Abstract
Objective
Blood vessel epicardial substance (BVES) is a tight junction-associated protein that regulates epithelial-mesenchymal states and is underexpressed in epithelial malignancy. However, the functional impact of BVES loss on tumorigenesis is unknown. Here we define the in vivo role of BVES in colitis-associated cancer (CAC), its cellular function, and its relevance to inflammatory bowel disease (IBD) patients.
Design
We determined BVES promoter methylation status using an Infinium HumanMethylation450 array screen of patients with ulcerative colitis with and without CAC. We also measured BVES mRNA levels in a tissue microarray consisting of normal colons and CAC samples. Bves−/− and wild-type mice (controls) were administered azoxymethane (AOM) and dextran sodium sulfate (DSS) to induce tumor formation. Lastly, we utilized a yeast two-hybrid screen to identify BVES interactors and performed mechanistic studies in multiple cell lines to define how BVES reduces c-Myc levels.
Results
BVES mRNA was reduced in tumors from patients with CAC via promoter hypermethylation. Importantly, BVES promoter hypermethylation was concurrently present in distant non-malignant appearing mucosa. As seen in human patients, Bves was underexpressed in experimental inflammatory carcinogenesis, and Bves−/− mice had increased tumor multiplicity and degree of dysplasia after AOM/DSS administration. Molecular analysis of Bves−/− tumors revealed Wnt activation and increased c-Myc levels. Mechanistically, we identified a new signaling pathway whereby BVES interacts with PR61α, a PP2A regulatory subunit, to mediate c-Myc destruction.
Conclusions
Loss of BVES promotes inflammatory tumorigenesis through dysregulation of Wnt signaling and the oncogene c-Myc. BVES promoter methylation status may serve as a CAC biomarker.
Keywords: Cancer, IBD, Ulcerative colitis, Colonic neoplasms, Colorectal cancer
INTRODUCTION
Chronic inflammation promotes the development of colorectal cancer (CRC)1,2. Patients with inflammatory bowel disease (IBD), for example, have an elevated risk of developing CRC3, particularly those who have extensive disease or long disease duration4. Although the pathogenesis of inflammatory carcinogenesis remains unclear, at least one component of malignant degeneration is thought to be disruption of intestinal epithelial function as a consequence of chronic inflammation5,6. Indeed, pathologic changes in adherens and tight junction proteins have been described in colitis and colitis-associated cancer (CAC)6–8. In addition to providing junctional integrity between cells, adherens and tight junctional complexes also transduce extracellular signals to direct intracellular programs (“outside-in” signaling9), such as those controlling cellular proliferation and differentiation. For example, E-cadherin can sequester β-catenin at the cell membrane, preventing its nuclear localization and transcriptional activity10. Given that dysregulation of junctional proteins commonly occurs in CAC, understanding their function in normal biology may yield clues to how their dysfunction promotes carcinogenesis.
Blood vessel epicardial substance (BVES/POPDC1) is a tight junction-associated protein often silenced in carcinomas secondary to promoter hypermethylation11–13. Restoring BVES expression in CRC cell lines promotes epithelial-like morphology and decreases proliferation, migration, invasion, xenograft tumor growth, and metastasis, together indicating broad regulatory capabilities11. Conversely, knockdown of BVES in epithelial-like cells induces a mesenchymal-like phenotype characterized by increased proliferation, altered morphology, and disorganized cell-cell contacts11. Yet how BVES regulates these phenotypes is incompletely understood. Indeed, while several BVES interacting proteins have been identified11, their known functions do not explain fully the role of BVES in maintaining epithelial phenotypes. Moreover, how BVES contributes to tumor development has not been tested using genetic approaches.
The transcription factor c-Myc is commonly overexpressed in cancer14,15 and regulates proliferation, differentiation, apoptosis, and epithelial-to-mesenchymal transition16. In mouse models of sporadic CRC, decreased c-Myc levels reduce Apc-driven tumorigenesis17. In IBD, c-Myc is overexpressed in both inflamed tissues and CAC tumors18, and network analysis of CAC samples indicated that c-Myc dysregulation functionally contributes to CAC progression19. c-Myc levels are also increased in experimental models of inflammatory carcinogenesis, such as the azoxymethane (AOM)/dextran sodium sulfate (DSS) mouse model of CAC20. Yet the processes responsible for c-Myc dysregulation in inflammatory carcinogenesis remain unidentified. To date, a complex network of proteins—including protein phosphatase 2A (PP2A), Axin1, and GSK3β—has been identified that regulates c-Myc protein levels by modifying the phosphorylation status of c-Myc at two residues, threonine 58 (T58) and serine 62 (S62)21. Ubiquitylation of c-Myc is initiated by phosphorylation at T58, leading to its ultimate degradation. Given the prominent role of c-Myc in driving oncogenic programs, understanding mechanisms that control PP2A dephosphorylation of c-Myc may identify new therapeutic targets in inflammatory carcinogenesis.
Here we report that BVES is an important regulator of inflammatory carcinogenesis programs and promotes c-Myc degradation through an interaction with the PR61α-PP2A complex. We observed that BVES is reduced in human CAC samples, and further that the BVES promoter was hypermethylated within the tumors and at distant unaffected mucosa, suggesting a field effect. Using the AOM/DSS inflammatory carcinogenesis model, we determined that Bves−/− mice demonstrate greater tumor incidence and multiplicity as well as a higher degree of dysplasia and intratumoral proliferation. Furthermore, molecular analysis of Bves−/− tumors revealed increased c-Myc protein and signaling activity. c-Myc protein was also elevated in intestinal crypts from Bves−/− mice. In line with in vivo results, knockdown of BVES in vitro increased c-Myc stability and consequently increased expression of key c-Myc targets ODC and CAD. Conversely, BVES overexpression reduced c-Myc stability and increased c-Myc ubiquitylation. Using a yeast two-hybrid (Y2H) screen, we identified PR61α, the PP2A regulatory subunit critical for c-Myc degradation, as a BVES-interacting protein, and show that this interaction is required for BVES to modulate cellular c-Myc levels. Thus, we demonstrate that BVES coordinates PR61α-containing PP2A phosphatase complexes to restrict c-Myc protein levels and that BVES is a key suppressor of inflammatory carcinogenesis whose promoter methylation status may define patients with ulcerative colitis (UC) at risk for colon cancer.
MATERIALS AND METHODS
Mice, treatments, and analysis
AOM and DSS were prepared as previously described22. Bves−/− mice have been previously described23. Detailed protocols can be found in the Supplementary Materials and Methods Section.
BVES promoter methylation analysis
Tissue samples were obtained from colectomy specimens from individuals without UC, individuals with UC but without dysplasia or cancer, and UC patients with high-grade dysplasia and/or colon cancer. Clinical information is described in online supplementary table 1. Detailed protocols regarding epithelial isolation, methylation array, and pyrosequencing can be found in the Supplementary Materials and Methods Section.
See Supplementary Materials and Methods for detailed methods regarding cell culture experiments, RNA scope, promoter methylation analyses, and mouse analysis.
