Abstract
The phylogeny of reptilian herpesviruses (HVs) relative to mammalian and avian HVs was investigated by using available gene sequences and by alignment of encoded amino acid sequences and derivation of trees by maximum-likelihood and Bayesian methods. Phylogenetic loci were obtained for green turtle HV (GTHV) primarily on the basis of DNA polymerase (POL) and DNA binding protein sequences, and for lung-eye-trachea disease-associated HV (LETV) primarily from its glycoprotein B sequence; both have nodes on the branch leading to recognized species in the Alphaherpesvirinae subfamily and should be regarded as new members of that subfamily. A similar but less well defined locus was obtained for an iguanid HV based on a partial POL sequence. On the basis of short POL sequences (around 60 amino acid residues), it appeared likely that GTHV and LETV belong to a private clade and that three HVs of gerrhosaurs (plated lizards) are associated with the iguanid HV. Based on phylogenetic branching patterns for mammalian HV lineages that mirror those of host lineages, we estimated a date for the HV tree's root of around 400 million years ago. Estimated dates for branching events in the development of reptilian, avian, and mammalian Alphaherpesvirinae lineages could plausibly be accounted for in part but not completely by ancient coevolution of these virus lines with reptilian lineages and with the development of birds and mammals from reptilian progenitors.
Herpesviruses (HVs) have a characteristic virion architecture, comprising an icosahedral capsid with a T = 16 arrangement of spikes, a surrounding proteinaceous tegument layer, and a bounding membrane with embedded protein species, with the overall diameter of the particle being around 200 nm. Historically, possession of this morphology was used to assign membership of the Herpesviridae family, and on this basis, HVs were defined that were associated with diseases in species across the animal kingdom, in mammals, birds, reptiles, amphibians, fish, and invertebrates (shellfish) (5, 20). Within the last two decades, sequence determination of HV genes and genomes has vastly improved our understanding of relationships among these viruses. Most sequencing studies have been concerned with mammalian HVs, and it is well established that these fall into three subfamilies (the Alpha-, Beta-, and Gammaherpesvirinae) which are all related by descent from a common ancestral HV species, as judged by extensive equivalences in their gene complements (13). The few avian HVs for which sequence data are available are related to the mammalian viruses, falling into two lineages in the Alphaherpesvirinae. However, amphibian and fish HVs comprise a separate grouping, which shows only a very marginal relationship in gene content to the mammalian and avian virus group, and the one characterized HV of an invertebrate (oyster) forms a third distinct group (5).
Over many years, HVs have been reported to be associated with diseases of reptiles, including species of snakes, lizards, and chelonians (i.e., turtles and tortoises); Wellehan et al. (28) have given an overview of the older literature on reptilian HVs. Until recently, such assignments depended on the criterion of virus particle morphology. On the basis of short DNA sequences obtained by PCR with primers for the HV DNA polymerase (POL) gene, Quackenbush et al. (16) reported that certain turtle HVs were related to the Alphaherpesvirinae. Further limited data have appeared for other HVs associated with chelonians, and longer sequences have now been published for the complete DNA polymerase and DNA binding protein (DBP) genes, plus parts of the UL28 and UL31 genes, of green turtle HV (GTHV) and for the glycoprotein B (gB) and protease/assembly protein genes of another HV of green turtle, lung-eye-trachea disease-associated HV (LETV). Limited sequences have also been described for HVs associated with lizards, including an iguanid HV (IgHV) and three HVs of gerrhosaurs (plated lizards) (GerHV1, GerHV2, and GerHV3). Accession numbers and references are listed in Table 1.
TABLE 1.
Gene sequences of reptilian HVs
| Virus | Gene | Sequence length (nucleotides) | Accession no. | Reference |
|---|---|---|---|---|
| GTHV | POL (complete) | 3,294 | AF239684 | 31 |
| UL28 (part) | 360 | 31 | ||
| UL31 (part) | 723 | 31 | ||
| DBP (complete) | 3,588 | AY332227 | 14 | |
| LETV | gB (complete) | 2,598 | AY124577 | 4 |
| Protease/assembly protein (complete) | 1,743 | AY124578 | 4 | |
| IgHV | POL (part) | 780 | AY236869 | 28 |
| Hawaiian GTHV | POL (part) | 483 | AF035003 | 16 |
| Florida GTHV | POL (part) | 483 | AF035004 | 16 |
| Loggerhead turtle HV | POL (part) | 483 | AF035005 | 16 |
| Olive Ridley turtle HV | POL (part) | 483 | AF049904 | 16 |
| Australian loggerhead turtle HV | POL (part) | 483 | AF299107 | 15 |
| Australian GTHV | POL (part) | 483 | AF299108 | 15 |
| Barbados GTHV | POL (part) | 483 | AF299110 | 15 |
| Loggerhead turtle HV | POL (part) | 181 | AF120208 | |
| Florida GTHV | POL (part) | 181 | AF120209 | |
| LETV | POL (part) | 181 | AY124579 | 4 |
| GerHV1 | POL (part) | 178 | AF416629 | 29 |
| GerHV2 | POL (part) | 178 | AF416628 | 29 |
| GerHV3 | POL (part) | 181 | AF416630 | 29 |
The phylogenetic status of reptilian HVs has remained rather ill defined, inasmuch as publications to date have reported only preliminary phylogenetic examinations of individual reptilian HVs. The purpose of the analyses reported in this paper was to assess the phylogenetic loci and relationships of reptilian HVs as fully as possible by using currently available sequences, to extend and integrate our understanding, and to explore emergent implications for herpesvirus evolution.
