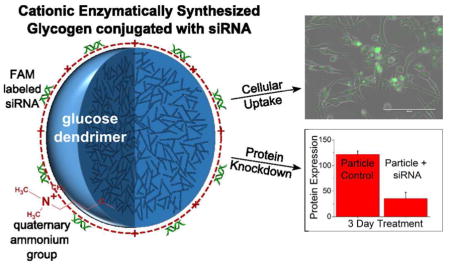
Abstract
Nanostructured starches are naturally derived nanomaterials that can be chemically modified to allow for the introduction of functional groups, enhancing their potential for drug delivery and other biotechnology applications. In this proof of concept study, we investigate chemically modified, enzymatically synthesized glycogen (ESG) nanodendrites as a biodegradable, biocompatible, siRNA delivery system. Commercially available ESG was modified using glycidyltrimethylammonium chloride (GTMA), introducing quaternary ammonium groups via an epoxide ring opening reaction. This cationic ESG (cESG) electrostatically bound siRNA and successfully knocked-down protein expression in an in vitro ovarian clear cell carcinoma model. The construct exhibited sustained siRNA delivery for up to six days while exhibiting less toxicity than a common liposome-based siRNA delivery reagent, Lipofectamine RNAiMAX. These promising results set the stage for use of dendritic starch as a cost-effective, easily modifiable nanoscale delivery system for a diverse range of cargo including nucleic acids and therapeutic compounds.
Graphical Abstract
Introduction
Biologically derived polysaccharides are attractive materials due to their biocompatibility, ease of purification, and low cost. Two examples are cellulose and chitosan, which have seen use in a plethora of applications ranging from drug delivery and tissue engineering to textiles, either “as is” or with chemical modifications.1–4 Another abundant natural polysaccharide with widespread use is starch. Beyond common use in the food industry, conventional starch has been adapted for pharmaceutical applications since the 1930s, primarily as a binding agent. More recently, various micro- and nano-particulate starch formulations have been investigated for the delivery of therapeutic molecules.5
Starch consists of repeated α-glucose units and can be found in several structural forms, such as linear amylose and branched amylopectin. The main difference between the two is in the glyosidic linkages. Amylose has α1→4 glycosidic bonds, whereas amylopectin also contains α1→6 bonds, leading to its branched structure.6 Glycogen, which is the main glucose storage form in mammals, has an even higher degree of α1→6 bonds (roughly one branch every 10 glucose units). The plant-based equivalent to glycogen, phytoglycogen, is a highly branched, high molecular weight α(1→4)(1→6) glucan that forms 30–100 nm nanodendrites. It can be extracted from various sources, including rice,7 barley,8 and algae,9 but is particularly abundant in sweet corn kernels.10 Depending on the source and maturation of plant seeds, size and branching density can vary.11 Compared to linear starch, phytoglycogen and branched starches tend to be more resistant to hydrolytic degradation in vivo.12
In vitro produced nanodendritic starch, herein referred to as enzymatically synthesized glycogen (ESG, Figure 1), mimics the structure of naturally occurring phytoglycogen. However, compared to phytoglycogen, ESG has a more uniform size distribution of 20–40 nm diameter and less variation in the lengths of its α(1→4) chains (narrower chain distribution) while retaining an average α(1→4) chain length of 9–12 glucosyl units.13,14 ESG is prepared from starch or dextran by treatment with isoamylase (EC 3.2.1.68) to produce short chain amylose followed by glycogen assembly using amylomaltase (EC 2.4.1.25) and branching enzyme (EC 2.4.1.18).15
Figure 1.
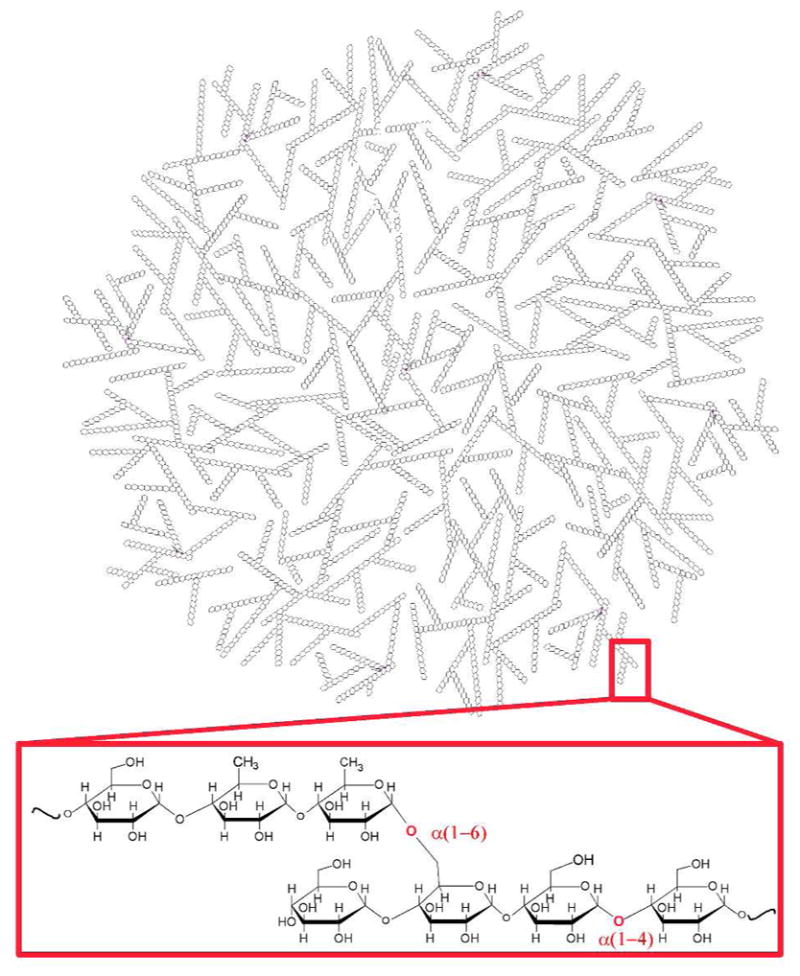
Schematic of ESG structure. Inset α(1→4) and α(1→6) glycosidic linkages of ESG
A safety evaluation of orally administered ESG in rats indicated a lack of toxicological and mutagenic response with the maximum dose of 2g/kg body weight/day tested, deeming ESG as a safe food ingredient.16 The biocompatibility of ESG, and the fact that it can be produced with high consistency at a large scale, makes it an interesting platform for adaptation in technical applications. Due to the relative ease in modifying their chemistries, ESG and similar branched dendrites have the potential to be developed as in vivo therapeutic delivery systems for a variety of cargo. For instance, phytoglycogen has been modified with octenyl succinate to introduce anionic charges and hydrophobicity, enabling the delivery of antigen-vaccines17 and improved efficacy of the antimicrobial peptide nisin.18 An area of interest to our group is the cationic modification of ESG to facilitate electrostatic binding and delivery of nucleic acids and small therapeutic molecules to mammalian cells. One approach to produce cationic carbohydrate carriers is to chemically introduce quaternary amine groups, as previously demonstrated for conventional starch,19 dextran,20 and cellulose.21 Both dextran and cellulose carriers were used for DNA plasmid condensation. While these complexes were shown to effectively enter cells, low transfection efficiency and translation of the plasmid was observed, presumably due to a strong DNA-polymer interaction that prohibited intracellular release of the plasmid.22 In contrast, we hypothesize that the release of cargo from ESG may be facilitated by metabolism of the glycolytic bonds by glycogen breakdown enzymes native to cells, such as glycogen debranching enzyme, glycogen phosphorylase, and phosphoglucomutase. In effect, these glycogen breakdown enzymes would provide an intrinsic mechanism that facilitates ESG cargo release once it has entered the cell.
