Abstract
Studies of Hepatitis C virus (HCV) RNA replication have become possible with the development of subgenomic replicons. This system allows the functional analysis of the essential components of the viral replication complex, which so far are poorly defined. In the present study we wanted to investigate whether lethal mutations in HCV nonstructural genes can be rescued by trans-complementation. Therefore, a series of replicon RNAs carrying mutations in NS3, NS4B, NS5A, and NS5B that abolish replication were transfected into Huh-7 hepatoma cells harboring autonomously replicating helper RNAs. Similar to data described for the Bovine viral diarrhea virus (C. W. Grassmann, O. Isken, N. Tautz, and S. E. Behrens, J. Virol. 75:7791-7802, 2001), we found that only NS5A mutants could be efficiently rescued. There was no evidence for RNA recombination between helper and mutant RNAs, and we did not observe reversions in the transfected mutants. Furthermore, we established a transient complementation assay based on the cotransfection of helper and mutant RNAs. Using this assay, we extended our results and demonstrated that (i) inactivating NS5A mutations affecting the amino-terminal amphipathic helix cannot be complemented in trans; (ii) replication of the helper RNA is not necessary to allow efficient trans-complementation; and (iii) the minimal sequence required for trans-complementation of lethal NS5A mutations is NS3 to -5A, whereas NS5A expressed alone does not restore RNA replication. In summary, our results provide the first insight into the functional organization of the HCV replication complex.
Hepatitis C virus (HCV) is an enveloped RNA virus that constitutes the genus Hepacivirus within the Flaviviridae family (53). A common feature of this virus group is a single-stranded, linear RNA genome of positive polarity. In the case of HCV the genome has a length of ∼9,600 nucleotides (nt), and it carries at the 5′ and 3′ ends short highly structured nontranslated regions (NTRs) that are important for RNA translation and replication (reviewed in reference 2). Located within the 5′ NTR is an internal ribosome entry site (IRES) directing translation of an approximately 3,000-amino-acid polyprotein that is co- and posttranslationally cleaved by cellular and viral proteinases into the following 10 products (listed from the N to the C terminus): core, envelope protein 1 (E1), E2, p7, nonstructural protein 2 (NS2), NS3, NS4A, NS4B, NS5A, and NS5B. The region from core to NS2 is processed by cellular signal peptidases and a signal peptide peptidase, whereas the proteins from NS3 to NS5B are generated by the NS2-3 and the NS3/4A viral proteinases. Defined functions have been ascribed to most of the nonstructural proteins. NS3 carries in the N-terminal domain a serine-type proteinase, whereas the C-terminal region possesses helicase and nucleoside triphosphatase activities (21, 24, 32). NS4A acts as a cofactor for the NS3 serine proteinase, and NS5B functions as RNA-dependent RNA polymerase (RdRp) (3, 5, 14, 40). The very hydrophobic NS4B protein appears to have no intrinsic enzymatic activity. However, it induces the formation of intracellular membranous vesicles, building up the so-called membranous web, which most likely is the site of HCV RNA replication (12, 20, 47). It is assumed that in analogy to other positive-strand RNA viruses, within the membranous web a multiprotein complex exists that contains, in addition to most if not all viral proteins, one or several host cell factors.
NS5A, which is presumed to be part of this replication complex (RC), is a highly phosphorylated polypeptide that associates with membranes via an N-terminal amphipathic α-helix (7, 13). Interestingly, a large number of cell culture-adaptive mutations that enhance RNA replication localize to the center of this molecule (6, 34). Several of them affect serine residues that are potential sites for phosphorylation by an as-yet-unknown cellular kinase(s) (28, 49). While single substitutions affecting these serine residues (S2197, S2202, and S2204) increase replication of HCV replicons to various extents, the combination of these mutations reduces or completely blocks RNA replication (38). It is not clear how these mutations enhance RNA replication, but their incompatibility suggests that they act via the same mechanism (38).
Several cellular proteins were found to interact with NS5A in vitro and in transfected cells (reviewed in reference 50). Some of these interactions, such as binding to and inhibition of the interferon-induced double-strand RNA-dependent protein kinase (PKR), are thought to play a role in the evasion by HCV of the interferon host defense system (16). In addition, NS5A was found to interact with growth factor receptor-bound protein 2 (Grb2) adaptor protein (48), karyopherin β3 (9), transcription factor SCRAP (18), p53 (42), cyclin-dependent kinase 1 (1), and tumor necrosis factor receptor-associated factor 2 (44). These interactions may perturb kinase signaling cascades or cause alterations in cell growth and cell signaling and thereby contribute to pathogenesis. More recently, an interaction of NS5A with two cellular proteins involved in intracellular vesicle transport, amphiphysin II (54) and human vesicle-associated membrane-protein associated protein A (hVAP-33, renamed hVAP-A) (52), was found. While interaction with the first seems to be dispensable for RNA replication, expression of a dominant negative hVAP-A mutant as well as an RNA interference-mediated knockdown of hVAP-A expression reduces RNA replication (17). This result suggests that the interaction of NS5A with this cellular protein is of functional importance and underscores the essential role of NS5A in HCV RNA replication. However, the exact mode of action is not known.
Virtually all positive-strand RNA viruses replicate their genomes on virus-induced vesicular membrane compartments. These membranous complexes are rather enclosed structures with a limited possibility for exchange of viral RNA and proteins by trans-complementation. For instance, in the case of Poliovirus (PV), only a very inefficient complementation of mutations in the proteins 2C and 3D (RdRp) was found (51). In the case of Kunjin virus (KV), efficient trans-complementation was reported for mutations affecting NS1, the NS3 helicase, or the NS5 RdRp (29-31, 37). For the Bovine viral diarrhea virus (BVDV), Grassmann and coworkers demonstrated that NS5A was the only nonstructural protein that could be complemented in trans (22).
With the aim of gaining insights into the functional organization of the HCV RC and the relationship between its individual components, in this study we established two alternative trans-complementation assays. The first one is based on the supertransfection of replicon cell clones with defective selectable subgenomic HCV RNAs, and the second approach is based on the cotransfection of defective and helper replicons into naive Huh-7 cells. Using these assays, we demonstrate that certain mutations in NS5A, but not in NS3, NS4B, or NS5B, can be complemented in trans. Coexpression of a minimal sequence from NS3 to NS5A is sufficient to rescue replication of mutants, whereas expression of NS5A alone does not restore functionality of defective replicon RNAs. Based on these results, we propose three alternative models for trans-complementation by HCV NS5A.
MATERIALS AND METHODS
Cell cultures and viruses.
Cell monolayers of the human hepatoma cell line Huh-7 were grown in Dulbecco's modified minimal essential medium (DMEM) (Life Technologies, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, penicillin at 100 U/ml, streptomycin at 100 μg/ml, and 10% fetal calf serum. G418 (Geneticin; Life Technologies) was added at a final concentration of 250 μg/ml to cell lines carrying HCV neomycin replicons. For generation of cell clones carrying subgenomic hygromycin replicons, 10 μg of in vitro-transcribed replicon RNAs hyg-ET and hyg-5.1 (Fig. 1A) was transfected into naive Huh-7 cells as described below. After selection for several weeks with hygromycin B at 50 μg/ml, single-cell colonies were isolated and expanded to obtain cell clones Huh-7-hyg/ET and Huh-7-hyg/5.1, respectively. Transient-replication assays were performed with a highly permissive Huh-7 cell clone that was generated as described recently (15).
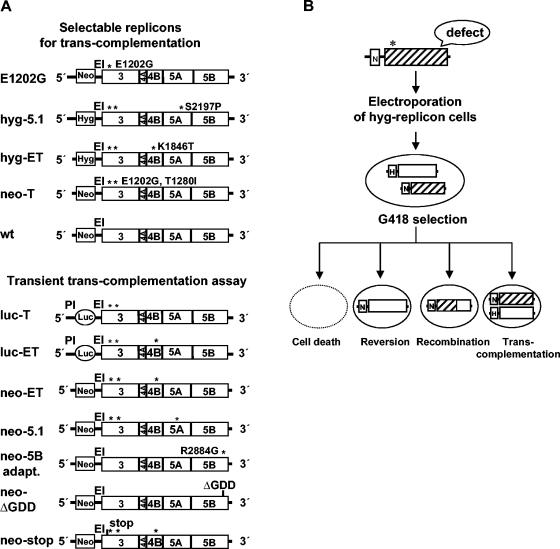
FIG. 1.
