Chemotaxis is the mechanism by which cells move in the direction of chemical gradients. The central circuit connecting basal movement and gradient sensing is unknown. Ras activation and F-actin form one coupled excitable system, which is the beating heart of cell movement in both the absence and presence of external cues.
Abstract
Many eukaryotic cells regulate their mobility by external cues. Genetic studies have identified >100 components that participate in chemotaxis, which hinders the identification of the conceptual framework of how cells sense and respond to shallow chemical gradients. The activation of Ras occurs during basal locomotion and is an essential connector between receptor and cytoskeleton during chemotaxis. Using a sensitive assay for activated Ras, we show here that activation of Ras and F-actin forms two excitable systems that are coupled through mutual positive feedback and memory. This coupled excitable system leads to short-lived patches of activated Ras and associated F-actin that precede the extension of protrusions. In buffer, excitability starts frequently with Ras activation in the back/side of the cell or with F-actin in the front of the cell. In a shallow gradient of chemoattractant, local Ras activation triggers full excitation of Ras and subsequently F-actin at the side of the cell facing the chemoattractant, leading to directed pseudopod extension and chemotaxis. A computational model shows that the coupled excitable Ras/F-actin system forms the driving heart for the ordered-stochastic extension of pseudopods in buffer and for efficient directional extension of pseudopods in chemotactic gradients.
INTRODUCTION
Chemotaxis, the capacity of cells to move directionally in gradients of chemicals, is critical for many biological processes, including the finding of food and sources of inflammation, but also during embryogenesis and wound healing (Artemenko et al., 2014). Cells in the absence of external cues extend protrusions in different directions, whereas in a gradient of chemoattractant, many protrusions are extended in the direction of the highest concentration of the chemical cues (Bosgraaf and Van Haastert, 2009a; Van Haastert, 2010; Insall, 2013). Protrusions in buffer and a gradient of chemoattractant are formed by local reshaping of the dendritic filamentous actin cytoskeleton (Krause and Gautreau, 2014). Key to understanding the molecular mechanism of chemotaxis is to determine how cells make protrusions and how chemoattractants bias the place and time where protrusions are formed.
Why do chemotactic cells make protrusions and move when there is no chemoattractant? Dennis Bray argued in his seminal perspective that “the high intrinsic activity serves to anticipate incoming stimuli” (Bray, 2014). An amoeboid cell is always moving, thereby constantly being responsive to internal and external signals that can bias this basal cell movement. With this concept, we proposed that basal cell movement is “prepared” to be biased by chemotactic signals. The molecular circuits that regulate basal pseudopod formation are constructed in such a way that they are easily biased by small spatial fluctuation of incoming signals. To understand chemotaxis at the lower limit of chemotactic detection, it is therefore essential to delineate the molecular mechanisms of how cells move in the absence of external stimuli and how tiny signals from activated chemoattractant receptors mix with this basal signaling, leading to a more localized response. Amoeboid cells move by extending pseudopodia. Cells in buffer do not extend pseudopodia in random directions but exhibit a rich repertoire of characteristic behavior (Andrew and Insall, 2007; Li et al., 2008; Bosgraaf and Van Haastert, 2009b). Most pseudopods are made at the side of an existing pseudopod (splitting), with a strong bias to do this alternatingly to the right and the left, leading to zigzag trajectories with persistence of direction. Alternatively, cells may extend pseudopods de novo from areas of the cell that have not extended pseudopodia in the recent past. These de novo pseudopodia are extended in random directions and interrupt the persistent zigzag trajectories of splittings (Li et al., 2008; Van Haastert and Bosgraaf, 2009a). Mutants in the cGMP/myosin pathway have been isolated that exhibit strongly enhanced de novo pseudopodia, and these cells move with nearly Brownian motion. On the other hand, starvation of wild-type cells strongly suppresses de novo pseudopodia, and these cells show very strong persistence, exploring large areas (Van Haastert and Bosgraaf, 2009a). In a shallow cAMP gradient, cells exhibit fewer de novo pseudopods, and splitting pseudopods have a small positional bias such that they are slightly better oriented toward the direction of the gradient than they would be in buffer (Andrew and Insall, 2007; Bosgraaf and Van Haastert, 2009a). At a molecular level, two circuits have been identified (Bretschneider et al., 2004, 2009; Ryan et al., 2012; Huang et al., 2013; Gerhardt et al., 2014; Tang et al., 2014; Bement et al., 2015; Gerisch and Ecke, 2016; Yang et al., 2016). The first circuit, called the cytoskeletal oscillatory network, is an excitable, actin-based cytoskeleton that can operate largely independently of external signals. This intrinsic network, consisting of kinases, GTP-binding proteins, and other regulatory molecules, modulates the actin-based cytoskeleton. Together these proteins determine how cells move in the absence of external stimuli. The second circuit, called the signal transduction excitable network, is the molecular network by which internal (myosin filaments for polarity) or external stimuli (chemoattractants) couple directly or indirectly to the cytoskeleton.
We estimate from genetic and biochemical studies that a few hundred proteins somehow are involved in chemotaxis or lead to altered chemotactic behavior when deleted or overexpressed (Fey et al., 2013; Basu et al., 2015). It will be difficult to completely dissect this complicated network. Uncovering these interactions and understanding details of chemotaxis will be facilitated if we can identify the signaling molecules that play a pivotal role at the interface of pseudopod formation and chemotactic signaling. Which signaling molecule is the critical component for basal pseudopod formation and receives spatial information from chemoattractant receptors? In Dictyostelium, Ras and Rac small GTP-binding proteins play a crucial role in chemotaxis. In the signaling cascade from chemoattractant receptor to F-actin, Ras is one of the first molecules that is activated much more strongly at the side of the cell facing the highest chemoattractant concentration than at the other side of the cell (Kae et al., 2004; Bolourani et al., 2006; Kortholt et al., 2011, 2013). Ras-GTP is frequently found in protrusions (Hecht et al., 2011; Kortholt et al., 2013). In addition, cells expressing an activated Ras in the absence of chemoattractants exhibit an increased number of F-actin–filled protrusions (Cai et al., 2010). This suggests that Ras is at a critical position, being part of both the basal and the chemoattractant-stimulated actin-regulated circuit. However, using the sensor Raf–Ras binding domain (RBD)–green fluorescent protein (GFP) to detect Ras activation, Huang et al. (2013) observed that cells in buffer make far fewer new Ras-GTP patches than pseudopods—only 0.6 Raf-RBD-GFP patch versus 3.8 pseudopods/min (Bosgraaf and Van Haastert, 2009b; Van Haastert and Bosgraaf, 2009a)—which argues against a fundamental role of Ras in basal pseudopod formation.
To address this discrepancy and investigate the role of Ras in basal cell movement in more detail, here we use a recently developed 10-fold–more-sensitive assay for the detection of activated Ras in live cells (Kortholt et al., 2013). Our data reveal as many as nine new Raf-RBD-GFP patches per minute. Simultaneously, detection of Ras activation, F-actin formation, and pseudopod extension in Dictyostelium cells in buffer and shallow gradients of cAMP show that all protrusions have strongly elevated levels of Ras-GTP, suggesting that Ras is part of both basal pseudopod formation and chemotactic signaling. Furthermore, we demonstrate that Ras activation and F-actin form excitable systems that are coupled through mutual positive feedback and memory. In buffer, this coupled excitable system induces the ordered-stochastic extension of pseudopods. In a shallow cAMP gradient, local Ras activation triggers full excitation of Ras and F-actin at the side of the highest cAMP concentration, leading to directed pseudopod extension and chemotaxis.
RESULTS
Cells in buffer contain multiple patches of active Ras
The RBD domain of mammalian Raf binds with high affinity to the active, GTP-bound state of Dictyostelium Ras but does not bind to the inactive, GDP-bound state. On Ras activation, the sensor RBD-Raf-GFP translocates from the cytoplasm to the plasma membrane. Although RBD-Raf-GFP has a high affinity for Ras-GTP, the translocation assay is not very sensitive because RBD-Raf-GFP in a boundary pixel not only is bound to active Ras at the membrane but is also present as soluble protein in the small cytosolic volume of the boundary pixels (Kortholt et al., 2013). Previously we showed that the sensitivity of this assay can be increased about 10-fold by coexpressing RBD-Raf-GFP and cytosolic red fluorescent protein (RFP; Bosgraaf et al., 2008). Cytosolic RFP in these boundary pixels is used to estimate the amount of cytosol in boundary pixels. Subtracting the RFP signal from the GFP signal gives a highly sensitive assay for active Ras. Data for each pixel are normalized by dividing by the average GFP signal in the cytosol and are presented as Ψ(i), which is directly proportional to the amount of activated Ras in that pixel (i; see Materials and Methods). Figure 1A shows a cell in buffer expressing both markers. In the green channel, two or three faint patches are visible at the boundary. However, after subtraction of the RFP signal, multiple patches are detectable. The fluorescence intensity at the boundary (Figure 1B) reveals at least five areas with increased Ras activation. Note that details on Ras activation in the green minus red signal at Ψ < 0.5 are undetectable in the GFP channel (see Materials and Methods).
