Abstract
The positional homologue in the infectious laryngotracheitis virus (ILTV) genome of the glycoprotein gJ gene of herpes simplex virus and the gp2 gene of equine herpesvirus 1 is expressed into four proteins of 85, 115, 160, and 200 kDa (J. Veits, B. Köllner, J. P. Teifke, H. Granzow, T. C. Mettenleiter, and W. Fuchs, Avian Dis. 47:330-342, 2003). RNA analyses revealed that these proteins are expressed from two different late (γ2) transcripts, an unspliced 5.5-kb and a spliced 4.3-kb mRNA that are translated into proteins of 985 and 611 amino acids, respectively. ILTV gJ is incorporated into virions and is modified by N- and O-linked glycosylation. After cotransfection of chicken cells with genomic DNA of a pathogenic ILTV strain and transfer plasmids, gJ-negative ILTV mutants could be isolated. In vitro growth studies demonstrated that deletion of the gJ gene has only minor effects on direct cell-to-cell spread as measured by plaque size. However, progeny virus titers of ILTV-ΔgJ were significantly reduced in comparison to those of the parental virus and a gJ rescue mutant. After experimental infection of chickens the gJ rescue mutant, like wild-type ILTV, caused severe disease and considerable mortality, whereas ILTV-ΔgJ was significantly attenuated. All immunized animals were protected against subsequent challenge infection with virulent ILTV. In sera collected after immunization with the gJ-rescue mutant or with wild-type ILTV, gJ-specific antibodies were detectable by immunofluorescence on cells that had been transfected with a gJ expression plasmid. As expected, no gJ-specific antibodies were found in sera obtained from chickens immunized with ILTV-ΔgJ. Thus, gJ deletion mutants of ILTV might be usable as attenuated live-virus vaccines. Furthermore, the gJ gene might constitute a reliable marker for serological discrimination between vaccinated and field virus-infected chickens.
Infectious laryngotracheitis virus (ILTV) causes a serious, worldwide-occurring respiratory disease of chickens which affects growth and egg production and may lead to death of the animals. The acute phase of infection lasts between 1 and 2 weeks and is often associated with clinical signs like gasping, coughing, expectoration of bloody mucus, and conjunctivitis. Subsequently, an asymptomatic latent infection of the central nervous system can be established. For prevention of disease chickens are immunized with attenuated live-virus vaccines that are suitable for mass application via eye drop, aerosol, or drinking water (reviewed in reference 2).
ILTV, also designated as gallid herpesvirus 1, has been classified as a member of the Alphaherpesvirinae subfamily of the Herpesviridae and represents the only species presently in the genus Infectious Laryngotracheitis-like Viruses (36). DNA restriction analyses (16, 28) and DNA sequencing (reviewed in reference 10) of the ca. 150-kbp genome of ILTV demonstrated that it consists of a long (UL) and a short (US) unique region and of inverted internal and terminal repeat sequences (IRs and TRs, respectively) flanking the US region (Fig. 1A). This genome structure corresponds to that of many other alphaherpesviruses, which are also similar to ILTV with respect to gene content and gene arrangement. Therefore, most gene and protein designations were adopted from herpes simplex virus type 1 (HSV-1) (33, 38) or from other alphaherpesviruses and used, e.g., for the ILTV homologues of the glycoproteins gB, gC, gD, gE, gG, gH, gI, gK, gL, gM, and gN (Fig. 1A).
FIG. 1.
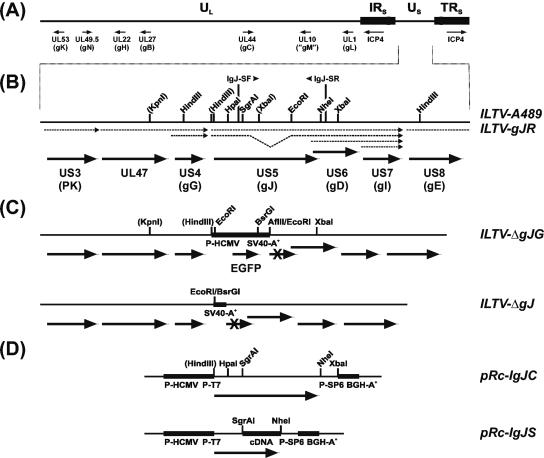
ILTV mutants and plasmids. (A) The ILTV genome consists of UL and US regions and of IRS and TRS flanking the US region. The positions of selected virus genes are indicated. (B) Enlarged section of the analyzed part of the US genome region of wild-type virus (ILTV-A489) and of a gJ rescue mutant (ILTV-gJR) with relevant restriction sites. Artificial sites are shown in parentheses. Viral transcripts are indicated by dotted arrows, and ORFs are drawn as bold arrows. Primers IgJ-SF and IgJ-SR were used for cDNA cloning. (C) ILTV-ΔgJG was derived from ILTV-A489 by substitution of gJ codons 1 to 729 by a reporter gene cassette encoding EGFP under control of P-HCMV. In ILTV-ΔgJ major parts of this cassette were removed, except the SV40 polyadenylation signal sequence (SV40-A+). (D) Plasmid pRc-IgJC contains the genomic gJ gene of ILTV, whereas in pRc-IgJS a part of the insert was replaced by a cDNA fragment of the spliced gJ mRNA species. The plasmids were used for in vitro transcription from bacteriophage promoters (P-T7 and P-SP6) and for transient expression in eukaryotic cells under control of P-HCMV and the bovine growth hormone polyadenylation signal (BGH-A+).
However, phylogenetic analyses of conserved genes indicated a considerable evolutionary distance between ILTV and other avian or mammalian alphaherpesviruses (17, 34). This was also emphasized by particularities of the ILTV genome, including the absence of a UL16 homologue (9), the translocation of the UL47 gene from the UL to the US region (58, 59), and an internal inversion within the UL region which encompasses the UL22 to UL44 genes (59). Interestingly, the recently published genome DNA sequence of psittacid herpesvirus 1 (PsHV-1) exhibits a similar inversion (GenBank accession number NC_005264) (48), and in the genome of pseudorabies virus (PrV) a related inversion ranging from UL27 to UL44 was found (24). Furthermore, PsHV-1 possesses a cluster of five open reading frames (ORFs) located upstream of the inversion in the UL region (48) that have homologues only in ILTV (53, 59). Other ILTV-specific or poorly conserved genes were identified close to both ends of the UL genome region (8, 19, 60) and within the US region and flanking parts of the IRS and TRS (58).
One of the poorly conserved ILTV genes is represented by ORF 5 in the US region (58), which encompasses 986 codons, and is located between the ORFs encoding the homologues of glycoproteins gG and gD (Fig. 1B). Positional homologues of ORF 5 are the US5 gene of HSV-1 and HSV-2 encoding gJ (6, 11, 32), gene 71 of equine herpesviruses (EHV) type 1 and 4 encoding gp2 (44, 46, 47, 55), and sORF2 of PsHV-1 (48). The deduced ORF 5 gene product of ILTV also exhibits characteristics of a membrane glycoprotein since it contains nine putative N-glycosylation sites and hydrophobic domains at its N and C termini, which may serve as signal sequence and as membrane anchor, respectively (58). The ILTV gene product shares only 24 to 26% identical amino acids with the respective proteins of EHV-1, EHV-4, and PsHV-1, and a significant homology to HSV-1 gJ was not detectable. Both gJ and gp2 have been shown to be dispensable for replication of the respective viruses (38, 43). Whereas no function has yet been assigned to gJ in HSV-1 or HSV-2, deletion of gp2 from EHV-1 was shown to impair virus entry and egress (39, 45).
Previous studies have shown that ORF 5 encodes a major target of ILTV-specific monoclonal antibodies (MAb), but controversial results were obtained with respect to protein size (25, 51). Although the calculated molecular mass of the ORF 5 translation product was 107 kDa (58), the viral gene product was initially described as a 60-kDa protein and, therefore, named gp60 (25). In contrast, we identified a set of four ORF 5-encoded proteins possessing apparent molecular masses of ca. 85, 115, 160, and 200 kDa and adopted the designation gJ from the positional HSV-1 homologue (51). Immunoelectron microscopy indicated an association of ILTV gJ with the envelope of mature virus particles, and indirect immunofluorescence (IIF) tests showed an accumulation of ILTV gJ in infected cell cultures and chicken tissues (51). IIF tests on cells transfected with gJ expression plasmids further revealed positive reactions not only with the majority of our MAb produced against purified ILT virions but also with most antisera from experimentally or naturally ILTV-infected chickens (51), demonstrating a high antigenicity of ILTV gJ. Thus, gJ-negative ILTV vaccines might be used to differentiate infected from vaccinated animals. Similar marker vaccines based on gE-deleted PrV have been used to successfully eradicate Aujeszky's disease from several countries including Germany and the United States (37, 49, 50). In a similar approach, gE-deleted bovine herpesvirus 1 (BHV-1) strains are currently being used in Europe for eradication of respiratory and genital infections caused by BHV-1 (21).