RESULTS
BVES is downregulated and its promoter is hypermethylated in CAC
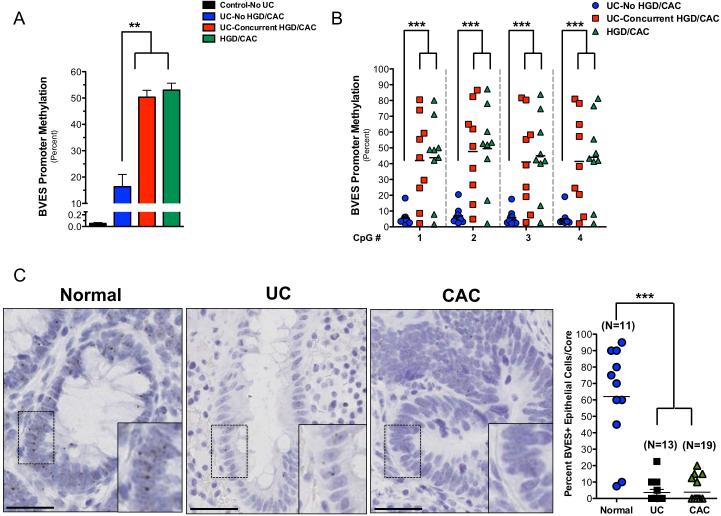
As BVES is underexpressed via promoter hypermethylation in CRC11, we asked whether the BVES promoter was also hypermethylated in CAC. Therefore, we analyzed BVES methylation status in an Infinium HumanMethylation450 array screen of IBD samples. The samples consisted of control patients (Control—No UC), patients with UC who did not have cancer (UC—no HGD/CAC), and two different types of samples from patients with UC who had colon cancer: the remote, non-malignant tissue (UC— concurrent HGD/CAC) and tissue with high-grade dysplasia and/or cancer (HGD/CAC). These analyses demonstrated that the BVES promoter was unmethylated in the controls–No UC (0.1% + 0.016%), moderately methylated in UC–no HGD/CAC (16% + 4.7%), and hypermethylated in the HGD/CAC among patients with colitis-associated carcinoma (HGD/CAC, 53% + 2.6%) (figure 1A). Furthermore, remote nonneoplastic, mucosal samples (UC-Concurrent HGD/CAC) from the same patients who had CAC (HGD/CAC) were hypermethylated (50% + 2.6%) to a similar degree as that observed in cancerous tissue. Interestingly, these results suggest that BVES promoter methylation may represent a field effect in CAC and that BVES promoter methylation status may identify UC patients with concurrent malignancy. To confirm the results derived from the HM450 methylation array studies, we pyrosequenced the BVES promoter in the same samples and again demonstrated low levels of methylation in the UC—no HGD/CAC cases, and higher methylation in both the UC—concurrent HGD/CAC and HGD/CAC cases (figure 1B).
Figure 1. A field effect of BVES promoter hypermethylation in colitis-associated cancer.
(A) Average BVES promoter methylation status in the indicated sample from the Infinium HumanMethylation 450 Array. Methylation was measured in four sample types: colon epithelia from patients who did not have UC (Control—No UC); colon epithelia from UC patients who did not have dysplasia or carcinoma (UC—no HGD/CAC); non-malignant colon epithelia from UC patients (UC—concurrent HGD/CAC) and malignant colon epithelia (HGD/CAC) from UC patients who had dysplasia/carcinoma. Control—No UC, n=17; UC—no HGD/CAC, n=11; UC—concurrent HGD/CAC, n=10; HGD/CAC, n=10. **p<0.01.
(B) Pyrosequencing at four sequential CpG dinucleotides in the BVES promoter. Each shape represents a separate individual, with mean methylation values depicted with black bars. ***p<0.001.
(C) Representative images of high-resolution in situ (RNAscope™) analysis of BVES message in normal colons (n=11), UC (n=13), and CAC (n=19). Right: Quantification of BVES expressing epithelial cells per tissue microarray core. Size standard=50 microns. ***p<0.001
It is possible that BVES promoter methylation, while increased, may not be sufficient to silence its expression. To determine whether BVES promoter methylation indeed reduced its transcription, we tested whether BVES mRNA was downregulated in CAC using high resolution in situ hybridization (RNAScope24) in a tissue microarray consisting of normal, UC, and CAC samples. BVES mRNA levels were low, but consistently present in normal colonic epithelial cells (figure 1C). In UC and CAC samples, however, BVES message was rarely detected and quantification of epithelial staining indicated a 5-fold decrease (p<0.001). Taken together, BVES RNA expression is downregulated in both UC and CAC, most likely due to promoter hypermethylation. Furthermore, as the BVES promoter is hypermethylated in both tumor and non-malignant mucosa in patients with CAC, BVES promoter methylation may serve as a biomarker associated with dysplasia or neoplasia in patients with IBD.
Bves loss promotes CAC development
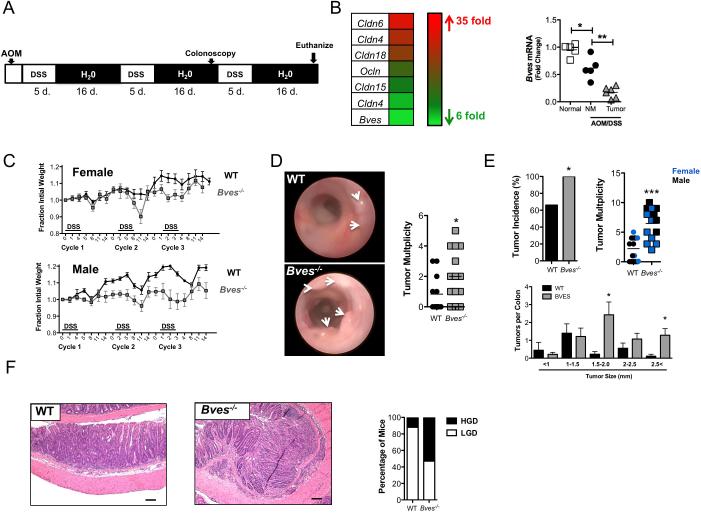
While BVES underexpression was consistently observed in human CAC, these studies do not establish whether BVES loss actively promotes tumorigenesis. Therefore, we used mouse genetic approaches combined with the AOM/DSS model (figure 2A) to determine if BVES loss contributed to inflammatory tumorigenesis. While Bves was expressed at baseline in the murine colon at both the RNA and protein levels (see online supplementary figure 1), transcriptome profiling of AOM/DSS-induced tumors in WT mice showed a 5-fold decrease in Bves transcripts (figure 2B), mirroring the results observed in human CAC. As expected, we also observed changes in other tight junction constituents, supporting previous reports of tight junctional dysregulation in colitis and CAC25. We confirmed that Bves message was decreased in AOM/DSS tumor tissue by qPCR in an independent sample set (figure 2B). Interestingly, Bves message also decreased in AOM/DSS treated non-malignant tissue compared to normal colons (figure 2B), again suggesting a field effect in inflammatory carcinogenesis. As a result, we hypothesized that complete loss of Bves might promote inflammatory carcinogenesis.
Figure 2. BVES modifies inflammatory carcinogenesis.
