IDH2. B and C: Expression of IDH2 protein in Eca109 and Eca9706 was downregulated after sh-IDH2-1 and sh-IDH2-2 transfection.
Downregulation of IDH2 inhibited ESCC cells proliferation and invasion
To ascertain whether IDH2 affects the proliferation of Eca109 and Eca9706 cells, we performed the CCK8 and colony formation assays. Compared with the sh-NC group, the OD450 values of the Eca109 and Eca9706 cells transfected with sh-IDH2-1 and sh-IDH2-2 were smaller (Figure 4A). At 144 h, the OD450 values in Eca109 transfected with sh-NC vs. sh-IDH2-1 or sh-IDH2-2 were: 1.02 ± 0.09 vs. 0.56 ± 0.06 and 0.66 ± 0.06, respectively, (all P<0.001) and in Eca9706 transfected with sh-NC vs. sh-IDH2-1 and sh-IDH2-2 were: 1.10 ± 0.11 vs. 0.63 ± 0.04 and 0.67 ± 0.07, respectively, (all P<0.001). Similarly, sh-IDH2 transfection groups of ESCC cells showed less colony formation than sh-NC groups (Figure 4B, 4C). The clone number in Eca109 transfected with sh-NC vs. sh-IDH2-1 and sh-IDH2-2 were: 124 ± 5 vs. 55 ± 7 and 60 ± 6, respectively, (all P<0.001) and in Eca9706 transfected with sh-NC vs. sh-IDH2-1 and sh-IDH2-2 were: 129 ± 7 vs. 70 ± 8 and 74 ± 9 respectively, (all P<0.001). To detect the effect IDH2 on the biological behaviors of Eca109 and Eca9706 cells, the transwell invasion assays were performed (Figure 4D, 4E). The number of invasive Eca109 cells of the sh-NC group vs. the sh-IDH2-1 and sh-IDH2-2 groups were: 431 ± 13 vs. 242 ± 18 and 230 ± 12, respectively, (all P<0.001), while the number of invasive Eca9706 cells of the sh-NC group vs. the sh-IDH2-1 and sh-IDH2-2 groups were: 447 ± 9 vs. 249 ± 12 and 258 ± 12, respectively, (all P<0.001). These results indicate that ESCC cells with higher level of IDH2 have stronger capability for invasion.
Figure 4.
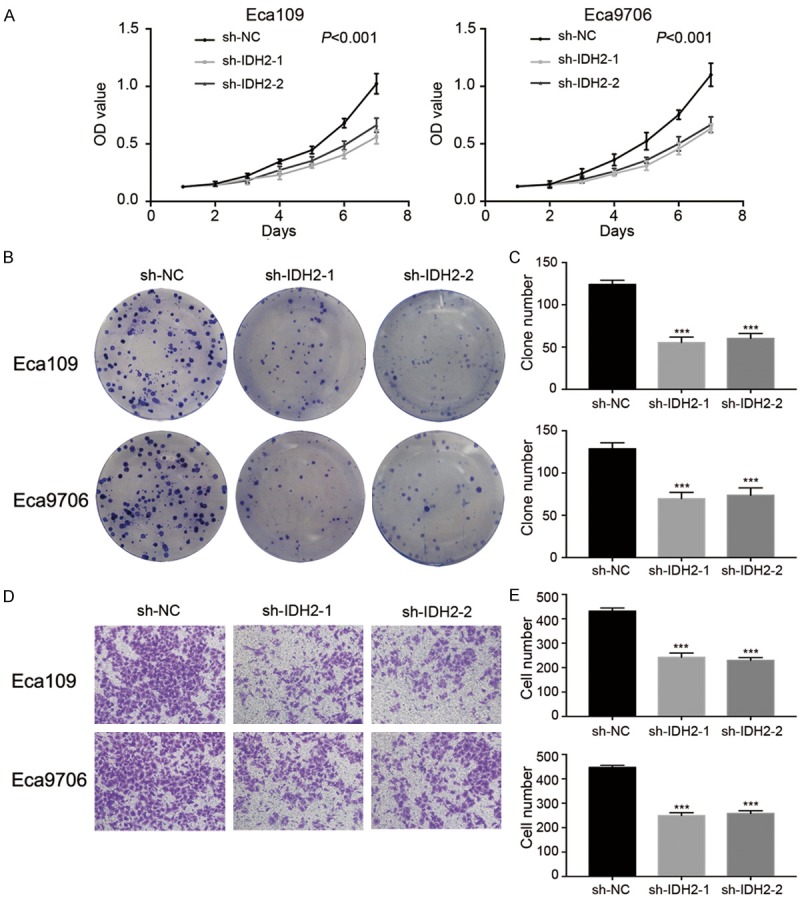
The effects of sh-IDH2-1 and sh-IDH2-2 on ESCC cells. A: The results of the CCK8 assays in Eca109 and Eca9706. B and C: The results of clonal efficiency assays in Eca109 and Eca9706. D and E: The results of cell invasion in Eca109 and Eca9706.