In this proof of principle study we investigated the feasibility of using cationically modified ESG (cESG) for binding and in vitro delivery of therapeutic short interfering ribonucleic acid (siRNA). cESG was synthesized by modifying free alcohol groups, conjugating these with glycidyltrimethylammonium chloride (GTMA). As a model system, we targeted the high expression of manganese superoxide dismutase (Sod2) in an ovarian clear cell carcinoma cell line, using Sod2-specific siRNA conjugated to cESG. We have previously shown that Sod2, a mitochondria localized antioxidant enzyme, is highly expressed in ovarian clear cell carcinomas and decreasing Sod2 expression may have therapeutic benefits by increasing the cancer cells susceptibility to chemotherapeutic-mediated redox damage (unpublished results). In comparison to a liposomal-based siRNA delivery vehicle, our work demonstrated that cESG is less cytotoxic, and shows a more sustained knockdown of Sod2 expression. Thus providing evidence that this dendritic starch can be modified and utilized as an effective nanoscale nucleic acid delivery vehicle to mammalian cells.
Results and Discussion
Synthesis and Characterization of cationic enzymatically synthesized glycogen (cESG)
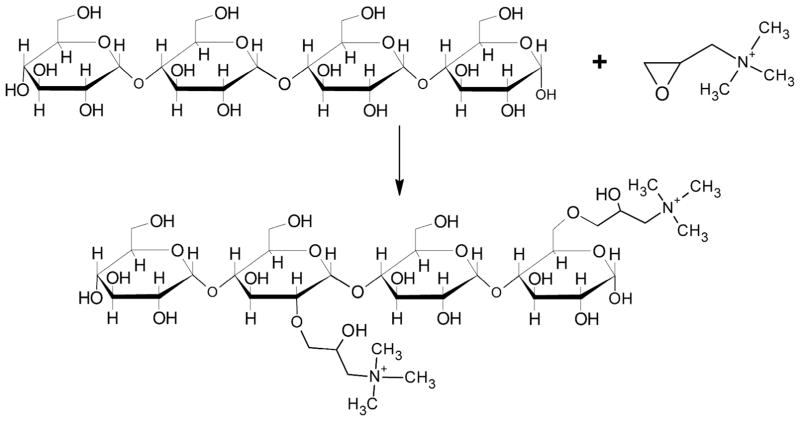
Established carbohydrate-epoxide chemistry was used to conjugate quaternary ammonium cations to commercially available ESG through a ring opening reaction (Scheme 1). Several synthesis conditions with molar ratios of anhydrous glucose units (AGU) to glycidyltrimethylammonium chloride (GTMA) were explored including 1:10, 1:5, 1:1, 2:1 and 10:1. Room temperature alkaline reaction conditions, pH 12.5, similar to those described by Thomas et al., were investigated.23 As a preliminary indicator of the conjugation reaction, zeta (ζ) potentials (the effective electrostatic potential of particles in colloidal suspension) were obtained to monitor the charge additions to cESG. It was found that a 1 to 10 molar ratio of AGU to GTMA resulted in a positive ζ-potential of 20.2 ± 2.9 mV, while reactions containing less GTMA resulted in cESG with considerably lower charge (Table 1). cESG had a similar hydrodynamic radius and appearance to unmodified particles, as measured by dynamic light scattering (DLS) and transmission electron microscopy (Figure S1). DLS indicated that the hydrodynamic radius was 32.4 ± 1.0 nm for the unmodified ESG and 31.9 ± 2.1 nm for cESG. It was found that extended dialysis against deionized water resulted in GTMA-modified ESG with greater than 20mV ζ-potential to fragment. However, adding 1–2 mM salt (sodium chloride) to the dialysis solution was sufficient to maintain particle integrity. The exact cause for this particle disintegration is unclear. The cESG synthesized at 1:10 AGU:GTMA molar ratio, with +20mV ζ-potential, was selected for continued elemental analysis, investigation of siRNA binding, and cellular delivery.
Scheme 1.
Epoxide ring opening reaction of GTMA with glucose residues.
Table 1.