The attenuated live-virus vaccines which are in use for prevention of ILT are genetically not precisely characterized and often exhibit residual virulence or increased pathogenicity after in vivo passage (13, 14). Genetically engineered ILTV mutants with defined deletions of nonessential genes could represent safer live-virus vaccines. However, up to now only very few ILTV recombinants were generated and tested in animals (10, 41, 52). Although these mutants, which lacked the ILTV-specific UL0 gene or the conserved genes encoding thymidine kinase (UL23) or dUTPase (UL50), were more or less attenuated, they were not suitable for use as vaccines to differentiate infected from vaccinated animals since the deletions did not affect immunogenic surface proteins.
In the present study, the transcription, translation, and processing of ILTV gJ were investigated to elucidate expression kinetics and the molecular basis for the discrepancies between expected and apparent protein sizes. Furthermore, gJ-negative ILTV mutants with and without an expression cassette for enhanced green fluorescent protein (EGFP), as well as a gJ rescue mutant were generated by cotransfection of chicken cells with virion DNA and transfer plasmids. In vitro growth properties of the ILTV recombinants were analyzed, and an animal trial was performed to determine the relevance of gJ for virulence in chickens and for establishment of protective immunity against challenge infection with pathogenic ILTV.
MATERIALS AND METHODS
Viruses and cells.
Virus recombinants were derived from the pathogenic ILTV strain A489 and propagated in primary chicken embryo kidney (CEK) cells (8). For transfection experiments a chicken hepatoma cell line (LMH) (23, 40) was used. Cells were grown in minimum essential medium supplemented with 10% fetal calf serum (Invitrogen, Karlsruhe, Germany).
Northern blot analyses and cDNA cloning.
CEK cells were infected with ILTV at a multiplicity of infection (MOI) of 5 PFU per cell and incubated in the presence of 100 μg of cycloheximide per ml for 6 h (α), in the presence of 250 μg of phosphonoacetic acid per ml for 16 h (β), or in the absence of any drugs for 6 h (γ1) or 16 h (γ2) at 37°C. Total RNA of infected and noninfected cells was prepared (4), separated in denaturing agarose gels, and hybridized with radiolabeled antisense cRNAs as described previously (8). For cDNA cloning of the gJ gene, synthetic oligonucleotide primers (MWG Biotech, Ebersberg, Germany) were deduced from the published DNA sequence of the US genome region of ILTV (GenBank accession no. U28832) (58). Five micrograms of late RNA (γ2) of ILTV-infected CEK cells was hybridized with 2.5 pmol of the primer IgJ-SR (reverse of nucleotides 11361 to 11379) and incubated for 1 h at 42°C with 200 U of reverse transcriptase (SuperScript II; Invitrogen). After inactivation for 15 min at 70°C, the template RNA was digested with RNases H and T1 for 30 min at 37°C. Then, an aliquot of the cDNA was amplified by PCR with primers IgJ-SR and IgJ-SF (nucleotides 9095 to 9113) by using Pfx DNA polymerase (Invitrogen). An initial denaturation step for 3 min at 95°C was followed by 30 cycles of 95°C for 15 s, 50°C for 30 s, and 72°C for 2 min, with a final elongation step at 72°C for 10 min (Primus 96 Thermocycler; MWG Biotech). After treatment with Klenow polymerase and polynucleotide kinase, the major 1,163-bp PCR product was purified from an agarose gel (QIAquick gel extraction kit; QIAGEN, Hilden, Germany), cloned into SmaI-digested plasmid pUC19, and analyzed by DNA sequencing.
Construction and applications of expression plasmids.
To permit in vitro transcription and transient expression of ILTV gJ in eukaryotic cells, the coding sequence (ORF 5) (58) was cloned into plasmid vector pRc-CMV (Invitrogen). For that purpose, the 5′-terminal 423 codons of ORF 5 were amplified from genomic ILTV DNA by PCR with primers IgJ-5F (GTGAAGCTTATTTCGCCGAGAGATGGGGAC) and IgJ-5R (GTTCTAGACAGTGTATTTTCTGACTCACCG), which contained artificial restriction sites (in italics) and the authentic start codon (underlined) of the gJ gene. The 1,301-bp amplification product was doubly digested with HindIII and XbaI and inserted into the appropriately cleaved vector. Subsequently, the 3′-terminal part of the cloned PCR product was replaced by an overlapping 3,070-bp HpaI-XbaI fragment of cloned ILT virion DNA. Thus, the resulting plasmid pRc-IgJC (Fig. 1D) contained the complete genomic gJ ORF of ILTV, which consists of 986 codons (58). To obtain an expression construct of the spliced gJ mRNA species, a 2,181-bp SgrAI-NheI fragment of pRc-IgJC was replaced by the corresponding 1,059-bp fragment of cloned gJ cDNA (see above). Consequently, in pRc-IgJS (Fig. 1D) the modified gJ ORF encompassed only 612 codons. The predicted gI gene homologue of ILTV (58) was cloned in expression plasmid pcDNA3 (Invitrogen) after PCR amplification of the entire ORF with primers IgI-F (CACAAGCTTCGAAAAGCATGGCATCG) and IgI-R (CACTCGAGCCACA TTCAGACTTAATCAC), followed by double digestion of the vector and the 1,130-bp PCR product with HindIII and XhoI (restriction sites are in italics and the start codon is underlined). To generate antisense cRNA probes for Northern blot hybridization, the resulting plasmid pcDNA-IgI, as well as pRc-IgJ and pRc-ICP4 (10) were transcribed with SP6 RNA polymerase (Roche) in the presence of [α-32P]UTP (MP Biochemicals, Eschwege, Germany). After transcription with T7 RNA polymerase, pRc-IgJC and pRc-IgJS were translated in vitro (TNT Coupled Reticulocyte Lysate System; Promega, Mannheim, Germany) in the presence of [35S]methionine (MP Biochemicals). Transient expression of gJ under control of the human cytomegalovirus immediate-early promoter (P-HCMV) was analyzed 24 to 48 h after calcium phosphate-mediated transfection (12) of LMH cells with plasmids pRc-IgJC or pRc-IgJS, respectively.
Generation of ILTV recombinants.
Plasmid pBl-GFP (9) which contains an EGFP expression cassette (Clontech, Heidelberg, Germany) inserted at the SmaI site within the polylinker of pBluescript SK(−) (Stratagene, Amsterdam, The Netherlands) was used for deletion of the gJ gene. To this end, a genomic 1,294-bp EcoRI-XbaI fragment containing the 3′ end of the ILTV gJ gene and downstream sequences was inserted downstream of the reporter gene cassette after digestion of pBl-GFP with AflII and XbaI and blunt-ending with Klenow polymerase, followed by double digestion with KpnI and HindIII to permit insertion of a 1,720-bp PCR product of ILTV DNA sequences from upstream of the gJ gene at the other side of the EGFP expression cassette. Amplification primer IΔgJ-F (AGGGTACCTGGCATGAGCGACATATAC) contained a vector-compatible KpnI-site (in italics), whereas the other terminus determined by primer IΔgJ-R (TTTACGGTTTCACCCGCAC) was fused to the HindIII site of the vector after Klenow treatment. LMH cells were cotransfected (12) with the resulting transfer plasmid pBl-ΔgJG, genomic DNA of ILTV-A489, and an expression plasmid encoding the UL48 homologue of ILTV which enhances the infectivity of virion DNA (10). EGFP-expressing ILTV recombinants were purified from transfection progeny, and a single plaque isolate, designated ILTV-ΔgJG (Fig. 1C), was further analyzed.
To generate a gJ deletion mutant of ILTV without reporter gene insertion, the EGFP ORF and the preceding promoter (P-HCMV) were removed from pBl-ΔgJG by double digestion with EcoRI and BsrGI, Klenow treatment, and religation. After cotransfection of cells with the resulting plasmid pBl-ΔgJ and virion DNA of ILTV-ΔgJG, the nonfluorescent virus recombinant ILTV-ΔgJ (Fig. 1C) could be plaque purified. In a similar manner, the gJ rescue mutant ILTV-gJR (Fig. 1B) was derived from ILTV-ΔgJG by using transfer plasmid pBl-IH6598, which contained a 6,598-bp HindIII fragment of wild-type ILTV DNA in pBluescript SK(−). All generated ILTV recombinants were characterized by restriction analyses and Southern blot hybridization of virion DNA (results not shown).
Growth kinetics and plaque assays.
CEK cells were inoculated at an MOI of 0.1, and growth kinetics were assayed as described (35, 53). Progeny virus plaques were visualized by IIF with an ILTV gC-specific MAb (51, 52) or autofluorescence (for ILTV-ΔgJG). For each time point after infection, mean progeny titers of two independent experiments were calculated. Furthermore, 30 plaques per virus mutant were measured microscopically, and average diameters as well as standard deviations were calculated.
Virion preparation, glycosidase and tunicamycin treatment, and Western blot analyses.