Overexpression of IDH2 promoted ESCC cells proliferation and invasion
To detect the gene expression level of IDH2 in control lentiviruses groups, IDH2W groups, and IDH2R groups. qRT-PCR and Western blot analysis were performed (Figure 5). IDH2 mRNA was successfully upregulated by IDH2W and IDH2R compared to control groups (Eca109: 0.96 ± 0.03 vs. 3.37 ± 0.22 and 3.33 ± 0.20, P<0.001, respectively; Eca9706: 0.96 ± 0.03 vs. 3.32 ± 0.31 and 3.40 ± 0.11, P<0.001, respectively). Western blot analysis confirmed the elevation of IDH2 expression level in IDH2W and IDH2R groups (Eca109: 0.79 ± 0.03 vs. 1.01 ± 0.02 and 1.02 ± 0.06, P<0.01 and 0.001, respectively; Eca9706: 0.81 ± 0.03 vs. 1.11 ± 0.04 and 1.13 ± 0.07, P<0.001, respectively). The effect of IDH2 upregulation on ESCC cells were showed in Figure 6. The OD450 values at 144 h were higher in IDH2W and IDH2R groups than in control groups (Eca109: 1.00 ± 0.10 vs. 1.22 ± 0.06 and 1.26 ± 0.09, P<0.05, respectively; Eca9706: 1.08 ± 0.07 vs. 1.27 ± 0.05 and 1.30 ± 0.06, P<0.05 and 0.01, respectively). IDH2W and IDH2R groups had more clone formation than control groups (Eca109: 579 ± 26 vs. 673 ± 40 and 671 ± 29, P<0.05, respectively; Eca9706: 572 ± 35 vs. 691 ± 39 and 682 ± 32, P<0.05, respectively). The invasive cell numbers were higher in IDH2W groups (Eca109: 352 ± 18 vs. 435 ± 15 and 425 ± 12, P<0.01, respectively; Eca9706: 335 ± 17 vs. 424 ± 19 and 422 ± 11, P<0.001 and 0.01, respectively) as well. These results demonstrated that the elevated expression of IDH2 promoted the proliferation and invasion of transducted cells compared with the control cells.
Figure 5.