Size and ζ-potential of cESG generated with different ratios of ESG to GTMA measured by DLS. (n=3 independent conjugation experiments. Each experiment was averaged over three measurements using 15 scans at 10 seconds each.) For all measurements cESG was dissolved in 1mM sodium phosphate buffer at pH 7.5.
| Molar Ratio (AGU:GTMA) | Size ± Std Dev. (nm) | ζ-potential ± Std Dev. (mV) |
|---|---|---|
| 1:10 | 31.9 ± 2.1 | +20.2 ± 2.9 |
| 1:5 | 31.1 ± 2.1 | +9.5 ± 4.1 |
| 1:1 | 32.5 ± 2.3 | +6.6 ± 8.4 |
| 2:1 | 31.7 ± 1.3 | 0.7 ± 1.0 |
| 10:1 | 32.2 ± 0.8 | −2.7 ± 0.9 |
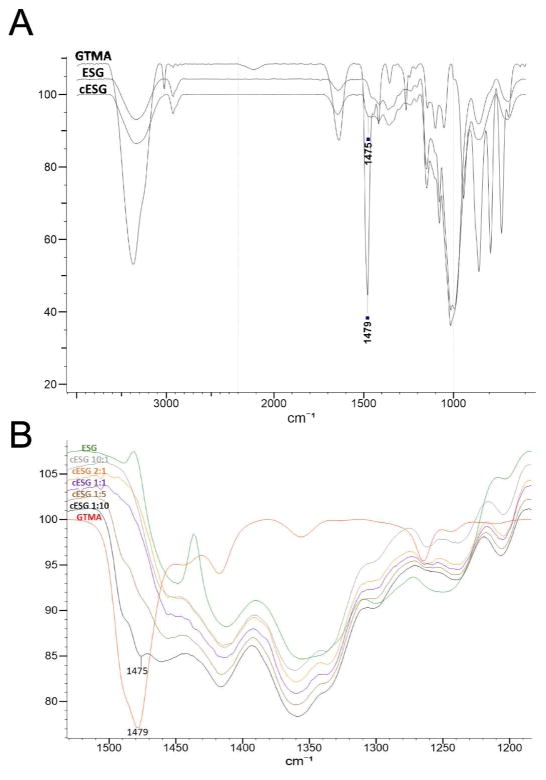
Incorporation of the quaternary amine group into cESG was verified using attenuated total reflectance Fourier transform infrared spectroscopy (ATR-FTIR) and proton nuclear magnetic resonance spectroscopy (1H NMR). ATR-FTIR on lyophilized cESG (Figure 2A) showed a peak at 1475 cm−1, assigned to C-H stretching of the quaternary amine group in GTMA. This peak was not present in unmodified ESG. Figure 2B shows a zoomed in overlay spectra of the products from the various AGU:GTMA ratios, with the 1475 cm−1 peak clearly visible in cESG synthesized using the 1:10, and 1:5 ratios. Similarly, proton NMR (Figure S3) of cESG synthesized at higher GTMA ratio show a quaternary amine peak at 3.116 ppm, corresponding to the 9 hydrogens of the methyl groups associated with nitrogen.
Figure 2.
ATR-FTIR shows incorporation of quaternary ammonium group in cESG A) ATR-FTIR spectra of ESG, cESG synthesized at 1:10 ratio (AGU:GTMA), and GTMA, where the peak at 1475 cm−1 in cESG is assigned to C-H stretching of methyl groups on the quaternary amine. See Figure S2 for additional cESG peak assignments. B) Zoomed in overlay spectra of the products from various AGU:GTMA ratios. The incorporation a quaternary amine, as indicated by the 1475 cm−1 peak, is clearly seen for the 1:10 and 1:5 samples.
X-ray photoelectron spectroscopy (XPS) and elemental analysis isotope ratio mass spectroscopy (EA-IRMS) were used to determine the degree of substitution of cESG (Table 2). XPS of cESG dried on silicon indicated a carbon to nitrogen (C/N) ratio of 37.9/1, equivalent to a degree of substitution of 0.188. No nitrogen was detected in the unmodified ESG (Figure S4). EA-IRMS was used to corroborate the XPS data, and showed a 0.28 degree of substitution, again with no nitrogen detected in the unmodified cESG. The above results represent approximately one cationic amino group per five AGU.
Table 2.
Degree of substitution of cESG according to XPS and EA-IRMS techniques. XPS degree of substitution from n=3 independent conjugation experiments. EA-IRMS, n=1 verification experiment.
| Method | C/N Ratio | Degree of substitution | AGU residues per cationic amino group |
|---|---|---|---|
| XPS | 37.9/1 | 0.188 ± 0.027 | 5.3 |
| EA-IRMS | 27.4/1 | 0.28 | 3.6 |
siRNA Binding of cESG
A fluorescein-labeled siRNA targeting Sod2 was incubated at various mass ratios with cESG and the binding of siRNA to cESG evaluated using a gel shift assay (Figure 3). During electrophoresis, unbound siRNA is free to migrate through an agarose gel matrix whereas siRNA bound to cESG (cESG-siRNA complex) either displays retarded or no migration. No siRNA was found to migrate through the gel when the cESG mass added exceeded that of siRNA (at least 2.5:1 w/w), indicating majority binding of the siRNA. The molecular weight of cESG (10,275,000 g/mol) was estimated based on the hydrodynamic radius using the DLS protein utilities software, and used for calculating the theoretical siRNA molecules per cESG. As the charge of the quaternary ammonium groups is not pKa dependent, the conjugation could be performed at biological pH 7.5 without affecting the intrinsic charge of cESG. All cESG-siRNA complexes that exhibited retention in the gel shift assay had the theoretical equivalent of 200 siRNA molecules or fewer per cESG and maintained a net positive ζ-potential of 2.5 ± 1 mV (Table S1). The slight positive charge on the resultant cESG-siRNA complexes may be beneficial for delivery to cells. For example, electrostatically condensed DNA polymer complexes that retain a net positive charge have demonstrated improved binding to cellular membranes, thereby facilitating cellular uptake.24 These complexes, with complete siRNA binding and positive charge, were further investigated for use in an in vitro cell culture model.
Figure 3.
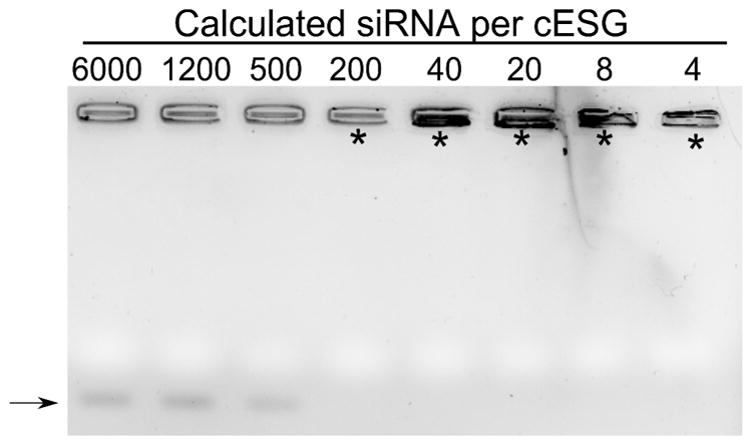
Gel shift assay of cESG-siRNA complexes confirms condensation of siRNA to cESG. Following condensation reactions, complexes were loaded into wells of 1% agarose gel, followed by electrophoresis. Migration of free siRNA (arrow) was observed in formulations with a higher numbers of siRNA per cESG. Impeded migration (asterisks), indicative of siRNA condensation, is observed in wells containing 200 siRNA per particle or less.
