A sequence from spikemoss uncovers the true extent of the Bowman-Birk Inhibitor family to be wider than cereals and legumes, solving a long-standing mystery about their origins.
Abstract
Bowman-Birk Inhibitors (BBIs) are a well-known family of plant protease inhibitors first described 70 years ago. BBIs are known only in the legume (Fabaceae) and cereal (Poaceae) families, but peptides that mimic their trypsin-inhibitory loops exist in sunflowers (Helianthus annuus) and frogs. The disparate biosynthetic origins and distant phylogenetic distribution implies these loops evolved independently, but their structural similarity suggests a common ancestor. Targeted bioinformatic searches for the BBI inhibitory loop discovered highly divergent BBI-like sequences in the seedless, vascular spikemoss Selaginella moellendorffii. Using de novo transcriptomics, we confirmed expression of five transcripts in S. moellendorffii whose encoded proteins share homology with BBI inhibitory loops. The most highly expressed, BBI3, encodes a protein that inhibits trypsin. We needed to mutate two lysine residues to abolish trypsin inhibition, suggesting BBI3’s mechanism of double-headed inhibition is shared with BBIs from angiosperms. As Selaginella belongs to the lycopod plant lineage, which diverged ∼200 to 230 million years before the common ancestor of angiosperms, its BBI-like proteins imply there was a common ancestor for legume and cereal BBIs. Indeed, we discovered BBI sequences in six angiosperm families outside the Fabaceae and Poaceae. These findings provide the evolutionary missing links between the well-known legume and cereal BBI gene families.
INTRODUCTION
Bowman-Birk Inhibitors (BBIs) were first described 70 years ago and were the subject of classical experiments in biochemistry (Bowman, 1946). BBIs are one of the many different families of plant protease inhibitors reviewed by Habib and Fazili (2007), who used the structural classifications provided by the MEROPS peptidase database (Rawlings et al., 2016). BBIs originally characterized from soybean (Glycine max) are dual inhibitors of both trypsin and chymotrypsin (Birk, 1961; Birk et al., 1963); this duality was later found to be the result of two separate inhibitory loops, each formed by a disulfide bridge (Odani and Ikenaka, 1973). This presence of two spatially separated loops is referred to as a “double-headed” structure (Figure 1). This double-headed structure is predicted to have arisen from an internal gene duplication of a single-headed ancestral inhibitor (Mello et al., 2003). The inhibitory loops are bound by proteases in a substrate-like conformation, a standard mechanism for protease inhibition. A double-headed BBI can bind two proteases simultaneously (Song et al., 1999). BBIs also typically possess many internal disulfide bonds between conserved Cys residues that provide structural stability and maintain the active conformation of the inhibitory loops. Highlighting this, a recent study showed that mutating a single Cys residue of a typical double-headed BBI abolished the activity attributed to both loops of the mutated BBI in seed extracts (Clemente et al., 2015).
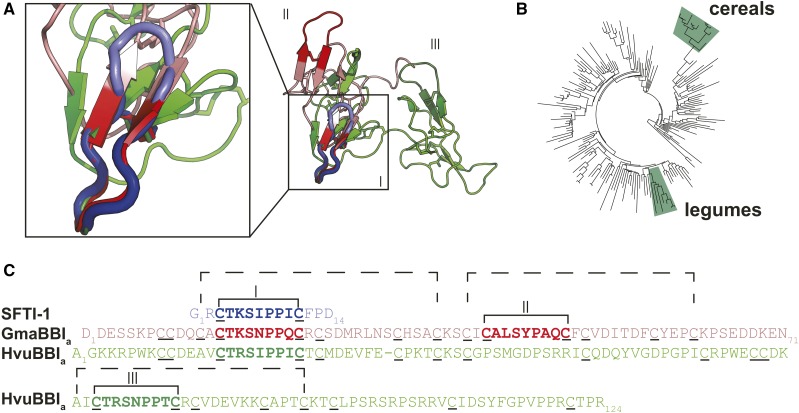
Figure 1.
The CTKSIPPIC Motif: Structures, Sequences, and Contexts.