MATERIALS AND METHODS
HV gene sequences.
Table 1 lists DNA sequences from reptilian HVs that were available as of late 2003. Sequences for mammalian and avian HV genes were obtained from public databases and from our own work; for the sake of brevity, only those that are discussed directly in the evaluation of the reptilian HVs are identified in this paper (but see reference 12).
General computational handling of sequences.
General sequence handling used the GCG package (Accelrys, Inc.). Amino acid sequence sets were aligned by using CLUSTAL W (27) or MAFFT (9). Positions in an alignment that had a gap in any sequence were removed, and any regions regarded as too diverged to align were also excised.
Inference of phylogenetic trees.
The initial procedure used to derive and evaluate phylogenetic trees was as follows. For a given alignment of amino acid sequences representing a single gene set, relationships were first evaluated by the rapid clustering method of neighbor joining with bootstrapping (6). Based on these results, operational taxonomic units were defined that consisted of securely associated groupings of species, and these were used to examine up to 104 trees with the maximum-likelihood program Protml (MOLPHY package) (1), with a single rate of change for all sites in each sequence. A set of the top-scoring trees was then evaluated by the maximum-likelihood program Codeml (PAML package, version 3.13) (30), with a distribution of rates across sites specified by a discrete gamma distribution. The output data from Codeml were assessed by Shimodaira's approximately unbiased (AU) test by using the CONSEL package (24, 25). This general approach has been criticized by Goldman et al. (7) as potentially vulnerable to excluding, in its early stages, trees that would have scored highly in the final stage. In practice, our early stage analyses were so broadly based as to make the possibility of any such error remote. In addition, we separately analyzed the alignments by a Bayesian approach with a Monte Carlo Markov process (MrBayes 3) (21), which generates a probability distribution of tree topologies contingent on the input data and which is not subject to the criticism of Goldman et al. (7); the two approaches yielded closely equivalent results in all cases. Evaluations of phylogenetic loci of reptilian HVs using alignments of very short sequences (i.e., representing a minor part of the POL gene) were attempted by three approaches: first, by simple comparisons of pairwise distances between aligned sequences; second, by attempting de novo construction of trees; and third, by computing maximum likelihoods for sets of trees in which a single test HV sequence was inserted in turn at every branch of the tree topology derived from the complete HV POL alignment. A Perl script (Treeadder) was written to generate such sets of tree topologies for testing.
Estimations of dates for phylogenetic events.
Dates for nodes within HV trees were estimated by two approaches. In both, a calibration was applied that equated paleontological dates in the host lineages with particular nodes in the HV trees. The first approach was to compute, by using Codeml, a molecular clock version of the tree under study that retained the previously obtained tree topology and, in addition, specified the branch on which the tree's root was to be located while enforcing a constant rate of change across all branches. The single rate for such a tree was then expressed from the calibration dates in terms of substitutions per amino acid site in a given time, and from this, estimates of dates were made for nodes of interest. The second approach used r8s, a program that takes previously estimated trees and aims to minimize differences in substitution rate for each branch by smoothing procedures, without imposing the global uniformity of the molecular clock approach (22, 23). We employed the penalized likelihood option of the program with quasi-newtonian optimization and scaling by specification of fixed dates.
RESULTS
Phylogenetic locus of GTHV.
