There is an intramolecular interaction in the lipid kinase Fab1 in which the upstream CCR domain contacts the Fab1 kinase region. Selected dominant-active alleles disrupt this interaction and result in elevated PI(3,5)P2. These findings suggest a regulatory mechanism that contributes to dynamic control of cellular PI(3,5)P2 synthesis.
Abstract
Phosphorylated phosphoinositide lipids (PPIs) are low-abundance signaling molecules that control signal transduction pathways and are necessary for cellular homeostasis. The PPI phosphatidylinositol (3,5)-bisphosphate (PI(3,5)P2) is essential in multiple organ systems. PI(3,5)P2 is generated from PI3P by the conserved lipid kinase Fab1/PIKfyve. Defects in the dynamic regulation of PI(3,5)P2 are linked to human diseases. However, few mechanisms that regulate PI(3,5)P2 have been identified. Here we report an intramolecular interaction between the yeast Fab1 kinase region and an upstream conserved cysteine-rich (CCR) domain. We identify mutations in the kinase domain that lead to elevated levels of PI(3,5)P2 and impair the interaction between the kinase and CCR domain. We also identify mutations in the CCR domain that lead to elevated levels of PI(3,5)P2. Together these findings reveal a regulatory mechanism that involves the CCR domain of Fab1 and contributes to dynamic control of cellular PI(3,5)P2 synthesis.
INTRODUCTION
Phosphatidylinositol (3,5)-bisphosphate (PI(3,5)P2) is a low-abundance signaling lipid (Dove et al., 1997; Whiteford et al., 1997), which constitutes ∼0.05–0.1% of total phosphoinositide lipids (Bonangelino et al., 2002; Duex et al., 2006a). That levels of this lipid are dynamically regulated suggests a role in adaptation to external stimuli and a role in the maintenance of cellular homeostasis (Dove et al., 1997; Sbrissa et al., 1999; Duex et al., 2006a; Bridges et al., 2012; Zolov et al., 2012).
PI(3,5)P2 is generated from phosphatidylinositol 3-phosphate (PI3P) by the conserved lipid kinase Fab1 (PIKfyve in mammals; Gary et al., 1998; Zolov et al., 2012). Fab1/PIKfyve is regulated via a protein complex with at least three proteins in mammalian cells—PIKfyve, Vac14, and Fig4—and five proteins in Saccharomyces cerevisiae—Fab1, Vac14, Fig4, Vac7, and Atg18 (Botelho et al., 2008; Jin et al., 2008). Little is known about how these proteins regulate Fab1.
Yeast Fab1 and mammalian PIKfyve are approximately fourfold longer than other phosphatidylinositol 5-kinases (Rao et al., 1998). The Fab1 kinase domain lies at the C-terminus and comprises less than one-fifth of the protein (Figure 1A). The N-terminal FYVE domain (Figure 1A) binds PI3P (Burd and Emr, 1998; Sbrissa et al., 1999), but the functions of additional conserved regions remain largely unknown. Fab1 residues 819–1550 encompass the conserved cysteine-rich (CCR) and chaperone-containing TCP1 (CCT) domain (Figure 1A). Based on sequence similarity between this region and the GroEL chaperonin, the CCR and CCT domains were proposed to associate with regulatory proteins (Efe et al., 2005; Michell et al., 2006; Botelho et al., 2008). The CCT domain is essential for the interaction of yeast Fab1 with the critical scaffold protein Vac14 (Botelho et al., 2008; Jin et al., 2008). Specific point mutations in the CCR domain were identified that result in greatly reduced PI(3,5)P2 levels (Botelho et al., 2008). However, the mechanisms that impair PI(3,5)P2 production in these CCR mutants are unknown.
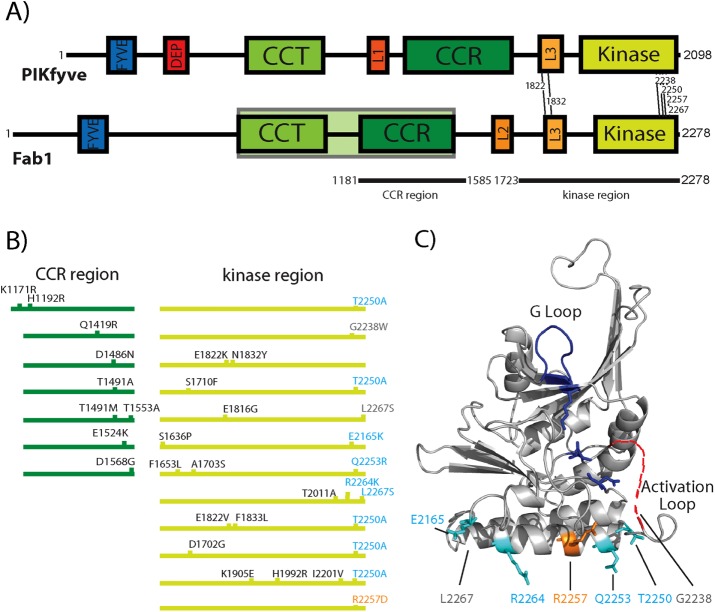
FIGURE 1:
Mutations in the kinase and CCR domains result in dominant-active Fab1 alleles. (A) Domain architecture of Fab1/PIKfyve. Amino acid residues of each domain. FYVE: Fab1 230–320, PIKfyve 143–234; DEP: not present in yeast Fab1, PIKfyve 368–438; CCT: Fab1 766–1039, PIKfyve 608–877; CCR: Fab1 1181–1500, PIKfyve 1157–1524; Kinase: Fab1 1900–2266, PIKfyve 1759–2084; L1: Not present in yeast Fab1, PIKfyve 1037–1090; L2: Fab1 1649–1700, Not present in PIKfyve; L3 Fab1 1821–1905, PIKfyve 1618–1693. The GroEL homology region corresponding to residues 819–1550 in yeast is shaded in green. Dominant-active mutations conserved between yeast Fab1 and mammalian PIKfyve are indicated with lines between the two schematics. The CCR and kinase regions (black lines below schematic) correspond to Y2H constructs used in this study. (B) Dominant-active alleles (CCR domain, dark green; kinase region, light green) with amino acid substitutions indicated. (C) Mapping putative causative dominant-active mutations onto a predicted structure of the kinase domain suggests a surface critical for the regulation of Fab1. Structure prediction, using Phyre2 (Kelley et al., 2015), based on the kinase domain of phosphatidylinositol phosphate kinase IIβ (Rao et al., 1998). Most of the previously identified dominant-active mutations (cyan) map to a surface of a predicted α-helix proximal to the activation loop. Surface residue R2257 is predicted to be part of the regulation of kinase activity and tested in Figure 2, A and B (orange). Residues G2238 and L2267 (gray) are outside of the predicted structure; approximate positions are indicated.