In Vitro mRNA Silencing by cESG-siRNA
Cellular uptake and cESG-siRNA mediated expression knockdown
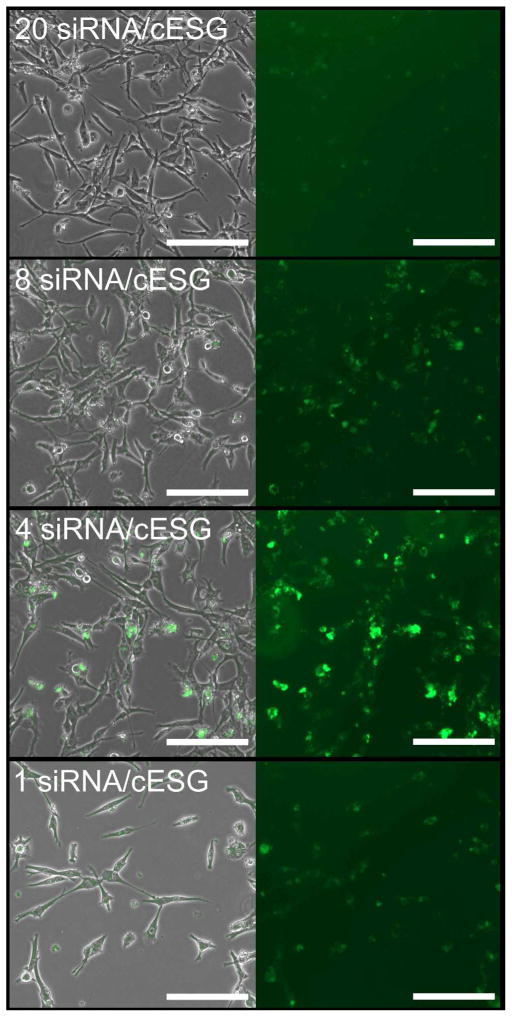
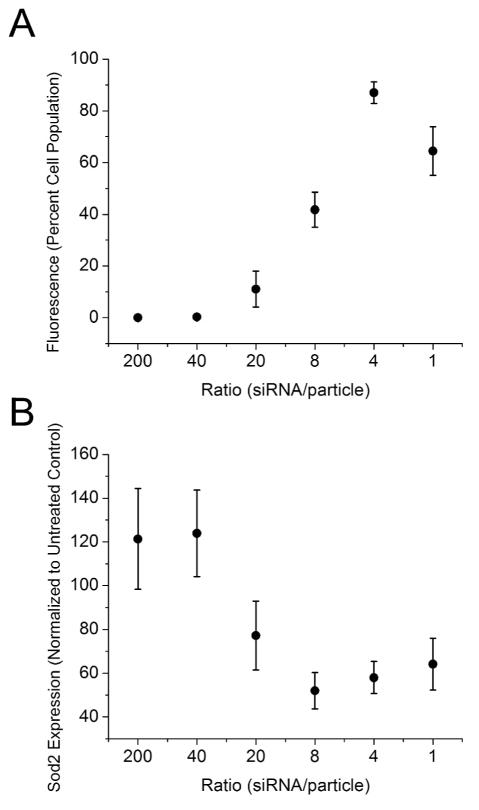
To evaluate cESG mediated siRNA delivery to cells, we chose to target Sod2 mRNA with a previously validated siRNA sequence, containing a 5′ fluorescein tag to visualize cellular uptake.25 The cESG-siRNA complexes with impeded migration in the gel shift assay (one to 200 siRNA per cESG) were added to cultures of ovarian clear cell carcinoma (ES-2) cells for 48 hrs. These cells exhibit high levels of Sod2 protein, making this a good model to test cESG-siRNA-mediated knockdown of Sod2 expression. For uptake studies, the concentration of siRNA transfected per well was held constant (100nM, approximately 7.5x108 siRNA per cell). Our lab has previously demonstrated that this siRNA concentration achieves optimal Sod2 expression knockdown using a commercially available liposomal-based siRNA transfection method (Lipofectamine RNAiMAX; LifeTechnologies) and is in the range of non-toxic siRNA concentrations commonly used in other nanoparticle delivery formulations.26–28 Monitoring uptake of cESG-siRNA by fluorescence microscopy revealed that lower siRNA to cESG particle ratios led to increased intracellular accumulation of complexes (Figure 4). Western blotting further confirmed that enhanced uptake of cESG-siRNA correlated well with knockdown of Sod2 protein expression (Figure 5). Since the siRNA concentration was held constant in all wells, cells were exposed to higher concentrations of cESG particles as the ratio of siRNA to cESG increased. For example, cells in Figure 2 pane 3 (4 siRNA/cESG) had fivefold more particles than Figure 2 pane 1 (20siRNA/cESG), but the same siRNA concentration. Thus the enhanced uptake of cESG-siRNA in these formulations and subsequent increase in efficiency of Sod2 knockdown may be an effect of the cells being exposed to a higher number of dendritic starch particles, in addition to potential effects from an optimal complex composition (i.e. number of siRNA per particle).
Figure 4.
Uptake of fluorescently labeled cESG-siRNA by ES-2 ovarian clear cell carcinoma cells increases as siRNA per cESG particle decreases. Fluorescence images (right) and overlay bright field images (left) of ES-2 cells incubated for 72hrs with cESG-siRNA complexes of various formulations were visualized by microscopy. Increased uptake was seen in complexes with fewer siRNA per cESG particle. Scale bar 200 μm.
Figure 5.
