Gps1 is a regulator of Rho GTPases during cell division. Cell cycle–regulated recruitment of Gps1 to the cell division site is under control of the conserved kinase Gin4 and the bud neck–associated protein Nba1. This biphasic recruitment is required for the spatiotemporal activation of Rho1 and inhibition of Cdc42.
Abstract
Cell cycle–dependent morphogenesis of unicellular organisms depends on the spatiotemporal control of cell polarity. Rho GTPases are the major players that guide cells through structural reorganizations such as cytokinesis (Rho1 dependent) and polarity establishment (Cdc42 dependent). In budding yeast, the protein Gps1 plays a pivotal role in both processes. Gps1 resides at the bud neck to maintain Rho1 localization and restrict Cdc42 activity during cytokinesis. Here we analyze how Gps1 is recruited to the bud neck during the cell cycle. We show that different regions of Gps1 and the septin-associated kinase Gin4 are involved in maintaining Gps1 at the bud neck from late G1 phase until midanaphase. From midanaphase, the targeting function of Gin4 is taken over by the bud neck–associated protein Nba1. Our data show that Gps1 is targeted to the cell division site in a biphasic manner, via Gin4 and Nba1, to control the spatiotemporal activation of Rho1 and inhibition of Cdc42.
INTRODUCTION
Cytokinesis is the process that physically divides one cell into two at the end of mitosis. In animal and yeast cells, a contractile actomyosin ring drives plasma membrane ingression to execute cytokinesis (Balasubramanian et al., 2004; Barr and Gruneberg, 2007; Green et al., 2012; Wloka and Bi, 2012; Meitinger and Palani, 2016). In budding yeast cells, cytokinesis is also driven by the formation of a septum composed of cell wall material.
During yeast cytokinesis, the establishment of polarity at the cell division site (also called the bud neck) drives a complex reorganization of the actin cytoskeleton that is required for the targeted secretion of cell material (Howell and Lew, 2012). Rho GTPases are the master regulators of cell polarity (Park and Bi, 2007). Rho1 plays a critical role in cytokinesis by promoting formin-dependent actin cable formation (Kohno et al., 1996), recruitment of the landmark protein Sec3 to the bud neck for targeted secretion (Guo et al., 2001), and activation of the glucan synthase Fks1, which is important for cell wall deposition between the dividing cells (Qadota et al., 1996).
Cdc42 is another Rho GTPase important for the establishment of cell polarity (Johnson and Pringle, 1990). Like Rho1, Cdc42 accumulates at the bud neck during cytokinesis. After cell division, Cdc42 establishes a new polarity site, where the new daughter cell (also named the bud) emerges (Park and Bi, 2007). The positioning of this site depends on signals emanating from the previous bud neck. In haploid cells, the new polarity site is placed adjacent to the bud neck (Casamayor and Snyder, 2002). Like Rho1, Cdc42 accumulates at the bud neck during cytokinesis. So far, no function for Cdc42 at the bud neck has been described. In fact, recent work suggested that Cdc42 has to be locally inactivated by GTPase-activating proteins (GAPs) to allow proper execution of cytokinesis (Tong et al., 2007; Atkins et al., 2013; Onishi et al., 2013).
The bud neck–associated protein Gps1 was shown to be an upstream regulator of Rho1 and Cdc42 (Meitinger et al., 2013, 2014). Gps1 directly interacts with Rho1 at the bud neck, where it maintains active Rho1 after the contraction of the actomyosin ring. This is important in order to maintain Rho1 effectors (Fks1, Sec3, and Bni1) at the bud neck for completion of septum formation. Consequently the deletion of Gps1 leads to cell wall (secondary septum) thinning and cell lyses during cell separation (Meitinger et al., 2013). In addition, Gps1 prevents reactivation of Cdc42 at the bud neck during and after completion of cytokinesis. This function of Gps1 is mediated by the protein Nba1, which prevents the formation of a new bud at the site where cytokinesis took place (Meitinger et al., 2014). In the absence of Gps1, cells recurrently rebud at the same site of cell division. This causes the narrowing of the bud neck and reduces replicative lifespan, most likely due to aberrant nuclear segregation (Meitinger et al., 2014). Gps1 is recruited to the bud neck by unknown mechanisms. Here we investigated the molecular players involved in this Gps1 recruitment. We show that the bud neck–associated kinase Gin4 and the integral membrane protein (Rax1 and Rax2)–associated Nba1 target Gps1 to the bud neck in different cell cycle phases. In addition, we identify three distinct regions of Gps1 required for its temporal localization. Our data suggest that Gps1 is recruited to the cell division site in a biphasic manner to provide proper Rho1 and Cdc42 regulation.