Here we provide insight into a function of the CCR domain. We report that the CCR domain contacts a C-terminal portion of Fab1, including the kinase domain, and that this interaction regulates Fab1 PI3 5-kinase activity. Point mutations in both the kinase and CCR domains result in elevation of PI(3,5)P2 levels. Furthermore, dominant-active mutations in the kinase domain impair a physical interaction between the kinase and CCR domains.
RESULTS AND DISCUSSION
Dominant-active mutations in the Fab1 kinase domain reveal a potential regulatory region
Several dominant-active Fab1 alleles with multiple point mutations were previously identified; however, in most cases, the causative point mutation was not determined (Duex et al., 2006b). Several of the mutations are at residues conserved between yeast and humans (Figure 1A). Ten of 11 alleles harbor at least one mutation at the C-terminal end of the kinase domain (Figure 1B, light green; Duex et al., 2006b). Intriguingly, the C-terminal mutations cluster on a single surface of a homology-based model of the kinase domain. Within this surface, three of the residues mutated in dominant-active alleles–T2250, Q2253, and R2264–reside on a predicted, solvent-exposed α-helix (Figure 1C).
On the basis of this model, we generated and characterized seven single–point mutation candidates. Six were mutations present in the original dominant-active alleles, whereas R2257D also resides on the putative solvent-exposed helix. In most cases, fragmented vacuoles correlate with elevated PI(3,5)P2 levels, whereas mutations that negatively affect PI(3,5)P2 levels lead to enlarged vacuoles (Duex et al., 2006b). Each of the seven mutants exhibited fragmented vacuoles indicative of elevated levels of PI(3,5)P2 (Figure 2A and Supplemental Figure S1A). These results support the model that this predicted surface of the kinase domain functions in the regulation of Fab1.
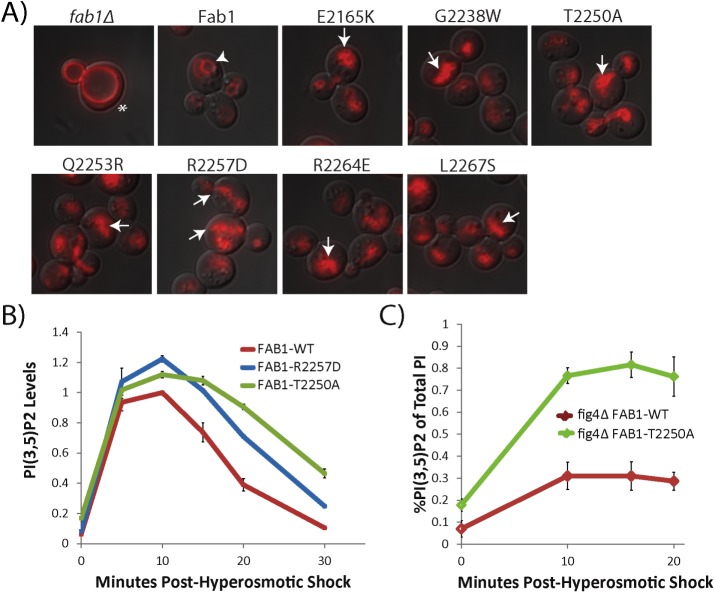
FIGURE 2:
Mutations in the C-terminal kinase domain result in dominant-active Fab1. (A) Single point mutations show a fragmented vacuole phenotype (arrows) indicative of elevated PI(3,5)P2 levels compared with WT Fab1 (arrowhead). The enlarged vacuole in fab1Δ (asterisk) is characteristic of reduced PI(3,5)P2 levels. Vacuoles labeled with FM4-64 (red). (B) During hyperosmotic shock, Fab1R2257D and Fab1T2250A produce more PI(3,5)P2, and the levels of this lipid decline more slowly than in wild-type yeast. Levels normalized to WT level at 10 min postshock within the same experiment. Statistical significance of observed differences between wild-type and mutant alleles over the shock time course was analyzed by two-way repeated-measures analysis of variance (ANOVA): p = 0.0013 for Fab1R2257D and p = 0.0001 for Fab1T2250A. (C) Expression of Fab1T2250A in a fig4Δ background results in higher PI(3,5)P2 levels than expression of Fab1WT under basal conditions, as well as during hyperosmotic shock. Statistical significance of observed difference between wild-type and Fab1T2250A over the shock time course was analyzed by two-way repeated-measures ANOVA: p = 0.0002. Error bars represent SD. Three independent experiments.