FIGURE 1:
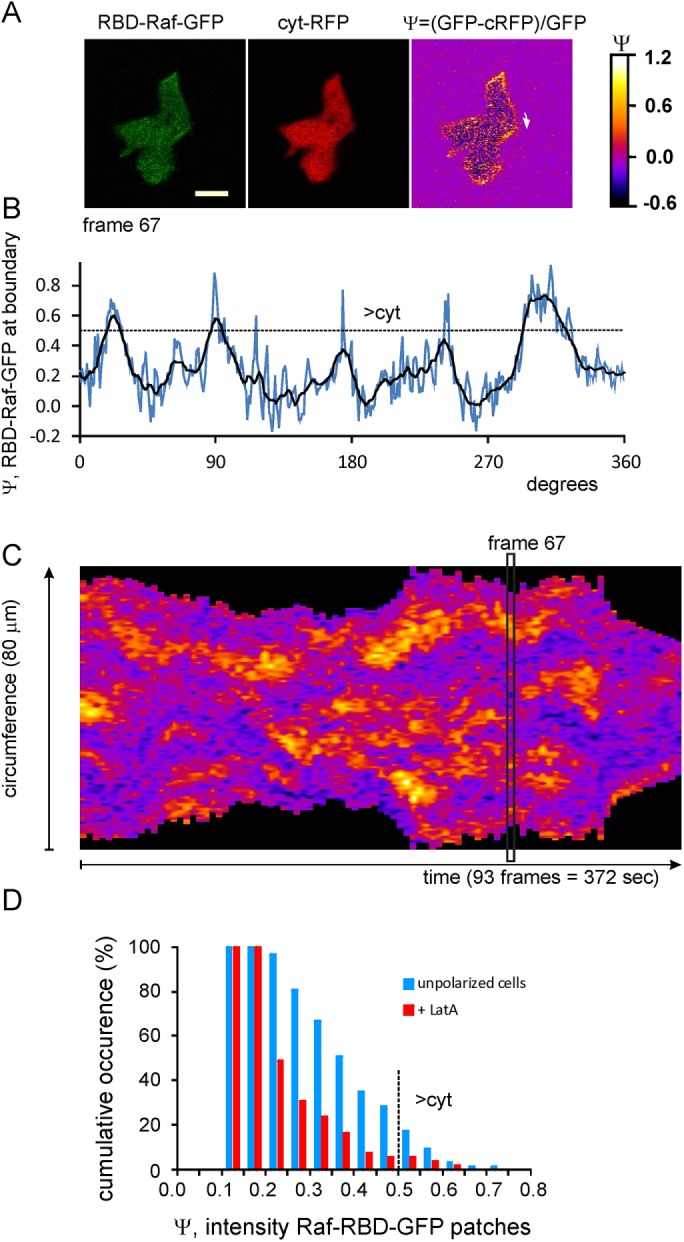
Ras-GTP patches in unpolarized Dictyostelium cells. Movies were made of cells in buffer expressing RBD-Raf-GFP and cytosolic (cyt) RFP (Supplemental Movies S1–S3). (A) Images of frame 67 in the green channel, the red channel, and the calculated GFP minus RFP signal Ψ, which is proportional to Ras-GTP (see Materials and Methods); scale bar, 5 μm. (B) Values of Ψ recorded in a line scan at a boundary 3 pixels wide (0.6 μm), starting at the arrow indicated in A. The dashed line at Ψ = 0.5 indicates that details below this line are not visible in the GFP channel. (C) Kymograph of the values of Ψ for the entire movie. This cell formed ∼53 Ras-GTP patches and extended 16 protrusions in the period of the movie. (D) Cumulative probability distribution of RAS-GTP patches in unpolarized cells with increasing intensity. The data represent 63 patches in buffer and 55 in LatA. Dashed line at Ψ = 0.5 indicates that only 10% of the patches in buffer and 5% of those in LatA are detectable in the GFP channel.
The kymograph of activated Ras at the boundary of an unpolarized cell moving in buffer reveals multiple small and large patches of activated Ras (Figure 1C). We define the minimal requirements for a patch of activated Ras as a group of pixels with an intensity of Ψ > 0.2, a width of at least 5 adjacent pixels (1 μm), and a duration of at least 2 pixels (8 s). According to this definition, the 372-s-long kymograph of Figure 1C shows 53 patches that have an average seize of 4.0 μm and an average lifetime of 24 s. On average, a cell has 3.6 ± 1.4 Ras-GTP patches (mean and SD, eight cells recorded for 5 min); with a lifetime of 24 s, cells extend a new Ras-GTP patch on average every ∼7 s (9.1 ± 1.0 new patches/min). About 90% of these patches have a relative fluorescence intensity Ψ < 0.5, because of which they were not easily detectable previously with less sensitive assays (Figure 1D). Indeed, previous experiments reported only 0.6 new Raf-RBD-GFP patch/min (Huang et al., 2013), consistent with present data showing ∼0.9 intense patch/min with Ψ > 0.5 (Figure 1D).
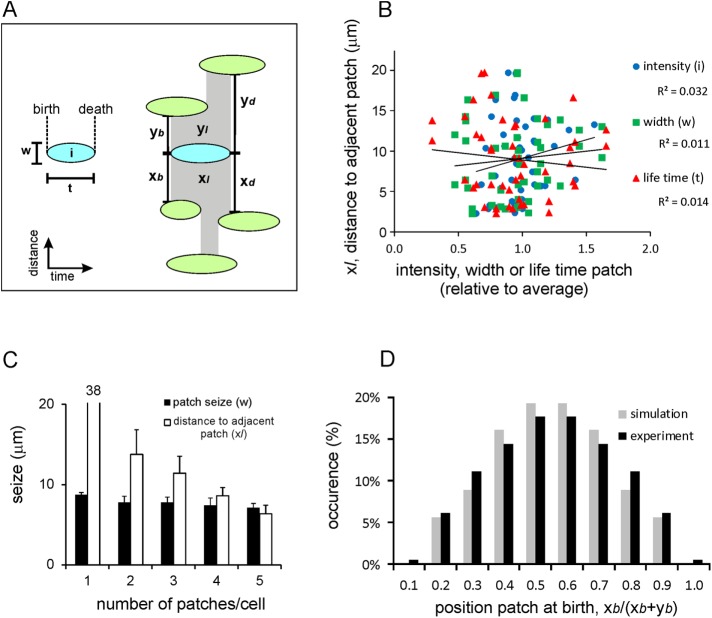
Many properties of a Raf-RBD-GFP patch appear to be independent of nearby patches (Figure 2). The lifetime, size, or intensity of a patch is indifferent to whether the two adjacent patches are very near or far away (Figure 2B) or whether the intensity of the adjacent patches is very high or very low (Supplemental Figure S1). In addition, the size of a patch is not affected by the number or intensity of other patches that are present simultaneously (Figure 2C). On the other hand, when a new patch appears between two existing patches, its location is not random but is more localized in the middle between the existing patches (Figure 2D). This may suggest that the existing patches produce a short-range inhibitor that reduces the probability that a new patch can arise near existing patches.
FIGURE 2:
Ras-GTP patches are not strongly influenced by neighboring patches. (A) Schematic of measurements. Left, a patch has a width w, lifetime t, intensity i, and given times of birth and death. Right, a patch is embedded by adjacent patches and has distances x and y to the adjacent patch to the right and left, respectively; these are measured at the times of birth (xb and yb) during life (xl and yl) and at the times of death (xd and yd) for the patch. (B) Lack of correlation between lifetime, width, or intensity of a patch and the distance to the adjacent patch (n = 23). All R2 values are very low and do not support a significant correlation. (C) A cell may have one or multiple Ras-GTP patches and patch-free spaces. The width of a Ras-GTP patch is independent of the number of other patches present; instead, patch-free spaces become smaller when more patches are present. Data are means and SEM from 50 patches. (D) Probability distribution of the appearance of a new Ras-GTP patch relative to the position of the two adjacent patches; at the moment of birth of a new patch, we measured distances xb and yb to the adjacent patches (data from 90 experimental Ras-GTP patches and 124 model R patches). The results show that a new Ras-GTP patch frequently starts somewhere in the middle of the open space between two patches. The data include cells with a single existing patch, where xb and yb represent the distances of the new patch to the right and left sides of that existing patch; the new patch preferentially appears at the opposite side of the existing patch. See Supplemental Figure S1 for other properties of Ras-GTP patches.
Patches of active Ras are associated with protruding activity
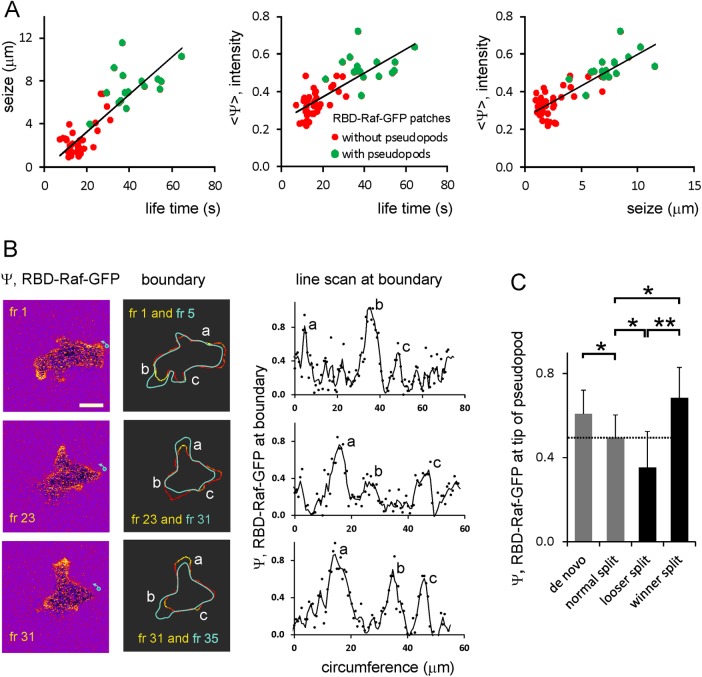
The cell presented in the kymograph of Figure 1B exhibits 53 Ras-GTP patches and has extended 16 protrusions during the 372 s of the recording. The average level of activated Ras at the entire boundary of the cell is Ψ = 0.28 ± 0.03 (eight cells); local patches have an intensity up to Ψ = 1.4. It appears that the membrane areas with protruding activity are highly associated with Ras-GTP patches. First, all protrusions occur in areas with Ras-GTP patches and never in Ras-GTP–free areas. Second, membrane areas with protruding activity contain Ras-GTP patches that are much larger, last longer, and have higher intensity than Ras-GTP patches that are not associated with protruding activity (Figure 3A). Third, when a cell has multiple Ras-GTP patches, a new protrusion starts at the patch with the highest intensity (Figure 3B). Fourth, occasionally an existing protrusion splits into two extending protrusions, which both contain a Ras-GTP patch. After 10–20 s, one protrusion dies, whereas the other survives; at the original moment of pseudopod splitting, the later winner already contains more Ras-GTP than the later loser protrusion (Figure 3C).
FIGURE 3:
Ras-GTP patches and pseudopod induction. (A) Correlation between size, lifetime, and intensity of Ras-GTP patches. Ras-GTP patches in pseudopodia are larger, live longer, and have a higher intensity than Ras-GTP patches in pseudopod-free areas of the cell. (B) Images (left) and outline (middle) and intensity at boundary (right) of Ras-GTP levels; scale bar, 5 μm. Top, frame 1, shows three major patches; the outline of frame 5 reveals that a protrusion was formed at patch b, which is the strongest Ras-GTP patch of frame 1. Middle, frame 23, shows pseudopod formation at the largest Ras-GTP patch a. Bottom, frame 31, shows a specific case; the strongest Ras-GTP patch a is present in a pseudopod that just stopped its extension, and a new pseudopod starts at the second-largest Ras-GTP patch b. (C) Ras-GFP levels in protrusions at the start of extension. De novo protrusions have more Ras-GTP than splitting protrusions. When a protrusion splits into two extending protrusions, a cell must retract one protrusion to prevent snapping; the data show that the winner protrusion had more Ras-GTP than the loser when they were formed. Data are means and SD of 12 determinations each; significant at *p < 0.05 or **p < 0.01.