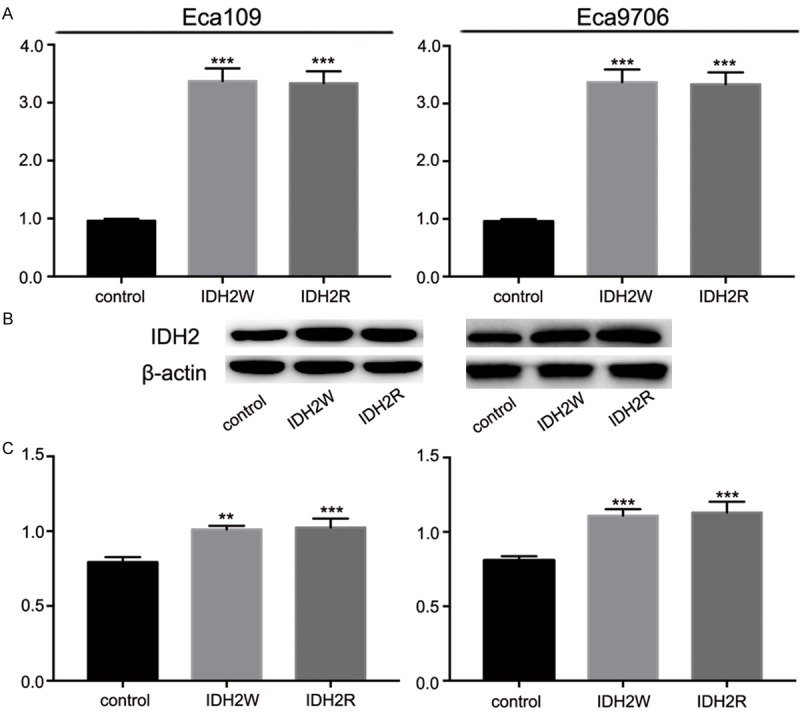
The upregulation effect of IDH2W and IDH2R. A: IDH2W and IDH2R increased IDH2 expression at mRNA level. B and C: IDH2W and IDH2R increased IDH2 expression at protein level.
Figure 6.
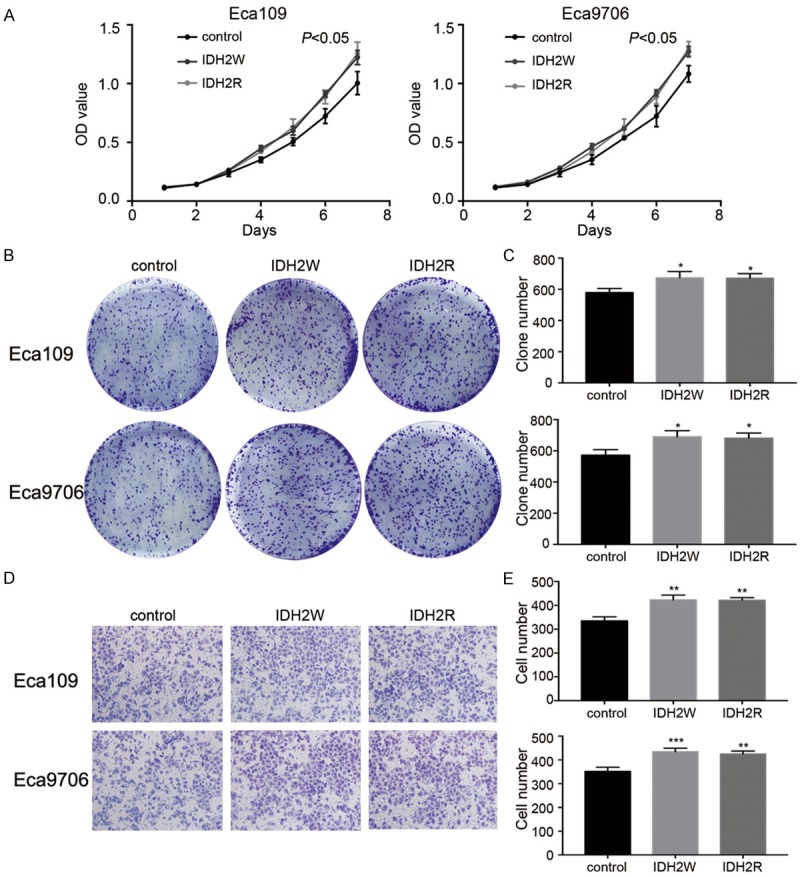
The effects of IDH2W and IDH2R on ESCC cells. A: CCK8 assays in Eca109 and Eca9706. B and C: Clonal efficiency assays in Eca109 and Eca9706. D and E: Invasion assays in Eca109 and Eca9706.