We directly measured PI(3,5)P2 levels of two alleles–Fab1-R2257D and Fab1-T2250A. In yeast, PI(3,5)P2 levels transiently change upon introduction into hyperosmotic media (Dove et al., 1997). Within 5 min, PI(3,5)P2 levels increase >15-fold, plateau for 10 min, and then rapidly return to basal levels (Duex et al., 2006a). Under basal conditions, both Fab1-T2250A and Fab1-R2257D display elevated PI(3,5)P2 levels compared with wild type (WT; Figure 2B). In addition, in response to a hyperosmotic stimulus, both mutants elevate PI(3,5)P2 levels and continue to display elevated levels at the 30-min time point compared with WT (Figure 2B). This indicates that the C-terminal portion of the kinase domain acts to control both basal levels of PI(3,5)P2 and production of PI(3,5)P2 in response to an extracellular stimulus.
The dominant-active mutation Fab1-T2250A elevates PI(3,5)P2 levels independently of the PI(3,5)P2 phosphatase Fig4
Fab1 exists in a complex with its opposing phosphatase, Fig4 (Botelho et al., 2008; Jin et al., 2008). Fig4 catalyzes PI(3,5)P2 turnover (Rudge et al., 2004) and only functions within the Fab1-Vac14-Fig4 complex (Duex et al., 2006b; Jin et al., 2008). Thus a Fab1 mutation that alters association of Fig4 with the Fab1-Vac14 complex could contribute to elevation of PI(3,5)P2 via a defect in turnover of PI(3,5)P2 rather than an increase in intrinsic Fab1 kinase activity. To test this possibility, we analyzed PI(3,5)P2 levels in cells expressing either Fab1-WT or Fab1-T2250A in a fig4Δ mutant. Under both basal and hyperosmotic shock conditions, Fab1-T2250A exhibits higher levels of PI(3,5)P2 relative to Fab1-WT even in the absence of Fig4 (Figure 2C). Thus Fab1-T2250A displays higher levels of PI(3,5)P2 relative to Fab1-WT independently of Fig4 function.
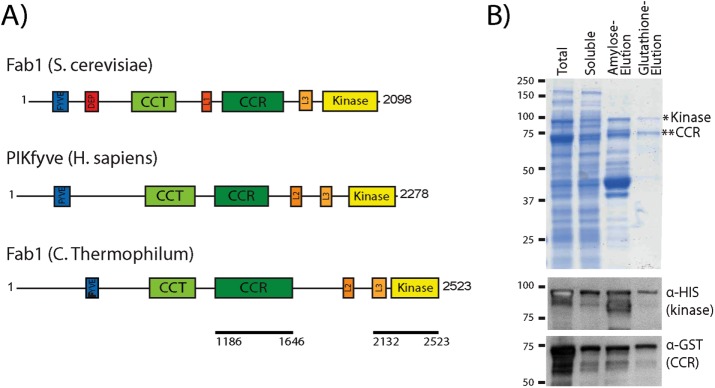
FIGURE 4:
C. thermophilum Fab1 kinase region interacts with the CCR domain in vitro. (A) Fab1 from C. thermophilum has similar domain architecture to S. cerevisiae Fab1 and human PIKfyve. Black lines indicate polypeptides used for in vitro studies. (B) Affinity copurification of a recombinant complex of C. thermophilum GST-CCR and 6xHis-MBP-kinase domains of Fab1. Domains were coexpressed in E. coli, and lysates were applied to amylose resin and eluted with maltose. Maltose eluates were applied to glutathione resin and eluted with reduced glutathione. Western blot analysis indicates that the Coomassie band at 89.5 kDa is the His-MBP-kinase region (Fab1 amino acids 2132–2523), and the Coomassie band at 75 kDa is the GST-CCR domain (Fab1 amino acids 1186–1646). Three independent experiments.