RESULTS AND DISCUSSION
Gps1 is targeted to the bud neck and cell cortex by different motifs
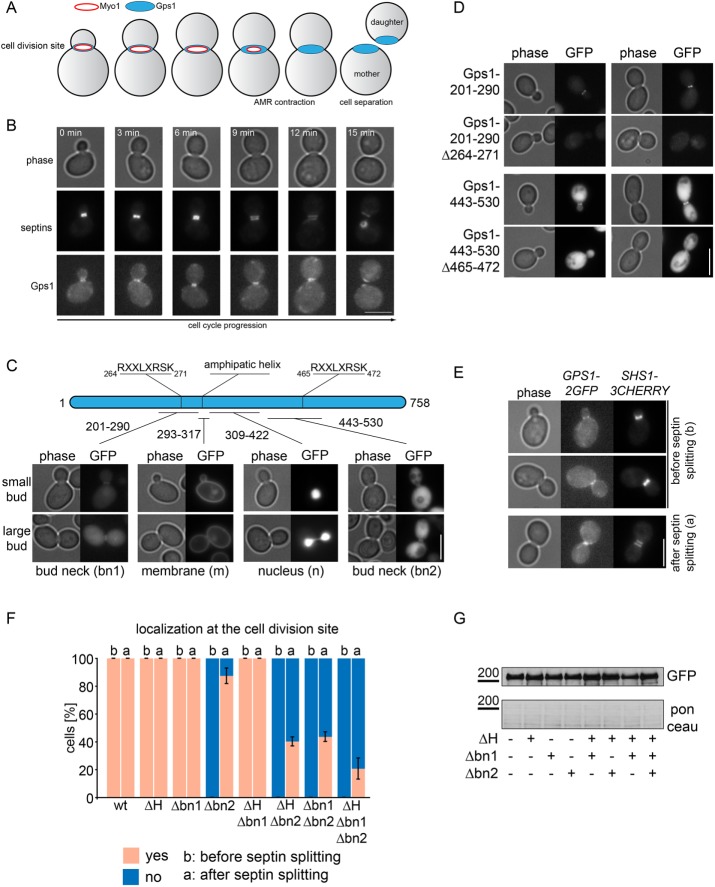
Gps1 localizes at the bud neck as a ring-like structure from G1 until the onset of cytokinesis (Figure 1, A and B; Meitinger et al., 2013; Wolken et al., 2014). During actomyosin ring contraction, Gps1 spreads over the closing bud neck and forms a disk-like structure at the daughter and mother side of the bud neck (Figure 1, A and B; Meitinger et al., 2013). Actomyosin ring contraction occurs after septin splitting, which is characterized by the reorganization of the septin structure from an hourglass shape into two distinct septin rings (Figure 1B). The process of septin splitting initiates during mitotic exit, which promotes mitotic cyclin-dependent kinase (Cdk) down-regulation (Lippincott et al., 2001). After septin splitting, the Gps1 disk becomes enclosed by the septin ring (Figure 1B; Meitinger et al., 2013).
FIGURE 1:
Gps1 localizes at the bud neck before and after septin splitting. (A) Schematic representation of Gps1 and Myo1 localization in cells at different stages of the cell cycle. (B) Still images of time-lapse series showing Gps1-GFP at the cell division site before and after septin splitting. The septin collar was stained by the septin Shs1-mCherry. Septin splitting (two distinct Shs1-mCherry collars) is observed at 9 min. Scale bar, 5 µm. (C) The fragments of Gps1 that can localize independently to the bud neck (bn1 and bn2), the membrane (m), and the nucleus (n) in gps1∆ cells and their corresponding localization as GFP fusions. The numbers indicate amino acid positions. Scale bar, 5 µm. (D) Localization of Gps1201–290 and Gps1443–530 depends on the RXXLXRSK motif in both fragments. The bud neck localization of Gps1201–290-GFP and Gps1443–530-GFP was analyzed before and after deletion of bn1 (codons 264–271) and bn2 (codons465–472), respectively. Scale bar, 5 µm. (E, F) The localization of Gps1 wild type and indicated Gps1 deletion mutants is quantified in F before and after septin splitting as indicated in E. Scale bar; 5 µm; 100 cells/strain. Error bars are SD of three independent experiments. (G) The immunoblot shows the protein levels Gps1-GFP wild type and indicated deletion mutants fused to GFP. The Ponceau S–stained membrane serves as a loading control.
To investigate how Gps1 associates with the bud neck before and after septin splitting, we analyzed the subcellular localization of a series of Gps1 truncation mutants (Supplemental Figure S1). We found that two fragments of Gps1, which comprise amino acids 201–290 and 443–530 (Supplemental Figure S1), respectively, localized to the bud neck in gps1∆ cells (Figure 1C). A Gps1 fragment with a predicted amphipathic helix (amino acids 293–317) strongly localized at the plasma membrane (Figure 1C), whereas amino acids 309–422 but not full-length Gps1 showed a strong nuclear localization (Figure 1C). Thus Gps1 carries a number of targeting signals that direct the protein to the bud neck and plasma membrane.
By comparing the amino acid composition of the Gps1 truncations 201–290 and 443–530, we found that both sequences carried a common feature defined by RXXLXRSK (where X is any amino acid). We named this motif bud neck 1 (bn1; amino acids 264–271) and 2 (bn2; amino acids 465–472). Deletion of any of these eight amino acids impaired bud neck localization of the Gps1aa201–290 and Gps1aa443–530 fragments (Figure 1D). This implies that Gps1 is targeted to the bud neck by the bn1 and bn2 motifs.
To investigate the importance of bn1 and bn2 motifs for Gps1 localization, we quantified cells with Gps1 at the bud neck before and after septin splitting (Figure 1E). We found that bn2 was important to target Gps1 to the bud neck mainly before septin splitting (Figure 1F). After septin splitting, deletion of bn2 decreased, but not completely abolished, Gps1 bud neck localization (Figure 1F). The deletion of bn1 or the predicted amphipathic helix (∆H) had no effect on Gps1 localization. However, both deletions increased the observed defect of the deletion of bn2 on Gps1 localization after septin splitting (Figure 1F). The expression of the analyzed Gps1 mutants was comparable to that for wild-type cells (Figure 1G), indicating that the loss of localization was not related to decrease in protein stability. Our data suggest that multiple motifs are responsible for targeting Gps1 to the bud neck before and after septin splitting. A genome-wide pattern search identified five proteins (www.yeastgenome.org/cgi-bin/PATMATCH/nph-patmatch; Gps1, Zds2, Mso1, Bpt1, and Nth1) containing the RXXLXRSK sequence. Three of these five proteins (Gps1, Zds2, and Mso1) were reported to localize at the bud neck (Rossio and Yoshida, 2011; Tkach et al., 2012). Thus the RXXLXRSK motif, or even a less stringent sequence, might be a more general bud neck–recruiting motif.
