Abstract
Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) is a specialized chaperone of phosphodiesterase 6, a key effector enzyme in the phototransduction cascade. The FKBP domain of AIPL1 is known to bind the farnesyl moiety of PDE6. Mutations in AIPL1, including many missense mutations in the FKBP domain, have been associated with Leber congenital amaurosis, a severe blinding disease. Here, we report the backbone and sidechain assignments of the N-terminal FKBPΔloop (with a loop deletion) of AIPL1 in complex with a farnesyl ligand. We also compare the predicted secondary structures of FKBPΔloop with those of a highly homologous AIP FKBP. These results show that the FKBP domains of AIP and AIPL1 have similar folds, but display subtle differences in structure and dynamics. Therefore, these assignments provide a framework for further elucidation of the mechanism of farnesyl binding and the function of AIPL1 FKBP.
Keywords: FKBP, AIPL1, chaperone, phosphodiesterase 6, AIP
Biological context
Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) was originally identified due to its association with one of the most severe forms of Leber congenital amaurosis (LCA), an early-onset, inherited retinopathy (Sohocki et al. 2000a). AIPL1 shares domain organization and 50% sequence identity with aryl hydrocarbon receptor-interacting protein (AIP) (Sohocki et al. 2000a). AIP is expressed in various tissues where it acts as a co-chaperone with HSP90 in the maturation of aryl hydrocarbon receptor and other nuclear receptors (Trivellin & Korbonits 2011). In contrast, AIPL1 is expressed exclusively in the retina and the pineal gland (Sohocki et al. 2000a, van der Spuy et al. 2002). AIPL1 is composed of an N-terminal FK506-binding protein (FKBP) domain and a C-terminal tetratricopeptide repeat (TPR) domain with three tetratricopeptide repeats (Sohocki et al. 2000a, Das et al. 1998). Distinctively, AIPL1 proteins in primates contain a third proline-rich region located C-terminally to the TPR domain.
Mutation-linked LCA are found in all three domains of AIPL1, but the FKBP domain appears to be a hot spot for the pathogenic mutations (Sohocki et al. 2000b, Stone 2007, Dharmaraj et al. 2004). AIPL1 in photoreceptor rods and cones serves as a specialized chaperone for cGMP-specific phosphodiesterase-6 (PDE6), the key effector enzyme in the phototransduction cascade (Ramamurthy et al. 2004, Liu et al. 2004, Gopalakrishna et al. 2016). AIPL1-knockout in mice revealed markedly reduced stability and activity of PDE6, which was followed by rapid retina degeneration (Ramamurthy et al. 2004). This phenotype parallels that of PDE6 mutations causing retinal degeneration and blindness in humans and animal models due to elevation of intracellular cGMP that triggers photoreceptor cell death (Farber & Lolley 1974, Bowes et al. 1990, Pittler & Baehr 1991). The mechanism of the chaperone/client relationships between AIPL1 and PDE6 is largely unknown. AIPL1 is critical for the proper assembly of rod PDE6. In this process, AIPL1 interacts with isoprenylated PDE6 catalytic subunits (Ramamurthy et al. 2003, Kolandaivelu et al. 2009), via direct binding of the PDE6 farnesyl or geranylgeranyl moiety to the FKBP domain (Majumder et al. 2013, Yadav et al. 2015).
Because proteins containing FKBP domains play essential roles in cellular signaling and protein folding, they have been a focus of intense research leading to a wealth of structural information. However, despite the critical significance of AIPL1 for the function and survival of photoreceptor cells, practically no structural information is available for its FKBP domain. NMR assignments for the AIPL1 FKBP is a first step in determining and understanding its unique structural features and interactions with farnesyl or geranylgeranyl ligand.
Methods and experiments
Cloning, protein expression and purification
For NMR assignments we utilized a construct of human AIPL1 FKBP (aa 2-161) where residues 111-132 were deleted and replaced with a short loop consisting of five glycines (AIPL1 FKBPΔ111-132). The AIPL1 FKBPΔ111-132 construct was designed based on the NMR structure of AIP FKBP (PDB ID 2LKN) where the corresponding region is unstructured and highly flexible (Linnert et al. 2013). Thus, thereafter we refer to AIPL1 FKBPΔ111-132 as AIPL1 FKBPΔloop. DNA coding AIPL1 FKBPΔloop was obtained in a two-step PCR procedure using plasmid for full-length human AIPL1 as template. During the first step, a DNA fragment was generated that codes the C-terminal portion of AIPL1 FKBPΔloop where residues 111–132 are replaced with five Gly. In the second PCR step, this fragment was paired with a primer such that the resulting AIPL1 FKBPΔloop construct had the N-terminal tag of MGHHHHHHG. This PCR construct was cloned into the pET15b vector using NcoI/NdeI sites, and the His6-tagged AIPL1 FKBPΔloop protein was expressed in BL21 (DE3) E. coli cells.