The dominant-active mutations in the kinase region impair an interaction with the Fab1 CCR domain
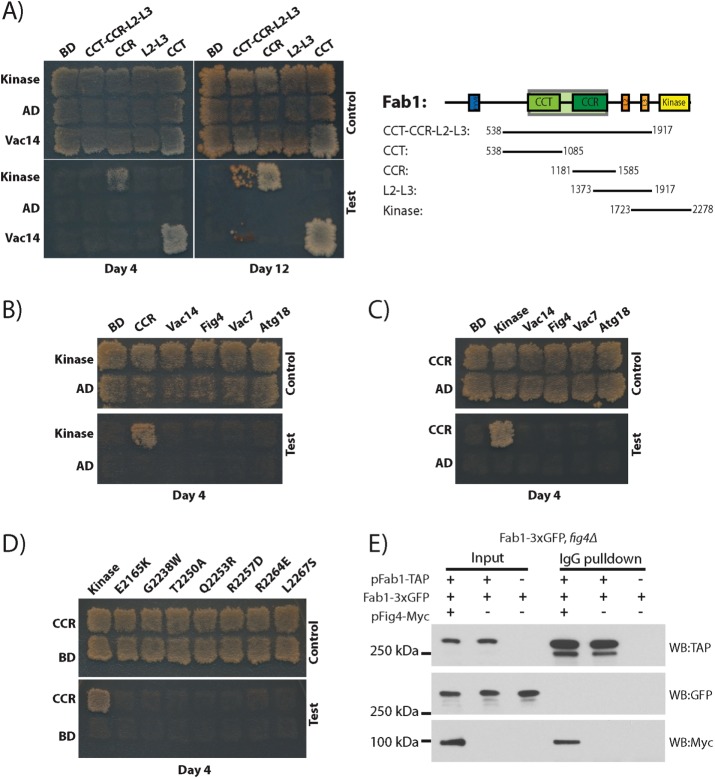
The clustering of dominant-active mutations to a conserved surface region of the Fab1 kinase domain led us to hypothesize that these mutations disrupt a protein–protein interaction that modulates kinase activity. As an initial test, we assayed for intramolecular interactions between the kinase domain and other regions of Fab1 using the yeast two-hybrid (Y2H) test (Figure 3A). The kinase region (residues 1723–2278; Kinase, Figure 1A) interacts with residues 1181–1585, which includes the CCR domain. There is a weaker interaction of the kinase region with a much larger Y2H construct spanning residues 538–1917 (CCT-CCR-L2-L3; Figure 1A), which also includes the CCR domain. In contrast, the CCT domain (residues 538–1085) and the L2 and L3 domains (residues 1373–1917) do not interact with the kinase region. Furthermore, neither the kinase domain nor the CCR domain interacts with other members of the Fab1 complex: Vac14, Fig4, Vac7, or Atg18 (Figure 3, B and C). These data suggest that the CCR domain and the kinase region interact with each other but not with other known regulators of Fab1. Strikingly, the interaction of the kinase domain with the CCR domain is impaired by every dominant-active kinase domain mutation tested (Figure 3D), suggesting that the CCR domain inhibits Fab1 activity.
FIGURE 3:
The Fab1 kinase region associates with the CCR region via an intramolecular interaction, and dominant-active mutations in the kinase region impair this interaction. For Y2H assay, transformants were patched onto SC-LEU-TRP and replica plated onto SC-LEU-TRP (control) or SC-LEU-TRP-ADE-HIS (test). CCR domain and kinase region constructs indicated in Figure 1A. (A) The Fab1 kinase region interacts with the Fab1-CCR domain. Gal4-AD fused with either the kinase region or empty vector tested against Gal4-BD fused with the indicated Fab1 fragments. (B) The kinase region interacts with the Fab1 CCR domain but not with other proteins in the Fab1 complex. Gal4–activation domain (AD) fused with either the kinase region or empty vector tested against Gal4–binding domain (BD) fused with the Fab1 CCR domain, Vac14, Fig4, Vac7 (amino acids 394–918), or Atg18. (C) The Fab1 CCR domain interacts with the Fab1-kinase region but not with other proteins in the Fab1 complex. Gal4-AD fused with the CCR region or empty vector tested with Gal4-BD fused with the kinase region, Vac14, Fig4, Vac7 (amino acids 394–918), or Atg18. (D) Dominant-active kinase domain mutants ablate the interaction with the Fab1-CCR domain. Gal4-BD fused with the Fab1 CCR region with Gal4-AD fused with either the kinase region of Fab1 or this region with one of the mutations E2165K, G2238W, T2250A, Q2253R, R2257D, R2264E, or L2267S. (A–D) All Y2H experiments were performed a minimum of six times, and the same results were obtained. (E) There is likely an intramolecular interaction between the kinase and CCR domains of Fab1 because Fab1 does not coimmunoprecipitate with itself in vivo. Coexpression of Fab1-TAP, Fab1-3xGFP, and Fig4-4xMyc in S. cerevisiae. Precipitation of Fab1-TAP with IgG pulls down a known Fab1 complex protein, Fig4-4XMyc, but not Fab1-3xGFP.
The interaction of the CCR domain with the kinase region could be intramolecular or intermolecular. To test for the ability of Fab1 to associate with other Fab1 molecules, we expressed Fab1-TAP in yeast harboring a Fab1–3x green fluorescent protein (GFP) genomic integrant. As a positive control for Fab1 complex assembly, Fig4-Myc was simultaneously expressed. As expected, Fab1-TAP pulls down Fig4-Myc, indicating that the Fab1 complex is intact. However, Fab1-TAP does not coimmunoprecipitate Fab1-3xGFP (Figure 3E), in agreement with previous reports that only one Fab1 molecule is present in each Fab1 complex (Botelho et al., 2008; Alghamdi et al., 2013). Thus the interaction between the Fab1 kinase region and CCR domain is likely intramolecular.
To assess further the interaction between the CCR domain and kinase region, we expressed the CCR domain and kinase regions in Escherichia coli. Owing to the instability of multiple S. cerevisiae Fab1 constructs tested, we turned to the thermophilic fungus Chaetomium thermophilum. Recombinant proteins from this organism are often more stable (Amlacher et al., 2011; Baker et al., 2015), and C. thermophilum Fab1 has a similar domain architecture to both S. cerevisiae Fab1 and human PIKfyve (Figure 4A). Of importance, the CCR domain and kinase region of Fab1, including residues that are mutated in S. cerevisiae dominant-active Fab1 alleles, are conserved in C. thermophilum (Supplemental Figure S2). We generated histidine (His) maltose-binding protein (MBP)–kinase and glutathione S-transferase (GST)–CCR fusion proteins (Figure 4A). Expression of either construct alone in E. coli yielded no soluble protein (unpublished data); however, as determined by SDS–PAGE and Western blot analysis, coexpression yielded soluble polypeptides for both the kinase region and CCR domain (Figure 4B). The recombinant polypeptides bound to each other, allowing copurification through two sequential affinity columns: amylose resin followed by glutathione resin, which bind alternatively to His-MBP-kinase and GST-CCR, respectively (Figure 4B). This provides in vitro evidence for a direct interaction between these domains.