(A) Schematic of AOM/DSS protocol and timeline. Mice were injected with 7.5 mg/kg of AOM and treated with 2.5% DSS at the indicated time.
(B) Left: Heat map of RNA-seq data derived from WT colons (n=3) and WT AOM/DSS tumors (n=3). Red indicates genes increased and green indicates genes decreased in tumors compared to normal colon. Right: qPCR of Bves message levels in normal colons (Normal, n=5), non-malignant AOM/DSS treated colon (NM, n=5) and AOM/DSS tumor (Tumor, n=6). Tissue harvested from WT mice after AOM/DSS treatment. ***p<0.001.
(C) Weights of Bves−/− and WT mice during AOM/DSS treatment. Weights are presented as fraction of initial weight. Bves−/− (male: n=8, female: n=7) and WT (male: n=5, female: n=10). *p<0.05, ***p<0.01, ***p<0.001.
(D) Representative colonoscopy images of WT and Bves−/− colons after the second cycle of DSS treatment. Right: Quantification of tumor multiplicity by endoscopy assessment.
(E) Tumor incidence, multiplicity, and size distribution at necropsy in WT and Bves−/− mice. Blue = female mice, black = male mice.*p<0.05, ***p<0.001.
(F) Left: Representative H&E stained sections demonstrating the histologic features of WT and Bves−/− tumors. Size standard=100 microns. Right: Blinded histological scoring of degree of dysplasia of tumors from WT and Bves−/− mice. Graph represents percentage of mice with intratumoral low or high-grade dysplasia.
To test the effect of Bves loss in CAC, we compared WT and Bves−/− mice subjected to the same inflammatory carcinogenesis protocol. We first observed that Bves−/− mice lost a greater fraction of body weight compared to WT mice, most notably during cycle 3 (figure 2C), suggesting increased sensitivity to AOM/DSS treatment. Indeed, endoscopy one-week prior to sacrifice demonstrated increased tumor multiplicity in Bves−/− mice (figure 2D) and this was confirmed at necropsy where we observed 100% tumor penetrance in Bves−/− mice compared to 60% in WT mice (figure 2E). Bves−/− mice also demonstrated increased tumor multiplicity (6.5 ± 0.6 tumors per Bves−/− mouse vs. 2.2±0.5 tumors per WT mouse, p<0.001), and tumor size (figure 2E). Furthermore, Bves−/− tumors exhibit more advanced dysplasia compared to WT tumors (figure 2F). Control mice treated with three cycles of DSS-only or a single AOM injection did not develop tumors during this time-frame (data not shown). Taken together, these results suggest that BVES underexpression in CAC functionally contributes to inflammatory carcinogenesis.
Increased proliferation and enhanced Wnt activation in Bves−/− tumors
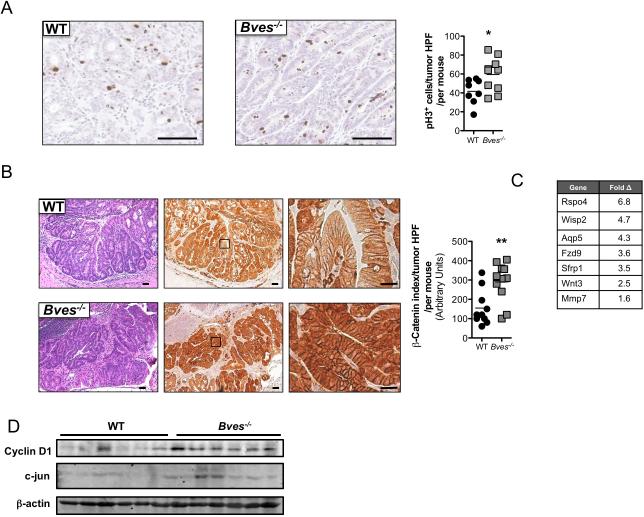
To identify BVES-directed mechanisms responsible for modifying tumorigenesis, we examined proliferation and apoptosis in the tumors of AOM/DSS treated Bves−/− mice. Proliferation, as measured by phospho-histone H3 staining, was increased in Bves−/− tumors (figure 3A). Conversely, staining for cleaved caspase-3 indicated no difference in intratumoral apoptosis between Bves−/− and WT mice (see online supplementary figure 2). As Wnt activation can drive proliferation, we postulated that Wnt signaling might be perturbed in Bves−/− tumors. β-catenin dysregulation is a key indicator of hyperactive Wnt signaling26, and β-catenin is also a mutational target in DMH/DSS inflammatory carcinogenesis, resulting in increased levels and altered subcellular distribution27. Therefore, we analyzed β-catenin by immunohistochemistry and observed excessive cytoplasmic and nuclear β-catenin localization in Bves−/− tumors compared to WT tumors(figure 3B). While these results suggested hyperactive Wnt signaling in Bves−/− tumors, we confirmed this by RNA-seq analysis, which indicated upregulation of established Wnt targets, such as Mmp7, Wisp2, and Rspo4 (figure 3C), in Bves−/− tumors. Ingenuity Pathway Analysis (IPA)28 of the RNA-seq data set also indicated hyperactive Wnt networks, such as β-catenin and Tcf. Finally, immunoblotting demonstrated greater expression of cyclin D1 and c-jun, two well-characterized Wnt target genes29,30, in Bves−/− tumors (figure 3D). While previous experiments demonstrated that BVES could regulate Wnt signaling using in vitro, cell-based assays11, these findings provide the first in vivo and genetic evidence supporting the hypothesis that BVES regulates Wnt activity.
Figure 3. Dysregulated Wnt signaling in Bves−/− tumors.
(A) Left: Representative images of phospho-histone H3 (pH3) immunohistochemistry in WT and Bves−/− tumors. Size standard=50 microns. Right: Quantification of pH3 positive cells per tumor high power field (HPF). Data is presented as the mean number of positive cells per tumor HPF per mouse. At least five HPF per mouse were scored. Student's t test, *p<0.05.
(B) Left: H&E stained sections, size standard=50 microns. Middle: Representative images of β-catenin immunohistochemistry, low magnification, size standard=50 microns. Right: β-catenin immunohistochemistry, high magnification, size standard=20 microns. Far right: Quantification of intratumoral β-catenin immunohistochemistry. An index was employed to quantify extent of nuclear and cytoplasmic β-catenin staining. This index is generated by multiplying the staining intensity (on a scale of 100-500) by percentage of the cells demonstrating nuclear staining. For example, a score of 500 indicates a field that demonstrated very intense nuclear β-catenin stain while a score of 100 indicates a field that has weak nuclear β-catenin staining. Data is presented as the mean score per tumor HPF per mouse. At least five HPF per mouse were scored.
(C) Wnt target genes upregulated in Bves−/− tumors identified in RNA-seq dataset (WT, n=3; Bves−/−, n=3).
(D) Immunoblot of Cyclin D1 and c-jun in Bves−/− and WT tumors. β-actin was used as a loading control.
