Calcium signals via calmodulin, myosin light chain kinase, and nonmuscle myosin II to mediate neuroepithelial apical-basal cell length during zebrafish brain morphogenesis.
Abstract
One of the first morphogenetic events in the vertebrate brain is the formation of the highly conserved midbrain–hindbrain boundary (MHB). Specific cell shape changes occur at the point of deepest constriction of the MHB, the midbrain–hindbrain boundary constriction (MHBC), and are critical for proper MHB formation. These cell shape changes are controlled by nonmuscle myosin II (NMII) motor proteins, which are tightly regulated via the phosphorylation of their associated myosin regulatory light chains (MRLCs). However, the upstream signaling pathways that initiate the regulation of NMII to mediate cell shape changes during MHB morphogenesis are not known. We show that intracellular calcium signals are critical for the regulation of cell shortening during initial MHB formation. We demonstrate that the MHB region is poised to respond to calcium transients that occur in the MHB at the onset of MHB morphogenesis and that calcium mediates phosphorylation of MRLC specifically in MHB tissue. Our results indicate that calmodulin 1a (calm1a), expressed specifically in the MHB, and myosin light chain kinase together mediate MHBC cell length. Our data suggest that modulation of NMII activity by calcium is critical for proper regulation of cell length to determine embryonic brain shape during development.
INTRODUCTION
Cell shape changes are central to the formation of tissue structure. One of the earliest and most highly conserved vertebrate brain structures that forms during development is the tissue fold at the midbrain–hindbrain boundary (MHB; Rhinn and Brand, 2001; Lowery and Sive, 2009; Hirth, 2010). Using the zebrafish MHB as a model to understand brain morphogenesis, we characterized the basic cell shape changes that are required to fold the neuroepithelium in this region (Gutzman et al., 2008, 2015). Zebrafish MHB formation is initiated at the 18-somite stage (ss) after the neural tube has formed. Cells at the point of deepest constriction at the MHB—the MHB constriction (MHBC)—shorten and narrow by 24 ss (Gutzman et al., 2015). By primordium-6 stage, cells at the MHBC constrict basally and expand apically, leading to an acute tissue angle and clear delineation of the midbrain from the hindbrain (Gutzman et al., 2008). The initial cell shape changes are differentially regulated by myosin motor proteins. Apical-basal cell length is specifically regulated by nonmuscle myosin IIA (NMIIA), and anterior-posterior cell width is specifically regulated by nonmuscle myosin IIB (NMIIB; Gutzman et al., 2015). However, the molecular pathways that activate NMII proteins to differentially mediate these cell shape changes remain unknown.
NMII activity can be regulated by several different kinase cascades via phosphorylation of myosin regulatory light chain (MRLC). Calcium (Ca2+) signaling plays a critical role in mediating phosphorylation of MRLC via myosin light chain kinase (MLCK; Berridge et al., 2000; Somlyo and Somlyo, 2003; Vicente-Manzanares et al., 2009) and is critical for many embryonic developmental processes (Webb and Miller, 2003; Slusarski and Pelegri, 2007). However, the specific role for Ca2+ signaling in brain morphogenesis is unknown. We hypothesized that Ca2+ functions as a morphogenetic signal to mediate cell shape changes at the MHB. We demonstrate that intracellular Ca2+ signals specifically determine cell length at the MHBC and that Ca2+ regulation of cell shape depends on NMII activity. We further demonstrate that proper regulation of MHBC cell shape requires calmodulin 1a (calm1a) and MLCK. Our data show for the first time that modulation of NMII function by Ca2+ is critical for regulation of MHBC cell length to determine embryonic brain shape during development.
RESULTS AND DISCUSSION
Ca2+ mediates cell length at the MHBC during brain development
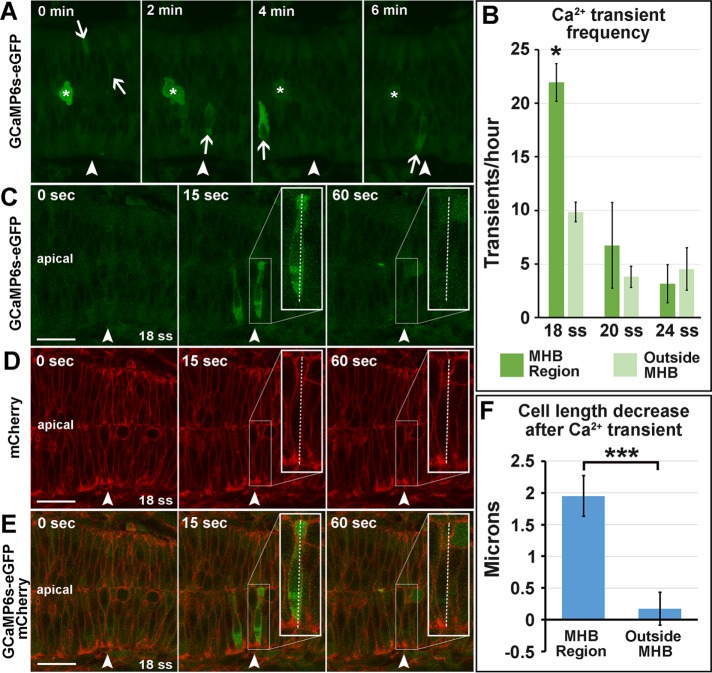
In zebrafish, Ca2+ signals precede morphological patterning in the brain (Webb and Miller, 2007), and apical-basal cell thinning in the enveloping layer cells depends on Ca2+ transients (Zhang et al., 2011). Ca2+ flashes have also been shown to drive apical constriction during neural tube closure (Christodoulou and Skourides, 2015). Therefore we investigated intracellular Ca2+ flashes in the MHB during morphogenesis. We used four-dimensional time-lapse imaging of GCaMP6s–green fluorescent protein (GFP) mRNA–injected embryos to determine whether there were Ca2+ flashes in MHB cells at the time of initial MHB formation. We found single-cell Ca2+ transients to be enriched in the MHB region compared with surrounding regions at 18 ss, during the initiation of MHB morphogenesis (Figure 1, A and B, and Supplemental Movie S1). We also found that apical-basal cell length decreased immediately after a Ca2+ transient specifically in the MHB region (Figure 1, C–F). Therefore we hypothesized that Ca2+ signals may initiate MHB morphogenesis and that cells in this region are poised to respond to Ca2+ transients.
