Abstract
Deerpox virus (DPV), an uncharacterized and unclassified member of the Poxviridae, has been isolated from North American free-ranging mule deer (Odocoileus hemionus) exhibiting mucocutaneous disease. Here we report the genomic sequence and comparative analysis of two pathogenic DPV isolates, W-848-83 (W83) and W-1170-84 (W84). The W83 and W84 genomes are 166 and 170 kbp, containing 169 and 170 putative genes, respectively. Nucleotide identity between DPVs is 95% over the central 157 kbp. W83 and W84 share similar gene orders and code for similar replicative, structural, virulence, and host range functions. DPV open reading frames (ORFs) with putative virulence and host range functions include those similar to cytokine receptors (R), including gamma interferon receptor (IFN-γR), interleukin 1 receptor (IL-1R), and type 8 CC-chemokine receptors; cytokine binding proteins (BP), including IL-18BP, IFN-α/βBP, and tumor necrosis factor binding protein (TNFBP); serpins; and homologues of vaccinia virus (VACV) E3L, K3L, and A52R proteins. DPVs also encode distinct forms of major histocompatibility complex class I, C-type lectin-like protein, and transforming growth factor β1 (TGF-β1), a protein not previously described in a mammalian chordopoxvirus. Notably, DPV encodes homologues of cellular endothelin 2 and IL-1R antagonist, novel poxviral genes also likely involved in the manipulation of host responses. W83 and W84 differ from each other by the presence or absence of five ORFs. Specifically, homologues of a CD30 TNFR family protein, swinepox virus SPV019, and VACV E11L core protein are absent in W83, and homologues of TGF-β1 and lumpy skin disease virus LSDV023 are absent in W84. Phylogenetic analysis indicates that DPVs are genetically distinct from viruses of other characterized poxviral genera and that they likely comprise a new genus within the subfamily Chordopoxvirinae.
Within the subfamily Chordopoxvirinae of the family Poxviridae, eight genera are currently recognized based primarily on morphological and biological characteristics (48). Viruses from seven genera infect mammalian species (Capripoxvirus, Leporipoxvirus, Molluscipoxvirus, Orthopoxvirus, Parapoxvirus, Suipoxvirus, and Yatapoxvirus), and one genus infects birds (Avipoxvirus). Comparative genome analysis has provided a genetic basis for poxviral genus classification (31, 43). Chordopoxvirus (ChPV) genomes range from 135 to 365 kb in size and contain 130 to 328 putative genes. Complete genomic sequences have been determined for representative and often multiple viruses from each ChPV genus, including the following viruses: sheeppox, goatpox, and lumpy skin disease viruses (Capripoxvirus) (61, 62); myxoma and rabbit (Shope) fibroma viruses (Leporipoxvirus) (14, 67); molluscum contagiosum virus (Molluscipoxvirus) (55); monkeypox, vaccinia, camelpox, variola, and ectromelia viruses (Orthopoxvirus) (4, 16, 28, 32, 42, 56); orf and bovine popular stomatitis viruses (Parapoxvirus) (17); swinepox virus (Suipoxvirus) (3); Yaba monkey tumor and Yaba-like disease viruses (Yatapoxvirus) (12, 41); and canarypox and fowlpox viruses (Avipoxvirus) (2, 60). Many poxviruses are presently unclassified, however, suggesting that greater phylogenetic breadth exists within the Chordopoxvirinae (48).
Genomic sequences, together with extensive genetic and reverse genetic studies of model poxviruses, have demonstrated that the chordopoxviral genome is organized into a large, central region containing genes involved in basic replicative mechanisms, including multistage viral transcription, viral genome replication, and virion assembly, and into terminal regions containing genes involved in virus-host interactions (45, 46, 63). Comparative genomic analysis has revealed that while gene content and gene order in the central regions are relatively well conserved among mammalian chordopoxviruses, terminal genomic regions are more variable, with distantly related viruses having greater differences in gene order and content (31, 55).
Natural and experimentally induced poxviral diseases have been reported for members of three subfamilies of cervids, including American deer (Odocoileinae), alces (Alcinae), and reindeer and caribou (Rangiferinae), and include diseases which resemble infections caused by parapoxvirus orf virus (8, 24, 40, 50, 68, 71). Deerpox viruses (DPVs) are poorly characterized viruses responsible for non-orf-like infections and are presently unclassified members of the Chordopoxvirinae. Reports of DPV-like infections in deer include a reindeer herd in the Metropolitan Toronto Zoo (8) and two mule deer (Odocoileus hemionus) a year apart in Bighorn Basin, Wyoming (68). The actual prevalence of infection and significance of DPV as a pathogen remain unknown. Clinical presentation of DPV infection includes keratoconjunctivitis and proliferative-ulcerative skin lesions on the face and feet. In the Wyoming cases, the disease was thought to be a significant factor in the death of the animals (68). Virions resembling vaccinia virus (VACV) were observed by electron microscopy upon examination of skin sections of DPV-infected animals (8, 68). Here we present genome analysis of two DPVs isolated in Wyoming. The data suggest that DPV represents a new genus within the Chordopoxvirinae (48).
MATERIALS AND METHODS
Virus strains, DNA isolation, cloning, sequencing, and sequence analysis.
DPVs W-848-83 (W83) and W-1170-84 (W84) were isolated in Basin, Wyoming, in 1983 and in Burlington, Wyoming, in 1984, respectively, from skin lesions of free-ranging mule deer. Viral genomic DNA was isolated from uncloned stocks as previously described (65) after three passages of W83 in fetal lamb kidney cells and W84 in Vero cells. Random DNA fragments were obtained by incomplete enzymatic digestion with Tsp509I endonuclease (New England Biolabs, Beverly, Mass.), and DNA fragments larger than 1.0 kbp were cloned and used in dideoxy sequencing reactions as previously described (2). Reaction products were analyzed on an ABI PRISM 3700 automated DNA sequencer (Applied Biosystems, Foster City, Calif.). Sequence data were assembled with the Phrap and CAP3 software programs (22, 36), and gaps were closed as described previously (1). Final DNA consensus sequences for W83 and W84 genomes represented on average 8.6- to 9.2-fold redundancy at each base position, with Consed estimated error rates of 0.3 and 0.9 per 10 kbp, respectively (22, 23, 30), and no significant genetic heterogeneity.
Genome DNA composition, structure, repeats, and restriction enzyme patterns were analyzed as previously described (1) by using the GCG version 10 software package (18). Pairwise genomic alignments were done by using WABA (Jim Kent; http://www.cse.ucsc.edu/∼kent/), and multiple genomic and protein alignments were done with DIALIGN (44) and/or CLUSTAL (58) alignment programs. Open reading frames (ORFs) longer than 30 codons were evaluated for coding potential as previously described (2). All ORFs with coding potential and ORFs greater than 60 codons were subjected to homology searches as previously described (1, 2). Based on these criteria, 172 ORFs were annotated as potential genes and numbered from left to right. Phylogenetic comparisons were performed on complete, concatenated datasets of 79 proteins encoded in conserved central core regions homologous to those located between VACV F17L and A24R. Alignment data were also manually edited with SEAVIEW to exclude ambiguously aligned gap and low-complexity regions prior to phylogenetic analysis (26). Phylogenetic analyses on unedited and edited protein alignments were done by using the PHYLO_WIN and TREE-PUZZLE version 5.2 software packages (26, 54).
Nucleotide sequence accession numbers.
The genome sequences of DPVs W83 and W84 have been deposited in GenBank under accession numbers AY689436 and AY689437, respectively.
RESULTS AND DISCUSSION
DPV genome organization.
Genomic sequences of DPV field isolates W83 and W84 were assembled into contiguous sequences of 166,259 and 170,560 bp, respectively, containing approximately 73% A+T. Terminal hairpin loops were not sequenced, but the assembled genome contained the putative telomeric resolution sequences at position 30 for W83 (ATTTATATACCTAAAAAAAAGATAAAAACA) and at position 122 for W84 (ATTTATATACCTTAAAAAAAAGATAAAACA), with the leftmost nucleotide of each assembled genome arbitrarily designated base 1. Like other poxviruses, DPV genomes contain a large, unique coding region (95% nucleotide identity between W83 and W84) bounded by two identical inverted terminal repeat (ITR) regions. Assembled ITRs of W83 and W84 are 5,012 and 7,061 bp, respectively, and contain significant differences in the lengths of tandem repeat regions (1.5 and 3.5 kbp, respectively). W83 contains 13 and 20 copies of a 39- and a 48-bp repeat, respectively, while W84 contains 109 and 2 copies of a 31- and a 48-bp repeat, respectively. All DPV repeats in this region share a 15-bp motif (GGGAAAGGGATAAAA).
W83 and W84 genomes contain 169 and 170 genes, respectively, coding for proteins of 53 to 1,953 amino acids and representing an approximate 96% coding density. The central DPV genomic region contains homologues of conserved poxviral genes involved in basic replicative mechanisms (including viral transcription, RNA modification, and DNA replication), virion structure, and assembly of intracellular mature and extracellular enveloped virions (Table 1) (45). DPV genomes also contain a complement of potential nucleotide metabolism genes similar to those of leporipox, capripox, swinepox, and yatapox viruses, including homologues of genes for thymidine kinase, dUTPase, and the small subunit of ribonucleotide reductase. A gene for the large subunit of ribonucleotide reductase is absent. DPV terminal genomic regions contain genes with functions likely affecting viral virulence, host range, and immune response modulation, many of which are members of gene families or have homologues in other poxviruses (Table 1).