That the kinase and CCR domains interact directly and that individual point mutations in the kinase region both enhance Fab1 kinase activity and impair the Y2H interaction between these domains suggest that the CCR domain inhibits Fab1 kinase activity through direct contact with the kinase region. Of note, another phosphoinositide kinase is hypothesized to be regulated via a protein–protein contact on the kinase domain (Budovskaya et al., 2002; Rostislavleva et al., 2015). The PI3-kinase p110α exhibits an intramolecular interaction between the adaptor-binding domain and the kinase domain (Huang et al., 2007, 2008). Oncogenic mutations in either region of p110α lead to elevated kinase activity (Zhao and Vogt, 2008) and were suggested to dissociate this interaction (Zhao and Vogt, 2008). Here we characterize a regulatory intramolecular interaction in Fab1. That this mode of regulation is seen in at least two lipid kinases makes it tempting to speculate that other phosphatidylinositol kinases and perhaps many protein kinases have similar types of regulation.
Dominant-active mutations in the CCR region of Fab1 indicate a regulatory role for this domain
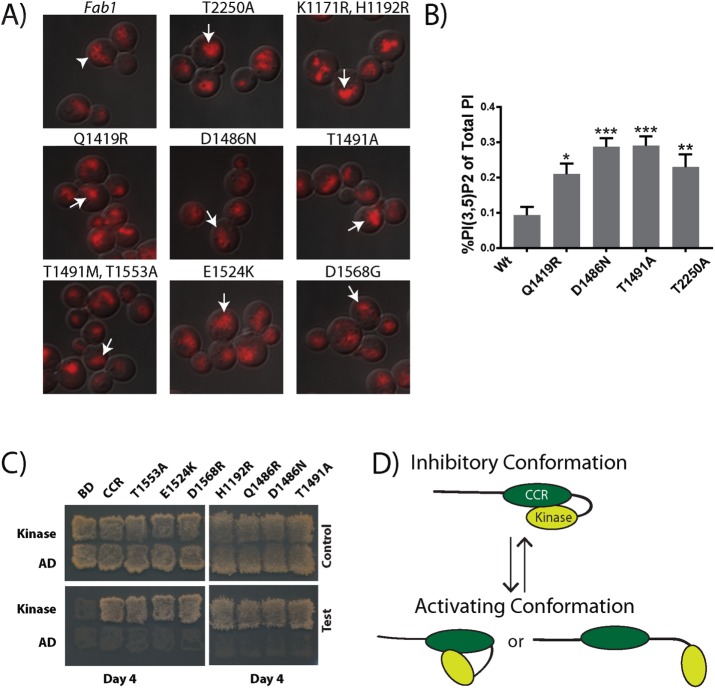
A model in which the CCR domain inhibits Fab1 kinase activity predicts that hyperactivation of Fab1 can be achieved through mutation of the CCR domain. To test this prediction, we performed random mutagenesis of the CCR domain and screened for colonies that bypass deletion of the Fab1 activator Vac7 (Supplemental Figure S3A). We isolated and sequenced 21 mutants representing seven unique alleles (Figure 1B, dark green). These mutants displayed fragmented vacuoles when expressed in WT or fab1Δ cells, indicative of elevated PI(3,5)P2 levels (Figure 5A and Supplemental Figure S3B). We directly measured PI(3,5)P2 levels for 3 of the CCR mutants (Fab1-Q1419R, Fab1-D1486N, and Fab1-T1491A) and found that each displayed a twofold to threefold elevation of PI(3,5)P2 compared with wild type (Figure 5B). The identification of hyperactive mutations in the CCR domain is consistent with an inhibitory role for this domain in regulating Fab1 kinase activity.
FIGURE 5:
Dominant active mutations in the CCR region of Fab1 indicate a regulatory role for this domain. (A) Selected CCR point mutants exhibit a fragmented vacuole phenotype (arrows) relative to the wild-type Fab1 control (arrowhead), suggesting an elevation in PI(3,5)P2 levels. Vacuoles labeled with FM4-64 (red). Three independent experiments. (B) Under basal conditions, each of the three CCR domain mutants tested (Fab1Q1419R, Fab1D1486N, and Fab1T1491A) displays elevated levels in PI(3,5)P2 relative to Fab1WT, similar to the kinase domain mutation Fab1T2250A. Statistical significance of observed differences between wild-type and the mutant alleles was analyzed by one-way ANOVA, p < 0.0001, with a Dunnett's multiple-comparison post hoc test: *p < 0.01, **p < 0.001, ***p < 0.0001. Error bars represent SD. Three independent experiments. (C) Dominant-active mutations identified in the CCR region do not block the Y2H interaction of the CCR region with the kinase region. Y2H tests of Gal4-AD fused with the kinase region (residues 1723–2278) with Gal4-BD fused with either the CCR region (residues 1181–1585) or this region with one of the mutations H1192R, Q1419R, D1486N, T1491A, T1491M, E1524K, T1553A, or D1568R. Six independent experiments. (D) Proposed regulatory intramolecular interaction within Fab1. The CCR domain interacts with the kinase region of Fab1 and reversibly inhibits its activity. Potential CCR inhibition of Fab1 kinase activity may be relieved by posttranslational modifications and/or altered interactions with other proteins.