ILT virions were purified from the supernatants of infected CEK cells by sucrose step-gradient centrifugation and incubated with neuraminidase and O-glycosidase or with N-glycosidase F (all enzymes purchased from Roche, Mannheim, Germany) (9). To inhibit N glycosylation, CEK and LMH cells were incubated for 24 h in the presence of 2 μg of tunicamycin (Sigma, Taufkirchen, Germany) per ml, which was added immediately after infection or transfection. Virion proteins (ca. 5 μg per lane), lysates of infected or transfected cells (ca. 104 cells per lane), and in vitro translation products were incubated in sample buffer containing 10% β-mercaptoethanol for 5 min at 95°C and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in discontinuous gels according to standard protocols (26). Proteins were transferred to nitrocellulose filters (Trans-Blot SD cell; Bio-Rad, München, Germany), and the blots were incubated with ILTV-specific MAb (51) against gJ (MAb 106, 1:5 dilution) and gC (MAb 201, 1:50 dilution) or with monospecific rabbit antisera against the UL31 (1:1,000 dilution), UL47 (1:100,000 dilution), and UL49.5 (1:5,000 dilution) gene products of ILTV (W. Fuchs and D. Wiesner, unpublished results). Antibody binding was detected with peroxidase-conjugated species-specific secondary antibodies (Dianova, Hamburg, Germany) and visualized by chemiluminescence (9).
Animal experiments.
White leghorn chickens were bred from specific pathogen-free eggs (Lohmann Tierzucht, Cuxhaven, Germany). At the age of 8 weeks, three groups of 12 chickens were infected intratracheally with ca. 2 × 104 PFU per animal of either wild-type ILTV-A489, ILTV-ΔgJ, or ILTV-gJR. For 10 days, chickens were observed daily for clinical symptoms. Clinical scores were determined by classifying the animals as healthy (0), slightly ill (1: occasional coughing, gasping or sneezing; general condition not affected), ill (2: permanent respiratory disorder, rhinitis; animals depressed), severely ill (3: marked dyspnea, expectoration of bloody mucus, open beaks; animals exhausted), or dead (4). For all animals of each group, the mean values were calculated for each day (daily scores) and for the period of days 1 to 10 after ILTV infection (total scores). For the determination of total scores, dead animals were considered until the end of the monitoring period but were no longer included in the daily scores after the day of death. At days 3 and 4 postinfection (p.i.), one animal of each group was necropsied and investigated for pathological alterations. Tracheal swabs were taken 3, 4, and 5 days p.i. and incubated in cell culture medium for 2 h at room temperature. After ultrasonic treatment and freeze-thawing, the suspensions were serially diluted, and virus titers were determined by plaque assays on CEK cells. Chicken sera collected before infection and at days 17 and 24 p.i. were tested by IIF tests for ILTV-specific antibodies on infected CEK cells (29) and for gC- and gJ-specific antibodies on LMH cells that had been fixed 48 h after transfection with pcDNA-gC (51) or pRc-IgJC. At day 28 p.i., all survivors, as well as five nonimmunized chickens, were intratracheally infected with 105 PFU of the virulent ILTV strain A489. Again, clinical signs were monitored for 10 days, and tracheal swabs were taken at days 3, 4, and 5 post-challenge infection (p.c.) to reisolate ILTV. Single animals of each group were necropsied and dissected at days 3 and 4 p.c., and after 28 days all remaining animals were killed and investigated for pathological alterations. For antibody detection sera were prepared at days 11 and 28 p.c.
RESULTS
Transcription patterns of the ILTV gJ gene.
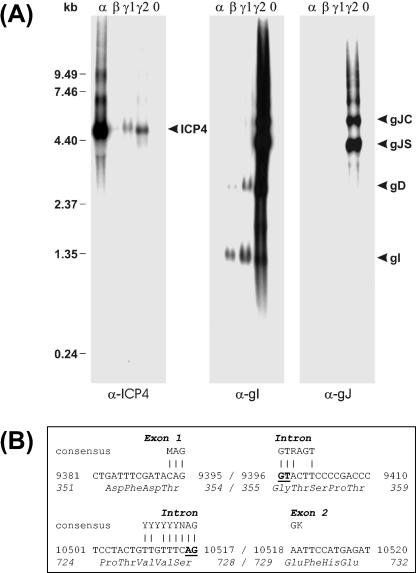
DNA sequence analysis of the US genome region of ILTV (58) indicated that the ORF 5 gene encoding gJ forms a 3′ coterminally transcribed gene cluster with the downstream gD and gI genes (Fig. 1B). The minimum transcript sizes calculated from the distance between the ATG initiation codons of the three genes and the next putative polyadenylation signal (AATAAA) immediately downstream of the gI ORF are listed in Table 1. To verify these predictions, Northern blot analyses were performed with total RNA from ILTV-infected cells (Fig. 2). To accumulate viral immediate-early transcripts (α) protein synthesis was blocked by incubation of the cells in the presence of cycloheximide. To identify early mRNAs (β) DNA replication was inhibited by phosphonoacetic acid. Late transcripts (γ1 and γ2) of ILTV could be detected in cells infected for 6 and 16 h without any drugs. As a control, one blot was hybridized with a labeled antisense cRNA of the gene encoding the ILTV homologue of the major immediate-early protein ICP4 of HSV-1 (18). In agreement with earlier results (60), the 4.5-kb ICP4 mRNA of ILTV was most abundantly expressed under immediate-early conditions (Fig. 2A, α-ICP4). In contrast, the viral transcripts detected by a labeled SP6 cRNA probe of pcDNA-IgI were not present in cycloheximide-treated cells (Fig. 2A, α-gI). Two transcripts of 1.3 and 2.8 kb, which fit the predicted sizes of coterminal gI and gD mRNAs (Table 1), were first detectable under early conditions but became more prominent when viral DNA replication was not inhibited (Fig. 2A, α-gI). Only at very late times after infection (16 h p.i.) were two strongly expressed ILTV-specific RNAs of 4.3 and 5.5 kb (gJS and gJC) also detected by the gI-specific probe (Fig. 2A, α-gI) and by a similarly prepared antisense cRNA of pRc-IgJC (Fig. 1D; Fig. 2A, α-gJ). Thus, both viral RNAs contained gJ coding sequences, but only the 5.5-kbp transcript had the required size to encode the complete gJ ORF (Table 1). The more abundant 4.3-kb RNA might be either the result of transcriptional initiation at a second, internal site or of mRNA splicing.
TABLE 1.
Properties of the analyzed mRNAs and gJ proteins of ILTVa
| Gene | gJ
|
gD | gI | |
|---|---|---|---|---|
| Unspliced | Spliced | |||
| mRNA size (kb) | ||||
| Calculated | >5.3 | >4.2 | >2.5 | >1.1 |
| Detected | 5.5 | 4.3 | 2.8 | 1.3 |
| Protein (no. of amino acids) | 985 | 611 | 434 | 362 |
| Molecular mass (kDa) | ||||
| Calculated | 106.52 | 66.82 | 48.47 | 39.74 |
| In vitro translation product | 140 | 62 | NI | NI |
| Plasmid-transfected cells | 200, 160, 115 | 85 | NI | NI |
| ILTV-infected cells | 200, 160, 115 | 85 | NI | NI |
| ILT virions | 200, 115 | 85 | NI | NI |
Calculated sizes of viral transcripts and of primary translation products were deducted from the published DNA sequence (58). Apparent mRNA sizes were determined by Northern blot hybridization, and apparent molecular masses of proteins were estimated by SDS-PAGE and Western blot analyses. The gD and gI proteins of ILTV have not yet been identified (NI).
FIG. 2.
RNA analyses. (A) CEK cells were infected at an MOI of 5 with ILTV-A489 and incubated for 6 h in the presence of cycloheximide (α), for 16 h in the presence of phosphonoacetic acid (β), and for 6 (γ1) or 16 h (γ2) without any drugs. Total RNA was separated in denaturing agarose gels, and Northern blots were hybridized with 32P-labeled antisense RNAs of the major immediate-early gene encoding ICP4 or of the gI and gJ genes. Positions of molecular mass markers and of detected viral transcripts are indicated. (B) Sequencing of a cloned cDNA of the smaller gJ mRNA (gJS) revealed removal of a 1,122-bp intron. Consensus sequences of eukaryotic splice donor and acceptor sites are shown, and completely conserved nucleotides are underlined (3). Nucleotide and amino acid numbering refers to the published DNA sequence (58).
To test the latter hypothesis, the viral DNA sequence (58) was analyzed for putative splice donor and splice acceptor sites (3), and the oligonucleotide primers IgJ-SF and IgJ-SR (Fig. 1B) were deduced that allowed reverse transcription and PCR amplification of the respective part of the gJ mRNAs from total late RNA of ILTV-infected cells. After PCR, only traces of the expected 2.3-kbp product were detectable, whereas a major portion of the synthesized DNA was 1.2 kbp in size. Cloning and sequencing of the latter product revealed removal of a 1,122-bp intron that contained highly conserved nucleotide doublets at its 5′ and 3′ends (Fig. 2B, underlined). However, the surrounding bases did not perfectly match the consensus sequences of splice donor and splice acceptor sites of higher eukaryotes (Fig. 2B) (3), which might explain why only a portion of the gJ mRNA was found to be spliced. Intron removal resulted in an in-frame deletion of amino acids 355 to 728 of the full-length gJ protein (Fig. 2B). Besides the spliced and unspliced gJ mRNAs (gJS and gJC), the Northern blot hybridization probes detected low amounts of even larger ILTV-specific RNAs (Fig. 2A). Most likely, these transcripts resulted from occasional read-through of the polyadenylation signals located downstream of the gG or gI genes, respectively (Fig. 1B). Thus, our analyses confirmed the predicted 3′-coterminal transcription of the gJ, gD, and gI genes and identified an additional spliced mRNA of the gJ gene.