TABLE 1.
Characterization of DPV ORFs
| ORF number | W83 position (length)a | W84 position (length)a | % Identityb | Best matchc
|
Description, putative function, and/or namef | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession no.d | Species and descriptiond | % Identity | LSDVe
|
SWPVe
|
VACVe
|
||||||||
| ORF | % Identityb | ORF | % Identityb | ORF | % Identityb | ||||||||
| DPV001 | 2176-1715 (154) | 4278-3817 | 95 | P18387 | SPPV T3A | 54 | LSDV001 | 53 | SPV001 | 54 | B15R | 35 | |
| DPV002 | 2754-2263 (164) | 4861-4364 (166) | 86 | LSDV002 | 43 | SPV002 | 38 | ||||||
| DPV003 | 4057-2975 (361) | 6176-5085 (364) | 88 | YLDV 149R | 29 | LSDV149 | 27 | SPV145 | 25 | C12L | 26 | Serpin-like protein | |
| DPV004 | 4788-4066 (241) | 6910-6185 (242) | 93 | LSDV003 | 46 | B9R | 42 | ER-localized apoptosis regulator | |||||
| DPV005 | 7351-7043 (103) | X94355 | CPXV C5L | 54 | A53R | 31 | CD30-like protein | ||||||
| DPV006 | 5312-5106 (69) | 7628-7422 | 67 | P22389 | Mus musculus endothelin 2 | 39 | Endothelin precursor | ||||||
| DPV007 | 6429-5365 (355) | 8745-7681 | 91 | LSDV007 | 46 | C10L | 35 | ||||||
| DPV008 | 7511-6492 (340) | 9808-8798 (337) | 79 | SPV003 | 57 | MHC-like TNF binding protein | |||||||
| DPV009 | 8278-7547 (244) | 10575-9844 | 92 | LSDV009 | 26 | SPV007 | 52 | C1L | 26 | ||||
| DPV010 | 9191-8376 (272) | 11488-10670 (273) | 86 | LSDV008 | 35 | SPV008 | 36 | B8R | 35 | Soluble IFN-γ receptor | |||
| DPV011 | 9972-9253 (240) | 12269-11604 (222) | 95 | LSDV009 | 41 | N2L | 30 | ||||||
| DPV012 | 10358-10017 (114) | 12632-12351 (94) | 97 | SPV009 | 40 | ||||||||
| DPV013 | 11331-10294 (346) | 13629-12592 | 94 | YLDV 7L | 50 | LSDV011 | 39 | SPV146 | 29 | CC-chemokine receptor-like protein | |||
| DPV014 | 12013-11387 (209) | 14313-13687 | 93 | LSDV012 | 48 | SPV142 | 28 | C19L | 23 | Ankyrin repeat protein | |||
| DPV015 | 12743-12066 (226) | 15045-14365 (227) | 68 | LSDV006 | 32 | B16R | 23 | IL-1 receptor-like protein | |||||
| DPV016 | 13391-12804 (196) | 15722-15126 (199) | 67 | AF012825 | ECTV EVM008 | 29 | Viral TNFR II-like C-terminal fragment | ||||||
| DPV017 | 14095-13418 (226) | 16432-15749 (228) | 86 | YLDV 9L | 40 | M2L | 29 | ||||||
| DPV018 | 15271-14126 (382) | 17612-16464 (383) | 94 | YLDV 10L | 44 | LSDV149 | 29 | SPV145 | 26 | K2L | 27 | Serpin-like protein | |
| DPV019 | 17220-15292 (643) | 19558-17630 | 93 | YLDV 11L | 52 | LSDV145 | 25 | SPV141 | 24 | B4R | 24 | Ankyrin repeat protein | |
| DPV020 | 17490-17224 (89) | 19828-19562 | 98 | LSDV014 | 56 | K3L | 45 | elF-2α-like PKR inhibitor | |||||
| DPV021 | 17959-17537 (141) | 20299-19880 (140) | 77 | YLDV 14L | 46 | LSDV015 | 40 | SPV011 | 39 | IL-18 binding protein | |||
| DPV022 | 18518-17982 (179) | 20859-20323 | 98 | YMTV 16L | 38 | LSDV017 | 34 | SPV012 | 32 | F1L | 27 | Antiapoptotic membrane protein | |
| DPV023 | 19002-18571 (144) | 21343-20912 | 97 | LSDV018 | 68 | SPV013 | 71 | F2L | 59 | dUTPase | |||
| DPV024 | 19424-19038 (129) | 21766-21380 | 92 | YLDV 18L | 40 | SPV014 | 33 | ||||||
| DPV025 | 21055-19469 (529) | 23397-21811 | 95 | LSDV019 | 40 | SPV015 | 44 | F3L | 23 | Kelch-like protein | |||
| DPV026 | 22092-21130 (321) | 24434-23472 | 97 | LSDV020 | 78 | SPV016 | 76 | F4L | 76 | Ribonucleotide reductase small subunit | |||
| DPV027 | 22407-22123 (95) | 24763-24479 | 86 | YMTV 21L | 28 | LSDV021 | 29 | SPV017 | 31 | ||||
| DPV028 | 22734-22459 (92) | 25090-24815 | 91 | LSDV022 | 60 | SPV018 | 28 | ||||||
| DPV029 | 22939-22745 (65) | LSDV023 | 63 | ||||||||||
| DPV030 | 25381-25139 (81) | SPV019 | 46 | ||||||||||
| DPV031 | 23655-23011 (215) | 26563-25919 | 95 | LSDV024 | 57 | SPV021 | 60 | F9L | 52 | ||||
| DPV032 | 24970-23639 (444) | 27878-26547 | 99 | LSDV025 | 81 | SPV022 | 82 | F10L | 72 | Serine/threonine protein kinase | |||
| DPV033 | 26136-24997 (380) | 29047-27905 (381) | 93 | LSDV026 | 31 | SPV023 | 43 | F11L | 32 | ||||
| DPV034 | 28123-26168 (652) | 31034-29079 | 95 | LSDV027 | 50 | SPV024 | 57 | F12L | 38 | EEV maturation protein | |||
| DPV035 | 29289-28165 (375) | 32201-31077 | 97 | LSDV028 | 75 | SPV025 | 72 | F13L | 55 | Palmitylated virion envelope protein | |||
| DPV036 | 29523-29314 (70) | 32435-32226 | 99 | AF012825 | ECTV EVM037 | 32 | SPV026 | 34 | F14L | 32 | |||
| DPV037 | 30238-29795 (148) | 33150-32707 | 97 | YLDV 29L | 66 | LSDV029 | 64 | SPV027 | 63 | F15L | 64 | ||
| DPV038 | 30964-30305 (220) | 33877-33218 | 94 | LSDV030 | 43 | SPV028 | 49 | F16L | 36 | ||||
| DPV039 | 31030-31353 (108) | 33946-34266 (107) | 97 | YLDV 31R | 72 | LSDV031 | 70 | SPV029 | 70 | F17L | 61 | DNA-binding virion protein | |
| DPV040 | 32765-31356 (470) | 35678-34269 | 99 | LSDV032 | 78 | SPV030 | 76 | E1L | 67 | Poly(A) polymerase large subunit | |||
| DPV041 | 34993-32798 (732) | 37906-35711 | 98 | LSDV033 | 53 | SPV031 | 60 | E2L | 43 | ||||
| DPV042 | 35662-35066 (199) | 38575-37979 | 91 | LSDV034 | 48 | SPV032 | 46 | E3L | 38 | dsRNA binding PKR inhibitor | |||
| DPV043 | 36446-35715 (244) | 39359-38628 | 99 | LSDV036 | 65 | SPV033 | 72 | E4L | 55 | RNA polymerase subunit RP030 | |||
| DPV044 | 36554-37795 (414) | 39467-40702 (412) | 93 | LSDV035 | 40 | E5R | 29 | ||||||
| DPV045 | 37833-39530 (566) | 40740-42437 | 99 | LSDV037 | 79 | SPV034 | 72 | E6R | 62 | ||||
| DPV046 | 39559-40359 (267) | 42466-43266 | 98 | LSDV038 | 83 | SPV035 | 79 | E8R | 69 | ||||
| DPV047 | 43395-40366 (1010) | 46305-43273 (1011) | 99 | MYXV m34L | 76 | LSDV039 | 75 | SPV036 | 78 | E9L | 67 | DNA polymerase | |
| DPV048 | 43433-43717 (95) | 46343-46627 | 94 | LSDV040 | 71 | SPV037 | 81 | E10R | 68 | IMV redox protein | |||
| DPV049 | 47025-46630 (132) | LSDV041 | 52 | E11L | 46 | Virion core protein | |||||||
| DPV050 | 45992-43956 (679) | 49048-47015 (678) | 97 | YLDV 42L | 49 | LSDV042 | 46 | SPV038 | 47 | O1L | 37 | ||
| DPV051 | 47089-46151 (313) | 50148-49210 | 98 | LSDV043 | 76 | SPV039 | 72 | I1L | 71 | DNA-binding virion core protein | |||
| DPV052 | 47321-47079 (81) | 50377-50135 | 93 | LSDV044 | 53 | SPV040 | 54 | I2L | 44 | ||||
| DPV053 | 48140-47325 (272) | 51196-50381 | 99 | LSDV045 | 63 | SPV041 | 70 | I3L | 53 | DNA-binding phosphoprotein | |||
| DPV054 | 48720-48226 (165) | 51774-51283 (164) | 89 | AB005148 | Bos taurus IL-1 receptor antagonist | 53 | IL-1 receptor antagonist | ||||||
| DPV055 | 48993-48760 (78) | 52042-51809 | 99 | YLDV 46L | 78 | LSDV046 | 64 | SPV043 | 61 | I5L | 47 | IMV membrane protein | |
| DPV056 | 50183-49017 (389) | 53226-52066 (387) | 97 | YMTV 47L | 54 | LSDV047 | 56 | SPV044 | 54 | I6L | 51 | ||