(A) Structures of replicon RNAs used for trans-complementation assays. Adaptive mutations localized in NS3 (E1202G and T1280I), NS4B (K1846T), NS5A (S2197P), and NS5B (R2884G) are indicated by asterisks. Numbers refer to amino acid positions of the HCV Con-1 complete polyprotein. In the case of the selectable replicons (upper panel), expression of the first cistron is mediated by the HCV IRES, whereas translation of the nonstructural proteins NS3 to -5B is directed by the EMCV IRES (EI). For the luciferase (luc) replicons, translation of this reporter is mediated by the PV IRES (PI), which was fused to the 3′ end of the HCV IRES. Translation of the HCV replicase is again mediated by the EMCV IRES. (B) Schematic drawing of the experimental approach used to analyze trans-complementation. Mutations were introduced into a subgenomic neo replicon carrying one weak adaptive mutation in NS3 (E1202G, indicated by an asterisk). Huh-7 cells containing hyg helper replicons were transfected with mutated RNAs and subjected to selection with G418. Possible outcomes are depicted at the bottom. G418-resistant colonies may arise in case of reversion, recombination, or trans-complementation. Neo or N, neomycin phosphotransferase; Hyg or H, hygromycin phosphotransferase; Luc, P. vulgaris luciferase gene; ΔGDD, deletion of the NS5B GDD motif.
Plasmid constructions.
All nucleotide and amino acid numbers refer to the Con-1 HCV genome (EMBL accession number AJ238799). Subgenomic replicons carrying the hygromycin phosphotransferase (hyg) gene were generated by replacing the selectable marker neo with the hyg gene in the subgenomic replicon pFK-rep-neo/5.1. For this purpose, the hyg gene was amplified from the plasmid pTKhyg by overlap PCR with primers that add AscI and PmeI restriction sites at the 5′ and 3′ ends of the amplicons, respectively, and that remove by silent mutation a ScaI site near the 3′ end of the gene. Upon restriction with AscI and PmeI, the fragment was inserted into pFK-rep-neo/5.1. Replicon hyg-ET was generated by transferring the SfiI fragment from replicon pFK-rep-luc-PI/ET into replicon hyg-5.1. The basic construct for selection assays, pFK-rep-neo/E1202G carrying one weak adaptive mutation in NS3 (Fig. 1A), has been described elsewhere (34). The plasmids carrying inactivating mutations in NS5A (Δ1977-1983, ins. 1980Ala, ins. 1983Ala, Δ2194-2207, Δ2200-2202, S2194A+S2197A+S2201A+S2204A, S2197A+S2201A, S2197A+S2204A, S2201A+S2204A, S2194A+S2201A+S2204A, and S2194A+S2200A+S2201A+S2202A) were generated by site-directed mutagenesis (25) with oligonucleotide primers carrying all necessary nucleotide alterations. EcoRI-XhoI (nt 6699 to 7186)- or HpaI-EcoRI (nt 5982 to 6699)-restricted PCR fragments were inserted into the parental replicon pFK-rep-neo/E1202G. For transient-replication assays the same PCR fragments were inserted into reporter replicons pFK-rep-luc-PI/ET and pFK-rep-luc-PI/E1202G+T1280I (38). These constructs contain the complete HCV 5′ NTR, a random 63-bp spacer element, the PV IRES, the luciferase gene of the firefly (Photinus vulgaris), the encephalomyocarditis virus (EMCV) IRES, the HCV nonstructural proteins NS3 to NS5B, and the HCV 3′ NTR (Fig. 1A). Plasmids harboring inactivating mutations or deletions in NS3 (A1027D and P1028H) or NS4B (V1897D, V1897K, V1897P, Δ1782-1821, Δ1822-1882, and Δ1905-1933) were generated by PCR-based mutagenesis and standard recombinant DNA technologies (46).
Plasmids pFK-rep-neo/wt, pFK-rep-neo/E1202G+T1280I, pFK-rep-neo/ΔGDD, pFK-rep-neo/5Badapt, pFK-rep-neo-stop, and pFK-rep-PI-luc/GND have been described recently (34, 38). The expression vector pTM was used to generate in vitro transcripts of NS3 to the 3′ NTR, NS3 to NS5A, and NS5A under translational control of the EMCV IRES. pTM/NS3-3′ plasmid variants containing different adaptive mutations (T, E1202G+T1280I; ET, E1202G+T1280I+K1846T; JT, and E1202G+T1280I+ΔS2202) were generated by transferring SfiI fragments (nt 3622 to 8499) from subgenomic adapted replicons into pTM/NS3-3′/wt (33).
Sequence analysis.
Nucleotide sequences of the final replicon constructs were confirmed by automated nucleotide sequencing with an ABI 310 sequencer (Applied Biosystems). Big Dye version 2.0 (Applied Biosystems) was used for cycle sequencing according to the instructions of the manufacturer.
In vitro transcription.
In vitro transcripts were generated by using the protocol described recently (41). In brief, plasmid DNA was restricted with AseI and ScaI in the case of replicon constructs or with XbaI and SpeI in the case of the pTM vectors. After extraction with phenol and chloroform, linearized plasmid DNA was precipitated with ethanol and dissolved in RNase-free water. In vitro transcription reaction mixtures contained 80 mM HEPES (pH 7.5), 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), 3.125 mM (each) nucleoside triphosphate, 1 U of RNasin (Promega, Mannheim, Germany) per μl, 0.1 μg of restricted plasmid DNA per μl, and 0.6 U of T7 RNA polymerase (Promega) per μl. After 2 h at 37 C, an additional 0.3 U of T7 RNA polymerase per μl was added, and the reaction mixture was incubated for another 2 h. Transcription was terminated by the addition of 1.2 U of RNase-free DNase (Promega) per μg of plasmid DNA and 30 min of incubation at 37°C. After one extraction with acidic phenol and chloroform, RNA was precipitated with isopropanol and dissolved in RNase-free water. The concentration was determined by measurement of the optical density at 260 nm, and RNA integrity was checked by denaturing agarose gel electrophoresis.
Electroporation and trans-complementation by coselection of replicons.
The conditions for electroporation have been described in detail elsewhere (34, 39). In pilot experiments, we found that the number of G418-resistant colonies obtained after transfection of naive Huh-7 and Huh-7-hyg cells depends on the amount of RNA transfected into the cells, the G418 concentration, and the duration of selection. Optimal results with respect to lowest colony formation of the mutants after transfection into naive cells and highest colony numbers when using Huh-7-hyg cells carrying the helper replicon were obtained with 250 μg of G418 per ml and 1 μg of mutant replicon RNA. Under these conditions, it took ∼6 months to establish a cell clone from one individual colony. Briefly, 1 μg of in vitro transcripts adjusted with total RNA from naive Huh-7 cells to a final amount of 10 μg was mixed with 400 μl of a suspension of 107 Huh-7 cells per ml. Electroporation conditions were 960 μF and 270 V with a Gene Pulser II system (Bio-Rad, Munich, Germany) and a cuvette with a gap width of 0.4 cm (Bio-Rad). Cells were immediately transferred to 7 ml of complete DMEM and seeded into a 10-cm-diameter cell culture dish. After 24 h, the medium was replaced by complete DMEM supplemented with 250 μg of G418 per ml. The medium was changed weekly, and at 5 to 8 weeks after electroporation, colonies were stained with Coomassie brilliant blue (0.6 g/liter in 50% methanol-10% acetic acid). For each replicon, about five independent transfections were performed.
Transient trans-complementation assay.
Huh-7 cells were cotransfected by electroporation as described above with 5 μg of a neo replicon as helper RNA and 1 μg of a replication-deficient luciferase reporter replicon. After addition of 12 ml of complete DMEM, 1-ml aliquots of the cell suspension were seeded into a 10-cm2 culture dish and harvested at the time points given in Results. For determination of luciferase activity, cells were washed three times with phosphate-buffered saline and scraped off the plate into 350 μl of ice-cold lysis buffer (1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, and 1 mM DTT). One hundred microliters of lysate was mixed with 360 μl of assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP, and 15 mM K2PO4 [pH 7.8]) and, after addition of 200 μl of a 200 μM luciferin stock solution, measured in a luminometer (Lumat LB9507; Berthold, Freiburg, Germany) for 20 s. Values obtained with cells harvested 4 h after electroporation were used to determine the transfection efficiency.
Amplification of RNA by RT-PCR and cloning of amplified DNA fragments.