To obtain uniformly 15N- and 13C-labeled AIPL1 FKBPΔloop, E. coli BL21 (DE3) cells were adapted on a minimal medium containing 15NH4Cl and 13C6-glucose. 0.5 mL of adapted BL21 (DE3) cells were inoculated in 50 mL of minimal media containing 15NH4Cl, 13C6-glucose and 100 μg/mL of ampicillin and incubated at 37°C with shaking until OD600 reached ~ 0.8–1.0. This culture was then added to 1 litre of minimal media containing 15NH4Cl, 13C6-glucose and 100 μg/mL of ampicillin, and cells were grown at 37°C to OD600 of ~0.6. Then protein expression was induced by adding 0.5 mM of IPTG, and cells were grown overnight at 20°C. The cell pellets were sonicated on ice (five 30s pulses) in a buffer containing 50 mM Tris-HCl (pH7.5), 100 mM NaCl, and 10 mM dithiothreitol (DTT) (buffer A), and protease inhibitor mixture (Roche Applied Science, Indianapolis, IN, USA). AIPL1 FKBPΔloop was purified over Ni-NTA resin (EMD Millipore: Billerica, MA, USA) using buffer A containing 250 mM imidazole for elution. AIPL1 FKBPΔloop was further purified by ion-exchange chromatography on a Mono Q5 column (Bio-Rad Laboratories, Hercules, CA, USA) and gel filtration chromatography on a HiLoad 16/600 Superdex 75 column (GE Healthcare, Pittsburgh, PA, USA) equilibrated with 25 mM Na2HPO4 (pH 7.5) and 10 mM of DTT.
NMR spectroscopy
NMR spectra were acquired on a 500 or 800 MHz Bruker Avance II NMR spectrometer at 25°C using 1.0 mM uniformly [13C,15N]-labeled AIPL1 FKBPΔloop or uniformly 15N-labeled AIPL1 FKBP wild type in complex with a farnesyl ligand of S-farnesyl-L-cysteine methyl ester (FC) in a buffer containing 25 mM sodium phosphate (pH 7.5) and 8 mM DTT either in 90% H2O / 10% D2O or 100% D2O. The AIPL1 FKBP/FC complexes were prepared by incubating 500 μl of 1.0 mM protein with 14 μl of 230 mM concentrated FC stock that was dissolved in deuterated DMSO overnight on a shaker. This represents a protein:FC molar ratio of 1:6.4 used in the incubation. After incubation overnight, the excess amount of FC which is not soluble in the aqueous buffer was removed by centrifugation before the NMR experiments were conducted. Therefore, the NMR samples should contain a protein:FC molar ratio of 1:1. The FC-binding to AIPL1 FKBP proteins was confirmed by comparing the 15N/1H HSQC spectra of the apo and complexed protein. A suite of triple resonance NMR experiments including HNCACB, HN(CO)CACB, HNCO, HN(CA)CO, HNCA, and HN(CO)CA experiments (Yamazaki et al. 1994) were collected for backbone assignments of the AIPL1 FKBPΔloop/FC complex. Side-chain assignments were obtained by acquiring C(CO)NH, H(CCO)NH, HBHA(CO)NH, HCCH-TOCSY, 15N-NOESY, 15N-TOCSY, and 13C-NOESY spectra (Clore & Gronenborn 1994). The collected data were processed using NMRPipe (Delaglio et al. 1995) and analyzed using NMRView (Johnson & Blevins 1994).
