Abstract
The pre-B cell receptor (pre-BCR) is an immature form of the BCR critical for early B lymphocyte development. It is composed of the membrane-bound immunoglobulin (Ig) heavy chain, surrogate light chain components, and the signaling subunits Igα and Igβ. We developed monovalent Quantum Dot (QD)-labeled probes specific for Igβ to study the behavior of pre-BCRs engaged in autonomous, ligand-independent signaling in live B cells. Single-particle tracking revealed that QD-labeled pre-BCRs engaged in transient, but frequent, homotypic interactions. Receptor motion was correlated at short separation distances, consistent with the formation of dimers and higher-order oligomers. Repeated encounters between diffusing pre-BCRs appeared to reflect transient co-confinement in plasma membrane domains. In human B cell precursor acute lymphoblastic leukemia (BCP-ALL) cells, we showed that frequent, short-lived, homotypic pre-BCR interactions stimulated survival signals, including expression of BCL6, which encodes a transcriptional repressor. These survival signals were blocked by inhibitory monovalent antigen-binding antibody fragments (Fabs) specific for the surrogate light chain components of the pre-BCR or by inhibitors of the tyrosine kinases Lyn and Syk. For comparison, we evaluated pre-BCR aggregation mediated by dimeric galectin-1, which has binding sites for carbohydrate and for the λ5 component of the surrogate light chain. Galectin-1 binding resulted in the formation of large, highly immobile pre-BCR aggregates, which was partially relieved by the addition of lactose to prevent the crosslinking of galectin-BCR complexes to other glycosylated membrane components. Analysis of the pre-BCR and its signaling partners suggested that they could be potential targets for combination therapy in BCP-ALL.
INTRODUCTION
The cell surface expression of the multimeric pre-B cell receptor (pre-BCR) complex is required for the survival and differentiation of B cell progenitors. This complex is composed of the immunoglobulin (Ig) heavy chain (surface μHC), two surrogate light chain components VpreB (also known as CD179a) and λ5 (also known as CD179b), and the signaling subunits Igα (also known as CD79a) and Igβ (also known as CD79b). Pre-BCR signaling stimulates the phosphorylation of Immunoreceptor Tyrosine-based Activation Motifs (ITAMs) in the cytoplasmic tails of both of the Igα and Igβ subunits by the tyrosine kinase Lyn (1). The phosphorylated ITAMs form binding sites for the dual Src homology 2 (SH2) domains of spleen tyrosine kinase (Syk), a critical component of downstream signaling cascades that determine cell fate decisions (2, 3).
The functional pre-BCR serves several crucial roles in B cell differentiation, including (i) initiating cell cycle exit (4, 5) and chromatin remodeling (6), which are critical for rearrangement of the genes encoding the light chains; (ii) regulating major changes in transcription (7, 8); (iii) providing quality control for Ig heavy chains (9); and (iv) balanced negative and positive selection of self-reactivity (10–12). Although it is broadly appreciated that the pre-BCR is positioned at a critical B cell developmental checkpoint, there remain some controversies regarding the primary signal initiating mechanism. Evidence exists for the following scenarios: Igα and Igβ signaling in the absence of external factors that might crosslink the Igα-Igβ heterodimer (13); pre-BCR recognition of self-antigens (14); involvement of μHC glycosylation at Asn46 (15); and pre-BCR aggregation by soluble or stromal-bound galectin-1, a lectin that has distinct binding sites for λ5 and for carbohydrates on glycoproteins and glycolipids (16). However, mutagenesis (17) and structural studies (18) suggest that autonomous signaling is driven by ligand-independent self-association of the λ5 components of the surrogate light chains. The weak signals attributed to these homotypic pre-BCR interactions have led some to refer to it as “tonic signaling” (19). The compelling evidence for homotypic pre-BCR associations motivates these questions: Can these interactions be directly measured and what is their lifetime? Do they engage in serial interactions (20)? How does the relative duration of pre-BCR homotypic interactions govern the strength and outcome of pre-BCR signaling? These questions can now be addressed through high-resolution imaging modalities, such as single particle tracking (SPT) methods that have captured the diffusion, clustering, and dynamics of the mature BCR (21, 22) and FcεRI (23, 24).
Our model system is B cell precursor acute lymphoblastic leukemia (BCP-ALL) (25), for which a substantial fraction of cases depend on pre-BCR autonomous signals for survival, expression of the gene encoding the transcriptional repressor BCL-6 (B-cell lymphoma 6 protein), and suppression of p53-mediated apoptosis (26). In these cases, there are parallels between the role of the BCR as a driver of or contributor to diffuse large B-cell lymphoma and chronic lymphocytic leukemia (27, 28). The knowledge that the BCR generates survival and proliferative signaling in mature B cell neoplasms provided motivation for successful clinical trials targeting Syk, Bruton’s tyrosine kinase (BTK), and other downstream signaling components (29). There is now strong interest in determining whether targeted agents against the pre-BCR and its signaling partners (such as Lyn and Syk) (30–32) will also have therapeutic potential for an important subset of BCP-ALL, particularly in the setting of adjuvant or combination therapy. New insights into the mechanisms governing pre-BCR activation also have profound implications for normal B cell development, as well as autoimmunity (33).
Here, we used state-of-the-art SPT methods to capture dimer events between pre-BCRs in real time. These studies provide evidence that pre-BCRs undergo homotypic interactions through their surrogate light chain components, and that these interactions stimulate autonomous signaling. We demonstrated that VpreB-specific monovalent Fab antibodies blocked self-association and inhibited autonomous signaling. Treatment with anti-VpreB Fabs resulted in an overall faster pre-BCR diffusion rate by eliminating the proportion of receptors that was engaged in dimerization. The clinical relevance of our findings was suggested by evidence that anti-VpreB Fabs sensitized BCP-ALL cells to low-dose chemotherapy. To complete our studies of pre-BCR oligomerization, we also evaluated receptor behavior on cells treated with the pre-BCR–crosslinking agent galectin-1. In addition to its well-known carbohydrate-binding properties (34), galectin-1 binds to the unique region of λ5 (35) and serves as a pre-BCR ligand (36). There is intense interest in galectin-mediated signaling in the leukemia microenvironment (37), in which galectin-1 can crosslink the pre-BCR with integrins and other glycoproteins at stromal–blast cell synapses (38). In addition to being produced by stromal cells, galectin-1 is secreted by BCP-ALL blasts for which it may be a soluble or cell-bound ligand for the pre-BCR (39).
We showed that short-lived autonomous signals from the pre-BCR were important to regulate BCL6 abundance, as a downstream readout of pre-BCR activity. BCL6 protein abundance was decreased after treatment with inhibitors that target the pre-BCR signaling partners Lyn, Syk, and SHIP-1 (SH2 domain-containing Inositol 5′-Phosphatase). The Janus-activated kinase (Jak) inhibitor tofacitinib enhanced accumulation of BCL6 protein, confirming reports that signal transducer and activator of transcription (STAT) actively represses BCL6 transcription in BCP-ALL (40). Completed and ongoing clinical trials for Jak inhibitors in BCP-ALL (NCT01914484, NCT01251965, and NCT01164163) provide motivation for further studies to evaluate whether de-repression of BCL6 and other targets of the pre-BCR pathway offer potential escape mechanisms. Studies of pre-BCR cell lines and patient-derived leukemia blasts (26, 30, 40, 41) suggest that predicting the therapeutic responses of individual patients to targeted inhibitors of the pre-BCR and Jak-STAT pathways may require case-by-case evaluation, the development of reliable biomarkers, and a systems level approach to understanding the complex crosstalk between both pathways.
RESULTS
SPT captures serial pre-BCR engagements
The first step in the experimental plan to track pre-BCR self-association dynamics was the design and production of monovalent Quantum Dot (QD) probes. We base our probes on the CB3-1 antibody to the Igβ (CD79b). As a positive control, we also generated probes based upon antibodies that recognize the Fc portion of the membrane-bound pre-BCR heavy chain (mIgμ). Both reagents have the advantage of not recognizing the VpreB and λ5 moieties of the surrogate light chain, which are proposed to mediate pre-BCR homotypic interactions (18). In brief, intact IgG antibodies were collected from hybridoma culture supernatants, which was followed by controlled pepsin cleavage to initially produce F(ab’)2 fragments. Anti-Igβ Fabs’ with exposed thiol groups (42) were generated by incubation in 2-mercaptoethylamine (MEA) containing EDTA, followed by covalent, maleimide-based coupling of the free cysteines to PEG2-biotin. Protein G beads were used to remove any contaminating intact IgG or Fc fragments. Monovalent Fab’-PEG2-biotin was purified by FPLC (Fast Protein Liquid Chromatography) and then mixed 1:1 with avidin-conjugated QD585 or QD655 for dual-color SPT. We then characterized the anti-Igβ Fab’ probe (fig. S1).
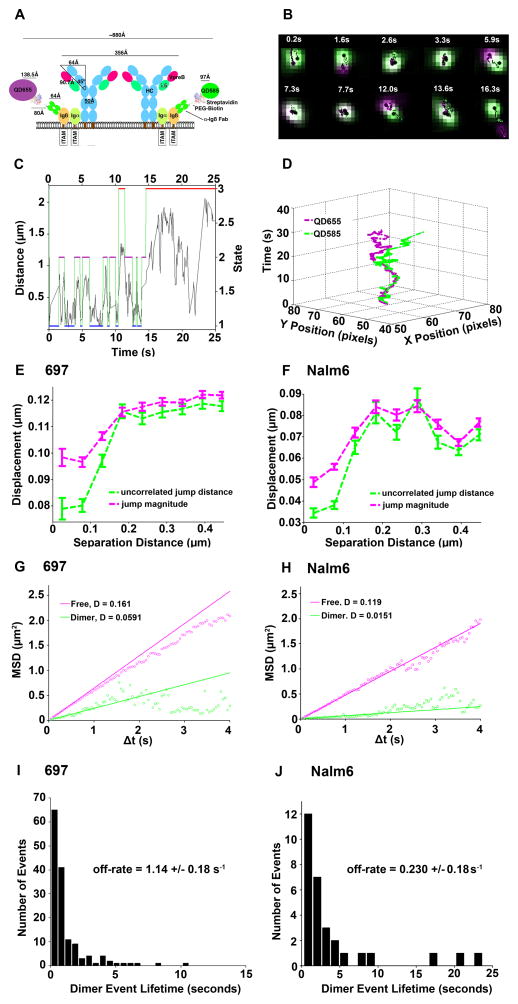
Our strategy for observing pre-BCR dimers by SPT involved tagging each multi-subunit pre-BCR with a different color of QD (QD585 or QD655) (Fig. 1A). Although there is limited structural information for the entire pre-BCR complex, if we assume a side-by-side orientation of all subunits, there would be approximately 80 to 100 nm between the two bound Fab’-QD probes in a dimer. Consistent with this, we found a best fit dimer distance between the QD probes of 100 nm with the Hidden Markov Model (HMM) to analyze large sets of two-color tracking data (43). Through SPT imaging (Fig. 1B), we captured pre-BCR dimerization in real time on the surface of live 697 cells [a cell line derived from a BCP-ALL patient (44)]. The two diffusing anti-Igβ Fab’-QD probes were distinguished by pseudo-coloring them as green or magenta dots throughout a 25-s time series. At 2.6 to 3.3 s into image acquisition, the dots were clearly overlaid, indicating the presence of dimers. The pair became segregated at 5.9 s, which was followed by two rebinding events (at 7.3 and 13.6 s) (Fig. 1B). The Viterbi plot (45) reported the state-specific transitions of these two receptors over time (Fig. 1C), using rate constants determined by the HMM as previously described in detail (43). The HMM distinguished three states for mobile receptors: (i) dimers; (ii) pairs that are transiently co-confined in the same region; and (iiii) pairs of receptors that are widely segregated (“free”). The two receptors cycled between dimer and co-confined states over a ~14.5-s time window, after which diffusion resulted in their separation by a distance of > 1 μm (Fig. 1C). For this pair, homotypic interaction lifetimes were short (<3 s).
Fig. 1. Direct evidence of pre-BCR dimerization on the surface of BCP-ALL cells.
(A) Experimental model with estimation of the interaction distance based upon the pre-BCR crystal structure (PDB ID: 2H32), streptavidin (PDB ID: 1STP), and the estimated QD radii. (B) Time-lapse imaging (20 frames/s) of a single cell under resting conditions. (C) Viterbi plot of the most likely state (dimer, domain, and free) between two QD interactions as derived from hidden Markov model (HMM) with separation distance as the observed parameter. (D) Two-channel 3D trajectory of anti-Igβ Fab’-QD655 (magenta) and Fab’-QD585 (green) bound to two receptors that serially engaged repeatedly over 14.5 s, with several instances of correlated motion before separating toward the end of acquisition. Data are representative of 35 independent experiments with 697 cells. (E and F) Analysis of displacement (magenta, jump magnitude) and degree of uncorrelated motion (green) as a function of separation distance (between 150 to 200 nm), observed with both 697 cells (E) and Nalm6 cells (F). (G and H) Diffusion by state, as determined by HMM analysis. (I and J) Distribution of dimer lifetimes in 697 and Nalm6 cells, with estimated off-rates as predicted by HMM analysis. Interaction distances in the HMM were set to 100 and 300 nm, respectively. Data in (E) to (J) represent 26,447 trajectories in 697 cells and 2,800 trajectories in Nalm6 cells.