One microgram of total RNA and 50 pmol of primer A9413 (CAG GAT GGC CTA TTG GCC TGG AG) were mixed in a total volume of 10.5 μl and denatured for 10 min at 65°C. Reverse transcription was performed with Expand-RT (Roche Biochemicals, Mannheim, Germany) in a total volume of 20 μl, and after 1 h at 42°C, different amounts of the reaction mixture were used for PCR with the Expand Long Template PCR system (Roche Biochemicals) and primers Sneo (CAA CGG GCG CGC CAT GAT TGA ACA A) and A9386Spe (CGT ACT AGT TTA GCT CCC CGT TCA TCG GTT GG). Cycle conditions were 2 min of initial denaturation at 94°C and 30 cycles of 10 s at 94°C, 90 s at 54°C, and 480 s at 68°C. After 10 cycles, the extension time was increased 10 s for each additional cycle. After a final 10-min incubation period at 68°C, PCR products were purified by preparative agarose gel electrophoresis, restricted with AscI and SpeI or SfiI, and inserted into the parental construct pFK-I389neo/NS3-3′/wt (41). To control for the specificity of the reverse transcriptase PCR (RT-PCR), 1 μg of total RNA from naive Huh-7 cells was mixed with 107 molecules of hyg or neo replicon in vitro transcript and RT-PCR was performed as described above. To control for PCR-mediated recombination, we mixed 107 molecules of hyg replicon with 107 molecules of a neo replicon lacking the sequence downstream of NS5A. Only in the case of the neo replicon alone did we obtain a PCR product with the correct size. All reactions performed in the absence of RT or with hyg or a shortened neo replicon as the template were negative (data not shown). Moreover, by using extension times of several minutes, we reduced the chance for PCR-mediated recombination.
Preparation of total RNA and Northern blot analysis.
Total RNA was prepared by a single-step isolation method (8), denatured by treatment with 5.9% glyoxal in a solution containing 50% dimethyl sulfoxide and 10 mM sodium phosphate buffer (pH 7.0), and analyzed after denaturing agarose gel electrophoresis by Northern blotting. Prior to hybridization, the membrane was stained with methylene blue and cut ∼0.5 cm below the 28S rRNA band. The upper strip containing the HCV replicon RNA was hybridized with a 32P-labeled negative-sense riboprobe complementary to the neo gene, the hyg gene, or the NS5A sequence. The lower strip, which was hybridized with a β-actin-specific antisense riboprobe, was used to correct for total RNA amounts loaded onto each lane of the gel (not shown).
RESULTS
Establishment of a trans-complementation assay by using selectable replicons.
In order to study which components of the HCV RC are exclusively cis active or can act in trans, we developed a trans-complementation assay that utilizes the transfection of selectable replication-deficient HCV replicons encoding the neomycin phosphotransferase (neo replicons) into Huh-7 cells that harbor replicons expressing the hygromycin phosphotransferase (hyg replicons) (Fig. 1B). Since the efficiency of trans-complementation can be affected both by the level of HCV proteins expressed from the helper RNA and by the compatibility of certain cell culture-adaptive mutations (see below), we used two different helper cell clones: Huh-7-hyg/ET, which harbors the hyg-ET-replicon, and Huh-7-hyg/5.1, which carries the hyg-5.1-replicon (Fig. 1A). These replicons have two adaptive mutations in NS3 in common but differ with respect to the highly adaptive mutation that resides in NS4B or NS5A (hyg-ET and hyg-5.1, respectively). To increase the efficiency of trans-complementation, transfected cells were cultured in the presence of only G418. Under these conditions, cells carrying only the hyg helper replicon are eliminated within about 1 week. The efficiency of trans-complementation was determined by the increase of G418-resistant cell clones obtained after transfection of the Huh-7-hyg cell clones in comparison to naive Huh-7 cells that lack the hyg helper RNA.
Three different mechanisms could account for the G418 resistance of a cell clone: (i) reversion or compensatory mutations in the defective neo replicon RNA, (ii) recombination between mutant and helper RNA, or (iii) trans-complementation (Fig. 1B). To differentiate between these possibilities, RNA present in G418-resistant cell clones was analyzed by Northern hybridization with neo- or hyg-specific probes and by sequence analysis of the mutated neo replicon RNA after amplification by RT-PCR and subcloning.
In the first set of experiments we compared the permissiveness of naive Huh-7 cells and the cells carrying the two different helper RNAs. This comparison was necessary because host cell permissiveness has a strong influence on the efficiency of RNA replication, and therefore, differences in the number of G418-resistant colonies obtained with different Huh-7 cells could merely reflect different levels of permissiveness rather than trans-complementation. To this end, we transfected a wild-type (wt) replicon or a replicon that carried two adaptive mutations in NS3 (E1202G and T1280I), enhancing RNA replication about two- to fivefold (replicon neo-T) (Fig. 1A). After G418 selection for 4 to 5 weeks, cells were fixed onto the plates and stained with Coomassie blue solution. As shown in Fig. 2, there was no difference in the number of G418-resistant colonies between naive Huh-7 cells and cells that carry the hyg helper RNAs, demonstrating the comparable permissiveness of the different Huh-7 cell cultures used for the trans-complementation assay.
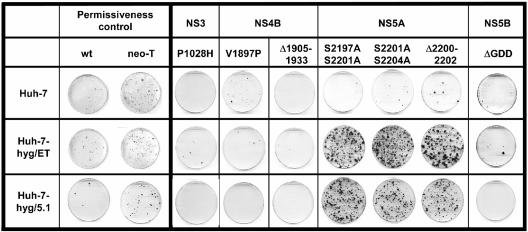
FIG. 2.
Representative results of trans-complementation experiments using selectable replicon mutants. Hygromycin replicon cells (Huh-7-hyg/ET and Huh-7-hyg/5.1) and naive Huh-7 cells were transfected with the neo replicons specified at the top. After G418 selection for 6 weeks, cells were fixed and colonies were stained with Coomassie blue solution.
Mutations in HCV NS5A can be complemented in trans.
To analyze whether HCV nonstructural proteins could be complemented in trans, we generated a panel of neo replicons that carried the adaptive mutation E1202G in NS3 and additional inactivating mutations in NS3, NS4B, NS5A, or NS5B that abolished RNA replication. The NS3 mutation served as a genetic marker of this RNA and would support RNA replication synergistically in the case of trans-complementation. For the inactivating NS3 mutation P1028H, we have recently shown that it has no influence on proteinase activity and therefore does not interfere with polyprotein processing (33). The exact reasons for the replication defects caused by the various mutations that we introduced into NS4B and NS5A are not known because thus far the exact functions of these proteins have not been firmly established. However, we analyzed all NS4B and NS5A mutants for their influence on polyprotein processing and found that in no case was it affected (reference 33 and data not shown). It is interesting that the amino acid substitutions S2197A, S2201A, and S2204A increased RNA replication when introduced individually into a replicon, whereas the combination of two or more of these mutations completely abolished RNA replication (N. Appel and R. Bartenschlager, unpublished results). In the case of NS5B, we inactivated the RdRp activity by deletion of 10 amino acids spanning the active-site GDD motif of the polymerase. Equal amounts of in vitro transcripts generated from these mutated template DNAs were introduced by electroporation into naive Huh-7 and the Huh-7-hyg cells. After 4 to 5 weeks of cultivation in medium containing only G418, cells were fixed, stained with Coomassie blue, and counted. In the case of mutations affecting NS3, NS4B, or NS5B, we found no difference in the number of G418-resistant colonies between naive Huh-7 cells and the two Huh-7-hyg cell clones (Fig. 2). The fact that a very low number of colonies were obtained after transfection of the NS5B mutant, which by definition is unable to replicate, argues that these colonies are most likely due to integration of a DNA fragment that carries the neo gene. Therefore, we considered this low number of G418-resistant colonies to be background. Furthermore, several repetitions of this experiment resulted in no colonies upon transfection of this NS5B mutant and G418 selection of transfected cells. In contrast to these results, about a 50- to 100-fold increase in the number of G418-resistant colonies was obtained upon transfection of Huh-7-hyg cells with RNAs that carried mutations in NS5A. This result suggested that only mutations in NS5A can be complemented in trans.