We further tested whether, as with dominant-active mutations in the kinase region, these mutations in the CCR domain disrupted the Y2H interaction of the CCR domain with the kinase region; none of these mutants did (Figure 5C). It is therefore unclear whether these CCR domain mutations enhance Fab1 kinase activity through the same mechanism as the dominant-active mutations identified in the kinase region. If the mechanisms are related, release of an inhibitory interaction between the CCR domain and kinase region may involve loss of key contacts rather than complete disruption of this interaction (Figure 5D). Alternatively, the enhanced kinase activity of the CCR mutants may be due to an independent mechanism, for instance, through enhanced association with additional positive regulators of Fab1.
These studies reveal that the Fab1 CCR domain regulates Fab1 kinase activity, and that this regulation at least in part, involves a physical interaction between these domains. Posttranslational modifications may reversibly stabilize or disrupt an inhibitory binding interaction between the CCR domain and the kinase region (Figure 5D) with or without the involvement of additional regulatory binding partners.
Structure–function studies that rely on loss-of-function mutants are often difficult to interpret: these mutations often impair protein folding or stability. These studies show that the generation and characterization of dominant-active mutations provide an effective approach to determine the function of specific protein domains. Characterization of dominant-active Fab1 alleles 1) identified a regulatory region on the putative surface of the C-terminal Fab1 kinase domain, which contains regulatory residues conserved among Fab1 proteins, and 2) identified an intramolecular interaction between this surface of the kinase domain and the upstream CCR domain (Figure 5E). Analogous genetic approaches with other genes that encode large proteins may be similarly informative. For instance, other proteins within the lipid kinase family, such as Stt4 and Pik1, have several domains of unknown function (Foti et al., 2001; Audhya and Emr, 2002; Strahl and Thorner, 2007; Baird et al., 2008). If dominant-active alleles could be generated, this would suggest that negative regulation occurs. Moreover, this type of mutant screen has the potential to reveal residues that are critical to the negative regulation of these kinases.
MATERIALS AND METHODS
Strains, plasmids, and media
Strains are listed in Supplemental Table S1. Strains were grown in either yeast extract/peptone/glucose (YEPD) or synthetic complete (SC) minimal medium. Plasmids are listed in Supplemental Table S2. Point mutations were generated as previously described (Weiner et al., 1993; Sawano and Miyawaki, 2000).
Fluorescence microscopy
Yeast cells were grown in the appropriate SC medium to an OD600 of 0.5 and then were labeled with FM4-64 (Vida and Emr, 1995). Images were acquired using the DeltaVision RT Restoration Microscopy System (Applied Precision, Issaquah, WA).
Phosphoinositide lipid labeling and quantification
Yeast [3H]inositol labeling and total cellular phosphatidylinositol extraction, deacylation, and measurements were performed as previously described (Bonangelino et al., 2002; Duex et al., 2006a). Cells were grown in the appropriate SC medium to an OD600 of 0.5. Cells (0.2 OD600 U) were washed three times with SC medium lacking inositol and then used to inoculate 5 ml of medium lacking inositol and containing 50 µCi of myo-[3H]inositol. Cells were labeled for between 12 and 16 h of shaking at 24°C, harvested by centrifugation, and resuspended in 100 µl of inositol-free medium. For hyperosmotic shock, 100 µl of inositol-free medium with 1.8 M NaCl was added to the 100 μl of sample and then stopped via the addition of ice-cold 4.5% perchloric acid. For basal conditions, 100 µl of inositol-free medium was added, followed by the addition of ice-cold 4.5% perchloric acid. Cells were lysed on a Beadbeater (Biospec) for 2 min at room temperature, followed by resting 2 min on ice. This was repeated two more times. Cell extracts were centrifuged at 14,000 rpm for 15 min at room temperature, and pellets were washed with 100 mM EDTA, pH 8.0, and then resuspended in 50 µl of distilled deionized water. The lipids were deacylated by treatment with methylamine, and then the samples were dried in a SpeedVac. Pellets were resuspended in 300 µl of distilled deionized water. After this, the samples were mixed with 300 µl of butanol/ethyl ether/formic acid ethyl ester (20:4:1), vortexed, and centrifuged at 14,000 rpm for 5 min, and then the aqueous phase was transferred to a fresh microcentrifuge tube. This sample extraction was performed twice. Samples were then dried on a SpeedVac, resuspended in 60 µl of distilled deionized water, and analyzed by high-performance liquid chromatography using an anion exchange, PartisphereSAX (Whatman), column, as previously described (Bonangelino et al., 2002). For comparison of phosphatidylinositol polyphosphate (PI) levels, the raw counts in each peak were expressed as a percentage of total phosphatidylinositol calculated from summation of the counts of the five detectable glycerol-inositol phosphate peaks (PI, PI3P, phosphatidylinositol 4-phosphate, PI(3,5)P2, and phosphatidylinositol 4,5-bisphosphate).