Targeting of Gps1 to the bud neck is essential for function
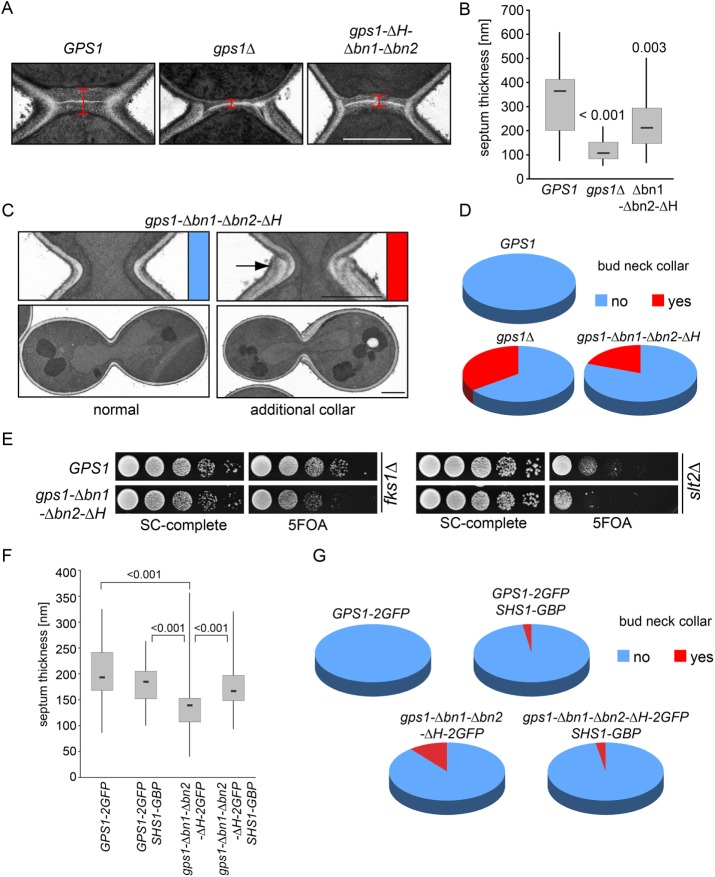
Gps1 is the factor that maintains Rho1 activity at the bud neck, which is necessary for secondary septum formation (Figure 2A; Meitinger et al., 2013, 2014). It also prevents Cdc42 activation at the bud neck and so ensures that cells always select a new bud site for each division (Meitinger et al., 2013, 2014). gps1∆ cells fail to suppress Cdc42 activation at the bud neck, resulting in the appearance of an additional collar around the bud neck that can be visualized by transmission electron microscopy (TEM; Meitinger et al., 2013, 2014). We therefore expected the bud-neck pool of Gps1 to control Rho1 or Cdc42 function. In line with this notion, we found that the mislocalization of Gps1 mutant protein lacking bn1, bn2, and the predicted amphipathic helix (Gps1-∆bn1-∆bn2-∆H) caused defects in the Rho1 (Figure 2, A and B) and Cdc42 pathways (Figure 2, C and D). Furthermore, like gps1∆, this mutant showed a synthetic growth defect with the cell wall mutant fks1∆ and the cell wall integrity pathway mutant slt2∆ (Figure 2E; Meitinger et al., 2013). These results indicate that Gps1-∆bn1-∆bn2-∆H has lost its function as a regulator of Rho1 and Cdc42.
FIGURE 2:
Gps1 localization at the bud neck is important for its function in the Rho1 and Cdc42 pathway. (A) Electron micrographs of the bud neck region of wild type, gps1∆, and gps1-∆bn1-∆bn2-∆H. Red bar, median thickness of the septum. Scale bar, 1 µm. (B) Box-and-whiskers plots show the quantification of the thickness of the septum of wild-type (n = 27), gps1∆ (n = 37), and gps1-∆bn1-∆bn2-∆H (n = 41) cells. The upper and lower boundaries in the boxes represent the third and first quartiles, respectively, and the lines within the boxes show the median values. (C) Electron micrographs of cells and their bud neck region with a normal bud neck (red) and an additional collar around the bud neck (blue, arrowhead). Scale bars, 1 µm. (D) Pie charts show the quantification of the phenotypes shown in C. (E) Genetic interactions of gps1-∆bn1-∆bn2-∆H with fks1∆ and slt2∆. Tenfold serial dilutions of strains with the indicated phenotype were spotted on SC-complete and 5-FOA plates (see Materials and Methods). (F, G) Box-and-whiskers plots show the quantification of the thickness of the septum (F) and the collar formation (G) of GPS1-2GFP (n = 59), GPS1-2GFP SHS1-GBP (n = 62), gps1-∆bn1-∆bn2-∆H-2GFP (n = 68), and gps1-∆bn1-∆bn2-∆H-2GFP SHS1-GBP (n = 42) cells.
Gps1-∆bn1-∆bn2-∆H may be nonfunctional because the deletions affect the folding of the protein. Alternatively, the mislocalization of Gps1-∆bn1-∆bn2-∆H may be the cause of the phenotype. In the latter case, artificial targeting of Gps1-∆bn1-∆bn2-∆H to the bud neck should be able to rescue Cdc42 and Rho1 regulation. To test this notion, we used a previously established strategy to constitutively target Gps1-∆bn1-∆bn2-∆H-GFP to the bud neck using a strain carrying the septin Shs1 fused to the green fluorescent protein (GFP)–binding protein (GBP; Meitinger et al., 2013). The binding between GFP and GBP allowed us to recruit Gps1-∆bn1-∆bn2-∆H-GFP to the bud neck independently of the bn1, bn2, and H motifs. We observed that the combination of Gps1-GFP and Shs1-GBP interfered only mildly with Rho1 and Cdc42 regulation at the bud neck (Figure 2, F and G). Of interest, the observed defects of bn1, bn2, and H deletion were rescued by targeting Gps1-∆bn1-∆bn2-∆H-GFP to the bud neck (Figure 2, F and G). This result clearly demonstrates that regions bn1, bn2, and H are essential for Gps1 targeting to the bud neck but not for Gps1 activity toward Rho1 and Cdc42. This data also show that Gps1 has to localize at the bud neck to spatially control the Rho1 and Cdc42 pathways.