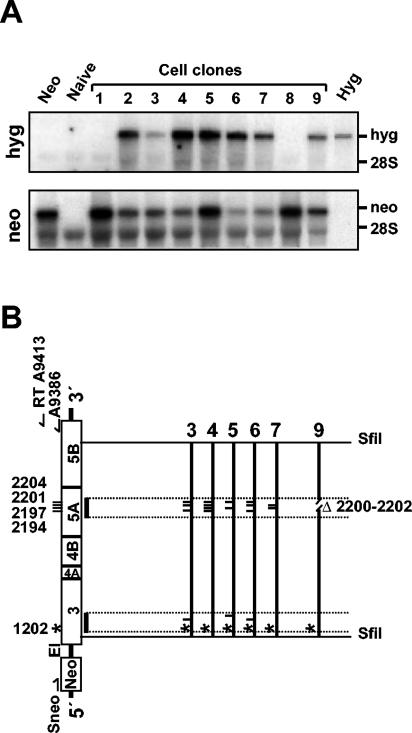
To further support this assumption, single-cell clones obtained after transfection of Huh-7-hyg cells with NS5A mutants were established, and replicon RNAs present in these cells were analyzed by Northern blot hybridization. To discriminate between the mutated and the helper RNAs, positive-strand-specific riboprobes that hybridized to the neo or the hyg gene, respectively, were used. Figure 3A shows the results of an analysis of nine independently generated cell clones. Except for two clones, both types of RNA were consistently detected in the cells. In total, we established 28 independent cell clones, of which 25 contained both RNA species, whereas for 3 cell clones we only detected the neo RNA. This argues that the hyg replicon indeed complemented the NS5A mutant, which in turn conferred G418 resistance to the cells. Alternatively, the mutated RNAs might have acquired reversions or second-site compensatory mutations or lost the inactivating mutations due to recombination with the helper RNA. In these cases, the neo replicon would be capable of self-replication. Although this possibility was not likely due to the rapid reduction of the hyg replicon in the absence of selective pressure (the cells were cultured in the presence of G418 only), we analyzed the sequences of the neo replicons in more detail. A neo-specific RT-PCR was performed to amplify almost the complete replicon sequence (see Materials and Methods) (Fig. 3B). After subcloning of an SfiI DNA fragment that spans most of the HCV coding region, at least two independent clones of each fragment were sequenced in the regions carrying the introduced mutations. We found that in all sequenced cell clones harboring both replicon RNA species, the inactivating mutations were still present in NS5A (Fig. 3B and Table 1). In addition, all of the replicons had preserved the adaptive mutation in NS3 at amino acid position 1202. Cell clones 3, 5, and 6 had one additional conserved mutation in NS3 (Table 1), whereas cell clones 4, 7, and 9 had no further mutations in this region. Sequence analysis of the complete SfiI fragment derived from cell clone 4 revealed that no further mutations were present in the HCV coding region, excluding the possibility that the NS5A mutations were compensated by amino acid substitutions elsewhere in the polyprotein. Moreover, we had no evidence for RNA recombination. This statement is based on the fact that the hyg helper replicon differed from the neo mutant RNA that was originally transfected into Huh-7 cells at 12 nucleotide positions. These were scattered throughout the coding sequence for NS3 to NS5A, with seven of them resulting in amino acid substitutions and the remaining ones being silent (34). The fact that none of these nucleotide substitutions was present in the neo mutant RNA amplified from cell clone 4 (and the other ones) clearly shows that our RT-PCR was specific for only this RNA and that RNA recombination did not occur in cells harboring both the mutant and the helper RNAs.
FIG. 3.
(A) Analysis of HCV RNA species in G418-resistant cell clones by Northern hybridization. Huh-7 cells carrying stably replicating hyg helper RNAs (hyg-ET or hyg-5.1) were transfected with neo replicon mutants carrying lethal mutations in NS5A. After several months of G418 selection, single-cell clones (clones 1 to 9) were established and analyzed for HCV RNAs by using neo- or hyg-specific (negative-sense) riboprobes. As controls for specificity, neo and hyg replicon transcripts (108 molecules) as well as total RNA of naive Huh-7 cells were analyzed in parallel. (B) Conserved mutations in neo replicons isolated from cell clones 3 to 7 and 9. After RT-PCR, two independent DNA clones were sequenced for each given cell clone. Conserved mutations are indicated by a asterisk in the case of the adaptive NS3 mutation and by vertical lines. The minimum region examined by sequence analysis is indicated by dotted lines. In the case of cell clone 4, the entire SfiI fragment was sequenced, and no further mutation was detected.
TABLE 1.
Sequence analysis of mutated neo replicon RNAs recovered from G418-resistant cell clones
| Cell clone | Input RNA | Rescued RNA |
|---|---|---|
| 3 | E1202G, S2194A, S2201A, S2204A | E1202G, G1388E, S2194A, S2201A, S2204A |
| 4 | E1202G, S2194A, S2197A, S2201A, S2204A | E1202G, S2194A, S2197A, S2201A, S2204A |
| 5 | E1202G, S2197A, S2204A | E1202G, S1408P, S2197A, S2204A |
| 6 | E1202G, S2194A, S2201A, S2204A | E1202G, G1388E, S2194A S2201A, S2204A |
| 7 | E1202G, S2197A, S2201A | E1202G, S2197A, S2201A |
| 8 | E1202G, Δ2194-2197 | E1202G, Δ2194-2197, I2430V |
| 9 | E1202G, Δ2200-2202 | E1202G, Δ2200-2202 |
In the case of cell clones harboring only the neo replicons (e.g., cell clones 1 and 8), the inactivating mutations in NS5A were also still present. However, further analysis of cell clone 8 revealed an additional conserved mutation (Table 1) that may have compensated for the defects in NS5A. Alternatively, replication of these neo mutant RNAs may be supported in a very minor subpopulation of Huh-7 cells. Irrespective of these possibilities, the absence of such mutations in at least one cell clone carrying both mutant and helper replicon RNAs strongly suggests that NS5A was complemented in trans. The fact that replication of the RNAs with inactivating mutations in the other nonstructural proteins could not be rescued indicated that only mutations in NS5A can be trans-complemented. This result implies that individual NS5A proteins rather than complete replicase protein complexes or RNA templates may become exchanged between helper and mutant RCs. Alternatively, complementing NS5A may provide a function outside the replicase complex and thereby indirectly rescue replication of the NS5A mutants.
Establishment of a transient trans-complementation assay.
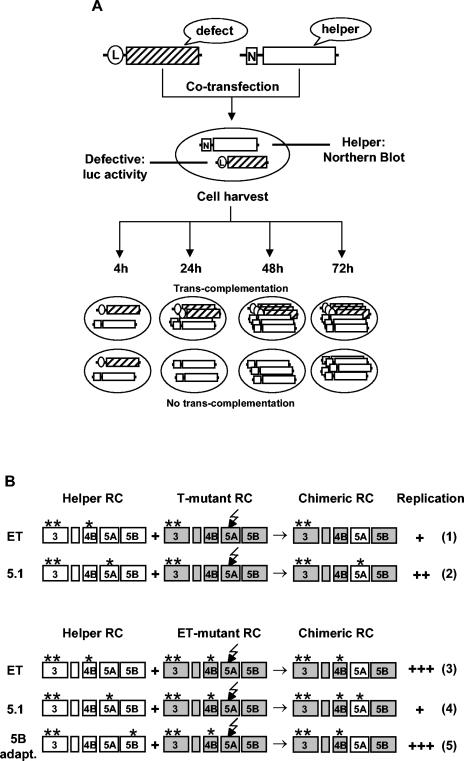
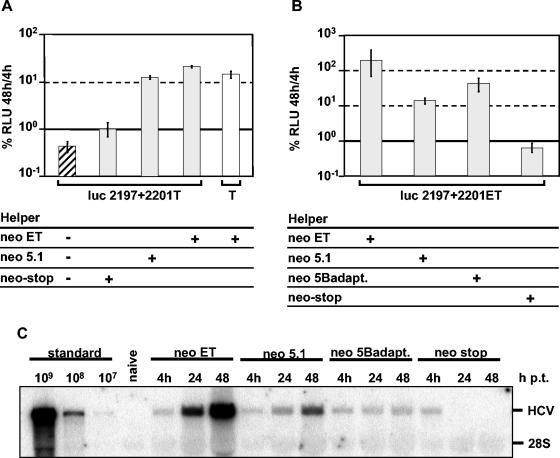
The results described thus far are based on trans-complementation of selectable replicon RNAs after supertransfection of helper replicon cells. Apart from the requirement to perform cell selection for several months, this approach has the risk of enriching additional mutations in the nonfunctional neo replicon during selection. These mutations may have a compensatory effect for the inactivating mutation and therefore circumvent trans-complementation by the helper RNA. To overcome this limitation and to screen a broader range of lethal mutations for rescue of RNA replication within a shorter time, we established a transient-complementation assay that was based on the cotransfection of mutant and helper RNAs. While the mutated replicon carried the firefly luciferase reporter gene, the replication-competent helper RNA carried the neo gene to allow discrimination between the two RNA species (Fig. 4). After cotransfection into a highly permissive naive Huh-7 cell clone (see Materials and Methods), replication of the mutant was determined at 4, 24, 48, and 72 h posttransfection by measurement of luciferase activity. These cells were chosen since they allow higher replication efficiencies in comparison to even highly permissive naive Huh-7 cell passages (38). In parallel, replication of the helper neo RNA was determined by Northern blot hybridization. In an attempt to achieve most efficient restoration of RNA replication, we used two different helper RNAs designated neo-ET and neo-5.1 (Fig. 1A). These replicons carry the same adaptive mutations in NS3 (E1202G and T1280I) but differ with respect to the additional highly adaptive mutation, which is K1846T in the case of neo-ET and S2197P in the case of neo-5.1. The inactivating NS5A mutations were introduced into the luc-T replicon, which harbors the same two adaptive mutations in NS3 (Fig. 1A). Based on our previous results indicating that only NS5A may be exchanged between different RCs, various combinations of cell culture-adaptive mutations were possible in trans-complemented RCs, depending on which mutant and helper RNAs were combined (Fig. 4B). In the first set of experiments we analyzed replication