Resonance assignments and data deposition
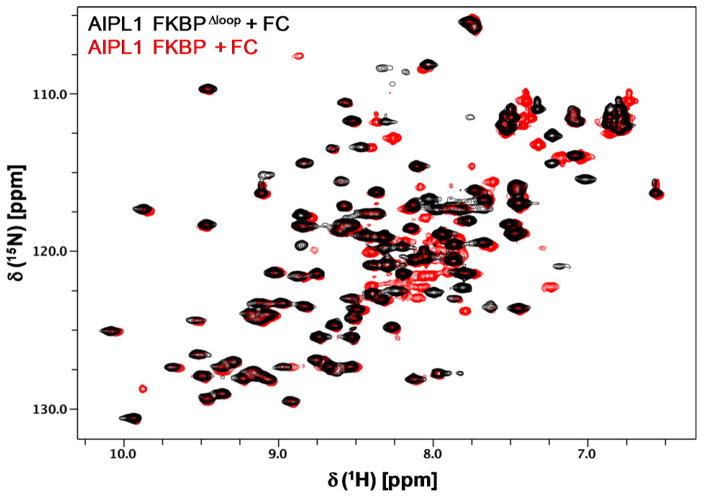
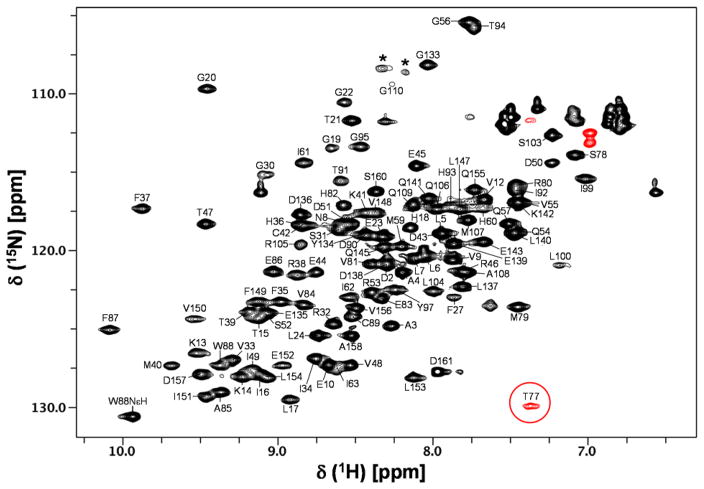
AIPL1 FKBP and AIP FKBP are highly homologous, sharing about 50% sequence identity and 70% sequence similarity and the loop residues 112–133 in AIP FKBP are known to be unstructured and highly flexible (Linnert et al. 2013). Therefore we performed NMR assignments of AIPL1 FKBP using a loop deletion construct (AIPL1 FKBPΔloop) where the corresponding loop residues 111–132 in AIPL1 FKBP were deleted and replaced with a short loop consisting of five glycines. The AIPL1 FKBPΔloop binds to FC with a similar affinity as the wild type protein, indicating that the deleted loop is not involved in the FC binding. Comparison of the 15N/1H HSQC spectra of AIPL1 FKBP wild type and AIPL1 FKBPΔloop in complex with FC (Fig. 1) clearly indicates that the two spectra are nearly identical, except for some expected changes including a few slightly shifted peaks (mostly located in α3 and α4 helices due to a short polyglycine linker in AIPL1 FKBPΔloop) and a dozen extra peaks present in the AIPL1 FKBP protein due to the presence of the loop residues 111–132. The peaks in the 15N/1H HSQC spectrum of AIPL1 FKBPΔloop in complex with FC are clearly well dispersed, indicating a well-folded protein (Fig. 2). The assigned backbone amides and Trp indole NεH are labeled using the wild type protein sequence numbering. All backbone amides are assigned except for G11, N26, I28-T29, G64-L76, V96, and S101-R102. Since all of the backbone peaks detected in the triple resonance experiments have been assigned, these missing backbone amides clustered nearby the FC-binding pocket must be broad beyond detection in solution in these experiments. T77 backbone amide (circled in red) is unique, having a highly upfield-shifted 15N chemical shift at 101.9 ppm (Fig. 2). The assigned chemical shifts were deposited in BioMagResBank (http://www.bmrb.wisc.edu) under the accession number 26947.
Fig. 1.
Overlay of 15N/1H HSQC spectra of AIPL1 FKBP and AIPL1 FKBPΔloop in complex with FC. These spectra are nearly identical, except for a few slightly shifted peaks and some extra peaks present in the AIPL1 FKBP protein due to the presence of the loop residues 111–132. It is noted that the folded negative peak of T77 amide is not shown in the figure.
Fig. 2.
15N/1H HSQC spectrum of AIPL1 FKBPΔloop in complex with FC. The assigned backbone amides and Trp indole NεH are labeled. Wild type protein sequence numbering is used in the labeling. The folded peaks are indicated in red. T77 backbone amide (circled in red) has a highly upfield-shifted 15N chemical shift at 101.9 ppm. Stars indicate peaks derived from the polyglycine linker.