Uptake of cESG-siRNA correlates with Sod2 protein expression knockdown. A) Uptake of cESG-siRNA was quantified as the percentage of cells which display fluorescence signals (at least 50 cells/image, n=3 images, analyzed and counted in three independent experiments, data represent mean +/− SEM). B) Sod2 protein expression was determined by western blotting 72 hrs after treatment with cESG-siRNA complexes (Figure S5). Expression was quantified by densitometry and normalization to the loading control β-actin and untreated control cells (n=3).
Subsequently, the cESG-siRNA complex that demonstrated significant knockdown of Sod2 (4 siRNA/cESG) was compared against the commonly used liposomal-based siRNA transfection reagent Lipofectamine RNAiMAX (LifeTechnologies). The amount of siRNA used for each delivery method remained constant (100 nM). ES-2 cells were incubated for 72 hrs with the respective complexes and then harvested immediately. Alternatively, the cells were incubated for 72 hrs with siRNA and left for an additional 72 hr recovery period in growth media before harvesting (6 day total). The level of siRNA-mediated Sod2 protein expression knockdown was analyzed by western blotting. Following a 72 hr incubation with cESG-siRNA, a decrease in Sod2 expression to 35% of untreated control was observed. At this time point, in comparison, an almost complete knockdown of Sod2 was observed in the Lipofectamine RNAiMAX-siRNA treated cells. After 6 days (3 days of treatment and 3 days recovery in media), extensive cell death was observed for Lipofectamine transfected cells, therefore Sod2 expression could not be analyzed. However, cESG-siRNA treated cells remained viable with sustained Sod2 knockdown (16% Sod2 expression) observed 6 days after the start of cESG-siRNA treatment (Figure 6 & 7).
Figure 6.
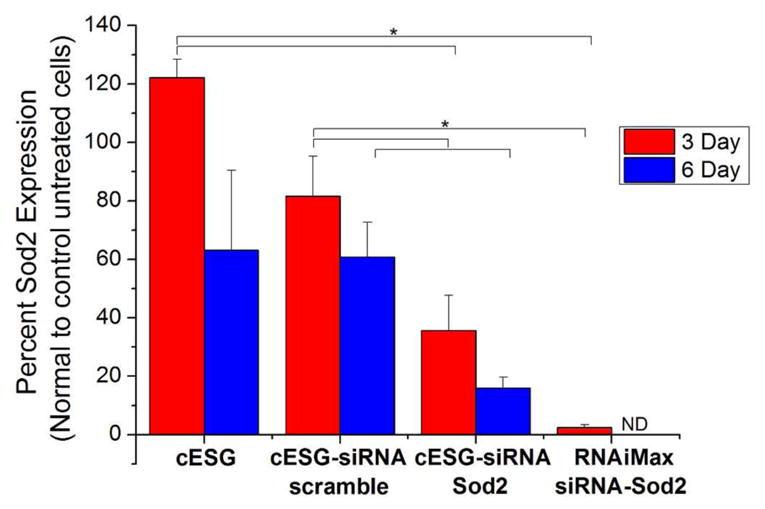
cESG-siRNA facilitates sustained protein expression knockdown. ES-2 cells were treated either with growth media (untreated control), cESG, cESG-siRNA-scramble, or cESG-siRNA-Sod2 for 72 hrs (3 Day) or allowed to recover in growth media for an additional 72hrs (6 Day) prior to assessment of Sod2 protein expression by western blotting (Figure S6). Western blot data was quantified by densitometric analysis and normalized to expression of the loading control β-Actin and expressed relative to Sod2 expression in untreated control cells. (n=3 independent experiments, mean ±SEM, * p<0.05, ANOVA with Tukey’s post-test.)
Figure 7.
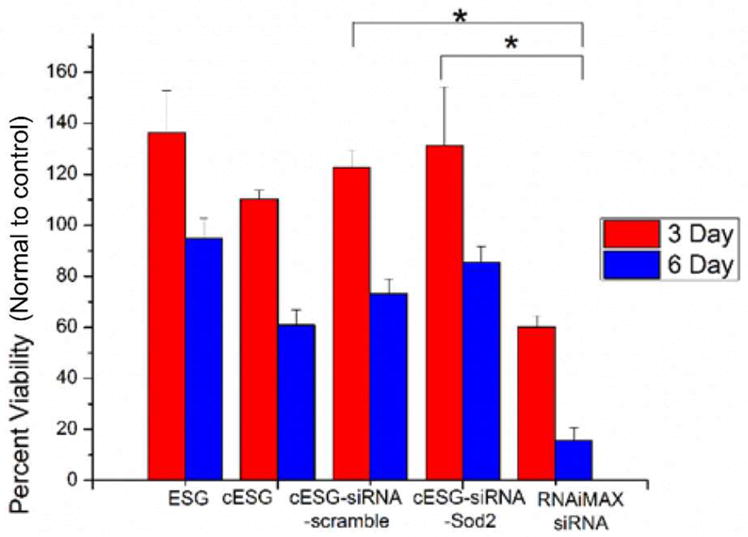
cESG-siRNA displays significantly less cytotoxicity than Lipofectamine RNAiMAX. Effects of cESG, cESG-siRNA and Lipofectamine RNAiMAX-siRNA complexes on ES-2 cell viability were assessed using a MTT Viability Assay. Treatments were conducted for 72 hrs (3 Day) or with an additional 72 hr recovery period in growth media (6 Day). MTT absorption values were normalized to control untreated cells. Representative data expressed as mean ± SEM; n=3 wells. *p<0.05 Controls compared to treatment with a student’s T-Tests
Cytotoxicity of cESG and cESG-siRNA
Given our observation that cESG-siRNA appears to be less toxic than Lipofectamine RNAiMAX transfection reagent, we further evaluated the cytotoxicity of cESG and cESG-siRNA using a MTT assay. Unmodified ESG, cESG and cESG-siRNA (bound to either scramble or Sod2 targeting sequence) displayed significantly less cytotoxicity after both 3 and 6 day treatments compared to Lipofectamine RNAiMAX (Figure 7). These data suggest that dendritic starch may be a valuable cellular delivery vehicle given its ability to effectively deliver nucleic acids and due to its lack of cytotoxicity compared to liposomal vectors.