| DPV057 | 51474-50179(432) | 54517-53222 | 99 | LSDV048 | 80 | SPV045 | 78 | I7L | 68 | Virion core protein | |||
| DPV058 | 51480-53528 (683) | 54523-56574 (684) | 97 | LSDV049 | 65 | SPV046 | 66 | I8R | 57 | RNA helicase | |||
| DPV059 | 55321-53531 (597) | 58367-56577 | 99 | LSDV050 | 66 | SPV047 | 67 | G1L | 56 | Metalloprotease | |||
| DPV060 | 55647-56309 (221) | 58693-59355 | 97 | LSDV051 | 56 | SPV048 | 53 | G2R | 45 | Transcriptional elongation factor | |||
| DPV061 | 55653-55321 (111) | 58699-58367 | 98 | AF170722 | SFV gp046L | 68 | LSDV052 | 58 | SPV049 | 59 | G3L | 45 | |
| DPV062 | 56656-56282 (125) | 59702-59328 | 100 | LSDV053 | 80 | SPV050 | 65 | G4L | 52 | Glutaredoxin | |||
| DPV063 | 56659-57960 (434) | 59705-61006 | 99 | LSDV054 | 64 | SPV051 | 63 | G5R | 45 | ||||
| DPV064 | 57964-58152 (63) | 61010-61198 | 98 | YLDV 55R | 85 | LSDV055 | 86 | SPV052 | 84 | G5.5R | 33 | RNA polymerase subunit RPO7 | |
| DPV065 | 58155-58658 (168) | 61201-61704 | 99 | LSDV056 | 61 | SPV053 | 63 | G6R | 45 | ||||
| DPV066 | 59806-58682 (375) | 62858-61734 | 98 | LSDV057 | 62 | SPV054 | 62 | G7L | 52 | Virion core protein | |||
| DPV067 | 59836-60615 (260) | 62888-63667 | 99 | LSDV058 | 93 | SPV055 | 92 | G8R | 84 | Late transcription factor VLTF-1 | |||
| DPV068 | 60655-61659 (335) | 63707-64711 | 98 | YLDV 59R | 64 | LSDV059 | 63 | SPV056 | 59 | G9R | 52 | Myristylated protein | |
| DPV069 | 61663-62409 (249) | 64715-65461 | 100 | LSDV060 | 87 | SPV057 | 84 | L1R | 70 | Myristylated IMV envelope protein | |||
| DPV070 | 62457-62741 (95) | 65509-65793 | 100 | LSDV061 | 53 | SPV058 | 56 | L2R | 31 | ||||
| DPV071 | 63719-62727 (331) | 66771-65779 | 99 | LSDV062 | 72 | SPV059 | 68 | L3L | 50 | ||||
| DPV072 | 63744-64499 (252) | 66796-67551 | 100 | LSDV063 | 81 | SPV060 | 80 | L4R | 64 | DNA-binding virion protein VP8 | |||
| DPV073 | 64522-64911 (130) | 67574-67963 | 99 | LSDV064 | 63 | SPV061 | 59 | L5R | 53 | Membrane protein | |||
| DPV074 | 64871-65320 (150) | 67923-68372 | 99 | LSDV065 | 72 | SPV062 | 64 | J1R | 59 | Virion protein | |||
| DPV075 | 65320-65892 (191) | 68372-68944 | 97 | LSDV066 | 67 | SPV063 | 69 | J2R | 67 | Thymidine kinase | |||
| DPV076 | 65871-66551 (227) | 69008-69604 (199) | 96 | LSDV067 | 58 | SPV064 | 47 | C7L | 35 | Host range protein | |||
| DPV077 | 66571-67611 (347) | 69623-70663 | 99 | LSDV068 | 82 | SPV065 | 80 | J3R | 74 | Poly(A) polymerase small subunit | |||
| DPV078 | 67529-68083 (185) | 70581-71135 | 99 | LSDV069 | 79 | SPV066 | 81 | J4R | 69 | RNA polymerase subunit RPO22 | |||
| DPV079 | 68503-68093 (137) | 71555-71145 | 100 | LSDV070 | 73 | SPV067 | 66 | J5L | 65 | ||||
| DPV080 | 68579-72436 (1286) | 71631-75488 | 99 | LSDV071 | 86 | SPV068 | 86 | J6R | 82 | RNA polymerase subunit RPO147 | |||
| DPV081 | 72974-72459 (172) | 76026-75511 | 99 | AF124517 | SPPV H1L | 83 | LSDV072 | 84 | SPV069 | 80 | H1L | 66 | Protein-tyrosine kinase, assembly |
| DPV082 | 72990-73559 (190) | 76042-76611 | 98 | LSDV073 | 74 | SPV070 | 73 | H2R | 65 | ||||
| DPV083 | 74548-73571 (326) | 77600-76623 | 100 | LSDV074 | 61 | SPV071 | 57 | H3L | 39 | IMV envelope protein p35 | |||
| DPV084 | 76948-74552 (799) | 80000-77604 | 99 | LSDV075 | 83 | SPV072 | 82 | H4L | 71 | RNA polymerase-associated RAP94 | |||
| DPV085 | 77116-77691 (192) | 80168-80743 | 99 | MYXV m73R | 55 | LSDV076 | 46 | SPV073 | 49 | H5R | 42 | Late transcription factor VLTF-4 | |
| DPV086 | 77734-78675 (314) | 80786-81727 | 99 | LSDV077 | 73 | SPV074 | 67 | H6R | 66 | DNA topoisomerase | |||
| DPV087 | 78699-79136 (146) | 81751-82188 | 99 | LSDV078 | 62 | SPV075 | 63 | H7R | 42 | ||||
| DPV088 | 79187-81715 (843) | 82239-84767 | 99 | LSDV079 | 73 | SPV076 | 72 | D1R | 66 | mRNA capping enzyme, large subunit | |||
| DPV089 | 82146-82889 (248) | 85198-85941 | 98 | LSDV081 | 45 | SPV078 | 41 | D3R | 36 | Virion protein | |||
| DPV090 | 82147-81680 (156) | 85199-84732 | 99 | LSDV080 | 45 | SPV077 | 45 | D2L | 39 | Virion protein | |||
| DPV091 | 82889-83542 (218) | 85941-86594 | 100 | MYXV m79R | 78 | LSDV082 | 77 | SPV079 | 76 | D4R | 68 | Uracil DNA glycosylase | |
| DPV092 | 83577-85934 (786) | 86629-88986 | 100 | YLDV 83R | 80 | LSDV083 | 78 | SPV080 | 80 | D5R | 69 | NTPase, DNA replication | |
| DPV093 | 85934-87838 (635) | 88986-90890 | 100 | LSDV084 | 89 | SPV081 | 91 | D6R | 82 | Early transcription factor VETFs | |||
| DPV094 | 87872-88363 (164) | 90924-91415 | 99 | LSDV085 | 83 | SPV082 | 80 | D7R | 67 | RNA polymerase subunit RPO18 | |||
| DPV095 | 88411-89043 (211) | 91463-92095 | 99 | LSDV086 | 70 | SPV083 | 65 | D9R | 59 | mutT motif | |||
| DPV096 | 89046-89795 (250) | 92098-92847 | 99 | YLDV 87R | 65 | LSDV087 | 67 | SPV084 | 64 | D10R | 48 | mutT motif | |
| DPV097 | 91724-89820 (635) | 94775-92871 | 100 | LSDV088 | 78 | SPV085 | 76 | D11L | 73 | NPH-I, transcription termination factor | |||
| DPV098 | 92623-91763 (287) | 95674-94814 | 100 | LSDV089 | 78 | SPV086 | 82 | D12L | 77 | mRNA capping enzyme, small subunit | |||
| DPV099 | 94305-92656 (550) | 97356-95707 | 100 | LSDV090 | 80 | SPV087 | 81 | D13L | 74 | Rifampin resistance protein | |||
| DPV100 | 94787-94335 (151) | 97838-97386 | 100 | MYXV m89L | 71 | LSDV091 | 68 | SPV088 | 64 | A1L | 64 | Late transcription factor VLTF-2 | |
| DPV101 | 95494-94823 (224) | 98545-97874 | 100 | AB015885 | YMTV Yb-B9L | 87 | LSDV092 | 88 | SPV089 | 88 | A2L | 86 | Late transcription factor VLTF-3 |
| DPV102 | 95721-95494 (76) | 98772-98545 | 99 | MYXV m91L | 75 | LSDV093 | 71 | SPV090 | 68 | A2.5L | 33 | ||
| DPV103 | 97700-95745 (652) | 100751-98796 | 100 | LSDV094 | 76 | SPV091 | 81 | A3L | 66 | Virion core protein P4b | |||
| DPV104 | 98213-97761 (151) | 101264-100812 | 99 | LSDV095 | 47 | SPV092 | 43 | A4L | 28 | Virion core protein, morphogenesis | |||
| DPV105 | 98253-98762 (170) | 101304-101810 (169) | 98 | LSDV096 | 68 | SPV093 | 63 | A5R | 64 | RNA polymerase subunit RPO19 | |||
| DPV106 | 99892-98771 (374) | 102940-101819 | 100 | LSDV097 | 77 | SPV094 | 76 | A6L | 56 | ||||
| DPV107 | 102066-99922 (715) | 105114-102970 | 99 | LSDV098 | 81 | SPV095 | 81 | A7L | 71 | Early transcription factor VETF | |||
| DPV108 | 102126-103007 (294) | 105174-106055 | 99 | MYXV m97R | 72 | LSDV099 | 70 | SPV096 | 70 | A8R | 63 | Intermediate transcription factor VITF-3 | |
| DPV109 | 103263-103021 (81) | 106311-106069 | 99 | LSDV100 | 79 | SPV097 | 82 | A9L | 71 | IMV membrane protein | |||