(A) Structural alignment of SFTI-1 (PDB code: 1JBL), a BBI from soybean (GmaBBIa; PDB code: 1BBI), and a BBI from barley (HvuBBIa; PDB code: 2FJ8). The trypsin inhibitory loop of all three sequences (boxed) share structural similarity.
(B) BBIs are common in the phylogenetically separate cereals and legumes marked on an angiosperm phylogeny of rbcL sequences (Elliott et al., 2014).
(C) Sequence alignment of SFTI-1, GmaBBI, and HvuBBI. The homologous inhibitory “heads” are indicated with dashed brackets. Inhibitory loops are indicated with Roman numerals and the corresponding loops are also labeled in (A). The soybean sequence exemplifies the “double-headed” structure, with two homologous inhibitory motifs, the second of which has been lost in cereal BBIs as a result of the loss of two Cys residues (inhibitory loop II). Many monocot BBIs have undergone internal gene duplications resulting in multiple inhibitory loops (e.g., inhibitory loop III).
Since their discovery, BBIs have been shown to be widely distributed in the legume (Fabaceae) and cereal (Poaceae) families (Mello et al., 2003). Although these two plant groups are distantly related (Figure 1B), the high sequence and structural similarity between their BBIs suggests they share a common ancestor (Mello et al., 2003). Cereal BBIs lack the double inhibitory-loop structure common in legume BBIs. This is due to a lack of inhibitory-loop-forming Cys residues in the region where the second loop is located in legumes, resulting in the second loop becoming nonfunctional (Park et al., 2004; Qi et al., 2005). Cereal BBIs differ in size from 8 to 20 kD, whereas legume BBIs are consistently around 8 kD in size. The wide size range for cereal BBIs was caused by internal gene duplication events. Although cereal BBIs lack the Cys residues required for a functional second inhibitory loop, the internal duplication events have resulted in multiple inhibitory loops for a single protein (Prakash et al., 1996; Song et al., 1999) (Figure 1).
The inhibitory loops of BBIs can function independently from the rest of the BBI protein, as demonstrated by analysis of synthetic peptides made for the nine-residue loop alone (Nishino et al., 1977). These small, synthetic peptides were also used to determine the essential residues necessary for specificity to trypsin or chymotrypsin (Terada et al., 1978). The sequences of the synthetic nonapeptides analyzed were CTKSNPPQC and CALSTPAQC, which correspond to the soybean BBI trypsin inhibitory and chymotrypsin inhibitory loops, respectively (Terada et al., 1978). By mutating the P1 residue (underlined; the first amino acid in the N-terminal direction from the cleaved bond following the notation described in Schechter and Berger, 1967), they showed that Lys or Arg at the P1 position confers specificity for trypsin, whereas a Tyr or Leu will confer specificity for chymotrypsin (Terada et al., 1978).
Since the first studies on synthetic BBI-loop peptides, every residue in the nonapeptide except the P5′ residue, which shows the weakest conservation, has been tested for its functional importance (McBride et al., 2002). Apart from the obviously critical P1 residue and loop-forming Cys residues (P3, P6′), the remaining residues also influence BBI structure and activity. For example, the hydroxyl and methyl group of the conserved Thr at position P2 directly interacts with trypsin to stabilize the active site, resulting in reduced hydrolysis (McBride et al., 1998). An Ile at P2′ is optimal for inhibition of trypsin (Gariani et al., 1999). The P3′ and P4′ Pro residues maintain a polyproline II conformation important for structure (Park et al., 2004). Furthermore, the P3′ Pro residue must be in a cis conformation for correct positioning of the P1 residue (Brauer et al., 2002). The P1′ position is a highly conserved Ser; however, substitution with Ala only slightly reduced BBI inhibitory activity (Brauer and Leatherbarrow, 2003). The low tolerance for substitution in these studies illustrates that the high conservation observed within BBI inhibitory loops is required for the optimal inhibition of trypsin.