Rescue of IDH2 knockdown phenotype by expression of shRNA-resistant IDH2
To further confirm the specificity of shRNA manipulations, rescue experiments using lentiviruses were carried out (Figure 7). ESCC cells with sh-IDH2-1 plasmids stably transfected were infected by IDH2W lentiviruses and IDH2R lentiviruses. The OD450 values at 144 h were higher in sh-IDH2-1 + IDH2W and sh-IDH2-1 + IDH2R groups than sh-IDH2-1 groups (0.65 ± 0.05 vs. 0.99 ± 0.11 and 1.33 ± 0.08 in Eca109 and 0.67 ± 0.06 vs. 1.04 ± 0.07 and 1.29 ± 0.07 in Eca9706, P<0.001, respectively). The clone numbers (281 ± 19 vs. 454 ± 28 and 580 ± 23 in Eca109 and 297 ± 14 vs. 475 ± 21 and 598 ± 23 in Eca9706, P<0.001, respectively) and invasive cell numbers (161 ± 18 vs. 291 ± 15 and 390 ± 15 in Eca109 and 134 ± 11 vs. 308 ± 19 and 397 ± 11 in Eca9706, P<0.001, respectively) showed the similar trend. Additionally, these results indicated that IDH2R had stronger ability of rescuing IDH2 knockdown phenotype than IDH2W (all P<0.05).
Figure 7.
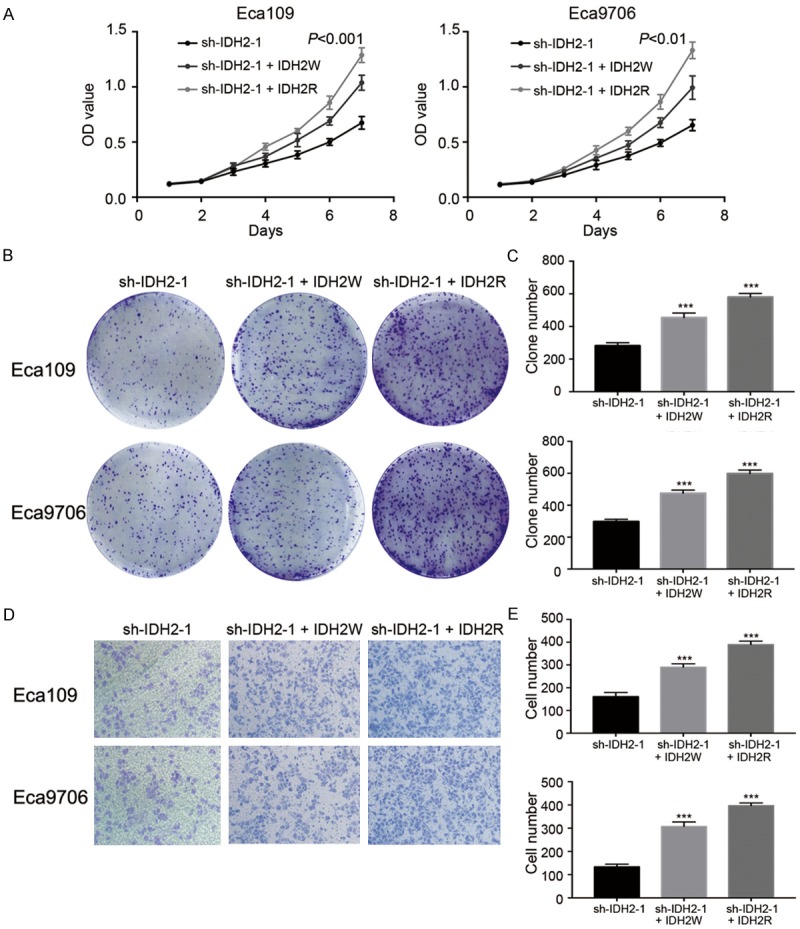
Rescue experiments of IDH2 knockdown phenotype. A: CCK8 assays in ESCC cells. B and C: Clonal efficiency assays in ESCC cells. D and E: Invasion assays in ESCC cells.