| DPV110 | 106011-103267 (915) | 109059-106315 | 99 | LSDV101 | 71 | SPV098 | 76 | A10L | 52 | Virion core protein P4a | |||
| DPV111 | 106026-106976 (317) | 109074-110024 | 99 | YLDV 102R | 78 | LSDV102 | 77 | SPV099 | 76 | A11R | 54 | ||
| DPV112 | 107555-106986 (190) | 110603-110034 | 97 | LSDV103 | 61 | SPV100 | 57 | A12L | 49 | Virion core protein | |||
| DPV113 | 107840-107622 (73) | 110887-110669 | 97 | LSDV104 | 63 | SPV101 | 53 | A13L | 36 | IMV membrane protein | |||
| DPV114 | 108203-107928 (92) | 111246-110971 | 100 | YLDV 105L | 86 | LSDV105 | 78 | SPV102 | 85 | A14L | 54 | IMV membrane protein | |
| DPV115 | 108381-108223 (53) | 111424-111266 | 100 | YLDV 106L | 84 | LSDV106 | 74 | SPV103 | 77 | A14.5L | 55 | Virulence factor | |
| DPV116 | 108655-108374 (94) | 111698-111417 | 100 | AB015885 | YMTV Yb-B23L | 54 | LSDV107 | 54 | SPV104 | 52 | A15L | 49 | |
| DPV117 | 109781-108642 (380) | 112827-111685 (381) | 98 | LSDV108 | 68 | SPV105 | 66 | A16L | 51 | Myristylated membrane protein | |||
| DPV118 | 110402-109812 (197) | 113449-112859 | 98 | YLDV 109L | 75 | LSDV109 | 66 | SPV106 | 73 | A17L | 41 | TMV membrane protein | |
| DPV119 | 110417-111862 (482) | 113464-114909 | 98 | LSDV110 | 59 | SPV107 | 64 | A18R | 54 | DNA helicase, elongation | |||
| DPV120 | 112073-111849 (75) | 115120-114896 | 100 | YLDV 111L | 76 | LSDV111 | 73 | SPV108 | 79 | A19L | 71 | ||
| DPV121 | 112420-113703 (428) | 115467-116750 | 99 | LSDV112 | 55 | SPV109 | 55 | A20R | 44 | DNA polymerase processivity factor | |||
| DPV122 | 112421-112077 (115) | 115468-115124 | 100 | LSDV113 | 64 | SPV110 | 68 | A21L | 59 | ||||
| DPV123 | 113687-114229 (181) | 116734-117276 | 100 | LSDV114 | 67 | SPV111 | 72 | A22R | 72 | Holliday junction resolvase | |||
| DPV124 | 114216-115394 (393) | 117263-118441 | 99 | LSDV115 | 65 | SPV112 | 64 | A23R | 62 | Intermediate transcription factor VITF-3 | |||
| DPV125 | 115423-118887 (1155) | 118470-121934 | 99 | LSDV116 | 91 | SPV113 | 89 | A24R | 83 | RNA polymerase subunit RPO132 | |||
| DPV126 | 119300-118890 (137) | 122344-121937 (136) | 95 | LSDV117 | 47 | SPV114 | 55 | A27L | 30 | Fusion protein | |||
| DPV127 | 119723-119304 (140) | 122767-122348 | 98 | AF170722 | SFV gp116L | 70 | LSDV118 | 69 | SPV115 | 71 | A28L | 59 | IMV protein |
| DPV128 | 120641-119742 (300) | 123685-122786 | 100 | LSDV119 | 69 | SPV116 | 68 | A29L | 61 | RNA polymerase subunit RPO35 | |||
| DPV129 | 120837-120613 (75) | 123881-123657 | 99 | AB018404 | YMTV Yb-D13L | 72 | LSDV120 | 72 | SPV117 | 67 | A30L | 57 | IMV, membrane |
| DPV130 | 121027-121476 (150) | 124069-124527 (153) | 95 | AF438165 | CMLV 150 | 47 | A31R | 53 | |||||
| DPV131 | 122253-121492 (254) | 125304-124543 | 100 | MYXV m120L | 88 | LSDV121 | 88 | SPV118 | 82 | A32L | 59 | DNA packaging, virus assembly | |
| DPV132 | 122383-122958 (192) | 125433-126008 | 96 | YLDV 122R | 48 | LSDV122 | 37 | SPV119 | 46 | A33R | 28 | EEV glycoprotein | |
| DPV133 | 122982-123485 (168) | 126032-126535 | 100 | LSDV123 | 56 | SPV120 | 69 | A34R | 51 | EEV protein | |||
| DPV134 | 123533-124075 (181) | 126583-127125 | 99 | LSDV124 | 43 | SPV121 | 45 | A35R | 37 | ||||
| DPV135 | 124111-124971 (287) | 127157-128014 (286) | 98 | LSDV125 | 45 | SPV122 | 48 | ||||||
| DPV136 | 125031-125675 (215) | 128074-128730 (219) | 93 | LSDV126 | 33 | SPV123 | 46 | A36R | 24 | EEV glycoprotein | |||
| DPV137 | 125741-126562 (274) | 128796-129617 | 99 | LSDV127 | 46 | SPV124 | 49 | A37R | 32 | ||||
| DPV138 | 127452-127883 (144) | 130507-130938 | 97 | YLDV 129R | 50 | Hypothetical protein | |||||||
| DPV139 | 127480-126584 (299) | 130535-129639 | 98 | MYXV m128L | 46 | LSDV128 | 40 | SPV125 | 44 | A38L | 26 | CD47-like protein | |
| DPV140 | 127918-128220 (101) | 130973-131317 (115) | 85 | LSDV129 | 30 | SPV126 | 23 | ||||||
| DPV141 | 128291-128536 (82) | 131384-131629 | 99 | LSDV130 | 53 | SPV127 | 47 | ||||||
| DPV142 | 129596-128550 (349) | 132691-131645 | 98 | SPV128 | 56 | A44L | 46 | Beta-hydroxysteroid dehydrogenase | |||||
| DPV143 | 129652-130143 (164) | 132747-133238 | 99 | LSDV131 | 64 | SPV129 | 62 | A45R | 36 | Superoxide dismutase-like protein | |||
| DPV144 | 131047-130400 (216) | 134133-133486 | 97 | AF320596 | Mus musculus C lectin-related protein | 52 | A40R | 27 | C-type lectin-like protein | ||||
| DPV145 | 131243-132928 (562) | 134328-136013 | 99 | LSDV133 | 64 | SPV130 | 67 | A50R | 53 | DNA ligase-like protein | |||
| DPV146 | 133038-138896 (1953) | 136122-141971 (1950) | 96 | LSDV134 | 53 | SPV131 | 52 | Variola virus B22R-like protein | |||||
| DPV147a | 138920-139969 (350) | 142021-142308 (96) | 81 | LSDV135 | 32 | SPV132 | 36 | B19R | 32 | IFN-α/β binding protein (fragment) | |||
| DPV147b | 142418-143050 (211) | 89 | YLDV 136R | 29 | LSDV135 | 30 | SPV132 | 31 | B19R | 34 | IFN-α/β binding protein fragment | ||
| DPV148 | 140001-140564 (188) | 143082-143645 | 96 | 45 | LSDV136 | 40 | SPV133 | 46 | K7R | 23 | |||
| DPV149 | 140620-141645 (342) | 143702-144727 | 99 | 47 | LSDV137 | 47 | SPV134 | 47 | A51R | 31 | |||
| DPV150 | 142576-141671 (302) | 145647-144748 (300) | 95 | AF030894 | MYXV α2,3-sialyltransferase | 44 | α2,3-sialyltransferase | ||||||
| DPV151 | 143540-142584 (319) | 146611-145655 | 99 | AJ010865 | Bos taurus MHC class I antigen | 27 | MHC class I-like protein | ||||||
| DPV152 | 143630-144205 (192) | 146701-147276 | 98 | MYXV m139R | 53 | SPV135 | 54 | A52R | 34 | IL-1R/TLR signaling inhibitor | |||
| DPV153 | 144262-144822 (187) | 147333-147893 | 96 | LSDV138 | 42 | A56R | 26 | Ig domain OX-2-like protein | |||||
| DPV154 | 144859-145803 (315) | 147930-148874 | 98 | LSDV139 | 66 | SPV137 | 63 | B1R | 49 | Serine/threonine protein kinase | |||
| DPV155 | 145827-146561 (245) | 148898-149632 | 99 | LSDV140 | 51 | SPV138 | 43 | N1R-like RING finger host range protein | |||||
| DPV156 | 146642-147526 (295) | 149713-150597 | 98 | LSDV141 | 41 | SPV139 | 53 | C3L | 36 | EEV host range protein | |||
| DPV157 | 147558-147971 (138) | 150629-151042 | 96 | LSDV142 | 39 | N1L | 42 | Virulence factor | |||||
| DPV158 | 148004-148933 (310) | 151075-152004 | 95 | LSDV143 | 53 | SPV140 | 54 | Tyrosine protein kinase-like protein | |||||
| DPV159 | 148966-149436 (157) | 152037-152507 | 99 | LSDV150 | 50 | A52R | 22 | ||||||
| DPV160 | 149486-151123 (546) | 152557-154194 | 96 | LSDV151 | 51 | SPV136 | 30 | A55R | 30 | Kelch-like protein | |||
| DPV161 | 151190-153112 (641) | 154261-156183 | 95 | LSDV145 | 46 | SPV141 | 50 | C9L | 23 | Ankyrin repeat protein | |||
