A donor knock-in vector optimized for the CRISPR/Cas9 system is constructed and a practical system developed that enables efficient disruption of cilia-related genes by exploiting homology-independent repair. A second version of the system can be used to reduce off-target cleavage frequency and increase versatility.
Abstract
The CRISPR/Cas9 system has revolutionized genome editing in virtually all organisms. Although the CRISPR/Cas9 system enables the targeted cleavage of genomic DNA, its use for gene knock-in remains challenging because levels of homologous recombination activity vary among various cells. In contrast, the efficiency of homology-independent DNA repair is relatively high in most cell types. Therefore the use of a homology-independent repair mechanism is a possible alternative for efficient genome editing. Here we constructed a donor knock-in vector optimized for the CRISPR/Cas9 system and developed a practical system that enables efficient disruption of target genes by exploiting homology-independent repair. Using this practical knock-in system, we successfully disrupted genes encoding proteins involved in ciliary protein trafficking, including IFT88 and IFT20, in hTERT-RPE1 cells, which have low homologous recombination activity. The most critical concern using the CRISPR/Cas9 system is off-target cleavage. To reduce the off-target cleavage frequency and increase the versatility of our knock-in system, we constructed a universal donor vector and an expression vector containing Cas9 with enhanced specificity and tandem sgRNA expression cassettes. We demonstrated that the second version of our system has improved usability.
INTRODUCTION
The development of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) technology has revolutionized genome editing in virtually all organisms (Doudna and Charpentier, 2014; Hsu et al., 2014). The programmable RNA-guided endonuclease Streptococcus pyogenes Cas9 (SpCas9) induces DNA double-strand breaks (DSBs) at target sites in the genome. These DSBs then trigger DNA repair mechanisms, which can be classified into two categories: homology-dependent and homology-independent repair (Jasin and Haber, 2016; Mladenov et al., 2016). Homology-dependent DNA repair occurs mainly through homologous recombination (HR), whereas homology-independent DNA repair (HIDR) occurs through nonhomologous end joining (NHEJ). These DNA repair systems have both advantages and disadvantages. Although HR can achieve precise repair, the repair rate is relatively low because it occurs in limited periods during the cell cycle, namely, the late S to G2 phases (which is approximately one-third of the period of the overall cell cycle). On the other hand, the rate of HIDR is high, although it is error-prone. One of the reasons for this high rate is that NHEJ can be performed during all phases of the cell cycle. Therefore DNA repair strategies should be selected in accordance with the actual purpose of genome editing.
hTERT-RPE1 (hereafter referred to as RPE1) is a cell line that was derived from human normal retinal pigment epithelial (RPE) cells, which were immortalized by the exogenous expression of human telomerase reverse transcriptase (hTERT; Bodnar et al., 1998; Jiang et al., 1999). Distinctive features of RPE1 cells are their high potential for ciliogenesis and normal karyotype. Because RPE1 cells efficiently form primary cilia under serum-starved conditions, these cells have been widely used for studies of ciliogenesis and ciliary function. Many culture cell lines, such as HeLa and HEK293T, are abnormal with respect to the number of chromosomes; recent sequencing of the HeLa genome demonstrated a high level of aneuploidy (Landry et al., 2013), and this can cause difficulties in genome editing because the copy number of the target genes is not always two. Therefore immortalized normal diploid cell lines, such as RPE1, are highly useful for genome editing.
HR activity varies among different cell lines, and, in general, it is lower in normal cell lines than in cancer cell lines. Because RPE1 cells retain characteristics of normal primary cells, HR activity is believed to be low in these cells. In fact, Miyamoto et al. (2015) reported that the establishment of biallelic KIF2A knockout (KO) lines from RPE1 cells was difficult, although not impossible; sequential gene targeting with two distinct targeting vectors containing the neomycin- and puromycin-resistance genes was required to establish the biallelic KO cell lines. Therefore the development of alternative genome editing methods for normal cell lines that do not require HR is highly anticipated. Examples of successful genome editing using the HIDR pathway have been reported for mammalian cells (Cristea et al., 2013; Maresca et al., 2013; Lackner et al., 2015; Geisinger et al., 2016), zebrafish (Auer and Del Bene, 2014; Auer et al., 2014; Kimura et al., 2014), and nematode (Katic et al., 2015). These studies reported efficient knock-in of a donor vector into the target genome DNA by concurrent cleavage of a donor plasmid and genomic DNA, followed by HIDR.
To enable the efficient KO of target genes in RPE1 cells, we developed a practical method that enables the efficient knock-in of a DNA cassette into target genes by a HIDR mechanism. We propose that our strategy is a promising alternative method for the establishment of KO cell lines.
RESULTS AND DISCUSSION
Design of a donor vector for the CRISPR/Cas9-mediated knock-in system
The hTERT-RPE1 cell line was originally established by immortalizing the RPE-340 cell line by transfection of the pGRN145 plasmid (ATCC MBA-141), which contains not only the hTERT gene but also the hygromycin B– and puromycin-resistance genes, followed by selection in the presence of hygromycin B. Therefore a conventional CRISPR/Cas9 protocol (Ran et al., 2013) involving transient puromycin selection is not applicable to RPE1 cells. Indeed, our attempts to establish IFT88 KO RPE1 cells by the conventional method using a high concentration of puromycin (15 µg/ml) for clonal selection were unsuccessful; as shown in Supplemental Figure S1B, lane 7, we could not detect any mobility-shifted bands indicative of indel mutations by heteroduplex mobility-shift assay of genomic DNA isolated from a mixture of surviving RPE1 cells.