Feedback loops between filamentous actin and Ras activation
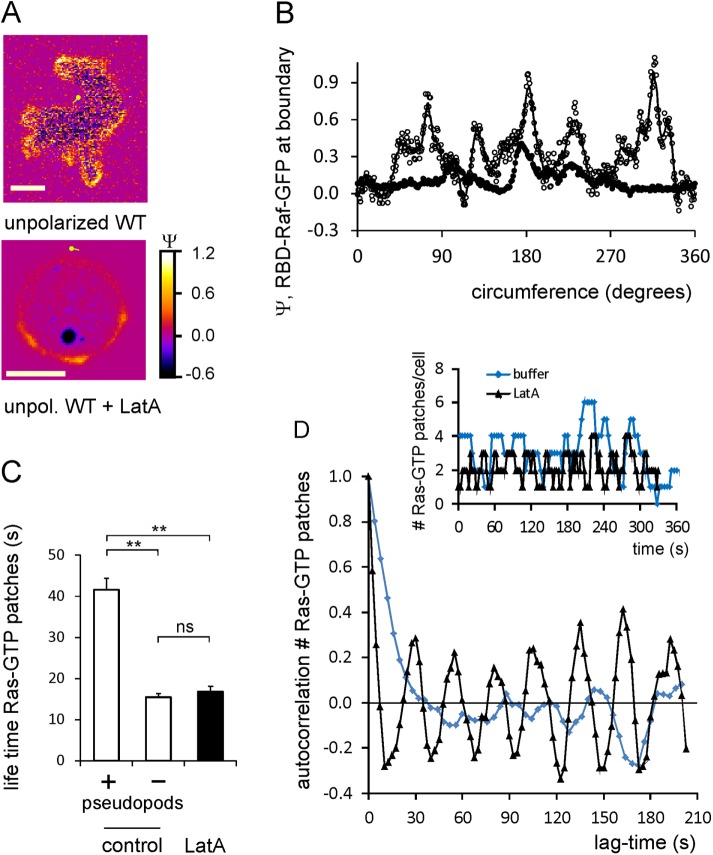
Unpolarized cells have an irregular shape with multiple Ras-GTP patches. On addition of the actin-polymerizing inhibitor latrunculin A (LatA), cells round up and protrusions disappear (Figure 4A). Ras activation in patches is still detectable but is reduced (Figure 4B); the mean Ras activation is <Ψ> = 0.31 ± 0.03 before LatA and <Ψ> = 0.13 ± 0.04 after addition of LatA (eight cells). The lifetime of Ras-GTP patches of control cells is 42 ± 11 s in protrusions and 16 ± 5 s outside protrusions; the lifetime of all Ras-GTP patches in LatA-treated cells is short, 16 ± 6 s (Figure 4C). In addition, the width and intensity of the Ras-GTP patches in LatA are essentially identical to the width and intensity of the subfraction of small Ras-GTP patches in control AX3 cells that are not associated to pseudopods (Supplemental Figure S2). The pronounced Ras-GTP patches that are associated with protrusions of control cells are not detectable in LatA-treated cells. On average, LatA-treated AX3 cells contains 2.2 ± 0.8 Ras-GTP patches/cell; with a lifetime of 16 s, this implies that cells extend a new Ras-GTP patch on average every ∼8 s (8.3 ± 1.5 in LatA vs. 9.1 ± 1.0 new patches/min without LatA, mean and SD; eight cells; not significantly different). Of importance, only 5% of the Ras-GTP patches have intensity Ψ > 0.5 (Figure 1D), consistent with the very scarce occurrence of 0.3 patch/min reported previously with the less sensitive assay (Huang et al., 2013).
FIGURE 4:
LatA inhibits Ras activation and uncovers a basal excitable Ras activation. (A) Ras-GTP levels of an unpolarized control cell and a cell in the presence of 5 μM F-actin inhibitor LatA; scale bar, 5 μm. (B) Line scan at the boundary of these cells. showing the levels of Ras-GTP; the control cell has approximately five Ras-GTP patches with Ψ ≈ 0.6, whereas the LatA-treated cell has only three Ras-GTP patches with Ψ ≈ 0.3. (C) Lifetime of Ras-GTP patches. In control cells, Ras-GTP patches in pseudopods live much longer than Ras-GTP patches in other areas of the cell. All Ras-GTP patches of LatA-treated cells have a short lifetime. Data show means and SEM with 16, 35, and 28 patches, respectively; significant at **p < 0.01; ns, not significant. (D) Autocorrelation analysis showing periodic Ras-GTP formation in LatA. Inset, number of Ras-GTP patches present in a typical control cell or a cell in LatA. Main graph, autocorrelation of this number at different lag times. Control cells have a relatively long autocorrelation time (time constant of exponential decay of autocorrelation at short lag times; t1/2 = 10.7 s) and no strong oscillations. Cells in LatA have a very short autocorrelation time of t1/2 = 3.4 s, and exhibit very strong oscillations with a periodicity of ∼28 ± 3 s. Data are derived from four cells for each condition.
The results suggest that F-actin stimulates Ras activation in protrusions of unpolarized cells in buffer. Because active Ras is known to induce the formation of F-actin (Cai et al., 2010), the combined data imply the presence of a positive feedback loop from F-actin to Ras-GTP and back to F-actin. In LatA, this positive feedback loop is interrupted, suggesting that the Ras-GTP patches observed in LatA are small, autonomous Ras-GTP patches formed in an excitable medium of Ras activation. To investigate the autonomous Ras activator further, we recorded the number of Ras-GTP patches per cell during several minutes in LatA-treated cells (inset, Figure 4D). Autocorrelation analysis of the variation in the number of Ras-GTP patches reveals that the autocorrelation time of Ras-GTP patches (the time period in which the autocorrelation decreases to zero) is much shorter for cells in LatA than for control cells (Figure 4D). This corresponds well with the measured shorter average lifetimes of 17 and 24 s for LatA and control cells, respectively. Of importance, autocorrelation of the number of Ras-GTP patches in LatA-treated cells exhibits very pronounced oscillations with a periodicity of 27 ± 3 s. This periodicity is consistent with the observation that about every 8 s, a LatA-treated cell makes a new Ras-GTP patch that has a lifetime of 17 s. Such a strong oscillatory behavior is not observed in control cells without LatA, probably because control cells have a mixture of small, short-lasting (16 s) Ras-GTP patches and large, long-lasting (42 s) Ras-GTP patches that are associated with protrusions.
Left/right asymmetry of pseudopod formation and Ras activation in polarized cells
Starved cells become elongated and polarized (Figure 5). The mean that Ras activation of polarized cells (<Ψ> = 0.31 ± 0.03) is not different from that for unpolarized cells (<Ψ> = 0.30 ± 0.02), but Ras-GTP is enriched in a strong patch at the leading edge, and several smaller Ras-GTP patches with lower intensity are present at the sides of the cell (Figure 5). Polarized Dictyostelium cells predominantly make protrusions by splitting of an existing protrusion at the front of the cell (Figure 5A). These splittings are very frequently alternating to the right and left, by which cells have a strong tendency to follow a persistent zigzag pattern (Li et al., 2008; Bosgraaf and Van Haastert, 2009b). Figure 5B shows Ras-GTP levels at the boundary of a subset of polarized cells that are all extending a protrusion to the left. In addition to the strong patch of activated Ras in the extending protrusion, a patch of activated Ras is present at the right of this protrusion, where the previous pseudopod was extended and the next protrusion is expected to be formed. The experiments thus reveal that moving polarized cells have a memory of pseudopod extension and Ras-GTP patches.
FIGURE 5:
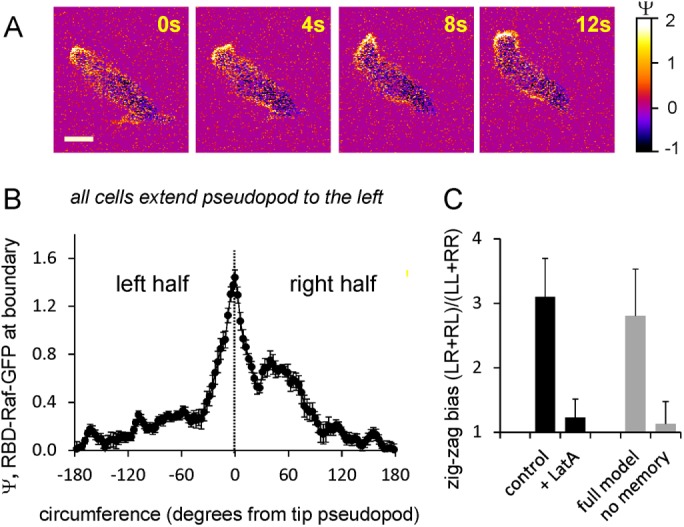
Left–right asymmetry of Ras activation and pseudopod formation. (A) Ras activation in a polarized cell moving to the upper left corner and then extending a splitting to the right. (B) Ras activation at the boundary of 19 cells all extending a pseudopod to the left. The cells have a pronounced Ras-GTP patch at the leading pseudopod but also elevated Ras-GTP levels at the right half of the cell at ∼55°, where the previous and the next pseudopod form. The data are means and SEM with n = 19. (C) Right/left bias. The formation of protrusions (control) or Ras-GTP patches (LatA) was recorded as being to the right or left relative to the previous one. For each triplet of protrusions or Ras-GTP patches, its configuration was identified as RR, RL, LR, or LL. In random movement, (RL + LR)/(RR + LL) is expected to be 1.0, whereas a value >1 indicates a preference for alternating R/L zigzag movement. The data are presented as means and SD of 12 cells each with a track of at least 20 protrusions. The gray bars represent the zigzag bias of P patches in model simulations, either the full model or no memory (k6 = 0). Data are means and SD with 20 simulations during 20 min.