| DPV162 | 153187-154434 (416) | 156252-157457 (402) | 68 | LSDV011 | 36 | SPV146 | 46 | CC-chemokine receptor-like protein | |||||
| DPV163 | 154548-155477 (310) | AF191297 | Cavia porcellus TGF-β | 28 | TGF-β1 | ||||||||
| DPV164 | 155544-157046 (501) | 157707-159209 | 93 | LSDV147 | 44 | SPV142 | 46 | B4R | 21 | Ankyrin repeat protein | |||
| DPV165 | 157088-158536 (483) | 159251-160699 | 94 | LSDV148 | 40 | SPV143 | 39 | C9L | 24 | Ankyrin repeat protein | |||
| DPV166 | 158557-160062 (502) | 160752-162230 (493) | 96 | LSDV152 | 39 | SPV144 | 36 | B4R | 26 | Ankyrin repeat protein | |||
| DPV167 | 160101-161105 (335) | 162261-163268 (336) | 92 | YLDV 149R | 48 | LSDV149 | 47 | SPV145 | 42 | C12L | 35 | Serpin-like protein | |
| DPV168 | 161118-161408 (97) | 163305-163586 (94) | 91 | LSDV153 | 45 | SPV147 | 52 | ||||||
| DPV169 | 161472-162194 (241) | 163651-164376 (242) | 93 | LSDV154 | 46 | B9R | 42 | ER-localized apoptosis regulator | |||||
| DPV170 | 162203-163285 (361) | 164385-165476 (364) | 88 | YLDV 149R | 29 | LSDV149 | 27 | SPV145 | 25 | C12L | 26 | Serpin-like protein | |
| DPV171 | 163506-163997 (164) | 165700-166197 (166) | 86 | LSDV155 | 43 | SPV149 | 38 | ||||||
| DPV172 | 164084-164545 (154) | 166283-166744 | 95 | P18387 | SPPV T3A | 54 | LSDV156 | 53 | SPV150 | 54 | B15R | 35 | |
Lengths of ORFs are in codons. W84 ORF lengths are presented only if differing from that of W83.
Percent amino acid identity was obtained by FASTA analysis.
Best scoring matches in BLAST analysis.
Accession numbers, species, and descriptions indicated are those different from lumpy skin disease virus (LSDV) and swinepox virus (SWPV). Other abbreviations are as follows: CPXV, cowpox virus; ECTV, ectromelia virus; MYXV, myxoma virus; SFV, rabbit (Shope) fibroma virus; SPPV, sheeppox virus; YLDV, Yaba-like disease virus; YMTV, yaba monkey tumor virus. GenBank database accession numbers are as follows: MYXV, AF170726; SFV, AF170722; and YLDV, AJ293568.
Best-matching ORFs from LSDV (accession no. AF325528), SWPV (accession no. AF410153), and VACV strain Copenhagen (accession no. M35027 and AF516337) genomes. Highlighted ORFs indicate best overall match to W84 in similarity searches.
Function was deduced from the degree of similarity to known genes and Prosite signatures. Abbreviations are as follows: IMV, intracellular mature virion; EEV, extracellular enveloped virion; eIF-2α, α subunit of eukaryotic initiation factor 2; dsRNA, double-stranded RNA.
Putative DPV virulence and host range proteins include those similar to secreted cytokine receptors (R) or binding proteins (BP), including gamma interferon receptor (IFN-γR; DPV010), interleukin-1 receptor (IL-1R; DPV015), IFN-α/βΒP (DPV147), IL-18BP (DPV021), major histocompatibility complex class I (MHC-I)-like tumor necrosis factor binding protein (TNFBP; DPV008), and two TNFR-like proteins (DPV016 and DPV005). DPV016 resembles a carboxyl-terminal fragment of viral TNFR-II, proteins present in several poxviral genera, and DPV005 resembles cellular CD30, a homologue of which has been found in orthopoxviruses cowpox virus, ectromelia virus, monkeypox virus, and variola virus (Table 1). Potential membrane-bound DPV immunomodulators include ORFs similar to cellular type 8 CC-chemokine receptor (DPV013 and DPV162), CD47 (DPV139), and OX-2 (DPV153). DPV proteins that are likely to inhibit intracellular signaling involved in immunological responses and/or apoptosis include homologues of VACV E3L and K3L (DPV042 and DPV020, respectively), myxoma virus M004 and M011R (DPV004 or DPV169 and DPV022, respectively), and serpins (DPV003, DPV018, DPV167, and DPV170). Notably, serpins DPV003 and DPV170, located in the ITR, are the least similar to known poxviral serpins but do contain the Asp P1 residue similar to poxvirus serpins known to affect inflammation, apoptosis, and virulence through inhibition of caspases 1 and 8 and granzyme B (57). DPV152 and DPV157 share similarity with VACV A52R and VACV N1L, respectively, proteins which affect intracellular signaling through IL-1R/Toll-like receptors and/or TNF superfamily receptors to affect viral virulence (10, 19, 33, 38).
DPV encodes six proteins containing ankyrin repeat motifs, two kelch-like proteins, and a protein similar to rabbit fibroma virus N1R (DPV155), proteins with homologues affecting poxviral virulence, host range, immunopathology, and/or apoptosis (Table 1) (11, 27, 37, 51). Other ORFs potentially affecting DPV-host interaction include homologues of poxvirus β-hydroxysteroid dehydrogenase (DPV142), superoxide dismutase (DPV143), α2,3-sialyltransferase (DPV150), and Tyr protein kinase-like protein (DPV158). Although many of these terminally located genes have similarity to those found in other poxviruses, this unique complement likely underlies DPV mechanisms of virulence and host range.
Notable host range and immunomodulatory genes.
DPVs contain several genes which are either completely novel within the Poxviridae or represent unique forms of cellular-like genes present in other poxviruses. Notably, some of these genes represent insertions in regions otherwise syntenic with other poxviruses (Table 1). These genes, likely involved in viral pathogenesis, encode proteins similar to cellular endothelin, IL-1R antagonist (IL-1Ra), transforming growth factor β1 (TGF-β1), C-type lectin-like receptors, and MHC-I.
DPV006 resembles endothelins (ETs), three potent vasoactive 21-amino-acid peptides (ET 1 to ET 3) with important roles in vascular homeostasis, and the structurally related snake venom sarafotoxins (Fig. 1A) (Table 1). ETs are synthesized as large precursors from which 40- to 90-amino-acid amino-terminal and 110- to 120-amino-acid carboxyl-terminal domains are sequentially removed by endopeptidases and endothelin-converting enzymes to yield biologically active ET peptides (49).
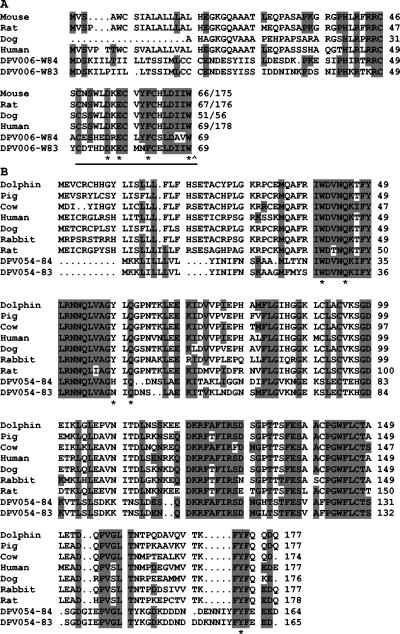
FIG. 1.
Multiple amino acid alignment of DPV006 with endothelins and DPV054 with secreted IL-1Ra (isoform 1). Amino acid positions are indicated on the right; / indicates truncation of the amino acid sequence, * indicates residues critical for receptor binding, and ^ indicates cleavage sites. (A) Alignment of DPV006 to endothelin homologues. ET peptide is underlined. Accession numbers are the following: P22389, mouse; P23943, rat; P12064, dog; and P20800, human. (B) Alignment of DPV054 to IL-1Ra. Accession numbers are the following: AB038268, dolphin; L38849, pig; AB005148, cow; P18510, human; AY026462, dog; P26890, rabbit; and P25086, rat.