Expression and processing of the gJ proteins.
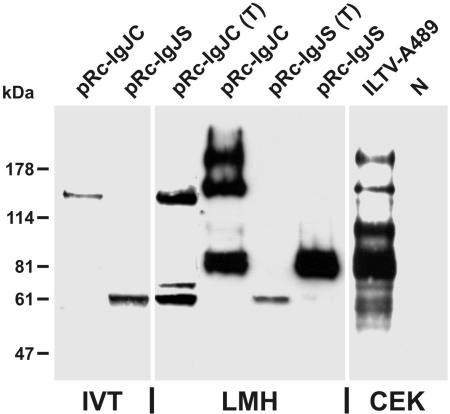
As described earlier, the protein products of the gJ gene detected in ILTV-infected cells exhibit molecular masses of 85, 115, 160, and 200 kDa (51). However, the deduced products of the two detected gJ mRNAs had sequences of 985 and 611 amino acids, corresponding to masses of 107 and 67 kDa, respectively (Table 1). For unknown reasons, the apparent molecular mass of the in vitro translation product of expression plasmid pRc-IgJC (Fig. 1D) containing the genomic gJ ORF was considerably larger than calculated, whereas the 62-kDa product of the spliced gJ RNA species cloned in pRc-IgJS (Fig. 1D) appeared slightly smaller than predicted (Fig. 3 and Table 1). A similar 62-kDa protein product was also detected with gJ-specific MAb (51) in Western blot analyses performed after transfection of LMH cells with pRc-IgJS in the presence of tunicamycin, which inhibits N glycosylation (Fig. 3). A second, weaker protein band at 70 kDa might have been caused by O glycosylation or other posttranslational modifications of the primary translation product. After transfection of tunicamycin-treated cells with pRc-IgJC, a 140-kDa protein corresponding to the in vitro translation product of the full-length gJ gene was additionally detected (Fig. 3). However, the concomitant presence of the smaller protein species indicated that the plasmid-derived gJ mRNA, like the viral transcript, was partially spliced in eukaryotic cells. In LMH cells transfected in the absence of drugs, the product of pRc-IgJS was further processed, leading to a diffuse protein band at ca. 85 kDa, which was similar to the major gJ-gene product in ILTV-infected CEK cells (Fig. 3 and Table 1). In untreated LMH cells transfected with pRc-IgJC, similar gJ gene products were detectable as in ILTV-infected CEK cells, although the 115-kDa species was less pronounced after transfection (Fig. 3 and Table 1). In transfected as well as in infected cells, traces of the 62-, 70-, and 140-kDa precursor proteins were also found after incubation in the absence of tunicamycin (Fig. 3). After tunicamycin treatment of infected CEK cells, viral gene expression overall was impaired, but mature input virus proteins were still detectable (results not shown).
FIG. 3.
In vitro translation (IVT) and expression of gJ in cell culture. Plasmids pRc-IgJC and pRc-IgJS were transcribed and translated in the presence of [35S]methionine. LMH cells transfected with pRc-IgJC or pRc-IgJS were incubated for 24 h in the presence (T) or absence of tunicamycin. CEK cells were also incubated for 24 h after infection with ILTV-A489 at an MOI of 2. Lysates of infected and noninfected (N) cells, transfected cells, and in vitro translation products were separated by SDS-PAGE and transferred to nitrocellulose filters. Binding of a gJ-specific MAb was visualized by a chemiluminescence reaction of peroxidase-conjugated secondary antibodies. The signals of radiolabeled proteins were enhanced by fluorography. Different exposures of the same blot were combined to achieve comparable signals. Locations of marker proteins are indicated.
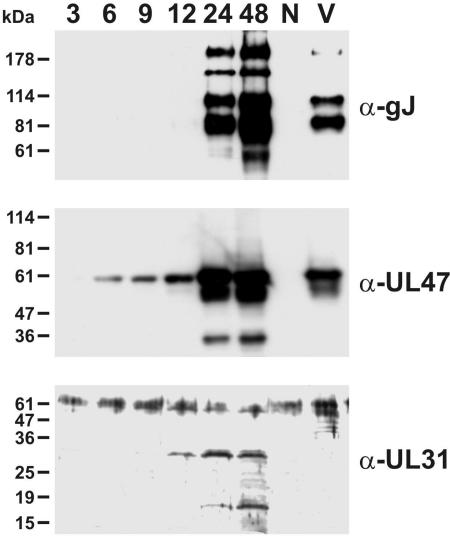
Western blot analyses of chicken cells harvested at different times after ILTV infection confirmed that gJ is a true late gene, since newly synthesized proteins were clearly detectable by specific MAb not before 24 h p.i. (Fig. 4, upper panel). In contrast, the UL47 and UL31 gene products of ILTV were detected by monospecific antisera (D. Wiesner, J. Veits, T. C. Mettenleiter, and W. Fuchs, unpublished results) from 6 or 12 h p.i., respectively (Fig. 4, lower panels). Like the UL47 protein, but unlike the UL31 gene product, different forms of gJ were present in sucrose gradient-purified ILT virions (Fig. 4). Apparently, only the 160-kDa gJ gene product was not incorporated into virus particles, indicating that this protein might represent an immature processing intermediate.
FIG. 4.
Expression kinetics of gJ. CEK cells were infected at an MOI of 2 with ILTV-A489. Proteins of cells harvested 3, 6, 9, 12, 24, and 48 h p.i., of noninfected cells (N), and of sucrose gradient-purified ILT virions (V) were separated by SDS-PAGE, and Western blots were analyzed with a gJ-specific MAb (α-gJ) or with monospecific antisera against a tegument protein (α-UL47) or a nonstructural protein (α-UL31). Locations of marker proteins are indicated.
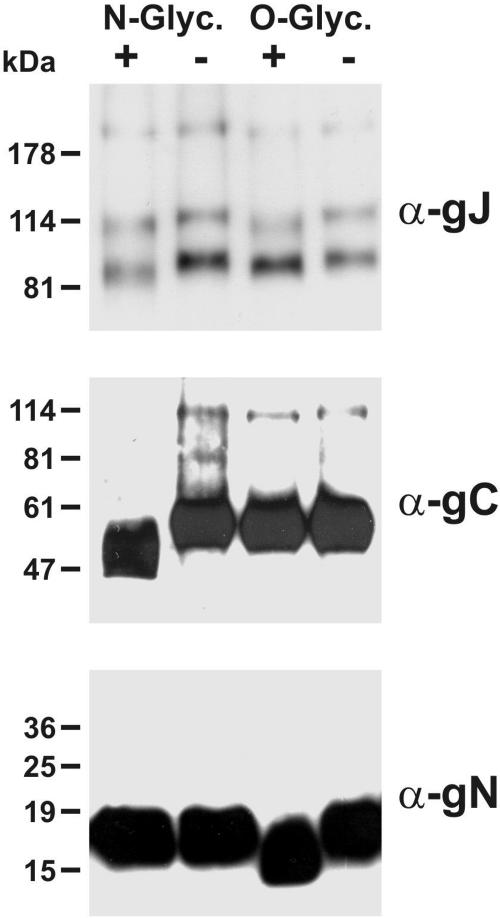
As already demonstrated by sensitivity to tunicamycin treatment, processing of ILTV gJ includes glycosylation. This could be verified by incubation of purified ILT virions with different glycosidases and subsequent Western blot analyses with a gJ-specific MAb. After treatment with N-glycosidase F, as well as with neuraminidase and O-glycosidase, the electrophoretic mobility of the 85- and 115-kDa gJ proteins was increased in comparison to the mobility in virions that were incubated without the enzymes (Fig. 5, upper panel). Thus, ILTV gJ contains asparagine, as well as serine- and/or threonine-linked sugar chains. The effects of glycosidases on the 200-kDa form of gJ were less evident, which might be explained in part by the size of the protein or by incomplete digestion. To test the enzyme activities, parallel blots were incubated with antibodies against gC (Fig. 5, middle panel), or gN (Fig. 5, lower panel). These proteins appeared to be only N or only O glycosylated, respectively (51; W. Fuchs and T. C. Mettenleiter, unpublished results).
FIG. 5.
Glycosidase treatment. Purified ILT virions were treated with N-glycosidase F or, consecutively, with neuraminidase and O-glycosidase (+). As controls, virions were similarly incubated without enzymes (−). Western blots were probed with a MAb against gJ, with a monospecific antiserum against gN, or with a gC-specific MAb. Locations of marker proteins are indicated.