Two decades after the first analysis of synthetic BBI inhibitory loops, a similar and naturally occurring peptide was discovered in sunflower (Helianthus annuus) seeds (Luckett et al., 1999). Sunflower Trypsin Inhibitor-1 (SFTI-1) shares the same function, a nearly identical amino acid sequence, and similar three-dimensional structural conformation with the trypsin inhibitory loop of BBIs; however, SFTI-1 is composed of only 14 residues and is a head-to-tail macrocycle (Figure 1). Originally thought to be the smallest member of the BBI family, SFTI-1 has an altogether different biosynthetic and evolutionary origin. SFTI-1 emerged from a protein that is also a precursor for a napin-type, seed storage albumin (Mylne et al., 2011). Evidence suggests SFTI-1 evolved de novo, in a stepwise manner within a standard preproalbumin gene, independently of BBIs (Elliott et al., 2014). A less potent, but similar, inhibitory loop also exists in ORB proteins from frogs (Li et al., 2007), implying BBI-like inhibitory loops have evolved independently multiple times for this function.
The existence of a highly conserved sequence in disparate proteins and protein contexts within distinct phylogenetic groups suggests the BBI inhibitory loop is a product of convergent evolution. Here, we investigated whether this trypsin inhibitory loop has appeared in additional protein contexts. We searched databases using the query sequence CTKSIPPIC, the sequence shared by SFTI-1 and BBIs, and found an identical match to a predicted protein from the ancient seedless, vascular plant Selaginella moellendorffii. S. moellendorffii belongs to the lycopod lineage, which is estimated to have diverged 400 million years ago, an estimated 200 to 230 million years before the evolution of angiosperms (Banks, 2009; Bell et al., 2010). Closer analysis revealed that rather than being a new protein context, the S. moellendorffii BBI-like protein represents an ancient member of the BBI protein family, as it shares conserved Cys residues outside the conserved inhibitory loops. Furthermore, a recombinant S. moellendorffii BBI inhibited trypsin, and mutating predicted P1 residues removed its trypsin inhibitory activity, suggesting it uses the same mechanism of inhibition as BBIs in seed plants. Consistent with the hypothesis that the S. moellendorffii BBI-like proteins share a common ancestor with legume and cereal BBIs, we discovered BBIs in six angiosperm families outside the Fabaceae and Poaceae plant families.
RESULTS
Identification of BBI-Like Genes in S. moellendorffii
Between SFTI-1 and BBIs, only the inhibitory loop is shared. As this loop is sufficient for inhibitory activity, we performed BLAST searches using the sequence CTKSIPPIC as bait to identify other proteins that possess this functional motif. We identified two sequences derived from the S. moellendorffii genome that contained highly similar motifs: BBI1 (NCBI accession: XM_002980953) and BBI2 (NCBI accession: XM_002980955).
The proteins encoded by BBI1 and BBI2 showed conservation to known BBIs within the inhibitory motifs, but less sequence similarity outside of these motifs (Figure 2). The translated sequences possessed a high number of Cys residues (13.7% of the complete primary amino acid sequence), which is a characteristic of BBIs. The top plant BLASTp hit in NCBI (NCBI accession: ADV40041) was a predicted BBI from the legume Lathyrus sativus that shares 25.4% identity and 44.9% similarity with BBI1, as calculated in a global alignment. Similarly, the most similar sequence to BBI2 by BLASTp (NCBI accession: NP_001236539) was a BBI from the legume G. max that shares 29.7% identity and 50.7% similarity in a global alignment.
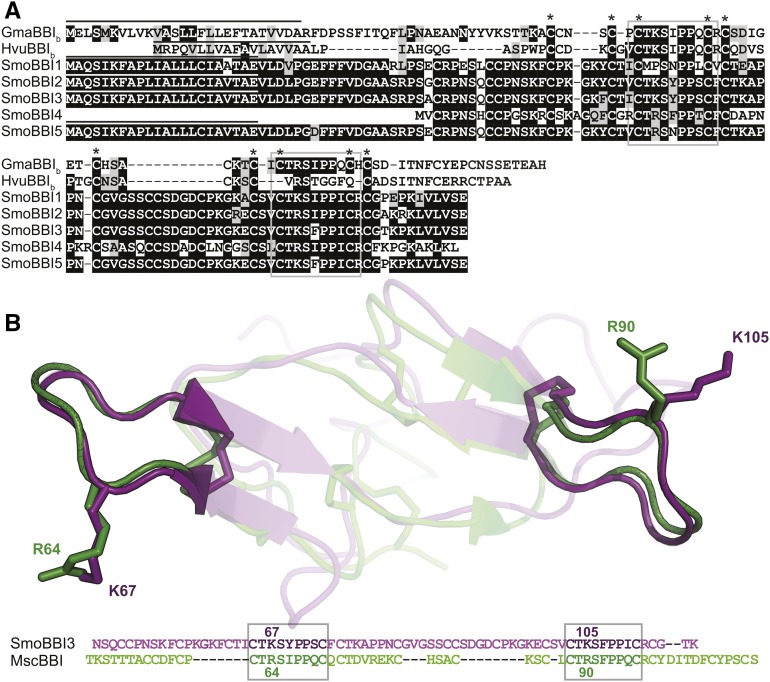
Figure 2.