rescue of a luc replicon carrying the NS5A double mutation S2197A+S2201A by neo-5.1 and neo-ET helper RNAs. To exclude nonspecific effects of these helper RNAs, we used as an additional negative control a neo replicon that carried a stop codon at the beginning of the NS3-coding region. This RNA (designated neo-stop) did not give rise to HCV proteins but otherwise was of the same structure as the other helper RNAs (Fig. 1A). A representative result of such a transient-replication assay is shown in Fig. 5A. Forty-eight hours after transfection, the luciferase activity observed with the luc-2197 + 2201-T mutant was below the detection limit. Cotransfection with the defective helper RNA neo-stop had no effect on the replication ability of the mutant. In contrast, cotransfection with either replication-competent helper RNA (neo-ET or neo-5.1) rescued the mutant to a level that was comparable to that for the parental replicon luc-T. Analysis of the neo helper replicons by Northern blot hybridization with a neo-specific probe revealed a much higher level of RNA replication in the case of neo-ET than in the case of the neo-5.1 replicon (Fig. 5C). Furthermore, using Western blot analysis, we found that the neo-ET helper RNA produces the highest levels of HCV proteins, arguing that one should expect a very high replication level of the mutant after cotransfection with this helper (data not shown). However, the neo-ET helper provides a wild-type NS5A protein in trans, and therefore, cotransfection with the luc-2197 + 2201-T mutant may lead to an RC that carries only a moderately adaptive mutation in NS3 (Fig. 4B, row 1). The neo-5.1 helper RNA produces an NS5A protein that carries a highly adaptive mutation in NS5A. Consequently, cotransfection with the luc-T mutant may restore a much more adapted RC (Fig. 4B, row 2), and therefore smaller amounts of NS5A are sufficient to rescue RNA replication of the mutant to the level obtained with the highly replicating neo-ET helper replicon (Fig. 5A and C). In agreement with this assumption, we found that increasing the amount of cotransfected helper RNA from 62 ng to 5 μg led to an increase of mutant RNA replication (data not shown). Based on these results, the highest level of rescue was expected for a combination of an efficiently replicating helper RNA (neo-ET) and an NS5A mutant that carried the same adaptive mutations (ET) (Fig. 4B, row 3). In contrast, the combination of the neo-5.1 helper RNA with the same mutant should lead to a lower degree of RNA replication rescue, because RCs in which NS4B and NS5A each carry a highly adaptive mutation may be generated (Fig. 4B, row 4). However, combination of highly adaptive mutations in NS4B, NS5A, or NS5B strongly reduces RNA replication (38, 39).
FIG. 4.
Transient trans-complementation assay. (A) A replication-deficient luciferase replicon (hatched box) was cotransfected with a neo helper RNA (open box) into cells of a highly permissive Huh-7 cell passage. Equal numbers of cells were seeded and harvested at given time points posttransfection. Replication of the mutant was monitored by luciferase assays, whereas replication of the helper RNA was controlled by Northern hybridization. (B) Possible outcomes of trans-complementation after cotransfection of various helper and mutant replicon RNAs. The jagged arrows indicate inactivating mutations in NS5A. Asterisks indicate the positions of adaptive mutations in NS3, NS4A, NS5A, or NS5B. Outcomes of trans-complementation based on the exchange of the NS5A protein and expected replication efficiencies are shown in the right. Numbers in parentheses indicate rows referred to in the text. +, poor replication; ++, moderate replication; +++ high replication.
FIG. 5.
trans-complementation of the replicon mutant luc-2197 + 2201 by different helper RNAs. (A) The inactivating mutations S2197A+S2201A were introduced into the luciferase replicon luc-T (Fig. 1A) carrying two weak adaptive mutations in NS3. After cotransfection with given helper RNAs, cells were harvested 4, 24, 48 and 72 h later and luciferase activities were determined. Values are the ratios of the values at 48 and 4 h, reflecting transfection efficiency. For comparison, the mutant was transfected without helper RNA (hatched box). Luciferase activities measured upon transfection of the parental replicon luc-T served as a positive control (open box). (B) trans-complementation of mutant luc-2197 + 2201-ET with given helper RNAs as described for panel A. Data are means and standard deviations from two independent experiments, each with duplicate measurements. (C) Analysis of the helper RNAs specified in panel B by Northern hybridization. Twenty micrograms of total RNA was analyzed with a neo-specific riboprobe. Total RNA of naive Huh-7 cells served as a negative control. Known amounts of replicon in vitro transcripts were used as positive controls. p.t., posttransfection.
To further substantiate this assumption, we performed an experiment in which we cotransfected the mutant luc-2197 + 2201-ET with helper replicons neo-ET, neo-5.1, and neo-5Badapt. As shown in Fig. 5B, the most efficient rescue was obtained with the neo-ET helper RNA, whereas an about 20-fold-lower rescue of the same mutant was found upon cotransfection of the neo-5.1 helper RNA. The fact that this helper did not support a higher rescue can be explained by the incompatibility of the highly adaptive mutations in NS4B (present in the mutant) with those in NS5A (present in the helper) (Fig. 4B) or by lower replication of the neo-5.1 helper RNA. To differentiate between these two possibilities, we cotransfected the same mutant with the neo-5Badapt. helper RNA. Although the replication efficiency of this helper RNA was lower than that of neo-5.1 (Fig. 5C), rescue of RNA replication was fivefold more efficient than that with neo-5.1 (Fig. 5B) due to the provision of wild-type NS5A in trans (Fig. 4B, row 5). Therefore, the lower production of NS5A (Fig. 5C) from the neo-5.1 helper plays only a minor role in trans-complementation, whereas the incompatibility of highly adaptive mutations seems to be a major determinant for the efficiency of trans-complementation. These results demonstrate that trans-complementation of NS5A is possible in transient-replication assays. Moreover, the data support the notion that either NS5A proteins can be exchanged between different RCs or NS5A provides a function outside the RC that indirectly rescues replication of the mutant. In the latter case, the lower trans-complementation efficiency of neo-5.1 RNA for luc-ET mutants may be due to a competition between the adapted proteins for the same replication-supporting function.
Mutations in the N-terminal amphipathic α-helix of NS5A cannot be complemented in trans.
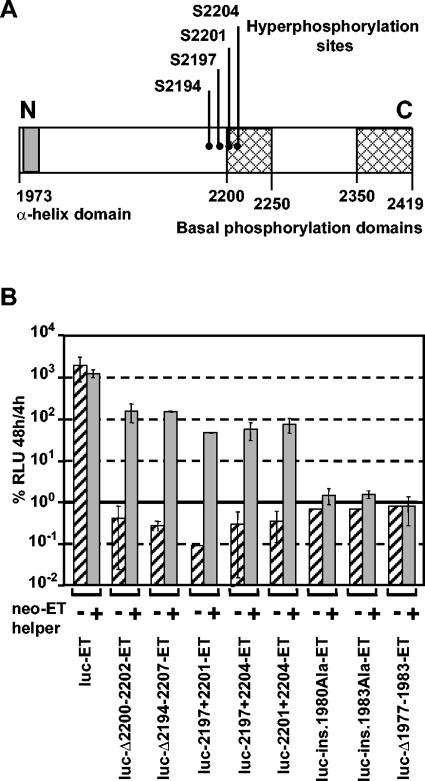
Having established a sensitive transient trans-complementation assay, we analyzed a comprehensive panel of replicons carrying mutations in different HCV proteins for their ability to be complemented in trans. To this end, a set of mutants was cotransfected with the neo-ET helper RNA. As summarized in Table 2, none of the analyzed mutations in NS3, NS4B, or NS5B, all of which reduced RNA replication to below the detection limit, could be complemented in trans. In contrast, all mutations in the central region of NS5A were rescued (Table 2 and Fig. 6). Among these were multiple mutations affecting potential phosphorylation sites as well as small deletions. In striking contrast, all replicons in which we had disturbed the structure of the N-terminal amphipathic helix by introducing a twist (ins. 1980Ala and ins. 1983Ala) or shortening its length (Δ1977-1983) could not be trans-complemented (Fig. 6B). Interestingly, these mutations not only disturb the proper folding of the helix but also cause a dramatic reduction of NS5A hyperphoshorylation, arguing that these mutations had a more general effect on the overall folding of the protein (45). In summary, the data show that neither mutations in NS3, NS4B, or NS5B nor mutations affecting the N-terminal membrane anchor of NS5A can be complemented in trans.