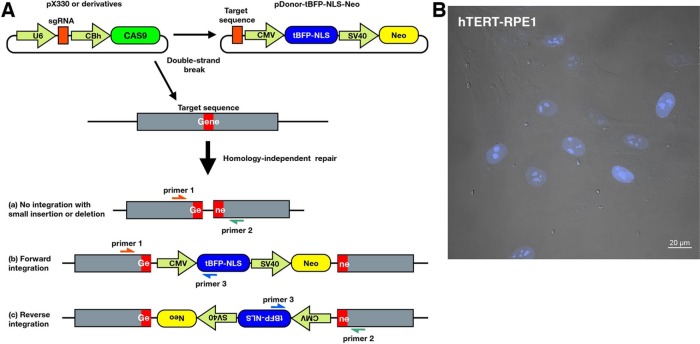
Therefore we first designed a donor vector for the CRISPR/Cas9-mediated knock-in system practically applicable to RPE1 cells. Because it was reported that the establishment of biallelic KO lines from RPE1 cells was difficult due to their low HR activity (Miyamoto et al., 2015), we sought to use an HIDR mechanism. The donor plasmid was created by modifying the pTagBFP-C1 vector because it contains two marker genes, one encoding blue fluorescent protein (tBFP) and the other encoding neomycin phosphotransferase (Neo; Figure 1A). We added cleavage sites for the restriction enzyme BbsI/BpiI upstream of the cytomegalovirus promoter to enable insertion of the target DNA sequence (pDonor-tBFP-NLS-Neo; Supplemental Figure S3A). The inserted target sequence before the protospacer adjacent motif (PAM) becomes a cleavage site for RNA-guided Cas9. Furthermore, a triplicated nuclear localization signal (NLS) was fused to tBFP to enable easy identification of knock-in cells with nuclear tBFP under a conventional fluorescence microscope (Figure 1B) without requirement for specialized equipment (i.e., fluorescence-activated cell sorter).
FIGURE 1:
Schematic overview of CRISPR/Cas9-mediated knock-in system. (A) RPE1 cells are transfected with an all-in-one vector, which contains spCas9 and an sgRNA expression cassette, together with a donor vector (pDonor-tBFP-NLS-Neo), which contains tBFP-3×NLS and Neo as selection markers. The RNA-guided Cas9 concurrently cleaves target sequences in the genome and donor vector. The DSBs trigger HIDR, in which the linearized donor vector can be integrated into the target locus in a forward or reverse direction. One of three types of donor integration can occur: (a) no integration of the donor vector, but with small indels; (b) forward integration of the donor vector; or (c) reverse integration of the donor vector. (B) Merged image of differential interference contrast and fluorescence microscopy. Signals of nuclear tBFP suggest successful knock-in of the donor vector. Scale bar, 20 μm.
An all-in-one vector for the expression of single-guide RNA (sgRNA) and Cas9, such as pX330 or its derivative, and the donor vector are cotransfected into the cells. RNA-guided Cas9 induces the concurrent cleavage of target sites in the donor plasmid and the genome, and the linearized donor plasmid is ligated into the genome by an HIDR mechanism (Figure 1A). As shown in Supplemental Figure S1B, lane 8, we could detect mobility-shifted bands by heteroduplex assay of genomic DNA from a mixture of surviving RPE1 cells after selection for several days in the presence of G418. Furthermore, PCR of the genomic DNA using three sets of primers confirmed integration of the donor vector (Figure 2A, lanes 5–7); the presence of PCR bands amplified with primer pair b (primers 1 and 3) and pair c (primers 2 and 3) indicate forward and reverse integration of the donor vector into the target locus (lanes 6 and 7, respectively). The presence of a band amplified with primer pair a (primers 1 and 2) indicates the absence of integration of the donor vector (lane 5; also see the band in lane 2, amplified from genomic DNA of wild-type [WT] RPE1 cells); however, there is the possibility that indels occur in the target locus by error-prone NHEJ. To confirm potential frameshift mutations by indels, Sanger sequencing of the PCR products is required after clonal selection.
FIGURE 2:
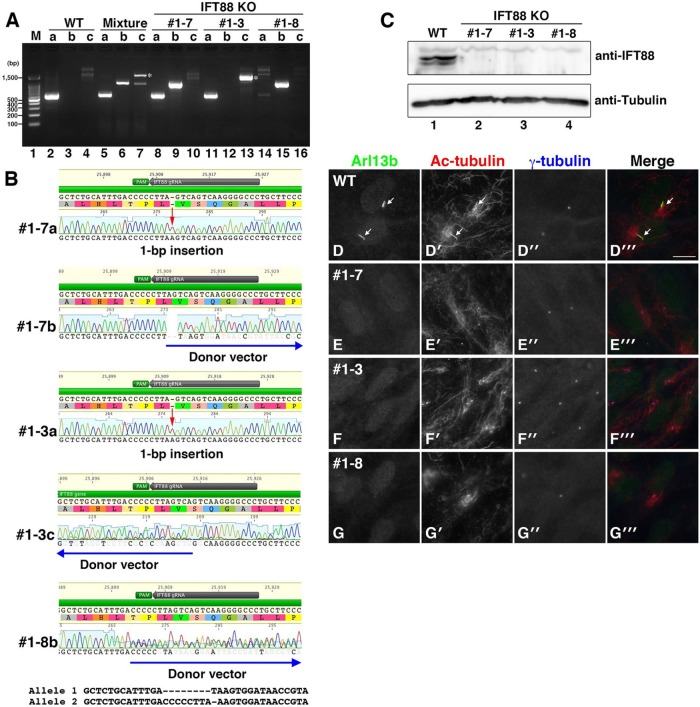
Establishment of IFT88-KO cell lines. (A) Integration of the donor vector was confirmed by genomic PCR using three sets of primers. The predicted sizes of the PCR products resulting from primer pairs a–c are ∼550, ∼1000, and ∼900 base pairs, respectively. Asterisks indicate PCR products containing an unexpectedly large insertion. (B) Indels and integration of the donor vector in IFT88-KO cell lines were confirmed by direct sequencing of the PCR products. Red arrows indicate a frameshift mutation by a 1–base pair insertion. Blue arrows indicate the integrated donor vector. The double peaks in the chromatogram of #1-8b were separated computationally. (C) Depletion of the IFT88 protein in IFT88-KO cell lines was confirmed by immunoblotting. (D) Ciliogenesis defect in IFT88-KO cell lines. Three IFT88-KO cell lines and WT RPE1 cells were serum starved for 24 h to induce ciliogenesis and triple immunostained for ARL13B (D–G), Ac-tubulin (D’–G’), and γ-tubulin (D”–G”). Merged images are shown in D”’–G”’. Arrows indicate primary cilia. Scale bar, 10 μm.