S. moellendorffii BBI3 Is Predicted to Be a BBI Based on Sequence Similarity at the Inhibitory Motifs and Shared Primary Protein Architecture.
(A) Boxshade alignment of S. moellendorffii BBI-like (SmoBBI) protein sequences with soybean BBI (GmaBBIb) and barley BBI (HvuBBIb) shows conservation of the trypsin inhibitory loop (gray box). All proteins, with the exception of SmoBBI4, share a similar protein architecture, with an ER signal (line above sequence) followed by Cys-rich sequence with two spatially distinct conserved inhibitory motifs. Cys residues that are conserved in all sequences are indicated with asterisks.
(B) Protein homology model of SmoBBI346-116 aligned with Medicago scutellata BBI (PDB code: 1MVZ) is shown along with the sequence alignment. The conserved inhibitory loops are shown as opaque, while the remaining protein is translucent and the corresponding inhibitory motifs are boxed in the alignment. The side chains of the labeled P1 residues (R64, K67, R90, and L105) and disulfide bonds are displayed in stick format.
A S. moellendorffii Transcriptome Identifies Five BBI-Like Sequences
Two BBI-like sequences were found through mining the published S. moellendorffii genome. To determine if these genes are expressed and to determine if BBI-like sequences are conserved within Selaginella, the transcriptomes of S. moellendorffii, S. kraussiana, and S. martensii were assembled from RNA-seq data. Searches within the S. martensii and S. kraussiana transcriptomes found no BBI-like sequences. A search of publicly available RNA-seq data for S. stauntoniana identified a sequence with two CTKSIPPIC-like motifs (CTMSYPPSC and CTKAPPNC). This suggests there are BBI-like sequences present in Selaginella species other than S. moellendorffii, despite being undetectable in S. kraussiana and S. martensii RNA-seq data sets.
In total, five BBI-like genes were identified as being expressed in S. moellendorffii (Figure 2; Supplemental Figure 1). All five genes were sequenced from both cDNA and genomic DNA and further validated by mapping RNA-seq reads to the complete open reading frames.
Using SignalP 4.1 (Petersen et al., 2011), we found that all encoded BBIs except BBI4 had a predicted endoplasmic reticulum (ER) localization signal (Figure 2). BBI4 is 39 residues shorter and shows the greatest sequence divergence (38–43% similarity) from the other four BBI-like sequences, which share >89% similarity. The shorter length was verified by 5′ RACE. Altogether, these data imply that BBI4, although expressed, probably does not code for a functional BBI and is a pseudogene.