FIGURE 1:
Calcium transients are enriched in the MHB at 18 ss and precede decreases in cell length. (A) Representative wild-type 18 ss time-lapse images of intracellular Ca2+ transients (panels from Supplemental Movie S1). Asterisks indicate mitotic cells, and arrows indicate Ca2+ transients analyzed. (B) Quantification of Ca2+ transients at 18, 20, and 24 ss. (C–E) Representative wild-type images at 18 ss analyzed for cell length changes after Ca2+ transients. (F) Quantification indicating the amount of decrease in cell length immediately after Ca2+ transients. For statistical analysis, one-way ANOVA with Tukey’s HSD post hoc test was conducted for Ca2+ transient frequency. *p < 0.05, mean ± SEM. Embryos analyzed: 18 ss, n = 11; 20 ss, n = 10; 24 ss, n = 8. Mann–Whitney U test was performed for analysis of cell length decrease after Ca2+ transients. ***p < 0.001, mean ± SEM. Embryos analyzed, n = 10; MHB region cells, 17; outside MHB cells, 6. Arrowheads indicate the MHBC. Scale bars, 25 μm.
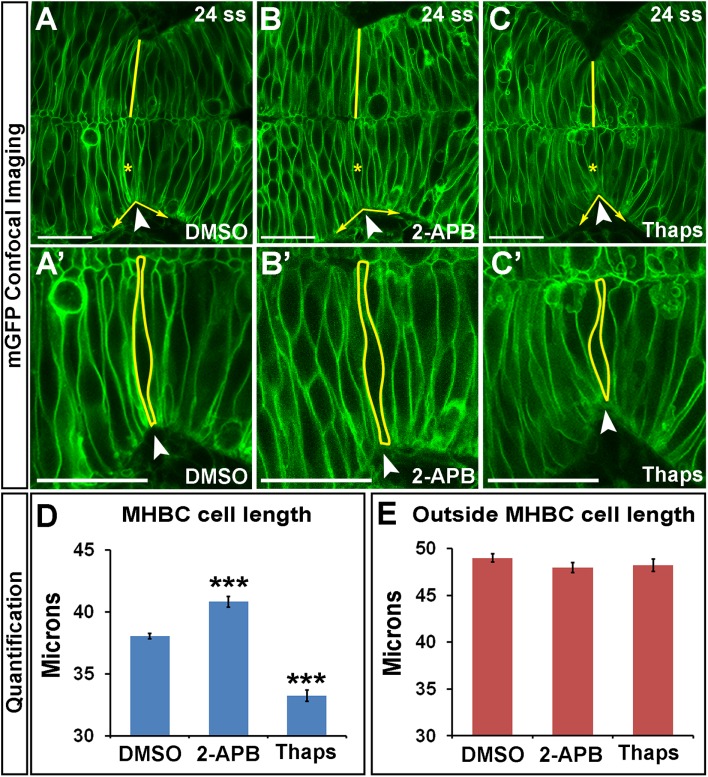
To investigate further the role of Ca2+ signals in regulating zebrafish MHB morphogenesis, we used pharmacological inhibitors to manipulate intracellular Ca2+ levels. We used 2-aminoethoxydiphenyl-borate (2-APB) to decrease intracellular Ca2+ levels (Bootman et al., 2002; Ashworth et al., 2007) and thapsigargin (Thaps) to increase intracellular Ca2+ levels (Kreiling et al., 2008; Zhang et al., 2011). Wild-type embryos were treated at 18 ss with dimethyl sulfoxide (DMSO), 2-APB, or Thaps. We analyzed cell shapes at 24 ss, after the initial morphogenetic events had occurred (Figure 2, A–C′). We found that embryos with decreased intracellular Ca2+ (2-APB treated) had significantly longer cells at the MHBC, whereas embryos with increased intracellular Ca2+ (Thaps treated) had significantly shorter cells at the MHBC (Figure 2D). Efficacy of 2-APB and Thaps was determined using the genetically encoded calcium indicator GCaMP6s-GFP (Supplemental Figure S2, A–D). Apoptosis and cell adhesion within the neuroepithelium, which are known to depend on intracellular Ca2+ homeostasis (Orrenius et al., 1992; Lagunowich et al., 1994), were not affected by these treatments at the doses used (Supplemental Figure S2, E–K).
FIGURE 2:
Calcium regulates cell length at the MHBC. (A–C′) Confocal images of embryos injected with mGFP and treated at 18 ss with (A, A′) DMSO, (B, B′) 100 μM 2-APB, or (C, C′) 2 μM Thaps. Embryos were washed, incubated, and live imaged at 24 ss. (A′–C′) Magnifications of A–C. Arrowheads indicate the MHBC. Asterisks in A–C indicate cell outlined in A′–C′. (D, E) Cell length quantification at the MHBC and 40 μm outside the MHBC. For statistical analysis, one-way ANOVA with Tukey’s HSD post hoc test was done. ***p < 0.001 compared with DMSO, mean ± SEM. For each measurement, DMSO (n = 29; 58 cells), 2-APB (n = 16; 32 cells), and Thaps (n = 14; 28 cells). Scale bars, 25 μm.
Quantification of Ca2+ effects on tissue shape showed that the MHBC angle was abnormal, as expected with changes in MHBC cell length, whereas MHB cell width was only slightly affected with Thaps treatment (Supplemental Figure S3, A and B). However, cells 40 μm posterior to the MHBC were not significantly different from controls after either treatment (Figure 2E), indicating a region-specific effect on cell shape with Ca2+ manipulation. Ca2+ has been previously reported to regulate cell shape in other cell types. Thaps treatment causes cell retraction, gap formation, and actin rearrangement in cultured endothelial cells (Moore et al., 1998), and in cardiac myocytes, Ca2+ regulates cellular contraction, causing shortening of cells in vitro (Bramlage et al., 2001).