TABLE 2.
trans-complementation of HCV replicon mutants
| Viral protein | Mutation | Complementationa |
|---|---|---|
| NS3 protease | A1027D | − (t) |
| P1028H | − (s) | |
| NS3 helicase; motif II | H1319A | − (t) |
| NS4B | V1897D | − (s) |
| V1897K | − (s) | |
| V1897P | − (s, t) | |
| Δ1782-1821 | − (s) | |
| Δ1905-1933 | − (s, t) | |
| NS5A | Δ1977-1983 | − (s, t) |
| ins. 1980 Ala | − (s, t) | |
| ins 1983 Ala | − (s, t) | |
| Δ2194-2197 | + (s) | |
| Δ2200-2202 | + (s, t) | |
| Δ2194-2207 | + (s, t) | |
| S2197A + S2201A | + (s, t) | |
| S2197A + S2204A | + (s, t) | |
| S2201A + S2204A | + (s, t) | |
| S2194A + S2197A + S2201A + S2204A | + (s, t) | |
| S2194A + S2200A + S2201A + S2202A | + (s, t) | |
| NS5B | ΔGDD/GND | − (s, t) |
| NS5BΔC12 | − (s, t) | |
| NS5BΔC21 | − (s, t) | |
| NS5B-LVL | − (s, t) | |
| NS5B-R568/570A | − (s, t) |
−, no trans-complementation; +, trans-complementation; s, trans-complementation with hyg replicon helper RNA; t, trans-complementation in transient-replication assay.
FIG. 6.
trans-complementation of NS5A mutants in a transient assay. (A) Schematic drawing of the NS5A protein structure. The region spanning the N-terminal amphipathic α-helix is shaded. Regions important for the basal phosphorylation of NS5A are indicated by crosshatched boxes. The major phosphoacceptor site (S2194) and the potential hyperphosphorylation sites (S2197, S2201, and S2204) are marked by black dots. (B) Transient replication of NS5A mutants after rescue with helper RNA neo-ET (Fig. 1A). Luciferase activities were determined as described in the legend to Fig. 5. Hatched bars represent the replication levels of the mutant in the absence of helper RNA. Bars are means and error ranges of quadruplicate results.
trans-complementation does not require replication of the helper RNA.
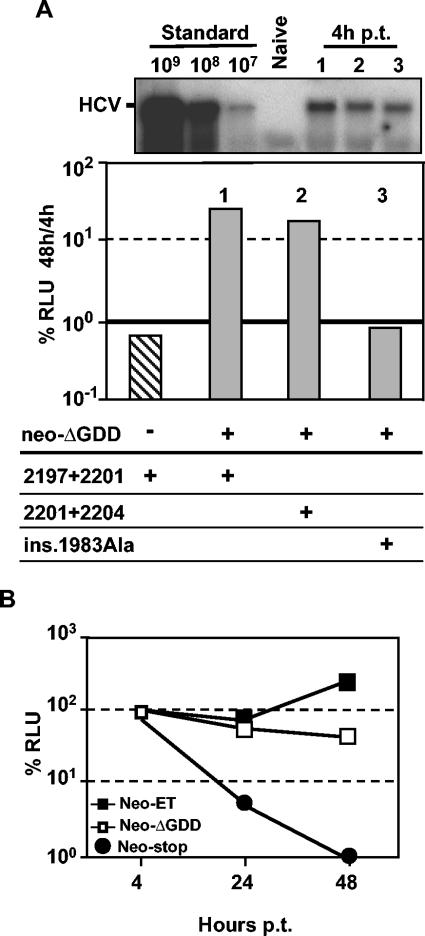
Encouraged by the successful complementation of replicon RNAs carrying mutations in the central domain of NS5A, we were interested to find out whether production of NS5A in the context of an active RC is required for trans-complementation. In the first set of experiments, we performed a transient-complementation assay with a helper RNA that cannot replicate due to a deletion spanning the GDD motif of the NS5B RdRp (neo-ΔGDD). As exemplified by the cotransfection of the NS5A mutants luc-2197 + 2201-ET and luc-2201 + 2204-ET, trans-complementation was possible to a level ∼10-fold above background (Fig. 7A). In contrast, the mutant with the altered N-terminal α-helix (ins. 1983Ala) was not rescued in spite of efficient transfection of the helper RNA as shown by Northern hybridization (Fig. 7A). This result demonstrates that trans-complementation does not require replication of the helper RNA. However, rescue of RNA replication of a mutant replicon was at least 10-fold higher when a replication-competent helper RNA (neo-ET) was used. In addition, by performing time course experiments we found that after cotransfection of nonreplicating helper RNA, luciferase activity, reflecting the replication efficiency of the trans-complemented luc mutant, steadily decreased, whereas an increase of luciferase activity over time was observed after cotransfection of replication-competent helper neo-ET (Fig. 7B). This increase correlates well with the RNA replication of the helper RNA. The 10-fold difference in rescue efficiency 48 h after transfection is therefore most likely due to the more continuous and higher expression of NS5A over time when a replication-competent helper is provided.
FIG. 7.
Rescue of NS5A mutants by a nonreplicating helper RNA. (A) Huh-7 cells were cotransfected with the indicated mutants and neo-ΔGDD as helper RNA. The transfection efficiency of the latter was confirmed by Northern hybridization with total RNA prepared from cells harvested 4 h posttransfection (p.t.) (upper panel). For further details, see the legend to Fig. 5C. (B) Huh-7 cells were cotransfected with the mutant luc-2197 + 2201-ET and given helper RNAs. Cells were harvested at 4, 24, and 48 h after transfection, and luciferase activities derived from the mutant were determined. Data were normalized for transfection efficiency by using the 4 h values. A representative result from three independent experiments is shown.
Expression of NS3 to NS5A is sufficient for trans-complementation.
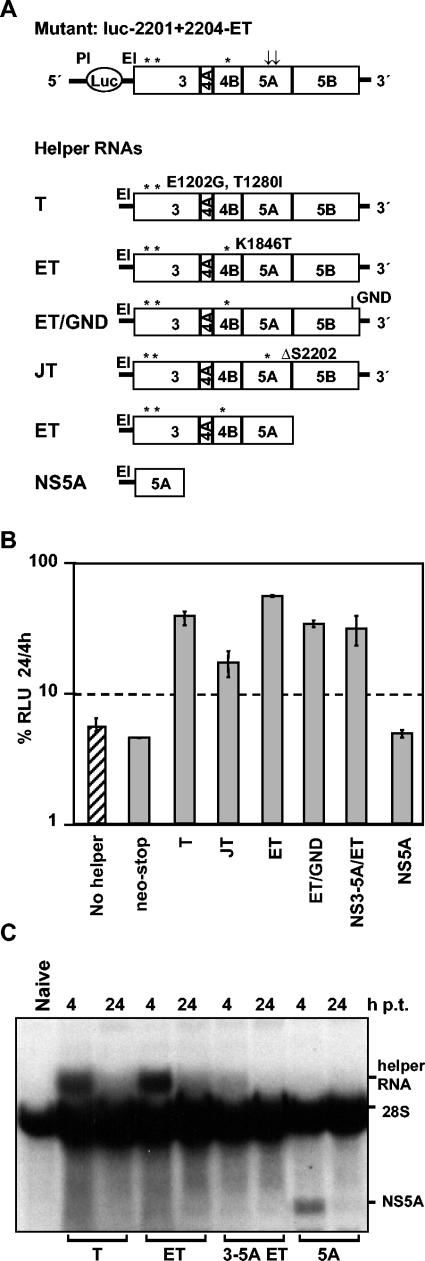
To further confirm that trans-complementation is possible without replication of the helper RNA and to map the minimal sequence sufficient for trans-complementation, we cotransfected the mutant luc-2201 + 2204-ET with RNAs encoding various fragments of the polyprotein encompassing NS3 to NS5B (Fig. 8A). Cells were harvested 4 and 24 h after transfection, and replication efficiency was determined by measuring luciferase activity. The results in Fig. 8B show that luciferase activity was more than 10-fold increased after coexpression of RNAs encoding a polyprotein encompassing NS3 to NS5B compared to the replicon mutant alone. Comparable to the results described above, reduced restoration efficiency was observed in the case of a helper RNA that carried a highly adaptive mutation in NS5A (Fig. 8A, JT). This result is most likely due to the incompatibility of the highly adaptive mutations in NS5A of the helper RNA (Δ2202) and NS4B of the mutant replicon (K1846T). Furthermore, cotransfection of an RNA that encodes only NS3 to NS5A was sufficient to restore replication of the mutant replicon to the same level as with the other helper RNAs. In contrast, when only NS5A was expressed, no rescue of RNA replication was observed. This was not due to low transfection efficiency, because the RNA was well detected 4 h after transfection, at a level comparable to those of the other helper RNAs (Fig. 8C), and the same was true for the NS5A protein as detected by Western blotting (data not shown). Taken together, these results show that the minimal region required for trans-complementation is a polyprotein fragment encompassing NS3 to NS5A.