The septin-associated kinase Gin4 recruits Gps1 to the bud neck
We next sought to identify upstream regulators of Gps1 bud neck recruitment. It was reported that septins recruit Gps1 to the bud neck (Wolken et al., 2014). Thus we first investigated whether the bud neck targeting of Gps1 required a functional septin ring. We analyzed the localization of Gps1 full-length (Gps1-FL, codons 1–758) and truncated forms Gps1-CT1 (codons 201–290), Gps1-CT2 (codons 443–530), and Gps1-CT3 (codons 293–318; Figure 3A) in the temperature-sensitive septin (cdc12-6) mutant cells. Cdc12 was essential for the binding of Gps1-FL or Gps1-CT2 to the bud neck but not for the plasma membrane binding of Gps1-CT3 (Figure 3, B and C). The signal of Gps1-CT1 at the bud neck was not detectable at 37°C and therefore was not analyzed. Our data indicate that Gps1 is recruited to the septin scaffold via amino acids 443–530. In contrast, plasma membrane binding of Gps1 (Gps1-CT3) was septin independent.
FIGURE 3:
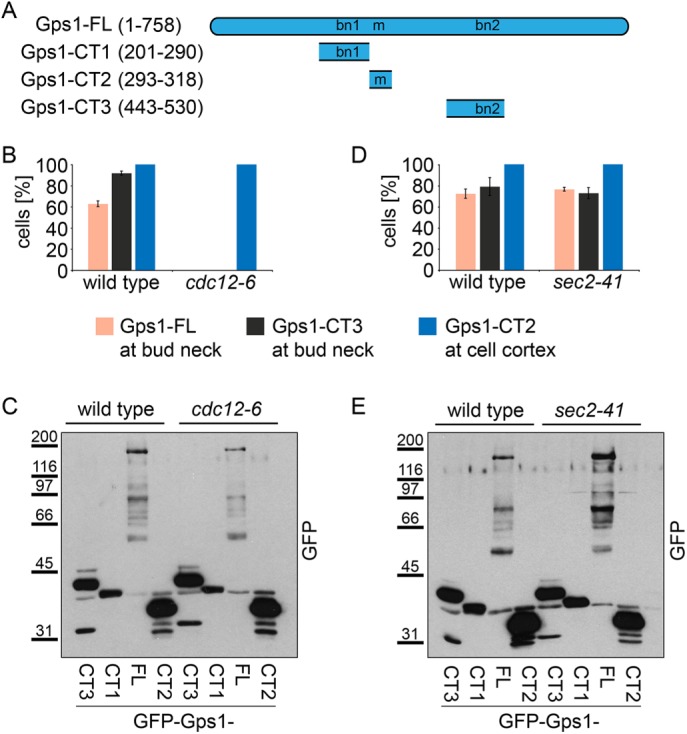
Gps1 localization at the bud neck depends on septins. (A) Schematic representation of Gps1 full-length and truncated constructs. Numbers indicate amino acid positions. bn1, bn2, bud neck regions 1 and 2; m, membrane-binding region. (B, D) Quantification of cells with the indicated localization of Gps1 full length and fragments fused to GFP in wild type and temperature-sensitive cdc12-6 (B) or sec2-41 (D) mutants. Cells were grown at 37°C for 3 h; 100–150 cells/strain; error bars are SD from three independent experiments. (C, E) Immunoblot shows the protein levels of GFP-GPS1, GFP-gps1-443-530, GFP-gps1-293-318, and GFP-gps1-201-290 in wild-type and cdc12-6 (C) or sec2-41 E) cells grown at 37°C for 3 h.
We next ask whether the septin scaffold could indirectly recruit Gps1 via its role in secretion (Barral et al., 2000). To address this question, we used the temperature-sensitive mutant sec2-41. Sec2 is an activator of the small GTPase Sec4, which is involved in targeted vesicular transport to the bud neck (Walch-Solimena et al., 1997). We found that the localization of Gps1-FL or Gps1-CT3 to the bud neck or Gps1-CT2 to the plasma membrane was Sec2 independent (Figure 3, D and E). This finding indicates that secretion is not required for Gps1 bud neck targeting. We thus postulate that septins may have a more direct role in Gps1 localization.