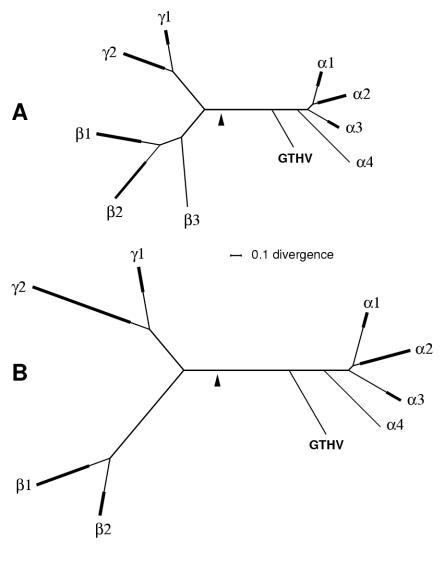
Phylogenetic trees were derived based on alignments of HV POL sequences (final alignment length, 809 amino acids, 45 species) and DBP sequences (final alignment length, 781 amino acids, 36 species). With each data set, the two methods employed (maximum likelihood by use of Codeml and Bayesian inference by use of MrBayes) gave closely comparable results, and one tree was identified as clearly top scoring. These top-scoring trees are shown in Fig. 1. The GTHV locus in each tree does not lie within any of the clades corresponding to recognized genera or genus-level groupings, and for our present purposes, this allows the trees to be usefully represented in a condensed format, focusing on genus-level clades rather than individual species. Figure 1 shows that with both POL and DBP trees, the GTHV lineage originates from the branch that connects the Alphaherpesvirinae to the Beta- and Gammaherpesvirinae, and in both cases, the node for the GTHV branch lies closer to nodes within the alpha subfamily than to any in the beta or gamma subfamilies. The best attainable estimate for the root of the HV tree is that it lies on the branch running between the alpha subfamily and the bifurcation of the lineages to the beta and gamma subfamilies (11). The estimated root loci are shown for Fig. 1 as the midpoint of the distance from the mean positions of branch tips in the alpha subfamily to the mean positions of branch tips in the beta and gamma subfamilies. By this criterion, the GTHV lineage in the POL and DBP trees forms a clade with the alpha HV lineages. We also investigated trees based on part of the UL31 gene (194 amino acids, 34 species); this smaller alignment gave an equivalent but noisier result (data not shown). The small part of the UL28 gene for which a GTHV sequence was available (Table 1) was not examined.
FIG. 1.
POL and DBP trees, including GTHV. The top-scoring trees are shown based on HV POL amino acid sequences (A) and HV DBP amino acid sequences (B). The trees are presented as unrooted and in a condensed format showing genus-level groupings rather than individual species, with the multiple-branch region of each genus-level clade represented by a single heavy line. In each tree, the estimated position of the root is indicated by a filled arrowhead, calculated as the midpoint of the distance from the mean positions of branch tips in the alpha subfamily to the mean positions of branch tips in the beta and gamma subfamilies. A common scale bar is indicated for divergence (i.e., substitutions per amino acid site). Genus-equivalent labels are as follows: α1, Simplexvirus; α2, Varicellovirus; α3, Mardivirus; α4, Iltovirus; β1, Cytomegalovirus (including Muromegalovirus and Tupaia HV); β2, Roseolovirus (including porcine cytomegalovirus); β3, elephant endothelial HV; γ1, Lymphocryptovirus; γ2, Rhadinovirus. (A) The POL tree contains 44 species in addition to GTHV, distributed as follows: α1, 4 species; α2, 7 species; α3, 3 species; α4, 1 species; β1, 8 species; β2, 3 species; β3, 1 species; γ1, 3 species; γ2, 14 species. (B) The DBP tree contains 35 species in addition to GTHV, distributed as follows: α1, 4 species; α2, 6 species; α3, 3 species; α4, 1 species; β1, 7 species; β2, 2 species; γ1, 3 species; γ2, 9 species.
Three separate gene trees thus gave a concordant result for the locus of GTHV in the phylogenetic tree of the Herpesviridae. The foremost interpretation of this analysis is that GTHV should be regarded as a member of the Alphaherpesvirinae, but for several reasons, this cannot be taken as an incontrovertible conclusion. First, inasmuch as the deep interior region of the tree from which the GTHV lineage springs is unexplored territory, it is conceivable that HVs mapping to this locus may turn out to be sufficiently distinct that they should be regarded taxonomically as a novel subfamily. Next, while the root assignments described above place GTHV on the alpha subfamily line of descent, the fact is that root placement in such trees is an estimation procedure that necessarily falls short of rigorous deduction, so that the root estimated may be significantly in error. An independent input to classifying GTHV is provided by information on the relative order and orientations of the UL28, DBP, POL, and UL31 genes in the GTHV genome: as pointed out by Nigro et al. (14), the GTHV arrangement matches that characteristic of the alpha subfamily and is distinct from the arrangements in the beta and gamma subfamilies. In cladistic terms, this common pattern may constitute either a shared derived state (so that GTHV would belong to the same clade as the recognized alphaherpesviruses) or the ancestral state (and so would not be informative on the relationship of GTHV to the alphaherpesviruses). Our position is that the available substantive evidence comprises a reasonably strong case for placing GTHV in the Alphaherpesvirinae, and this would meet present pragmatic norms of virus taxonomic practice.
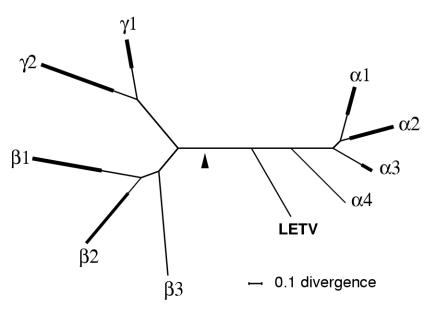
Phylogenetic locus of LETV.