Isolation and in vitro growth properties of gJ mutants of ILTV.
To analyze the functions of the ILTV gJ protein, two deletion mutants were generated by homologous recombination between virion DNA and transfer plasmids in transfected chicken cells. Isolation of the first virus recombinant (Fig. 1C, ILTV-ΔgJG) was facilitated by concomitant insertion of a reporter gene cassette encoding EGFP under control of a strong promoter (P-HCMV) and subsequent plaque purification from green fluorescent cells. However, since previous studies had demonstrated that overexpression of foreign proteins including EGFP might negatively affect replication of ILTV (10, 52, 53), a second recombinant was derived from ILTV-ΔgJG by purification of nonfluorescent virus plaques. The resulting virus (Fig. 1C, ILTV-ΔgJ) lacked P-HCMV and the EGFP gene but retained a foreign DNA fragment of 272 bp that represented the polyadenylation signal of simian virus 40 (SV40-A+) and served as a positive genome marker of the desired ILTV recombinant. To verify that possible replication defects of the deletion mutants were really caused by the absence of gJ, a rescue mutant (Fig. 1B, ILTV-gJR) was also derived from ILTV-ΔgJG.
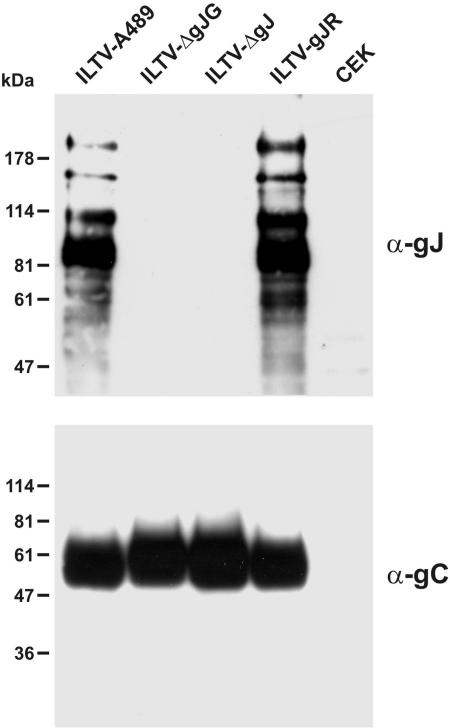
Virus mutants ILTV-ΔgJG and ILTV-ΔgJ exhibited identical deletions of ORF 5 codons 1 to 729. The 3′-terminal part of the gJ gene was preserved since it overlaps with the promoter region and coding sequences of the gD gene (Fig. 1B) (58). Although the remaining part of the ORF contained three in-frame ATG codons, expression of functional proteins appeared very unlikely. In Western blot analyses of infected cell lysates of ILTV-ΔgJG and ILTV-ΔgJ with gJ-specific MAb (Fig. 6, upper panel), neither full-length nor truncated gene products were detectable, whereas the gJ proteins of ILTV-gJR were indistinguishable from those of wild-type ILTV-A489. Incubation of a parallel blot with a gC-specific MAb (Fig. 6, lower panel) confirmed that comparable amounts of protein were loaded in all lanes and that viral gene expression was not generally affected in the absence of gJ.
FIG. 6.
Protein expression of gJ mutants. CEK cells were harvested 24 h p.i. (MOI of 2) with ILTV-A489, ILTV-ΔgJG, ILTV-ΔgJ, or ILTV-gJR and separated by SDS-PAGE. Western blots were probed with MAb against gJ or gC. Marker proteins are indicated on the left.
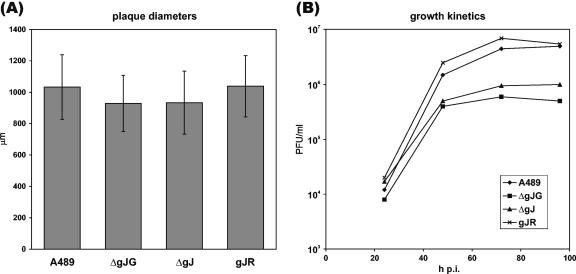
Successful isolation of gJ-negative ILTV mutants had already demonstrated that gJ is dispensable for virus replication in cell culture. More detailed studies revealed that deletion of gJ had only minor effects on the cell-to-cell spread of ILTV, since average plaque diameters of ILTV-ΔgJG and ILTV-ΔgJ on CEK cells were reduced by only ca. 10% compared to ILTV-gJR and wild-type virus (Fig. 7A). However, gJ appears to be more relevant for formation of infectious virus particles, since multistep growth studies reproducibly showed that maximum virus titers of ILTV-ΔgJG and ILTV-ΔgJ were ca. 10-fold lower than those of ILTV-A489 and ILTV-gJR (Fig. 7B). Because of the low titers of the investigated deletion mutants, one-step growth kinetics could not be determined.
FIG. 7.
In vitro growth properties of gJ mutants. (A) For analysis of cell-to-spread, CEK cells infected with ILTV-A489, ILTV-ΔgJG, ILTV-ΔgJ, and ILTV-gJR were incubated for 48 h under semisolid medium. Virus plaques were visualized either by GFP-autofluorescence or by indirect immunofluorescence reactions of a gC-specific MAb, and average diameters of 30 plaques per virus, as well as standard deviations, were calculated. (B) For analysis of growth kinetics, cells were infected at an MOI of 0.1 and harvested together with the supernatant after 24, 48, 72, and 96 h. Mean progeny virus titers of two experiments were determined by plaque assays.
Relevance of gJ for virulence of ILTV in chickens.
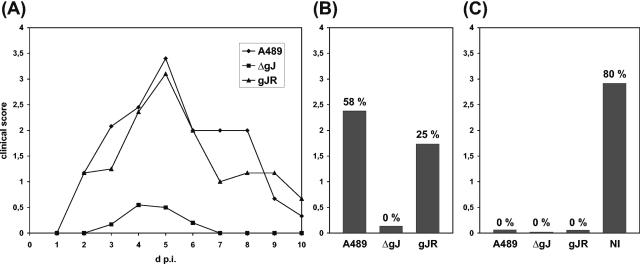
Since it could be detected that in vitro replication of ILTV was affected in the absence of gJ, this protein might also be required for efficient virus replication in its natural host. To test this hypothesis, three groups of 12 8-week-old chickens were intratracheally infected with ca. 2 × 104 PFU per animal of either wild-type ILTV-A489, deletion mutant ILTV-ΔgJ, or rescue mutant ILTV-gJR. From day 1 to 10 after infection, clinical signs of ILT, such as respiratory disorder and depression, were monitored and scored (Table 2 and Fig. 8A and B). All animals infected with wild-type virus or ILTV-gJR developed severe disease leading to mortality rates of 58 or 25%, respectively (Fig. 8A and B). In contrast, all chickens survived infection with ILTV-ΔgJ, and only 7 of 12 animals were slightly ill for a few days, whereas the others remained healthy (Table 2 and Fig. 8A). The different clinical scores correlated with the severity of lesions observed in larynx, trachea, and lung of dissected animals (results not shown). However, in tracheal swabs that were taken at days 3, 4, and 5 p.i., virus could be reisolated from almost all animals of each group, although the mean titer of shed ILTV-ΔgJ was ca. 10-fold lower than that of ILTV-gJR and ILTV-A489 (Table 2).
TABLE 2.
Summary of animal experimentsa
| Parameter | Time scale (days) | No. of chicks scoring/no. tested | ||||||
|---|---|---|---|---|---|---|---|---|
| Primary infection | 0 | ILTV-A489 | ILTV-ΔgJ | ILTV-gJR | Control | |||
| PFU/animal | 2 × 104 | 2 × 104 | 2 × 104 | |||||
| Morbidity | 1-10 p.i. | 12/12 | 7/12 | 12/12 | 0/5 | |||
| Clinical scores | 2.38 | 0.14 | 1.74 | |||||
| Mortality | 3-8 p.i. | 7/12 | 0/12 | 3/12 | 0/5 | |||
| Virus shedding | 3-5 p.i. | 9/9 | 8/10 | 9/9 | NT | |||
| PFU/ml | 4.3 × 103 | 4.5 × 102 | 6.6 × 103 | |||||
| ILTV-specific Ab | 17 p.i. | 3/3 | 9/10 | 7/7 | NT | |||
| +++ | + | +++ | ||||||
| 24 p.i. | 3/3 | 9/10 | 7/7 | NT | ||||
| +++ | ++ | +++ | ||||||
| gC-specific Ab | 24 p.i. | 3/3 | 8/10 | 7/7 | NT | |||
| ++ | + | ++ | ||||||
| gJ-specific Ab | 24 p.i. | 3/3 | 0/10 | 7/7 | NT | |||
| +++ | +++ | |||||||
| Challenge infection | 28 p.i. | ILTV-A489
|
||||||
| PFU/animal | 1 × 105
|
|||||||
| Morbidity | 1-10 p.c. | 1/3 | 1/10 | 2/7 | 5/5 | |||
| Clinical scores | 0.07 | 0.02 | 0.06 | 2.92 | ||||
| Mortality | 4-7 p.c. | 0/3 | 0/10 | 0/7 | 4/5 | |||
| Virus shedding | 3-5 p.c. | 0/3 | 0/8 | 0/6 | 4/4 | |||
| PFU/ml | 1.8 × 103 | |||||||
| ILTV-specific Ab | 11 p.c. | 3/3 | 8/8 | 6/6 | 1/1 | |||
| +++ | +++ | +++ | ++ | |||||
| 28 p.c. | 3/3 | 8/8 | 6/6 | 1/1 | ||||
| +++ | +++ | +++ | +++ | |||||
| gC-specific Ab | 11 p.c. | 3/3 | 8/8 | 6/6 | NT | |||
| ++ | ++ | ++ | ||||||
| gJ-specific Ab | 11 p.c. | 3/3 | 6/8 | 6/6 | NT | |||
| +++ | + | +++ | ||||||
Chickens were intratracheally infected with ILTV-A489, ILTV-ΔgJ, or ILTV-gJR. After 28 days immunized and nonimmunized control animals were challenged with a high dose of ILTV-A489. For 10 days after immunization (p.i.), and after challenge infection (p.c.), animals were examined daily to determine morbidity and mortality rates, as well as clinical scores. Shedding of ILTV was detected by plaque assays of tracheal swabs on CEK cells, and mean virus titers were calculated. At the indicated times sera were analysed by IIF tests for antibodies (Ab) against total ILTV proteins, or glycoproteins gC and gJ, respectively. Signal strengths were roughly quantified by 1 to 3 plus signs. Some parameters were not tested (NT) in all groups.