Candidate knock-in cells were then selected on the basis of the presence of nuclear tBFP signals (Figure 1B). Because the distinct blue fluorescence in the nucleus does not affect the subsequent procedures, such as immunofluorescence microscopy for organellar proteins, nuclear tBFP is an ideal marker for the selection of knock-in cells.
Establishment of IFT88-KO cell lines
To examine the efficiency of our system, we sought to establish IFT88 KO cell lines using RPE1 cells. IFT88 is a subunit of the IFT-B complex, which is composed of 16 subunits (Katoh et al., 2016; Taschner et al., 2016). Because the IFT-B complex plays a crucial role in anterograde trafficking of ciliary proteins, including tubulins, disruption of the IFT88 gene results in a severe ciliogenesis defect (Pazour et al., 2000; Taulman et al., 2001; Huangfu et al., 2003). We used the CRISPR design (Ran et al., 2013) and CRISPRscan (Moreno-Mateos et al., 2015) sgRNA design programs to select efficient sgRNA for the IFT88 gene and for the following experiments (see Materials and Methods and Supplemental Table S1). Double-stranded oligonucleotides for the targeting of IFT88 were inserted into both the all-in-one vector pSpCas9(BB)-2A-Puro (PX459) (Ran et al., 2013) and the donor vector pDonor-tBFP-NLS-Neo. RPE1 cells were cotransfected with these vectors, selected in medium containing G418 (600 µg/ml) for ∼14 d, and further cultured for ∼14 d for colony formation. Isolated colonies with nuclear tBFP, as well as a mixture of nonisolated cells, were used for the following experiments.
To identify RPE1 clones with a disrupted IFT88 gene, we subjected 15 isolated clones to genotyping. As shown in Figure 2A, PCR analysis of genomic DNA from a mixture of nonisolated cells using three sets of primers (Figure 1A) suggested integration of the donor vector in both forward (lane 6) and reverse (lane 7) directions. After the initial test, we obtained four independent IFT88 KO clones (summarized in Table 1). Among them, we selected three clones (#1-3, 1-7, and 1-8) for further analyses because these clones had distinct genetic modifications: monoallelic forward integration of the donor vector (clone #1-7; lanes 8–10), monoallelic reverse integration of the donor vector (clone #1-3; lanes 11–13), and biallelic forward integration of the donor vector (clone #1-8; lanes 14–16). The predicted sizes of the PCR products using primer pairs a–c are ∼550, 1000, and 900 base pairs, respectively. Note that a band of ∼1600 base pairs indicated by asterisks was amplified using primer pair c in the nonisolated clone mixture (lane 7) and clone #1-3 (lane 13); direct sequencing of the ∼1600–base pair PCR product revealed an unexpectedly large insertion of the donor vector. Sequencing of the PCR products demonstrated a 1–base pair insertion in one allele of clones #1-7 and 1-3 and a forward and reverse integration, respectively, of the donor vector in the other allele (Figure 2B). The double peaks found in the sequencing profile of #1-8b were separated using CRISP-ID (Dehairs et al., 2016), which is a web-based application for identifying indels.
TABLE 1:
Summary of KO cell lines established by our knock-in systems.
| Gene | gRNA | KI system | Genotyping | KO clones (mono-KI: bi-KI) | KO clones/genotyping (%) | Independent KO (mono-KI: bi-KI) | Independent KO/genotyping (%) |
|---|---|---|---|---|---|---|---|
| IFT88 | #1 | Version 1 | 15 | 6 (5:1) | 40.0 | 4 (3:1) | 26.7 |
| IFT20 | #1 | Version 1 | 6 | 1 (1:0) | 16.7 | 1 (1:0) | 16.7 |
| IFT20 | #2 | Version 1 | 6 | 3 (3:0) | 50.0 | 3 (3:0) | 50.0 |
| IFT56 | #1 | Version 1 | 17 | 5 (5:0) | 29.4 | 3 (3:0) | 17.6 |
| IFT56 | #2 | Version 1 | 14 | 3 (3:0) | 21.4 | 2 (2:0) | 14.3 |
| IFT139 | #2 | Version 1 | 9 | 8 (7:1) | 88.9 | 3 (2:1) | 33.3 |
| IFT144 | #2 | Version 1 | 4 | 3 (3:0) | 75.0 | 3 (3:0) | 75.0 |
| ARL13B | #1 | Version 1 | 7 | 5 (5:0) | 71.4 | 5 (5:0) | 71.4 |
| ARL13B | #2 | Version 1 | 6 | 4 (4:0) | 66.7 | 4 (4:0) | 66.7 |
| IFT88 | #1 | Version 2 | 9 | 5 (4:1) | 55.6 | 3 (2:1) | 33.3 |
| IFT22 | #2 | Version 2 | 12 | 10 (10:0) | 83.3 | 3 (3:0) | 25.0 |
| IFT27 | #2 | Version 2 | 4 | 2 (2:0) | 50.0 | 1 (1:0) | 25.0 |
RPE1 cells were transfected with an expression vector for Cas9 and sgRNA together with the donor vector. After drug selection, single colonies expressing nuclear tBFP were isolated. PCR and Sanger sequencing were performed on genomic DNA of the isolated clones. Genotyping column shows the number of clones checked by genomic PCR and Sanger sequencing. KO clones column shows the number of biallelic KO clones confirmed by Sanger sequencing. Numbers enclosed in parentheses indicate the numbers of monoallelic and biallelic knock-in clones. In monoallelic knock-in clones, small indels on another allele causing gene disruption were confirmed by Sanger sequencing. Independent KO column shows the number of KO clones each having different genotypes.
We next confirmed depletion of the IFT88 protein in the established KO cell lines. Immunoblotting analysis of cell lysates using antibodies against IFT88 and α-tubulin demonstrated specific depletion of the IFT88 protein in the three KO cell lines (Figure 2C, lanes 2–4).