Our three-dimensional (3D) view of the same pre-BCR dimer pair (Fig. 1D) illustrated the relative X and Y positions of each anti-Igβ Fab’-QD probe over time. Based upon their close proximity and correlated motion, the two pre-BCRs became dimerized at the beginning of the imaging. Their dissociation at 5.9 s was readily apparent, which was followed by a short re-engagement period and then dissociation and diffusion in separate directions toward the end of the image acquisition period. Additional examples of pre-BCR dimer events were analyzed (fig. S2 and movie S1). We applied two other criteria to evaluate dimerization (Fig. 1, E and F), which examine whether pairs of molecules in the entire data set move independently (“uncorrelated motion”, with dissimilar jump magnitudes) or together (correlated motion with similar jump magnitudes). These plots show characteristic drops in pre-BCR uncorrelated motion (green lines) and jump magnitude (magenta lines) that were a function of close separation distances, an expected requirement for interactions. These analyses were performed for 697 cells (Fig. 1E), as well as for Nalm6 cells (46) (Fig. 1F). These data provide statistically rigorous validation that pre-BCR dimerization occurred frequently on the surface of two distinct BCP-ALL cell lines. Analysis of the cytogenetics and the cell surface expression of BCP-ALL markers for both cell lines was also performed (table S1).
We then compared the relative mobilities of pre-BCR monomers and dimers on the surface of both cell lines (Fig. 1, G and H). In the free state, pre-BCRs had a relatively fast diffusion coefficient (0.16 and 0.12 μm2/s on 697 cells and Nalm6 cells, respectively). By comparison, the diffusion rates for interacting pairs of pre-BCRs (representing jump distributions only during the intervals when the receptors were identified as being dimerized) were markedly slower at 0.06 and 0.015 μm2/s on 697 cells and Nalm6 cells, respectively. When considering the whole population of receptors (that is, not state-specific), the ensemble Mean Square Displacement (MSD) plots for pre-BCRs on these cells were similar when either the anti-Igβ or the anti-mIgμ QD probes were used (fig. S3). The mean diffusion coefficients for the pre-BCR on the surfaces of 697 and Nalm6 cells were 0.13 and 0.09 μm2/s, respectively (table S2). We also summarized the number of trajectories tracked under each experimental condition in this work (table S2).
The dissociation of individual receptors from dimers was previously shown to be a stochastic process, resulting in a characteristic range of dimer lifetimes that represent all of the binding and unbinding events within a data set (43, 47). For 697 cells, most (84%) of lifetimes of the 146 bound events were less than 2 s, with an off-rate of 1.14 +/− 0.18/s calculated by the HMM (Fig 1I, fig. S4, and table S3). Homotypic interactions of the pre-BCR were slightly more stable on Nalm6 cells, as evidenced by a higher percentage of longer lifetime binding events (43% > 2 s) and an off-rate of 0.230 +/− 0.08/s (Fig 1J, fig. S4, and table S3). Note the correlation between the slower overall diffusion rate of the pre-BCR in Nalm6 cells and the slower off-rates for homotypic interactions.
Because lateral diffusion can be restricted by barriers and protein-protein interactions in a non-linear fashion (48), the observation of slower pre-BCR diffusion rates on Nalm6 cells may reflect intrinsic differences in the cortical cytoskeleton, the increased size of pre-BCR complexes and their associated adaptor proteins, or both. The possibility of self-associated pre-BCR aggregates has been suggested previously (18, 19); their side-by-side alignment would orient them in the form of chains. A potential outcome of the five-fold slower off-rate (Fig. 1J and fig. S4) for self-engaged pre-BCRs on Nalm6 cells, compared to that of pre-BCRs on 697 cells, may be the greater likelihood of them forming pre-BCR oligomers larger than dimers. We captured a pair of QD probes undergoing correlated motion on the surface of Nalm6 cells that were further apart than the 80 to 100 nm theoretical distance consistent with receptor dimers and their probes (movie S2). We interpret this movie as evidence for the diffusion of homotypic pre-BCR trimers or tetramers with the two Igβ Fab’-QD probes bound to the ends. We found that a median distance of 227 nm was maintained by the two probes undergoing correlated motion before blinking of the QD655 occurred (fig. S5, magenta).
Galectin leads to the formation of complex pre-BCR–glycoprotein aggregates with slow diffusion rates
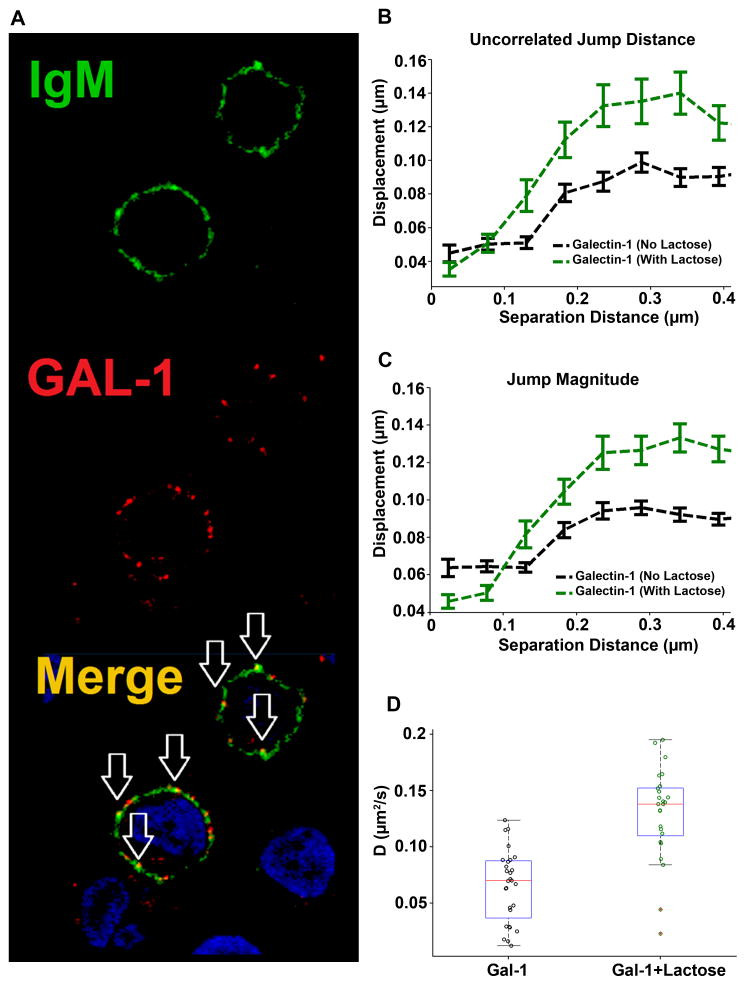
Galectin-1 binds to λ5 with high specificity (Ka =2 × 106 M−1) (36). We next evaluated the effects of exogenous galectin-1 on the diffusion and aggregation dynamics of pre-BCRs. We first verified the purity of the protein used in our experiments (fig. S6A) and took note of a structural model for monomeric galectin-1 (fig. S6B). Incubation of 697 cells with Alexa 555–conjugated galectin-1 (red) led to the marked clustering of the pre-BCR (Fig. 2A, green) within 5 min. The merged image shows substantial colocalization (Fig. 2A, yellow) of galectin-1 and the pre-BCR in these clusters. For these experiments, we used 10 μM galectin-1, a concentration at which soluble galectin-1 should be >90% dimerized (49).
Fig. 2. Galectin-1 stabilizes receptor dimerization and promotes the formation of higher-order complexes.
(A) Analysis of the binding of labeled galectin-1 (red) to the pre-BCR (green) in 697 cells. Arrows indicate cluster formation and the colocalization of galectin-1 with the pre-BCR. Images are representative of three experiments. (B and C) Blocking the carbohydrate-binding domain of galectin-1 with 10 mM lactose revealed differences in displacements for the uncorrelated jump distance and the jump magnitude of distinct anti-Igβ probes at different separation distances. (D) Analysis of the diffusion of the pre-BCR in 697 cells treated with galectin-1 (GAL-1) in the absence or presence of 10 mM lactose. Plots represent cumulative data from 1491 (galectin) and 1560 (galectin + lactose) trajectories tracked over five independent experiments.
We performed SPT with the anti-Igβ Fab’-QD probes to evaluate the effects of galectin-1 on pre-BCR diffusion. We observed a marked decrease in both jump magnitude and uncorrelated motion for pre-BCRs on the surface of galectin-1–treated 697 cells (Fig. 2, B and C). A large fraction of pre-BCR essentially became immobile (Fig. 2D and movie S3), which is best explained by the formation of complex aggregates containing pre-BCR, galectin-1, and other cell surface glycoproteins. To provide evidence that the marked slowdown of the pre-BCR bound to galectin-1 could be attributed in part to carbohydrate-mediated lattices with other glycoproteins (37), we also performed experiments in the presence of excess lactose (10 mM) to block the lectin-binding site of galectin-1 (fig. S6B). Under these conditions, we observed larger overall jump distributions for diffusing receptors and a shorter interaction distance for the onset of correlated motion (Fig. 2, B and C, green dashed lines). The overall diffusion rate of galectin-1–crosslinked pre-BCRs was markedly faster in the presence of lactose (Fig. 2D), approaching values for pre-BCRs under autonomous signaling conditions (table S2). Note that controls were performed with lactose alone (see fig. S6C) to ensure that the addition of lactose in the absence of galectin-1 did not alter the average diffusion behavior of the pre-BCR. Additional controls included the treatment of cells with 2 μM galectin-1, a concentration at which galectin-1 monomers predominate, which failed to markedly slow the diffusion rate of the pre-BCR (fig. S6D). Thus, these data suggest that soluble galectin-1 dimers are required to provide a bivalent ligand that binds to λ5 and promotes the aggregation of cell surface pre-BCRs (36).
Fabs against VpreB block the self-association of pre-BCRs
Our next goal was to test the effects of dimer-disrupting and dimer-stabilizing reagents upon the overall diffusive behavior of pre-BCRs. We reasoned that monovalent antibody fragments specific for the surrogate light chains of the pre-BCR would have potential as dimer-disrupting reagents. As described in the Materials and Methods, we screened the ConCIRT Synthetic Library to identify 16 phage-displayed clones that bound to recombinant VpreB1 polypeptide. A high-affinity clone was selected for the production of recombinant human IgG, and we then prepared anti-VpreB1 Fabs. The parent IgG bound to the pre-BCR on the surface of 697 cells with subnanomolar affinity (fig. S7).
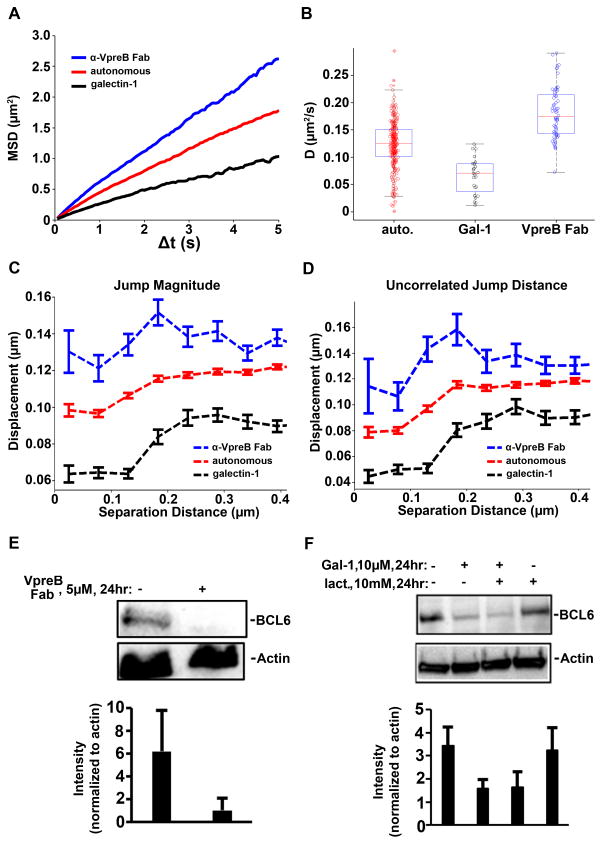
We then generated MSD plots (Fig. 3A) for the pre-BCR in the absence and presence of a saturating concentration of anti-VpreB1 Fabs (1 μM). In these experiments, Fabs were added to 697 cells for 10 min at 37°C before we performed SPT. We observed that targeting the pre-BCR with these monovalent blocking reagents markedly increased the overall diffusion rate of the pre-BCR. The overall diffusion coefficient for the pre-BCR on untreated 697 cells was 0.129 μm2/s, which was slower than the rate of 0.172 μm2/s that was observed when anti-VpreB Fabs blocked pre-BCR self-association (Fig. 3B and table S2). For comparison, we also plotted the values for pre-BCR diffusion in 697 cells after treatment with galectin-1 (Fig. 3A, black lines). After crosslinking with galectin-1, a greater percentage of pre-BCRs approached an immobile state, with an ensemble MSD-derived diffusion coefficient of 0.069 μm2/s (table S2 and movie S3). We also performed an overall comparison of the changes in jump magnitude (Fig. 3C) and uncorrelated motion (Fig. 3D) for pairs of pre-BCRs tracked on the surface of untreated cells (red lines; autonomous signaling) compared to those of cells treated with the anti-VpreB Fabs (blue lines) or galectin-1 (black lines). These data underscore our observations that pre-BCR self-association was prevented by monovalent blocking antibodies specific for the surrogate light chain and that galectin-1 induced the formation of large-scale pre-BCR aggregates.
Fig. 3. Comparison of the effects of VpreB Fabs and galectin-1 on pre-BCR diffusion and dimerization.
(A and B) Ensemble MSD values (A) and diffusion coefficients (B) revealed changes in pre-BCRs after 10 min of treatment with anti-VpreB Fabs (1 μM, blue) or galectin-1 (10 μM, black). The two-tailed t test from MATLAB was used to confirm statistical significance (see table S2). (C and D) Changes in uncorrelated jump distance and jump magnitude after 10 min of treatment with either 1 μM anti-VpreB Fab (blue dotted lines) or 10 μM galectin-1 (black dotted lines). (E and F) 697 cells were left untreated or were treated for 24 hours with 5 μM anti-VpreB Fab (E) or with 10 μM galectin-1 in the presence or absence of 10 mM lactose (F) before being subjected to Western blotting analysis with antibodies against the indicated proteins. Each blot is representative of three or more independent experiments. Bar graphs report total densitometry data for the abundances of the indicated proteins normalized to the abundance of actin.