Calcium signals to nonmuscle myosin II to mediate cell shape at the MHB
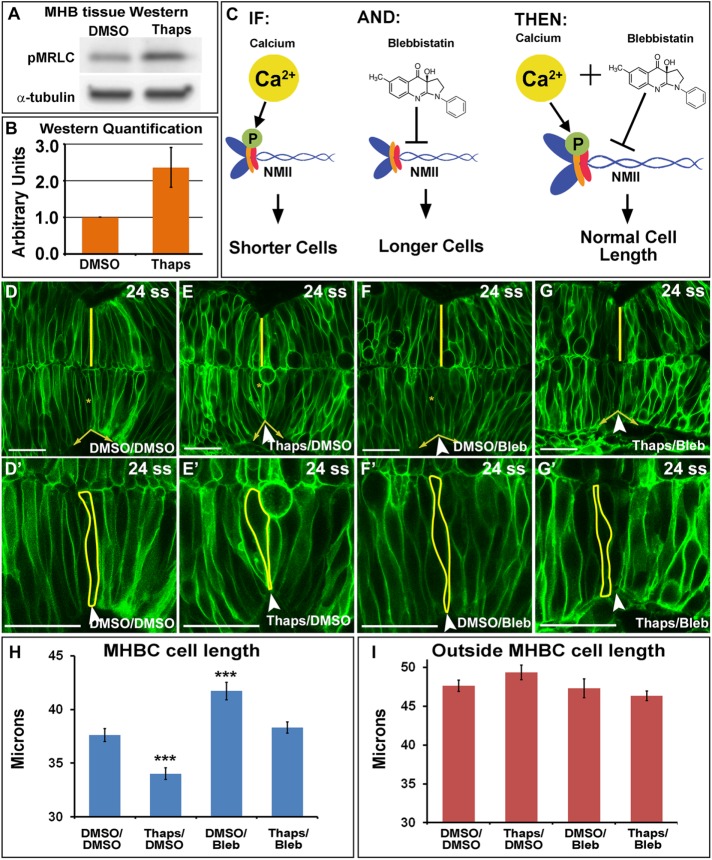
On the basis of our previous work demonstrating that NMII proteins are critical mediators of cell shape at the MHB (Gutzman et al., 2015) and the known role for Ca2+ in regulation of NMII activity (Somlyo and Somlyo, 2003), we hypothesized that Ca2+ may mediate its effects on MHB cell shape via downstream activation of NMII. To test this hypothesis, we first determined whether increasing Ca2+ in the MHB would modulate phosphorylation of MRLC (pMRLC), an indicator of NMII activity (Vicente-Manzanares et al., 2009). We treated wild-type embryos with DMSO or Thaps and microdissected MHB tissue for Western analysis of pMRLC. MHB dissections were confirmed to be specific using reverse transcription-PCR (RT-PCR) with tissue-specific markers (Supplemental Figure S4, A–D). Western analysis of MHB protein showed that pMRLC levels increased approximately twofold after Thaps treatment (Figure 3, A and B), suggesting that increased intracellular Ca2+ leads to increased NMII activity. We were unable to detect a change in pMRLC with 2-APB treatment. This is consistent with the observed difference in the magnitude of effects on Ca2+ between 2-APB and Thaps. Specifically, 2-APB decreases Ca2+ by half, whereas Thaps increases Ca2+ by fivefold (Supplemental Figure S2, A–D).
FIGURE 3:
Calcium signals to NMII at the MHB to modulate MHBC cell length. (A) Representative Western blot for pMRLC in MHB-specific tissue dissected after DMSO or 2 μM Thaps treatment. (B) pMRLC Western quantification using α-tubulin as a control (n = 4). (C) Hypothesized role of Ca2+ and NMII interactions. IF, increase in Ca2+ leads to increased pMRLC to activate NMII and causes shorter cells at the MHBC; AND, Bleb inhibits NMII function; THEN, Thaps treatment to increase Ca2+ leading to shorter cells can be rescued with NMII inhibition by Bleb. (D–G′) Confocal images of mGFP-injected embryos treated with (D, D′) DMSO/DMSO, (E, E′) Thaps/DMSO, (F, F′) DMSO/Bleb, or (G, G′) Thaps/Bleb. (D′–G′) Magnifications of D–G. Arrowheads indicate the MHBC. Asterisks in D–G indicate cell outlined in D′–G′. (H, I) Cell length quantification at the MHBC and 40 μm outside the MHBC. For statistical analysis, one-way ANOVA with Tukey’s HSD post hoc test was done. ***p < 0.001 compared with DMSO/DMSO; mean ± SEM. For each measurement, DMSO/DMSO (n = 9; 18 cells), Thaps/DMSO (n = 9; 18 cells), DMSO/Bleb (n = 7; 14 cells), and Thaps/Bleb (n = 8; 16 cells). Scale bars, 25 μm.
Next we tested our hypothesis that Ca2+ mediates cell shape via NMII protein function, using two complementary rescue experiments that coupled modulation of intracellular Ca2+ levels with manipulation of NMII function. We used 2-APB or Thaps treatment to manipulate Ca2+ levels, as described in Figure 2. NMII activity was manipulated using the pharmacological reagent blebbistatin (Bleb) or with mypt1 gene knockdown to inhibit or activate NMII, respectively. Bleb is a well-established myosin II inhibitor (Kovacs et al., 2004), and mypt1 encodes the regulatory subunit of myosin phosphatase and is required to inactivate pMRLC. Therefore mypt1 knockdown leads to an increase in pMRLC, resulting in NMII overactivation (Ito et al., 2004; Gutzman and Sive, 2010).