DPV006 encodes an ET precursor-like protein including an amino-terminal signal peptide and a highly conserved Arg/Lys-Arg-Cys tripeptide endopeptidase cleavage site (positions 47 to 49) (Fig. 1A). The lack of a carboxyl-terminal domain in DPV006 suggests that endothelin-converting enzyme-mediated cleavage is not required for activation (52). Although W83 and W84 ET-like peptides are only 52% identical, both peptides contain two predicted disulfide bonds and conserved residues which are important for ET 1 and 2 receptor binding and biological activity (Fig. 1A) (49). Upstream nucleotide sequences resembling early poxviral promoters suggest that DPV006 is expressed as an early gene.
ETs are produced primarily by endothelial cells, but also by epithelial cells and neurons, and exert their actions in a paracrine-autocrine fashion by interacting with G protein-coupled receptors expressed in vascular smooth muscle cells, endothelial cells, and, to a lesser extent, other cell types (29). Mammalian ETs have been implicated in a number of airway, pulmonary vascular, and cardiovascular disorders and in chronic and acute inflammatory diseases (5, 29, 34). ET 1 binding to smooth muscle cell receptors leads to vasoconstriction, cytokine production, cell growth, and inflammatory cell recruitment, while binding to endothelial receptors has been associated with nitric oxide release and prevention of apoptosis (5, 34). DPV ETs may have similar functions in the host, conceivably contributing to the marked proliferative and necrotizing character of DPV-induced lesions (68). Alternatively, DPV006 may function as an ET antagonist, interfering with normal host ET functions. DPV006 represents a second poxviral gene with similarity to host genes primarily associated with vascular physiology and, like parapoxvirus vascular endothelial growth factor, may have a significant role in virus virulence (53).
DPV054 is similar to cellular IL-1Ra, an IL-1-like molecule which acts as a competitive inhibitor of IL-1 and antagonizes IL-1R signaling (Table 1) (Fig. 1B). DPV054 in W83 and W84 are 89% identical and contain a predicted amino-terminal signal peptide, indicating that DPV054, similar to mammalian secreted IL-1Ra isoforms, is secreted. Although overall identity between DPV and mammalian IL-1Ra is 41 to 53%, a region between residues 27 and 48 of DPV054 is 76 to 90% identical to mammalian IL-1Ra and contains 3 of 5 residues involved in the binding of IL-1Ra to IL-1R. A fourth residue involved in binding is also conserved in DPV054 (Tyr159) (21).
The balance between IL-1 and IL-1Ra is known to influence the course of many inflammatory and viral diseases (6). For instance, elevated IL-1Ra levels relative to IL-1β levels in human immunodeficiency virus-infected patients may reflect direct stimulation of monocyte IL-1Ra production by human immunodeficiency virus (39). Correlation of increased IL-1Ra levels during rhinovirus infection with peak symptomatology and onset of clinical resolution has led to the suggestion that IL-1Ra may play a role in the resolution of this respiratory infection (70). Poxviruses inhibit proinflammatory IL-1β activity, often through multiple strategies, as evidenced in DPV, which encodes homologues of viral serpins, IL-1R, and an intracellular IL-1R/Toll-like receptor inhibitor, which affect IL-1 maturation or signaling (Table 1) (46). To our knowledge, DPV054 encodes the first viral protein with similarity to IL-1Ra, thus adding an additional poxviral strategy to block host IL-1β-mediated responses.
DPV163, present only in W83, is similar to TGF-β1 (Table 1). Although multiple copies of distantly related TGF-β homologues are present in avian poxviruses, this is the first observation of a TGF-β1-like gene in a mammalian chordopoxvirus (2). DPV163 encodes a 310-amino-acid protein that contains most of the TGF-β1 propeptide region and the TGF-β1 chain, including a TGF-β1 prosite motif and all 10 Cys residues necessary for disulfide bridge formation. As with avian poxviral TGF homologues, DPV163 is most similar to cellular TGF-β1 in the TGF-β1 chain region (50% amino acid identity between DPV163 residues 214 to 310).
DPV163 lacks features associated with the amino-terminal propeptide of eukaryotic TGF-β1, including 36 amino acids containing the predicted signal peptide, an Arg-Gly-Asp cell attachment site, and the Arg-His-Arg-Arg cleavage site (DPV163 amino acids 210 to 214) necessary for removal of the propeptide and subsequent activation of TGF-β1. Notably, DPV163 contains an Ile-Asn-Met-Pro motif (DPV163 amino acids 262 to 265) instead of the Trp-Ser-Leu-Asp motif important for the interaction of mammalian TGF-β1 with its receptor, for growth inhibition of epithelial cells, and for growth stimulation of fibroblasts (35). Divergence in the propeptide region, lack of the cleavage site needed for release of the mature peptide, and substitutions at significant sites suggest that processing or specificities of DPV163 may be distinct from cellular TGFs.
TGF-β1 suppresses multiple immune functions, including polyclonal antibody production, cytotoxic T lymphocytes, natural killer (NK) and lymphokine-activated killer cell activity, macrophage activation, and IL-1R expression (20). At the site of injury, TGF-β induces production of inflammatory cytokines IL-1, TNF, and IL-6 (20). TGF-β also affects cell growth, stimulating connective tissue cell growth and differentiation during neovascularization and wound healing while suppressing proliferation in most other cell types, including T and B lymphocytes, monocytes, and macrophages (7, 9, 15, 20, 47). DPV163 may affect similar host responses.
DPV144 encodes a protein with similarity to members of a glycoprotein gene superfamily which exhibit a C-type animal lectin domain (Table 1). DPV144 in W83 and W84 are 97% identical and are most similar to proteins encoded by the NK gene complex (NKC) and related cell receptors (40 to 60% amino acid identity). Similar to NKC proteins, DPV144 is a predicted type II integral membrane protein, containing four conserved Trp residues and two of the three Cys pairs believed to form intrachain disulfide bonds within the lectin-like domain (69). DPV144 also resembles viral lectin-like proteins encoded by rat cytomegalovirus (45% amino acid identity), fowlpox virus (FPV239; 36% amino acid identity), and VACV (A40R; 27% amino acid identity). These rat cytomegalovirus and VACV proteins are not essential for virus growth in vitro (64, 66), and disruption of A40R attenuates VACV strain WR following intradermal but not intranasal inoculation of mice (59, 64). Although poxviral C-type lectin-like proteins share sequence similarity to NK cell receptors, evidence for a role of these proteins in NK cell activation or modulation is lacking.
DPV151 is most similar (27% identity over 187 amino acids) to cellular HLA class I histocompatibility antigen α chain precursors, containing putative extracellular α1, α2, and α3 domains, connecting peptide, transmembrane domains, and four Cys residues necessary for disulfide bond formation (Table 1). DPV151 lacks amino-terminal signal peptide and carboxyl-terminal cytoplasmic domains homologous to cellular MHC-I, and the α 1 domain is not well conserved (data not shown). DPV151 is less similar to the MHC-I homologue from molluscum contagiosum virus (16% identity over 201 amino acids to MC080R) and to homologues of the MHC-I-like TNFBP of Tanapox virus and its homologues in DPV (DPV008), Yaba-like disease virus, and swinepox virus (21% identity over 254 amino acids to SPV003) (13). Notably, an MHC-I homologue encoded by murine cytomegalovirus (m144 gene) functions to protect against NK-mediated clearance of virus-infected cells (25). A similar function has not been demonstrated for poxviral MHC-I, but it is tempting to speculate that DPV151 could have a role in interfering with NK-mediated antiviral immunity.
Comparison of DPVs and other ChPV genera.
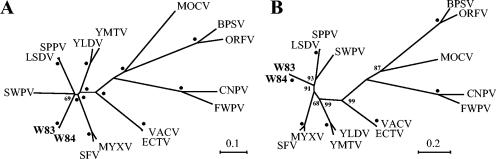
DPVs are most similar to viruses of the capripoxvirus, suipoxvirus, leporipoxvirus, and yatapoxvirus (CSLY) genera, grouping with these viruses by phylogenetic analysis (Fig. 2). In addition, DPV and CSLY share distinctive genomic features, such as the insertion of the VACV C7L homologue (DPV076) between homologues of VACV J2R and J3R, the absence of A-type inclusion protein genes (VACV A25L/A26L), and more extensive gene colinearity (Table 1 and Fig. 2). Phylogenetic analysis also suggests that DPVs, capripoxviruses, and swinepox virus are monophyletic (Fig. 2). However, data indicate that DPV is a group as distinct as other ChPV genera are from each other (Fig. 2). Maximum likelihood analysis of whole genome sequences reveals distance estimates between DPV and other CSLY genera (0.654 to 0.754) on the same order of magnitude as those between established CSLY genera (0.505 to 0.725). Other genomic features distinguish DPV from other CSLY viruses, including the presence of DPV-specific genes and a homologue of VACV A31R, a gene otherwise present only in orthopoxviruses and avipoxviruses. Taken together, these data indicate that DPV represents a new poxvirus genus.
FIG. 2.