Conclusion
In this study, we investigated the possibility to use cationic carbohydrate nanodendrimers (cESG) for siRNA delivery. Chemical modification using GTMA was used to incorporate one positive amino group for every five AGU. Multiple formulations of cESG-siRNA were produced via electrostatic condensation and demonstrated to successfully be taken up by ES-2 clear cell carcinoma cells while exhibiting low cytotoxicity. This non-covalent condensation to cESG may be investigated further for co-delivery of small therapeutic molecules. The system is of interest for sustained delivery of siRNA to cancer, where the leaky vasculature of tumors gives rise to the enhanced permeability and retention (EPR) effect allowing for size dependent particle accumulation within the tumor compared to neighboring normal cells.29 Of specific interest is the finding that cESG-siRNA maintained sustained knockdown of the protein target for 6 days after initiating treatment. This may indicate that siRNA is slowly released from the particle, which is a particularly attractive feature for therapeutic delivery. The contribution of enzymes within the target cell that may aid in this cargo release, by specific breakdown of glycogen bonds, remain to be investigated. However, the observation that enhanced expression of enzymes involved in glycogen breakdown are induced by the hypoxic tumor microenvironment30,31 indicates the degradation of cESG in cancers may also be enhanced, facilitating release of bound material. The uptake and degradation behavior of cESG for delivery of adjuvant therapies to ovarian cancer is the subject of future studies with this new nanoparticle system.
Experimental Procedures
Materials
Enzymatically synthesized glycogen (ESG) (Bioglycogen™ lot 100526) was purchased from Glico Nutrition Co. Ltd. Glycidyltrimethylammonium chloride (GTMA) was purchased from Sigma-Aldrich Chemicals Co. A 5′ fluorescein 6-FAM-labeled, previously validated siRNA targeting Sod2 was purchased from Dharmacon (On-Target Plus 5′-CAACAGGCCUUAUUCCACU-3′). A scramble oligonucleotide sequence was used as a non-targeting control (Dharmacon, OnTarget Plus Control siRNA Nontargeting siRNA #1). Antibodies were obtained from Cell Signaling Technology (Boston, MA) or Abcam (Cambridge, MA). Synthesis experiments were conducted using 18.2 MOhm-cm, RNAse free water, cell treatments were prepared in sterile, nuclease free water (Qiagen). All experiments were conducted with reagent grade chemicals.
Synthesis of cESG
100 mg of ESG were dissolved in 10 mL of 0.1M sodium phosphate buffer of pH 12.5 (0.6mmols AGU). While stirring GTMA was added dropwise to the solution at the following molar ratios AGU:GTMA 1:10, 1:5, 1:1, 2:1, and 10:1. The reaction was stirred at room temperature for 24 hrs. The products were purified by 48 hrs of dialysis against 4 L of 1mM sodium chloride, with 3 changes, using a 10,000 kDa MWCO regenerated cellulose membrane (ThermoScientific). The dialyzed ESG-GTMA product (cESG) was lyophilized on a Labconco Freezone 2.5 Plus before further analysis.
Dynamic Light Scattering (DLS)
Zeta (ζ) potential and mean particle diameter were collected in 1mM sodium phosphate buffer pH 7.5 (5mg lyophilized compound/mL) using DLS (Malvern Zetasizer Nano, Worcestershire, UK). Mean particle diameter measurements were averaged over three measurements using 15 scans at 10 seconds each. The particle diameters reported were calculated by the percent volume algorithm of the Malvern Zetasizer collection software. Zeta potential measurements were averaged over three measurements of 60 scans each.
Fourier Transform Infrared Spectroscopy (FTIR)
FTIR spectra of samples were taken of lyophilized compounds using a Bruker Tensor27 spectrometer with a Pike Miracle single bounce attenuated total reflectance (ATR) attachment. Spectra were collected over a range of 800 – 4000 cm−1 and averaged over 32 scans at 4 cm−1 resolution.
Nuclear Magnetic Resonance (NMR)
NMR spectra were obtained of lyophilized sample dissolved in deuterium oxide (100mg/mL) at room temperature using a 400 MHz Bruker Avance spectrometer with Topspin 2.1 software. Spectra were averaged over 64 scans at 400.13 MHz with a pulse of 1200 μs and a 1s relaxation delay.
X-Ray Photoelectron Spectrometry (XPS)
For XPS preparation, approximately 0.5 μM ESG or cESG complexes were prepared in 1 mM sodium phosphate buffer and 100 μL added onto an acetone cleaned 1cm2 silicon substrate. Samples were dried overnight under nitrogen. The samples were analyzed on a Thermo VG Scientific Theta Probe X-ray Photoelectron Spectroscopy (XPS) at 125 eV detector pass energy with a stepsize of 0.1eV and a dwell time of 50ms. Each high resolution spectra was averaged over 30 scans. Fresh samples were used for each analysis.
Transmission Electron Microscopy (TEM)
TEM specimens were prepared on formvar coated gold grids using the drop method. Briefly, 10 μL of 0.1 μM solution of ESG or cESG was dropped onto a grid for 30 seconds prior to the solution being wicked away. A 10 uL drop of methylamine tungstate dye (NanoW, Nanoprobes) was dropped on the grid and the specimen allowed to dry overnight under nitrogen. Microscopy was done in a JEOL 2010 field emission transmission electron microscope or JEOL 2010 with thermionic LaB6 emitter operating at 200kV in brightfield mode. ImageJ software was used to determine particle dimensions.
Elemental Analysis Isotope Ratio Mass Spectroscopy (EA-IRMS)
EA-IRMS was conducted using a Carlo Erba NA 1500 elemental analyzer (Milano, IT) coupled to a VG Isochrom continuous flow IRMS (Isoprime Inc., Manchester, UK) and an integrated thermal conductivity detector (TCD) for the determination of carbon and nitrogen content. Analysis was performed by EcoCore Analytical Services (Colorado State University, Fort Collins CO).
Degree of Substitution
Degree of substitution (DS) calculations were performed from the molar C/N ratio using the equation:
Condensation of cESG-siRNA
Lyophilized cESG was dissolved in 1mM NaPO4 buffer pH 7.5 at a concentration of 3 μM ESG. 10 μM fluorescein-labeled siRNA with a Sod2-specific targeting sequence was prepared in distilled water. The particle and siRNA were then mixed at various mass ratios (1:2, 1:12.5, 1:25, 1:60, 1:125, 1:500), at a constant siRNA concentration of 1 μM, in distilled water. Solutions were vortexed and centrifuged for 15 seconds every 5 minutes, for a total 40 minute incubation period.