In our first rescue experiments, we increased Ca2+ levels and rescued the cell-length phenotype with inhibition of NMII. We hypothesized that we could rescue Thaps-induced short cells by inhibiting NMII with Bleb (Figure 3C). As predicted, we found that Thaps decreased cell length and Bleb increased cell length at the MHBC, and treatment of embryos with Thaps followed by Bleb rescued the abnormal-cell-length phenotypes specifically at the MHBC (Figure 3, D–I). These data suggest that the Ca2+ signal affects cell length via modulation of NMII function. Quantification of tissue shape showed that the MHBC angle was abnormal, as expected with changes in MHBC cell length, but was also partially rescued with Thaps and Bleb treatment together. Bleb had a slight effect on MHB cell width, which is expected because Bleb inhibits both NMIIA and NMIIB, and we know that NMIIB is essential in mediating cell width (Supplemental Figure S3, C and D; Gutzman et al., 2015). Cell length outside of the MHBC was not significantly different from controls for any treatment (Figure 3I), suggesting that MHBC cells are poised to respond to slight changes in intracellular Ca2+ levels.
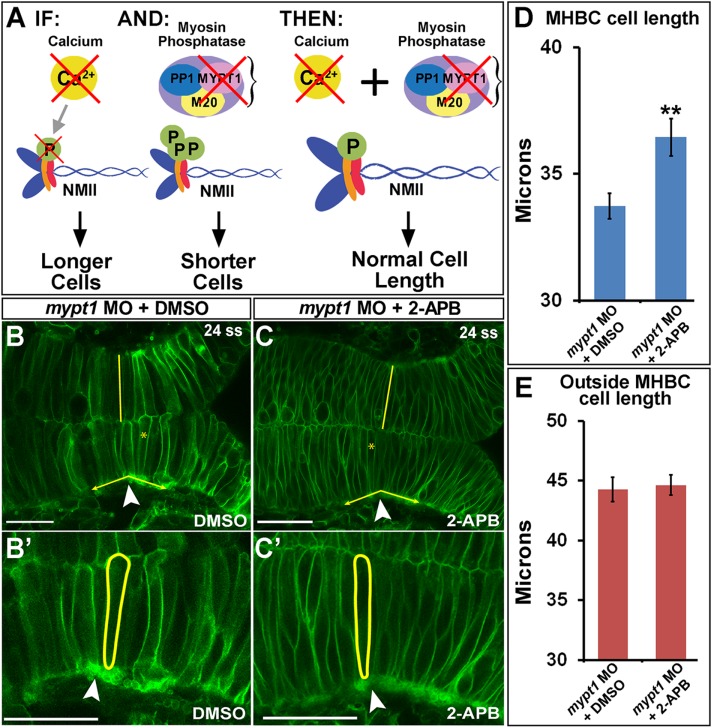
In a complementary set of experiments, we overactivated NMII and rescued the cell-length phenotype by decreasing Ca2+ levels. We hypothesized that decreasing Ca2+ with 2-APB would specifically rescue short cells induced by NMII overactivation (Figure 4A). The mypt1 morphants revealed cells that were both shorter and wider than controls, as we previously reported (Figure 4B; Gutzman and Sive, 2010; Gutzman et al., 2015). As predicted, cell length in mypt1 morphants treated with 2-APB was rescued compared with mypt1 morphant controls (Figure 4, B–D). No other significant differences were observed (Figure 4E and Supplemental Figure S3, E and F). We did not observe a rescue of the MHB tissue angle in these experiments, which is expected because we know that cell width also contributes to normal MHB tissue angle (Supplemental Figure S3, E and F).
FIGURE 4:
Calcium inhibition rescues NMII overactivation at the MHBC. (A) Hypothesized role of Ca2+ and NMII interactions. IF, inhibition of Ca2+ by 2-APB results in longer MHBC cells; AND, mypt1 knockdown results in overactivation of MRLC, causing shorter and wider MHB cells; THEN, inhibition of Ca2+ in mypt1-knockdown embryos would rescue MHBC cell length. (B, C) Confocal images of 24-ss embryos coinjected with mGFP and mypt1 MO and treated with (B) DMSO or (C) 2-APB. (B′, C′) Magnified images from B and C. Arrowheads indicate MHBC. (D, E) Cell length quantification at the MHBC and 40 μm outside the MHBC. For statistical analysis, the Mann–Whitney U test was done. **p < 0.01, mean ± SEM. For each measurement, mypt1 MO plus DMSO (n = 11; 22 cells) and mypt1 MO plus 2-APB (n = 13; 26 cells). Scale bars, 25 μm.
Taking the results together, we showed in vivo that MHBC cell length is regulated by Ca2+ and NMII activity. These complementary rescue experiments further suggest an important correlation between regulation of cell length and NMII. In our previous work, we determined that NMIIA specifically regulates cell length at the MHBC (Gutzman et al., 2015). Here 2-APB rescues only the cell-length phenotype in mypt1-knockdown embryos, without the rescue of cell width. These results lead us to speculate that Ca2+ might signal through intermediate molecules specifically to NMIIA to differentially mediate cell length at the MHBC. Additional experiments are needed to specifically address this possibility.
Calmodulin and MLCK regulate cell shape at the MHB
Because Ca2+ is a ubiquitous ion and additional manipulations of Ca2+ levels are not region specific, the question remains as to how Ca2+ transients might lead to modulation of cell shape specifically at the MHBC. Ca2+ interacts with calcium-binding proteins to initiate downstream signaling cascades (Berridge et al., 2000). Therefore one possible mechanism for a region-specific Ca2+ response to transient activity would be through tissue specific expression of a calcium-binding protein required to mediate cell shape changes. We hypothesized that the calcium-binding partner calmodulin, a highly conserved multifunctional Ca2+ sensor and signal transducer (Crivici and Ikura, 1995; Tidow and Nissen, 2013), might play a role in allowing MHBC cells to respond specifically to Ca2+.