A similar analysis was carried out for an alignment of gB sequences (final alignment length, 616 amino acids, 61 species) that included LETV and an unambiguous top-scoring tree identified. As shown in Fig. 2, the gB tree gave a locus for LETV that was comparable to that found for GTHV. The shorter protease data set sequences (final alignment length, 188 amino acids, 38 species) gave a tree with an equivalent locus for LETV, but this was of lower value for our purposes, inasmuch as it did not include a sequence for infectious laryngotracheitis virus (which comprises the α4 clade in the trees shown in Fig. 1 and 2); the protease tree is not shown. Coberley et al. (4) have demonstrated that the relative order and orientations of the LETV genes for gB and the protease/assembly protein are characteristic of the alpha subfamily. Overall then, as for GTHV, there is a reasonable case for assigning LETV to the Alphaherpesvirinae.
FIG. 2.
gB tree, including LETV. The top-scoring tree is shown based on HV gB amino acid sequences, obtained by two separate methods (Codeml and MrBayes). The format is as described for Fig. 1. The gB tree contains 60 species in addition to LETV, distributed as follows: α1, 10 species; α2, 13 species; α3, 3 species; α4, 1 species; β1, 9 species; β2, 3 species; β3, 1 species; γ1, 6 species; γ2, 14 species.
The question then arises of the detail of the phylogenetic relationship between GTHV and LETV. Because their tree loci were derived by using disjunct sets of genes, the trees obtained are not informative on whether GTHV and LETV belong to a single clade branching from the main Alphaherpesvirinae lineage or to separate lineages with distinct points of divergence from the main Alphaherpesvirinae lineage. However, the only presently available sequence that may facilitate a direct comparison of GTHV and LETV is a short section (181 nucleotides) of the LETV POL gene (Table 1). We postpone treatment of this until the section below on analysis with short sequence fragments.
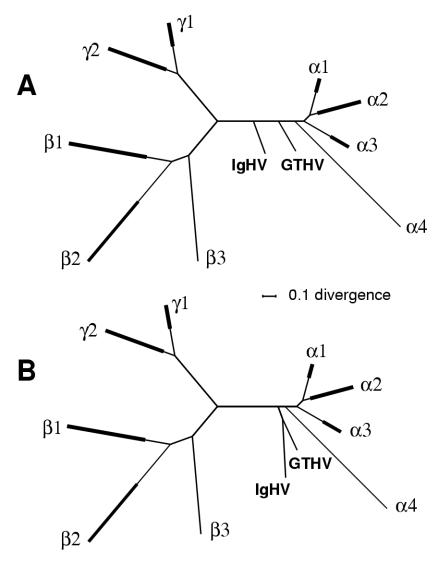
Phylogenetic locus of IgHV.
Wellehan et al. (28) have described a partial POL gene sequence of 780 nucleotides for IgHV. We incorporated the corresponding amino acid sequence into an alignment containing 46 species and with a final length of 226 amino acids, i.e., comprising 28% of the complete POL alignment length used to examine the GTHV locus. The locus for IgHV was primarily analyzed by starting with the tree topology previously obtained from the whole POL alignment for 45 species, interpolating IgHV at each branch in turn, and evaluating the resulting set of 83 trees with Codeml. Two top-scoring trees were identified, as shown in Fig. 3. In tree A, the IgHV lineage branches from the lineage leading to the Alphaherpesvirinae deeper in the tree than the GTHV branch, whereas in tree B, IgHV is placed in a clade with GTHV. Trees A and B had closely equivalent AU scores (0.62 and 0.55, respectively), so we cannot discriminate between these two possible loci for IgHV with the available data.
FIG. 3.
Trees based on a part of POL, including IgHV. The two top equal trees are shown based on a 226-amino-acid alignment of a section of POL. The alignment included 46 species: 45 as described for Fig. 1A plus IgHV. The format is as described for Fig. 1.
Evaluation of phylogenetic relationships from short sequence fragments.
Other available sequences for reptilian HVs are short, representing fragments obtained by PCR (Table 1). The sequences available included seven 483-nucleotide and two 181-nucleotide POL gene fragments of turtle HVs that were all very closely related to the GTHV complete POL sequence; these evidently represented strains of the same virus, or closely related viruses, and were not examined further. The four other available sequences comprised POL gene fragments for LETV, GerHV1, GerHV2, and GerHV3 (29). The amino acid sequences (59 or 60 residues) translated from these sequences were aligned with the 46 known HV POL amino acid sequences (including the partial IgHV sequence). We found that the section of POL represented in the short sequences did not enable an alignment of high quality. The N-terminal region of 22 residues is strongly conserved and has 7 residues invariant across the 50 input sequences. However, much of the C-terminal portion is quite diverse and has a level of insertion-deletion differences among sequences that limits its usefulness. Removing gapped loci and a highly diverse section reduced the alignment, for alphaherpesvirus and test sequences only, to 52 residues. Other versions of reduced alignment that included beta- and gammaherpesvirus sequences were also produced, with lengths of 31 to 40 residues. While these alignments were the best achievable, we regarded them as of indifferent overall quality.