Other studies suggest that early B cells are governed by a balance of finely tuned thresholds that govern early B cell checkpoints on self-reactivity and survival (40, 41, 50). We investigated the abundance of BCL6 protein as a readout of pre-BCR signaling under cell autonomous and galectin-mediated crosslinking conditions. We found that BCL6 protein abundance in 697 cells was greatest under autonomous signaling conditions (Fig. 3, E and F, left lanes). BCL6 abundance was decreased after overnight treatment with anti-VpreB Fabs, which inhibited homotypic interactions (Fig. 3E, right lane). Galectin-1–mediated crosslinking of the pre-BCR, in the absence, but not presence, of lactose, led to substantially reduced amounts of BCL6 (Fig. 3F).
Tyrosine phosphorylation and leukemia cell survival are finely tuned by autonomous pre-BCR Signaling
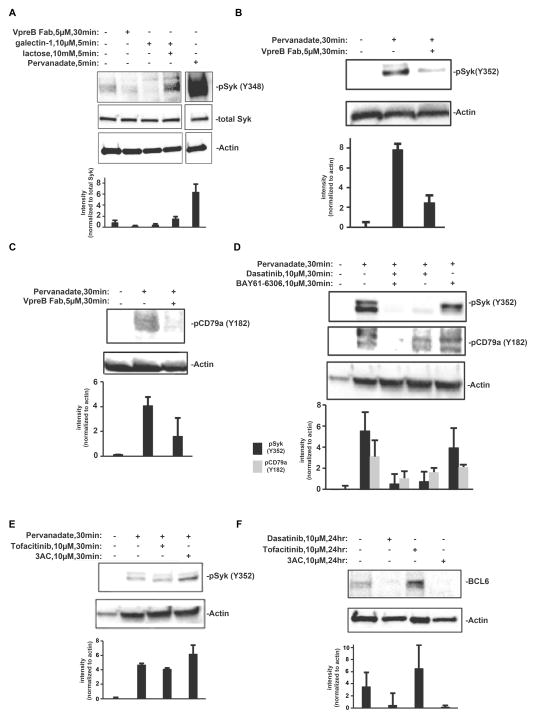
We next evaluated the extent of Syk and CD79a phosphorylation under each of these experimental conditions (Fig. 4). We found that Syk phosphorylation at Tyr348 was detectable in lysates prepared from 3 × 107 Nalm6 cells under autonomous signaling conditions (Fig. 4A, left lane). Furthermore, treatment of cells for 30 min with anti-VpreB Fabs reduced the amount of Syk phosphorylated at Tyr348 by 40%. In addition, cells were treated for 5 min with galectin-1 either alone or in combination with lactose to block the carbohydrate-binding site of galectin-1. Addition of galectin-1 alone decreased the amount of Syk phosphorylated at Tyr348 (Fig. 4A), whereas crosslinking in the presence of lactose led to enhanced Syk phosphorylation on this site. This finding suggests that the inclusion of other glycoproteins and their signaling partners in the galectin-1–pre-BCR aggregate rapidly inhibits Syk-mediated signaling. Because the surface of BCP-ALL blasts is rich with ITIM-containing inhibitory receptors (50), co-aggregation of pre-BCRs with these glycoproteins might explain the reduced phosphorylation of Syk in response to galectin-1 alone. In our hands, detection of phosphorylated Syk (pSyk) under autonomous signaling conditions often required the overexposure of Western blots. Wienands et al. previously showed that treatment of B cells with the pan phosphatase inhibitor pervanadate reveals kinase activity that is constitutively opposed by phosphatase activity (51). We found a ~7-fold enhancement in the phosphorylation of Syk at Tyr348 after a 30-min treatment of Nalm6 cells with pervanadate (Fig. 4A).
Fig. 4. Western blotting analysis of pre-BCR signaling.
(A) Western blotting analysis of Nalm6 cell lysates (3 × 107cells) for Syk phosphorylation at Tyr348 (pSyk Y348) after the indicated treatments. Blots were stripped and incubated with antibodies against total Syk and actin. Note that the pervanadate lane is shown after gray scale adjustment of the digital image, to adjust for the intensity of the signal for pSykY348 compared to that in resting cells and after treatment with inhibitors. (B and C) Western blotting analysis of 697 cells (1.0 × 106) with antibodies specific for pSyk and pCD79a (Igα) after the cells were treated for 30 min with anti-VpreB Fab in the presence of pervanadate. (D) Western blotting analysis of 697 cells (1.0 × 106) with antibodies against pSyk and pCD79a (Igα) after the cells were treatment with BAY61-3606 (Syk inhibitor) or dasatinib (Lyn and Abl inhibitor) in the presence of pervanadate. (E) Western blotting analysis of 697 cells (1.0 × 106) with antibody specific for pSyk after the cells were treated for 30 min with 3AC (SHIP inhibitor) or tofacitinib (Jak inhibitor) in the presence of pervanadate. (F) Western blotting analysis of 697 cells (1.0 × 106) with antibody specific for total BCL6 protein after the cells were treated for 24 hours with dasatinib, tofacitinib, or 3AC. All blots are representative of three or more independent experiments. Accompanying bar graphs report total densitometry data for all experiments for a given condition, with the abundance of the protein of interest normalized to that of actin or total Syk.
Because phosphorylation detection under basal conditions required large quantities of cells, we took advantage of pervanadate to enhance the amounts of phosphorylated Igα (CD79a) ITAM and pSyk (Y182, Y352) (Fig. 4, B to D). We interpreted the presence of two or more bands corresponding to phosphorylated CD79a (Fig. 4C, lane 2) as differential mobility caused by the unequal phosphorylation of tyrosine residues in the cytoplasmic tail of CD79a. Treatment of cells with the combination of pervanadate and anti-VpreB Fabs resulted in the near-complete ablation of phosphorylation of both Syk Tyr182 and CD79a. Thus, the increased phosphorylation of these proteins, revealed by inhibiting phosphatase activity, was completely dependent on homotypic pre-BCR interactions that stimulated autonomous signaling. Receptor phosphorylation by the Src family kinase Lyn precedes the recruitment and activation of Syk in the canonical ITAM signaling pathway (19). There is also evidence that once Syk is recruited to the receptor, it also participates in the phosphorylation of ITAMs (52). We next evaluated these early steps in the autonomous pre-BCR signaling pathway in experiments with kinase-selective, cell-permeable inhibitors. Dasitinib is a cell-permeable inhibitor of the tyrosine kinases Src and Abl (53). Consistent with our earlier findings and those of others (26), we found that the addition of dasatinib to pervanadate-treated 697 cells blocked the Lyn-mediated phosphorylation of the pre-BCR, as detected by anti-CD79a-pY182 antibodies (Fig. 4D). The combination of the Syk inhibitor BAY61-3606 and dasatinib abolished the accumulation of phosphorylated CD79a during phosphatase inhibition, suggesting that trans-phosphorylation by Syk combined with Lyn activity was required for the robust phosphorylation of CD79a. The Syk inhibitor also resulted in a 20% reduction in the amount of pSyk phosphorylated at Tyr352, consistent with this tyrosine residue being a substrate of both Lyn and Syk (2). Treatment of 697 cells with the Jak inhibitor tofacitinib (54) did not substantially alter Syk phosphorylation in the presence of pervanadate (Fig. 4E).
We also demonstrated the contribution of the pre-BCR–Lyn-Syk axis in autonomous signaling to BCL6 production, because treating 697 cells overnight with dasatinib resulted in reduced BCL6 abundance (Fig. 4F). Overnight treatment of 697 cells with tofacitinib increased the abundance of BCL6 protein, which we attributed to relief from a Jak-STAT pathway that represses BCL6 transcription (Fig. 4F) (26, 55). Genetic deletion of SH2-containing inositol-5 phosphatase-1 (SHIP1, also known as INPP5D) or inhibition of its phosphatase activity with the small molecule inhibitor 3AC (3-a-aminocholestane) leads to cell death in BCR-ABL1+ acute lymphoblastic leukemia cells (50). We also evaluated the effects of 3AC on 697 cells. As reported previously (50), 3AC slightly increased Syk phosphorylation in these cells (here performed in the presence of pervanadate) (Fig. 4E). In addition, overnight treatment with 3AC alone resulted in a decrease in BCL6 protein abundance (Fig. 4F).
Inhibitors of tyrosine kinases and inositol phosphatases differentially affect the survival of BCP-ALL cells
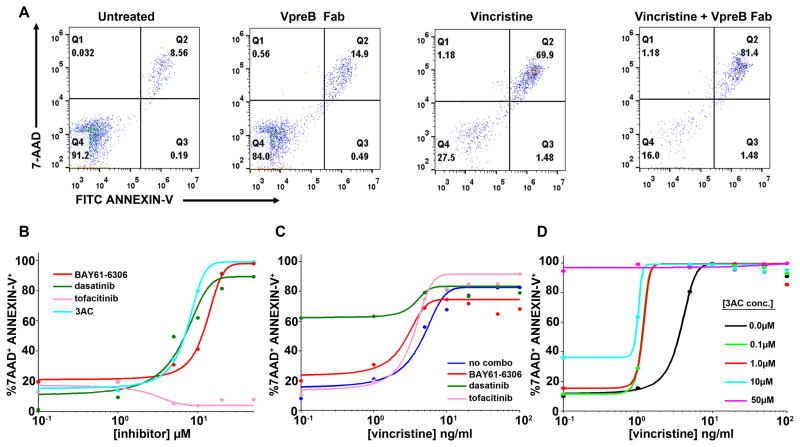
We next evaluated whether anti-VpreB Fabs or our panel of pharmacologic inhibitors could induce apoptosis in 697 cells, either alone or in combination with low concentrations of the chemotherapeutic agent vincristine. Overnight incubation of the cells with VpreB Fabs slightly enhanced the labeling of the cells with both 7-AAD and Annexin-V (14.9% compared to 8.5% of untreated cells, Fig. 5A). The combination of VpreB Fabs with vincristine (10 ng/ml) also slightly enhanced apoptosis (81.4% apoptotic cells compared to 69.9% for vincristine alone). At least for 697 cells, which lack Jak mutations known to be present in a minor subset of BCP-ALL cases (56), treatment with tofacitinib did not directly induce apoptosis (Fig. 5B). By comparison, both dasatinib (a Lyn inhibitor) and BAY61-3606 (a Syk inhibitor) resulted in increased 7-AAD labeling of non-viable cells (Fig. 5B). All three inhibitors (tofacitinib, dasatinib, and BAY61-3606; each at 10 μM) modestly potentiated apoptosis in 697 cells treated with vincristine over a range of 0.1 to 2 ng/ml (Fig. 5C). When used at 1 μM, 3AC sensitized cells to low concentrations of vincristine (0.1 to 2 ng/ml) (Fig. 5D). Higher concentrations of 3AC (10 to 50 μM) were required to induce direct killing of 697 cells, as determined by analysis of 7-AAD labeling (Fig. 5D and figs. S8 to S10).
Fig. 5. Analysis of apoptosis and viability in BCP-ALL cell lines after treatment with pre-BCR pathway inhibitors.
(A) Annexin V (FITC-conjugated) and 7-AAD labeling of 697 cells after 3 days of culture in the absence (autonomous) and presence of anti-VpreB Fabs or vincristine, as indicated. (B) 7-AAD and Annexin-V labeling of 697 cells treated for 3 days with a range of concentrations (0.1 to 100 μM) of tyrosine kinase inhibitors (BAY61-6306, dasatinib, tofacitinib) or inositol phosphatase (3AC). (C) 7-AAD and Annexin-V labeling of 697 cells treated for 3 days with or without a single dose (10 μM) of tyrosine kinase inhibitors (BAY61-6306, dasatinib, tofacitinib) in combination with increasing concentrations (0.1 to 100 ng/ml) of vincristine. (D) 7-AAD and Annexin-V labeling of 697 cells incubated for 3 days with or without 3AC (0.1 to 50 μM) in combination with increasing doses of vincristine (0.1 to 100 ng/ml). In all plots, the percentage of cells that were 7AAD+Annexin-V+ is plotted against increasing concentrations of vincristine or inhibitor and fit with a sigmoidal dose-response curve. All data were acquired by flow cytometric analysis of at least 10,000 events for each condition and are representative of xxx independent experiments.
Tracking of pre-BCRs in primary cells reveals differential effects of anti-VpreB Fabs and Lyn inhibition
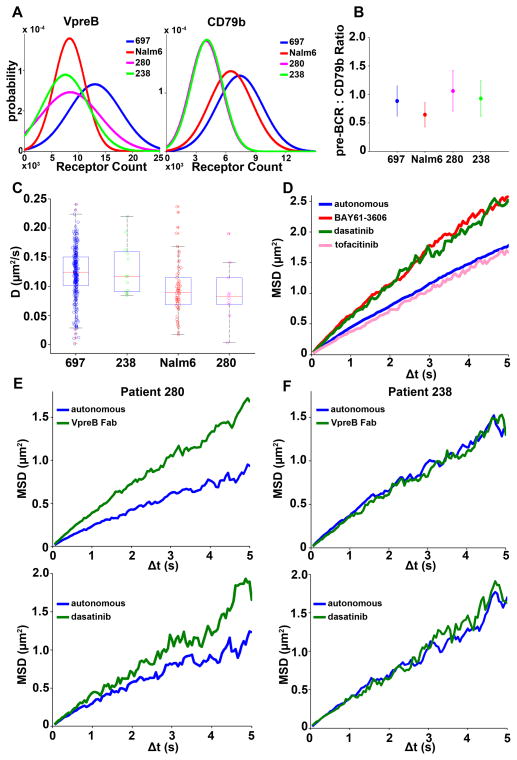
The pre-BCR is absent from BCR-ABL1+ acute lymphoblastic leukemia cells (50); however, its presence in other BCP-ALL subsets likely defines those patients for which therapies that target autonomous signaling are of potential value (26). We identified two distinct BCP-ALL patient samples (#280 and #238) that had measurable amounts of pre-BCR on the cell surface (Fig. 6). To enable repeated measurements to be made of primary cells to validate pre-BCR diffusion and drug sensitivity studies, the patient samples were cryopreserved on the day of acquisition and then were either placed in tissue culture immediately after thawing or were passaged through Nod-SCID-γ2–deficient mice (a necessary step to expand primary leukemia blasts, which have a limited lifespan in culture). The cell surface abundance of the pre-BCR, based on quantitative binding of anti-VpreB antibodies (fig. S11), for these two patient samples ranged from ~4000 to 15,000 copies per cell, with average values of 6000 to 7000 (Fig. 6A). These values were very similar to those determined from experiments with Nalm6 cells and are equivalent to about half of the abundance of the pre-BCR on 697 cells. The pre-BCR complex is commonly assumed to have a uniform stoichiometry of two mIgμ: VpreB:λ5 complexes to one Igα–Igβ heterodimer; however, our flow cytometry–based quantitative measurements suggest that both sets of patient cells had an average of 4000 Igβ molecules, whereas there were ~6500 copies of Igβ on Nalm6 cells and ~8000 copies on Igβ on 697 cells. We found that there were slightly differing ratios (0.7 to 1.1) of mIgμ:Igβ for each cell type (Fig. 6B). We speculate that this variability may be explained by some of the pre-BCRs being composed of homodimers of Igβ or Igα.