BVES regulates c-Myc degradation
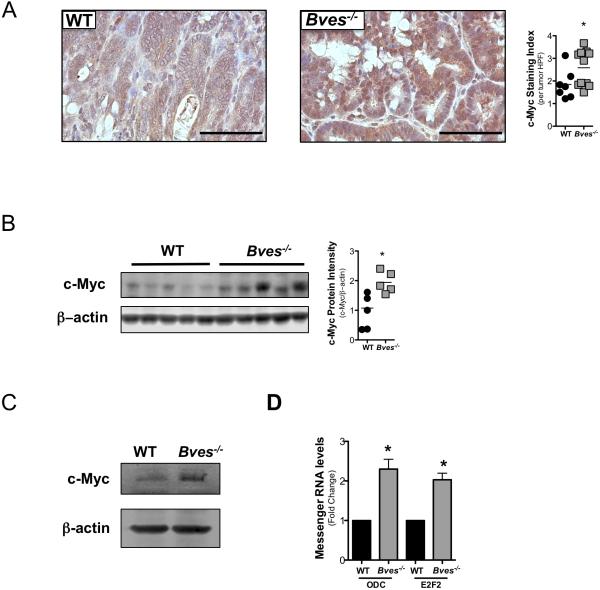
As c-Myc is a bona-fide Wnt transcriptional target17, has been identified as a potential biomarker in patients with IBD at risk for CAC19, and is overexpressed in AOM/DSS tumors20, we postulated that c-Myc was dysregulated in Bves−/− tumors. Indeed, IPA analysis of intratumoral transcriptomes identified causal dysregulation28 of c-Myc networks (see online supplementary figure 3A). While analysis of RNA-seq datasets showed only a modest increase in c-Myc transcripts in Bves−/− tumors compared to WT tumors (see online supplementary figure 3B), immunohistochemical staining for c-Myc demonstrated an increase in both total c-Myc protein (figure 4A) and transcriptionally active, phosphorylated serine 62 c-Myc species in Bves−/− tumors (see online supplementary figure 4). Interestingly, immunoblotting in tumor-adjacent mucosa also showed higher c-Myc levels in Bves−/− colons, which suggested c-Myc was increased prior to tumor formation and that BVES might regulate c-Myc levels in the gut at baseline (figure 4B). To test this, we isolated crypts from untreated Bves−/− and WT mice and observed greater c-Myc protein in Bves−/− samples (figure 4C). Consistent with elevated c-Myc, qPCR for Ornithine decarboxylase (Odc), a c-Myc transcriptional target, indicated a 4-fold increase in Bves−/− colons (see online supplementary figure 5). We also observed increased mRNA of c-Myc targets Odc and E2f transcription factor 2 (E2f2) (figure 4D) in “mini-gut” 3D cultures, demonstrating that BVES regulation of c-Myc activity was epithelial cell-autonomous.
Figure 4. c-Myc signaling is dysregulated in Bves−/− mice in inflammatory carcinogenesis.
(A) Left: Representative images of immunohistochemistry for intratumoral c-Myc. Right: quantification of c-Myc positive cells per tumor high power field (HPF). HPFs were scored according to an index from 1-4 (a score of 1 denotes less than 25% of positive cells per HPF; a score of 2 denotes 25-50%; a score of 3 denotes 50-75%; a score of 4 denotes 80-100%). Data is presented as the mean score per tumor HPF per mouse. At least five HPF per mouse were scored. Student's t test, *p<0.05. Size standard=50 microns
(B) Immunoblot of c-Myc in WT and Bves−/− normal adjacent colons. Blots were imaged using the LiCor Odyssey system and quantified using Image Studio analysis. Student's t test, p<0.05.
(C) Immunoblot of c-Myc in WT (n=3) and Bves−/− (n=3) intestinal crypts.
(D) qPCR for Odc and E2f2 in enteroid cultures Student's t test, *p<0.05.
In all western blots, β-actin served as loading control.
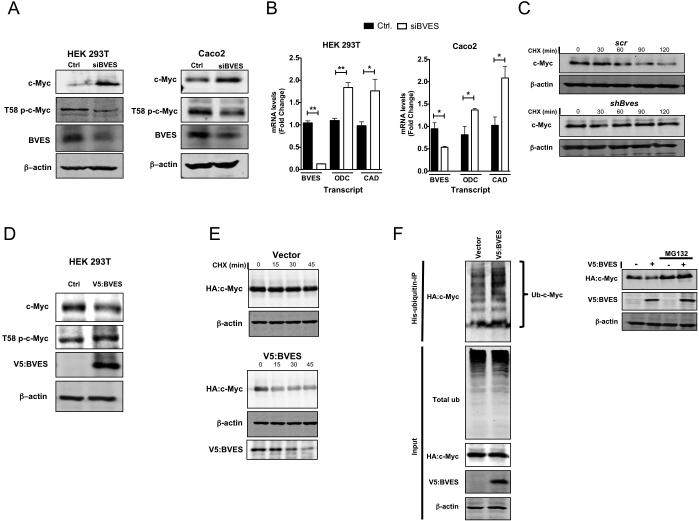
As we observed increased c-Myc protein in Bves−/− tumors, we postulated that BVES could regulate c-Myc protein stability. Three cell lines—HEK 293T (non-malignant cell line), Caco2 (CRC cell line that can form a polarized epithelium), and HCE (Human Corneal Epithelial)—which all express BVES (supplementary figure 6)11 were used for BVES knockdown experiments. In all three cell lines, BVES RNAi increased c-Myc protein levels (figure 5A and supplementary figure 7). In addition to increasing total c-Myc protein, we also observed that BVES knockdown reduced T58 phosphorylation, a key post-translational modification which signals for c-Myc degradation by the ubiquitin-proteasome system (figure 5A). This increase in c-Myc was functionally relevant as transcript levels of c-Myc targets ODC and Carbamoyl-Phosphate Synthetase 2 Aspartate Transcarbamylase and Dihydroorotase (CAD) were increased with BVES knockdown (figure 5B). Furthermore, knockdown of BVES doubled c-Myc half-life compared to non-targeting control samples (figure 5C). Conversely, overexpressing BVES reduced c-Myc protein levels, increased T58 c-Myc species (figure 5D), dampened c-Myc transcriptional activation of the c-Myc responsive E2F2 reporter (see online supplementary figure 8), and decreased c-Myc protein half-life (figure 5E, lower panel). We then tested whether BVES could regulate c-Myc ubiquitylation, a central post-translational event targeting its destruction. Indeed, by overexpressing BVES we observed increased c-Myc polyubiquitylation (figure 5F).Moreover, inhibiting the proteasome using MG132 blocked BVES-induced reduction of c-Myc (figure 5F). Hence, our results suggest that BVES promotes the post-translational degradation of c-Myc.
Figure 5. BVES regulates c-Myc stability and activity.
(A) c-Myc and T58 phospho-c-Myc protein levels after BVES knockdown in HEK 293T (left) or Caco2 (right) cells after 48 hr serum starvation.
(B) qRT-PCR assay for c-Myc targets ODC and CAD following BVES knockdown in the indicated cell lines. Data are presented as mean ±SEM and in triplicates. *p<0.05, **p<0.01.
(C) Cycloheximide treatment (100 μg/ml) of HEK 293T cells with and without Bves knockdown followed by immunoblotting for c-Myc.