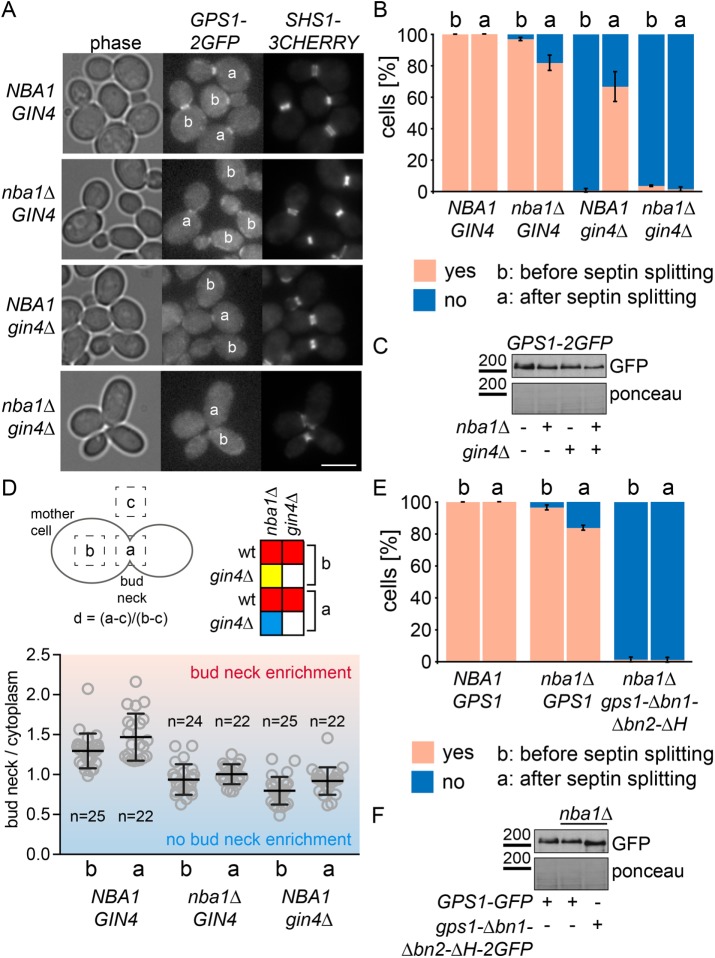
Several protein complexes are anchored to the bud neck via binding to the septin scaffold (Oh and Bi, 2010). Because Gps1 and septins did not interact in the yeast two-hybrid system (unpublished data), we considered the possibility that Gps1 binds indirectly to the septin scaffold via a septin-interacting protein. To identify such binding mediators, we determined Gps1 localization in strains carrying gene deletions of septin-associated components. Deletion of most genes, including MYO1 and BNI5, coding for proteins with scaffolding functions at the bud neck (Fang et al., 2010; Wloka et al., 2013), did not affect Gps1 bud neck localization (Supplemental Figure S2). In striking contrast, the septin-associated kinase GIN4 (Carroll et al., 1998; Mortensen et al., 2002) was essential for the recruitment of Gps1-FL, as well as of the Gps1-CT1 or Gps1-CT3 fragments, to the bud neck before septin splitting (Supplemental Figure S2 and Figure 4, A–C). Consistently, the kinase Elm1, which is an upstream activator of Gin4 (Asano et al., 2006), also influenced Gps1 bud neck recruitment (Supplemental Figure S2). This indicates that the Elm1-Gin4 pathway targets Gps1 to the bud neck at early stages of mitosis. However, after septin splitting, the bud neck–associated protein Nba1 became important to maintain Gps1 at this location (Figure 4, A–C). Because the proportion of cells with split septin rings are rare in a growing cell population, the role of Nba1 in Gps1 localization has been previously overlooked (Meitinger et al., 2014). The role of Nba1 in Gps1 localization became even more apparent in the gin4∆ nba1∆ double mutant cells, where Gps1 completely failed to bind to the bud neck (Figure 4, A–C).
FIGURE 4:
Gps1 is targeted to the bud neck by Gin4 and Nba1. (A) Examples of wild-type, nba1∆, gin4∆, and nba1∆ gin4∆ cells expressing GPS1-2GFP SHS1-Cherry before (b) or after (a) septin splitting. Scale bar, 5 µm. (B) Quantification of Gps1-GFP at the bud neck before and after septin splitting in wild-type, gin4∆, nba1∆, and gin4∆ nba1∆ cells (n > 100 for each condition; error bars are SD from three independent experiments). (C) Immunoblot shows the protein levels of GPS1-GFP in wild-type, gin4∆, nba1∆, and gin4∆ nba1∆ cells. The Ponceau S–stained membrane serves as a loading control. (D) Quantification of Gps1-2GFP bud neck enrichment (ratio bud neck/cytoplasm) before (b) or after (a) septin splitting. Graph shows mean values; error bars are SD; red, p < 0.001; yellow, p < 0.01; blue, p = 0.07. (E) Quantification of cells with Gps1-GFP and Gps1-∆bn1-∆bn2-∆H-GFP at the bud neck before and after septin splitting in wild-type and nba1∆ cells (n > 100 for each condition; error bars are SD from three independent experiments). (F) Immunoblot shows the proteins levels of GPS1-GFP and gps1-∆bn1-∆bn2-∆H-GFP in wild-type and nba1∆ cells. The Ponceau S–stained membrane serves as a loading control.
Although the majority of nba1∆ cells showed Gps1-2GFP at the bud neck before septin splitting, the level of bud neck–enriched Gps1-2GFP was significantly lower in nba1∆ than in wild-type cells (Figure 4, A and D). In gin4∆ cells, the levels of Gps1 at the bud neck were strongly reduced before septin splitting (no Gps1 enrichment visible) and less strongly reduced after septin splitting (Gps1 enrichment visible; Figure 4, A and D). These data indicate that both Gin4 and Nba1 are required for maximal levels of Gps1 bud neck recruitment irrespective of cell cycle stage.
Mislocalization of Gps1 in gin4∆ nba1∆ cells was similar to the localization of the Gps1-∆bn1-∆bn2-∆H protein in GIN4 nba1∆ cells (Figure 4, B, E, and F). In addition, we observed an interaction between Gps1 and Gin4 in the yeast two-hybrid system. This interaction was lost in mutants lacking the bn1 and bn2 regions (Figure 5A). The known interactions of Gps1 with Rho1, Cdc42, Nba1, Nap1, and Nis1 (Meitinger et al., 2013, 2014) were not affected upon deletion of bn1 and bn2 (Figure 5A). Thus it is likely that Gin4 specifically recruits Gps1 via bn1 and bn2 to the bud neck.