Fig. 6. Expression profile and single particle tracking of pre-BCRs on different primary BCP-ALL cells.
(A) Comparison of the cell surface abundances of VpreB and CD79b (Igβ) on 697 cells, Nalm6 cells, and blasts from patients #238 and #280, as a readout of pre-BCR on the membrane of BCP-ALL cells. Cells were incubated with saturating concentrations of labeled fluorophore-conjugated anti-VpreB or anti-CD79b antibodies and binding values were quantified by flow cytometry. Results were calibrated by saturation binding of each antibody to Simply Cellular anti-mouse IgG microspheres. Data are representative of at least three independent experiments. (B) Comparison of the mean fluorescent values after anti-VpreB and anti-CD79b labeling of the indicated cell types, plotted as a ratio of pre-BCR (2 VpreB = 1 pre-BCR) to CD79b. (C) Comparison of the diffusion coefficient (μm2/s) of the pre-BCR in 697 cells (n = 26,447 trajectories) and Nalm6 cells (n = 3,573 trajectories) to primary cells from patient 238 (304 trajectories) and patient 280 (353 trajectories); single particle tracking was performed on untreated cells in all cases and performed over multiple days. (D) Ensemble MSD plots derived from SPT of the pre-BCR on 697 cells with or without a 10-min treatment with the indicated tyrosine kinase inhibitors. (E) Ensemble MSD plots derived from SPT of the pre-BCR on the surface of primary cells from patient 280 with and without a 10-min treatment with dasatinib (1 μM) or anti-VpreB Fabs (1 μM). (F) Ensemble MSD plots derived from SPT of the pre-BCR on the surface of primary cells from patient 238 with and without a 10-min treatment with dasatinib (1 μM) or anti-VpreB Fabs (1 μM). Data in (D) to (F) are representative of xxx independent experiments.
We next performed SPT with the anti-Igβ Fab’-QD probes to evaluate the diffusion of pre-BCRs on untreated primary cells. We found that the overall diffusion coefficient for pre-BCRs on the plasma membrane of B cells from patient #238 was similar to that of 697 cells (Fig. 6C). The range of the observed values, as well as the overall diffusion coefficient for pre-BCRs on cells from patient #280, matched well with the diffusion characteristics of pre-BCRs on Nalm6 cells. As an extension of the drug profiling reported earlier, we also treated 697 cells with each of the three tyrosine kinase inhibitors and performed SPT experiments. Treatment of 697 cells with the Lyn and Syk inhibitors (dasatinib and BAY61-3606, respectively) increased pre-BCR mobility (Fig. 6D), as was expected given that ITAM phosphorylation generates docking sites for signaling partners that slow receptor diffusion. Incubation of B cells from patient #280 with either anti-VpreB Fab or dasatinib resulted in a marked increase in pre-BCR mobility (Fig. 6E). In contrast, there was little change in pre-BCR diffusion in cells from patient #238 after either treatment (Fig. 6F).
Together, our earlier findings related to the diffusion and dimerization properties of pre-BCRs (Figs. 3 and 6) suggest that these receptors fall into at least two distinct categories. To address potential explanations for these observations, we isolated mRNA from each of the two cell lines, as well as from the two patient samples. After RT-PCR amplification, we performed sequencing analysis for comparison to the reference sequence for VpreB (UniProt #P12018). We found that a substantial fraction of VpreB sequences expressed in each cell source differed from the reference sequence (fig. S12). Furthermore, the two groups with distinct diffusion profiles also sorted together based upon single nucleotide polymorphisms (SNPs). For 697 cells and those from patient #238, which had the fastest pre-BCR diffusion rates, we documented substitutions of lysine for glutamic acid at position 132 and glutamic acid for lysine at position 77. For Nalm6 cells and those from patient #280, which showed the slowest overall pre-BCR diffusion rates, we found either a 33 or 80% frequency, respectively, in substitution of asparagine for the aspartic acid at position 76. This latter change results in a putative N-linked glycosylation site, which is worth pursuing further because there are no N-linked glycosylation sites in the other forms of VpreB. With the exception of the K77E variant, all of these variants are listed on the UniProt site as natural variants.
Primary BCP-ALL cells show differential sensitivity to blockade of autonomous pre-BCR signaling
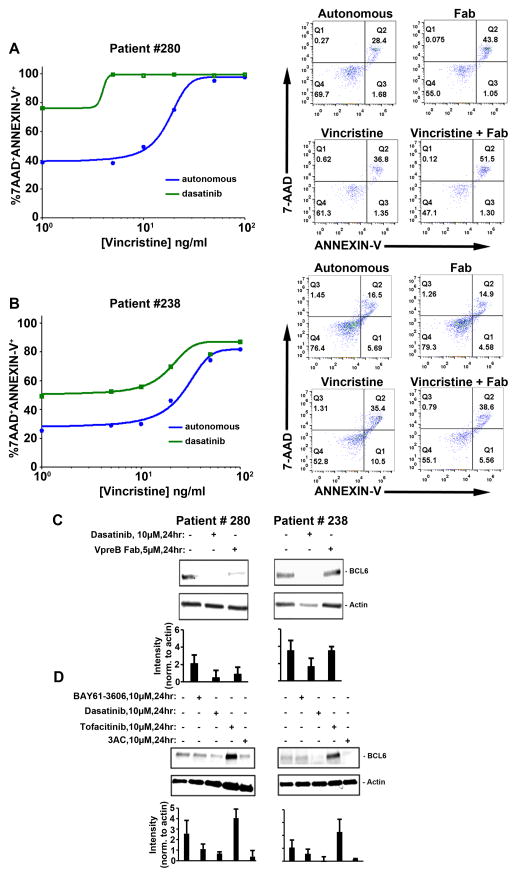
Given our observations that BCP-ALL cells fall into at least two distinct classes with respect to pre-BCR mobility and their possible linkage to VpreB polymorphisms, we evaluated whether blockade of homotypic pre-BCR self-associations might differentially sensitize the two primary patient samples to vincristine. We found that incubation of cells from patient #280 with anti-VpreB Fabs enhanced apoptosis, whether administered alone or in combination with a low concentration of vincristine (Fig. 7A). Blasts from patient #280 were also very sensitive to dasatinib as a single agent; 80% of cells were strained with 7-AAD and Annexin-V after 24 hours of culture. Longer incubations were not feasible, because the cells from this patient were 40% 7-AAD–positive when incubated with no drug or in the presence of up to 5 ng/ml vincristinel (Fig. 7A, blue line).
Fig. 7. Flow cytometric and Western blot analyses of primary BCP-ALL cells after treatment with tyrosine kinase inhibitors or anti-VpreB Fab.
(A and B) Left: Plots show Annexin V (FITC-conjugated) and 7-AAD labeling of primary cells from patient #280 (A) and patient #238 (B) after 24 hours of culture with or without anti-VPreB Fabs (5 μM) or vincristine (1 to 100 ng/ml) as indicated. Right: Scatter plots report flow-based measurements of primary cells from patient 280 (A) and patient 238 (B) that were double labeled for 7-AAD and Annexin-V after 24 hours of culture with or without a single dose of anti-VpreB Fab (5 μM), vincristine (10 ng/ml), or both, as indicated. All data in (A) and (B) were acquired by flow cytometric analysis of at least 10,000 events for each condition and are representative of xxx independent experiments. (C) Western blotting analysis of cell lysates (1.0 × 106 primary cells each) for patients 280 or 238 to compare the relative abundance of BCL6 protein after the cells were treated for 24 hours with dasatinib (10 μM) or anti-VpreB Fab (5 μM). (D) Western blotting analysis of cell lysates (1.0 × 106 cells) from patients 280 and 238 to compare the relative abundance of BCL6 protein after the cells were treated for 24 hours with BAY61-6306, dasatinib, tofacitinib, or 3AC. Each blot is representative of three or more independent experiments. Bar graphs show total densitometry data for all experiments for a given condition, with the abundance of the protein of interest normalized to that of actin.
Blasts from patient #238, which is a high-risk case with MLL-rearrangement (table S1), were relatively resistant to vincristine alone up to concentrations of 10 ng/ml (Fig. 7B). Anti-VpreB Fabs did not potentiate low-dose vincristine treatment of cells from patient #238; however, we observed a ~50% increase in 7-AAD labeling of patient #238 cells when dasatinib was added over a range of concentrations of vincristine (1 to 100 ng/ml). Thus, the combination of dasatinib and vincristine was effective in killing cells from both patients, but higher concentrations of dasatinib were required to kill the cells from patient #238. Finally, basal amounts of BCL6 protein were slightly greater in patient #280 cells compared to those in cells from patient #238 (Fig. 7C). The addition of VpreB Fab to block autonomous signaling decreased the abundance of BCL6 only in the cells from patient #280 (Fig. 7C). Furthermore, BCL6 abundance was reduced or became undetectable in cells from either patient when treated with either dasatinib or 3AC (Fig. 7D). Because the cells from patient #238 cells harbor an MLL-AF4 fusion, they fall into a category of BCP-ALL that is not frequently associated with the “Ph-like” transcriptional signature (57). Nevertheless, based upon the inhibitor studies, the basal activities of Lyn and SHIP (and possibly Abl) apparently contributed to the survival of the cells from patient #238. As for 697 cells (Fig. 4D), BCL6 abundance in both sets of primary cells was markedly increased in response to the Jak inhibitor tofacitinib, which suggests that BCL6 expression was repressed by the Jak-STAT pathway. Together, our results suggest that agents targeting the pre-BCR or its downstream partners lead to lymphoblast killing in vitro. However, the extent to which this will apply in a clinical setting will require further evaluation, including analysis of surrogate light chain polymorphisms as well as of the abnormal activation of alternative tyrosine kinase signaling pathways linked to specific mutations in non-pre-BCR pathways.
DISCUSSION
Seminal studies over 20 years ago led to the discovery of the surrogate light chain components, VpreB and λ5, which proved to be critical for the cell surface expression of the μH chain in early B cells (58). The assembly of Igμ with surrogate light chain components generates a functional cell surface receptor that signals the successful recombination of the heavy chain, initiates a burst of pre-B-cell proliferation, and then sets the stage for cell cycle arrest and the recombination of light chain–encoding genes (59). Reviews have covered critical aspects of the pre-B cell checkpoint, including the proximal events associated with pre-BCR signaling (19) and complicated transcriptional regulation, which are important to control B cell development and avoid autoreactivity (60). These checkpoints are often overridden in the pre-B leukemia setting (25).
We chose to study the pre-BCR in the context of BCP-ALL, which is one of the most prevalent neoplasms in children (61) and an aggressive cancer in adolescents and adults (62). Although therapies have markedly increased the number of relapse-free patients in the past 40 years, there remain inherent concerns with existing treatments (63). Among these include the association of early chemotherapeutic administration with serious effects that alter early and late life stages (64). Considering these factors and the fact that high-risk leukemia outcome appears to have generally plateaued with conventional therapy (65), a push for new therapeutic approaches and agents is of rising importance. Based on success in mature B cell neoplasms (66–68), the pre-BCR and its signaling partners (31) may also offer new promise for targeted therapy in BCP-ALL.
We focused here on the earliest events in pre-BCR signaling: the homo-oligomerization that drives autonomous signaling. We applied state-of-the-art imaging techniques to demonstrate the highly transient nature of homotypical pre-BCR interactions. These interactions influence pre-BCR diffusion in the plasma membrane of early lineage B cells. We found that pre-BCR diffusion in 697 cells and blasts from patient #238 was relatively fast at ~0.12 μm2/s (table S2 and Fig. 6C). HMM analysis of >50,000 trajectories in 697 cells enabled these trajectories to be further divided into 2 groups: The fast group, traveling at 0.16 μm2/s, were considered “free” QD-tagged receptors and assumed to be mostly monomers, with the caveat that interactions with dark receptors cannot be captured under these SPT imaging conditions. The slow group, traveling at 0.059 μm2/s, represents QD-tagged pairs during the intervals in which they are engaged in homotypic interactions (Fig. 1G). It is likely that the slow population also reflects the transient residency of the pre-BCR in protein islands, which is already described as a mechanism to cluster the TCR and BCR in lymphocyte membranes (69, 70). Our results, which capture repeated interactions between the same pair of pre-BCRs (Fig. 1C), are consistent with the transient co-confinement of pre-BCRs in these specialized membrane microdomains and within cytoskeletal corrals.