FIG. 8.
Rescue of an NS5A mutant by coexpression of NS3 to NS5A in trans. (A) The structure of the mutant is shown at the top, and the those of the different helper RNAs are given below the mutant structure. All RNAs carry the EMCV IRES at the 5′ end to allow efficient protein expression. Helper RNAs expressing NS3 to NS5Bcontain the HCV 3′ NTR, which is not present with RNAs encoding for NS3 to NS5A or NS5A alone. GND indicates the position of the inactivating amino acid substitution in the NS5B RdRp. Adaptive mutations in NS3, NS4B, and NS5A are marked by asterisks. (B) Huh-7 cells were cotransfected with mutant luc-2201 + 2204-ET and a given helper RNA carrying adaptive mutations as shown in panel A. Cells were harvested at 4, 24 and 48 h after transfection. Shown are the 24-h values normalized to the 4-h value. Data are means and standard deviations from at least three independent experiments. The hatched bar represents the replication efficiency of the mutant in the absence of the helper RNA. (C) Detection of helper RNAs by Northern hybridization. Positions of helper RNAs are indicated on the right. As a negative control, total RNA from naive Huh-7 cells was used (left lane). Note that the NS3 to NS5A helper RNA had a size similar to that of the 28S RNA and therefore could not be detected in this analysis. p.t., posttransfection.
DISCUSSION
In this study we analyzed the conditions necessary for trans-complementation of HCV proteins that constitute the viral RC. This analysis became possible by the establishment of two alternative assays. The first one is based on the supertransfection of nonreplicating selectable neo replicons into Huh-7 cells that carry a functional helper replicon. The disadvantages of this assay are the length of time required to establish cell clones, the low success in recovery of viable cell clones, and the possibility for accumulation of second-site compensatory mutations or reversions in the replicon mutant. On the other hand, this approach has the advantage that stable cell clones can be examined in detail for the replicating RNA species. In this way we found that only mutations in NS5A could be rescued and that in all complemented cell clones that were analyzed, the mutants still carried the inactivating substitutions in NS5A. Complete sequence analysis of the HCV coding region of one cell clone revealed no further mutations, suggesting that trans-complementation took place. In 3 out of 28 cases, only the mutated neo replicons were detected. Analysis of one of these clones revealed that the neo RNAs had acquired an additional mutation that may have compensated for the defects. Another possibility is that we selected by chance for a more permissive host cell that supports replication of this RNA. Further studies will be required to differentiate between these two possibilities.
Surprisingly, in no case was RNA recombination observed, although naturally occurring recombinant HCV isolates have been described (10, 11, 27). Very recently, a model was proposed in which homologous recombination occurs during minus-strand RNA synthesis by template switching of the polymerase that is facilitated by conserved hairpin structures (26). For members of the Picornaviridae family, recombination is a well-known phenomenon. For instance, in the case of PV it was reported that up to 79% of virus strains secreted after vaccination are recombinants (11). RNA recombination was thought to occur by a copy choice mechanism. More recently, two independent groups reported the generation of recombinant genomes of PV (19) and BVDV (4, 16a) by a nonreplicative recombination mechanism in vivo.
The second assay allowed us to monitor transient replication of an inactivated replicon after cotransfection with a helper RNA. This assay has two advantages over the selection approach. First, trans-complementation can be measured within a very short time and does not depend on the selection of cell clones, and the problem of accumulation of compensatory mutations is circumvented. Second, nonreplicating helper RNAs can be used, which is not possible with the coselection approach. In fact, by using this assay we found that expression of a polyprotein encompassing NS3 to NS5A was sufficient to rescue replication of an NS5A replicon mutant, whereas NS5A expressed on its own did not support trans-complementation. These data suggest that the other nonstructural proteins, NS3, NS4A, and NS4B, affect production of a functional NS5A protein. This result is in remarkable agreement with the previous observation that proper phosphorylation of NS5A requires expression of this protein in the context of the same polyprotein (33). Such a requirement may reflect the contribution of the region from NS3 to NS4B to the folding of NS5A in a highly ordered complex, with only properly folded NS5A being able to rescue RNA replication in trans. In turn, a misfolded NS5A that is part of an RC might be difficult to complement because of possible tethering to components of the complex in a nonproductive way. Such a scenario may explain why mutations affecting the overall folding or the length of the N-terminal amphipathic helix could not be rescued. Interestingly, these mutations also caused a dramatic reduction in NS5A hyperphosphorylation, indicating a more profound effect on the conformation of the protein, which in turn probably interferes with its proper phosphorylation (45). We assume that in addition to its function as a membrane anchor, the N terminus of NS5A is required for correct folding and incorporation of the protein into the RC. Mutations in this domain may therefore result in an NS5A that is nonproductively associated with other components of the RC, making trans-complementation very difficult.
Our results for HCV are in line with a recent report by Grassmann and coworkers, who studied the conditions for trans-complementation of the pestivirus BVDV (22). It was found that only mutations in NS5A, and not mutations in NS3, NS4A, NS4B, and NS5B, can be rescued by trans-complementation. However, in that study complementation only was possible when mutants were transfected into cell clones that carried stably replicating replicons, whereas cotransfection of helper RNAs or infection with helper viruses did not restore replication of the mutants. In contrast, we also achieved trans-complementation with nonreplicating helper RNAs. The reason for this discrepancy is not known, but it may be due to a higher sensitivity of our transient trans-complementation assay.
trans-complementation has also been described for the two flaviviruses Yellow fever virus and KV. In both cases, trans-complementation of NS1 was possible after coexpression of this protein by noncytopathic Sindbis virus replicons with the NS1 mutant (30, 36). For KV, it was also possible to complement NS5 polymerase mutants by expression of isolated NS5, although rescue in trans was more efficient when NS5 was expressed as part of a polyprotein encompassing NS1 to NS5 (29, 30). Furthermore, trans-complementation was demonstrated for the NS3 helicase region of KV (37). In contrast, for both HCV and BVDV, only mutations in NS5A can be complemented in trans. This result suggests that NS3 (presumably as a NS3/4A complex), NS4B, and NS5B are strictly cis acting within a higher-order complex. This result is in agreement with the general notion that the RC of HCV has a rather closed conformation. For instance, it was found that in vitro, isolated RCs are unable to switch to an exogenous template (35). Furthermore, RNA present within this complex is fairly resistant to nucleases, as are the viral proteins that are actively engaged in RNA replication (35, 43). Like for virtually all other positive-strand RNA viruses, HCV RNA replication takes place within a membranous compartment that probably limits access for exogenous RNAs and proteins but, on the other side, protects these viral factors from degradation and reduces the induction of antiviral defense mechanisms in the host cell. The formation of such a compartment may limit the possibility for trans-complementation.
In view of a model for trans-complementation, three possibilities can be envisaged. In the first model, expression of functional NS5A may confer an increased permissiveness to the host cell, for instance, by interfering with a cellular inhibitor, which would result in an overall enhancement of HCV RNA replication. In this case NS5A would have the capacity to support the defective replicon by performing a function outside the RC. However, we found that replication of low-efficiency replicons such as the wild type or a replicon carrying two weakly adaptive mutations in NS3 was not enhanced in trans-complementation assays. Neither in the coselection approach nor in the transient assay was an increase in replication of such RNAs found, suggesting that an indirect function of NS5A that enhances host cell permissiveness and/or RNA replication in general is less likely. A second model is based on the exchange of the complete polyprotein complex encompassing NS3 to NS5B between the functional and the inactive RCs. This model seems to be less likely, since trans-complementation due to a template switch should also be possible for proteins other than NS5A. In the third model, trans-complementation is mediated by an exchange of the functional NS5A protein into the altered RC. Three lines of evidence support this notion. First, trans-complementation was observed only for NS5A and not for the other NS proteins. Second, complementation efficiency was significantly reduced with mutants carrying a highly adaptive mutation in NS4B upon cotransfection with a helper RNA that delivered an adapted NS5A protein. This result suggests that a chimeric RC carrying two proteins with highly adaptive mutations was generated. In this case the reduction of replication is due to the combination of proteins with highly adaptive mutations that are antagonistic when present in the same RC (38). Third, NS5A is anchored to endoplasmic reticulum membranes by an amphipathic α-helix but not by a transmembrane domain, as is the case for NS4B, for NS5B, and, via NS4A, for the NS3/4A complex. It is therefore tempting to speculate that NS5A is less tightly associated with membranes that provide the scaffold of the RC. This looser association may facilitate trans-complementation.