Memory plays a critical role in persistent cell movement and efficient chemotaxis (Li et al., 2008; Bosgraaf and Van Haastert, 2009a,b; Cooper et al., 2012; Skoge et al., 2014). Does the basic excitable system for Ras activation has memory leading to memory of protrusions, or is the memory of Ras-GTP patches the consequence of memory in F-actin/protrusions? Control cells exhibit a strong left (L)/right (R) bias (Figure 5C; (RL + LR)/(LL + RR) = 3.1; random would be 1.0). We measured in LatA-treated cells the position where a new Ras-GTP patch started in relation to the position where the previous patch disappeared. A new Ras-GTP patch appears equally probable at the left or right side of the previous Ras-GTP patch, leading to a very low L/R bias (Figure 5C; (RL + LR)/(LL + RR) = 1.2). We also measured the actual distance where a new Ras-GTP patch starts relative to where previous Ras-GTP patches extinguish. With a circumference of 32 μm of LatA-treated cells, the expected random distance is 8 μm to the left or right of the previous Ras-GTP patch; if memory is present, the distance is expected to be ≪8 μm. We observed that the distance between a new and old patch is 7.8 ± 1.2 μm (mean and SEM, n = 31), which is statistically not different from random. These data thus suggest that the basic Ras-excitable system has no detectable memory.
Either F-actin or Ras-GTP can be the trigger for a new protrusion
Our data show that relatively high levels of activated Ras are present locally at the membrane where a protrusion starts (Figure 3), supporting current models that Ras-GTP acts upstream of the machinery that induces local F-actin polymerization (Sasaki et al., 2004; Bolourani et al., 2006). However, our data also show that F-actin strongly enhances excitable Ras activation (Figure 4; Sasaki et al., 2007). This suggests that Ras activation and F-actin form a positive feedback loop in extending protrusions. With such a positive feedback loop, activated Ras may induce F-actin/protrusions, F-actin/protrusions may induce Ras activation, or both may occur. What is the trigger for protrusions, Ras-GTP or F-actin?
To untangle this F-actin/Ras positive-feedback loop, we made careful kinetic analysis of the local activation of Ras and formation of F-actin in individual protrusions. We performed experiments in wild-type cells expressing RBD-Raf-GFP and LifeAct-RFP, which exhibit similar rate constants of dissociation from Ras-GTP and F-actin, respectively (see Materials and Methods). We recorded the moment and position at the membrane where a protrusion started. Subsequently we determined back in time when F-actin and activated Ras were beginning to increase at that location (see Supplemental Figure S3 for the procedure). The analysis of seven de novo pseudopods revealed that RBD-Raf-GFP started 2.0 ± 0.3 s and LifeAct-RFP 0.8 ± 0.3 s before the protrusion. In all seven de novo pseudopods, Ras activation started first, ∼1.3 s before F-actin formation (Figure 6). Of interest, the analysis of 21 splitting pseudopods revealed the opposite: in 76% of the cases, F-actin started before Ras activation. The increase of LifeAct-RFP preceded the protrusion by 1.9 ± 0.5 s, whereas RBD-Raf-GFP started to increase 0.7 ± 0.4 s before the protrusion; thus Ras activation started 1.2 s after F-actin formation (mean and SD of all 21 splitting pseudopods).
FIGURE 6:
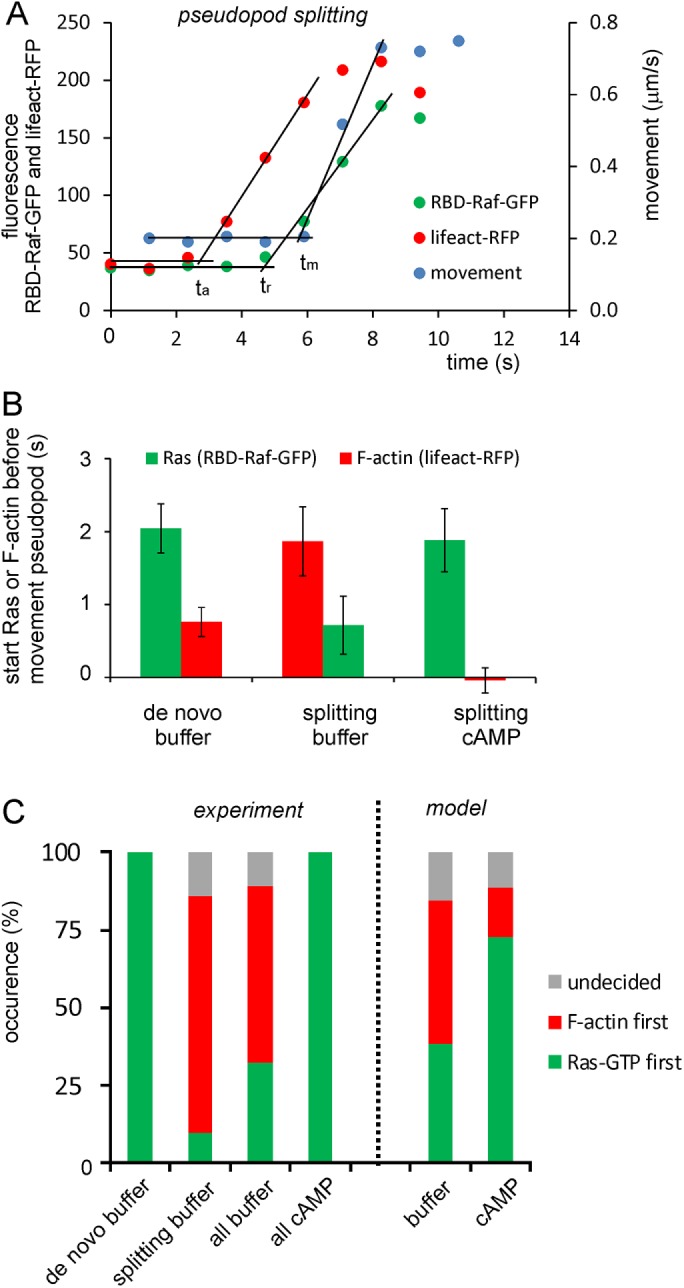
Kinetics of Ras activation, F-actin formation, and the extension of pseudopods. Confocal movies of cells expressing RBD-Raf-GFP and LifeAct-RFP to detect Ras-GTP and F-actin, respectively, were recorded at 1.18 s/frame. For seven de novo pseudopods and 22 splitting pseudopods, the frame number was identified when the extension of the protrusions started, and then in several frames before and after that moment the exact position of the tip of the pseudopod and the fluorescence intensity of RBD-Raf-GFP and LifeAct-RFP around that position were determined. (A) Raw data of one splitting pseudopod; three data sets with initially low values that increase and then level off at a high level. Each data set was fitted with two linear regression lines—a horizontal line for the data points before the increase and a line for the increasing data points. The selection of the data points belonging to these two lines was optimized using the lowest sum of residual squares of the two regression lines. The intersection of these two lines represents the time point (tx) where the data set started its increase. The increase of LifeAct-RFP (ta) and RBD-Raf-GFP (tr) started 3.11 and 1.17 s, respectively, before the start of the moving pseudopod (tm). Raw data and its analysis for a de novo pseudopod are presented in Supplemental Figure S3. (B) Combined means and SD of seven de novo and 22 splitting pseudopods in buffer. Movies were also recorded in a gradient of 0.1 nM/μm cAMP. Chemotaxis cells extend mainly splitting pseudopods (Bosgraaf and Van Haastert, 2009a). The data show the means and SD of 12 splitting pseudopods. (C) Percentage of pseudopods in which RBD-Raf-GFP (green) or LifeAct-RFP (red) started at least 1 s before the other; the gray bar represents cases in which this difference is <1 s. The data are derived from the same experiment as in B. The data for the model present the percentage of cases in which R (green) or P (red) started at least 1 s before the other; the gray bar represents cases in which this difference is <1 s (Figure 7D).
The chemoattractant induces pseudopods by Ras activation
The chemoattractant cAMP induces Ras activation, which is strongest at the side of the cell facing the highest concentration of chemoattractant (Sasaki et al., 2004; Kortholt et al., 2013). We analyzed the position and kinetics of Ras activation and F-actin formation in shallow cAMP gradients (Figure 6). As mentioned, splitting pseudopods in buffer begin with F-actin formation, followed after ∼1 s by Ras activation, and still 1 s later, the extension of the protrusion starts. In contrast, in cells moving in a shallow cAMP gradient, nearly all protrusions are preceded by Ras activation, which is followed after ∼2 s by both F-actin formation and the extension of the protrusion. This strongly suggests that in a cAMP gradient, the splitting pseudopod is not triggered by the endogenous F-actin memory but by the cAMP-induced Ras activation. We therefore postulate that cAMP-induced Ras activation “mixes” with the endogenous repertoire of activated Ras/F-actin of unstimulated cells described earlier. Protrusions are still made at areas with the strongest Ras activation, but the position of the activated Ras patch has a positional bias that depends on the direction and steepness of the cAMP gradient.
A computational model for coupled Ras/F-actin activation
The experiments suggest that in buffer, Ras activation occurs in an excitable medium, leading to multiple Ras-GTP patches with specific distributions of number, width, lifetime, and intensity. We designed a computation model to investigate three aspects: 1) the coupling between Ras-GTP patches, F-actin, and pseudopod formation; 2) the role of memory in the system; and 3) the bias by local external gradients of cAMP. The model combines components of previously published models on patches, coupled oscillators, local excitation global inhibition (LEGI) for stimulation by cAMP, and memory for alternating right/left extension of pseudopods (Hecht et al., 2010, 2011; Cooper et al., 2012; Yan et al., 2012; Huang et al., 2013; Shi et al., 2013). Our model does not include deformable cells (Hecht et al., 2011; Neilson et al., 2011; Shi et al., 2013) and polarity other than by memory, such as a front–rear gradient of myosin filaments in starved elongated cells (Nakajima et al., 2014).