S. moellendorffii BBI3 Is a Functional Trypsin Inhibitor
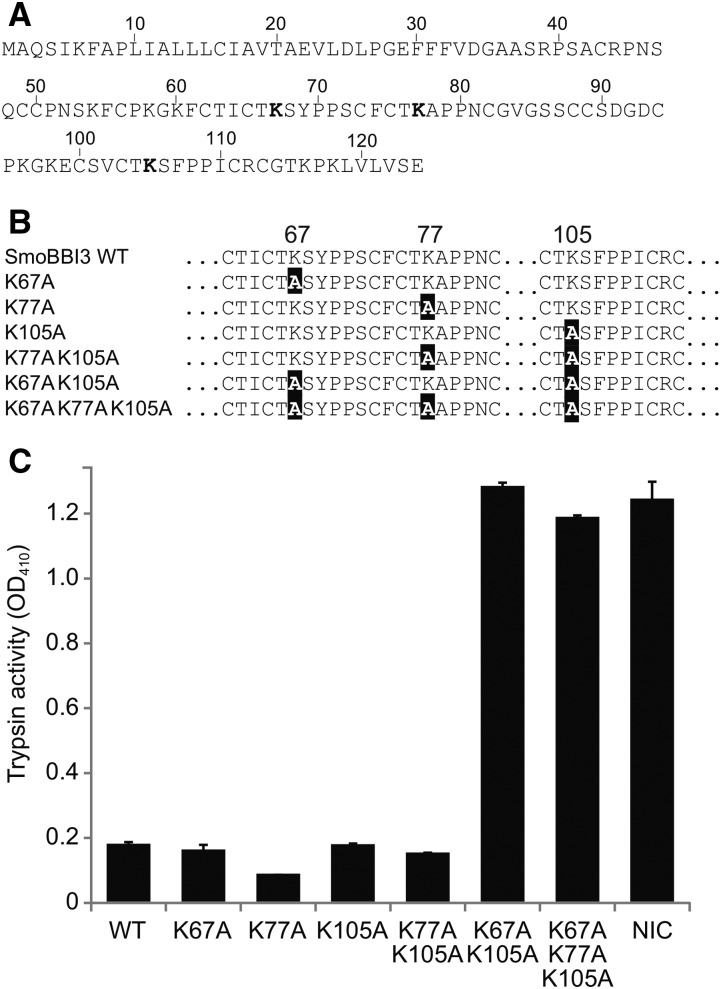
To test if the S. moellendorffii proteins share a similar function with characterized BBIs, we produced recombinant BBI3 in Escherichia coli and purified it for trypsin inhibition assays (Supplemental Figure 2). BBI3 was chosen because it had the highest number of reads mapped to its open reading frame (ORF), suggesting it is the most highly expressed of the five BBI transcripts (Table 1). Trypsin inhibitory ability was determined by end-point analysis after 45 min, assuming that the reaction had reached equilibrium at this stage. Therefore, the results will not indicate any intermediate levels of inhibition due to the inhibitor being in molar excess. Comparing BBI3 to soybean BBI in trypsin inhibition assays showed that both were effective trypsin inhibitors, reducing activity by 99.0 and 99.5%, respectively (Supplemental Figure 3). A protein model of BBI3 predicted a double-headed structure similar to that of legume BBIs (Figure 2). Based on the protein model generated, Lys-67 and Lys-105 were identified as the putative P1 Lys residues (Figure 2). However, a third region within the BBI3 sequence shares similarity with the BBI inhibitory motif and included a third potential inhibitory P1 residue, Lys-77. In order to provide support to our protein model and to test which predicted P1 residues contribute to the inhibition by BBI3, we generated a series of BBI3 mutants (Figure 3). Mutating Lys-67, Lys-77, or Lys-105 to Ala in isolation had no effect on the ability of BBI3 to inhibit trypsin. Similarly, a double mutant containing Lys77Ala and Lys105Ala did not reduce inhibition of trypsin activity compared with the wild-type control. The inhibitory activity of the Lys67Ala/Lys105Ala BBI3 double mutant, however, was completely abolished (Figure 3). Furthermore, the triple mutant showed no difference in inhibition compared with the Lys67Ala/Lys105Ala double mutant (Figure 3). Taken together, these data suggest that a combination of Lys67Ala and Lys105Ala mutations are sufficient to completely abolish inhibitory activity to a level equivalent to that of a negative control and that Lys-77 is not an inhibitory residue.
Table 1. SmoBBI mRNA Expression.
| Gene | Mapped Reads | Average Coverage |
|---|---|---|
| SmoBBI1 | 77 | 20.7 |
| SmoBBI2 | 1539 | 413.2 |
| SmoBBI3 | 2383 | 639.7 |
| SmoBBI4 | 30 | 11.7 |
| SmoBBI5 | 589 | 158.1 |
The number of RNA-seq clean reads mapping to each SmoBBI open reading frame and the average coverage, which is indicative of expression level. Average coverage represents the average number of reads mapping to each position of the reference gene. Mapped reads are shown as a graphical representation in Supplemental Figure 1.