FIG. 8.
Virulence of gJ mutants. Three groups of chickens were intratracheally infected with ca. 2 × 104 PFU of ILTV-A489, ILTV-ΔgJ, or ILTV-gJR. After 4 weeks the immunized chickens and nonimmunized (NI) control animals were challenged with 105 PFU of ILTV-A489. From day 1 to 10 after each infection, the animals were daily classified as healthy (0), slightly ill (1), ill (2), severely ill (3), or dead (4) (for details, see text). The mean clinical scores of each group were calculated. The daily scores after primary infection (A), as well as the cumulative scores after primary infection (B) and after challenge infection (C) were plotted. Mortality rates are indicated above the bars.
Previous studies indicated that gJ might be a dominant antigen for the humoral immune response against ILTV (51). Therefore, sera were collected from all animals at days 17 and 24 p.i. with the gJ-negative and gJ-positive viruses and analyzed in parallel by IIF tests. On fixed monolayers of wild-type ILTV-infected CEK cells, the fluorescence intensities observed with sera from ILTV-ΔgJ-infected animals were generally lower than those of sera from chickens infected with ILTV-gJR or ILTV-A489 (Table 2). Nevertheless, 90% of the ILTV-ΔgJ-specific sera exhibited unequivocally positive reactions. Similar results were obtained by IIF tests on LMH cells transfected with a gC expression plasmid (51), indicating that this glycoprotein is also an important antigen of ILTV (Table 2). As expected, serum antibodies of chickens infected with ILTV-ΔgJ did not bind to pRc-IgJC transfected cells, whereas the serum samples of all other animals showed strong reactions with the transiently expressed gJ protein (Table 2).
To test whether the immune response induced by gJ-negative ILTV was sufficient to confer protection against subsequent infection with virulent wild-type virus, immunized chickens and five nonvaccinated control animals were intratracheally challenged with high doses (105 PFU per animal) of ILTV-A489 at day 28 after primary infection. As expected, all nonvaccinated chickens developed severe clinical symptoms, shed high amounts of virus, and four of five animals died within 7 days p.i. (Table 2 and Fig. 8C). In contrast, the chickens that had been previously immunized with either ILTV-A489, ILTV-gJR, or ILTV-ΔgJ showed no or only minor clinical signs; all animals survived the infection, and no shedding of challenge virus could be detected (Table 2 and Fig. 8C). Concordantly, no significant pathological alterations were detectable in respiratory tissues of vaccinated animals that were necropsied and dissected at days 3 and 4 p.c., whereas control animals exhibited hemorrhagic inflammations of the tracheas (data not shown). Antisera prepared at days 11 and 24 p.c. from chickens immunized with ILTV-498 or ILTV-gJR showed similar reactions in IIF tests with ILTV-infected cells or transiently expressed gC and gJ as after vaccination (Table 2). In contrast, after wild-type virus challenge of chickens immunized with ILTV-ΔgJ, serum reactions were enhanced with respect to infected cells and gC, and 80% of the animals additionally produced detectable amounts of gJ-specific antibodies (Table 2).
In summary, the animal experiment demonstrated that gJ-negative ILTV is attenuated in chickens but able to confer protective immunity after live-virus vaccination. Furthermore, the gJ protein might be suitable as a marker that permits serological differentiation of field virus-infected and vaccinated animals.
DISCUSSION
In this study we investigated expression, structure, and in vitro as well as in vivo functions of the products of the ORF 5 gene of the US genome region of ILTV (58). Positional homologues of this gene were found only in several alphaherpesvirus genomes, including those of HSV-1, HSV-2, EHV-1, EHV-4, and PsHV-1 (6, 32, 46, 47, 48). Other alphaherpesviruses like varicella zoster virus, PrV, and BHV-1 do not possess corresponding genes (5, 24, 42). The deduced products of all ORF 5 homologues exhibit characteristics of membrane glycoproteins, but their sizes range from 92 amino acids in herpes simplex viruses to 985 amino acids in ILTV, and sequence conservation is hardly detectable. The gene product of HSV-1, a minor N-glycosylated virion protein, was named gJ (11, 38). The respective gene product of EHV-1, gp2, is one of the most immunogenic envelope proteins (1, 44, 55) that is heavily O glycosylated and partially cleaved by proteases during processing (27, 55, 56, 57). The 85-, 115-, 160-, and 180-kDa ORF 5 gene products of ILTV also represent immunodominant proteins (51), which contain both N-, and O-linked carbohydrate chains.
Since the major 85-kDa form of ILTV gJ appeared significantly smaller than calculated, gJ, like its EHV-1 homologue, might be processed by proteolytic cleavage. However, the amino acid sequence around the cleavage site of gp2 is not conserved in gJ. Northern blot hybridization and cDNA analyses demonstrated the presence of two gJ-specific transcripts. The larger one contained the genomic ORF 5, whereas the smaller one lacked a 1,122-bp intron sequence leading to an in-frame deletion of codons 355 to 728. Posttranscriptional modification by mRNA splicing is common in higher eukaryotes but only rarely used by herpesviruses. In HSV-1, splicing has been described only for the mRNAs of the terminase encoded by UL15, of the immediate early proteins ICP0, ICP22, and ICP47, and for the latency-associated transcripts (38). However, mRNA splicing has not been detected for the gJ mRNA of HSV-1 or for its homologue in EHV-1 and EHV-4.
Previously described split genes of ILTV include the homologue of HSV-1 UL15 and the duplicated positional counterparts of the ICP0 gene, UL0 and UL[-1] (8, 60). When compared to these genes, gJ is peculiar in the simultaneous presence of spliced and unspliced mRNAs that are translated into different proteins. Expression of the plasmid-cloned gJ gene in cultured chicken cells indicated that the partial mRNA splicing and subsequent processing of the translation products are independent of other viral factors. Thus, incomplete processing of the gJ mRNA by cellular spliceosomes is presumably the consequence of suboptimal splice donor and acceptor sites which do not perfectly match known consensus sequences (3).
Transfection of cells with a plasmid containing the spliced gJ cDNA demonstrated that it only encodes the major 85-kDa form of the protein. Therefore, the 115-, 160-, and 200-kDa proteins are modified translation products of the unspliced mRNA. Remarkably, the 115-kDa protein is smaller than the in vitro translation product of the genomic ORF, indicating that in addition to mRNA splicing, cleavage by a cellular protease at a yet unknown site might also contribute to the complex expression pattern of ILTV gJ. An almost complete inhibition of gJ processing was achieved by the incubation of plasmid-transfected cells in the presence of tunicamycin, an inhibitor of N glycosylation (31). Under these conditions, the two major gene products appeared similar to the in vitro translation products of spliced and unspliced mRNAs, and no mature proteins were detectable. This indicates that N glycosylation in the endoplasmic reticulum might be a prerequisite for gJ transport to the Golgi apparatus, where other modifications including O glycosylation and, possibly, proteolytic cleavage can occur. Remarkably, treatment with either N- or O-glycosidases significantly increased the electrophoretic mobility of the mature 85- and 115-kDa forms of gJ but had only minor effects on the 200-kDa protein. Either a part of the sugar chains are not accessible to the enzymes in this protein, or it contains other uncharacterized modifications.