Fab1-TAP-tag pull down
A fig4Δ, FAB1-3xGFP strain was transformed with plasmids expressing Fab1-TAP and Fig4-Myc or empty vectors. We harvested 25 OD600 U of log-phase cells, lysed them in immunoprecipitation (IP) buffer (50 mM Tris, pH 7.5, 120 mM NaCl, 10 mM EDTA, 1 mM ethylene glycol tetraacetic acid, 5 mM 2-glycerophosphate, 1× Roche Complete inhibitor cocktail, and 3 mM benzamidine) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), 3 µg/ml leupeptin, 5 mg/ml aprotonin, and 18 µg/ml chymostatin, and removed debris by centrifugation for 5 min at 500 × g. The supernatant was mixed with 5% octyl-glucoside (Sigma-Aldrich) in lysis buffer for a final concentration of 0.5% octyl-glucoside and incubated for 30 min. Octyl-glucoside–solubilized lysate was cleared by spinning at 13,000 × g for 10 min. The supernatant was incubated for 1 h with 20 µl of immunoglobulin G (IgG) Sepharose beads (GE Healthcare). IgG beads were washed three times with 500 µl of IP buffer containing 0.5% octyl-glucoside. Bound protein was eluted by heating IgG beads with 25 μl of sample buffer (50 mM Tris, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 10% glycerol, 0.01% bromophenol blue) at 80°C for 5 min. SDS–PAGE and Western blot analysis were used to detect Fab1-TAP, Fab1-3xGFP, and Fig4–Myc.
Protein purification
Protein expression plasmids were generated using the pQlink plasmid vectors (Scheich et al., 2007). Proteins were expressed in E. coli BL21 star (DE3). Cells were grown in modified terrific broth (1.2% [wt/vol] tryptone, 2.4% [wt/vol] yeast extract, 4% glycerol, 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5) with 0.05 mg/ml ampicillin at 37°C to an OD600 of 0.4–0.5. After 18 h of induction with 0.2 mM isopropyl-β-d-thiogalactoside at 20°C, cells were harvested via centrifugation at 5000 × g for 15 min at 4°C. Cells were lysed via sonication in 1 g/10 ml of lysis buffer (25 mM NaPO4, pH 7.5, 100 mM NaCl, 100 μl/100 ml Sigma-Aldrich protease inhibitor cocktail, one tablet/100 ml of cOmplete EDTA-free Protease Inhibitor, 0.1 mg/ml PMSF, 5 mM MgCl2, 100 μl [100 U]/100 ml RQ1 RNase-free DNase from Promega). After a 25,000 × g spin at 4°C for 35 min, proteins in the supernatant were purified with amylose resin (New England Biolabs, Ipswich, MA), washed with wash buffer (25 mM NaPO4, pH 7.5, 100 mM NaCl, 100 μl/100 ml of Sigma-Aldrich protease inhibitor cocktail, one tablet/100 ml of cOmplete EDTA-free Protease Inhibitor Tablet, 0.1 mg/ml PMSF), eluted with wash buffer plus 20 mM Maltose, and then purified on glutathione–Sepharose resin (GE Healthcare, Little Chalfont, England) and eluted with wash buffer plus 25 mM reduced glutathione.
Yeast two-hybrid assay
The indicated pGAD and pGBD plasmids (see Supplemental Table S2) were cotransformed into the yeast strain PJ69-4A as previously described (Jin et al., 2008). Transformants were plated onto SC-LEU-TRP, replica plated onto SC-LEU-TRP (control), SC-LEU-TRP-ADE-HIS+3AT (stringent test), and SC-LEU-TRP-ADE-HIS (test), and grown for 4–12 d at 24°C. Yeast strain PJ69-4A and the pGAD and pGBD vectors were described previously (James et al., 1996).
Coomassie stain and Western blot analysis
Samples were diluted from their original concentrations as follows. Total and soluble protein, 1:50 dilution; amylose elution and glutathione elution, 1:2 dilution. Samples were heated in sample buffer for 10 min at 70°C and run on a 10% SDS–polyacrylamide gel at 70 V until the dye front ran off the gel (2–3 h). Gels were stained with GelCode Blue Stain Reagent (Thermo Scientific) for 1 h at room temperature and then washed with water to destain and image. Images were processed using ImageJ, version 1.6.0_24 (National Institutes of Health).
For Western blot analysis, samples were run on an SDS–PAGE gel, followed by an 850 V-h transfer to polyvinylidene fluoride membrane. Membranes were blocked with 2.5% milk in Tris-buffered saline/Tween-20, probed with primary antibody, washed, probed with secondary antibody, and imaged on a Typhoon 9410 Molecular Imager (GE Amersham Molecular Dynamics). Images were processed using ImageJ, version 1.6.0_24. Antibody dilutions were as follows: anti-hexahistidine (552565; BD Pharmingen), 1:3000; anti-GST (901601; BioLegend), 1:3000; anti-GFP (Roche), 1:1000; anti-TAP (Pierce), 1:5000; and anti–Myc clone 9E10 (EMD Millipore), 1:1000.
Screen for dominant-active CCR domain alleles
This screen is adapted from a previously published dominant-active screen of the kinase domain of Fab1 (Duex et al., 2006b; Supplemental Figure S3A). A pRS416-Fab1 plasmid was gapped with a Pflm1 restriction enzyme digest at 37°C overnight, which cut out a region encoding amino acids 1014–1575 of Fab1. The gapped plasmid was purified by agarose electrophoresis followed by DNA purification (Qiagen). After this, primers MJL299_PflM1_FWD (5-CATTTCTGTGGATAAGTTGGCTACG-3) and MJL300_PflM1_REV (5-CATGAGTCATAGATATACCTGTTCCAC-3) were used to reamplify this region along with 100 base pairs of overhang on either side of the cut site using Taq polymerase with the recommended buffer (Invitrogen). Pflm1-gapped pRS416-Fab1 and the Taq-amplified gapped region were then cotransformed into an exponentially growing culture of vac7Δ (LWY2054). One hundred fifty single colonies that grew faster than pRS416-Fab1 WT control were picked and struck to single colonies. Of the original 150 mutants, 79 were able to grow at 37°C. These 79 candidate mutant Fab1 plasmids were extracted from yeast, amplified in E. coli, and retransformed into vac7Δ. Twenty-one candidate mutant plasmids of the original 150 rescued the 37°C vac7Δ growth defect. Visualization of the vacuole with FM4-64 and fluorescence microscopy indicated that all 21 mutant plasmids displayed a fragmented vacuolar phenotype in a fab1Δ mutant, indicative of elevated PI(3,5)P2 levels. Sanger sequencing of candidate plasmids revealed eight unique alleles.