Discussion
In the present study, the result of qRT-PCR showed no difference of mRNA expression between these two groups (P>0.05). mRNA level of one gene is not always interrelated with protein level. In addition to transcription and mRNA stability, translation and protein degradation are also essential factors for protein expression [24]. The protein expression levels of IDH2 in cancerous tissues were higher than those in paracancerous tissues (IHC: P<0.001; Western: P<0.001), despite the range of expression levels from patient to patient. The study suggests that IDH2 protein may be a good biomarker for ESCC patients. The biological behavior of carcinoma and molecular pathogenesis differ with respect to different sites of origin and histological types. Unlike the results in this study on ESCC, IDH2 is downregulated in HCC cells, GC cells and the early phase of colon cancer [19,20,23]. The upregulation of IDH2 in endometrial cancer, prostate and testicular cancer, as well as advanced phase of colon cancer is in line with the IDH2 overexpression in ESCC [21-23]. Here, the analysis of the correlations of IDH2 expression with clinicopathological characteristics revealed that the expression level of IDH2 was related to the T stage, but not to other factors, which may be due to the heterogeneity and insufficient number of samples evaluated.
Additionally, the prognostic value of IDH2 was investigated in ESCC. The Kaplan-Meier curve revealed that IDH2 overexpression was correlated to poor outcomes in patients with ESCC, including worse OS and PFS. Univariate analysis showed that IDH2 may be an independent prognostic marker for OS and PFS. Multivariate analysis further confirmed this result. The ROC curve was used for judging the prognostic value of IDH2 and the AUC was 0.638 (P=0.014) for OS and 0.638 (P=0.030) for PFS. This indicated that IDH2 had highly sensitive and specific prognostic value. Furthermore, the T stage, N stage and adjuvant therapy were relevant to the outcomes of ESCC patients as well. These outcomes raised the possibility of IDH2 as an oncogene in ESCC.
To verify the cancer promotion effect of IDH2 in vitro, we performed CCK8, clonal efficiency, and invasion assays. The OD values, number of colonies formed and the number of invasive cells were lower in the sh-IDH2 transfected groups. These results indicated that IDH2 inhibition decreased the proliferation and invasion abilities of ESCC cells. The overexpression of IDH2 showed an opposite effect of sh-IDH2 on ESCC cells. Furthermore, the specificity of sh-IDH2-1 was verified by rescue experiment. Thus, IDH2 can act as an adverse prognostic biomarker for the postoperative outcomes of ESCC patients.
The mechanisms of regulation of IDH2 and its effects on cancer have not been elucidated. IDH2 plays an effective role in producing GSH reductase and regenerating thioredoxin (Trx) by supplying NADPH [25]. GSH and Trx are critically important antioxidative systems for protecting cells against oxidative damage and xenobiotic toxicity by scavenging the reactive oxygen species (ROS) [26,27]. Furthermore, GSH and Trx systems also promote cancer cells growth and cause immune response suppression [28]. The redox status changes the survival and proliferation of cancer cells by modifying the transcriptional and posttranscriptional status of proteins involved in cell cycle control, such as apoptosis signal-regulating kinase 1 (ASK1), c-Jun N-terminal kinase, and p38 mitogen-activated protein kinase [29,30].
In addition to the significant role of IDH2 in the antioxidative system, the effect of 2-HG on epigenetic modification is another important mechanism. Mutations in the active site’s arginine residues IDH2 R172 and IDH2 R140 have been shown to result in the conversion of NADPH and α-KG to NADP+ and 2-HG [31,32]. 2-HG inhibits differentiation and facilitates oncogenesis by promoting hypermethylation of global DNA and histone [33]. Numerous kinds of cancer cells are characterized by ectopic DNA methylation [34], and epigenetic silencing is regarded as a third pathway satisfying the hypothesis of two-hit that at least two classes of gene mutations are responsible for the silencing of tumor-suppressor genes (TSGs) [35]. Several TSGs, such as EDNRB, TIP30, TPEF and ECRG4 have been reported to be silenced through promoter hypermethylation during the development of ESCC [36-39]. Interestingly, high levels of 2-HG occur not only in cancer cells with mutant IDH but even in cancer cells with wild-type IDH in hypoxic microenvironment. In hypoxic cells, upregulated reductive metabolism of wild-type mitochondrial IDH2 could also increase the reductive carboxylation of α-KG to citrate and lead to the accumulation of 2-HG [40].