Evaluations of the phylogenetic loci of LETV and the GerHVs were then attempted by three approaches, as described in Materials and Methods. Analyses based on such short sequences are intrinsically limited and can be expected to yield definite answers only in favorable cases, in particular, where the query sequence is closely similar to an already well-characterized instance. None of the short reptilian HV sequences were close to each other, nor to any other sequence, and the results obtained with them were judged indicative but not precise or robust; they are not described here in detail. In summary, LETV and GTHV appeared to be each other's closest relative, consistent with them belonging to the same lineage, while GerHV2 and GerHV3 appeared to be related to IgHV, with GerHV1 perhaps also belonging to this grouping.
Assessment of the evolution of the Herpesviridae.
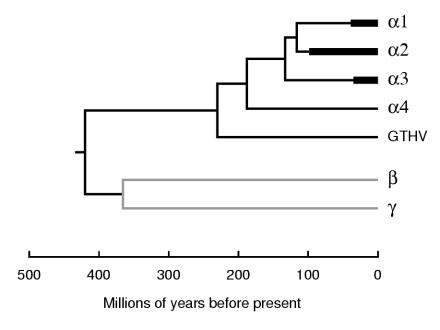
In this section, we treat evolutionary implications of the finding that a group of reptilian HVs of some diversity forms a clade with the avian and mammalian HVs of the Alphaherpesvirinae; in this context, we refer to the whole clade as the alphaherpesvirus lineage. Our laboratory previously observed that many elements of the branching patterns for mammalian HVs in each subfamily of the Herpesviridae show congruence with the tree for corresponding lineages of mammalian host species, suggesting a prominent component of coevolution of hosts and viruses (10-12). The question thus arose as to whether this phenomenon of coevolution may extend also to reptilian and avian HVs, on a deeper timescale. To examine this, we estimated dates of nodes in the GTHV- and LETV-containing trees reported above by two approaches, namely application of a molecular clock and use of a rate-smoothing program.
Molecular clock trees are constructed to impose a constant rate across all branches. This may represent oversimplification of the data but allows a calibration of timescale from one or more nodes specified as having known dates. We used Codeml to compute versions of trees that retained the previously identified topologies but imposed the constraints of a molecular clock and placed the tree's root on the branch connecting the alphaherpesvirus lineage to the beta- plus gammaherpesvirus lineages. For this purpose, the DBP and POL datasets were concatenated to give a single alignment of 1,590 amino acids. We wished to infer a timescale for each tree by using the same calibration system for all trees and also to employ only calibration points from the alpha subfamily, as this was the most appropriate for our present purpose. The calibration used was therefore based on nodes in the α2 lineage, taking the divergence between bovine HV 1 and pseudorabies virus (suid HV 1) and the divergence between these artiodactyl viruses and equine HVs 1 and 4 as corresponding to the paleontological dates for separation of the ruminant and pig lineages and of artiodactyl and perissodactyl lineages, respectively. The values applied were 63.8 and 82.1 millions of years ago (Ma), respectively (26). The divergence rate for each HV data set was calculated as the mean value for the two data points in terms of substitutions/amino acid site/lineage/109 years, and date estimates for nodes of interest were then obtained. In addition to HV trees from the present study, we included a longer data set that comprised seven genes plus a single exon for 19 species (12). Table 2 summarizes the trees examined and dates estimated, and Fig. 4 shows the molecular clock tree for the DBP plus POL tree with the timescale applied. Estimates of dates for the trees containing turtle HV species are in good agreement with available corresponding estimates for the tree based on the large, 19-species alignment, with values for the date of the HV tree's root falling in the range of 374 to 420 Ma in the three trees examined. We consider the two estimates for nodes leading to turtle HVs (230 Ma for GTHV and 248 Ma for LETV) to be indistinguishable given the limitations of the data on which they are based.
TABLE 2.
Date estimates from molecular clock trees
| Parameter | Result for:
|
||
|---|---|---|---|
| 7 genes + 1 exona | DBP + POL | gB | |
| No. of species | 19 | 36 | 61 |
| Alignment length (amino acid residues) | 4,580 | 1,590 | 616 |
| Estimated substitution rateb | 2.87 | 3.50 | 3.36 |
| Date of node for α1 + α2 cladec | 114 | 116 | 122 |
| Date of node for α3 lineage | 133 | 134 | |
| Date of node for α4 lineage | 188 | 187 | |
| Date of node for GTHV lineage | 230 | ||
| Date of node for LETV lineage | 248 | ||
| Date of node for β + γ clade | 351 | 366 | 331 |
| Date for root of tree | 413 | 420 | 374 |
Data are from reference 12.
Rates are substitutions/amino acid site/lineage/109 years.
Dates are millions of years before the present.
FIG. 4.