Phylogenetic analysis of DPV proteins. Seventy-nine conserved ORFs between DPV039 and DPV125 were concatenated from W83 and W84 and aligned with similarly concatenated ORF sets from other ChPVs with DIALIGN. Unrooted trees were generated by neighbor-joining analysis with Poisson correction for multiple substitutions and 500 bootstrap replicates as implemented in PHYLO_WIN (A) and maximum likelihood analysis with JTT correction for multiple substitutions and 1,000 quartet puzzling steps as implemented in TREE-PUZZLE (B). Bootstrap (A) or support (B) values of 100% are marked with dots; values less than 100% are presented at appropriate nodes. Homologous protein sequences from the following viruses and accession numbers were compared: bovine popular stomatitis virus (BPSV), AY386265; canarypox virus (CNPV), AY318871; ectromelia virus (ECTV), AF012825; fowlpox virus (FWPV), AF198100; lumpy skin disease virus (LSDV), AF325528; molluscum contagiosum virus (MOCV), MCU60315; myxoma virus (MYXV), AF170726; orf virus (ORFV), AY386264; rabbit (Shope) fibroma virus (SFV), AF170722; sheeppox virus (SPPV), AY077833; swinepox virus (SWPV), AF410153; vaccinia virus (VACV), M35027; Yaba-like disease virus (YLDV), AJ293568; and Yaba monkey tumor virus (YMTV), AY386371. Similar results were obtained by using an alignment manually edited to include only unambiguously aligned sites (20,132 of 30,019 sites) and using alignments generated with CLUSTAL W (data not shown).
Despite the high degree of similarity between W83 and W84 genomes relative to other ChPV genera (Table 1 and Fig. 2), significant differences between these DPVs exist. While centrally located ORFs (DPV020 to DPV160) are the most conserved between DPVs (97% average amino acid identity), terminally located ORFs are less similar (88% average amino acid identity [Table 1]). Whole genome maximum likelihood distances between W83 and W84 (0.042) are less than distances between both sequenced viruses of the genus leporipoxvirus (0.166) but greater than distances between eight sequenced viruses of the genus capripoxvirus (0.023 to 0.034). Although W83 and W84 have similar gene orders and contents, in W84 two genes are absent (DPV030 and DPV163) and one gene is fragmented into two ORFs (DPV147a and DPV147b) by an in-frame stop, and in W83 three genes are absent (DPV005, DPV031, and DPV051). With the exception of DPV147, genomic indels of 165 to 860 bp are responsible for differences in gene content between W83 and W84. These include CD30-like, TGF-β-like, and IFN-α/βBP genes, which conceivably could impart virus-specific host range and virulence functions to each DPV. These genomic differences suggest that W83 and W84 are distinct viruses within the genus.
Conclusions.
Genome sequences of W83 and W84 provide the first view of DPV genomics. A unique complement of DPV virulence and host range genes predicts novel mechanisms underlying virus-cervid host interactions in infection and immunity. Genomic analysis indicates that DPV represents a new genus within the Chordopoxvirinae.
Acknowledgments
We thank A. Lakowitz and C. Balinsky for providing excellent technical assistance.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, E. Oma, G. F. Kutish, and D. L. Rock. 1999. The genome of Melanoplus sanguinipes entomopoxvirus. J. Virol. 73:533-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2000. The genome of fowlpox virus. J. Virol. 74:3815-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, F. A. Osorio, C. Balinsky, G. F. Kutish, and D. L. Rock. 2002. The genome of swinepox virus. J. Virol. 76:783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, N. T. Sandybaev, U. Z. Kerembekova, V. L. Zaitsev, G. F. Kutish, and D. L. Rock. 2002. The genome of camelpox virus. Virology 295:1-9. [DOI] [PubMed] [Google Scholar]
- 5.Alonso, D. M. W., and M. W. Radomski. 2003. The nitric oxide-endothelin-1 connection. Heart Fail. Rev. 8:107-115. [DOI] [PubMed] [Google Scholar]
- 6.Arend, W. P. 2002. The balance between IL-1 and IL-1Ra in disease. Cytokine Growth Factor Rev. 13:323-340. [DOI] [PubMed] [Google Scholar]
- 7.Ashcroft, G. S. 1999. Bidirectional regulation of macrophage function by TGF-β. Microbes Infect. 1:1275-1282. [DOI] [PubMed] [Google Scholar]
- 8.Barker, I. K., K. G. Mehren, W. A. Rapley, and A. N. Cagnon. 1980. Keratoconjunctivitis and oral cutaneous lesions associated with poxvirus infection in reindeer, p. 171-177. In R. J. Montali and G. Migaki (ed.), The comparative pathology of zoo animals: proceedings of a symposium held at the National Zoological Park, Smithsonian Institution. Smithsonian Institution Press, Washington, D.C.
- 9.Blobe, G. C., W. P. Schiemann, and H. F. Lodish. 2000. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 342:1350-1358. [DOI] [PubMed] [Google Scholar]
- 10.Bowie, A., E. Kiss-Toth, J. A. Symons, G. L. Smith, S. K. Dower, and L. A. O'Neill. 2000. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc. Natl. Acad. Sci. USA 97:10162-10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brick, D. J., R. D. Burke, L. Schiff, and C. Upton. 1998. Shope fibroma virus RING finger protein N1R binds DNA and inhibits apoptosis. Virology 249:42-51. [DOI] [PubMed] [Google Scholar]
- 12.Brunetti, C. R., H. Amano, Y. Ueda, J. Qin, T. Miyamura, T. Suzuki, X. Li, J. W. Barrett, and G. McFadden. 2003. Complete genomic sequence and comparative analysis of the tumorigenic poxvirus Yaba monkey tumor virus. J. Virol. 77:13335-13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunetti, C. R., M. Paulose-Murphy, R. Sing, J. Qin, J. W. Barrett, A. Tardivel, P. Schneider, K. Essani, and G. McFadden. 2003. A secreted high-affinity inhibitor of human TNF from Tanapox virus. Proc. Natl. Acad. Sci. USA 100:4831-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cameron, C., S. Hota-Mitchell, L. Chen, J. Barrett, J. X. Cao, C. Macaulay, D. Willer, D. Evans, and G. McFadden. 1999. The complete DNA sequence of myxoma virus. Virology 264:298-318. [DOI] [PubMed] [Google Scholar]
- 15.Cerwenka, A., and S. L. Swain. 1999. TGF-β1: immunosuppressant and viability factor for T lymphocytes. Microbes Infect. 1:1291-1296. [DOI] [PubMed] [Google Scholar]
- 16.Chen, N., M. I. Danila, Z. Feng, R. M. Buller, C. Wang, X. Han, E. J. Lefkowitz, and C. Upton. 2003. The genomic sequence of ectromelia virus, the causative agent of mousepox. Virology 317:165-186. [DOI] [PubMed] [Google Scholar]
- 17.Delhon, G., E. R. Tulman, C. L. Afonso, Z. Lu, A. de la Concha-Bermejillo, H. D. Lehmkuhl, M. E. Piccone, G. F. Kutish, and D. L. Rock. 2004. Genomes of the parapoxviruses ORF virus and bovine papular stomatitis virus. J. Virol. 78:168-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiPerna, G., J. Stack, A. G. Bowie, A. Boyd, G. Kotwal, Z. Zhang, S. Arvikar, E. Latz, K. A. Fitzgerald, and W. L. Marshall. 2004. Poxvirus protein N1L targets the I-kappaB kinase complex, inhibits signaling to NF-kappaB by the tumor necrosis factor superfamily of receptors, and inhibits NF-kappaB and IRF3 signaling by toll-like receptors. J. Biol. Chem. 279:36570-36578. [DOI] [PubMed] [Google Scholar]
- 20.Durum, S. K., and J. J. Oppenheim. 1993. Proinflammatory cytokines and immunity, p. 801-835. In W. E. Paul (ed.), Fundamental immunology, 3rd ed. Raven Press, Ltd., New York, N.Y.