Northern and Western blot analyses demonstrated that the gJ proteins are encoded by a true late gene and incorporated into virus particles. Virion incorporation and membrane association of ILTV gJ had previously been shown by immunoelectron microscopy (51), but our present studies additionally revealed that ILTV particles contain the 85-kDa product of the spliced, as well as the 115- and 200-kDa products of the unspliced mRNA species. This was not surprising, since both primary translation products contain identical signal sequences and transmembrane domains at their N and C termini, respectively. The 160-kDa form of gJ most likely represents an immature precursor protein since it was only found in infected cells. Presence of the 115-kDa protein in virions indicates that it might be truncated in the N-terminal part but retains the C-terminal membrane anchor. On the other hand, direct membrane association might not be required for all forms of gJ if they are linked in a protein complex with each other or with different envelope proteins. Complex formation has been described for many herpesvirus membrane proteins including homodimers of gB and gC and heterodimers of gE and gI, gH and gL, or gM and gN (15, 20, 38). However, if ILTV gJ forms or is part of a multimeric protein complex, its components are apparently not linked by intermolecular disulfide bonds, since the protein patterns observed after electrophoresis under reducing or nonreducing conditions were very similar (results not shown).
The HSV-1 gJ and EHV-1 gp2 genes are dispensable for virus replication in cell culture (38, 43), as is ORF 5 of ILTV. Thus, ORF 5 is the 10th nonessential ILTV gene to be described after the conserved thymidine kinase, dUTPase, and UL10 genes and the specific ORF A to ORF E and UL0 genes (9, 10, 41, 52, 53). Deletion of ILTV gJ did not significantly affect virus spread in chicken cell monolayers, but led to a ca. 10-fold reduction of maximum virus titers. Similar growth deficiencies were described for gp2-negative EHV-1 and attributed to both impaired virus entry and less efficient viral egress (39, 45). Immunoelectron microscopic analyses of ILTV-ΔgJ did not show any conspicuous defects in virion morphogenesis (results not shown). The role of ILTV gJ during virus entry remains to be investigated. It is also unclear whether the products of the spliced and unspliced gJ mRNAs possess different functions during virus replication.
EHV-1 mutants which completely lacked gp2 or expressed a truncated form of this protein were apathogenic in a mouse model (30, 54). We show that gJ of ILTV is also a virulence factor, since a gJ-negative mutant was strongly attenuated in chickens, the natural host of ILTV. Thus, gJ-negative ILTV mutants might be suitable as live vaccines. Remarkably, avirulence of ILTV-ΔgJ was not associated with a dramatic reduction of virus shedding as found after immunization with attenuated, UL0-deleted ILTV recombinants (52). The more efficient in vivo replication of gJ-negative ILTV might enhance the risk of unwanted virus spread from vaccinated to nonvaccinated animals, but, on the other hand, might facilitate successful vaccination of huge chicken flocks after mass application of low virus doses per animal via aerosol or drinking water. Some conventional ILTV live-virus vaccines are suitable for this mode of administration (2), but it remains to be tested for ILTV-ΔgJ.
Unlike the other known nonessential proteins of ILTV, gJ represents a major surface protein, which induces specific antibodies in experimentally or naturally infected animals (51). In our previous experiments, all tested chicken sera exhibiting positive IIF reactions with ILTV-infected cells also reacted with cells transfected with a gJ expression plasmid. Thus, ILTV-ΔgJ might be used as a vaccine that permits serological discrimination of vaccinated and field virus-infected animals, since, as expected, chickens immunized with the deletion mutant did not produce gJ-specific antibodies. In IIF tests on ILTV-infected cells, the sera of most chickens immunized with ILTV-ΔgJ showed positive reactions although reactivity was generally weaker than in sera of animals immunized with wild-type virus. Presumably, the absence of an immunodominant antigen, as well as the attenuation of the deletion mutant, contributed to this phenomenon. Nevertheless, chickens immunized with gJ-negative ILTV were completely protected against challenge infection with a high dose of pathogenic ILTV. Neither clinical signs nor shedding of the challenge virus could be observed. Thus, gJ-specific antibodies are apparently not required for virus neutralization. This is in agreement with previous results obtained with bursectomized chickens that indicated that the cellular immune response is probably more relevant for protection against ILTV than antibodies (7).
After wild-type challenge the intensity of IIF reactions of antisera collected from chickens immunized with ILTV-ΔgJ increased, and in most animals a gJ-specific antibody response became detectable. Due to limited replication of the challenge virus, the amounts of gJ-specific antibodies were very low, and it is conceivable that in several cases also field virus infections of chickens that were previously immunized with a ΔgJ marker vaccine would remain unrecognized. However, sensitivity of detection of gJ-specific antibodies in chicken sera might be improved by development of blocking enzyme-linked immunosorbent assays using available MAb against gJ (51). Similar tests were successfully applied in combination with gE-negative vaccines of PrV and BHV-1 (22, 49). Thus, our previously described gJ-specific MAb, together with the gJ-negative ILTV recombinants presented in this study, might become useful tools for control of ILT and eventual eradication of ILTV.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (Fu 395).
The authors thank D. Lütticken for providing the parental ILTV strain, R. Riebe for preparation of chicken cell cultures, and H. Granzow for electron microscopic investigations. The technical assistance of C. Ehrlich, P. Meyer, and M. Voss is greatly appreciated.
REFERENCES
- 1.Allen, G. P., and M. R. Yeargan. 1987. Use of λgt11 and monoclonal antibodies to map the genes for the six major glycoproteins of equine herpesvirus 1. J. Virol. 61:2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagust, T. J., and J. S. Guy. 1997. Laryngotracheitis, p. 527-539. In B. W. Calnek, H. J. Barnes, C. W. Beard, L. R. McDougald, and Y. M. Saif (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames, Iowa.
- 3.Breathnach, R., and P. Chambon. 1981. Organization and expression of eucaryotic split genes coding for proteins. Annu. Rev. Biochem. 50:349-383. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 6.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahey, K. J., and J. J. York. 1990. The role of mucosal antibody in immunity to infectious laryngotracheitis virus in chickens. J. Gen. Virol. 71:2401-2405. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs, W., and T. C. Mettenleiter. 1996. DNA sequence and transcriptional analysis of the UL1 to UL5 gene cluster of infectious laryngotracheitis virus. J. Gen. Virol. 77:2221-2229. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs, W., and T. C. Mettenleiter. 1999. DNA sequence of the UL6 to UL20 genes of infectious laryngotracheitis virus and characterization of the UL10 gene product as a nonglycosylated and nonessential virion protein. J. Gen. Virol. 80:2173-2182. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs, W., K. Ziemann, J. P. Teifke, O. Werner, and T. C. Mettenleiter. 2000. The non-essential UL50 gene of avian infectious laryngotracheitis virus encodes a functional dUTPase which is not a virulence factor. J. Gen. Virol. 81:627-638. [DOI] [PubMed] [Google Scholar]
- 11.Ghiasi, H., A. B. Nesburn, S. Cai, and S. L. Wechsler. 1998. The US5 open reading frame of herpes simplex virus type 1 does encode a glycoprotein (gJ). Intervirology 41:91-97. [DOI] [PubMed] [Google Scholar]
- 12.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 13.Guy, J. S., H. J. Barnes, and L. M. Morgan. 1990. Virulence of infectious laryngotracheitis viruses: comparison of modified-live vaccine viruses and North Carolina field isolates. Avian Dis. 34:106-113. [PubMed] [Google Scholar]
- 14.Guy, J. S., H. J. Barnes, and L. Smith. 1991. Increased virulence of modified-live infectious laryngotracheitis vaccine virus following bird-to-bird passage. Avian Dis. 35:348-355. [PubMed] [Google Scholar]