We then examined the phenotype of the IFT88-KO cell lines. Three IFT88-KO cell lines, as well as WT RPE1 cells, were cultured for 24 h under serum-starved conditions to induce ciliogenesis, followed by triple immunostaining for ARL13B (Figure 2, D–G) and acetylated α-tubulin (Ac-tubulin; Figure 2, D’–G’), which are markers for primary cilia, and γ-tubulin (Figure 2, D”–G”), a marker protein for the basal body. Approximately 80% of WT cells formed primary cilia under serum-starved conditions. In striking contrast, none of the IFT88-KO cell lines formed primary cilia (Figure 2, E–G; also see Figure 3F). The phenotype of IFT88-KO RPE1 cells is consistent with that previously reported of cells derived from IFT88-deficient mice (Pazour et al., 2000; Taulman et al., 2001; Huangfu et al., 2003). Taking these results together, we concluded that we successfully established IFT88-KO cell lines by our newly established CRISPR/Cas9-mediated knock-in system.
FIGURE 3:
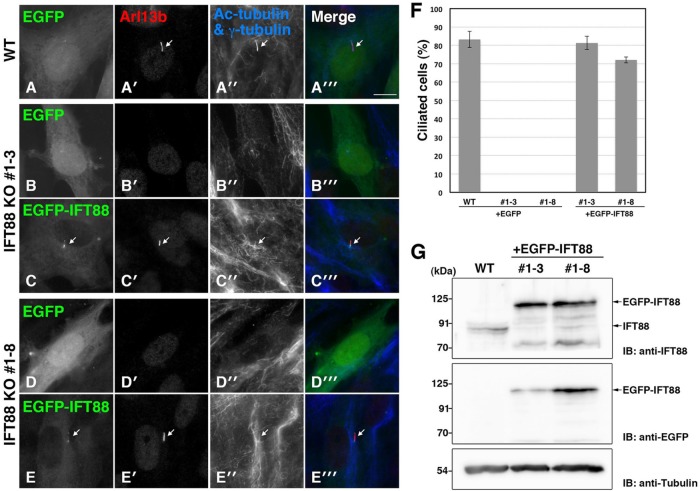
Rescue experiments of IFT88-KO cell lines. WT RPE1 cells (A) and IFT88-KO cell lines #1-3 (B, C) and #1-8 (D, E) were transfected with pEBMulti-Ble-EGFP (A, B, D) or pEBMulti-Ble-EGFP-IFT88 (C, E). The transfected cells were cultured under serum-starved conditions and immunostained for Arl13b (A’–E’), Ac-tubulin, and γ-tubulin (A”–E”). Merged images are shown in A”’–E”’. Arrows indicate primary cilia. Scale bar, 10 μm. WT cells and IFT88-KO cells exogenously expressing EGFP-IFT88 were ciliated, whereas IFT88-KO cells expressing EGFP were not. (F) Ciliated cells in the experiments shown in A–E were counted, and percentages of ciliated cells are represented as bar graphs. Values are means ± SE (error bars) of three independent experiments. In each experiment, 40–80 cells with EGFP signals were examined. (G) Confirmation of exogenously expressed EGFP-IFT88 in IFT88-KO cell lines by immunoblotting. Lysates from WT RPE1 cells and from IFT88-KO clones #1-3 and 1-8 expressing EGFP-IFT88 were immunoblotted with antibodies against IFT88 and GFP. Tubulin was analyzed as a loading control.
Rescue experiments of IFT88-KO cell lines
We next performed rescue experiments to analyze the possibility of off-target cleavage events causing unexpected effects. For rescue experiments of IFT88-KO cells, we used an episomal expression vector system. The pEBMulti-Ble vector harbors a replication origin, oriP, and Epstein–Barr virus nuclear antigen 1, as well as a selection marker, the bleomycin/Zeocin-resistance gene. These components enable episomal replication of the plasmid and its distribution to daughter cells.
WT RPE1 cells and two IFT88-KO cell lines were transfected with pEBMulti-Ble–enhanced green fluorescent protein (EGFP) or pEBMulti-Ble-EGFP-IFT88 (see Materials and Methods) and selected in medium containing Zeocin (75 µg/ml) for ∼7 d. The cells were then cultured for 24 h under serum-starved conditions to induce ciliogenesis. Defective ciliogenesis of clones #1-3 and 1-8 was rescued by the exogenous expression of EGFP-IFT88 (Figure 3, C–C”’ and E–E”’, respectively) but not by that of EGFP (Figure 3, B–B”’ and D–D”’, respectively). Quantification of ciliated cells demonstrated that the exogenous EGFP-IFT88 expression in IFT88-KO cells increased the percentage of ciliated cells to levels similar to that of WT cells (Figure 3F). As shown in Figure 3G, immunoblotting analysis demonstrated that the amount of exogenously expressed EGFP-IFT88 protein was comparable to that of endogenous IFT88. These results confirmed that defective ciliogenesis is the true phenotype caused by disruption of the IFT88 gene.
Establishment of other KO cell lines
To demonstrate the general application of our method, we then sought to establish RPE1 cell lines defective in IFT20, another IFT-B subunit. Using two different target sequences, we obtained four independent IFT20-KO cell lines from 12 clones subjected to genotyping, as summarized in Table 1. Among them, two cell lines obtained using different target sequences (#1-3 and 2-6) were selected for the following analyses. As shown in Supplemental Figure S2, A and B, clone #1-3 has a 1–base pair insertion in one allele and a reverse integration of the donor vector in the other allele (lanes 5–7), and clone #2-6 has a 1–base pair insertion in one allele and a forward donor vector integration in the other allele (8–10). When WT RPE1 and IFT20-KO cells were stained with an anti-IFT20 antibody, Golgi-like staining typical of IFT20 (Follit et al., 2006) was abolished in the two IFT20-KO cell lines (Supplemental Figure S2, C–E). Furthermore, these IFT20-KO cell lines demonstrated defective ciliogenesis (Supplemental Figure S2, C’–E’), consistent with previous reports on the phenotypes of IFT20-knockdown cells and IFT20-conditional KO mice (Follit et al., 2006; Jonassen et al., 2008).