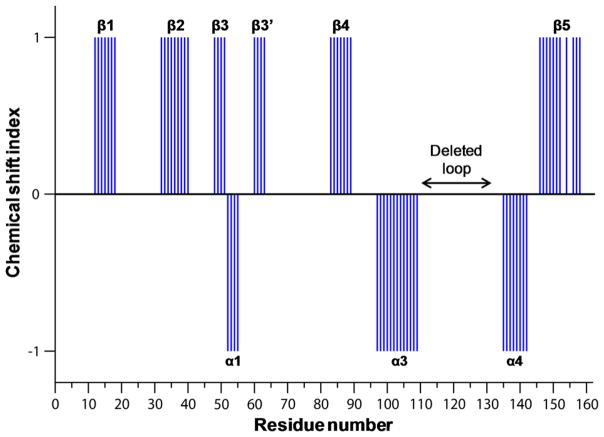
Figure 3 shows the plot of chemical shift index (CSI) as a function of the residue number of AIPL1 FKBPΔloop. The secondary structures of AIPL1 FKBPΔloop obtained from the assigned backbone using CSI and TALOS+ (Shen et al. 2009) are labeled (Fig. 3). Clearly, 5 β-strands are present in AIPL1 FKBPΔloop as in AIP FKBP. The 3rd β-strand consisting of a short β3 and a short β3′ is interrupted by a short α1 helix in both AIPL1 and AIP FKBP proteins. Analyses of 13C-NOESY spectra indicate that the β-strands have similar registry and form an antiparallel β-sheet as observed in AIP FKBP. Helices α3 and α4 are also present in both proteins. However, the N-terminal α0 helix present in AIP FKBP (residues 4–11) is completely absent in AIPL1 FKBPΔloop (corresponding residues 3–10). Moreover, the residues G65-C78 in AIP FKBP are well behaved in solution and contain a short loop (G65-P71 that links β3′ and α2) and an α2 helix consisting of residues V72-C78 (Linnert et al. 2013, Linnert et al. 2012). However, the backbone amides of the corresponding residues G64-L76 in AIPL1 FKBPΔloop were broad beyond detection. We only detected the backbone amide of the last residue (T77) on the α2 helix in AIPL1 FKBPΔloop. These data suggest that the α2 helix and/or its preceding loop in AIPL1 FKBPΔloop exhibit heterogeneous conformations, in direct contrast to the AIP FKBP protein.
Fig. 3.
Plot of chemical shift index as a function of residue number of AIPL1 FKBPΔloop. Wild type AIPL1 FKBP protein sequence numbering is used here. The deleted loop residues in our AIPL1 FKBPΔloop construct are indicated. The secondary structures obtained from the analyses of the assigned backbone of the AIPL1 FKBPΔloop are also labeled in the figure. Interestingly, the N-terminal α0 helix present in AIP FKBP is absent in AIPL1 FKBPΔloop. Furthermore, the backbone amides of G64-L76 in AIPL1 FKBPΔloop were broad beyond detection, and the corresponding residues in AIP FKBP contain a short loop and an α2 helix.
In summary, despite of the high sequence homology between the FKBP domains of AIP and AIPL1, AIPLI FKBP binds to FC with a high affinity (Majumder et al. 2013), but AIP FKBP does not bind to FC at all. Our NMR assignments and analyses reported here suggest that these two proteins have similar folds, but exhibit subtle differences in structure and dynamics. In future studies, we will use these NMR assignments to fully elucidate the mechanism of farnesyl binding and the function of AIPL1 FKBP.
Acknowledgments
This work was supported by the National Institutes of Health grant EY-10843 to N.O.A.
Footnotes
Conflict of interest: the authors declare that they have no conflicts of interest.
Ethical standards: all of our experiments comply with accepted ethical standards.