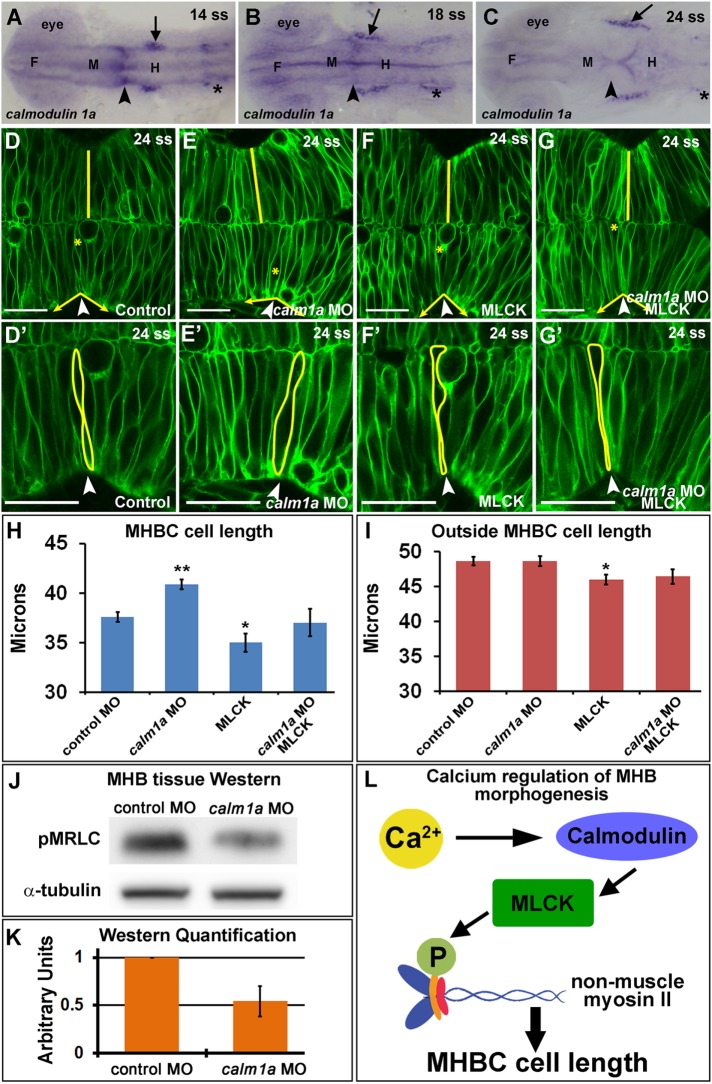
Mammalian vertebrates express three calmodulin genes, whereas zebrafish have six calmodulin genes due to genome duplication that all encode the same calmodulin protein (Friedberg and Taliaferro, 2005). In zebrafish, calmodulin 3b is ubiquitously expressed in the brain during development, whereas other calmodulin genes have region-specific expression (Thisse et al., 2001; Thisse and Thisse, 2004), and the protein exhibits cell type–specific subcellular localization (Caceres et al., 1983; Barreda and Avila, 2011). Of particular interest is calmodulin 1a (calm1a), which was shown to have potential MHB-specific expression during development (Thisse et al., 2001). Differential expression patterns of calmodulin have been observed in the eye and nervous system in other vertebrates (Friedberg and Rhoads, 2001; Thut et al., 2001; Kobayashi et al., 2015); however, comparative expression patterns during neural tube formation and brain morphogenesis at the time points examined here have not been reported. We used in situ hybridization to confirm the MHB localized expression of calm1a during brain morphogenesis. We found that calm1a is expressed specifically at the MHB before and during the initiation of MHB morphogenesis, with other specific expression detectable in the trigeminal ganglia and otic vesicles (Figure 5, A–C). Therefore, we hypothesized that the spatial localization of calm1a expression may provide an MHBC region-specific response to the Ca2+ transients. To test this, we knocked down calm1a using morpholino antisense oligonucleotides. We chose a knockdown method rather than a mutant approach because we hypothesize that the multiple calmodulin genes, all encoding the same protein, would lead to gene compensation, masking any specific role for calm1a in MHB tissue (Rossi et al., 2015). The calm1a morpholino (MO) effectiveness and specificity were confirmed using RT-PCR, MHB tissue-specific Western analysis, and calm1a mRNA rescue (Supplemental Figure S4). The morpholino resulted in a deletion of exon 2 and an early stop codon as well as an approximate 50% reduction in calmodulin protein within the MHB region (Supplemental Figure S4, E–G). We found that calm1a knockdown resulted in longer MHBC cells but had no effect on cell length outside of the MHBC (Figure 5, D–E′ and H–I). The MHBC cell length defect was rescued by calm1a mRNA expression (Supplemental Figure S4, H–L). The calm1a knockdown also had a dramatic effect on MHBC angle, as expected with the cell-length phenotype, but had no effect on MHB cell width (Supplemental Figure S3, G and H). These results indicate a requirement for calm1a in mediating cell length at the MHBC.
FIGURE 5:
Calmodulin 1a and MLCK mediate cell length at the MHBC. (A–C) calm1a gene expression by in situ hybridization. Arrowheads indicate MHBC, and arrows indicate trigeminal ganglia; asterisks indicate otic vesicle. F, forebrain; M, midbrain; H, hindbrain. (D–G′) Confocal images of embryos injected with mGFP and (D, D′) control MO, (E, E′) calm1a MO, (F, F′) MLCK mRNA, or (G, G′) calm1a MO and MLCK mRNA. (D′–G′) Magnification of D–G. Arrowheads indicate the MHBC. Asterisks in D–G indicate cell outlined in D′–G′. (H, I) Cell length quantification at the MHBC and 40 μm outside the MHBC. (J) Representative Western blots for pMRLC in MHB-specific tissue dissected after control MO or calm1a MO injection. (K) pMRLC Western quantification using α-tubulin as a control (n = 5). (L) Proposed signaling pathway for Ca2+ regulation of cell length at the MHBC. For statistical analysis, one-way ANOVA with Tukey’s HSD post hoc test was done. **p < 0.01 and *p < 0.05 compared with control, mean ± SEM. Control MO (n = 17; 34 cells), calm1a MO (n = 19; 38 cells), MLCK mRNA (n = 13; 26 cells), and calm1a MO plus MLCK (n = 9; 18 cells). Scale bars, 25 μm.