Our in vitro assays confirmed the effects of IDH2 on ESCC. By obtaining more information about IDH2, we will better understand how it affects the biological behaviors of ESCC. Further studies will be conducted to clarify in more details the mechanisms of action of IDH2 and its mutation status in ESCC patients. Additionally, ROS are important mediators of radiotherapy and chemotherapy for damaging DNA of cancer cells. Accordingly, the expression level of IDH2 may be an effective predictive marker for chemoradiotherapy, and targeting IDH2 may sensitize their effect in addition to interfering with other biological behaviors of ESCC.
Conclusion
In summary, IDH2 is significantly upregulated in cancerous tissues of patients with ESCC. IDH2 could act as a promising biomarker for identifying ESCC patients with poor outcomes.
Acknowledgements
This study was supported by the grants from the National Natural Science Foundation of China (No. 81572958).
Disclosure of conflict of interest
None.
References
- 1.Napier KJ, Scheerer M, Misra S. Esophageal cancer: a review of epidemiology, pathogenesis, staging workup and treatment modalities. World J Gastrointest Oncol. 2014;6:112–120. doi: 10.4251/wjgo.v6.i5.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Metzger R, Schneider PM, Warnecke-Eberz U, Brabender J, Holscher AH. Molecular biology of esophageal cancer. Onkologie. 2004;27:200–206. doi: 10.1159/000076913. [DOI] [PubMed] [Google Scholar]
- 4.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 5.Mandard AM, Hainaut P, Hollstein M. Genetic steps in the development of squamous cell carcinoma of the esophagus. Mutat Res. 2000;462:335–342. doi: 10.1016/s1383-5742(00)00019-3. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Leonardi R, Subramanian C, Jackowski S, Rock CO. Cancer-associated isocitrate dehydrogenase mutations inactivate NADPH-dependent reductive carboxylation. J Biol Chem. 2012;287:14615–14620. doi: 10.1074/jbc.C112.353946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skiold S, Azimzadeh O, Merl-Pham J, Naslund I, Wersall P, Lidbrink E, Tapio S, Harms-Ringdahl M, Haghdoost S. Unique proteomic signature for radiation sensitive patients; a comparative study between normo-sensitive and radiation sensitive breast cancer patients. Mutat Res. 2015;776:128–135. doi: 10.1016/j.mrfmmm.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia. 2017;31:272–281. doi: 10.1038/leu.2016.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol. 2016;27:599–608. doi: 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- 11.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O’Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM, Kwak EL, Clark JW, Ryan DP, Deshpande V, Dias-Santagata D, Ellisen LW, Zhu AX, Iafrate AJ. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O’Donnell P, Grigoriadis A, Diss T, Eskandarpour M, Presneau N, Hogendoorn PC, Futreal A, Tirabosco R, Flanagan AM. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 17.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, Spath D, Kayser S, Zucknick M, Gotze K, Horst HA, Germing U, Dohner H, Dohner K. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J. Clin. Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 18.Houillier C, Wang X, Kaloshi G, Mokhtari K, Guillevin R, Laffaire J, Paris S, Boisselier B, Idbaih A, Laigle-Donadey F, Hoang-Xuan K, Sanson M, Delattre JY. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75:1560–1566. doi: 10.1212/WNL.0b013e3181f96282. [DOI] [PubMed] [Google Scholar]
- 19.Tian GY, Zang SF, Wang L, Luo Y, Shi JP, Lou GQ. Isocitrate dehydrogenase 2 suppresses the invasion of hepatocellular carcinoma cells via matrix metalloproteinase 9. Cell Physiol Biochem. 2015;37:2405–2414. doi: 10.1159/000438593. [DOI] [PubMed] [Google Scholar]