Molecular clock tree based on DBP plus POL sequences. A concatenated alignment for DBP and POL amino acid sequences of 36 species was used to produce a molecular clock, and a timescale was applied based on the correlation of divergence in the α2 lineage with paleontological dates as described in the text. Regions in the α1, α2, and α3 lineages occupied by multiple branches are shown as heavy lines, and the reduced outlines for the beta and gamma lineages are shown in gray.
We also applied the program r8s to estimating dates in the alphaherpesvirus portions of the DBP plus POL tree and the gB tree. The r8s program aims to estimate rates and dates in phylogenetic trees without imposing a molecular clock, by using smoothing procedures to find solutions that optimize compatibility among the branch lengths provided as input. In these analyses, the beta- and gammaherpesvirus portions in each tree served only to provide the root locus. The same calibration points were specified as for the molecular clock analysis. Both datasets required large smoothing factors, indicating that the trees approximated molecular clock behavior. The dates obtained, shown in Table 3, are similar to those obtained via molecular clock trees. While for both methods precision of the estimates would improve with data for more genes of turtle HVs, we consider that the overall consistency among the data sets gives confidence in employing their dates in discussion of large-scale evolutionary scenarios.
TABLE 3.
Date estimates from rate-smoothing analysisa
| Tree locus | Date for:
|
|
|---|---|---|
| DBP + POL | gB | |
| Node for α1 + α2 clade | 118 | 119 |
| Node for α3 lineage | 135 | 135 |
| Node for α4 lineage | 180 | 186 |
| Node for GTHV lineage | 246 | |
| Node for LETV lineage | 263 | |
Dates are millions of years before the present.
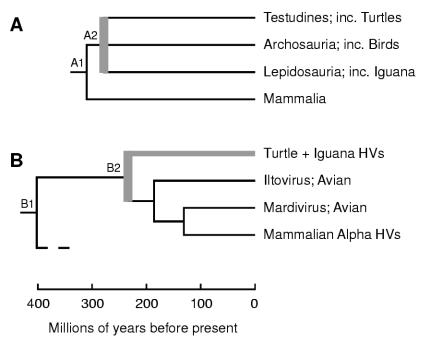
Understanding of patterns in reptile evolution, which is required for our analysis, is presently incomplete but maturing rapidly. The earliest major split in reptilian lineages, estimated to have occurred 310 Ma, was into lines distinguished by patterns of cranial temporal aperture: most modern reptiles, and also birds, evolved from the diapsid lineage (with two temporal openings) while mammals eventually arose from the synapsid lineage (with one temporal opening) (2). Chelonians are anapsids, with no temporal aperture; however, recent morphological and sequence-based analyses are pointing to this condition as secondarily derived from an earlier diapsid state (3, 8, 17-19, 32). Recent molecular phylogenetic analyses have placed chelonians (order Testudines) as associated with either the Archosauria (extant members birds and crocodiles) or the Lepidosauria (including lizards and snakes) (3, 8, 17, 32). From these published analyses we can, as shown in Fig. 5A, draw a reasonably well-founded consensus tree that presents lineages for the Testudines, Archosauria, and Lepidosauria as an unresolved polytomy originating at around 270 to 285 Ma, with the root of the tree as the diapsid-synapsid split at 310 Ma. This turns out to provide sufficient detail for our present purpose. The corresponding HV tree is shown in Fig. 5B, drawn to indicate incomplete resolution of details for GTHV, LETV, and IgHV.
FIG. 5.
Comparison of host and alphaherpesvirus trees. (A) Consensus tree for reptilian, avian, and mammalian lineages, derived from published data, with branch names chosen for comparison with the HV tree. The gray bar represents multiple branchings treated as unresolved. (B) Summary tree for alphaherpesvirus lineages, based on the gB tree and DBP plus POL molecular clock tree (Table 2). The gray bar and branch represent unresolved branching details for GTHV, LETV, and IgHV. In both trees, nodes that are discussed in the text are labeled. A common timescale is shown at the foot. inc., includes.
Our estimated date for the most recent common ancestor of alpha-, beta-, and gammaherpesviruses (node B1 in Fig. 5) is earlier than the date for the diapsid-synapsid divergence (node A1), so our discussion here focuses on the alphaherpesvirus lineage. The alphaherpesvirus lineage is now seen to comprise viruses whose hosts come from highly diverged reptilian groups (chelonians and lizards) plus reptile-derived groups (birds and mammals) and to have developed from a common ancestor on a timescale that approximates that of the major reptilian lineages plus avian and mammalian lineages. We should thus treat the reptilian, avian, and mammalian alphaherpesviruses as all of comparable significance in considering the evolution of the lineage. While neither the host tree nor the alphaherpesvirus tree (as depicted in Fig. 5) has yet been fully resolved, it is clear that they do not show global congruence. There are, however, two possible but mutually exclusive scenarios that would economically relate the two trees, both based on the date for node B2, the divergence of reptilian HVs from avian and mammalian HVs. The first is that node B2 represents the counterpart of the diapsid-synapsid divergence, i.e., node A1 in the host tree. In this scenario, reptilian and mammalian HV lines could have each coevolved with their host lineages, but the two avian HV lines would have arisen by transfer of HVs from synapsid reptilian or mammalian hosts. The second scenario is that node B2 corresponds to node A2, the radiation of the diapsids, so that reptilian and avian HV lines could each have coevolved with their hosts, but the mammalian HV clade would have arisen by a transfer mechanism. We regard both of these as plausible and attractive interpretations. There is no cogent reason to prefer one on the basis of the trees in Fig. 5, although from a wider perspective, the fact that the beta- and gammaherpesvirus clades both consist of mammalian viruses with no known avian members could be taken as lending support to the first scenario. Our overall evaluation is that there was major ancient involvement of reptilian hosts in the evolution of the alphaherpesvirus lineage but that unresolved complexities remain in the details of the lineage's development.