One known downstream target of calmodulin is activation of MLCK (Holzapfel et al., 1983), and activated MLCK is known to phosphorylate MRLC (Somlyo and Somlyo, 2003). Therefore we investigated the role for MLCK in regulation of cell shape at the MHBC. We overexpressed MLCK and found that cells at the MHBC and outside the MHBC were slightly shorter than control cells, indicating that overexpression of MLCK increased cell shortening throughout the region (Figure 5, F, F’, and H–I). MLCK expression had no effect on overall MHB angle because all cells had a cell length defect (Supplemental Figure S3, G and H). From these results, we hypothesized that calm1a and MLCK may function in the same pathway to mediate MHBC cell length. To test this, we coinjected embryos with calm1a MO and MLCK mRNA, which rescued the MHBC cell-length phenotype (Figure 5, G–I), further suggesting that calmodulin works in conjunction with MLCK to regulate cell shape specifically at the MHBC. In addition, we hypothesized that calm1a knockdown would influence activation of NMII. Therefore we analyzed phosphorylated MRLC levels in MHB tissue microdissected from control and calm1a MO–injected embryos by Western blot, as in Figure 2. Western analysis of MHB protein demonstrated that pMRLC levels decreased ∼50% with calm1a knockdown (Figure 5, J and K), further indicating a requirement for calm1a at the MHB for NMII activity.
Together our results suggest a model in which Ca2+/calmodulin specifically activates MLCK at the MHBC to mediate cell length via activation of NMII (Figure 5L). This is the first time a role for Ca2+-mediated regulation of cell length during brain morphogenesis has been demonstrated. We hypothesize that the specific expression of calm1a in this region is what allows the MHBC neuroepithelial cells to respond to Ca2+ transients that occur in the MHB region at the onset of morphogenesis.
MATERIALS AND METHODS
Zebrafish maintenance and husbandry
Zebrafish maintenance, husbandry procedures, and staging were followed as per Kimmel et al. (1995) and Westerfield (2000). Wild-type (AB and EK) zebrafish embryos were used for all experiments. Somites were counted to establish specific and consistent staging and eliminate concerns regarding potential developmental delay for all experiments. This study was approved by the University of Wisconsin–Milwaukee Institutional Animal Care and Use Committee.
Live imaging of calcium transients and analysis
For imaging of Ca2+ transients, wild-type single-cell embryos were injected with 100 ng of GCaMP6s-GFP mRNA encoding the Ca2+ indicator, a gift from Philipp Keller and Yinan Wan (Howard Hughes Medical Institute, Janelia Research Campus), and 100 ng mCherry mRNA. Live time-lapse imaging was conducted beginning at 18, 20, or 24 ss using a Nikon CS2 scanning confocal microscope. Time-lapse data sets were acquired as single slices or 5-μm z-stacks and taken ∼15–20 μm into the neural tube tissue from the dorsal surface at a frame rate of 10–30 s per image. Overall capture time was between 10 and 120 min, with the average total time being 35 min per embryo. Ca2+ transients observed were quantified spatially within the MHB region (∼10 μm on either side of the MHBC, ∼20 μm total) and the outside MHB region (∼30 μm on either side of the MHB region, ∼60 μm total). Therefore the MHB region accounted for 25% of the total area analyzed, and the outside MHB region accounted for 75% of the total area analyzed. To determine Ca2+ transient frequency, we normalized the number of Ca2+ transients observed during each time-lapse experiment to the number of transients per hour at an acquisition rate of one image per 10 s. Each region was normalized to represent an equal area of the neural tube tissue for comparison. To examine change in cell shape after Ca2+ transients, we measured the apical-basal length of single cells using the mCherry cell outline images. Single cells were measured during the Ca2+ transient, and the same cell was measured again immediately after the transient. Only cells in the MHBC region demonstrated decreases in cell length after Ca2+ transients. All confocal images were analyzed using the Nikon Imaging Systems (NIS) Elements software.
Drug treatments
Wild-type embryos were injected at the one-cell stage with 200 ng/μl membrane GFP (mGFP) mRNA (CAAX-eGFP). At 18 ss, embryos were dechorionated and treated for 10 min with 100 μM 2-APB (D9754-1G; Sigma-Aldrich), an inositol triphosphate receptor (IP3R) antagonist that blocks Ca2+ release from the endoplasmic reticulum, to decrease intracellular Ca2+ levels (Bootman et al., 2002; Ashworth et al., 2007). Alternatively, embryos were treated for 15 min with 2 μM Thaps (T9033-1MG; Sigma-Aldrich), a sarcoplasmic and endoplasmic reticulum Ca2+ ATPase (SERCA) inhibitor that depletes Ca2+ stores, to increase intracellular Ca2+ levels (Kreiling et al., 2008; Zhang et al., 2011). After the treatment times, embryos were washed and allowed to mature to 24 ss in 1% agarose–lined Petri dishes at 28°C and imaged using live confocal microscopy. DMSO control treatments were conducted using the same volume percentage as other treatment groups in each experiment. For Thaps/Bleb combined drug treatment rescue experiments, wild-type embryos were injected at the one-cell stage with mGFP mRNA. At 18 ss, embryos were separated into two treatment groups: DMSO or 2 μM Thaps. After treatment, embryos were washed and incubated at 28°C until 20–21 ss. At 20–21 ss, embryos from each treatment group were separated into two additional treatment groups: DMSO or 50 μM Bleb (B0560-1MG; Sigma-Aldrich). Finally, each group was incubated at 28°C until 24 ss and imaged using live confocal microscopy.
Live confocal imaging and cell shape analysis
All mGFP live confocal imaging was conducted as previously described (Graeden and Sive, 2009; Gutzman et al., 2015) using a Nikon CS2 scanning confocal microscope. Live confocal images presented in each figure are single slices taken from a z-series of images ∼15–20 μm into the tissue from the dorsal surface. All confocal images were processed using Nikon Imaging Systems (NIS) Elements software and Photoshop (Adobe). Cell shape analysis was performed as described in Gutzman et al. (2015). Briefly, cell length was determined using the NIS-Elements software measurement tool by measuring a single cell spanning the neuroepithelium from apical to basal in the region described, either directly at the MHBC or 40 μm posterior to the MHBC (outside the MHBC) on the hindbrain side.