In addition, using our system, we succeeded in establishing RPE1 cells defective in IFT56 (an IFT-B subunit), IFT139 (an IFT-A subunit), IFT144 (an IFT-A subunit), and ARL13B (a ciliary small GTPase; summarized in Table 1), all of which are involved in ciliary protein trafficking and/or ciliary assembly (Madhivanan and Aguilar, 2014; Mourão et al., 2016; Taschner and Lorentzen, 2016). It is noteworthy that all of the defects in ciliogenesis and/or ciliary protein trafficking observed in these KO cells can be restored by exogenous expression of corresponding proteins; the characterization of these KO cells has been reported in other studies (Funabashi et al., 2017; Hirano et al., 2017; Nozaki et al., 2017). Of course, our strategy is also applicable to more widely used cell lines. Indeed, we recently reported that HeLa cells defective in ATP9A, a phospholipid flippase, were successfully established using our knock-in system (Tanaka et al., 2016). These examples demonstrate the utility of our system.
Improved CRISPR/Cas9-mediated knock-in system
One of the most critical concerns with genome editing using the CRISPR/Cas9 system is off-target cleavage. The original SpCas9 can cleave off-target sites that are not exactly complementary to the guide RNA. To overcome this problem, improved SpCas9s, called eSpCas9(1.1) and SpCas9-HF1, have recently been developed by the groups of Zhang and Joung, respectively (Kleinstiver et al., 2016; Slaymaker et al., 2016). The two groups showed that these improved Cas9s dramatically decreased levels of off-target cleavage activity while retaining high levels of on-target cleavage activity in mammalian cells. We therefore exploited one of these Cas9 vectors for the improvement of our knock-in system, namely, to reduce the frequency of off-target cleavage events and increase the versatility of the donor vector.
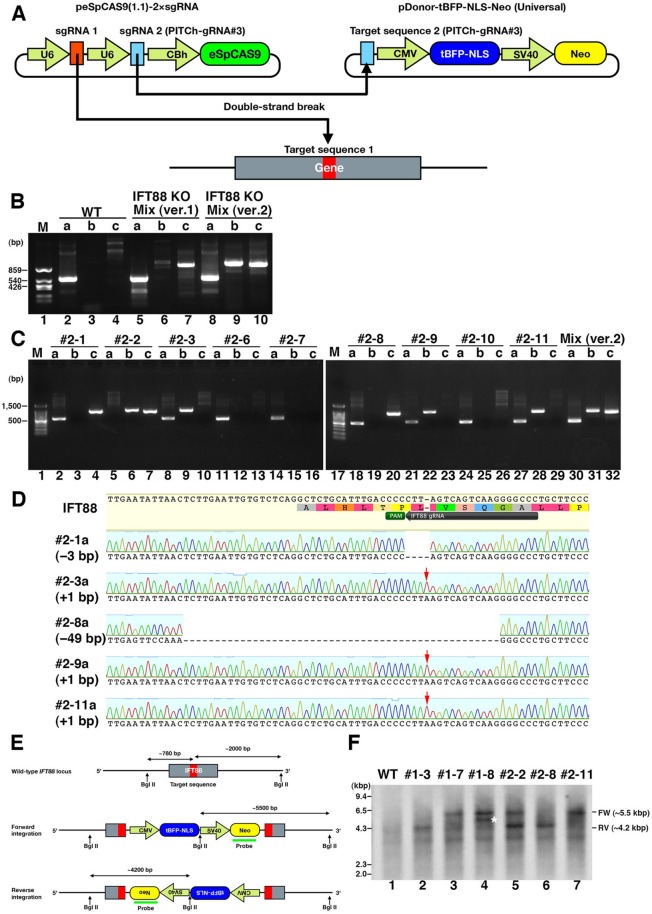
The second version of our knock-in system is shown schematically in Figure 4A. We selected the eSpCas9(1.1) plasmid as the starting material because its framework is the same as those of pX330 and its derivatives. To enable multisite cleavage, we added a second sgRNA expression cassette after the first sgRNA cassette in the plasmid. The second sgRNA (sgRNA2) was designed to enable cleavage of the donor vector at target sequence 2. Note that target sequences 1 and 2 are not necessarily identical, as we exploit homology-independent mechanism to knock-in the donor vector. As the second sgRNA and target sequence 2, we selected the PITCh-gRNA#3 sequence (see Materials and Methods and Supplemental Figure S2B), which was designed and confirmed experimentally by Sakuma et al. (2015) to reduce off-target cleavage events in mammalian cells. The plasmid containing the PITCh-gRNA#3 targeting sequence, pDonor-tBFP-NLS-Neo (Universal), can be used as a universal donor vector by combining its use with the peSpCas9(1.1)-2×sgRNA vector. Therefore, to disrupt the target gene in the improved system (Figure 4A), all one has to do is to construct the all-in-one vector, peSpCas9(1.1)-2×sgRNA, containing the sgRNA sequence for the corresponding target sequence.
FIGURE 4:
Our novel improved CRISPR/Cas9-mediated knock-in system. (A) Schematic illustration of the improved knock-in system. The all-in-one vector peSpCAS9(1.1)-2×sgRNA has a Cas9 with enhanced specificity (eSpCas9) and tandem expression cassettes of sgRNAs. The first sgRNA targets the genome; the second one targets the donor vector. The universal donor vector pDonor-tBFP-NLS-Neo (Universal) contains an artificial sequence (PITCh-gRNA#3) designed by Sakuma et al. (2015). (B) Integration of the donor vector in a mixture of nonisolated cells was confirmed by genomic PCR using three sets of primers (Figure 1A). (C) Integration of donor vector in isolated clones was confirmed by genomic PCR using three sets of primers. (D) Small indels of nonintegrated alleles in isolated cell lines (#2-1, 2-3, 2-8, 2-9, 2-11) were confirmed by direct sequencing of PCR products that were amplified using the primer pair a. Red arrows indicate 1–base pair insertion. (E) Schematic illustration of the IFT88 locus with the integrated donor vector. (F) Southern blot analysis. Genomic DNA from WT RPE1 cells and six IFT88 KO clones indicated were digested with BglII. Southern blotting was carried out using a probe for the Neo gene in the donor vector. Approximately 5.5- and 4.2-kbp bands were detected when the donor vector was integrated into the IFT88 locus in forward and reverse directions, respectively. An asterisk indicates a band of unexpected size.