Agarose gel retardation studies
10 μL of cESG-siRNA solutions, containing 60 ng of siRNA, were mixed with 2 μL of 10x gel loading dye. 10 μL of the solution was loaded into a 1% Agarose gel. Electrophoresis was run in 1x Tris-acetate-EDTA (TAE) buffer at 100 V for 20 minutes. Ethidium bromide bath staining was conducted for 15 min, followed by imaging on a FluorChem E imager (Protein Simple).
Cell culture
ES-2 clear cell ovarian carcinoma cells (purchased from ATCC) were maintained in McCoy’s 5A media supplemented with 10% fetal bovine serum and cultured at 37 °C under 5% CO2.
Analysis of cESG-siRNA cellular uptake and transfection efficiency
ES-2 cells were seeded into 6-well plates at a density of 80,000 cells per well. After 24 hrs cells were incubated with fresh cESG-siRNA complexes (100 nM siRNA/well) in growth media (2 mL). For fluorescent imaging, media was replaced with PBS after indicated treatment times and FAM-labeled siRNA-cESG within cells detected using an EVOSfl fluorescent microscope (AMG). Bright field images were taken simultaneously and the number of fluorescing to total number of cells quantified. Cells were washed and collected on ice in 50 μL RIPA buffer,32 containing proteinase inhibitors, for western blotting analysis. For comparison to Lipofectamine transfection ES-2 cells were cultured as above and incubated for 72 hrs in growth media containing 100 nM siRNA freshly prepared as either cESG-siRNA or Lipofectamine RNAiMAX-siRNA complexes (prepared according to the manufacturer’s directions). After 72 hrs cells were either (1) immediately replaced with PBS for fluorescence imaging and collected on ice in RIPA buffer or (2) incubated in fresh growth media for an additional 72 hrs before fluorescent imaging and harvesting for western blot analysis. Controls (including growth media alone, cESG and cESGScramble- siRNA) were treated in the same manner.
Western Blotting Protocol
Protein concentration was determined using a BCA assay (Pierce) and samples were loaded into a mini-Protean gel (BioRad) along with a Precision Plus Protein™ dual color standard ladder (BioRad), and run in 1x Tris-Glycine-SDS for 45 min at 160 V. Samples were transferred to a polyvinylidene fluoride (PVDF) membrane using a TurboBlot transfer unit (Bio-Rad) and blocked for 1 hr in 5% powdered milk in Tris-buffered saline-0.1% Tween 20 (TTBS). The membrane was incubated with primary antibody to Sod2 or β-Actin control overnight and rinsed prior to a 1-hr incubation with secondary anti-mouse or anti-rabbit antibody respectively. West femto substrate (Thermo Scientific) was used for band visualization in a ChemiDoc MP imaging system (Bio-Rad). Band intensities were quantified using ImageJ software. Band intensities were standardized to the average band intensity of the blot, followed by normalization of the Sod2 band intensity to the corresponding β-Actin controls.
Statistical Analysis
All data presented are representative of at least three independent experiments (unless otherwise noted) and expressed as mean ± SEM. Statistical data analysis ANOVA with Tukey’s post test or student t-test was performed using OriginPro Software v8.5.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the SUNY Research Fund and NIH for funding (NIH/NCI grant R00CA143229 to NH). We are grateful to Vijay Jain Bharamaiah Jeevendrakumar and Bushra Alam for their support in the collection of XPS and TEM analysis.
Abbreviations
- ESG
Enzymatically synthesized glycogen
- cESG
cationic enzymatically synthesized glycogen
- siRNA
short interfering RNA
- GTMA
glycidyltrimethylammonium chloride
- Sod2
manganese superoxide dismutase
- DLS
dynamic light scattering
- EA-IRMS
elemental analysis – isotope ratio mass spectroscopy
- XPS
X-Ray Photoelectron Spectroscopy
- AGU
anhydrous glucose units
Footnotes
Conflict of Interest Disclosure: The authors declare no competing financial interest.
Supporting Information Available: Transmission electron micrographs, FTIR peak assignments, NMR and XPS spectra, and representative western blots. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Shukla SK, Mishra AK, Arotiba OA, Mamba BB. Chitosan-based nanomaterials: A state-of-the-art review. Int J Biol Macromol. 2013;59:46–58. doi: 10.1016/j.ijbiomac.2013.04.043. [DOI] [PubMed] [Google Scholar]
- 2.Dash M, Chiellini F, Ottenbrite RM, Chiellini E. Chitosan—A versatile semi-synthetic polymer in biomedical applications. Prog Polym Sci. 2011;36:981–1014. [Google Scholar]
- 3.Kong M, Chen XG, Xing K, Park HJ. Antimicrobial properties of chitosan and mode of action: A state of the art review. Int J Food Microbiol. 2010;144:51–63. doi: 10.1016/j.ijfoodmicro.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Kalia S, Dufresne A, Cherian BM, Kaith B, Avérous L, Njuguna J, Nassiopoulos E. Cellulose-based bio-and nanocomposites: a review. Int J Polym Sci. 2011;2011:35. [Google Scholar]
- 5.Mateescu MA, Ispas-Szabo P, Assaad E. Controlled Drug Delivery. Chapter 2. Woodhead Publishing; Cambridge: 2015. Starch and derivatives as pharmaceutical excipients: From nature to pharmacy; pp. 21–84. [Google Scholar]
- 6.Rodrigues A, Emeje M. Recent applications of starch derivatives in nanodrug delivery. Carbohydr Polym. 2012;87:987–994. [Google Scholar]
- 7.Wong KS, Kubo A, Jane J-l, Harada K, Satoh H, Nakamura Y. Structures and Properties of Amylopectin and Phytoglycogen in the Endosperm of sugary-1 Mutants of Rice. J Cereal Sci. 2003;37:139–149. [Google Scholar]
- 8.Burton RA, Jenner H, Carrangis L, Fahy B, Fincher GB, Hylton C, Laurie DA, Parker M, Waite D, Van Wegen S. Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. The Plant J. 2002;31:97–112. doi: 10.1046/j.1365-313x.2002.01339.x. [DOI] [PubMed] [Google Scholar]