Antisense MO oligonucleotide and mRNA injections
All knockdown experiments were performed using splice site–blocking MO antisense oligonucleotides. MO details are as follows: mypt1 MO (5′-ATTTTTTGTGACTTACTCAGCGATG-3′; Gutzman et al., 2008, 2015), calmodulin 1a (ENSDART00000034580), and calm1a MO (5′-CCACAAACAGACTGCCTTACCTGCA-3′). Zebrafish p53 MO (5′-GCGCCATTGCTTTTGCAAGAATTG-3′) was used in conjunction with the calm1a MO at equal concentration. Standard control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′). All MOs are from Gene Tools. Morpholinos were injected into one-cell-stage embryos either alone or in conjunction with mGFP mRNA as indicated. MO concentrations were 5 ng mypt1 MO, 2 ng calm1a MO, and 2 ng p53 MO. Control MO concentration was equal to the highest concentration of any MO used in that experiment. For MLCK mRNA expression experiments, 50 ng of MLCK mRNA (zfMLCK fused to the globin 3′ untranslated region, kindly provided by Erez Raz, University of Munster, Munster, Germany), was injected with 200 ng/μl mGFP mRNA separately or combined with calm1a MO or control MO. All mRNA was in vitro transcribed for injections using the mMessage mMachine Transcription Kit (Ambion). Embryos were incubated at 28°C to 24 ss and imaged using live confocal microscopy.
In situ hybridization
In situ hybridization was conducted according to standard procedures. For the EST/cDNA clone, calm1a, cb617, from ZFIN was used to produce the calmodulin 1a in situ probe. Bright-field imaging was conducted using an Olympus SZX12 stereomicroscope with an Olympus DP72 camera.
Western blot analysis with MHB-specific tissue
Embryos were dechorionated and treated as described with either DMSO or Thaps at 18 ss. Alternatively, embryos were injected with calm1a MO or control MO. MHB tissue was dissected from 18 to 24 ss for protein isolation and analysis (see Supplemental Figure S4 for details). Primary antibodies were pMLC2 (pMRLC; 3671; Cell Signaling Technology) at 1:500 (Gutzman and Sive, 2010) and α-tubulin (T6199; Sigma-Aldrich) at 1:1000. Secondary antibodies were anti-mouse horseradish peroxidase (HRP; 7076S; Cell Signaling Technology) and anti-rabbit HRP (7074S; Cell Signaling Technology) at 1:2000. Blots were imaged on a UVP Biospectrum Imaging System.
Statistical analyses
Statistical analysis between two groups was carried out using the Mann–Whitney U test; p values denoting significance are reported in each figure legend. Statistical analysis for comparisons between more than two treatment groups was carried out by one-way analysis of variance (ANOVA). For ANOVA p < 0.05, Tukey’s honest significant difference (HSD) post hoc tests were performed to determine significance between control treatment and experimental treatment groups. The p values for post hoc comparisons are presented in each figure legend. All ANOVA and Tukey’s HSD post hoc analyses were carried out using R-3.1.2. At least three independent experiments were conducted for all data presented; n indicates the total number of embryos analyzed, and numbers of single cells analyzed are indicated where necessary.
Supplementary Material
Acknowledgments
We thank Ava Udvadia (University of Wisconsin–Milwaukee) for helpful discussion and comments on the manuscript and Elizabeth Falat for assistance in making the calmodulin 1a rescue construct. We acknowledge Philipp Keller and Yinan Wan from the Howard Hughes Medical Institute, Janelia Research Campus for kindly providing the previously unpublished pCS2+GCaMP6s-GFP construct. We thank Erez Raz from the Institute of Cell Biology at the University of Munster for providing the zebrafish MLCK expression construct. This work was funded in part by the University of Wisconsin–Milwaukee Research Growth Initiative.
Abbreviations used:
- MHB
midbrain–hindbrain boundary
- MHBC
midbrain–hindbrain boundary constriction
- MLCK
myosin light chain kinase
- MRLC
myosin regulatory light chain
- NMII
nonmuscle myosin II.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-08-0561) on February 1, 2017.
REFERENCES
- Ashworth R, Devogelaere B, Fabes J, Tunwell RE, Koh KR, De Smedt H, Patel S. Molecular and functional characterization of inositol trisphosphate receptors during early zebrafish development. J Biol Chem. 2007;282:13984–13993. doi: 10.1074/jbc.M700940200. [DOI] [PubMed] [Google Scholar]
- Barreda EG, Avila J. Tau regulates the subcellular localization of calmodulin. Biochem Biophys Res Commun. 2011;408:500–504. doi: 10.1016/j.bbrc.2011.04.082. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Bootman MD, Collins TJ, Mackenzie L, Roderick HL, Berridge MJ, Peppiatt CM. 2-aminoethoxydiphenyl borate (2-APB) is a reliable blocker of store-operated Ca2+ entry but an inconsistent inhibitor of InsP3-induced Ca2+ release. FASEB J. 2002;16:1145–1150. doi: 10.1096/fj.02-0037rev. [DOI] [PubMed] [Google Scholar]
- Bramlage P, Joss G, Staudt A, Jarrin A, Podlowski S, Baumann G, Stangl K, Felix SB, Stangl V. Computer-aided measurement of cell shortening and calcium transients in adult cardiac myocytes. Biotechnol Prog. 2001;17:929–934. doi: 10.1021/bp010080q. [DOI] [PubMed] [Google Scholar]
- Caceres A, Bender P, Snavely L, Rebhun LI, Steward O. Distribution and subcellular localization of calmodulin in adult and developing brain tissue. Neuroscience. 1983;10:449–461. doi: 10.1016/0306-4522(83)90145-8. [DOI] [PubMed] [Google Scholar]
- Christodoulou N, Skourides PA. Cell-autonomous Ca(2+) flashes elicit pulsed contractions of an apical actin network to drive apical constriction during neural tube closure. Cell Rep. 2015;13:2189–2202. doi: 10.1016/j.celrep.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Crivici A, Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- Friedberg F, Rhoads AR. Evolutionary aspects of calmodulin. IUBMB Life. 2001;51:215–221. doi: 10.1080/152165401753311753. [DOI] [PubMed] [Google Scholar]