We applied the second version of the knock-in system to knock out the IFT88 gene and compared the efficacy of the first and second knock-in systems. As shown in Figure 4B, genomic PCR analysis of nonisolated cell mixtures using three sets of primers (Figure 1A) demonstrated that both systems succeeded in gene knock-in. Genomic PCR analysis of nine clones established by the second system demonstrated that one clone (#2-2) had biallelic knock-in and five clones (#2-1, 2-3, 2-8, 2-9, and 2-11) had monoallelic forward or reverse integration of the donor vector (Figure 4C). Direct sequencing of the PCR products amplified from the monoallelic knock-in clones using primer pair a (Figure 1A) confirmed the presence of indels (Figure 4D), although one clone had a 3–base pair deletion without a frameshift, and three clones had the same 1–base pair insertion. As summarized in Table 1, by using the first and second versions of the knock-in system, we succeeded in establishing independent IFT88 KO clones in 4 of 15 isolated clones (26.7%) and 3 of 9 isolated clones (33.3%), respectively. In addition, using our improved system, we succeeded in establishing RPE1 cells defective in IFT22 and IFT27 (summarized in Table 1), both of which are subunits of the IFT-B complex; the characterization of these KO cells will be described elsewhere in detail. Thus, despite the second version of the knock-in system being more practical and versatile than the first system, the knockout efficiency was comparable between the two methods.
Finally, we performed Southern blot analysis of genomic DNA from the IFT88-KO cells established by the first and second knock-in systems to examine whether the donor vector was integrated into off-target sites (Figure 4, E and F). When the genomic DNA is digested with BglII and probed with a Neo fragment, the forward and reverse integration of the donor vector into the IFT88 locus is expected to yield bands of ∼5.5 and ∼4.2 kbp, respectively (Figure 4E). For clones #1-3 and 1-7 established by the first knock-in system, only an ∼4.2- or ∼5.5-kbp band was detected (Figure 4F, lanes 2 and 3), indicating integration of the donor vector into the expected site. In contrast, another band in addition to the ∼5.5-kbp band was detected for clone #1-8 (lane 4; band indicated by an asterisk), suggesting integration of the donor vector into an unexpected site in addition to the expected site. It is noteworthy, however, that the ciliary assembly in clone #1-8 cells was restored by exogenous IFT88 expression (Figure 3, D–D”’, E–E”’, and F), excluding the possibility that the donor vector integration into an off-target site gave rise to the ciliogenesis defect.
On the other hand, only a band(s) of expected size(s) were detected for clones #2-2, 2-8, and 2-11 established by the second system (Figure 4F, lanes 5–7), indicating integration of the donor vector into the expected site(s) in the IFT88 locus. However, even with the use of an improved Cas9, off-target cleavage is not completely abolished. Rescue experiments will hence be essential to confirm the correlation between the genotype and phenotype in KO cell lines.
Recently the effectiveness of homology-independent genome-editing strategy was reported. He et al. (2016) reported that knock-in of a donor plasmid through HIDR occurs at a higher efficiency than through homology-dependent repair in several human cell lines. Lackner et al. (2015) reported that precise gene tagging by a HIDR mechanism is possible in mammalian culture cells. More recently, Suzuki et al. (2016) reported that homology-independent targeted integration enables precise genome editing in vitro and in vivo. Therefore our results and recent reports show that the strategy of using the HIDR mechanism is a promising approach for efficient genomic editing.
MATERIALS AND METHODS
Plasmids
A customizable donor vector containing BbsI/BpiI cleavage sites for oligonucleotide insertion was constructed from pTagBFP-C1 (Evrogen) by insertion of the BbsI/BpiI cleavage site sequences. The resulting donor vector, named pDonor-tBFP-NLS-Neo (Supplemental Figure S2A), has been deposited in Addgene (ID 80766). A universal donor vector with an artificially designed sequence, PITCh-gRNA #3 (5′-GCATCGTACGCGTACGTGTT-3′), was constructed from pDonor-tBFP-NLS-Neo by inserting the corresponding sequence. The resulting universal donor vector, named pDonor-tBFP-NLS-Neo (Universal) (Supplemental Figure S2B), has been deposited in Addgene (ID 80767). An eSpCas9(1.1)-based expression vector with a second sgRNA expression cassette was constructed by inserting the PITCh-gRNA #3 sequence into eSpCas9(1.1) (Addgene plasmid #71814) by a SLiCE cloning method (Motohashi, 2015). The tandem sgRNA expression vector, named peSpCAS9(1.1)-2×sgRNA (Supplemental Figure S2C), has been deposited in Addgene (ID 80768).
Cell culture
hTERT-RPE1 cells (CRL-4000; American Type Cell Collection [ATCC]) were grown in DMEM/Ham’s F-12 (Nacalai Tesque) supplemented with 10% fetal bovine serum (FBS) and 0.348% sodium bicarbonate at 37°C in 5% CO2.
Establishment of IFT88- and IFT20-KO cell lines
The sgRNA sequences targeting human IFT88 (5′-GGGCCCCTTGACTGACTAAG-3′) and IFT20 (5′-GCCTTCAGCCTCTATGGTCTGC-3′ and 5′-GTGATGAACTGAACAAGCTGA-3′) were designed using CRISPR design (crispr.mit.edu) and CRISPRscan (www.crisprscan.org). These sequences were separately inserted into pSpCas9(BB)-2A-Puro (PX459; Addgene plasmid #48139) and peSpCAS9(1.1)-2 × sgRNA. The cleavage activities of these Cas9 vectors were tested as described by Mashiko et al. (2013). Briefly, HEK293T cells were cotransfected with pSpCas9(BB)-2A-Puro (PX459) containing the IFT88 sgRNA and pCAG-EGxxFP containing the IFT88 cDNA. After 48 h of transfection, the expression level of EGFP was examined by fluorescence microscopy. Cleavage activities for IFT20 were tested using pSpCas9(BB)-2A-Puro (PX459) containing IFT20 sgRNA and pCAG-EGxxFP containing the IFT20 cDNA in the same way.