Diffusion of the pre-BCR in the plasma membranes of Nalm6 cells and patient #280 blasts was slightly slower overall at ~0.09 μm2/s, with values of 0.12 μm2/s for free receptors and 0.015 μm2/s for receptors engaged in homotypic interactions. The slower diffusion rates for Nalm6 receptors in the bound state might reflect the five-fold slower off-rate for pre-BCR in Nalm6 compared to 697 cells (Fig. 1J and fig. S4), and therefore a greater propensity to form slow-moving, higher-order oligomers. We observed several instances in Nalm6 cells in which interacting pre-BCR pairs had an interaction distance >100 nm (fig. S5 and movie S2). Similarly, single-molecule imaging studies of the BCR demonstrated that higher affinity interactions support an enhanced ability of the receptor to oligomerize (71). We do not currently have a definitive explanation for the two distinct off-rates that we observed for the homotypic pre-BCR interactions on different BCP-ALL cells, but we speculate that it could be because of the observed VpreB SNPs that we documented for this group (fig. S12A). Of particular interest is the SNP variant that introduces a putative N-glycosylation site (substitution of asparagine for aspartic acid at position 76). A 3D structural model based upon the solved NMR and X-ray crystal structures of the pre-BCR (fig. S12B) reveals that this amino acid (and any putative posttranslational modification) is at a protein surface that is exposed to be solvent. Differential N-glycosylation of Asn46 of the pre-BCR heavy chain was previously noted to have a major influence on pre-BCR autonomous signaling, possibly by neutralizing an inhibitory influence imposed by the arginine-rich λ5 tail of the surrogate light chain (15). The substitution of lysine for glutamic acid at position 132, seen in those 697 cells with a faster off-rate for pre-BCR homotypic interactions, could also reflect the change in charge near the putative VpreB dimerization site (17).
The BCRs of naïve and mature B cells have been the subject of impactful, high-resolution imaging studies (22, 70–73) that provide a context for the current work. BCR diffusion rates in the absence of ligand are generally slower than those of the pre-BCR, with variability amongst B cell sources and across imaging conditions. For example, BCR diffusion in J558L B cells was measured in TIRF mode on antigen-free lipid bilayers to have a range from 0.039 to 0.069 μm2/s (71). Up to 20% of BCRs are immobile under those conditions (D < 0.01 μm2/s). On the surface of A20 B cells, wild-type IgM BCR travels at an average rate of 0.031 μm2/s, with the caveat that those IgM BCRs that are outside actin-rich regions diffuse at a faster rate (0.108 μm2/s) than that of BCRs within actin-rich membrane (0.039 μm2/s) (22). The heavy chain moiety of BCR is very influential, because the IgD BCR exhibits very slow diffusion (0.005 μm2/s) (22). The cellular context also matters, particularly the influence of the actin cortical cytoskeleton, which forms corrals to restrict receptor motion (24). The BCR undergoes faster diffusion in B cells treated with latrunculin A (22) or in cells stimulated with ligands for Toll-like receptors, which induce cofilin-mediated severing of actin filaments (73). We speculate that variability in the cortical actin cytoskeleton of Nalm6 and 697 cells and in cells from the two primary patients could contribute to our observed differences in pre-BCR diffusion rates.
Our experimental design was inspired by mutagenesis and structural studies that implicate the unique regions of λ5 in pre-BCR homotypic oligomerization for ligand-independent signaling (17, 18). We reasoned that Fabs directed at the VpreB component of the surrogate light chain would sterically block λ5-mediated homodimerization. This strategy was successful, because incubation with these monomeric antibody fragments resulted in faster pre-BCR diffusion rates and a marked reduction in observed dimerization events. Furthermore, the Fabs also blocked the accumulation of pCD79a and pSyk in the presence of pervanadate and H2O2, a strategy that reveals the close control of autonomous signaling by phosphatases (51). Kinetic proofreading is proposed as a generalized schema by which the number of receptors occupied and the duration of receptor occupancy translate into functional signals (74). In the absence of high-affinity interactions, frequent rebinding events associated with spatial confinement appear to provide the TCR a mechanism to propagate even weak signals (75). We speculate that frequent, low-affinity rebinding events also provide exactly the right threshold needed for the pre-BCR to maintain the narrow window for early B survival.
Galectin-1 binds to the λ5-UR domain of the pre-BCR (35) and, although this interaction is independent of the glycan-binding site of galectin, engagement with the pre-BCR induces local conformational changes that alter the affinity of galectin-1 for carbohydrates (37). Thus, it was important to evaluate the behavior of pre-BCRs that were crosslinked with soluble, dimeric galectin-1 in the presence and absence of lactose to distinguish “lattice-like” complexes, which include other cell-surface glycans, from dimers and chains of pre-BCRs and galectin-1 alone. We showed that the pre-BCR rapidly became immobile after the addition of galectin-1 alone (Fig. 3B and movie S3), forming clusters that were large enough to observe by standard epifluorescence techniques (Fig. 2A). Inclusion of lactose during galectin-1–mediated crosslinking disrupted complex, glycan-mediated aggregation, which was observed as a substantial change in the values for uncorrelated jump distance (Fig. 2B) and jump magnitude (Fig. 2C), as well as in overall diffusion (Fig. 2D). It is remarkable that galectin-mediated crosslinking in either context did not support accumulation of BCL6 protein. The need for a perfectly tuned autonomous pre-BCR signal was previously described as the “Goldilocks” l threshold that optimally supports the PI3K-Akt pro-survival pathway (76, 77). The role for galectin-1–mediated crosslinking of the pre-BCR in ALL deserves further study. There may be a difference between galectin-1–mediated pre-BCR signaling when galectin-1 is produced as a soluble autocrine factor (a notable feature of MLL-rearranged BCP-ALL cells) (39) versus galectin presentation on the surface of stromal cells (78). It has been proposed that early B cells occupy different bone marrow niches as they progress from dependency on IL-7 to stromal presentation of galectin (16).
One challenge will be to identify those BCP-ALL patients for which pre-BCR targeted therapy is appropriate, because there are multiple mechanisms that enable leukemia cells to bypass the pre-BCR checkpoint (25). A recent study of 830 cases from four clinical trials identified 13.5% of patients as being particularly reliant on autonomous pre-BCR signaling (26). On face value, the two patient samples (#280 and #238) that we studied here would fall within this class, because they expressed pre-BCR on the cell surface and were sensitive to dasatinib (Fig. 7). However, only one patient sample (#280) exhibited faster diffusion of the pre-BCR after exposure to anti-VpreB Fabs, which was accompanied by an increase in the fraction of apoptotic cells. This indicates that the cell surface expression of the pre-BCR alone does not predict reliance on autonomous signaling for survival.
Important BCP-ALL subsets rely on aberrant Abl or Jak signaling pathways, indicating that they are arrested very early in the B cell developmental process. Twenty-five percent of BCP-ALL cases express BCR-ABL because of a 9:22 translocation (Philadelphia chromosome, Ph+), and their responses to BCR-ABL kinase inhibitors can be short-lived (79). A “Ph-like” transcriptional signature has been linked to poor outcome in 10% of childhood ALL and 27% of adult ALL cases; the novel translocations and activating mutations associated with Ph-like ALL involve a number of cytokine receptors (FTL3, IL-7R, CRLF2, EPOR, PDGFRB, TSLF) and their associated signaling partners (kinases of the Jak and Abl families) (80). It appears that the ITAM-Syk and the ITIM-phosphatase axes are operating in the background of these oncogenic signaling pathways, because inhibitors of both pathways can stimulate pre-existing B cell programs for cell death (50). ITIM-bearing receptors (including, PECAM1, CD300A, LAIR1, and CD22/SIGLEC-2) are expressed in BCP-ALL cells, where they serve critical roles in recruiting the tyrosine and inositol phosphatases SHP1 (PTPN6) and SHIP-1 (INPP5D), respectively (50, 81). Whereas SHIP-1 acts as a mediator of negative signaling from ITIM-bearing receptors in immune cells, SHIP-1 is important in negative regulatory loops downstream of the BCR (82). SHIP-1 can be engaged under conditions that promote the incomplete phosphorylation of Igα and Igβ ITAMs, which favors the recruitment of single SH2 domain–bearing phosphatases over the recruitment of the dual SH2 domain–bearing kinase Syk (83). The Lyn–SHIP-1–PTEN–SHP1 negative regulatory axis is an essential and constitutively engaged regulator of the BCR, particularly in the context of anergy (83, 84).
The complexities of the pre-BCR and its downstream partners (Syk, SLP65, and BCL6) alternatively serving as oncogenes or tumor suppressors have been reviewed (25). Central to understanding this complexity will be accurate maps of the stage-specific transcription factor network, which normally coordinates B lymphoid progression, and how disruption at key checkpoints leads to oncogenesis (85). We expect that improved understanding of pre-BCR dynamics and signaling outcomes, begun here with the current study, will be particularly important as tyrosine kinase inhibitors are incorporated into BCP-ALL therapy. Because of complex regulatory feed-forward and feed-back loops in early B cell development (26, 41, 85), it is critical to understand if low-level pre-BCR signaling events offer potential sources of drug resistance when the strongly proliferative signals from BCR-ABL and mutant Jak-STAT signaling pathways are suppressed. The exquisite sensitivity of BCP-ALL cells to inhibitors of Lyn, Syk, and SHIP-1, as shown here, underscores that a substantial number of BCP-ALL cases are likely to benefit when chemotherapy and kinase inhibitors are administered in combination This concept has already been demonstrated in an animal model of Ph+ ALL, combining dasatinib (which inhibits Abl and Lyn) with ruxolitinib (which inhibits Jak) (86). Serial administration or immunotherapy may be possible if toxicity is a barrier when chemotherapeutics and kinase inhibitors are co-administered.
MATERIALS AND METHODS
Cell culture and treatments
Suspension cultures of BCP-ALL cell lines (697 and Nalm6) were cultured in RPMI 1640 with phenol red (Sigma-Aldrich, Saint Louis, MO), with 10% (v/v) heat-inactivated fetal bovine serum (HI FBS), 1% Penicillin/Streptomycin (Gibco, Gaithersburg, MD), and 2 mM L-Glutamine. Patient samples were cultured in IMDM (Iscove’s Modified Dulbecco’s Medium), GlutaMAX (Gibco) supplemented with 20% (v/v) HI FBS, 1% Penicillin/Streptomycin, 1x Insulin, transferrin, and selenium (Gibco), 1 mM sodium pyruvate (Gibco), and 55 μM 2-Mercaptoethanol (Gibco). Before experiments were performed, cells were washed twice with Tyrode’s solution. Unless otherwise stated, cells were pretreated for 10 min with BAY61-3606 (1 μM), dasatinib (1 μM), tofacitinib (1 μM), anti-VpreB Fab (1 μM), or recombinant galectin-1 (10 μM) in the presence or absence of lactose in Tyrode’s solution, 0.1% bovine serum albumin (BSA), and 20 mM glucose. For SPT experiments, galectin-1 was purchased from Peprotech (Rocky Hill, NJ). Inhibitors were purchased from Santa Cruz Biotechnology (Dallas, TX), Selleck Chemicals (Houston, TX), and Echelon Biosciences (Salt Lake City, UT).
Recombinant anti-VpreB antibody screening and production
The Contextual Combinatorial Immune Repertoire (ConCIRT) Synthetic Library (87), consisting of 56 billion synthetically constructed human antibodies arrayed in over 100 phage-displayed sub-libraries, was screened against recombinant “surrobodies.” Surrobodies consist of a functional human IgG1 heavy chain isolated from an anti-influenza H5N1 hemagglutinin antibody and paired with either surrogate light chain components (VpreB1, λ5) or a chimeric polypeptide that is a product of the fusion of human VpreB1 and λ5. Phage bound to surrobodies coated on a 96-well microtiter plate were eluted and amplified for use in a subsequent round of screening. After four rounds of phage screening, individual clones from enriched pools were analyzed for specific binding by ELISA. Testing for binding specificity to VpreB1 was performed on ELISA plates coated with a chimeric VpreB1-λ5 polypeptide. Detection was quantified with HRP-conjugated anti-myc antibodies, which was followed by ELISA-based hit identification. After positive identification, 16 phage-derived clones with an affinity for VpreB in the range 0.85 to 250 nM were sequenced. Clone 2460B04 was selected for integration into a mammalian expression plasmid, which was followed by transient transfection of HEK 293-F cells for the production of recombinant IgG antibodies. Intact IgG was reduced with 2MEA (2-mercaptoethylamine-HCl) to produce anti-VpreB1 Fab fragments. The reactivity of anti-VpreB1 Fabs against live 697 cells was evaluated by flow cytometry (fig. S7).
Anti-CD79b IgG purification
CB3-1 hybridoma cells (generously provided by Dr. Max D. Cooper, Emory University) were cultured in RPMI 1640 with phenol red (Sigma-Aldrich) containing 10% (v/v) HI FBS, 1% Penicillin/Streptomycin (ThermoFisher Scientific), 2 mM L-Glutamine (Gibco) and 1x β-mercaptoethanol (Gibco). Flask cultures were expanded by 2-week culture in 250 ml in RPMI media without FBS, which was followed by media collection, clarification by centrifugation at 560g, and filtration through a 0.22-μm vacuum filter. The culture medium was circulated overnight (4°C) on a protein A/G affinity column and the IgG was eluted with glycine buffer (pH 2.5 to 3.0). Eluted 1-ml fractions were neutralized with 1 M TRIS buffer (pH 9.0).