DISCUSSION
Our analyses located GTHV and LETV lineages as both originating from the lineage leading to the recognized members of the Alphaherpesvirinae, and on presently minimal evidence, it seems likely that GTHV and LETV belong to a private clade. The IgHV lineage originates in the same region of the HV tree, but its relationship to the GTHV and LETV lineages is unresolved. We expect that a modest additional amount of comparative data will serve to solidify the GTHV-LETV relationship, for instance, either the GTHV gB gene or LETV POL gene sequences. Resolving the locus for IgHV could well prove a more demanding undertaking, given that the present analysis based on 28% of the complete POL alignment gave a result nicely balanced between two possibilities, and we speculate that full resolution may require a set of several gene sequences for IgHV, GTHV, and LETV. The fact that these reptilian HV lineages originate in a central region of the HV tree remote from other species can be expected to contribute to the difficulty of fully resolving details of their phylogeny. For the GerHV species, their relationships to each other and to IgHV may well resolve with, say, a complete POL sequence for each, contingent on the preliminary indication obtained proving correct.
Our conclusions bear on arrangements for the taxonomy of the Herpesviridae. GTHV and LETV should now be included in the Alphaherpesvirinae. However, the deep points of origin of their lineages would require definition of either one or two new genera to accommodate them, and clearly, this action must await clarification of their relationship. For IgHV, we suggest that membership of the Alphaherpesvirinae should not be considered until more sequence data have been applied to phylogenetic analysis; data on gene arrangement would also be useful.
Our laboratory's first attempts at analyzing the apparent occurrence of coevolution of host and HV lineages, carried out a decade ago (10, 11), produced an HV family tree of estimated depth around 200 million years; that work depended on the straightforward but limited neighbor-joining method of tree construction. However, our subsequent modeling of the HV tree and possible host-virus coevolution by using computing-intensive maximum-likelihood methods and revised, deeper estimates of host paleontological dates has yielded a tree with its root at around 400 Ma (see reference 12 and the present paper). The detection of reptilian HV lines in the alphaherpesvirus lineage together with the increased estimate for the antiquity of the HV tree's root have now acted to revise our perspective on early evolution of the HV family since divergence from the most recent common ancestor. We consider that the most important outcome of this paper is its case that reptilian HV lineages be brought to the forefront in considering evolution of the family. The two possibilities described for coevolutionary development of early alphaherpesviruses with hosts remain a tentative sketch that should be developed with more data. A novel point is that the dates obtained for the timescale of the HV tree suggest that in principle there may exist, as yet undetected, HVs of reptiles or birds whose lineages originate from nodes deep in the trees of the beta and gamma subfamilies.
Acknowledgments
This work was supported by the United Kingdom Medical Research Council.
We thank J. Wellehan for early sight of data and A. Davison and R. Bowden for critical reading of the manuscript.
REFERENCES
- 1.Adachi, J., and M. Hasegawa. 1994. The MOLPHY 2.2 package. Institute of Statistical Mathematics, Tokyo, Japan.
- 2.Benton, M. J. 1997. Vertebrate palaeontology. Chapman & Hall, London, United Kingdom.
- 3.Cao, Y., M. D. Sorenson, Y. Kumasawa, D. P. Mindell, and M. Hasegawa. 2000. Phylogenetic position of turtles among amniotes: evidence from mitochondrial and nuclear genes. Gene 259:139-148. [DOI] [PubMed] [Google Scholar]
- 4.Coberley, S. S., R. C. Condit, L. H. Herbst, and P. A. Klein. 2002. Identification and expression of immunogenic proteins of a disease-associated marine turtle herpesvirus. J. Virol. 76:10553-10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 7.Goldman, N., J. P. Anderson, and A. G. Rodrigo. 2000. Likelihood-based tests of topologies in phylogenetics. Syst. Biol. 49:652-670. [DOI] [PubMed] [Google Scholar]
- 8.Hedges, S. B., and L. L. Poling. 1999. A molecular phylogeny of reptiles. Science 283:998-1001. [DOI] [PubMed] [Google Scholar]
- 9.Katoh, K., K. Misawa, K. Kuma, and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGeoch, D. J., and S. Cook. 1994. Molecular phylogeny of the Alphaherpesvirinae subfamily and a proposed evolutionary timescale. J. Mol. Biol. 238:9-22. [DOI] [PubMed] [Google Scholar]
- 11.McGeoch, D. J., S. Cook, A. Dolan, F. E. Jamieson, and E. A. R. Telford. 1995. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247:443-458. [DOI] [PubMed] [Google Scholar]
- 12.McGeoch, D. J., A. Dolan, and A. C. Ralph. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 74:10401-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minson, A. C., A. J. Davison, R. C. Desrosiers, B. Fleckenstein, D. J. McGeoch, P. E. Pellett, R. Roizman, and D. M. J. Studdert. 2000. Herpesviridae, p. 203-225. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Academic Press, New York, N.Y.