- 15.Handler, C. G., R. J. Eisenberg, and G. H. Cohen. 1996. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J. Virol. 70:6067-6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, M. A., C. T. Prideaux, K. Kongsuwan, M. Sheppard, and K J. Fahey. 1991. Gallid herpesvirus 1 (infectious laryngotracheitis virus): cloning and physical maps of the SA-2 strain. Arch. Virol. 119:181-198. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, M. A., and S. G. Tyack. 1995. Molecular evolution of infectious laryngotracheitis virus (ILTV; gallid herpesvirus 1): an ancient example of the Alphaherpesviridae? Vet. Microbiol. 46:221-231. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, M. A., S. G. Tyack, C. T. Prideaux, K. Kongsuwan, and M. Sheppard. 1995. Nucleotide sequence of infectious laryngotracheitis virus (gallid herpesvirus 1) ICP4 gene. Virus Res. 35:193-204. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, M. A., S. G. Tyack, C. T. Prideaux, K. Kongsuwan, and M. Sheppard. 1997. Nucleotide sequence of the left-terminus of infectious laryngotracheitis virus (Gallid herpesvirus 1) SA-2 strain. Arch. Virol. 142:1903-1910. [DOI] [PubMed] [Google Scholar]
- 20.Jöns, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaashoek, M. J., A. Moerman, J. Madic, F. A. Rijsewijk, J. Quak, A. L. Gielkens, and J. T. van Oirschot. 1994. A conventionally attenuated glycoprotein E-negative strain of bovine herpesvirus type 1 is an efficacious and safe vaccine. Vaccine 12:439-444. [DOI] [PubMed] [Google Scholar]
- 22.Kaashoek, M. J., A. Moerman, J. Madic, K. Weerdmeester, M. Maris-Veldhuis, F. A. Rijsewijk, and J. T. van Oirschot. 1995. An inactivated vaccine based on a glycoprotein E-negative strain of bovine herpesvirus 1 induces protective immunity and allows serological differentiation. Vaccine 13:342-346. [DOI] [PubMed] [Google Scholar]
- 23.Kawaguchi, T., K. Nomura, Y. Hirayama, and T. Kitagawa. 1987. Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res. 47:4460-4464. [PubMed] [Google Scholar]
- 24.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78:424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kongsuwan, K., M. A. Johnson, C. T. Prideaux, and M. Sheppard. 1993. Use of λgt11 and monoclonal antibodies to map the gene for the 60,000 dalton glycoprotein of infectious laryngotracheitis virus. Virus Genes 7:297-303. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Learmonth, G. S., D. N. Love, J. E. Wellington, J. R. Gilkerson, and J. M. Whalley. 2002. The C-terminal regions of the envelope glycoprotein gp2 of equine herpesviruses 1 and 4 are antigenically distinct. Arch. Virol. 147:607-615. [DOI] [PubMed] [Google Scholar]
- 28.Leib, D. A., J. M. Bradbury, C. A. Hart, and K. McCarthy. 1987. Genome isomerism in two alphaherpesviruses: herpesvirus saimiri-1 (herpesvirus tamarinus) and avian infectious laryngotracheitis virus. Arch. Virol. 93:287-294. [DOI] [PubMed] [Google Scholar]
- 29.Lüschow, D., O. Werner, T. C. Mettenleiter, and W. Fuchs. 2001. Protection of chickens from lethal avian influenza A virus infection by live-virus vaccination with infectious laryngotracheitis virus recombinants expressing the hemagglutinin (H5) gene. Vaccine 19:4249-4259. [DOI] [PubMed] [Google Scholar]
- 30.Marshall, K. R., Y. Sun, S. M. Brown, and H. J. Field. 1997. An equine herpesvirus-1 gene 71 deletant is attenuated and elicits a protective immune response in mice. Virology 231:20-27. [DOI] [PubMed] [Google Scholar]
- 31.McDowell, W., and R. T. Schwarz. 1988. Dissecting glycoprotein biosynthesis by the use of specific inhibitors. Biochimie 70:1535-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGeoch, D. J., A. Dolan, S. Donald, and F. J. Rixon. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 181:1-13. [DOI] [PubMed] [Google Scholar]
- 33.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 34.McGeoch, D. J., A. Dolan, and A. C. Ralph. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 74:10401-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623-625. [DOI] [PubMed] [Google Scholar]
- 36.Minson, A. C., A. Davison, R. Eberle, R. C. Desrosiers, B. Fleckenstein, D. J. McGeoch, P. E. Pellet, B. Roizman, and D. M. J. Studdert. 2000. Family Herpesviridae, p. 203-225. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 37.Müller, T., H.-J. Bätza, H. Schlüter, F. J. Conraths, and T. C. Mettenleiter. 2003. Eradication of Aujeszky's disease in Germany. J. Vet. Med. B 50:207-213. [DOI] [PubMed] [Google Scholar]
- 38.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 39.Rudolph, J., and N. Osterrieder. 2002. Equine herpesvirus type 1 devoid of gM and gp2 is severely impaired in virus egress but not direct cell-to-cell spread. Virology 293:356-367. [DOI] [PubMed] [Google Scholar]
- 40.Schnitzlein, W. M., J. Radzevicius, and D. N. Tripathy. 1994. Propagation of infectious laryngotracheitis virus in an avian liver cell line. Avian Dis. 38:211-217. [PubMed] [Google Scholar]
- 41.Schnitzlein, W. M., R. Winans, S. Ellsworth, and D. N. Tripathy. 1995. Generation of thymidine kinase-deficient mutants of infectious laryngotracheitis virus. Virology 209:304-314. [DOI] [PubMed] [Google Scholar]
- 42.Schwyzer, M., and M. Ackermann. 1996. Molecular virology of ruminant herpesviruses. Vet. Microbiol. 53:17-29. [DOI] [PubMed] [Google Scholar]
- 43.Sun, Y., and S. M: Brown. 1994. The open reading frames 1, 2, 71, and 75 are nonessential for the replication of equine herpesvirus type 1 in vitro. Virology 199:448-452. [DOI] [PubMed] [Google Scholar]
- 44.Sun, Y., A. R. MacLean, D. Dargan, and S. M. Brown. 1994. Identification and characterization of the protein product of gene 71 in equine herpesvirus 1. J. Gen. Virol. 75:3117-3126. [DOI] [PubMed] [Google Scholar]
- 45.Sun, Y., A. R. MacLean, J. D. Aitken, and S. M. Brown. 1996. The role of the gene 71 product in the life cycle of equine herpesvirus 1. J. Gen. Virol. 77:493-500. [DOI] [PubMed] [Google Scholar]
- 46.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 47.Telford, E. A., M. S. Watson, J. Perry, A. A. Cullinane, and A. J. Davison. 1998. The DNA sequence of equine herpesvirus-4. J. Gen. Virol. 79:1197-1203. [DOI] [PubMed] [Google Scholar]
- 48.Thureen D. R., C. L. Keeler, Jr., and M. Dolan. Unpublished results.
- 49.van Oirschot, J. T., H. J. Rziha, P. J. Moonen, J. M. Pol, and D. van Zaane. 1986. Differentiation of serum antibodies from pigs vaccinated or infected with Aujeszky's disease virus by a competitive enzyme immunoassay. J. Gen. Virol. 67:1179-1182. [DOI] [PubMed] [Google Scholar]
- 50.van Oirschot, J. T. 1999. Diva vaccines that reduce virus transmission. J. Biotechnol. 73:195-205. [DOI] [PubMed] [Google Scholar]
- 51.Veits, J., B. Köllner, J. P. Teifke, H. Granzow, T. C. Mettenleiter, and W. Fuchs. 2003. Isolation and characterization of monoclonal antibodies against structural proteins of infectious laryngotracheitis virus. Avian Dis. 47:330-342. [DOI] [PubMed] [Google Scholar]
- 52.Veits, J., D. Lüschow, K. Kindermann, O. Werner, J. P. Teifke, T. C. Mettenleiter, and W. Fuchs. 2003. Deletion of the nonessential UL0 gene of infectious laryngotracheitis (ILT) virus leads to attenuation in chickens, and UL0 mutants expressing influenza virus hemagglutinin (H7) protect against ILT and fowl plague. J. Gen. Virol. 84:3343-3352. [DOI] [PubMed] [Google Scholar]
- 53.Veits, J., T. C. Mettenleiter, and W. Fuchs. 2003. Five unique open reading frames of infectious laryngotracheitis virus are expressed during infection but are dispensable for virus replication in cell culture. J. Gen. Virol. 84:1415-1425. [DOI] [PubMed] [Google Scholar]
- 54.von Einem, J., J. Wellington, J. M. Whalley, K. Osterrieder, D. J. O'Callaghan, and N. Osterrieder. 2004. The truncated form of glycoprotein gp2 of equine herpesvirus 1 (EHV-1) vaccine strain KyA is not functionally equivalent to full-length gp2 encoded by EHV-1 wild-type strain RacL11. J. Virol. 78:3003-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wellington, J. E., G. P. Allen, A. A. Gooley, D. N. Love, N. H. Packer, J. X. Yan, and J. M. Whalley. 1996. The highly O-glycosylated glycoprotein gp2 of equine herpesvirus 1 is encoded by gene 71. J. Virol. 70:8195-8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wellington, J. E., A. A. Gooley, D. N. Love, and J. M. Whalley. 1996. N-terminal sequence analysis of equine herpesvirus 1 glycoproteins D and B and evidence for internal cleavage of the gene 71 product. J. Gen. Virol. 77:75-82. [DOI] [PubMed] [Google Scholar]
- 57.Whittaker, G. R., L. A. Wheldon, L. E. Giles, J. M. Stocks, I. W. Halliburton, R. A. Killington, and D. M. Meredith. 1990. Characterization of the high Mr glycoprotein (gP300) of equine herpesvirus type 1 as a novel glycoprotein with extensive O-linked carbohydrate. J. Gen. Virol. 71:2407-2416. [DOI] [PubMed] [Google Scholar]
- 58.Wild, M. A., S. Cook, and M. Cochran. 1996. A genomic map of infectious laryngotracheitis virus and the sequence and organization of genes present in the unique short and flanking regions. Virus Genes 12:107-116. [DOI] [PubMed] [Google Scholar]
- 59.Ziemann, K., T. C. Mettenleiter, and W. Fuchs. 1998. Gene arrangement within the unique long genome region of infectious laryngotracheitis virus is distinct from that of other alphaherpesviruses. J. Virol. 72:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ziemann, K., T. C. Mettenleiter, and W. Fuchs. 1998. Infectious laryngotracheitis herpesvirus expresses a related pair of unique nuclear proteins which are encoded by split genes located at the right end of the UL genome region. J. Virol. 72:6867-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]