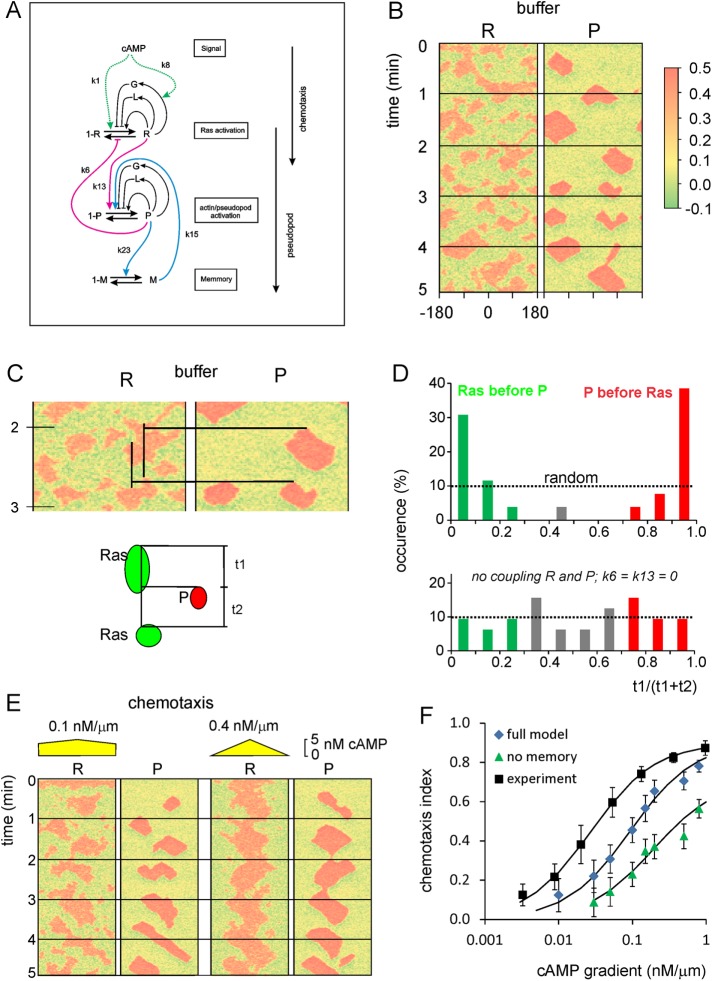
A simple reaction-diffusion model for excitable Ras-GTP patches consists of a local activator and a global inhibitor that are both enhanced by active Ras-GTP (R). Because new patches are not likely being formed close to existing patches but are otherwise independent of other Ras-GTP patches, we included a short-range local inhibitor that is induced by activated Ras (Figure 7A). We formulated a similar model for a hypothetical excitable molecule (P) that induces protrusions. The molecular identity of P is not specified; it is not the protrusion itself, but it includes molecules such as Rac-GTP, activated Scar, Arp2/3, and F-actin, which induce the beginning of a protrusion. These two excitable loops are coupled to each other by two reactions, R stimulating the formation of P, and P inhibiting the degradation of R. Furthermore, the chemoattractant regulates the formation of Ras-GTP in a LEGI-type scheme by stimulating both the formation of R and, somewhat more slowly, the formation of the global inhibitor of R. Finally, we include memory (M); experiments reveal memory in pseudopod formation but not in basic Ras activation (Figure 4). Therefore the formation of M is stimulated by P; M degrades slowly and stimulates the formation of new P patches (Figure 7A). In the equations describing this model, we include Gaussian white noise for R and P (Materials and Methods). Parameters for diffusion rate constants and kinetic reactions were chosen to yield the multiple patches of R matching experimental properties with respect to number, width, and duration of activated Ras and to yield patches of P matching the number and duration of protrusions (see Supplemental Figure S4 for details on parameter selection). Simulations were performed in one dimension with 120 grid points on a circle with radius 6.25 µm; the model does not include cell deformation.
FIGURE 7:
Computational model for the coupled excitable activation of Ras and inducer of protrusions. (A) Scheme of the model. Ras-GTP is represented by R, which is formed by GEFs that are autocatalytically activated by R or by cAMP and are inhibited by a global inhibitor and a local inhibitor; R is degraded by GAPs. The inducer of protrusions, which could be Rac-GTP, Scar, or F-actin, is represented by P and is regulated in a similar way as R. The excitable R and P are coupled (red lines). Finally, experiments reveal that the system has memory, which resides in protrusions but not in Ras-GTP; it is modeled as a slowly degrading M that is formed in response to P and stimulates the formation of P. The model contains Gaussian white noise, which is slightly larger for R than for P, and by which patches of R are formed more frequently than patches of P. Simulations are performed on a circle with 120 grid points. (B) Kymograph of R and P for a typical simulation in buffer. (C, D) Coupling of R and P. The schematic shows a red patch of P. At the grid point of the start of P, the time intervals t1 and t2 were determined at which in that grid point the previous and the next patch of R started. The kymograph in C shows two cases; the upper P patch is closely preceded by an R patch, and the next R patch follows much later; the lower P patch shows the opposite. The data from 58 P patches are summarized in D; bottom, coupling constants k6 and k13 are set to zero. (E) Kymograph of R and P for typical simulations in a shallow (left) and steep (right) gradient of cAMP. (F) The chemotactic index was calculated using the position where each P patch started relative to the highest cAMP concentration (Supplemental Figure S5). Data are means and SD of 100 simulations for each cAMP gradient for the full model and the model lacking the memory (k15 = 0). The experimental data were collected before using micropipettes (Kortholt et al., 2013).
Figure 7B shows the outcome of a 5-min simulation in buffer, revealing multiple patches of (R) with a range of sizes and durations and only a few patches of P. The noise in R and P generates a new outcome of the model for each simulation. We varied parameter settings and analyzed ∼100 simulations for each condition. The model successfully predicts many experimental observations: new patches of R start preferentially in the middle between existing patches (Figure 2D), and patches of P are formed alternating to the right and left, depending on M (Figure 5C).
We investigated the properties of the model with respect to the coupling between R and P and how this is biased by a gradient of cAMP. We identified the grid point where a new P patch begins and then identified for that grid point the times t1 and t2 that the previous or next R patch started, respectively (Figure 7C). The results reveal that R and P patches are strongly coupled; with the chosen settings of the coupling constants k6 and k13, ∼40% of the P patches start just after an R patch, and 45% of the P patches start just before an R patch (Figure 7D). When the coupling constants k6 and k13 are set to zero, P patches start any time relative to R patches, essentially as expected for random uncoupled formation of P patches (Figure 7D, bottom). In a gradient of cAMP, which induces the activation of R patches, significantly more P patches start immediately after R than in buffer, as also observed experimentally (Figure 6C).
As mentioned earlier, the model parameter settings were chosen to produce more R patches than P patches, as observed experimentally. Thus, although R and P patches are coupled, only a fraction of the R patches overlap with a P patch. This was also observed experimentally; only about half of the Ras-GTP-patches are associated with protrusions, whereas other Ras-GTP patches are formed in pseudopod-free areas of the cell. It appears that the width, lifetime, and intensity are significantly larger in R patches associated with a P patch than in R patches not associated with a P patch, as observed experimentally (Supplemental Figure S2). We modeled LatA-treated cells by setting the coupling constant k6 between P and R to zero (i.e., the protrusion does not stimulate Ras-GTP formation). The results reveal that all R patches are small with respect to size, lifetime, and intensity, exactly as observed experimentally (Supplemental Figure S2). We conclude that the coupled excitable R–P model successfully describes many observations on Ras activation, F-actin formation, and cell movement in buffer.
Next we investigated chemotaxis in the model cell; we kept all parameters the same, except for a small stimulation of R by cAMP. In a gradient of cAMP, more R patches are formed at the side of the model cell facing the highest cAMP concentration, and consequently also more P patches are formed at that side. To model cell movement, we assume that a protrusion starts at the position of the first grid point of an new patch of P and that this protrusion is 5 µm long and perpendicular to the cell surface (Van Haastert and Bosgraaf, 2009b; Supplemental Figure S5A). The derived tracks show that cells in buffer move in all directions. In a cAMP gradient, the P patches are formed mainly at the side of the cell facing the highest cAMP concentration (Figure 7E). As a consequence, the protrusions point in the direction of cAMP (Supplemental Figure S5E). From these model tracks, a chemotaxis index can be calculated and compared with experimentally observed data (Figure 7F). The results reveal that experimental data are only slightly better than the chemotaxis index predicted by the model.
Finally, we investigated the role of memory on the model cell. Living cells exhibit a right/left bias of pseudopod extension with approximately threefold preference for alternating LR + LR extensions relative to consecutive RR or LL extensions (Figure 5C). The tracks of the model cell (Supplemental Figure S5) show many alternating zigzag segments with a right/left bias of 2.8 ± 0.7. Deletion of M from the model (i.e., k15 = 0) yields tracks with a right/left bias of 1.1 ± 0.3, which is statistically not different from 1.0 for random (Figure 5C). Cell movement of amoeboid cells is characterized by a correlated random walk (CRW): the direction of the next pseudopod is not random but correlated with the direction of the previous step, leading to persistence of movement. The analysis of the mean square displacement as a function of the number of steps provides a very sensitive method to uncover the mode of movement. Dictyostelium cells exhibit a persistence of 3.4 min, equivalent to approximately seven protrusions (Bosgraaf and Van Haastert, 2009b). The model cells also exhibits a CRW with a persistence of 6.1 steps (Supplemental Figure S5D). Removing M from the model does not alter the speed of the model cell but strongly reduces the persistence to 1.2 steps. Finally, in a gradient of cAMP, the trajectories of model cells are relatively straight in the direction of cAMP, but this is less obvious in model cells without memory (Supplemental Figure S5F), leading to a strongly reduced chemotaxis index (Figure 7F). Thus memory contributes to alternating right/left pseudopod extensions, persistence of cell movement, and improvement of chemotaxis.
DISCUSSION
Our new data provide detailed information on the role of Ras in basal pseudopod formation. First, the guanine exchange factors (GEFs) and GTPase-activating proteins (GAPs) form an excitable medium for Ras activation to produce Ras-GTP patches at a high frequency. Second, Ras-GTP and F-actin form a positive feedback loop by which Ras activation is amplified in protrusions. Third, protrusions are initiated by Ras activation or F-actin formation.
The data strongly suggest that the F-actin/Ras coupled excitable system is a critical component in buffer to induce protrusions and that it receives information from the signaling pathways to induce chemotaxis. Previously it was difficult to define the role of activated Ras for excitable F-actin and basal pseudopod formation because the less sensitive assay for activated Ras could detect only the strongest Ras-GTP patches, which prevented the characterization of the high-frequency excitable Ras system (Hecht et al., 2011; Huang et al., 2013; Kortholt et al., 2013). Our data now show that Ras is at the heart of chemotaxis, being part of both a coupled F-actin/Ras excitable system for basal pseudopod formation and a receptor-stimulated signaling pathway mediating directional information.