(D) c-Myc and T58 phospho-c-Myc protein levels after overexpression of V5:BVES in HEK 293T cells.
(E) HEK 293T cells co-transfected with HA:c-Myc and V5:BVES were then treated with cycloheximide (100 μg/ml) followed by immunoblotting for the indicated protein.
(F) Left: His:Ubiquitin and HA:c-Myc were co-transfected into HEK 293T cells along with V5:BVES. Cells were treated with proteasome inhibitor MG132 (20 μm) for 4 hours before His:Ubiquitin complexes were immunoprecipitated and resolved by SDS-PAGE. Ubiquitylated HA:c-Myc complexes were visualized by immunoblotting (Ub-c-Myc). Total ubiquitylated protein (Total ub) was examined as a control. Right: HEK 293T cells co-transfected with HA:c-Myc and V5:BVES were treated with proteasome inhibitor MG132 (20 μm) for 4 hours. Whole cell lysates were analyzed for HA:c-Myc expression. In all immunoblots, β-actin was used as a loading control. All experiments were replicated three times.
BVES interacts with PR61α, PP2Ac, and c-Myc
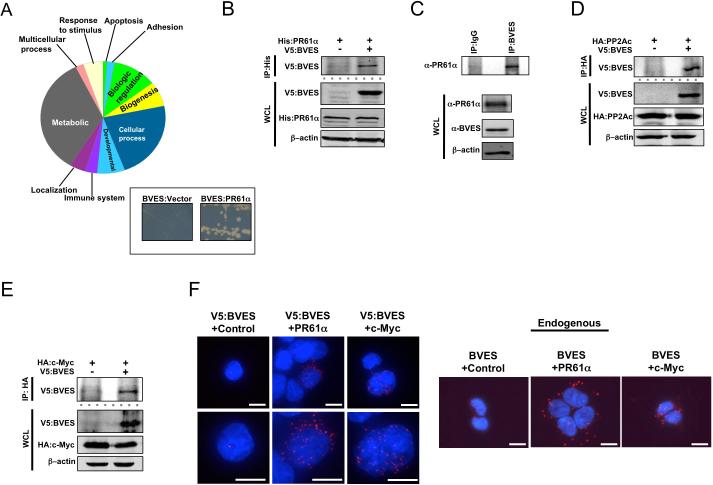
To identify a molecular mechanism by which BVES orchestrates c-Myc degradation, we conducted a Y2H screen to define the BVES interactome. Characterization of this interactome using the PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System31 identified a number of biologic processes influenced by BVES (figure 6A). Interestingly, the screen identified that BVES interacted with four of the five members of the B’ family of PP2A regulatory subunits (PPP2R5A, PPP2R5B, PPP2R5D, and PPP2R5E). PPP2R5A, also known as PR61α, is a key regulator of PP2A mediated c-Myc dephosphorylation. PR61α directs the heterotrimeric PP2A complex, consisting of a regulatory, catalytic, and structural subunit, to associate with doubly phosphorylated (T58/S62) c-Myc and dephosphorylate S62, resulting in increased levels of monophosphorylated T58 c-Myc species, which signals c-Myc to be degraded by the proteaosome32.
Figure 6. BVES interacts with PR61α, PP2A, and c-Myc.
(A) PANTHER Biologic Process Analysis of BVES interactome. Inset: Directed yeast two-hybrid of BVES and PR61α.
(B) Co-immunoprecipitation of exogenous and (C) endogenous PR61α:BVES complexes in HEK 293T cells.
(D) Co-immunoprecipitation of V5:BVES and HA:PP2Ac or (E) HA:c-Myc.
(F) Left: Proximity ligation assay in HEK 293T cells transfected with V5:BVES. Left: control, middle: α-PR61α, right: α-c-Myc. Right: Proximity ligation assay in HEK 293T cells for endogenous protein interactions. Left: control, middle: α-PR61α, right: α-c-Myc. Anti-HA was used as a control in both exogenous and endogenous localization. Scale bar denotes 10 μm. Red staining is positive signal from the PLA interaction, and blue staining is DAPI. In all immunoblots blots, β-actin was used to ensure loading consistency. All experiments were repeated at least two times.
The BVES:PR61α interaction was then confirmed by directed Y2H (figure 6A) and by exogenous and endogenous co-immunoprecipitation in HEK 293T cells (figure 6B and C). If BVES were interacting with PR61α to promote c-Myc degradation, we hypothesized that BVES would complex with both the PP2A catalytic subunit (PP2Ac) and c-Myc, which we then demonstrated by co-immunoprecipitation (figure 6D and E and see online supplementary figure 9). We further used the proximity ligation assay (PLA) and confirmed interaction of both exogenous and endogenous BVES with endogenous PR61α and c-Myc (figure 6F). Overall, these data indicate that BVES complexes with c-Myc, PR61α, and the PP2A catalytic subunit.
BVES is essential for PR61α-mediated c-Myc degradation
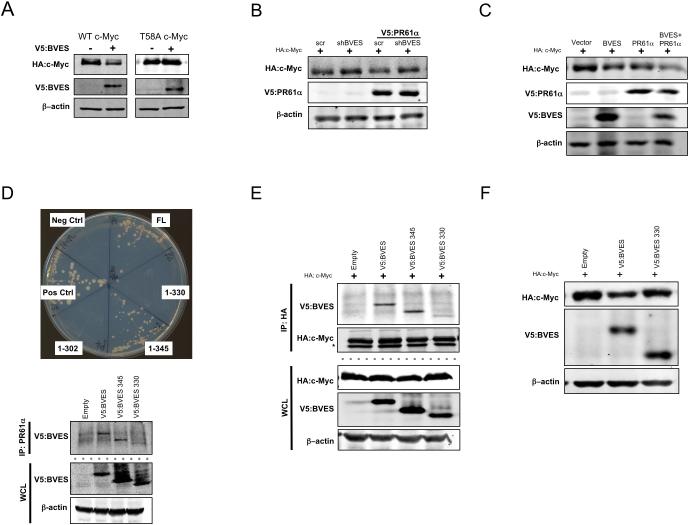
PP2A dephosphorylation of S62 requires c-Myc to be phosphorylated at residue T5833. If BVES reduces c-Myc through PP2A, we reasoned c-MycT58A, a c-Myc mutant resistant to T58 phosphorylation, would escape BVES-induced degradation. Indeed, BVES expression consistently reduced c-MycWT but had no effect on c-MycT58A (figure 7A). We next hypothesized that knockdown of BVES would ablate PR61α-PP2A induced c-Myc degradation. Overexpression of PR61α reduced c-Myc protein subtly but consistently as previously reported32 (figure 7B; compare lane 1 and 3). Knocking down BVES, however, rescued PR61α-induced degradation (figure 7B; compare lanes 3 and 4). We then tested whether BVES could enhance PR61α-mediated c-Myc degradation, and indeed, overexpression of BVES and PR61α substantially reduced c-Myc protein compared to PR61α or BVES alone (figure 7C; compare lane 4 to 2 or 3).
Figure 7. The BVES:PR61α interaction is required to promote c-Myc degradation.