Fab’ generation, biotin conjugation, and QD labeling
Fab’-biotin probes were prepared as previously described (42). IgG derived from the CB3-1 hybridoma or anti-IgM (Jackson Immuno Research, West Grove, PA) was dialyzed into sodium acetate solution (pH 4.5) to prepare for addition to pepsin-conjugated agarose (ThermoFisher Scientific, Waltham, MA) according to the manufacturer’s instructions. IgG digestion was performed for 5 h at 37°C, and the eluate was dialyzed at 4°C into 100 mM phosphate, 5 mM EDTA, (pH 6.0) to prepare for reduction of the interchain disulfides of F(ab’)2 by 2-mercaptoethylamine (2-MEA, ThermoFisher Scientific). The sample was incubated in 2-MEA (50 mM) for 90 min at 37°C and then dialyzed into storage buffer [100 mM phosphate, 5 mM EDTA, (pH 6.5 to 7.0)] for 4 hours at 4°C, exchanging buffer multiple times. The exposed thiol groups, generated from the reduction of interchain disulfides, were selectively conjugated to biotin with the EZ-Link Maleimide-PEG2-Biotin (ThermoFisher Scientific) and resolved by SDS-PAGE. To remove higher molecular weight bands (F(ab’)2 and undigested IgG, digested fragments were recovered at room temperature by FPLC size-exclusion chromatography with Superdex 75 10/300 column (GE Healthcare Life Sciences, Marlborough, MA) at a flow rate of 0.5 ml/min in phosphate-buffered saline (PBS) (fig. S1A). Residual intact Fc fragments were removed by batch processing with protein A/G agarose beads (ThermoFisher Scientific). The purity of the anti-CD79b Fab’-maleimide biotin conjugates was determined by Coomassie stain and Western blotting analysis (fig. S1, B and C) after the samples were resolved by SDS-PAGE under nonreducing conditions. The biotin to Fab’ molar ratio was determined with the FluoReporter biotective green reagent (ThermoFisher Scientific) with a reported mean biotin:Fab molar ratio of 1:1. Anti-CD79b Fab’-Biotin or Anti-IgM Fab’-Biotin were mixed at a 1:1 ratio with Qdot655-strepavidin or Qdot585-streptavidin in PBS containing 1% (w/v) BSA to generate stock solutions.
QD labeling of live cells
Lab-Tek imaging chambers were coated with poly-l-lysine hydrobromide (Sigma-Aldrich) at 1 mg/ml in sterile water for 20 min at room temperature, which was followed by three washes with water. BCP-ALL cells were serum-starved in RPMI 1640 without FBS for 2 hours before being added to coated wells and incubated for 15 min at 37°C. QD-Fab’-CD79b or QD-Fab’-IgM were added at a final concentration of 100 pM in Tyrodes’s solution with 0.1% (w/v) BSA and 20 mM glucose and were incubated for 10 to 15 min at 37°C.
Single particle tracking and analysis
All corresponding GPU SPT and track elongation, squared displacement, mean square displacement, correlated motion, fiducial data acquisition and image registration, and three-state hidden Markov Model used in this study were described in detail previously (43).
Galectin-1 cloning, expression, and purification
MGC human galectin-1–encoding cDNA (LGALS1, accession# BC001693) was PCR-amplified with the CACC forward primer (FWD: 5′-CAC CAT GGC TTG TGG TCT GG-3′) and reverse primer (REV: 5′-TCA GTC AAA GGC CAC ACA TTT GAT CT-3′). The amplified product was subcloned into pET101 by directional TOPO expression. The cloned product was transformed into One SHOT TOP10 Escherichia coli by heat-shock. Cells were grown overnight on ampicillin. Resistant cultures were selected and were used for DNA isolation. BL21 StarTM One Shot cells were transformed with the pET101 vector with LGALS1 by heat-shock, brief outgrowth in SOC medium, followed by transfer to 10 ml of LB containing ampicillin, and the cultures were grown overnight at 37°C while shaking. The next day, 50 ml of LB containing ampicillin was inoculated with 1 ml of the overnight culture. The culture was grown at 37°C with shaking (225 to 250 rpm) for 2 to 3 hours. IPTG (1 mM) was added to induce the expression of galectin-1 for 3 to 4 hours. The cells were then harvested by centrifugation at 3000g for 10 min at 4°C. The cells were purified by α-lactose/agarose (Sigma-Aldrich) as detailed previously (88).
Western blotting
After the cells were incubated in the presence or absence of the appropriate inhibitors or crosslinking agents, they were washed with ice-cold PBS and incubated for 15 min on ice in Tris-based lysis buffer containing 1% NP-40 and protease and phosphatase inhibitors (Thermo Scientific). To examine the effects of inhibitors on BCL6 abundance, treatments were performed for 24 hours. Cell lysates were clarified by centrifugation at 14,000g for 10 min at 4°C and then were added to Laemmli’s Reducing buffer (final 2x). Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. After the blots were blocked with Tris-buffered saline containing 5% BSA, they were incubated with antibodies specific for pCD79a-Tyr182 (CST, Danver, MA), pSyk-Tyr348 (Novus Biologicals, Littleton, CO), pSyk-Tyr348 (Abcam, Cambridge, MA), pSyk-Tyr352 (CST), or BCL6 (Santa Cruz Biotechnology). Antibody specific for β-actin (Sigma-Aldrich) was used to confirm equivalent loading of gels.
Pervanadate treatment
The use of pervanadate to enhance signaling in various cell types has been reported previously (89–91). Here, all BCP-ALL cells were treated with freshly prepared pervanadate solution (0.2 mM sodium orthovanadate and 1 mM hydrogen peroxide) in the presence or absence of inhibitors and then treated for the times indicated in the figure legends.
Chemosensitization
To establish an effective concentration range for the potential synergy of targeted inhibitors with vincristine, 697 cells (50,000 cells per condition) were incubated for 3 days at 37°C with a range of concentrations of vincristine (0.1 to 100.0 ng/ml) (Sigma-Aldrich). Controls are indicated in the figures, including vincristine alone and vehicle at the highest DMSO concentration used (0.2%). Curves were fit with a sigmoidal dose-response curve with the curve fitting toolbox of MATLAB.
Flow cytometry assays
Leukemia cell apoptosis was determined based on measurement of the binding of Annexin-V-FITC (BioLegend, San Diego, CA) or Pacific Blue Annexin-V (BioLegend), whereas loss of cell viability was measured by 7-AAD labeling (BioLegend), according to the manufacturer’s procedures. For receptor-binding experiments, cells were incubated under saturating conditions for 1.5 hours on ice in PBS with anti-CD79b-APC antibody (BioLegend) or anti-VpreB-PE antibody (BioLegend). Flow cytometry data were calibrated with QuantumTM Simply Cellular beads (Bangs Laboratories, Fishers, IN) with anti-mouse Fc antibody according to the manufacturer’s instructions. Dissociation constants were estimated with nonlinear regression analysis under KD-controlled conditions as described previously (92). All flow cytometry data collection was conducted on HyperCyt (93) or BD LSRFortessa flow cytometers and analyzed with the FlowJo Software Suite (Tree Star, Ashland, OR).
RT-PCR mRNA sequence analysis
Total RNA was extracted from the 697 and Nalm6 cell lines and from patient samples #238 and #280 with NucleoSpin RNA XS (Macherey-Nagel, Duren, Germany). cDNA synthesis was performed with both random hexamers and d(T)20 primers with SuperScript First-Strand Synthesis System (ThermoFisher Scientific). Primers were designed for the 5′-UTR (FWD: 5′-GAGCTCTGCATGTCTGCACC-3′) and 3′-UTR (REV: 5′-CGTGCCTCTGCTGTCTTCAG-3′) based upon the sequence of the reference VPREB1 mRNA (NM_007128.3) sequence. PCR reactions used the Platinum PCR SuperMix (ThermoFisher Scientific). The Thermo Cycler set-up was as follows: 94°C for 2 min to denature the DNA and activate the enzyme; 35 cycles of PCR amplification followed by 30 s at 94°C, 30 s at 55°C, and 72°C for 1 min. All PCR amplicons were agarose gel–purified (Macherey-Nagel) and subcloned into the pCR-BluntII-TOPO vector according to the guidelines of the Zero Blunt TOPO PCR cloning kit (ThermoFisher Scientific). One Shot Competent E. coli were then transformed with the plasmids, and 10 to 20 colonies were selected on LB plates with kanamycin (50 μg/ml) and grown in 5 ml of LB containing kanamycin overnight. Plasmids were purified with the NucleoSpin Plasmid Miniprep (Macherey-Nagel). Purified plasmids were sequenced (Genewiz, Cambridge, Massachusetts) using their predefined Sp6 and T7 primers. Only sequences that passed Genewiz quality control were analyzed. Each DNA sequence was compared to the reference sequence (NM_007128.3) by NCBI BLAST search. All multiple protein sequence alignments were performed with the MUSCLE server (94) and were compared to the reference sequence (UniProt #P12018). Prediction of the N-glycosylation of the reference and variant sequences was assessed with the NetNGlyc 1.0 Server.
Composite 3D model of preBCR Fab-like arms and Fcμ
Because the Fab-like X-Ray crystal structure of the preBCR (PDB: 2H32) does not contain the unique region, we used the MODELLER software suite (95) to build a homology model including the surrogate light chain unique region (UR) components. These surrogate light chain λ5-UR and VpreB-UR structural components were derived from the NMR solution structure (PDB: 2LKQ) and de novo peptide design PEP-FOLD online server (96), respectively. The composite 3D model shows the Fab-like arms extended from the Fcμ region derived from NMR and X-Ray structures (PDB: 4JVU, 4BA8, and 4JVW). Residues implicated in polymorphism were analyzed and highlighted with the VMD software package (97).
Supplementary Material
Fig. S1. Generation, purification, and characterization of anti-Igβ Fab’ probes.
Fig. S2. Viterbi plots of states observed for pairs of QD-bound receptors under autonomous signaling conditions.
Fig. S3. Comparison of MSD values for pre-BCRs tracked under different conditions.
Fig. S4. Comparison of off-rates for homotypic interactions between pre-BCRs in two different cell lines.
Fig. S5. 3D plot for pairs of QD probes showing correlated motion at wide separation distances.
Fig. S6. Galectin-1 purification, homology model, and diffusion profile with varying concentrations.
Fig. S7. Characterization of the binding of the anti-VpreB antibody to 697 cells.
Fig. S8. Apoptotic response to single-agent treatment with kinase inhibitors targeting the pre-BCR pathway.
Fig. S9. Apoptotic response to combination treatment with vincristine and inhibitors of the pre-BCR pathway.
Fig. S10. Apoptotic response to combination treatment with vincristine and a SHIP inhibitor.
Fig. S11. Quantitation of the amounts of VpreB and Igβ on the surface of different BCP-ALL cells.
Fig. S12. Sequence alignment reveals polymorphism among different cell lines and patient samples.
Table S1. Characteristics of BCP ALL cells used in this study.
Table S2. SPT receptor diffusion statistics.
Table S3. SPT receptor dimer statistics.
Movie S1. Two-color SPT analysis of 697 cells depicting correlated motion and serial engagement.
Movie S2. Two-color SPT analysis of Nalm6 cells depicting correlated motion at wide (> 200 nm) distances.
Movie S3. SPT analysis showing pre-BCR diffusion in 697 cells in the presence or absence of exogenous galectin-1.
Acknowledgments
Use of the UNM Cancer Center microscopy and flow cytometry facilities, and NIH P30CA118100 support for these cores, is gratefully acknowledged. We thank the clinical staff in UNM Pediatric Oncology for participating in the HRRC 05-235 study and members of the New Mexico Spatiotemporal Modeling Center for technical advice and assistance, particularly Drs. Mara Steinkamp and Cedric Cleyrat.
Funding: This work was supported by NIH grants P50GM085273 (to B.W.), F31CA192848 (to M.F.E.), and R01BM100114 (to D.L.).
Footnotes
Author contributions: M.F.E. performed most of the experiments; K.M. and I.K. assisted in the culture and development of hybridomas and patient-derived cells; A.M. assisted in development of biochemical assays; L.X. and M.H. screened for and produced the anti-VpreB antibodies; S.W. was clinical coordinator for the study; D.L. developed SPT approaches and supervised data collection and analysis; and B.W. developed the study strategy, supervised biochemical and cell-based studies, reviewed results, and prepared the manuscript with M.F.E. and D.L.
Competing interests: B.W. and colleagues disclose intellectual property related to pre-BCR immunotherapy initiatives.