Excitable Ras activation
Cells form Ras-GTP patches at a high frequency of approximately nine new patches per minute. This is about 15 times faster than could be observed previously with less sensitive methods. Of importance, cells make approximately two to three times more Ras-GTP patches than pseudopods. Because hyperactivation of Ras induces pseudopods (Cai et al., 2010), and in wild-type cells, all pseudopods contain elevated levels of Ras-GTP (Figure 3), we conclude that activated Ras is a critical component of basal pseudopod formation. The strong correlation between size, intensity, and lifetime strongly suggests that the Ras-GTP patches are self-amplifying, a critical property of excitability. We observed that existing patches do not influence each other, suggesting that the self-amplification within a patch does not extend to adjacent patches. Ras excitability for new Ras patches is not uniform at the membrane but low close to existing patches and maximal in the middle between existing patches. The model with a local inhibitor produced by Ras-GTP patches successfully reproduces the observed landscape of excitability.
Coupled excitable Ras and F-actin
In the absence of F-actin, the intrinsic Ras module leads to small Ras patches. In the presence of F-actin, Ras activation is strongly enhanced in F-actin filled protrusions, and, conversely, activated Ras enhances the formation of F-actin; together these lead to a crucial positive feedback loop between F-actin and Ras. Several pathways, including TORC2, phosphatidylinositol 3-kinase, and the small GTP-binding proteins Rap1 and Rac, may contribute to Ras-stimulated actin polymerization (Sasaki et al., 2004; Charest and Firtel, 2007; Charest et al., 2010; Cai et al., 2010; Kortholt et al., 2011; Liu et al., 2016). The second component of the positive feedback loop—the molecular mechanism by which F-actin regulates Ras activation—is not well known. Filamentous actin is present in the entire cortex of the Dictyostelium cells as relatively stable parallel bundles, whereas in protrusion, F-actin is present more as a dynamic dendritic network (Gerisch and Ecke, 2016). Most likely, the dendritic bundles are instrumental in both Ras activation and pseudopod formation. Ras activation may be mediated by F-actin–binding RasGEFs (Kortholt et al., 2013; Liu et al., 2016; Senoo et al., 2016). Alternatively, Ras activation by F-actin polymers could be indirect (e.g., a F-actin–binding protein stimulates a RasGEF), or it could be through coincidental activation (e.g., TORC2 regulates both Ras and F-actin; Charest et al., 2010; Kortholt et al., 2011). In addition, cytoskeletal components such as myosin filaments and IQGAP/cortexillin may inhibit Ras activation indirectly (Lee et al., 2010) because they inhibit the formation of dendritic F-actin in the rear and at the sides of the cell. Many reports have shown that actin filament formation occurs in an excitable medium (Bretschneider et al., 2004, 2009; Ryan et al., 2012; Huang et al., 2013; Gerhardt et al., 2014; Tang et al., 2014; Bement et al., 2015; Gerisch and Ecke, 2016; Yang et al., 2016). Of importance, the Ras and F-actin excitable systems are coupled because dendritic F-actin stimulates Ras activation, and active Ras leads to F-actin polymerization. Coupled excitable systems are very sensitive to small fluctuations of internal or external regulators. Such internal factors could be cell shape, membrane tension, local differences in diffusion, position of the nucleus, or some form of memory.
Protrusions are triggered by activation of Ras or F-actin
In a coupled excitable system, stochastic variation in the concentration of one of the components (noise) can bring the coupled system easily to an excited state. Dictyostelium cells can extend either of two types of pseudopod: splitting of an existing pseudopod or formation of a de novo pseudopod in an area that was devoid of pseudopod activity for some time. De novo pseudopodia are formed at the side of the cell where the formation of dendritic F-actin is suppressed by myosin filaments and IQGap/cortexillin (Bosgraaf and Van Haastert, 2009b; Lee et al., 2010). Therefore the probability that the coupled system can start a de novo protrusion with F-actin is low. Indeed, we observed that all de novo protrusions started with an increase of Ras-GTP. On the other hand, pseudopod splitting occurs in an environment of existing dendritic F-actin, and we saw new protrusions triggered by F-actin rather than activated Ras.
Coupled excitable systems are also very sensitive to small fluctuations of an external regulator, such as a chemoattractant gradient that stimulates one of the components of the coupled system. In Dictyostelium, the chemoattractant cAMP stimulates Ras activation. Of importance, almost all protrusions in a cAMP gradient start with a local patch of activated Ras, whereas F-actin is formed later, simultaneous with the extension of the pseudopod. Thus the cAMP-induced local Ras activation increases the probability that the coupled excitable system starts amplification to a full-grown Ras-GTP patch at the place of the initial cAMP-induced Ras activation, and this leads subsequently to an F-actin patch making protrusion in the direction of the cAMP gradient. Our model of this coupled excitable module successfully explains many experimental observations on cell movement in both the absence and presence of chemoattractants. The coupled excitable system is not only very efficient for chemotaxis, but also may allow other forms of coordinated movement, such as collective movement, by which cell–cell contact may trigger Ras activation, or electrotaxis, by which an electric field may trigger mobility or activation of Ras.
Although Ras and F-actin are entangled in a positive feedback loop, and therefore all pseudopods have elevated levels of activated Ras, note that pseudopod formation is still possible in the absence of Ras activation (Bolourani et al., 2006) with the F-actin excitable system alone. Thus rasC/G-null cells can form protrusions and move in buffer, but they cannot chemotax efficiently to cAMP because Ras is an important intermediate between receptor and the cytoskeleton.
Memory
In addition to the coupled Ras/F-actin–excitable system, the system contains memory: cells exhibit a bias toward alternating right/left extension of pseudopods. Our experiment reveal that the basal excitable Ras activation has no memory, suggesting that memory of directional movement is located in some component of the pseudopod cytoskeleton that slowly fades away after the pseudopod has stopped. The role of the memory can be deduced from experiments with mutant cells, revealing two main functions. In buffer, alternating right/left movement allows the cell to move in a persistent manner, thereby exploring a much larger area in the same amount of time (Van Haastert and Bosgraaf, 2009a). During chemotaxis, memory serves as an integrator of directional information by which cells chemotax much more efficiently in shallow gradients (Bosgraaf and Van Haastert, 2009a). The proposed roles for right/left memory is well replicated by M in the computational model, showing that model cells without memory lose the right/left bias, lose persistence in buffer, and exhibit impaired chemotaxis. Our computer model was designed to investigate the kinetic interaction between Ras and pseudopod-inducing activity and its activation by chemoattractant gradients. The model did not incorporate components that could further improve gradient sensing and directional movement. Cell deformation will lead to a cell that is elongated in the direction of the gradient, thereby sensing a larger difference between front and rear (Tweedy et al., 2013). Starved cells are polarized due to myosin filaments in the rear that inhibit pseudopods in the rear, thereby increasing the accuracy of movement in gradients (Bosgraaf and Van Haastert, 2009a; Shi et al., 2013).
A model for chemotaxis
In our present view of chemotaxis, the coupled excitable Ras/F-actin module forms the beating heart of basal and directed cell locomotion. In the absence of external signals, local stochastic fluctuations of Ras-GTP, F-actin, or one of their regulators triggers excitation and pseudopod formation. In a chemoattractant gradient, local stimulation by chemoattractant receptors is easily sufficient to trigger local excitation of Ras. In this view, a simple signaling pathway from chemoattractant receptors to Ras activation is essential and sufficient for chemotaxis, as we reported previously (Kortholt et al., 2011). However, nature is not simple, and many additional signaling pathways have been reported that improve chemotaxis, none of them being essential. For Dictyostelium, these pathways include phosphoinositide 3-kinase , PLA2, TORC2, and guanylyl cyclase, which probably enhance signaling efficiency between Ras and F-actin (Veltman et al., 2008; Bosgraaf and Van Haastert, 2009a; Kortholt et al., 2013). Besides enhancing chemotaxis in shallower gradients, the additional signaling pathways may also contribute to improved directed cell movement in complex environment with irregular chemoattractant gradients and complex three-dimensional surfaces (Skoge et al., 2016).
Basal cell movement and chemotaxis of mammalian cells may be regulated by a similar coupled excitable system composed of a small GTP-binding protein and F-actin. In neutrophils, the GTP-binding protein is most likely cdc42, which possesses excitable properties in buffer and is stimulated by the chemoattractant (Yang et al., 2016), similar to excitable Ras in Dictyostelium. Therefore the coupled excitable model presented here for Dictyostelium is likely to be applicable for mammalian cells as well but with different identities of the excitable R and P.
Why do chemotactic cells move when there is no chemoattractant? We showed here that the central Ras/F-actin excitable module is so sensitive to shallow gradients of chemoattractants because the module is active in the absence of chemoattractants. Because cells move in the absence of chemoattractant, there is no threshold of chemoattractant for movement or chemotaxis. Thus each cAMP molecule that binds to the receptor induces some Ras activation and contributes to a positional bias of excitable Ras activation. At the lower limit of chemotaxis, the cell does not need a compass for navigation that determines where it is to go. Instead, the protrusion formed by the coupled excitable system in buffer is also formed in a shallow gradient but with a small positional bias. The limit for directional movement then depends only on the statistical probability that more chemoattractant receptors are activated at the side of the cell facing the highest concentration (Van Haastert and Postma, 2007). Some cells, such as human neutrophils and Dictyostelium, need optimal chemotaxis for survival of the species and operate at the physical limit of chemotaxis. We postulate that these cells have a machinery for movement in buffer that is optimized for chemotaxis. In such cells, the characterization of cell behavior in buffer is instrumental for understanding how these cells sense and respond to external cues (Bray, 2014).
MATERIALS AND METHODS
Cell culture and preparation
To study Ras activation, we coexpressed RBD-Raf-GFP (amino acids 50–134 of RAF1) with cytosolic mRFP in wild-type AX3 cells from a single plasmid as described (Bosgraaf et al., 2008; Veltman et al., 2009; Kortholt et al., 2013). To visualize simultaneously F-actin and Ras-GTP, we replaced mRFP of the foregoing vector by LifeAct-RFP. Cells were grown in HL5-C medium including glucose (Formedium) containing 50 μg/ml hygromycin B (Invitrogen) for selection. Cells were collected and starved on nonnutrient agar for 2 h (unpolarized cells) or 5 h (polarized cells). Cells were then harvested, suspended in 10 mM KH2PO4/Na2HPO4, pH 6.5 (PB), and used in experiments. Confocal images were recorded using a Zeiss LSM 510 META-NLO confocal laser scanning microscope equipped with a Zeiss Plan-Apochromatic 63×/ numerical aperture 1.4 objective.