(A) WT HA:c-Myc or phospho-mutant HA:T58A c-Myc levels after either empty vector (negative control) or V5:BVES transfection in HEK 293T cells.
(B) Immunoblotting for HA:c-Myc levels following PR61α overexpression in the setting of BVES knockdown or (C) when both PR61α and BVES are present HEK 293T cells.
(D) Top: Mapping the PR61α BVES binding interface via directed yeast two-hybrid (Full length BVES, residues 1-345, 1-330, 1-302, negative control (Neg Ctrl), and positive control (Pos Ctrl)). Below: Co-immunoprecipitation of the indicated BVES mutants and PR61α in HEK 293T cells..
(E) Co-immunoprecipitation of the indicated BVES mutants and HA:c-Myc in HEK 293T cells.
(F) HA:c-Myc protein levels after transfection of the indicated BVES construct in HEK 293T cells.
In all immunoblots, β-actin was used as a loading control. All experiments were repeated at least two times.
We then sought to determine whether BVES requires PR61α to degrade c-Myc. For these experiments we first mapped the BVES:PR61α interaction domain by serial deletions to the carboxy-terminus of BVES. Deleting the carboxy-terminal 30 residues, but not the last 15 residues, disrupted the BVES:PR61α interaction as demonstrated by Y2H and by co-IP, thus mapping the interaction domain to residues 330-345 (figure 7D). Importantly, this uncoupling mutant (BVES-330) demonstrated reduced affinity for c-Myc (figure 7E) and was unable to reduce c-Myc levels (figure 7F), indicating BVES indeed requires interaction with PR61α to regulate c-Myc. Overall, our results demonstrate that BVES, through PR61α, promotes c-Myc dephosphorylation, destabilization, and destruction, and thus provides mechanistic insight into one manner by which BVES may contribute to inflammatory carcinogenesis.
DISCUSSION
We, and others, have shown that BVES is underexpressed in gastrointestinal cancers and that restoration of BVES in cancer cell lines induces epithelial features. Here we provide the first genetic evidence that BVES modifies cancer phenotypes, as we demonstrate that mice lacking Bves have increased tumor multiplicity and dysplasia after establishment of inflammatory carcinogenesis. Further, we show Bves−/− tumors have increased c-Myc protein resulting in activation of c-Myc regulated networks. Moreover, we identify that BVES interacts with PR61α, a key regulatory subunit of the PP2A phosphatase complex, and promotes PP2A-mediated c-Myc dephosphorylation leading to c-Myc degradation. Uncoupling the BVES:PR61α interaction blocks BVES-dependent reduction of cellular c-Myc levels. To our knowledge, this is the first junctional-associated protein identified that regulates post-translational c-Myc status. The potential clinical relevance is demonstrated as we observed that BVES is downregulated in CAC likely secondary to promoter methylation. However, perhaps more importantly, we establish that the BVES promoter is also aberrantly methylated in distant, normal appearing tissues in patients with CAC/HGD—suggesting a field effect. Thus, our findings not only reveal that deletion of BVES promotes CAC, but also that BVES promoter methylation status may be a clinically important surrogate marker of colitis-associated dysplasia or CAC in IBD patients.
Chronic colitis has been shown to accelerate genome-wide methylation changes34; it has been hypothesized that this greater rate of methylation contributes to the increased cancer risk in patients with colitis by silencing tumor suppressors. Understanding how methylation broadly affects inflammatory tumorigenesis is important to design therapies and screening strategies for IBD patients. Our report specifically identifies that the BVES promoter is hypermethylated in UC patients who have CAC. Interestingly, BVES promoter hypermethylation is observed not only in the cancerous tissue, but also in the non-malignant mucosa in these patients. Currently, the standard method of cancer screening in IBD patients, who are at up to a 10-fold elevated risk of developing CAC1, is surveillance colonoscopy performed with the hope that cancer will be detected at an early, treatable stage. Yet the detection of neoplasia in the colon can be challenging in individuals with IBD, as the lesions are frequently flat and difficult to detect in a background of acute and chronic inflammatory changes. Our data suggest that aberrant BVES promoter methylation may be a useful biomarker for the presence of CAC, or even dysplasia, and that measuring BVES promoter methylation status could serve as a clinically useful tool to identify patients at risk for colon dysplasia or cancer.
While the molecular pathogenesis of CAC remains incompletely understood, recent work has shown the importance of NF-κB signaling35, the intestinal microbiota36, the tumor microenvironment37, and the innate immune system38 in regulating inflammatory tumorigenesis. A growing body of evidence also supports the important role of epithelial junctional constituents in inflammation and CRC. For example, mice expressing a dominant negative N-cadherin display disrupted adherens junctions and develop severe inflammation and colitis-associated dysplasia39. Likewise, knocking out Junctional adhesion molecule (Jam-A) results in a dramatic increase in susceptibility to DSS-induced colitis8. Here we show that deletion of Bves, a tight junction-associated protein, augments inflammatory carcinogenesis. Indeed, loss of BVES appears to increase tumor initiation and progression. We postulate that this is likely due to dual regulation of Wnt signaling and c-Myc protein degradation. Our results further strengthen the concept that junctional constituents are important regulators of colitis-induced tumor initiation and progression.
In the last decade, BVES has been shown to regulate a variety of cellular processes. For example, a Y2H screen of a mouse heart library identified an interaction between BVES and GEFT, a guanine nucleotide exchange factor40. Indeed, it was shown that expression of BVES modulated cell shape and locomotion, thus linking BVES to Rho-family GTPase signaling40. BVES has also been shown, via an interaction with ZO-1, to regulate GEF-H1-mediated RhoA activity11. More recently, it was reported that BVES plays a regulatory role in cardiac pacemaking through binding of cAMP and interacting with potassium channel TREK-141. Further, BVES interacts with CAV3, a caveolin expressed in the muscle tissue, and cardiac myocytes in Bves−/− mice have altered calveolar number and size42. Thus, BVES, through scaffolding with protein complexes, regulates a wide variety of basic, yet essential, cellular processes.
Our results now expand the known regulatory roles of BVES to include maintaining appropriate c-Myc protein levels. We show that BVES, through its interaction with the PR61α-containing PP2A phosphatase complex, can promote c-Myc degradation and that silencing BVES prevents PR61α-induced degradation of c-Myc. Moreover, mutating BVES so that it is unable to associate with PR61α renders BVES unable to initiate c-Myc destruction. The post-translational regulation of c-Myc requires coordination of numerous proteins to modify its phosphorylation and ubiquitylation status21. Precisely how BVES coordinates the PR61α-PP2A complex remains to be understood, but given that analysis of BVES structure shows no apparent enzymatic motifs in BVES, it is likely that BVES acts as a scaffold allowing for complex formation, similar to AXIN121. Interestingly, in addition to the membranous staining of the BVES:PR61α complex, there also appears to be peri-nuclear and cytoplasmic localization (figure 6F), which is consistent with previous reports describing the dynamic subcellular localization of BVES and its family members43. The PP2A family has been associated with tight junctional complexes regulating cellular permeability, but their exact role remains controversial44. BVES may bridge PP2A complexes to tight junctions and our report adds a new molecular mechanism for “outside-in” signaling in the epithelium.