FIGURE 5:
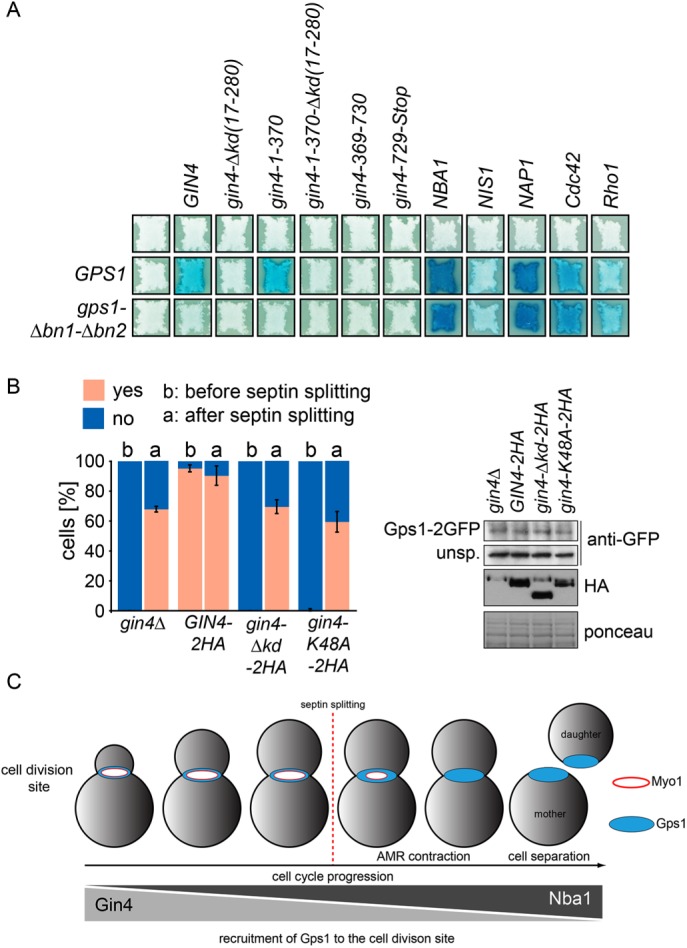
Gin4 interacts with and recruits Gps1 to the bud neck in a kinase-dependent manner. (A) Yeast two-hybrid assay between Gps1 or Gps1-∆bn1-∆bn2 and the indicated interaction partners. Blue indicates interaction. (B) Quantification of cells with Gps1-GFP at the bud neck before (b) and after (a) septin splitting in gin4∆ cells expressing GIN4-2HA, gin4-∆kd-2HA, or gin4-K48A-2HA (n > 100 for each strain; error bars are SD from three independent experiments). Immunoblot shows the protein levels of GIN4-2HA, gin4-∆kd-2HA, and gin4-K48A-2HA. (C) Schematic representation of Gps1 localization at the bud neck during the cell cycle. During early phases of the cell cycle, Gps1 recruitment to the bud neck mainly depends on Gin4. After mitotic exit and septin splitting, Gps1 localization at the bud neck mainly depends on Nba1. Both Gin4 and Nba1 are important, however, to maintain high levels of Gps1 at the bud neck.
The Gin4 kinase domain is important to recruit Gps1 to the bud neck
Gin4 consists of an N-terminal kinase domain and a C-terminal tail (Iwase and Toh-e, 2004; Buttery et al., 2012). Because Gin4 has been shown to have kinase-dependent and -independent functions at the bud neck (Longtine et al., 1998; Buttery et al., 2012), we asked whether the catalytic domain of Gin4 is required for Gps1 localization. To test this notion, we first analyzed the interaction of Gps1 with truncations of Gin4. We found that the interaction between Gin4 and Gps1 was specific to the kinase domain (Figure 5A). Next, we analyzed the localization of Gps1 in a Gin4 kinase-dead mutant (Gin4-K48A) and in a Gin4 mutant lacking the kinase domain (Gin4-∆kd; Altman and Kellogg, 1997; Iwase and Toh-e, 2004; Buttery et al., 2012). In line with our interaction studies, we found that the localization of Gps1 at the bud neck was lost in gin4-∆kd and gin4-K48A mutants (Figure 5B). In conclusion, our data show that Gin4 recruits Gps1 to the bud neck in a manner that depends on its kinase activity and the Gps1 bud neck–targeting motifs bn1 and bn2. Gin4 may phosphorylate either Gps1 directly or a bud neck–associated factor that tethers Gps1 to the septin complex. This targeting mechanism is important to control Rho1 and Cdc42 activity at the bud neck.
In conclusion, we established two mechanisms that control the targeting of Gps1 to the bud neck. At initial phases of the cell cycle, mainly the kinase Gin4 promotes the association of Gps1 with the septin ring (Figure 5C). In late anaphase, after septin splitting and actomyosin ring contraction, the ring-like Gps1 signal spreads over the closing bud neck, thereby losing its association with septins. During this phase, Nba1 and the predicted amphipathic helix region of Gps1 become essential to stabilize Gps1 at the plasma membrane as a disk-like structure. We propose that similar to Myo1 (Fang et al., 2010), Gps1 is targeted to the bud neck in a biphasic manner. This might be a general principle to target proteins to the bud neck before and after mitotic exit.
MATERIALS AND METHODS
Yeast strains and plasmid construction
Yeast strains and plasmids used in this study are listed in Supplemental Tables S1 and S2, respectively. Genes were expressed from their endogenous promoter and locus unless specified otherwise. Gene deletions and epitope tagging were performed using PCR-based methods (Knop et al., 1999; Janke et al., 2004). Gps1 wild type or mutants fused to 2GFP-kanMX6 were integrated into the genome. For this, the corresponding plasmids (pMF650, 755, 793, 794, 795, 796, 798, and 799) were first digested with NheI and NotI. The fragments were purified and transformed into MFY1588. After selection on G418-containing plates, positive clones were selected by microscope and confirmed by immunoblotting. Afterward, the wild type containing URA3-based plasmid was removed by negative selection on 5FOA plates. For localization studies shown in Figure 2, A and B, and Supplemental Figure S1, GPS1 fragments were under the control of the Gal1 promoter and expressed from a CEN plasmid in a gps1∆ strain. GIN4-2HA, gin4-∆kinase-2HA (Iwase and Toh-e, 2004), and gin4-K48A-2HA were expressed from a CEN plasmid in a gin4∆ strain. For yeast two-hybrid assay, the genes and gene fragments were cloned into pMM5 or pMM6 (Schramm et al., 2001).