- 14.Nigro, O., G. Yu, A. A. Aguirre, and Y. Lu. 2004. Sequencing and characterization of the full-length gene encoding the single-stranded DNA binding protein of a novel Chelonian herpesvirus. Arch. Virol. 149:337-347. [DOI] [PubMed] [Google Scholar]
- 15.Quackenbush, S. L., R. N. Casey, R. J. Murcek, T. A. Paul, T. M. Work, C. J. Limpus, A. Chaves, L. duToit, J. V. Perez, A. A. Aguirre, T. R. Spraker, J. A. Horrocks, L. A. Vermeer, G. H. Balazs, and J. W. Casey. 2001. Quantitative analysis of herpesvirus sequences from normal tissue and fibropapillomas of marine turtle with real-time PCR. Virology 287:105-111. [DOI] [PubMed] [Google Scholar]
- 16.Quackenbush, S. L., T. M. Work, G. H. Balazs, R. N. Casey, J. Rovnak, A. Chaves, L. duToit, J. D. Baines, C. R. Parrish, P. R. Bowser, and J. W. Casey. 1998. Three closely related herpesviruses are associated with fibropapillomatosis in marine turtles. Virology 246:392-399. [DOI] [PubMed] [Google Scholar]
- 17.Rest, J. S., J. A. Ast, C. C. Austin, P. J. Waddell, E. A. Tibbetts, J. A. Hay, and D. P. Mindell. 2003. Molecular systematics of primary reptilian lineages and the tuatara mitochondrial genome. Mol. Phylogenet. Evol. 29:289-297. [DOI] [PubMed] [Google Scholar]
- 18.Rieppel, O., and M. deBraga. 1996. Turtles as diapsid reptiles. Nature 384:453-455. [Google Scholar]
- 19.Rieppel, O., and R. R. Reisz. 1999. The origin and early evolution of turtles. Annu. Rev. Ecol. Syst. 30:1-22. [Google Scholar]
- 20.Roizman, B., R. C. Desrosiers, B. Fleckenstein, C. Lopez, A. C. Minson, and M. J. Studdert. 1992. The family Herpesviridae: an update. Arch. Virol. 123:425-449. [DOI] [PubMed] [Google Scholar]
- 21.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian inference under mixed methods. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 22.Sanderson, M. J. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19:101-109. [DOI] [PubMed] [Google Scholar]
- 23.Sanderson, M. J. 2003. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19:301-302. [DOI] [PubMed] [Google Scholar]
- 24.Shimodaira, H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51:492-508. [DOI] [PubMed] [Google Scholar]
- 25.Shimodaira, H., and M. Hasegawa. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246-1247. [DOI] [PubMed] [Google Scholar]
- 26.Springer, M. S., W. J. Murphy, E. Eizirik, and S. J. O'Brien. 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. USA 100:1056-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. J. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wellehan, J. F. X., J. L. Jarchow, C. Reggiardo, and E. R. Jacobson. 2003. A novel herpesvirus associated with hepatic necrosis in a San Esteban chuckwalla, Sauromalus varius. J. Herpetol. Med. Surg. 13:15-19. [Google Scholar]
- 29.Wellehan, J. F. X., D. K. Nichols, L.-L. Li, and V. Kapur. 2004. Three novel herpesviruses associated with stomatitis in Sudan plated lizards (Gerrhosaurus major) and a black-lined plated lizard (Gerrhosaurus nigrolineatus). J. Zoo Wildl. Med. 35:50-54. [DOI] [PubMed] [Google Scholar]
- 30.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 31.Yu, Q., N. Hu, Y. Lu, V. R. Nerurkar, and R. Yanagihara. 2001. Rapid acquisition of entire DNA polymerase gene of a novel herpesvirus from green turtle fibropapilloma by a genomic walking technique. J. Virol. Methods 91:183-195. [DOI] [PubMed] [Google Scholar]
- 32.Zardoya, R., and A. Meyer. 2000. Mitochondrial evidence on the phylogenetic position of caecilians (Amphibia: Gymnophiona). Genetics 155:765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]