- 9.Dauvillée D, Colleoni C, Mouille G, Buléon A, Gallant DJ, Bouchet B, Morell MK, d’Hulst C, Myers AM, Ball SG. Two loci control phytoglycogen production in the monocellular green alga Chlamydomonas reinhardtii. Plant Physiol. 2001;125:1710–1722. doi: 10.1104/pp.125.4.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris DL, Morris CT. Glycogen in the Seed of Zea Mays (Variety Golden Bantam) J Biol Chem. 1939;130:535–544. [Google Scholar]
- 11.Tateishi K, Nakano A. Effects of Degree of Branching on Dispersion Stability of Phytoglycogen in Aqueous Solution. Biosci Biotech Biochem. 1997;61:455–458. [Google Scholar]
- 12.Huang L, Yao Y. Particulate structure of phytoglycogen nanoparticles probed using amyloglucosidase. Carbohydr Polym. 2011;83:1665–1671. [Google Scholar]
- 13.Takata H, Kajiura H, Furuyashiki T, Kakutani R, Kuriki T. Fine structural properties of natural and synthetic glycogens. Carbohydr Res. 2009;344:654–659. doi: 10.1016/j.carres.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Kajiura H, Takata H, Kuriki T, Kitamura S. Structure and solution properties of enzymatically synthesized glycogen. Carbohydr Res. 2010;345:817–824. doi: 10.1016/j.carres.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Kajiura H, Kakutani R, Akiyama T, Takata H, Kuriki T. A novel enzymatic process for glycogen production. Biocatal Biotransform. 2008;26:133–140. [Google Scholar]
- 16.Tafazoli S, Wong AW, Kajiura H, Kakutani R, Furuyashiki T, Takata H, Kuriki T. Safety evaluation of an enzymatically-synthesized glycogen (ESG) Regulatory tox and pharm. 2010;57:210– 219. doi: 10.1016/j.yrtph.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Lu F, Mencia A, Bi L, Taylor A, Yao Y, HogenEsch H. Dendrimer-like alpha-d-glucan nanoparticles activate dendritic cells and are effective vaccine adjuvants. J Controlled Release. 2015;204:51–59. doi: 10.1016/j.jconrel.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Bi L, Yang L, Narsimhan G, Bhunia AK, Yao Y. Designing carbohydrate nanoparticles for prolonged efficacy of antimicrobial peptide. J Controlled Release. 2011;150:150–156. doi: 10.1016/j.jconrel.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 19.Amar-Lewis E, Azagury A, Chintakunta R, Goldbart R, Traitel T, Prestwood J, Landesman-Milo D, Peer D, Kost J. Quaternized starch-based carrier for siRNA delivery: from cellular uptake to gene silencing. J Controlled Release. 2014;185:109–120. doi: 10.1016/j.jconrel.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 20.Yudovin-farber I, Yanay C, Azzam T, Linial M, Domb AJ. Quaternary Ammonium Polysaccharides for Gene Delivery. Bioconjugate Chem. 2005;16:1196–1203. doi: 10.1021/bc050066p. [DOI] [PubMed] [Google Scholar]
- 21.Fayazpour F, Lucas B, Alvarez-Lorenzo C, Sanders NN, Demeester J, De Smedt SC. Physicochemical and transfection properties of cationic Hydroxyethylcellulose/DNA nanoparticles. Biomacromolecules. 2006;7:2856–2862. doi: 10.1021/bm060474b. [DOI] [PubMed] [Google Scholar]
- 22.Song Y, Wang H, Zeng X, Sun Y, Zhang X, Zhou J, Zhang L. Effect of Molecular Weight and Degree of Substitution of Quaternized Cellulose on the Efficiency of Gene Transfection. Bioconjugate Chem. 2010;21:1271–1279. doi: 10.1021/bc100068f. [DOI] [PubMed] [Google Scholar]
- 23.Thomas JJ, Rekha MR, Sharma CP. Dextran-glycidyltrimethylammonium chloride conjugate/DNA nanoplex: A potential non-viral and haemocompatible gene delivery system. Int J Pharm. 2010;389:195–206. doi: 10.1016/j.ijpharm.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Kabanov AV. Taking polycation gene delivery systems from in vitro to in vivo. Pharm Sci Technolo Today. 1999;2:365–372. doi: 10.1016/s1461-5347(99)00186-8. [DOI] [PubMed] [Google Scholar]
- 25.Dier U, Shin D-H, Hemachandra LPMP, Uusitalo LM, Hempel N. Bioenergetic analysis of ovarian cancer cell lines: profiling of histological subtypes and identification of a mitochondria-defective cell line. PLoS One. 2014:9. doi: 10.1371/journal.pone.0098479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taratula O, Garbuzenko OB, Kirkpatrick P, Pandya I, Savla R, Pozharov VP, He H, Minko T. Surface-engineered targeted PPI dendrimer for efficient intracellular and intratumoral siRNA delivery. J Controlled Release. 2009;140:284–293. doi: 10.1016/j.jconrel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Florinas S, Nam HY, Kim SW. Enhanced siRNA delivery using a combination of an arginine-grafted bioreducible polymer, ultrasound, and microbubbles in cancer cells. Mol Pharm. 2013;10:2021–2030. doi: 10.1021/mp400048p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krebs MD, Jeon O, Alsberg E. Localized and sustained delivery of silencing RNA from macroscopic biopolymer hydrogels. J Am Chem Soc. 2009;131:9204–9206. doi: 10.1021/ja9037615. [DOI] [PubMed] [Google Scholar]
- 29.Torchilin V. Tumor delivery of macromolecular drugs based on the EPR effect. Adv Drug Deliv Rev. 2011;63:131–135. doi: 10.1016/j.addr.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Favaro E, Bensaad K, Chong MG, Tennant DA, Ferguson DJ, Snell C, Steers G, Turley H, Li JL, Gunther UL, et al. Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metab. 2012;16:751–764. doi: 10.1016/j.cmet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 31.Zois CE, Favaro E, Harris AL. Glycogen metabolism in cancer. Biochem Pharmacol. 2014;92:3–11. doi: 10.1016/j.bcp.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Gómez-Sánchez R, Pizarro-Estrella E, Yakhine-Diop SMS, Rodríguez-Arribas M, Bravo-San Pedro JM, Fuentes JM, González-Polo RA. Routine Western blot to check autophagic flux: Cautions and recommendations. Anal Biochem. 2015;477:13–20. doi: 10.1016/j.ab.2015.02.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.