To generate the plasmid pMF650, GPS1 together with promoter and terminator region was amplified from genomic DNA (strain ESM356) using primers GCGGGATCCCTTCTACTTCTGTCTGTTGC and TCCGGGTACCGAATAAATGGACGAGCTAGC. The PCR product was digested with BamHI/KpnI and cloned into pRS316 (Sikorski and Hieter, 1989). The GPS1-2GFP fusion was generated by gap repair (Sherman, 1991). For this, the plasmid pRS316-GPS1 was digested with SnaBI/EcoRI and transformed into the yeast strain ESM356 GPS1-2GFP-kanMX6. The resulting plasmid was purified from a single yeast clone and confirmed by sequencing.
We used PCR to delete regions of GPS1 in pMF650, pMF726, and pMF736 to create pMF755, pMF774, pMF775, and pMF793-pMF799. We used the primers CCTCATTGGGAATCAAGATACTGTTTTCATATAATGTTCA and TGAACATTATATGAAAACAGTATCTTGATTCCCAATGAGG (to delete residues 293–299), GCAGTAAATCAAGTAGCAGTGCTATTAGATGTAAGGGCGG and CCGCCCTTACATCTAATAGCACTGCTACTTGATTTACTGC (to delete residues 264–217), and GCAACAGCAGCATGTACTCGCCATCGACGGCCCTAAATGC and GCATTTAGGGCCGTCGATGGCGAGTACATGCTGCTGTTGC (to delete residues 465–472).
GPS1 full length and truncations were cloned by PCR into pRS315-pGal1-GFP (gift from Michael Knop, University of Heidelberg, Heidelberg, Germany) to create plasmids pMF726, pMF730, pMF736, and pMF739. Plasmid pMF726 was cloned with primers GCGGGATCCATATGAACTCCTTCATTTCTGTACCG and TCCGGTCGACAGGCGATATCGCCATTAAGG (BamHI/SalI), and three stop codons were subsequently introduced after residue 530 with primers TGCTCATCTGATTATTAATGATAATCATCTGAAAGCTAC and GTAGCTTTCAGATGATTATCATTAATAATCAGATGAGCA. Plasmid pMF730 was cloned with primers AAAGGATCCCAGGAATGATCATCAGAGCTCC and TCCGCCCGGGGAATAAATGGACGAGCTAGC (BamHI/XmaI). Plasmid pMF736 was cloned with primers GCGGGATCCATATGCAAGAAGCCAAACCGATGAC and TCCGCCCGGGAGGCGATATCGCCATTAAGG (BamHI/XmaI), and three stop codons were subsequently introduced after residue 290 with primers TTGGGAATCAAGATATGATAATAATTACGGAAATTACGG and CCGTAATTTCCGTAATTATTATCATATCTTGATTCCCAA. Plasmid pMF739 was cloned with primers GCGGGATCCATATGAAGTTACGGAAATTACGGCTAG and TCCGGTCGACTTAATTCGTGGCGGAGGGGAC (BamHI/SalI). Primers CAAGAGGCGGCAGTTGCGGTAATATCAAAAGCAG and CTGCTTTTGATATTACCGCAACTGCCGCCTCTTG were used to introduce the mutation K48A in GIN4 (pMF1328).
Yeast growth conditions
Yeast strains were grown in yeast/peptone/dextrose medium containing 0.1 mg/l adenine (YPAD) as described (Sherman, 1991). The temperature-sensitive yeast strains cdc12-6 (kind gift of Daniel Lew, Duke University, Durham, NC) and sec2-41 (kind gift of Anne Spang, Biocenter, University of Basel, Basel, Switzerland) were grown at 23°C until they reached OD600 = 0.5 and then shifted to 37°C for 3 h before inspection. Strains carrying plasmids were grown in synthetic complete (SC) medium lacking the corresponding amino acids.
Genetic interactions based on growth
Synthetic growth defects were analyzed using the plasmid shuffle strategy. Therefore mutant strains containing an URA3-based plasmid (pRS316) with the corresponding wild-type gene were subjected to a growth test on 5-fluoroorotic acid (5-FOA) plates (selection against URA3). For each mutant, six or more individual transformants were analyzed. Tenfold serial dilutions of wild type (ESM356-1) and one representative mutant with the indicated genotype were spotted onto SC and 5-FOA plates. Mutants were complemented by pRS316-GPS1 (Figures 3E). Plates were incubated for 2 d at 30°C.
Protein detection methods and quantifications
Western blotting of yeast extracts was performed as described (Janke et al., 2004). Primary antibodies were rabbit anti-GFP, mouse anti-hemagglutinin (clone 12CA5; Sigma-Aldrich), and mouse anti-Myc (clone 9E10; Sigma-Aldrich). Secondary antibodies were goat anti-mouse and goat anti-rabbit immunoglobulins G coupled to horseradish peroxidase (Jackson ImmunoResearch Laboratories).
Yeast two-hybrid assay
Plasmids pMM5 (fusion to LexA) and pMM6 (fusion to Gal4) containing the indicated genes and gene fragments were transformed into the yeast strains YPH500 (MATalpha) and SGY37 (MATa), respectively. The plasmids containing YPH500 and SGY37 strains were matted, and the yeast two-hybrid assay was performed as described (Schramm et al., 2001). Interacting proteins allow the expression of β-galactosidase, which converts X-Gal into a blue-detectable product.
Microscopy techniques
Cells were fixed in 4% formaldehyde for 20 min and washed with phosphate-buffered saline before inspection of fluorescence signals (GFP or Cherry). Live-cell imaging and quantification of fluorescence still images were performed as described (Meitinger et al., 2011). Briefly, for the quantification and image preparation, three to five z-sections of the region of interest were averaged or maximum projected, respectively. At least 100 cells were counted for each strain, experiment, and each time point shown in Figures 1F, 3, B and D, 4, B and E, and 5B. Specimens for electron microscopy were prepared as described previously (Meitinger and Pereira, 2016).