Supplementary Material
Acknowledgments
We thank Richard Baker for his gift of C. thermophilum DNA and protein expression constructs. This work was supported by National Institutes of Health Grants R01-NS064015 and R01-GM050403 to L.S.W. M.J.L. was supported in part by a Bradley Merrill Patten Fellowship from the Department of Cell and Developmental Biology, University of Michigan. B.S.S. was supported in part by a Jane Coffin Childs Memorial Fund Fellowship and National Institutes of Health Grant K99-GM-120511.
Abbreviations used:
- CCR
conserved cysteine-rich domain
- CCT
chaperone-containing TCP1 domain
- PI(3,5)P2
phosphatidylinositol (3,5)-bisphosphate
- PI3P
phosphatidylinositol 3-phosphate
- Y2H
yeast two-hybrid.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-06-0390) on February 1, 2017.
REFERENCES
- Alghamdi TA, Ho CY, Mrakovic A, Taylor D, Mao D, Botelho RJ. Vac14 protein multimerization is a prerequisite step for Fab1 protein complex assembly and function. J Biol Chem. 2013;288:9363–9372. doi: 10.1074/jbc.M113.453712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- Baird D, Stefan C, Audhya A, Weys S, Emr SD. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J Cell Biol. 2008;183:1061–1074. doi: 10.1083/jcb.200804003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, Hughson FM. A direct role for the Sec 1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 2015;349:1111–1114. doi: 10.1126/science.aac7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Nau JJ, Duex JE, Brinkman M, Wurmser AE, Gary JD, Emr SD, Weisman LS. Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J Cell Biol. 2002;156:1015–1028. doi: 10.1083/jcb.200201002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho RJ, Efe JA, Teis D, Emr SD. Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell. 2008;19:4273–4286. doi: 10.1091/mbc.E08-04-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D, Ma JT, Park S, Inoki K, Weisman LS, Saltiel AR. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell. 2012;23:2955–2962. doi: 10.1091/mbc.E11-12-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Hama H, DeWald DB, Herman PK. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J Biol Chem. 2002;277:287–294. doi: 10.1074/jbc.M109263200. [DOI] [PubMed] [Google Scholar]
- Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- Duex JE, Nau JJ, Kauffman EJ, Weisman LS. Phosphoinositide 5-phosphatase Fig4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell. 2006a;5:723–731. doi: 10.1128/EC.5.4.723-731.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Tang F, Weisman LS. The Vac14p-Fig4p complex acts independently of Vac7p and couples PI(3,5)2 synthesis and turnover. J Cell Biol. 2006b;172:693–704. doi: 10.1083/jcb.200512105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe JA, Botelho RJ, Emr SD. The Fab1 phosphatidylinositol kinase pathway in the regulation of vacuole morphology. Curr Opin Cell Biol. 2005;17:402–408. doi: 10.1016/j.ceb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell. 2001;12:2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Mandelker D, Gabelli SB, Amzel LM. Insights into the oncogenic effects of PIK3CA mutations from the structure of p110alpha/p85alpha. Cell Cycle. 2008;7:1151–1156. doi: 10.4161/cc.7.9.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang Y, Park S, Duex JE, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michell RH, Heath VL, Lemmon MA, Dove SK. Phosphatidylinositol 3,5-bisphosphate: metabolism and cellular functions. Trends Biochem Sci. 2006;31:52–63. doi: 10.1016/j.tibs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Rao VD, Misra S, Boronenkov IV, Anderson RA, Hurley JH. Structure of type IIbeta phosphatidylinositol phosphate kinase: a protein kinase fold flattened for interfacial phosphorylation. Cell. 1998;94:829–839. doi: 10.1016/s0092-8674(00)81741-9. [DOI] [PubMed] [Google Scholar]
- Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, Williams RL. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350:aac7365. doi: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge SA, Anderson DM, Emr SD. Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol Biol Cell. 2004;15:24–36. doi: 10.1091/mbc.E03-05-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Shisheva A. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J Biol Chem. 1999;274:21589–21597. doi: 10.1074/jbc.274.31.21589. [DOI] [PubMed] [Google Scholar]
- Scheich C, Kummel D, Soumailakakis D, Heinemann U, Bussow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MP, Felts KA, Simcox TG, Braman JC. A method for the site-directed mono- and multi-mutagenesis of double-stranded DNA. Gene. 1993;126:35–41. doi: 10.1016/0378-1119(93)90587-s. [DOI] [PubMed] [Google Scholar]
- Whiteford CC, Brearley CA, Ulug ET. Phosphatidylinositol 3,5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem J. 1997;323:597–601. doi: 10.1042/bj3230597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolov SN, Bridges D, Zhang Y, Lee WW, Riehle E, Verma R, Lenk GM, Converso-Baran K, Weide T, Albin RL, et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc Natl Acad Sci USA. 2012;109:17472–17477. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.