RPE1 cells grown to ∼3.0 × 105 cells on a 12-well plate were transfected with 1 µg of all-in-one vector and 0.25 µg of the donor vector using X-tremeGENE9 DNA Transfection Reagent (Roche Applied Science) and selected in medium containing G418 (600 µg/ml) for ∼14 d. Subsequent culturing for ∼14 d was required for colony formation. The colonies with nuclear tBFP fluorescence were isolated and subjected to extraction of genome DNA using MagExtractor Genome (TOYOBO) according to the manufacturer’s instructions. The extracted genome DNA was subjected to PCR using KOD FX Neo DNA polymerase (TOYOBO) and three sets of primers (Supplemental Table S1) to distinguish among the following three states of integration of the donor vector: (a) no integration with small insertions or deletions (indels), (b) forward integration of the donor vector, and (c) reverse integration of the donor vector (Figure 1A). The PCR products were treated with Illustra ExoProStar (GE Healthcare Life Sciences) to remove unincorporated primers and nucleotides. Biallelic disruption of the target gene was confirmed by direct sequencing of the PCR products. Comparison of the DNA sequences was performed using Geneious Pro (Biomatters). Heteroduplex mobility assay was performed as described by Ota et al. (2013).
Rescue experiments
The human IFT88 cDNA was cloned into the pEBMulti-Ble-EGFP episomal vector (Wako Pure Chemical Industries). WT and IFT88 KO cell lines transfected with pEBMulti-Ble-EGFP or pEBMulti-Ble-EGFP-IFT88 were selected in medium containing Zeocin (75 µg/ml) for ∼7 d. These cells were used for the following immunofluorescence and immunoblotting analyses.
Immunofluorescence microscopy
RPE1 cells were incubated in serum-free Opti-MEM (Invitrogen) supplemented with 0.2% bovine serum albumin for 24 h to induce ciliogenesis. The cells were fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 5 min, permeabilized with methanol at −20°C for 5 min, and incubated in PBS containing 10% FBS at room temperature for 1 h. The cells were then incubated with primary antibodies at room temperature for 1 h, washed three times with PBS, and incubated with secondary antibodies at room temperature for 1 h. Coverslips were mounted in Mowiol (Calbiochem), and the cells were observed using an Axiovert 200 M microscope (Carl Zeiss). Merged images of differential interference and fluorescence were obtained using a confocal laser-scanning microscope (Nikon A1R-MP).
Immunoblotting
RPE1 cells were lysed in lysis buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid–KOH, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, and 10% glycerol) containing a protease inhibitor cocktail (Nacalai Tesque). After 15 min on ice, the cell lysates were centrifuged at 16,100 × g for 15 min at 4°C in a microcentrifuge. Proteins (30 µg) in cell lysates were separated by SDS–PAGE and electroblotted onto an Immobilon-P transfer membrane (Millipore EMD). The membrane was blocked with 5% skimmed milk and incubated sequentially with primary and horseradish peroxidase–conjugated secondary antibodies. Detection was carried out using a Chemi-Lumi One L Kit (Nacalai Tesque).
Antibodies
The following antibodies were obtained from the indicated vendors: monoclonal mouse anti–Ac-tubulin (6-11B-1; Sigma-Aldrich), anti–γ-tubulin (GTU-88; Sigma-Aldrich), and anti-GFP (JL-8; BD Biosciences); monoclonal rat anti–α-tubulin (YL1/2; Abcam); polyclonal rabbit anti-ARL13B (17711-1-AP; Proteintech), anti-IFT88 (13967-1-AP; Proteintech), and anti-IFT20 (13615-1-AP; Proteintech); Alexa Fluor 555–conjugated goat anti-rabbit immunoglobulin G (IgG; A21429, Invitrogen); Alexa Fluor 488–conjugated goat anti-rabbit IgG (A11034; Invitrogen); Alexa Fluor 555–conjugated goat anti-mouse IgG2b (A21147; Invitrogen); Alexa Fluor 647–conjugated goat anti-mouse IgG1 (A21240; Invitrogen); DyLight649-conjugated goat anti-mouse IgG (115-495-209; Jackson ImmunoResearch Laboratories); and horseradish peroxidase–conjugated secondary antibodies (115-035-166 and 111-035-144; Jackson ImmunoResearch Laboratories).
Southern blotting
The extracted genome DNA was digested with BglII, electrophoresed on a 0.7% agarose gel., and transferred onto a GeneScreen Plus membrane (PerkinElmer) by the alkaline transfer method. A DNA fragment covering the neomycin phosphotransferase gene was amplified from the donor vector by PCR and labeled using an AlkPhos Direct kit (GE Healthcare). Hybridization of the probe was performed according to the manufacturer’s instructions. Luminescence generated by a CDP-Star Detection reagent (GE Healthcare) was detected using LAS-3000mini (Fuji Film).
Supplementary Material
Acknowledgments
We thank Feng Zhang for providing pSpCas9(BB)-2A-Puro (PX459) and eSpCas9(1.1) and Helena Akiko Popiel for critical reading of the manuscript. This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas “Cilia and Centrosome” from the Ministry of Education, Culture, Sports, Science and Technology, Japan (Grants 25113514 and 15H01211 to K.N.); the Japan Society for the Promotion of Science (Grants 22390013 and 15H04370 to K.N. and 25860044 and 15K07929 to Y.K.); and grants from the Uehara Memorial Foundation and from the Astellas Foundation for Research on Metabolic Disorders to K.N. and from the Takeda Science Foundation to Y.K. S.N. was supported by the JSPS Research Fellowship for Young Scientists.
Abbreviations used:
- Cas9
CRISPR-associated 9
- CRISPR
clustered regularly interspaced short palindromic repeats
- DSB
double-strand break
- EGFP
enhanced green fluorescent protein
- eSpCas9
enhanced specificity Cas9
- FBS
fetal bovine serum
- HIDR
homology-independent DNA repair
- HR
homologous recombination
- hTERT
human telomerase reverse transcriptase
- IFT
intraflagellar transport
- KO
knockout
- NHEJ
nonhomologous end joining
- NLS
nuclear localization signal
- PAM
protospacer adjacent motif
- PBS
phosphate-buffered saline
- RPE
retinal pigment epithelial
- sgRNA
single-guide RNA
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-01-0051) on February 8, 2017.