Supplementary Material
Acknowledgments
We thank David Pellman, Anne Spang, Michael Knop, and Daniel Lew for reagents; Birgit Hub, Karsten Richter, and the German Cancer Research Center Electron Microscopy Facility for support with electron microscopy; Astrid Hofmann for technical support; and members of G.P.’s lab for comments on the manuscript. The work of G.P. is financed by the Heisenberg Programme of the German Research Council (PE1883-3) and German Research Council Collaborative Research Grant SFB1036.
Abbreviations used:
- GBP
GFP-binding protein
- TEM
transmission electron microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-09-0687) on February 1, 2017.
REFERENCES
- Altman R, Kellogg D. Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol. 1997;138:119–130. doi: 10.1083/jcb.138.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano S, Park JE, Yu LR, Zhou M, Sakchaisri K, Park CJ, Kang YH, Thorner J, Veenstra TD, Lee KS. Direct phosphorylation and activation of a Nim1-related kinase Gin4 by Elm1 in budding yeast. J Biol Chem. 2006;281:27090–27098. doi: 10.1074/jbc.M601483200. [DOI] [PubMed] [Google Scholar]
- Atkins BD, Yoshida S, Saito K, Wu CF, Lew DJ, Pellman D. Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J Cell Biol. 2013;202:231–240. doi: 10.1083/jcb.201301090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806–R818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Barral Y, Mermall V, Mooseker MS, Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- Buttery SM, Kono K, Stokasimov E, Pellman D. Regulation of the formin Bnr1 by septins and a MARK/Par1-family septin-associated kinase. Mol Biol Cell. 2012;23:4041–4053. doi: 10.1091/mbc.E12-05-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Altman R, Schieltz D, Yates JR, Kellogg D. The septins are required for the mitosis-specific activation of the Gin4 kinase. J Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A, Snyder M. Bud-site selection and cell polarity in budding yeast. Curr Opin Microbiol. 2002;5:179–186. doi: 10.1016/s1369-5274(02)00300-4. [DOI] [PubMed] [Google Scholar]
- Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, Iwase M, Vallen EA, Bi E. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J Cell Biol. 2010;191:1333–1350. doi: 10.1083/jcb.201005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Guo W, Tamanoi F, Novick P. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat Cell Biol. 2001;3:353–360. doi: 10.1038/35070029. [DOI] [PubMed] [Google Scholar]
- Howell AS, Lew DJ. Morphogenesis and the cell cycle. Genetics. 2012;190:51–77. doi: 10.1534/genetics.111.128314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M, Toh-e A. Ybr267w is a new cytoplasmic protein belonging to the mitotic signaling network of Saccharomyces cerevisiae. Cell Struct Funct. 2004;29:1–15. doi: 10.1247/csf.29.1. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, et al. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Shannon KB, Shou W, Deshaies RJ, Li R. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J Cell Sci. 2001;114:1379–1386. doi: 10.1242/jcs.114.7.1379. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Fares H, Pringle JR. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Gen Dev. 2011;25:875–888. doi: 10.1101/gad.622411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Khmelinskii A, Morlot S, Kurtulmus B, Palani S, Andres-Pons A, Hub B, Knop M, Charvin G, Pereira G. A memory system of negative polarity cues prevents replicative aging. Cell. 2014;159:1056–1069. doi: 10.1016/j.cell.2014.10.014. [DOI] [PubMed] [Google Scholar]
- Meitinger F, Palani S. Actomyosin ring driven cytokinesis in budding yeast. Semin Cell Dev Biol. 2016;53:19–27. doi: 10.1016/j.semcdb.2016.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Pereira G. Visualization of cytokinesis events in budding yeast by transmission electron microscopy. Methods Mol Biol. 2016;1369:87–95. doi: 10.1007/978-1-4939-3145-3_7. [DOI] [PubMed] [Google Scholar]
- Meitinger F, Richter H, Heisel S, Hub B, Seufert W, Pereira G. A safeguard mechanism regulates Rho GTPases to coordinate cytokinesis with the establishment of cell polarity. PLoS Biol. 2013;11:e1001495. doi: 10.1371/journal.pbio.1001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen EM, McDonald H, Yates J, 3rd, Kellogg DR. Cell cycle-dependent assembly of a Gin4-septin complex. Mol Biol Cell. 2002;13:2091–2105. doi: 10.1091/mbc.01-10-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Bi E. Septin structure and function in yeast and beyond. Trends Cell Biol. 2010;21:141–148. doi: 10.1016/j.tcb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Ko N, Nishihama R, Pringle JR. Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J Cell Biol. 2013;202:311–329. doi: 10.1083/jcb.201302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Python CP, Inoue SB, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin DE, Ohya Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- Rossio V, Yoshida S. Spatial regulation of Cdc55-PP2A by Zds1/Zds2 controls mitotic entry and mitotic exit in budding yeast. J Cell Biol. 2011;193:445–454. doi: 10.1083/jcb.201101134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm C, Janke C, Schiebel E. Molecular dissection of yeast spindle pole bodies by two hybrid, in vitro binding, and co-purification. Methods Cell Biol. 2001;67:71–94. doi: 10.1016/s0091-679x(01)67006-7. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C, et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z, Gao XD, Howell AS, Bose I, Lew DJ, Bi E. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J Cell Biol. 2007;179:1375–1384. doi: 10.1083/jcb.200705160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Collins RN, Novick PJ. Sec 2p mediates nucleotide exchange on Sec 4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C, Bi E. Mechanisms of cytokinesis in budding yeast. Cytoskeleton. 2012;69:710–726. doi: 10.1002/cm.21046. [DOI] [PubMed] [Google Scholar]
- Wloka C, Vallen EA, The L, Fang X, Oh Y, Bi E. Immobile myosin-II plays a scaffolding role during cytokinesis in budding yeast. J Cell Biol. 2013;200:271–286. doi: 10.1083/jcb.201208030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolken DM, McInnes J, Pon LA. Aim44p regulates phosphorylation of Hof1p to promote contractile ring closure during cytokinesis in budding yeast. Mol Biol Cell. 2014;25:753–762. doi: 10.1091/mbc.E13-06-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.