References
- Bowes C, Li T, Danciger M, Baxter LC, Applebury ML, Farber DB. Retinal degeneration in the rd mouse is caused by a defect in the beta subunit of rod cGMP-phosphodiesterase. Nature. 1990;347:677–680. doi: 10.1038/347677a0. [DOI] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods in enzymology. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dharmaraj S, Leroy BP, Sohocki MM, et al. The phenotype of Leber congenital amaurosis in patients with AIPL1 mutations. Archives of Ophthalmology. 2004;122:1029–1037. doi: 10.1001/archopht.122.7.1029. [DOI] [PubMed] [Google Scholar]
- Farber DB, Lolley RN. Cyclic guanosine monophosphate: elevation in degenerating photoreceptor cells of the C3H mouse retina. Science. 1974;186:449–451. doi: 10.1126/science.186.4162.449. [DOI] [PubMed] [Google Scholar]
- Gopalakrishna KN, Boyd K, Yadav RP, Artemyev NO. Aryl Hydrocarbon Receptor-interacting Protein-like 1 Is an Obligate Chaperone of Phosphodiesterase 6 and Is Assisted by the gamma-Subunit of Its Client. The Journal of biological chemistry. 2016;291:16282–16291. doi: 10.1074/jbc.M116.737593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. Journal of biomolecular NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kolandaivelu S, Huang J, Hurley JB, Ramamurthy V. AIPL1, a protein associated with childhood blindness, interacts with alpha-subunit of rod phosphodiesterase (PDE6) and is essential for its proper assembly. Journal of Biological Chemistry. 2009;284:30853–30861. doi: 10.1074/jbc.M109.036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnert M, Haupt K, Lin YJ, Kissing S, Paschke AK, Fischer G, Weiwad M, Lucke C. NMR assignments of the FKBP-type PPIase domain of the human aryl-hydrocarbon receptor-interacting protein (AIP) Biomolecular NMR assignments. 2012;6:209–212. doi: 10.1007/s12104-012-9359-0. [DOI] [PubMed] [Google Scholar]
- Linnert M, Lin YJ, Manns A, Haupt K, Paschke AK, Fischer G, Weiwad M, Lucke C. The FKBP-type domain of the human aryl hydrocarbon receptor-interacting protein reveals an unusual Hsp90 interaction. Biochemistry. 2013;52:2097–2107. doi: 10.1021/bi301649m. [DOI] [PubMed] [Google Scholar]
- Liu X, Bulgakov OV, Wen XH, et al. AIPL1, the protein that is defective in Leber congenital amaurosis, is essential for the biosynthesis of retinal rod cGMP phosphodiesterase. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13903–13908. doi: 10.1073/pnas.0405160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder A, Gopalakrishna KN, Cheguru P, Gakhar L, Artemyev NO. Interaction of aryl hydrocarbon receptor-interacting protein-like 1 with the farnesyl moiety. The Journal of biological chemistry. 2013;288:21320–21328. doi: 10.1074/jbc.M113.476242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittler SJ, Baehr W. Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase beta-subunit gene of the rd mouse. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:8322–8326. doi: 10.1073/pnas.88.19.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy V, Niemi GA, Reh TA, Hurley JB. Leber congenital amaurosis linked to AIPL1: a mouse model reveals destabilization of cGMP phosphodiesterase. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13897–13902. doi: 10.1073/pnas.0404197101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy V, Roberts M, van den Akker F, Niemi G, Reh TA, Hurley JB. AIPL1, a protein implicated in Leber’s congenital amaurosis, interacts with and aids in processing of farnesylated proteins. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12630–12635. doi: 10.1073/pnas.2134194100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of biomolecular NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohocki MM, Bowne SJ, Sullivan LS, et al. Mutations in a new photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Nat Genet. 2000a;24:79–83. doi: 10.1038/71732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohocki MM, Perrault I, Leroy BP, et al. Prevalence of AIPL1 mutations in inherited retinal degenerative disease. Molecular Genetics and Metabolism. 2000b;70:142–150. doi: 10.1006/mgme.2000.3001. [DOI] [PubMed] [Google Scholar]
- Stone EM. Leber congenital amaurosis - a model for efficient genetic testing of heterogeneous disorders: LXIV Edward Jackson Memorial Lecture. American Journal of Ophthalmology. 2007;144:791–811. doi: 10.1016/j.ajo.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Trivellin G, Korbonits M. AIP and its interacting partners. The Journal of endocrinology. 2011;210:137–155. doi: 10.1530/JOE-11-0054. [DOI] [PubMed] [Google Scholar]
- van der Spuy J, Chapple JP, Clark BJ, Luthert PJ, Sethi CS, Cheetham ME. The Leber congenital amaurosis gene product AIPL1 is localized exclusively in rod photoreceptors of the adult human retina. Human molecular genetics. 2002;11:823–831. doi: 10.1093/hmg/11.7.823. [DOI] [PubMed] [Google Scholar]
- Yadav RP, Majumder A, Gakhar L, Artemyev NO. Extended conformation of the proline-rich domain of human aryl hydrocarbon receptor-interacting protein-like 1: implications for retina disease. Journal of neurochemistry. 2015;135:165–175. doi: 10.1111/jnc.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR, Kay LE. A suite of triple-resonance NMR experiments for the backbone assignment of 15N, 13C, 2H-labeled proteins with high sensitivity. J Am Chem Soc. 1994;116:11655–11666. [Google Scholar]