REFERENCES
- Auer TO, Del Bene F. CRISPR/Cas9 and TALEN-mediated knock-in approaches in zebrafish. Methods. 2014;69:142–150. doi: 10.1016/j.ymeth.2014.03.027. [DOI] [PubMed] [Google Scholar]
- Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Cristea S, Freyvert Y, Santiago Y, Holmes MC, Urnov FD, Gregory PD, Cost GJ. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol Bioeng. 2013;110:871–880. doi: 10.1002/bit.24733. [DOI] [PubMed] [Google Scholar]
- Dehairs J, Talebi A, Cherifi Y, Swinnen JV. CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Sci Rep. 2016;6:28973. doi: 10.1038/srep28973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi T, Katoh Y, Michisaka S, Terada M, Sugawa M, Nakayama K. Ciliary entry of KIF17 is dependent on its binding to the IFT-B complex via IFT46–IFT56 as well as on its nuclear localization signal. Mol Biol Cell. 2017;28:624–633. doi: 10.1091/mbc.E16-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger JM, Turan S, Hernandez S, Spector LP, Calos MP. In vivo blunt-end cloning through CRISPR/Cas9-facilitated non-homologous end-joining. Nucleic Acids Res. 2016;44:e76. doi: 10.1093/nar/gkv1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Tan C, Wang F, Wang Y, Zhou R, Cui D, You W, Zhao H, Ren J, Feng B. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016;44:e85. doi: 10.1093/nar/gkw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Katoh Y, Nakayama K. Intraflagellar transport-A complex mediates ciliary entry as well as retrograde trafficking of ciliary G protein-coupled receptors. Mol Biol Cell. 2017;28:429–439. doi: 10.1091/mbc.E16-11-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Jasin M, Haber JE. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair (Amst) 2016;44:6–16. doi: 10.1016/j.dnarep.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang XR, Jimenez G, Chang E, Frolkis M, Kusler B, Sage M, Beeche M, Bodnar AG, Wahl GM, Tlsty TD, et al. Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat Genet. 1999;21:111–114. doi: 10.1038/5056. [DOI] [PubMed] [Google Scholar]
- Jonassen JA, San Agustin J, Follit JA, Pazour GJ. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J Cell Biol. 2008;183:377–384. doi: 10.1083/jcb.200808137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I, Xu L, Ciosk R. CRISPR/Cas9 Genome editing in caenorhabditis elegans: evaluation of templates for homology-mediated repair and knock-ins by homology-independent DNA Repair. G3 (Bethesda) 2015;5:1649–1656. doi: 10.1534/g3.115.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Terada M, Nishijima Y, Takei R, Nozaki S, Hamada H, Nakayama K. Overall architecture of the intraflagellar transport (IFT)-B complex containing Cluap1/IFT38 as an essential component of the IFT-B peripheral subcomplex. J Biol Chem. 2016;291:10962–10975. doi: 10.1074/jbc.M116.713883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima S-I. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Carré A, Guzzardo PM, Banning C, Mangena R, Henley T, Oberndorfer S, Gapp BV, Nijman SMB, Brummelkamp TR, et al. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun. 2015;6:10237. doi: 10.1038/ncomms10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JJM, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stütz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 2013;3:1213–1224. doi: 10.1534/g3.113.005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhivanan K, Aguilar RC. Ciliopathies: the trafficking connection. Traffic. 2014;15:1031–1056. doi: 10.1111/tra.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca M, Lin VG, Guo N, Yang Y. Obligate Ligation-Gated Recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013;23:539–546. doi: 10.1101/gr.145441.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep. 2013;3:3355. doi: 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Hosoba K, Ochiai H, Royba E, Izumi H, Sakuma T, Yamamoto T, Dynlacht BD, Matsuura S. The microtubule-depolymerizing activity of a mitotic kinesin protein KIF2A drives primary cilia disassembly coupled with cell proliferation. Cell Rep. 2015;10:664–673. doi: 10.1016/j.celrep.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov E, Magin S, Soni A, Iliakis G. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: Cell cycle and proliferation-dependent regulation. Semin Cancer Biol. 2016;37-38:51–64. doi: 10.1016/j.semcancer.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin J-D, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K. A simple and efficient seamless DNA cloning method using SLiCE from Escherichia coli laboratory strains and its application to SLiP site-directed mutagenesis. BMC Biotechnol. 2015;15:47. doi: 10.1186/s12896-015-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourão A, Christensen ST, Lorentzen E. The intraflagellar transport machinery in ciliary signaling. Curr Opin Struct Biol. 2016;41:98–108. doi: 10.1016/j.sbi.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Nozaki S, Katoh Y, Terada M, Michisaka S, Funabashi T, Takahashi S, Kontani K, Nakayama K. Regulation of ciliary retrograde protein trafficking by Joubert syndrome proteins ARL13B and INPP5E. J Cell Sci. 2017;130:563–576. doi: 10.1242/jcs.197004. [DOI] [PubMed] [Google Scholar]
- Ota S, Hisano Y, Muraki M, Hoshijima K, Dahlem TJ, Grunwald DJ, Okada Y, Kawahara A. Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes Cells. 2013;18:450–458. doi: 10.1111/gtc.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Nakade S, Sakane Y, Suzuki KT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs andCRISPR-Cas9 with the PITCh systems. Nat Protoc. 2015;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Ono N, Shima T, Tanaka G, Katoh Y, Nakayama K, Takatsu H, Shin H-W. The phospholipid flippase ATP9A is required for recycling pathway from endosomes to the plasma membrane. Mol Biol Cell. 2016;27:3883–3893. doi: 10.1091/mbc.E16-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Lorentzen E. The intraflagellar transport machinery. Cold Spring Harb Perspect Biol. 2016;8:a028092. doi: 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Weber K, Mourão A, Vetter M, Awasthi M, Stiegler M, Bhogaraju S, Lorentzen E. Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 2016;35:773–790. doi: 10.15252/embj.201593164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulman PD, Haycraft CJ, Balkovetz DF, Yoder BK. Polaris, a protein involved in left-right axis patterning, localizes to basal bodies and cilia. Mol Biol Cell. 2001;12:589–599. doi: 10.1091/mbc.12.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.