REFERENCES AND NOTES
- 1.Bannish G, Fuentes-Panana EM, Cambier JC, Pear WS, Monroe JG. Ligand-independent signaling functions for the B lymphocyte antigen receptor and their role in positive selection during B lymphopoiesis. The Journal of experimental medicine. 2001;194:1583–1596. doi: 10.1084/jem.194.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geahlen RL. Syk and pTyr’d: Signaling through the B cell antigen receptor. Biochim Biophys Acta. 2009;1793:1115–1127. doi: 10.1016/j.bbamcr.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurosaki T, Johnson SA, Pao L, Sada K, Yamamura H, Cambier JC. Role of the Syk autophosphorylation site and SH2 domains in B cell antigen receptor signaling. J Exp Med. 1995;182:1815–1823. doi: 10.1084/jem.182.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nahar R, Ramezani-Rad P, Mossner M, Duy C, Cerchietti L, Geng H, Dovat S, Jumaa H, Ye BH, Melnick A, Muschen M. Pre-B cell receptor-mediated activation of BCL6 induces pre-B cell quiescence through transcriptional repression of MYC. Blood. 2011;118:4174–4178. doi: 10.1182/blood-2011-01-331181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandal M, Powers SE, Ochiai K, Georgopoulos K, Kee BL, Singh H, Clark MR. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat Immunol. 2009;10:1110–1117. doi: 10.1038/ni.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stadhouders R, de Bruijn MJ, Rother MB, Yuvaraj S, Ribeiro de Almeida C, Kolovos P, Van Zelm MC, van Ijcken W, Grosveld F, Soler E, Hendriks RW. Pre-B cell receptor signaling induces immunoglobulin kappa locus accessibility by functional redistribution of enhancer-mediated chromatin interactions. PLoS Biol. 2014;12:e1001791. doi: 10.1371/journal.pbio.1001791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuh W, Meister S, Herrmann K, Bradl H, Jack HM. Transcriptome analysis in primary B lymphoid precursors following induction of the pre-B cell receptor. Mol Immunol. 2008;45:362–375. doi: 10.1016/j.molimm.2007.06.154. [DOI] [PubMed] [Google Scholar]
- 8.Anbazhagan K, Rabbind Singh A, Isabelle P, Stella I, Celine AD, Bissac E, Bertrand B, Remy N, Naomi T, Vincent F, Rochette J, Lassoued K. Human pre-B cell receptor signal transduction: evidence for distinct roles of PI3kinase and MAP-kinase signalling pathways. Immun Inflamm Dis. 2013;1:26–36. doi: 10.1002/iid3.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawano Y, Yoshikawa S, Minegishi Y, Karasuyama H. Pre-B cell receptor assesses the quality of IgH chains and tunes the pre-B cell repertoire by delivering differential signals. J Immunol. 2006;177:2242–2249. doi: 10.4049/jimmunol.177.4.2242. [DOI] [PubMed] [Google Scholar]
- 10.Almqvist N, I, Martensson L. The pre-B cell receptor; selecting for or against autoreactivity. Scand J Immunol. 2012;76:256–262. doi: 10.1111/j.1365-3083.2012.02751.x. [DOI] [PubMed] [Google Scholar]
- 11.Zouali M. Transcriptional and metabolic pre-B cell receptor-mediated checkpoints: implications for autoimmune diseases. Mol Immunol. 2014;62:315–320. doi: 10.1016/j.molimm.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Grimsholm O, Ren W, Bernardi AI, Chen H, Park G, Camponeschi A, Chen D, Bergmann B, Hook N, Andersson S, Stromberg A, Gjertsson I, Cardell S, Yrlid U, De Riva A, Martensson IL. Absence of surrogate light chain results in spontaneous autoreactive germinal centres expanding V(H)81X-expressing B cells. Nat Commun. 2015;6:7077. doi: 10.1038/ncomms8077. [DOI] [PubMed] [Google Scholar]
- 13.Fuentes-Panana EM, Bannish G, Shah N, Monroe JG. Basal Igalpha/Igbeta signals trigger the coordinated initiation of pre-B cell antigen receptor-dependent processes. J Immunol. 2004;173:1000–1011. doi: 10.4049/jimmunol.173.2.1000. [DOI] [PubMed] [Google Scholar]
- 14.Kohler F, Hug E, Eschbach C, Meixlsperger S, Hobeika E, Kofer J, Wardemann H, Jumaa H. Autoreactive B cell receptors mimic autonomous pre-B cell receptor signaling and induce proliferation of early B cells. Immunity. 2008;29:912–921. doi: 10.1016/j.immuni.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 15.Ubelhart R, Bach MP, Eschbach C, Wossning T, Reth M, Jumaa H. N-linked glycosylation selectively regulates autonomous precursor BCR function. Nat Immunol. 2010;11:759–765. doi: 10.1038/ni.1903. [DOI] [PubMed] [Google Scholar]
- 16.Mourcin F, Breton C, Tellier J, Narang P, Chasson L, Jorquera A, Coles M, Schiff C, Mancini SJ. Galectin-1-expressing stromal cells constitute a specific niche for pre-BII cell development in mouse bone marrow. Blood. 2011;117:6552–6561. doi: 10.1182/blood-2010-12-323113. [DOI] [PubMed] [Google Scholar]
- 17.Ohnishi K, Melchers F. The nonimmunoglobulin portion of lambda5 mediates cell-autonomous pre-B cell receptor signaling. Nat Immunol. 2003;4:849–856. doi: 10.1038/ni959. [DOI] [PubMed] [Google Scholar]
- 18.Bankovich AJ, Raunser S, Juo ZS, Walz T, Davis MM, Garcia KC. Structural insight into pre-B cell receptor function. Science. 2007;316:291–294. doi: 10.1126/science.1139412. [DOI] [PubMed] [Google Scholar]
- 19.Monroe JG. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat Rev Immunol. 2006;6:283–294. doi: 10.1038/nri1808. [DOI] [PubMed] [Google Scholar]
- 20.Tsourkas PK, Liu W, Das SC, Pierce SK, Raychaudhuri S. Discrimination of membrane antigen affinity by B cells requires dominance of kinetic proofreading over serial engagement. Cell Mol Immunol. 2012;9:62–74. doi: 10.1038/cmi.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tolar P, Meckel T. Imaging B-cell receptor signaling by single-molecule techniques. Methods Mol Biol. 2009;571:437–453. doi: 10.1007/978-1-60761-198-1_29. [DOI] [PubMed] [Google Scholar]
- 22.Treanor B, Depoil D, Gonzalez-Granja A, Barral P, Weber M, Dushek O, Bruckbauer A, Batista FD. The membrane skeleton controls diffusion dynamics and signaling through the B cell receptor. Immunity. 2010;32:187–199. doi: 10.1016/j.immuni.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrews NL, Pfeiffer JR, Martinez AM, Haaland DM, Davis RW, Kawakami T, Oliver JM, Wilson BS, Lidke DS. Small, mobile FcepsilonRI receptor aggregates are signaling competent. Immunity. 2009;31:469–479. doi: 10.1016/j.immuni.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews NL, Lidke KA, Pfeiffer JR, Burns AR, Wilson BS, Oliver JM, Lidke DS. Actin restricts FcepsilonRI diffusion and facilitates antigen-induced receptor immobilization. Nat Cell Biol. 2008;10:955–963. doi: 10.1038/ncb1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eswaran J, Sinclair P, Heidenreich O, Irving J, Russell LJ, Hall A, Calado DP, Harrison CJ, Vormoor J. The pre-B-cell receptor checkpoint in acute lymphoblastic leukaemia. Leukemia. 2015;29:1623–1631. doi: 10.1038/leu.2015.113. [DOI] [PubMed] [Google Scholar]
- 26.Geng H, Hurtz C, Lenz KB, Chen Z, Baumjohann D, Thompson S, Goloviznina NA, Chen WY, Huan J, LaTocha D, Ballabio E, Xiao G, Lee JW, Deucher A, Qi Z, Park E, Huang C, Nahar R, Kweon SM, Shojaee S, Chan LN, Yu J, Kornblau SM, Bijl JJ, Ye BH, Ansel KM, Paietta E, Melnick A, Hunger SP, Kurre P, Tyner JW, Loh ML, Roeder RG, Druker BJ, Burger JA, Milne TA, Chang BH, Muschen M. Self-enforcing feedback activation between BCL6 and pre-B cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer cell. 2015;27:409–425. doi: 10.1016/j.ccell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis RE, V, Ngo N, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, Xu W, Shaffer AL, Wright G, Xiao W, Powell J, Jiang JK, Thomas CJ, Rosenwald A, Ott G, Muller-Hermelink HK, Gascoyne RD, Connors JM, Johnson NA, Rimsza LM, Campo E, Jaffe ES, Wilson WH, Delabie J, Smeland EB, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Pierce SK, Staudt LM. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson FK, Krysov S, Davies AJ, Steele AJ, Packham G. B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2011;118:4313–4320. doi: 10.1182/blood-2011-06-338855. [DOI] [PubMed] [Google Scholar]
- 29.Niemann CU, Wiestner A. B-cell receptor signaling as a driver of lymphoma development and evolution. Semin Cancer Biol. 2013;23:410–421. doi: 10.1016/j.semcancer.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bicocca VT, Chang BH, Masouleh BK, Muschen M, Loriaux MM, Druker BJ, Tyner JW. Crosstalk between ROR1 and the Pre-B cell receptor promotes survival of t(1;19) acute lymphoblastic leukemia. Cancer cell. 2012;22:656–667. doi: 10.1016/j.ccr.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perova T, Grandal I, Nutter LM, Papp E, Matei IR, Beyene J, Kowalski PE, Hitzler JK, Minden MD, Guidos CJ, Danska JS. Therapeutic potential of spleen tyrosine kinase inhibition for treating high-risk precursor B cell acute lymphoblastic leukemia. Sci Transl Med. 2014;6:236ra262. doi: 10.1126/scitranslmed.3008661. [DOI] [PubMed] [Google Scholar]
- 32.Muschen M. Rationale for targeting the pre-B-cell receptor signaling pathway in acute lymphoblastic leukemia. Blood. 2015;125:3688–3693. doi: 10.1182/blood-2015-01-567842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ubelhart R, Jumaa H. Autoreactivity and the positive selection of B cells. European journal of immunology. 2015 doi: 10.1002/eji.201444622. [DOI] [PubMed] [Google Scholar]
- 34.Barondes SH, Cooper DN, Gitt MA, Leffler H. Galectins. Structure and function of a large family of animal lectins. J Biol Chem. 1994;269:20807–20810. [PubMed] [Google Scholar]
- 35.Elantak L, Espeli M, Boned A, Bornet O, Bonzi J, Gauthier L, Feracci M, Roche P, Guerlesquin F, Schiff C. Structural basis for galectin-1-dependent pre-B cell receptor (pre-BCR) activation. J Biol Chem. 2012;287:44703–44713. doi: 10.1074/jbc.M112.395152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gauthier L, Rossi B, Roux F, Termine E, Schiff C. Galectin-1 is a stromal cell ligand of the pre-B cell receptor (BCR) implicated in synapse formation between pre-B and stromal cells and in pre-BCR triggering. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13014–13019. doi: 10.1073/pnas.202323999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonzi J, Bornet O, Betzi S, Kasper BT, Mahal LK, Mancini SJ, Schiff C, Sebban-Kreuzer C, Guerlesquin F, Elantak L. Pre-B cell receptor binding to galectin-1 modifies galectin-1/carbohydrate affinity to modulate specific galectin-1/glycan lattice interactions. Nat Commun. 2015;6:6194. doi: 10.1038/ncomms7194. [DOI] [PubMed] [Google Scholar]
- 38.Rossi B, Espeli M, Schiff C, Gauthier L. Clustering of pre-B cell integrins induces galectin-1-dependent pre-B cell receptor relocalization and activation. J Immunol. 2006;177:796–803. doi: 10.4049/jimmunol.177.2.796. [DOI] [PubMed] [Google Scholar]
- 39.Juszczynski P, Rodig SJ, Ouyang J, O’Donnell E, Takeyama K, Mlynarski W, Mycko K, Szczepanski T, Gaworczyk A, Krivtsov A, Faber J, Sinha AU, Rabinovich GA, Armstrong SA, Kutok JL, Shipp MA. MLL-rearranged B lymphoblastic leukemias selectively express the immunoregulatory carbohydrate-binding protein galectin-1. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:2122–2130. doi: 10.1158/1078-0432.CCR-09-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S, Klemm L, Kweon SM, Nahar R, Braig M, Park E, Kim YM, Hofmann WK, Herzog S, Jumaa H, Koeffler HP, Yu JJ, Heisterkamp N, Graeber TG, Wu H, Ye BH, Melnick A, Muschen M. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shojaee S, Caeser R, Buchner M, Park E, Swaminathan S, Hurtz C, Geng H, Chan LN, Klemm L, Hofmann WK, Qiu YH, Zhang N, Coombes KR, Paietta E, Molkentin J, Koeffler HP, Willman CL, Hunger SP, Melnick A, Kornblau SM, Muschen M. Erk Negative Feedback Control Enables Pre-B Cell Transformation and Represents a Therapeutic Target in Acute Lymphoblastic Leukemia. Cancer cell. 2015;28:114–128. doi: 10.1016/j.ccell.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peluso P, Wilson DS, Do D, Tran H, Venkatasubbaiah M, Quincy D, Heidecker B, Poindexter K, Tolani N, Phelan M, Witte K, Jung LS, Wagner P, Nock S. Optimizing antibody immobilization strategies for the construction of protein microarrays. Anal Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 43.Low-Nam ST, Lidke KA, Cutler PJ, Roovers RC, van Bergen en Henegouwen PM, Wilson BS, Lidke DS. ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat Struct Mol Biol. 2011;18:1244–1249. doi: 10.1038/nsmb.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Findley HW, Jr, Cooper MD, Kim TH, Alvarado C, Ragab AH. Two new acute lymphoblastic leukemia cell lines with early B-cell phenotypes. Blood. 1982;60:1305–1309. [PubMed] [Google Scholar]
- 45.Forney G. The Viterbi algorithm. Proc IEEE. 1973;61:268–278. [Google Scholar]
- 46.Hurwitz R, Hozier J, LeBien T, Minowada J, Gajl-Peczalska K, Kubonishi I, Kersey J. Characterization of a leukemic cell line of the pre-B phenotype. Int J Cancer. 1979;23:174–180. doi: 10.1002/ijc.2910230206. [DOI] [PubMed] [Google Scholar]
- 47.Steinkamp MP, Low-Nam ST, Yang S, Lidke KA, Lidke DS, Wilson BS. erbB3 Is an Active Tyrosine Kinase Capable of Homo- and Heterointeractions. Mol Cell Biol. 2014;34:965–977. doi: 10.1128/MCB.01605-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F, Crise B, Su B, Hou Y, Rose JK, Bothwell A, Jacobson K. Lateral diffusion of membrane-spanning and glycosylphosphatidylinositol-linked proteins: toward establishing rules governing the lateral mobility of membrane proteins. The Journal of cell biology. 1991;115:75–84. doi: 10.1083/jcb.115.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stowell SR, Cho M, Feasley CL, Arthur CM, Song X, Colucci JK, Karmakar S, Mehta P, Dias-Baruffi M, McEver RP, Cummings RD. Ligand reduces galectin-1 sensitivity to oxidative inactivation by enhancing dimer formation. J Biol Chem. 2009;284:4989–4999. doi: 10.1074/jbc.M808925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Z, Shojaee S, Buchner M, Geng H, Lee JW, Klemm L, Titz B, Graeber TG, Park E, Tan YX, Satterthwaite A, Paietta E, Hunger SP, Willman CL, Melnick A, Loh ML, Jung JU, Coligan JE, Bolland S, Mak TW, Limnander A, Jumaa H, Reth M, Weiss A, Lowell CA, Muschen M. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature. 2015;521:357–361. doi: 10.1038/nature14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wienands J, Larbolette O, Reth M. Evidence for a preformed transducer complex organized by the B cell antigen receptor. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7865–7870. doi: 10.1073/pnas.93.15.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rolli V, Gallwitz M, Wossning T, Flemming A, Schamel WW, Zurn C, Reth M. Amplification of B cell antigen receptor signaling by a Syk/ITAM positive feedback loop. Mol Cell. 2002;10:1057–1069. doi: 10.1016/s1097-2765(02)00739-6. [DOI] [PubMed] [Google Scholar]