- 20.Wu D. Isocitrate dehydrogenase 2 inhibits gastric cancer cell invasion via matrix metalloproteinase 7. Tumour Biol. 2016;37:5225–5230. doi: 10.1007/s13277-015-4358-2. [DOI] [PubMed] [Google Scholar]
- 21.Guirguis A, Elishaev E, Oh SH, Tseng GC, Zorn K, DeLoia JA. Use of gene expression profiles to stage concurrent endometrioid tumors of the endometrium and ovary. Gynecol Oncol. 2008;108:370–376. doi: 10.1016/j.ygyno.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Lv Q, Xing S, Li Z, Li J, Gong P, Xu X, Chang L, Jin X, Gao F, Li W, Zhang G, Yang J, Zhang X. Altered expression levels of IDH2 are involved in the development of colon cancer. Exp Ther Med. 2012;4:801–806. doi: 10.3892/etm.2012.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jo SH, Son MK, Koh HJ, Lee SM, Song IH, Kim YO, Lee YS, Jeong KS, Kim WB, Park JW, Song BJ, Huh TL. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J Biol Chem. 2001;276:16168–16176. doi: 10.1074/jbc.M010120200. [DOI] [PubMed] [Google Scholar]
- 26.Lee SM, Koh HJ, Park DC, Song BJ, Huh TL, Park JW. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med. 2002;32:1185–1196. doi: 10.1016/s0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Holmgren A. Thioredoxin system in cell death progression. Antioxid Redox Signal. 2012;17:1738–1747. doi: 10.1089/ars.2012.4650. [DOI] [PubMed] [Google Scholar]
- 28.Benhar M, Shytaj IL, Stamler JS, Savarino A. Dual targeting of the thioredoxin and glutathione systems in cancer and HIV. J Clin Invest. 2016;126:1630–1639. doi: 10.1172/JCI85339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sainz RM, Lombo F, Mayo JC. Radical decisions in cancer: redox control of cell growth and death. Cancers (Basel) 2012;4:442–474. doi: 10.3390/cancers4020442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueda S, Masutani H, Nakamura H, Tanaka T, Ueno M, Yodoi J. Redox control of cell death. Antioxid Redox Signal. 2002;4:405–414. doi: 10.1089/15230860260196209. [DOI] [PubMed] [Google Scholar]
- 31.Ward PS, Cross JR, Lu C, Weigert O, Abel-Wahab O, Levine RL, Weinstock DM, Sharp KA, Thompson CB. Identification of additional IDH mutations associated with oncometabolite R(-)-2-hydroxyglutarate production. Oncogene. 2012;31:2491–2498. doi: 10.1038/onc.2011.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Gilbert MR, Kyprianou N, Rangnekar VM, Horbinski C. The tumor suppressor prostate apoptosis response-4 (Par-4) is regulated by mutant IDH1 and kills glioma stem cells. Acta Neuropathol. 2014;128:723–732. doi: 10.1007/s00401-014-1334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baylin SB, Jones PA. A decade of exploring the cancer epigenome-biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 36.Zhao BJ, Sun DG, Zhang M, Tan SN, Ma X. Identification of aberrant promoter methylation of EDNRB gene in esophageal squamous cell carcinoma. Dis Esophagus. 2009;22:55–61. doi: 10.1111/j.1442-2050.2008.00848.x. [DOI] [PubMed] [Google Scholar]
- 37.Dong W, Shen R, Cheng S. Reduction of TIP30 in esophageal squamous cell carcinoma cells involves promoter methylation and microRNA-10b. Biochem Biophys Res Commun. 2014;453:772–777. doi: 10.1016/j.bbrc.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 38.Zhao BJ, Tan SN, Cui Y, Sun DG, Ma X. Aberrant promoter methylation of the TPEF gene in esophageal squamous cell carcinoma. Dis Esophagus. 2008;21:582–588. doi: 10.1111/j.1442-2050.2007.00808.x. [DOI] [PubMed] [Google Scholar]
- 39.Li LW, Yu XY, Yang Y, Zhang CP, Guo LP, Lu SH. Expression of esophageal cancer related gene 4 (ECRG4), a novel tumor suppressor gene, in esophageal cancer and its inhibitory effect on the tumor growth in vitro and in vivo. Int J Cancer. 2009;125:1505–1513. doi: 10.1002/ijc.24513. [DOI] [PubMed] [Google Scholar]
- 40.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]