A sensitive assay for Ras activation at the cell boundary
In Dictyostelium, Ras is located uniformly at the plasma membrane, where it can shuttle between an inactive, GDP-bound state and an active, GTP-bound state. GEFs activate Ras proteins, whereas GAPs catalyze the inactivation of Ras-GTP (Kortholt and van Haastert, 2008). Stimulation of cells with cAMP does not change the localization of Ras but converts Ras from the inactive, Ras-GDP state to the active, Ras-GTP state (Sasaki et al., 2004; Sasaki and Firtel, 2009). The RBD domain or mammalian Raf binds specifically to the GTP form of Ras. On cAMP stimulation, RBD-Raf-GFP translocates from the cytoplasm to the cell boundary. Assays measuring the activation of a membrane protein using the translocation of a cytosolic marker to the cell boundary are fundamentally insensitive because a boundary pixel contains membrane and an unknown amount of cytosol. By coexpressing RBD-Raf-GFP and cytosolic RFP from one plasmid, we use the RFP signal to estimate the cytosolic volume, which allows us to calculate the amount of RBD-Raf-GFP that specifically binds to Ras-GTP at the membrane (Bosgraaf et al., 2008; Kortholt et al., 2013). Calculations used the following steps. First, the images of the movie are corrected if needed for bleaching using the EMBL ImageJ plug-in Bleach corrector; usually, the RFP signal shows 10–30% bleaching during 5 min. Then the mean background fluorescence intensity in the red and green channels outside the cells is determined and subtracted from all pixels of the movie. Subsequently individual cells are analyzed. To correct for the difference in expression levels of the two markers within one cell, large areas of the cytoplasm are selected (excluding nucleus and vacuoles), yielding the mean fluorescence intensity in the cytoplasm of the red (<Rc>) and green channels (<Gc>), respectively. This provides the correction factor, c = <Gc>/<Rc>, and all pixels in the red channel are multiplied by c. Then, for each pixel i of that cell, we calculate the difference of green and corrected red signals and normalize this by dividing by the average fluorescence intensity of GFP in the cytoplasm. Thus the amount of RBD-Raf-GFP that specifically binds to Ras-GTP at the membrane in pixel i is given by Ψ(i) = (Gi – cRi)/<Gc>. Previous analyses with a PIP3 detector (Bosgraaf et al., 2008) and the current Ras-GTP detector (Kortholt et al., 2013) reveal that this method provides an ∼10-fold increase of sensitivity for detecting local activated Ras. Note that a value of Ψ = 0.4 is highly significant and easily detectable using the information from the green and red channels but is nearly undetectable as an increase RBD-Raf-GFP at the cell boundary in the green channel only. Thus all experiments yielding values of Ψ between 0 and ∼0.5 give new information on Ras activation not found previously. The value of Ψ is a direct indicator for the amount of RBD-Raf-GFP bound to Ras-GTP at the membrane and is therefore proportional to the fraction of Ras at the membrane in the activated state. We can estimate the proportionality p between Ψ and the fraction of Ras only in the activated state (Ras-GTP/total-Ras = pΨ). The highest values of Ψ frequently observed is ∼3; because Ras-GTP/total Ras is maximally 1, this means that p is maximally 0.33. Unstimulated Dictyostelium cells have <Ψ> = 0.31 ± 0.03; with p = 0.33, this would mean that in unstimulated cells, ∼10% of Ras is in the activated, Ras-GTP state, which is consistent with the observed fraction of Ras activation in many cell types (Markevich et al., 2004).
Chemotaxis assays
To investigate how Ras and/or F-actin are locally activated during chemotaxis, we stimulated cells expressing RBD-Raf-GFP and cRFP or cells expressing RBD-Raf-GFP and LifeAct-RFP with a micropipette releasing cAMP. Using pipettes with different cAMP concentrations and recordings at different distances from the pipette, we exposed cells to shallow or steep cAMP gradients (Postma and van Haastert, 2009). The chemotaxis index is the distance moved in the direction of the gradient divided by the total distance moved during intervals of 30 s.
Kinetics of Ras activation F-actin formation and pseudopod extension
We used the sensor pair RBD-Raf-GFP and LifeAct-RFP to detect the relative kinetics of Ras activation and F-actin formation. Of importance, these sensors have similar off-rates: single-molecule experiments show that the interaction between RBD-Raf and Ras-GTP has an off-rate of ∼3 s–1 (Hibino et al., 2009). FRAP experiments of LifeAct-GFP bound to F-actin show half-maximal recovery after 0.2–0.3 s, indicating an off-rate between 2 and 3 s–1 (Riedl et al., 2008; Riedl, 2010). Movies of cells coexpressing RBD-Raf-GFP and LifeAct-RFP from one plasmid were recorded on a confocal microscope at 1.18 s/frame. The extension of a new pseudopod was determined using Quimp3 as described (Bosgraaf and Van Haastert, 2010). This provided the first frame (t0) and starting position at the cell boundary of a new protrusion. A line scan (width 660 nm) was drawn perpendicular to the cell surface at this position (Supplemental Figure S3B), and several frames before and after t0 were analyzed using this line segment. The tip of the pseudopod was identified as the x, y-position along this line segment with the half-maximal increase of the GFP signal; the distance of this tip position in a specific frame relative to the tip position in the previous frame, divided by the frame interval, represents the speed of movement of the pseudopod in this frame. Using the fluorescence intensities along the line segment, we determined the time of onset of the increase of the intensity of RBD-Raf-GFP (tr) and LifeAct-RFP (ta) as follows (Supplemental Figure S3). For the GFP signal, we first subtracted the intensity value of cytosolic pixels and then determined the sum of the intensities of those pixels at the cell boundary that have an intensity above the intensity of cytosolic pixels. For the RFP signal, we used a similar procedure to determine the sum of intensities in the cortex. GFP data were analyzed by linear regression using a model in which the level of Ras-GTP is low and constant before tr and increases linearly after tr. Thus tr is the time coordinate of the intersection point of the two regression lines. The values of ta and tm (start point of movement) were estimated in the same way, using data from the RFP channel and data on movement of the protrusion, respectively.
Computational model for coupled excitable Ras activation and pseudopod extension
The model consists of four parts: stimulation of Ras activation by cAMP (S), excitable Ras activation (R), excitable pseudopod inducer (P), and pseudopod memory (M). The cell is described as a circle with a radius of 6.25 µm (circumference 39.3 µm). Simulations were performed in a one-dimensional model with 120 grid points.
The cAMP concentration (A in nM) is zero, uniform at the specified concentration, or a linear gradient with the highest concentration at grid point 60. cAMP binds to the receptor with dissociation constant 30 nM to provide the stimulus (S),
|
|
Ras activation and pseudopod initiation contain a noise term, which was calculated in the numerical simulations as follows:
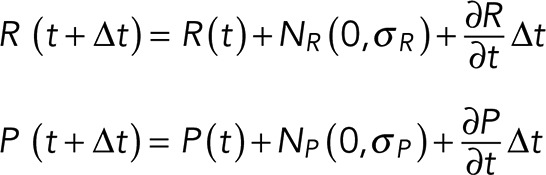 (2) (2)
|
where N(0, σ) is Gaussian white noise with mean 0 and variance σ2 and Δt is the integration time.
The partial differential equations are
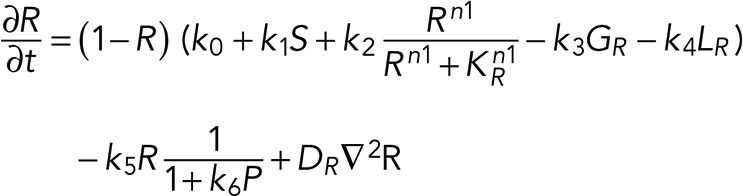 (3) (3)
|
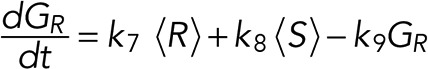 (4) (4)
|
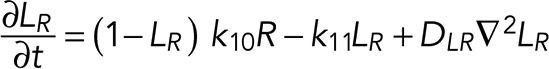 (5) (5)
|
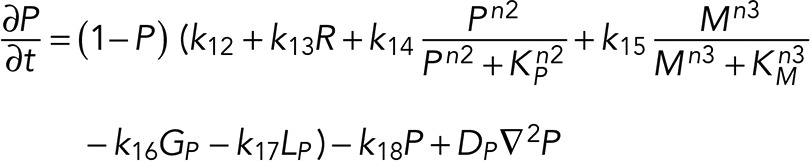 (6) (6)
|
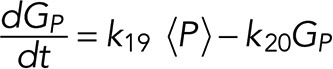 (7) (7)
|
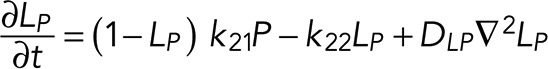 (8) (8)
|
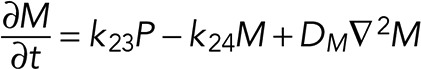 (9) (9)
|
See Supplemental Table SI for the meaning and value of the parameters and constants and Supplemental Figure S4 for parameter sensitivity, selection, and optimization.
Supplementary Material
Acknowledgments
This work was supported by a Netherlands Organization for Scientific Research NWO-VIDI grant to A.K.
Abbreviations used:
- CRW
correlated random walk
- GAP
GTPase-activating protein
- GEF
guanine exchange factor
- LatA
latrunculin A
- LEGI
local excitation global inhibition
- RBD
Ras-binding domain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-10-0733) on February 1, 2017.