Figure 3.
Trypsin Inhibition by Wild-Type and Mutant S. moellendorffii BBI3.
(A) Full-length wild-type BBI3 showing predicted inhibitory residues (bold).
(B) Partial BBI3 sequences showing mutations (black highlight).
(C) Inhibitors were incubated with trypsin and a colorogenic substrate of trypsin whose activity is measurable as OD410. Error bars represent sd for three technical replicates in a single microtiter plate. Trypsin incubated with the colorogenic substrate but no inhibitor was used as a control (NIC).
BBIs Are Widely Distributed in the Plant Kingdom
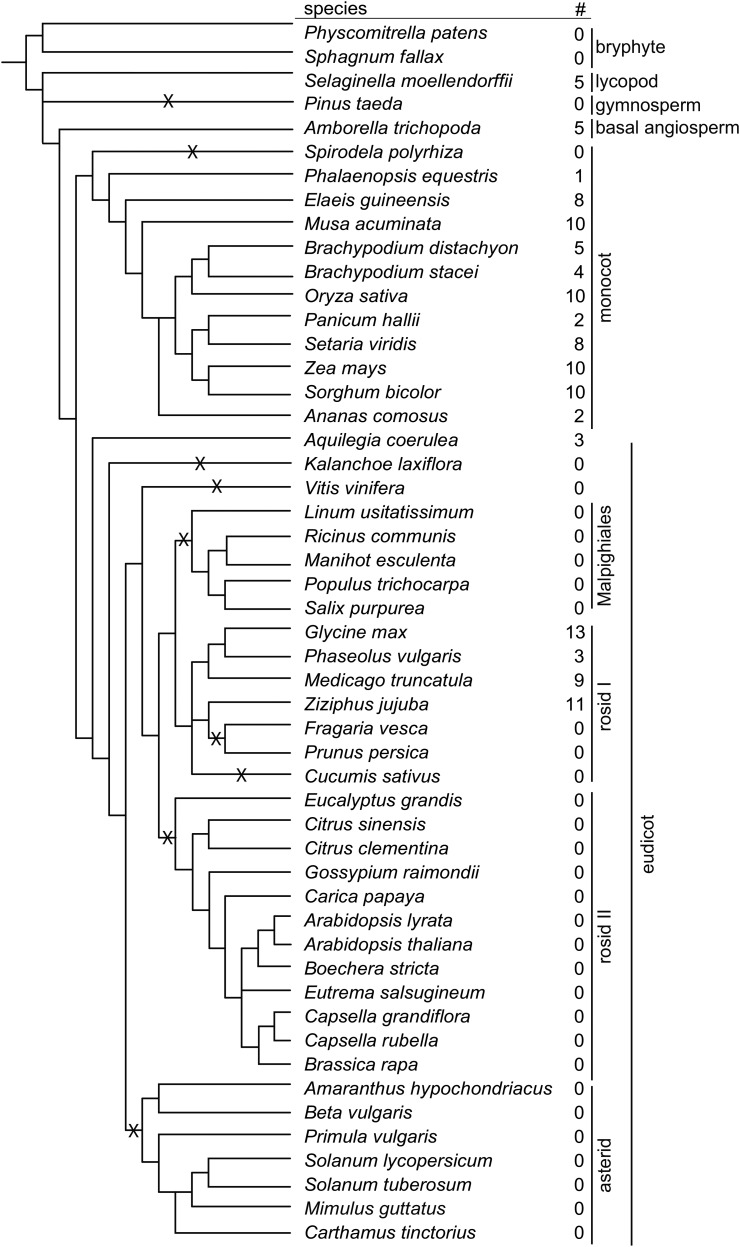
The discovery of BBI-like sequences in S. moellendorffii suggests there was an ancient common ancestor for BBIs in legumes and cereals. If so, BBIs should be more widely distributed in the plant kingdom. Comprehensive BLAST searches in published plant genomes identified BBI-like sequences in plant lineages outside legumes and cereals (Figure 4). BBI-like sequences were not identified in the genomes of the two bryophyte species searched. At the time the BLAST searches were completed, there were no published genomes for monilophytes (ferns and horsetails), which diverged after the evolution of lycopods and prior to the evolution of angiosperms. We did not find any sequence in the genome of loblolly pine (Pinus taeda), the only gymnosperm genome searched. All plant genomes searched are included in Figure 4 along with the number of BBIs identified in each genome.