- Friedberg F, Taliaferro L. Calmodulin genes in zebrafish (revisited) Mol Biol Rep. 2005;32:55–60. doi: 10.1007/s11033-004-2891-y. [DOI] [PubMed] [Google Scholar]
- Graeden E, Sive H. Live imaging of the zebrafish embryonic brain by confocal microscopy. J Vis Exp 2009(26), 1217. 2009 doi: 10.3791/1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzman JH, Graeden EG, Lowery LA, Holley HS, Sive H. Formation of the zebrafish midbrain-hindbrain boundary constriction requires laminin-dependent basal constriction. Mech Dev. 2008;125:974–983. doi: 10.1016/j.mod.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzman JH, Sahu SU, Kwas C. Non-muscle myosin IIA and IIB differentially regulate cell shape changes during zebrafish brain morphogenesis. Dev Biol. 2015;397:103–115. doi: 10.1016/j.ydbio.2014.10.017. [DOI] [PubMed] [Google Scholar]
- Gutzman JH, Sive H. Epithelial relaxation mediated by the myosin phosphatase regulator Mypt1 is required for brain ventricle lumen expansion and hindbrain morphogenesis. Development. 2010;137:795–804. doi: 10.1242/dev.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirth F. On the origin and evolution of the tripartite brain. Brain Behav Evol. 2010;76:3–10. doi: 10.1159/000320218. [DOI] [PubMed] [Google Scholar]
- Holzapfel G, Wehland J, Weber K. Calcium control of actin-myosin based contraction in triton models of mouse 3T3 fibroblasts is mediated by the myosin light chain kinase (MLCK)-calmodulin complex. Exp Cell Res. 1983;148:117–126. doi: 10.1016/0014-4827(83)90192-1. [DOI] [PubMed] [Google Scholar]
- Ito M, Nakano T, Erdodi F, Hartshorne DJ. Myosin phosphatase: structure, regulation and function. Mol Cell Biochem. 2004;259:197–209. doi: 10.1023/b:mcbi.0000021373.14288.00. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Saragai S, Naito A, Ichio K, Kawauchi D, Murakami F. Calm1 signaling pathway is essential for the migration of mouse precerebellar neurons. Development. 2015;142:375–384. doi: 10.1242/dev.112680. [DOI] [PubMed] [Google Scholar]
- Kovacs M, Toth J, Hetenyi C, Malnasi-Csizmadia A, Sellers JR. Mechanism of blebbistatin inhibition of myosin II. J Biol Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- Kreiling JA, Balantac ZL, Crawford AR, Ren Y, Toure J, Zchut S, Kochilas L, Creton R. Suppression of the endoplasmic reticulum calcium pump during zebrafish gastrulation affects left-right asymmetry of the heart and brain. Mech Dev. 2008;125:396–410. doi: 10.1016/j.mod.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Lagunowich LA, Stein AP, Reuhl KR. N-cadherin in normal and abnormal brain development. Neurotoxicology. 1994;15:123–132. [PubMed] [Google Scholar]
- Lowery LA, Sive H. Totally tubular: the mystery behind function and origin of the brain ventricular system. Bioessays. 2009;31:446–458. doi: 10.1002/bies.200800207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TM, Brough GH, Babal P, Kelly JJ, Li M, Stevens T. Store-operated calcium entry promotes shape change in pulmonary endothelial cells expressing Trp1. Am J Physiol. 1998;275:L574–L582. doi: 10.1152/ajplung.1998.275.3.L574. [DOI] [PubMed] [Google Scholar]
- Orrenius S, McCabe MJ, Nicotera P. Ca(2+)-dependent mechanisms of cytotoxicity and programmed cell death. Toxicol Lett. 1992;64–65:357–364. doi: 10.1016/0378-4274(92)90208-2. (Spec No) [DOI] [PubMed] [Google Scholar]
- Rhinn M, Brand M. The midbrain–hindbrain boundary organizer. Curr Opin Neurobiol. 2001;11:34–42. doi: 10.1016/s0959-4388(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- Slusarski DC, Pelegri F. Calcium signaling in vertebrate embryonic patterning and morphogenesis. Dev Biol. 2007;307:1–13. doi: 10.1016/j.ydbio.2007.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somlyo AP, Somlyo AV. Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: modulated by G proteins, kinases, and myosin phosphatase. Physiol Rev. 2003;83:1325–1358. doi: 10.1152/physrev.00023.2003. [DOI] [PubMed] [Google Scholar]
- Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. Expression of the zebrafish genome during embryogenesis. ZFIN Direct Data Submission. 2001 . Available at https://zfin.org/ZDB-PUB-010810-1 (accessed 13 February 2013) [Google Scholar]
- Thisse B, Thisse C. Fast release clones: A high throughput expression analysis. ZFIN Direct Data Submission. 2004 https://zfin.org/ZDB-PUB-040907-1 (accessed 13 February 2013) [Google Scholar]
- Thut CJ, Rountree RB, Hwa M, Kingsley DM. A large-scale in situ screen provides molecular evidence for the induction of eye anterior segment structures by the developing lens. Dev Biol. 2001;231:63–76. doi: 10.1006/dbio.2000.0140. [DOI] [PubMed] [Google Scholar]
- Tidow H, Nissen P. Structural diversity of calmodulin binding to its target sites. FEBS J. 2013;280:5551–5565. doi: 10.1111/febs.12296. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb SE, Miller AL. Calcium signalling during embryonic development. Nat Rev. 2003;4:539–551. doi: 10.1038/nrm1149. [DOI] [PubMed] [Google Scholar]
- Webb SE, Miller AL. Ca2+ signalling and early embryonic patterning during zebrafish development. Clin Exp Pharmacol Physiol. 2007;34:897–904. doi: 10.1111/j.1440-1681.2007.04709.x. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene: University of Oregon Press; 2000. [Google Scholar]
- Zhang J, Webb SE, Ma LH, Chan CM, Miller AL. Necessary role for intracellular Ca2+ transients in initiating the apical-basolateral thinning of enveloping layer cells during the early blastula period of zebrafish development. Dev Growth Differ. 2011;53:679–696. doi: 10.1111/j.1440-169X.2011.01275.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.