REFERENCES
- Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- Artemenko Y, Lampert TJ, Devreotes PN. Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell Mol Life Sci. 2014;71:3711–3747. doi: 10.1007/s00018-014-1638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Fey P, Jimenez-Morales D, Dodson RJ, Chisholm RL. dictyBase 2015: Expanding data and annotations in a new software environment. Genesis. 2015;53:523–534. doi: 10.1002/dvg.22867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement WM, Leda M, Moe AM, Kita AM, Larson ME, Golding AE, Pfeuti C, Su KC, Miller AL, Goryachev AB, von Dassow G. Activator-inhibitor coupling between Rho signalling and actin assembly makes the cell cortex an excitable medium. Nat Cell Biol. 2015;17:1471–1483. doi: 10.1038/ncb3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolourani P, Spiegelman GB, Weeks G. Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol Biol Cell. 2006;17:4543–4550. doi: 10.1091/mbc.E05-11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosgraaf L, Keizer-Gunnink I, Van Haastert PJM. PI3-kinase signaling contributes to orientation in shallow gradients and enhances speed in steep chemoattractant gradients. J Cell Sci. 2008;121:3589–3597. doi: 10.1242/jcs.031781. [DOI] [PubMed] [Google Scholar]
- Bosgraaf L, Van Haastert PJM. Navigation of chemotactic cells by parallel signaling to pseudopod persistence and orientation. PLoS One. 2009a;4:e6842. doi: 10.1371/journal.pone.0006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosgraaf L, Van Haastert PJM. The ordered extension of pseudopodia by amoeboid cells in the absence of external cues. PLoS One. 2009b;4:e5253. doi: 10.1371/journal.pone.0005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosgraaf L, Van Haastert PJM. Quimp3, an automated pseudopod-tracking algorithm. Cell Adh Migr. 2010;4:46–55. doi: 10.4161/cam.4.1.9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D. Intrinsic activity in cells and the brain. Mol Biol Cell. 2014;25:737–738. doi: 10.1091/mbc.E13-12-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider T, Anderson K, Ecke M, Müller-Taubenberger A, Schroth-Diez B, Ishikawa-Ankerhold HC, Gerisch G. The three-dimensional dynamics of actin waves, a model of cytoskeletal self-organization. Biophys J. 2009;96:2888–2900. doi: 10.1016/j.bpj.2008.12.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider T, Diez S, Anderson K, Heuser J, Clarke M, Müller-Taubenberger A, Köhler J, Gerisch G. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Cai H, Das S, Kamimura Y, Long Y, Parent CA, Devreotes PN. Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J Cell Biol. 2010;190:233–245. doi: 10.1083/jcb.201001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest P, Firtel R. Big roles for small GTPases in the control of directed cell movement. Biochem J. 2007;401:377–390. doi: 10.1042/BJ20061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest PG, Shen Z, Lakoduk A, Sasaki AT, Briggs SP, Firtel RA. A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev Cell. 2010;18:737–749. doi: 10.1016/j.devcel.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RM, Wingreen NS, Cox EC. An excitable cortex and memory model successfully predicts new pseudopod dynamics. PLoS One. 2012;7:e33528. doi: 10.1371/journal.pone.0033528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P, Dodson RJ, Basu S, Chisholm RL. One stop shop for everything Dictyostelium: dictyBase and the Dicty Stock Center in 2012. Methods Mol Biol. 2013;983:59–92. doi: 10.1007/978-1-62703-302-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt M, Ecke M, Walz M, Stengl A, Beta C, Gerisch G. Actin and PIP3 waves in giant cells reveal the inherent length scale of an excited state. J Cell Sci. 2014;127:4507–4517. doi: 10.1242/jcs.156000. [DOI] [PubMed] [Google Scholar]
- Gerisch G, Ecke M. Wave patterns in cell membrane and actin cortex uncoupled from chemotactic signals. Methods Mol Biol. 2016;1407:79–96. doi: 10.1007/978-1-4939-3480-5_6. [DOI] [PubMed] [Google Scholar]
- Hecht I, Kessler DA, Levine H. Transient localized patterns in noise-driven reaction-diffusion systems. Phys Rev Lett. 2010;104:158301. doi: 10.1103/PhysRevLett.104.158301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht I, Skoge ML, Charest PG, Ben-Jacob E, Firtel RA, Loomis WF, Levine H, Rappel W-J. Activated membrane patches guide chemotactic cell motility. PLoS Comput Biol. 2011;7:e1002044. doi: 10.1371/journal.pcbi.1002044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino K, Shibata T, Yanagida T, Sako Y. A RasGTP-induced conformational change in C-RAF is essential for accurate molecular recognition. Biophys J. 2009;97:1277–1287. doi: 10.1016/j.bpj.2009.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-H, Tang M, Shi C, Iglesias PA, Devreotes PN. An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat Cell Biol. 2013;15:1307–1316. doi: 10.1038/ncb2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall R ( The interaction between pseudopods and extracellular signalling during chemotaxis and directed migration. Curr Opin Cell Biol. 2013;25:526–531. doi: 10.1016/j.ceb.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Kae H, Lim CJ, Spiegelman GB, Weeks G. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 2004;5:602–606. doi: 10.1038/sj.embor.7400151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortholt A, Kataria R, Keizer-Gunnink I, Van Egmond WN, Khanna A, Van Haastert PJM. Dictyostelium chemotaxis: essential Ras activation and accessory signalling pathways for amplification. EMBO Rep. 2011;12:1273–1279. doi: 10.1038/embor.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortholt A, Keizer-Gunnink I, Kataria R, Van Haastert PJM. Ras activation and symmetry breaking during Dictyostelium chemotaxis. J Cell Sci. 2013;126:4502–4513. doi: 10.1242/jcs.132340. [DOI] [PubMed] [Google Scholar]
- Kortholt A, van Haastert PJM. Highlighting the role of Ras and Rap during Dictyostelium chemotaxis. Cell Signal. 2008;20:1415–1422. doi: 10.1016/j.cellsig.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- Lee S, Shen Z, Robinson DN, Briggs S, Firtel RA. Involvement of the cytoskeleton in controlling leading-edge function during chemotaxis. Mol Biol Cell. 2010;21:1810–1824. doi: 10.1091/mbc.E10-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Nørrelykke SF, Cox EC. Persistent cell motion in the absence of external signals: a search strategy for eukaryotic cells. PLoS One. 2008;3:e2093. doi: 10.1371/journal.pone.0002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lacal J, Veltman DM, Fusetti F, van Haastert PJM, Firtel RA, Kortholt A. A Gα-stimulated RapGEF is a receptor-proximal regulator of dictyostelium chemotaxis. Dev Cell. 2016;37:458–472. doi: 10.1016/j.devcel.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Markevich NI, Moehren G, Demin OV, Kiyatkin A, Hoek JB, Kholodenko BN. Signal processing at the Ras circuit: what shapes Ras activation patterns. Syst Biol (Stevenage) 2004;1:104–113. doi: 10.1049/sb:20045003. [DOI] [PubMed] [Google Scholar]
- Nakajima A, Ishihara S, Imoto D, Sawai S. Rectified directional sensing in long-range cell migration. Nat Commun. 2014;5:5367. doi: 10.1038/ncomms6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson MP, Veltman DM, van Haastert PJM, Webb SD, Mackenzie JA, Insall RH. Chemotaxis: a feedback-based computational model robustly predicts multiple aspects of real cell behaviour. PLoS Biol. 2011;9:e1000618. doi: 10.1371/journal.pbio.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma M, van Haastert PJM. Mathematics of experimentally generated chemoattractant gradients. Methods Mol Biol. 2009;571:473–488. doi: 10.1007/978-1-60761-198-1_31. [DOI] [PubMed] [Google Scholar]
- Riedl J. Munich, Germany: University of Munich; 2010. Development and Characterization of Lifeact-A Versatile Marker for the Visualization of F-Actin. PhD Thesis. [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan GL, Petroccia HM, Watanabe N, Vavylonis D. Excitable actin dynamics in lamellipodial protrusion and retraction. Biophys J. 2012;102:1493–1502. doi: 10.1016/j.bpj.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki AT, Firtel RA. Spatiotemporal regulation of Ras-GTPases during chemotaxis. Methods Mol Biol. 2009;571:333–348. doi: 10.1007/978-1-60761-198-1_23. [DOI] [PubMed] [Google Scholar]
- Sasaki AT, Janetopoulos C, Lee S, Charest PG, Takeda K, Sundheimer LW, Meili R, Devreotes PN, Firtel RA. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol. 2007;178:185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo H, Cai H, Wang Y, Sesaki H, Lijima M. The novel race binding protein GflB sharpens ras activity at the leading edge of migrating cells. Mol Biol Cell. 2016;27:1596–1605. doi: 10.1091/mbc.E15-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Huang C-H, Devreotes PN, Iglesias PA. Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells. PLoS Comput Biol. 2013;9:e1003122. doi: 10.1371/journal.pcbi.1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoge M, Wong E, Hamza B, Bae A, Martel J, Kataria R, Keizer-Gunnink I, Kortholt A, Van Haastert PJ, Charras G, et al. A worldwide competition to compare the speed and chemotactic accuracy of neutrophil-like cells. PLoS One. 2016;11:e0154491. doi: 10.1371/journal.pone.0154491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoge M, Yue H, Erickstad M, Bae A, Levine H, Groisman A, Loomis WF, Rappel W-J. Cellular memory in eukaryotic chemotaxis. Proc Natl Acad Sci USA. 2014;111:14448–14453. doi: 10.1073/pnas.1412197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Wang M, Shi C, Iglesias PA, Devreotes PN, Huang C-H. Evolutionarily conserved coupling of adaptive and excitable networks mediates eukaryotic chemotaxis. Nat Commun. 2014;5:5175. doi: 10.1038/ncomms6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedy L, Meier B, Stephan J, Heinrich D, Endres RG. Distinct cell shapes determine accurate chemotaxis. Sci Rep. 2013;3:2606. doi: 10.1038/srep02606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert PJM ( Chemotaxis: insights from the extending pseudopod. J Cell Sci. 2010;123:3031–3037. doi: 10.1242/jcs.071118. [DOI] [PubMed] [Google Scholar]
- Van Haastert PJM, Bosgraaf L. Food searching strategy of amoeboid cells by starvation induced run length extension. PLoS One. 2009a;4:e6814. doi: 10.1371/journal.pone.0006814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert PJM, Bosgraaf L. The local cell curvature guides pseudopodia towards chemoattractants. HFSP J. 2009b;3:282–286. doi: 10.2976/1.3185725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert PJM, Postma M. Biased random walk by stochastic fluctuations of chemoattractant-receptor interactions at the lower limit of detection. Biophys J. 2007;93:1787–1796. doi: 10.1529/biophysj.107.104356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman DM, Akar G, Bosgraaf L, Van Haastert PJM. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 2009;61:110–118. doi: 10.1016/j.plasmid.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Keizer-Gunnik I, Van Haastert PJM. Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum. J Cell Biol. 2008;180:747–753. doi: 10.1083/jcb.200709180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Mihaylov V, Xu X, Brzostowski JA, Li H, Liu L, Veenstra TD, Parent CA, Jin T. A Gβγ effector, ElmoE, transduces GPCR signaling to the actin network during chemotaxis. Dev Cell. 2012;22:92–103. doi: 10.1016/j.devcel.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Collins SR, Meyer T. Locally excitable Cdc42 signals steer cells during chemotaxis. Nat Cell Biol. 2016;18:191–201. doi: 10.1038/ncb3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.