While this paper was under review, Graziani and Paonessa described a dominant negative effect of wt NS5A on subgenomic replicons carrying adaptive mutations in NS5A (23). The inhibition was observed upon expression of wt NS5A in the context of a polyprotein encompassing NS3 to NS5A or from a wt- or NS5B-adapted replicon, and it reduced replication of an NS5A-adapted replicon to about 10%, whereas replicons with wt NS5A were not affected. The authors proposed two models to explain their results. In one of these, NS5A is engaged in protein-protein interactions, e.g., between NS5A and NS5B. This hypothesis implies that the dominant negative effect of wt NS5A is due to its interaction with NS5B, encoded by another (the NS5A-adapted) replicon. In fact, our observation that NS5A mutants can be complemented in trans is compatible with such an exchange of proteins encoded by different replicons. Another similarity is the observation that a dominant negative effect is exerted only when NS5A is expressed as part of a polyprotein encompassing NS3 to NS5A, whereas expression of NS5A alone or together with other individual nonstructural proteins has no effect. Likewise we found that efficient trans-complementation depends on the expression of NS5A as part of a polyprotein encompassing NS3 to NS5A. Finally, Graziani and Paonessa observed that an adapted NS5A protein neither has a dominant negative phenotype nor enhances replication of wt replicons (23). We also did not observe enhanced replication of wt or weakly adapted replicons in our trans-complementation assays. This result is difficult to understand, because an exchange of proteins or an indirect function of NS5A outside the RC should increase RNA replication of weakly replicating RNAs. Further studies will be required to clarify the underlying mechanism.
In summary, our results constitute the first report describing the trans-complementation of an HCV nonstructural protein. The assays reported here may help us to better understand the role of NS5A in the viral life cycle.
Acknowledgments
We thank Michael Frese, Kerry Mills, Thomas Pietschmann, Matthias Reiss, and Marc Windisch for critical reading of the manuscript.
This work was supported by grants from the Sonderforschungsbereich 638 (Teilprojekt A5), the European Union (QLK2-CT-2002-01329), and the Bristol-Myers Squibb Foundation.
REFERENCES
- 1.Arima, N., C. Y. Kao, T. Licht, R. Padmanabhan, Y. Sasaguri, and R. Padmanabhan. 2001. Modulation of cell growth by the hepatitis C virus nonstructural protein NS5A. J Biol. Chem. 276:12675-12684. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., V. Lohmann, T. Wilkinson, and J. O. Koch. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69:7519-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becher, P., M. Orlich, M. König, and H. J. Thiel. 1999. Nonhomologous RNA recombination in bovine viral diarrhea virus: molecular characterization of a variety of subgenomic RNAs isolated during an outbreak of fatal mucosal disease. J. Virol. 73:5646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens, S. E., L. Tomei, and R. de Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Brass, V., E. Bieck, R. Montserret, B. Wölk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 9.Chung, K. M., J. Lee, J. E. Kim, O. K. Song, S. Cho, J. Lim, M. Seedorf, B. Hahm, and S. K. Jang. 2000. Nonstructural protein 5A of hepatitis C virus inhibits the function of karyopherin β3. J. Virol. 74:5233-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colina, R., D. Casane, S. Vasquez, L. Garcia-Aguirre, A. Chunga, H. Romero, B. Khan, and J. Cristina. 2004. Evidence of intratypic recombination in natural populations of hepatitis C virus. J. Gen. Virol. 85:31-37. [DOI] [PubMed] [Google Scholar]
- 11.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egger, D., B. Wölk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elazar, M., K. H. Cheong, P. Liu, H. B. Greenberg, C. M. Rice, and J. S. Glenn. 2003. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J. Virol. 77:6055-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Failla, C., L. Tomei, and R. de Francesco. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 68:3753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. A kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gale, M. J., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 16a.Gallei, A., A. Pankraz, H.-J. Thiel, and P. Becher. 2004. RNA recombination in vivo in the absence of viral replication. J. Virol. 78:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, L., H. Aizaki, J. W. He, and M. M. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh, A. K., M. Majumder, R. Steele, P. Yaciuk, J. Chrivia, R. Ray, and R. B. Ray. 2000. Hepatitis C virus NS5A protein modulates transcription through a novel cellular transcription factor SRCAP. J. Biol. Chem. 275:7184-7188. [DOI] [PubMed] [Google Scholar]
- 19.Gmyl, A. P., E. V. Belousov, S. V. Maslova, E. V. Khitrina, A. B. Chetverin, and V. I. Agol. 1999. Nonreplicative RNA recombination in poliovirus. J. Virol. 73:8958-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grassmann, C. W., O. Isken, N. Tautz, and S. E. Behrens. 2001. Genetic analysis of the pestivirus nonstructural coding region: defects in the NS5A unit can be complemented in trans. J. Virol. 75:7791-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graziani, R., and G. Paonessa. 2004. Dominant negative effect of wild-type NS5A on NS5A-adapted subgenomic hepatitis C virus RNA replicon. J. Gen. Virol. 85:1867-1875. [DOI] [PubMed] [Google Scholar]
- 24.Gwack, Y., D. W. Kim, J. H. Han, and J. Choe. 1995. NTPase activity of hepatitis C virus NS3 protein expressed in insect cells. Molecules Cells. 5:171-175. [Google Scholar]
- 25.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 26.Kalinina, O., H. Norder, and L. O. Magnius. 2004. Full-length open reading frame of a recombinant hepatitis C virus strain from St Petersburg: proposed mechanism for its formation. J. Gen. Virol. 85:1853-1857. [DOI] [PubMed] [Google Scholar]
- 27.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko, T., Y. Tanji, S. Satoh, M. Hijikata, S. Asabe, K. Kimura, and K. Shimotohno. 1994. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem. Biophys. Res. Commun. 205:320-326. [DOI] [PubMed] [Google Scholar]
- 29.Khromykh, A. A., M. T. Kenney, and E. G. Westaway. 1998. trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 72:7270-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khromykh, A. A., P. L. Sedlak, K. J. Guyatt, R. A. Hall, and E. G. Westaway. 1999. Efficient trans-complementation of the flavivirus kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J. Virol. 73:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 1999. trans-complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J. Virol. 73:9247-9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, D. W., Y. Gwack, J. H. Han, and J. Choe. 1995. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 215:160-166. [DOI] [PubMed] [Google Scholar]
- 33.Koch, J. O., and R. Bartenschlager. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 73:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai, V. C., S. Dempsey, J. Y. Lau, Z. Hong, and W. Zhong. 2003. In vitro RNA replication directed by replicase complexes isolated from the subgenomic replicon cells of hepatitis C virus. J. Virol. 77:2295-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindenbach, B. D., and C. M. Rice. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, W. J., P. L. Sedlak, N. Kondratieva, and A. A. Khromykh. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J. Virol. 76:10766-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohmann, V., F. Körner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 42.Majumder, M., A. K. Ghosh, R. Steele, R. Ray, and R. B. Ray. 2001. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J. Virol. 75:1401-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyanari, Y., M. Hijikata, M. Yamaji, M. Hosaka, H. Takahashi, and K. Shimotohno. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J. Biol. Chem. 278:50301-50308. [DOI] [PubMed] [Google Scholar]
- 44.Park, K. J., S. H. Choi, D. H. Choi, J. M. Park, S. W. Yie, S. Y. Lee, and S. B. Hwang. 2003. Hepatitis C virus NS5A protein modulates c-Jun N-terminal kinase through interaction with tumor necrosis factor receptor-associated factor 2. J. Biol. Chem. 278:30711-30718. [DOI] [PubMed] [Google Scholar]
- 45.Penin, F., V. Brass, N. Appel, S. Ramboarina, R. Montserret, D. Ficheux, H. E. Blum, R. Bartenschlager, and D. Moradpour. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. [DOI] [PubMed]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan, S. L., H. Nakao, Y. He, S. Vijaysri, P. Neddermann, B. L. Jacobs, B. J. Mayer, and M. G. Katze. 1999. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc. Natl. Acad. Sci. USA 96:5533-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanji, Y., T. Kaneko, S. Satoh, and K. Shimotohno. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tellinghuisen, T. L., and C. M. Rice. 2002. Interaction between hepatitis C virus proteins and host cell factors. Curr. Opin. Microbiol. 5:419-427. [DOI] [PubMed] [Google Scholar]
- 51.Teterina, N. L., W. D. Zhou, M. W. Cho, and E. Ehrenfeld. 1995. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J. Virol. 69:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. M. Wen, A. E. Gorbalenya, S. B. Hwang, and M. C. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 53.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the VIIth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 54.Zech, B., A. Kurtenbach, N. Krieger, D. Strand, S. Blencke, M. Morbitzer, K. Salassidis, M. Cotten, J. Wissing, S. Obert, R. Bartenschlager, T. Herget, and H. Daub. 2003. Identification and characterization of amphiphysin II as a novel cellular interaction partner of the hepatitis C virus NS5A protein. J. Gen. Virol. 84:555-560. [DOI] [PubMed] [Google Scholar]