Because c-Myc regulates thousands of genes, even subtle changes in c-Myc expression can have profound effects on cellular transcriptomes that promote tumorigenesis45. Indeed, strict regulation of c-Myc is an important component of homeostasis, and this is particularly true in the intestine. Acute expression of c-Myc, for example, dramatically expands the intestinal crypts and results in loss of differentiated cells46. Moreover, it has been shown that c-Myc is essential for Apc-mediated intestinal tumorigenesis17. Thus, BVES may serve as an important suppressor of inflammatory tumorigenesis via attenuating excessive c-Myc levels. More broadly, BVES could act as a regulator of c-Myc in a variety of tissues, as BVES is expressed in most epithelial tissues, such as lung, stomach, and breast, and its downregulation or promoter hypermethylation has been documented in diverse epithelium11–13.
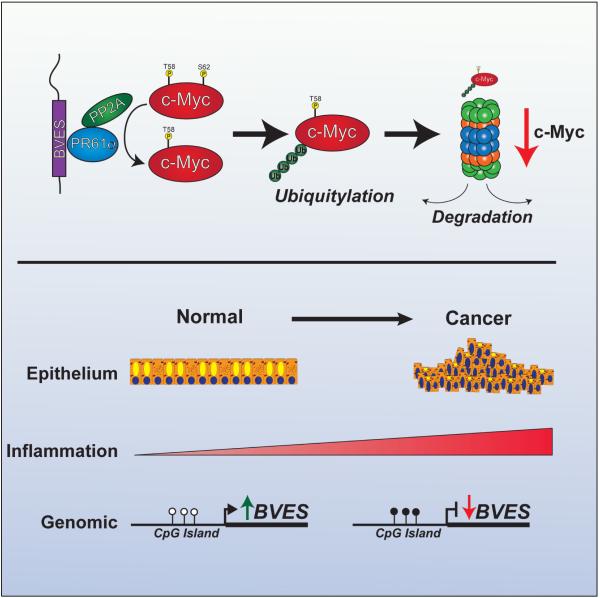
Taking our data together, one can envision a model in which chronic inflammation leads to BVES promoter hypermethylation, resulting in suppression of BVES transcription and reduced cellular protein levels. Loss of BVES impairs PR61α directed PP2A dephosphorylation of c-Myc, thus favoring increased cellular pools of c-Myc, a potent oncogene, likely, in cooperation with other oncogenic events, contributing to tumor progression (figure 8).
Figure 8.
Working model of the role of BVES in regulating c-Myc degradation and colitis-associated cancer development.
Supplementary Material
SUMMARY BOX.
What is already known about this subject?
➢ Patients with ulcerative colitis are at greater risk for developing colon cancer.
➢ Blood vessel epicardial substance (BVES) is a tight junction protein that regulates epithelial-to-mesenchymal transition in vitro.
➢ c-Myc is an oncogene overexpressed in 50% of all malignancies, including colitis-associated cancer (CAC).
➢ What are the new findings?
➢ BVES promoter hypermethylation is present in CAC and distant uninvolved mucosa.
➢ BVES is underexpressed in patients with CAC compared to normal colonic tissue.
➢ Deletion of Bves promotes colitis-associated tumor multiplicity and dysplasia.
➢ BVES directs the PR61α-PP2A complex to target c-Myc for proteasomal destruction.
How might it impact on clinical practice in the foreseeable future?
➢ BVES promoter hypermethylation status is a potential biomarker to identify UC patients at risk for cancer.
➢ Our studies demonstrate a new mechanism for regulation of c-Myc, an oncogene that is dysregulated in numerous malignancies.
➢ BVES plays a key role in maintaining the integrity of the colonic mucosa and protecting from inflammatory carcinogenesis, and may represent a therapeutic target in CAC.
ACKNOWLEDGEMENTS
We would like to thank the members of the Williams lab who helped discuss and review the manuscript. We would also like to thank Dr. Brian Grieb and Dr. Joseph Roland for their helpful suggestions. Finally, we appreciate the advice and counsel from Dr. R. Daniel Beauchamp and Dr. Barbara Fingelton in preparing the manuscript.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors have no conflicts of interest to disclose.
Author Contributions: BP and AMK contributed equally to the work in the manuscript. WMG and CSW are co-corresponding authors. BP, AMK, CWB, and SPS developed the hypothesis, designed experiments, analyzed the data, and wrote the manuscript. BP, AMK, CWB, SPS, CEK, WN, MKM, RDN, FLR performed experiments. BP, AMK, CWB, SPS, MKW, JJS, XC, KTW, TAB, DMB, WPT, RC, TAB, WMG, CSW contributed to experimental design, generation of the reagents, and manuscript editing. BP, AMK, WMG, CSW conceived and supervised the project.
REFERENCES
- 1.Mantovani A A, Allavena P, et al. Cancer-related inflammation. Nature. 2008;454(7203):436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 2.Danese S, Mantovani A. Inflammatory bowel disease and intestinal cancer: a paradigm of the Yin-Yang interplay between inflammation and cancer. Oncogene. 2010;29(23):3313–3323. doi: 10.1038/onc.2010.109. [DOI] [PubMed] [Google Scholar]
- 3.Terzić J, Grivennikov S, Karin E, et al. Inflammation and colon cancer. Gastroenterology. 2010;138(6):2101–2114. e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 4.Jess T, Simonsen J, Jørgensen KT, et al. Decreasing risk of colorectal cancer in patients with inflammatory bowel disease over 30 years. Gastroenterology. 2012;143(2):375–81. doi: 10.1053/j.gastro.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 5.Schmitz H, Barmeyer C, Fromm M, et al. Altered tight junction structure contributes to the impaired epithelial barrier function in ulcerative colitis. Gastroenterology. 1999;116(2):301–309. doi: 10.1016/s0016-5085(99)70126-5. [DOI] [PubMed] [Google Scholar]
- 6.Gibson P, Rosella O, Nov R, et al. Colonic epithelium is diffusely abnormal in ulcerative colitis and colorectal cancer. Gut. 1995;36(6):857–863. doi: 10.1136/gut.36.6.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karayiannakis AJ, Syrigos KN, Efstathiou J, et al. Expression of catenins and E-cadherin during epithelial restitution in inflammatory bowel disease. J Pathol. 1998;185(4):413–418. doi: 10.1002/(SICI)1096-9896(199808)185:4<413::AID-PATH125>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 8.Vetrano S, Rescigno M, Rosaria Cera M, et al. Unique role of junctional adhesion molecule-A in maintaining mucosal homeostasis in inflammatory bowel disease. Gastroenterology. 2008;135(1):173–184. doi: 10.1053/j.gastro.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Severson EA, Parkos CA. Mechanisms of outside-in signaling at the tight junction by junctional adhesion molecule A. Ann N Y Acad Sci. 2009;1165:10–18. doi: 10.1111/j.1749-6632.2009.04034.x. [DOI] [PubMed] [Google Scholar]
- 10.Orsulic S, Huber O, Aberle H, et al. E-cadherin binding prevents β-catenin nuclear localization and β-catenin / LEF- 1-mediated transactivation. J Cell Sci. 1999;1245:1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- 11.Williams CS, Zhang B, Smith JJ, et al. BVES regulates EMT in human corneal and colon cancer cells and is silenced via promoter methylation in human colorectal carcinoma. J Clin Invest. 2011;121(10):4056–4069. doi: 10.1172/JCI44228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Q, Hawes SE, Stern JE, et al. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17(3):645–654. doi: 10.1158/1055-9965.EPI-07-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim M, Jang HR, Haam K, et al. Frequent silencing of popeye domain-containing genes, BVES and POPDC3, is associated with promoter hypermethylation in gastric cancer. Carcinogenesis. 2010;31(9):1685–1693. doi: 10.1093/carcin/bgq144. [DOI] [PubMed] [Google Scholar]