- 53.Finn RS, Dering J, Ginther C, Wilson CA, Glaspy P, Tchekmedyian N, Slamon DJ. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/”triple-negative” breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105:319–326. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- 54.Degryse S, Cools J. JAK kinase inhibitors for the treatment of acute lymphoblastic leukemia. Journal of hematology & oncology. 2015;8:91. doi: 10.1186/s13045-015-0192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swaminathan S, Klemm L, Park E, Papaemmanuil E, Ford A, Kweon SM, Trageser D, Hasselfeld B, Henke N, Mooster J, Geng H, Schwarz K, Kogan SC, Casellas R, Schatz DG, Lieber MR, Greaves MF, Muschen M. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16:766–774. doi: 10.1038/ni.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Mullighan CG, Harvey RC, Wu G, Chen X, Edmonson M, Buetow KH, Carroll WL, Chen IM, Devidas M, Gerhard DS, Loh ML, Reaman GH, Relling MV, Camitta BM, Bowman WP, Smith MA, Willman CL, Downing JR, Hunger SP. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2011;118:3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Veer A, Waanders E, Pieters R, Willemse ME, Van Reijmersdal SV, Russell LJ, Harrison CJ, Evans WE, van der Velden VH, Hoogerbrugge PM, Van Leeuwen F, Escherich G, Horstmann MA, Mohammadi Khankahdani L, Rizopoulos D, De Groot-Kruseman HA, Sonneveld E, Kuiper RP, Den Boer ML. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 2013;122:2622–2629. doi: 10.1182/blood-2012-10-462358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conley ME, Burrows PD. Plugging the leaky pre-B cell receptor. J Immunol. 2010;184:1127–1129. doi: 10.4049/jimmunol.0990113. [DOI] [PubMed] [Google Scholar]
- 59.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 60.von Boehmer H, Melchers F. Checkpoints in lymphocyte development and autoimmune disease. Nat Immunol. 2010;11:14–20. doi: 10.1038/ni.1794. [DOI] [PubMed] [Google Scholar]
- 61.Winick NJ, Carroll WL, Hunger SP. Childhood leukemia--new advances and challenges. The New England journal of medicine. 2004;351:601–603. doi: 10.1056/NEJMe048154. [DOI] [PubMed] [Google Scholar]
- 62.Douer D. Adult acute lymphoblastic leukemia: a cancer with no standard of care. Acta Haematol. 2013;130:196–198. doi: 10.1159/000351601. [DOI] [PubMed] [Google Scholar]
- 63.Gaynon PS, Angiolillo AL, Carroll WL, Nachman JB, Trigg ME, Sather HN, Hunger SP, Devidas M. Long-term results of the children’s cancer group studies for childhood acute lymphoblastic leukemia 1983–2002: a Children’s Oncology Group Report. Leukemia. 2010;24:285–297. doi: 10.1038/leu.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Asner S, Ammann RA, Ozsahin H, Beck-Popovic M, von der Weid NX. Obesity in long-term survivors of childhood acute lymphoblastic leukemia. Pediatric blood & cancer. 2008;51:118–122. doi: 10.1002/pbc.21496. [DOI] [PubMed] [Google Scholar]
- 65.Winter SS. Pediatric acute leukemia therapies informed by molecular analysis of highrisk disease. Hematology Am Soc Hematol Educ Program. 2011;2011:366–373. doi: 10.1182/asheducation-2011.1.366. [DOI] [PubMed] [Google Scholar]
- 66.Awan FT, Byrd JC. New strategies in chronic lymphocytic leukemia: shifting treatment paradigms. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:5869–5874. doi: 10.1158/1078-0432.CCR-14-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharman J, Hawkins M, Kolibaba K, Boxer M, Klein L, Wu M, Hu J, Abella S, Yasenchak C. An open-label phase 2 trial of entospletinib (GS-9973), a selective spleen tyrosine kinase inhibitor, in chronic lymphocytic leukemia. Blood. 2015;125:2336–2343. doi: 10.1182/blood-2014-08-595934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puri KD, Di Paolo JA, Gold MR. B-cell receptor signaling inhibitors for treatment of autoimmune inflammatory diseases and B-cell malignancies. Int Rev Immunol. 2013;32:397–427. doi: 10.3109/08830185.2013.818140. [DOI] [PubMed] [Google Scholar]
- 69.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maity PC, Blount A, Jumaa H, Ronneberger O, Lillemeier BF, Reth M. B cell antigen receptors of the IgM and IgD classes are clustered in different protein islands that are altered during B cell activation. Sci Signal. 2015;8:ra93. doi: 10.1126/scisignal.2005887. [DOI] [PubMed] [Google Scholar]
- 71.Sohn HW, Tolar P, Pierce SK. Membrane heterogeneities in the formation of B cell receptor-Lyn kinase microclusters and the immune synapse. J Cell Biol. 2008;182:367–379. doi: 10.1083/jcb.200802007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, Carter R, Justement LB, Bruckbauer A, Batista FD. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 2013;38:461–474. doi: 10.1016/j.immuni.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 73.Freeman SA, Jaumouille V, Choi K, Hsu BE, Wong HS, Abraham L, Graves ML, Coombs D, Roskelley CD, Das R, Grinstein S, Gold MR. Toll-like receptor ligands sensitize B-cell receptor signalling by reducing actin-dependent spatial confinement of the receptor. Nat Commun. 2015;6:6168. doi: 10.1038/ncomms7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hlavacek WS, Redondo A, Metzger H, Wofsy C, Goldstein B. Kinetic proofreading models for cell signaling predict ways to escape kinetic proofreading. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:7295–7300. doi: 10.1073/pnas.121172298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aleksic M, Dushek O, Zhang H, Shenderov E, Chen JL, Cerundolo V, Coombs D, van der Merwe PA. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity. 2010;32:163–174. doi: 10.1016/j.immuni.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCarthy N. Tumour suppressors: Turning a negative into a positive. Nat Rev Cancer. 2016;16:272–273. doi: 10.1038/nrc.2016.39. [DOI] [PubMed] [Google Scholar]
- 77.Shojaee S, Chan LN, Buchner M, Cazzaniga V, Cosgun KN, Geng H, Qiu YH, von Minden MD, Ernst T, Hochhaus A, Cazzaniga G, Melnick A, Kornblau SM, Graeber TG, Wu H, Jumaa H, Muschen M. PTEN opposes negative selection and enables oncogenic transformation of pre-B cells. Nat Med. 2016;22:379–387. doi: 10.1038/nm.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fei F, Joo EJ, Tarighat SS, Schiffer I, Paz H, Fabbri M, Abdel-Azim H, Groffen J, Heisterkamp N. B-cell precursor acute lymphoblastic leukemia and stromal cells communicate through Galectin-3. Oncotarget. 2015;6:11378–11394. doi: 10.18632/oncotarget.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fielding AK, Rowe JM, Buck G, Foroni L, Gerrard G, Litzow MR, Lazarus H, Luger SM, Marks DI, McMillan AK, Moorman AV, Patel B, Paietta E, Tallman MS, Goldstone AH. UKALLXII/ECOG2993: addition of imatinib to a standard treatment regimen enhances long-term outcomes in Philadelphia positive acute lymphoblastic leukemia. Blood. 2014;123:843–850. doi: 10.1182/blood-2013-09-529008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, McCastlain K, Ding L, Lu C, Song G, Ma J, Becksfort J, Rusch M, Chen SC, Easton J, Cheng J, Boggs K, Santiago-Morales N, Iacobucci I, Fulton RS, Wen J, Valentine M, Cheng C, Paugh SW, Devidas M, Chen IM, Reshmi S, Smith A, Hedlund E, Gupta P, Nagahawatte P, Wu G, Chen X, Yergeau D, Vadodaria B, Mulder H, Winick NJ, Larsen EC, Carroll WL, Heerema NA, Carroll AJ, Grayson G, Tasian SK, Moore AS, Keller F, Frei-Jones M, Whitlock JA, Raetz EA, White DL, Hughes TP, Guidry Auvil JM, Smith MA, Marcucci G, Bloomfield CD, Mrozek K, Kohlschmidt J, Stock W, Kornblau SM, Konopleva M, Paietta E, Pui CH, Jeha S, Relling MV, Evans WE, Gerhard DS, Gastier-Foster JM, Mardis E, Wilson RK, Loh ML, Downing JR, Hunger SP, Willman CL, Zhang J, Mullighan CG. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. The New England journal of medicine. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shah NN, Stevenson MS, Yuan CM, Richards K, Delbrook C, Kreitman RJ, Pastan I, Wayne AS. Characterization of CD22 expression in acute lymphoblastic leukemia. Pediatric blood & cancer. 2015;62:964–969. doi: 10.1002/pbc.25410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Daeron M, Jaeger S, Du Pasquier L, Vivier E. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunological reviews. 2008;224:11–43. doi: 10.1111/j.1600-065X.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- 83.O’Neill SK, Getahun A, Gauld SB, Merrell KT, Tamir I, Smith MJ, Dal Porto JM, Li QZ, Cambier JC. Monophosphorylation of CD79a and CD79b ITAM motifs initiates a SHIP-1 phosphatase-mediated inhibitory signaling cascade required for B cell anergy. Immunity. 2011;35:746–756. doi: 10.1016/j.immuni.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Getahun A, Beavers NA, Larson SR, Shlomchik MJ, Cambier JC. Continuous inhibitory signaling by both SHP-1 and SHIP-1 pathways is required to maintain unresponsiveness of anergic B cells. J Exp Med. 2016;213:751–769. doi: 10.1084/jem.20150537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Somasundaram R, Prasad MA, Ungerback J, Sigvardsson M. Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood. 2015;126:144–152. doi: 10.1182/blood-2014-12-575688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Appelmann I, Rillahan CD, de Stanchina E, Carbonetti G, Chen C, Lowe SW, Sherr CJ. Janus kinase inhibition by ruxolitinib extends dasatinib- and dexamethasone-induced remissions in a mouse model of Ph+ ALL. Blood. 2015;125:1444–1451. doi: 10.1182/blood-2014-09-601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu L, Yee H, Chan C, Kashyap AK, Horowitz L, Horowitz M, Bhatt RR, Lerner RA. Combinatorial surrobody libraries. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10756–10761. doi: 10.1073/pnas.0805293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carlow DA, Williams MJ, Ziltener HJ. Modulation of O-glycans and N-glycans on murine CD8 T cells fails to alter annexin V ligand induction by galectin 1. J Immunol. 2003;171:5100–5106. doi: 10.4049/jimmunol.171.10.5100. [DOI] [PubMed] [Google Scholar]
- 89.Imbert V, Peyron JF, Farahi Far D, Mari B, Auberger P, Rossi B. Induction of tyrosine phosphorylation and T-cell activation by vanadate peroxide, an inhibitor of protein tyrosine phosphatases. Biochem J. 1994;297(Pt 1):163–173. doi: 10.1042/bj2970163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smrz D, Lebduska P, Draberova L, Korb J, Draber P. Engagement of phospholipid scramblase 1 in activated cells: implication for phosphatidylserine externalization and exocytosis. J Biol Chem. 2008;283:10904–10918. doi: 10.1074/jbc.M710386200. [DOI] [PubMed] [Google Scholar]
- 91.Klasener K, Maity PC, Hobeika E, Yang J, Reth M. B cell activation involves nanoscale receptor reorganizations and inside-out signaling by Syk. Elife. 2014;3:e02069. doi: 10.7554/eLife.02069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drake AW, Klakamp SL. A rigorous multiple independent binding site model for determining cell-based equilibrium dissociation constants. J Immunol Methods. 2007;318:147–152. doi: 10.1016/j.jim.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 93.Edwards BS, Young SM, Ivnitsky-Steele I, Ye RD, Prossnitz ER, Sklar LA. High-content screening: flow cytometry analysis. Methods Mol Biol. 2009;486:151–165. doi: 10.1007/978-1-60327-545-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 96.Shen Y, Maupetit J, Derreumaux P, Tuffery P. Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J Chem Theory Comput. 2014;10:4745–4758. doi: 10.1021/ct500592m. [DOI] [PubMed] [Google Scholar]
- 97.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Generation, purification, and characterization of anti-Igβ Fab’ probes.
Fig. S2. Viterbi plots of states observed for pairs of QD-bound receptors under autonomous signaling conditions.
Fig. S3. Comparison of MSD values for pre-BCRs tracked under different conditions.
Fig. S4. Comparison of off-rates for homotypic interactions between pre-BCRs in two different cell lines.
Fig. S5. 3D plot for pairs of QD probes showing correlated motion at wide separation distances.
Fig. S6. Galectin-1 purification, homology model, and diffusion profile with varying concentrations.
Fig. S7. Characterization of the binding of the anti-VpreB antibody to 697 cells.
Fig. S8. Apoptotic response to single-agent treatment with kinase inhibitors targeting the pre-BCR pathway.
Fig. S9. Apoptotic response to combination treatment with vincristine and inhibitors of the pre-BCR pathway.
Fig. S10. Apoptotic response to combination treatment with vincristine and a SHIP inhibitor.
Fig. S11. Quantitation of the amounts of VpreB and Igβ on the surface of different BCP-ALL cells.
Fig. S12. Sequence alignment reveals polymorphism among different cell lines and patient samples.
Table S1. Characteristics of BCP ALL cells used in this study.
Table S2. SPT receptor diffusion statistics.
Table S3. SPT receptor dimer statistics.
Movie S1. Two-color SPT analysis of 697 cells depicting correlated motion and serial engagement.
Movie S2. Two-color SPT analysis of Nalm6 cells depicting correlated motion at wide (> 200 nm) distances.
Movie S3. SPT analysis showing pre-BCR diffusion in 697 cells in the presence or absence of exogenous galectin-1.