- 21.Evans, R. J., J. Bray, J. D. Childs, G. P. A. Vigers, B. J. Brandhuber, J. J. Skalicky, R. C. Thompson, and S. P. Eisenberg. 1995. Mapping receptor binding sites in interleukin (IL)-1 receptor antagonist and IL-1B by site-directed mutagenesis. J. Biol. Chem. 270:11477-11483. [DOI] [PubMed] [Google Scholar]
- 22.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 23.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 24.Falk, E. S. 1978. Parapoxvirus infections of reindeer and musk ox associated with unusual human infections. Br. J. Dermatol. 99:647-654. [DOI] [PubMed] [Google Scholar]
- 25.Farrell, H. E., N. J. Davis-Poynter, D. M. Andrews, and M. A. Degli-Esposti. 2002. Function of CMV-encoded MHC class I homologues. Curr. Top. Microbiol. Immunol. 269:131-151. [DOI] [PubMed] [Google Scholar]
- 26.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-554. [DOI] [PubMed] [Google Scholar]
- 27.Gillard, S., D. Spehner, R. Drillien, and A. Kirn. 1986. Localization and sequence of a vaccinia virus gene required for multiplication in human cells. Proc. Natl. Acad. Sci. USA 83:5573-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247-266. [DOI] [PubMed] [Google Scholar]
- 29.Goraca, A. 2002. New views on the role of endothelin. Endocr. Regul. 36:161-167. [PubMed] [Google Scholar]
- 30.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:192-202. [DOI] [PubMed] [Google Scholar]
- 31.Gubser, C., S. Hue, P. Kellam, and G. L. Smith. 2004. Poxvirus genomes: a phylogenetic analysis. J. Gen. Virol. 85:105-117. [DOI] [PubMed] [Google Scholar]
- 32.Gubser, C., and G. L. Smith. 2002. The sequence of camelpox virus shows it is most closely related to variola virus, the cause of smallpox. J. Gen. Virol. 83:855-872. [DOI] [PubMed] [Google Scholar]
- 33.Harte, M. T., I. R. Haga, G. Maloney, P. Gray, P. C. Reading, N. W. Bartlett, G. L. Smith, A. Bowie, and L. A. O'Neill. 2003. The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J. Exp. Med. 197:343-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hocher, B., A. Schwarz, K. A. Fagan, C. Thone-Reineke, K. El-Hag, H. Kusserow, S. Elitok, C. Bauer, H. H. Neumayer, D. M. Rodman, and F. Theuring. 2000. Pulmonary fibrosis and chronic lung inflammation in ET-1 transgenic mice. Am. J. Respir. Cell Mol. Biol. 23:19-26. [DOI] [PubMed] [Google Scholar]
- 35.Huang, S. S., M. Zhou, F. E. Johnson, H. S. Shieh, and J. S. Huang. 1999. An active site of transforming growth factor-β(1) for growth inhibition and stimulation. J. Biol. Chem. 274:27754-27758. [DOI] [PubMed] [Google Scholar]
- 36.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ink, B. S., C. S. Gilbert, and G. I. Evan. 1995. Delay of vaccinia virus-induced apoptosis in nonpermissive Chinese hamster ovary cells by the cowpox virus CHOhr and adenovirus E1B 19K genes. J. Virol. 69:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotwal, G. J., A. W. Hugin, and B. Moss. 1989. Mapping and insertional mutagenesis of a vaccinia virus gene encoding a 13,800-Da secreted protein. Virology 171:579-587. [DOI] [PubMed] [Google Scholar]
- 39.Kreuzer, K. A., J. M. Dayer, J. K. Rockstroh, T. Sauerbruch, and U. Spengler. 1997. The IL-1 system in HIV infection: peripheral concentrations of IL-1β, IL-1 receptor antagonist and soluble IL-1 receptor type II. Clin. Exp. Immunol. 109:54-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kummeneje, K. 1979. Contagious ecthyma (orf) in reindeer (Rangifer tarandus). Vet. Rec. 105:60-61. [DOI] [PubMed] [Google Scholar]
- 41.Lee, H. J., K. Essani, G. L. Smith, F. Jeanmougin, and D. G. Higgins. 2001. The genome sequence of Yaba-like disease virus, a yatapoxvirus. Virology 281:170-192. [DOI] [PubMed] [Google Scholar]
- 42.Massung, R. F., L.-I. Liu, J. Qi, J. C. Knight, T. E. Yuran, A. R. Kerlavage, J. M. Parsons, J. C. Venter, and J. J. Esposito. 1994. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology 201:215-240. [DOI] [PubMed] [Google Scholar]
- 43.McLysaght, A., P. F. Baldi, and B. S. Gaut. 2003. Extensive gene gain associated with adaptive evolution of poxviruses. Proc. Natl. Acad. Sci. USA 100:15655-15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgenstern, B., K. Frech, A. Dress, and T. Werner. 1998. DIALIGN: finding local similarities by multiple sequence alignment. Bioinformatics 14:290-294. [DOI] [PubMed] [Google Scholar]
- 45.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 46.Moss, B., and J. L. Shisler. 2001. Immunology 101 at poxvirus U: immune evasion genes. Semin. Immunol. 13:59-66. [DOI] [PubMed] [Google Scholar]
- 47.Moustakas, A., K. Pardali, A. Gaal, and C. H. Heldin. 2002. Mechanisms of TGF-β signaling in regulation of cell growth and differentiation. Immunol. Lett. 82:85-91. [DOI] [PubMed] [Google Scholar]
- 48.Moyer, R. W., B. Arif, D. N. Black, D. B. Boyle, R. M. Buller, K. R. Dumbell, J. J. Esposito, G. McFadden, B. Moss, A. Mercer, S. Ropp, D. N. Tripathy, and C. Upton. 2000. Family Poxviridae, p. 137-147. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Academic Press, New York, N.Y.
- 49.Nakajima, K., S. Kubo, S. Kumagaye, H. Nishio, M. Tsunemi, T. Inui, H. Kuroda, N. Chino, T. X. Watanabe, T. Kimura, and S. Sakakibara. 1989. Structure-activity relationship of endothelin: importance of charged groups. Biochem. Biophys. Res. Commun. 163:424-429. [DOI] [PubMed] [Google Scholar]
- 50.Patton, J. F., R. W. Nordhausen, L. W. Woods, and N. J. MacLachlan. 1996. Isolation of a poxvirus from a black-tailed deer (Odocoileus hemionus columbianus). J. Wildl. Dis. 32:531-533. [DOI] [PubMed] [Google Scholar]
- 51.Pires de Miranda, M., P. C. Reading, D. C. Tscharke, B. J. Murphy, and G. L. Smith. 2003. The vaccinia virus kelch-like protein C2L affects calcium-independent adhesion to the extracellular matrix and inflammation in a murine intradermal model. J. Gen. Virol. 84:2459-2471. [DOI] [PubMed] [Google Scholar]
- 52.Rubanyi, G. M., and L. H. Parker-Botelho. 1991. Endothelins. FASEB J. 5:2713-2720. [DOI] [PubMed] [Google Scholar]
- 53.Savory, L. J., S. A. Stacker, S. B. Fleming, B. E. Niven, and A. A. Mercer. 2000. Viral vascular endothelial growth factor plays a critical role in orf virus infection. J. Virol. 74:10699-10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 55.Senkevich, T. G., E. V. Koonin, J. J. Bugert, G. Darai, and B. Moss. 1997. The genome of Molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology 233:19-42. [DOI] [PubMed] [Google Scholar]
- 56.Shchelkunov, S. N., A. V. Totmenin, P. F. Safronov, M. V. Mikheev, V. V. Gutorov, O. I. Ryazankina, N. A. Petrov, I. V. Babkin, E. A. Uvarova, L. S. Sandakhchiev, J. R. Sisler, J. J. Esposito, I. K. Damon, P. B. Jahrling, and B. Moss. 2002. Analysis of the monkeypox virus genome. Virology 297:172-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shisler, J. L., and B. Moss. 2001. Immunology 102 at poxvirus U: avoiding apoptosis. Semin. Immunol. 13:67-72. [DOI] [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tscharke, D. C., P. C. Reading, and G. L. Smith. 2002. Dermal infection with vaccinia virus reveals roles for virus proteins not seen using other inoculation routes. J. Gen. Virol. 83:1977-1986. [DOI] [PubMed] [Google Scholar]
- 60.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2004. The genome of canarypox virus. J. Virol. 78:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2001. Genome of lumpy skin disease virus. J. Virol. 75:7122-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, J. H. Sur, N. T. Sandybaev, U. Z. Kerembekova, V. L. Zaitsev, G. F. Kutish, and D. L. Rock. 2002. The genomes of sheeppox and goatpox viruses. J. Virol. 76:6054-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turner, P. C., and R. W. Moyer. 2002. Poxvirus immune modulators: functional insights from animal models. Virus Res. 88:35-53. [DOI] [PubMed] [Google Scholar]
- 64.Voigt, S., G. R. Sandford, L. Ding, and W. H. Burns. 2001. Identification and characterization of a spliced C-type lectin-like gene encoded by rat cytomegalovirus. J. Virol. 75:603-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wesley, R. D., and A. E. Tuthill. 1984. Genome relatedness among African swine fever virus field isolates by restriction endonuclease analysis. Prev. Vet. Med. 2:53-62. [Google Scholar]
- 66.Wilcock, D., S. A. Duncan, P. Traktman, W.-H. Zhang, and G. L. Smith. 1999. The vaccinia virus A40R gene product is a nonstructural, type II membrane glycoprotein that is expressed at the cell surface. J. Gen. Virol. 80:2137-2148. [DOI] [PubMed] [Google Scholar]
- 67.Willer, D. O., G. McFadden, and D. H. Evans. 1999. The complete genome sequence of shope (rabbit) fibroma virus. Virology 264:319-343. [DOI] [PubMed] [Google Scholar]
- 68.Williams, E. S., V. M. Becerra, E. T. Thorne, T. J. Graham, M. J. Owens, and C. E. Nunamaker. 1985. Spontaneous poxviral dermatitis and keratoconjunctivitis in free-ranging mule deer (Odocoileus hemionus) in Wyoming. J. Wildl. Dis. 21:430-433. [DOI] [PubMed] [Google Scholar]
- 69.Yokoyama, W. M., and B. F. M. Plougastel. 2003. Immune functions encoded by the natural killer gene complex. Nat. Rev. Immunol. 3:304-316. [DOI] [PubMed] [Google Scholar]
- 70.Yoon, H. J., Z. Zhu, J. M. Gwaltney, Jr., and J. A. Elias. 1999. Rhinovirus regulation of IL-1 receptor antagonist in vivo and in vitro: a potential mechanism of symptom resolution. J. Immunol. 162:7461-7469. [PubMed] [Google Scholar]
- 71.Zarnke, R. L., R. A. Dieterich, K. A. Neiland, and G. Ranglack. 1983. Serologic and experimental investigations of contagious ecthyma in Alaska. J. Wildl. Dis. 19:170-174. [DOI] [PubMed] [Google Scholar]