- 14.Toon CW, Chou A, Clarkson A, et al. Immunohistochemistry for Myc predicts survival in colorectal cancer. PLoS One. 2014;9(2):e87456. doi: 10.1371/journal.pone.0087456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koo SH, Kwon KC, Shin SY, et al. Genetic alterations of gastric cancer: comparative genomic hybridization and fluorescence in situ hybridization studies. Cancer Genet Cytogenet. 2000;117(2):97–103. doi: 10.1016/s0165-4608(99)00152-1. [DOI] [PubMed] [Google Scholar]
- 16.Cowling VH, Cole MD. E-cadherin repression contributes to c-Myc-induced epithelial cell transformation. Oncogene. 2007;26(24):3582–3586. doi: 10.1038/sj.onc.1210132. [DOI] [PubMed] [Google Scholar]
- 17.Yekkala K, Baudino TA. Inhibition of intestinal polyposis with reduced angiogenesis in ApcMin/+ mice due to decreases in c-Myc expression. Mol Cancer Res. 2007;5(12):1296–1303. doi: 10.1158/1541-7786.MCR-07-0232. [DOI] [PubMed] [Google Scholar]
- 18.Ciclitira PJ, Macartney JC, Evan G. Expression of c-myc in non-malignant and pre-malignant gastrointestinal disorders. J Pathol. 1987;151(4):293–296. doi: 10.1002/path.1711510409. [DOI] [PubMed] [Google Scholar]
- 19.Brentnall TA, Pan S, Bronner MP, et al. Proteins that underlie neoplastic progression of ulcerative colitis. Proteomics - Clin Appl. 2009;3(11):1326–1337. doi: 10.1002/prca.200900061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki R, Miyamoto S, Yasui Y, et al. Global gene expression analysis of the mouse colonic mucosa treated with azoxymethane and dextran sodium sulfate. BMC Cancer. 2007;7:84. doi: 10.1186/1471-2407-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnold HK, Zhang X, Daniel CJ, et al. The Axin1 scaffold protein promotes formation of a degradation complex for c-Myc. EMBO J. 2009;28(5):500–512. doi: 10.1038/emboj.2008.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett CW, Fingleton B, Williams A, et al. MTGR1 is required for tumorigenesis in the murine AOM/DSS colitis-associated carcinoma model. Cancer Res. 2011;71(4):1302–1312. doi: 10.1158/0008-5472.CAN-10-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrée B, Fleige A, Arnold HH, et al. Mouse Pop1 is required for muscle regeneration in adult skeletal muscle. Mol Cell Biol. 2002;22(5):1504–1512. doi: 10.1128/mcb.22.5.1504-1512.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, Flanagan J, Su N, et al. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagnostics. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber CR, Nalle SC, Tretiakova M, et al. Claudin-1 and claudin-2 expression is elevated in inflammatory bowel disease and may contribute to early neoplastic transformation. Lab Invest. 2008;88(10):1110–1120. doi: 10.1038/labinvest.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korinek V, Barker N, Morin PJ, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275(5307):1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 27.Kohno H, Suzuki R, Sugie S, et al. B-catenin mutations in a mouse model of inflammation-related colon carcinogenesis induced by 1,2-dimethylhydrazine and dextran sodium sulfate. Cancer Sci. 2005;96(2):69–76. doi: 10.1111/j.1349-7006.2005.00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krämer A, Green J, Pollard J, et al. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398(6726):422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 30.Mann B, Gelos M, Siedow Q, et al. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci U S A. 1999;96(4):1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41(D1):D377–86. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold HK, Sears RC. Protein Phosphatase 2A regulatory subunit B56α associates with c-Myc and negatively regulates c-Myc accumulation protein. Mol Cell Biol. 2006;26(7):2832–2844. doi: 10.1128/MCB.26.7.2832-2844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh E, Cunningham M, Arnold H, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6(4):308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 34.Issa JJ, Ahuja N, Toyota M. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001:3573–3577. [PubMed] [Google Scholar]
- 35.Grivennikov S, Karin E, Terzic J, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15(2):103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arthur JC, Perez-Chanona F, Muhlbauger M, et al. Intestinal inflammation targets cancer-inducing actvity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katoh H, Wang D, Daikoku T, et al. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell. 2013;24(5):631–644. doi: 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukata M, Chen A, Vamadevan AS, et al. Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology. 2007;133(6):1869–81. doi: 10.1053/j.gastro.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hermiston ML, Gordon JI. Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-Cadherin. Science. 1995;270(5239):1203–1207. doi: 10.1126/science.270.5239.1203. [DOI] [PubMed] [Google Scholar]
- 40.Smith TK, Hager H, Francis R, et al. Bves directly interacts with GEFT, and controls cell shape and movement through regulation fo Rac1/Cdc42 activity. Proc Natl Acad Sci U S A. 2008;105(24):8298–8303. doi: 10.1073/pnas.0802345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Froese A, Breher SS, Waldeyer C, et al. Popeye domain containin proteins are essential for stress-mediated modulation of cardiac pacemaking in mice. J Clin Invest. 2012;122(3):1119–1130. doi: 10.1172/JCI59410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alcalay Y, Hochhauser E, Kliminski V, et al. Popeye domain containing 1 (Popdc1/Bves) is a caveolae-associated protein involved in ischemia tolerance. PLoS One. 2013;8(9):e71100. doi: 10.1371/journal.pone.0071100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osler ME, Chang MS, Bader DM. Bves modulates epithelial integrity through an interaction at the tight junction. J Cell Sci. 2005;118(20):4667–4678. doi: 10.1242/jcs.02588. [DOI] [PubMed] [Google Scholar]
- 44.Dunagan M, Chaudhry K, Samak G, et al. Acetaldehyde disrupts tight junctions in Caco-2 cell monolayers by a protein phosphatase 2A-dependent mechanism. AJP Gastrointest Liver Physiol. 2012;303(12):G1356–64. doi: 10.1152/ajpgi.00526.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J, Woo AJ, Chu J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finch AJ, Soucek L, Junttila MR, et al. Acute overexpression of Myc in intestinal epithelium recapitulates some but not all the changes elicited by Wnt/beta-catenin pathway activation. Mol Cell Biol. 2009;29(19):5306–5315. doi: 10.1128/MCB.01745-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.