Figure 4.
Distribution of BBIs in Land Plants.
The number of BBI-like sequences identified (#) in each plant genome searched. Accession numbers for all sequences are included in Supplemental Data Set 1. Predicted gene loss events are indicated with an X. The phylogeny of the plant genomes investigated is derived from species trees available from Cogepedia and APG III.
To provide experimental support of the publicly available data, we chose to confirm the presence of three of the BBI-like sequences from banana (Musa acuminata), which does not belong to the legume or cereal families. Sequences that matched the publicly available sequences were cloned from banana leaf genomic DNA (Figure 5). This supports the bioinformatics finding that angiosperm-type BBIs are found in plant lineages outside legumes and cereals. As we did not identify BBI-like sequences in the other Selaginella species, S. kraussiana and S. martensii, we assembled a transcriptome of an available lycopod, Isoetes drummondii, and found a BBI-like sequence. The I. drummondii BBI-like sequence was verified by cloning and sequencing the full-length cDNA (Figure 5). Therefore, despite not being expressed in S. kraussiana and S. martensii, BBIs likely are present in the common ancestor of Selaginella and Isoetes.
Figure 5.
BBI-Like Sequences Identified from Quillwort and Banana.
Alignment of translated sequences of genes cloned from genomic DNA from banana (M. acuminata; Mac) and cDNA from quillwort (I. drummondii, Idr). Sequences are aligned with a BBI encoded by a gene from soybean (GmaBBIb) to illustrate conserved regions including the inhibitory loops (gray boxes). Conserved Cys residues are indicated by asterisks.
The number of BBIs identified in each plant genome searched was mapped onto a phylogenetic reconstruction derived from trees from Cogepedia and Angiosperm Phylogeny Group III (Figure 4) (http://genomevolution.org/wiki/index.php/Sequenced_plant_genomes; Angiosperm Phylogeny Group, 2016). BBIs appear to be maintained throughout monocot evolution, with examples of BBI-like sequences found in all monocot species genomes searched with the exception of duckweed (Spirodela polyrhiza). In dicots, the presence and absence of genes indicate a complex pattern of gene loss. The identification of a BBI-like sequence in the basal eudicot, the Colorado blue columbine (Aquilegia coerulea), suggests that dicot BBIs share a common ancestor followed by several independent loss events. BBI-like sequences were not found in any asterid species searched, suggesting there was a loss event prior to their divergence. This is true for Malphigiales and rosid clade II as well. The pattern of loss events in rosid clade II is more complex, with BBI-like sequences found in Fabaceae and Rhamnaceae but apparently lost in all other rosid clade II species searched.
All dicot sequences analyzed had the typical double-headed BBI motif with the exception of one sequence from jujube (Ziziphus jujuba) (XP_015882735.1), which has lost two inhibitory loop forming Cys residues and another that appears to have lost one of the two loop-forming residues (XP_015882689.1). As predicted, most monocot sequences lack the conserved Cys residues forming the second inhibitory loop. Three of the 10 M. acuminata sequences had a similar sequence to dicot BBIs, with a double-headed structure. Two of double-headed sequences were confirmed by PCR cloning and sequencing. One single-headed M. acuminata sequence was also confirmed by PCR cloning and sequencing (Figure 5). This suggests that the double-headed sequences could represent the ancestral form of monocot BBIs prior to the predicted loss of the two Cys residues of the second BBI loop. One of the BBI-like sequences from Amborella trichopoda is predicted by sequence homology to possess the same double-headed motif as dicots, suggesting this is likely the ancestral structure of angiosperm BBIs. However, four of the five A. trichopoda BBI-like sequences have an additional Cys residue within their first BBI loop, suggesting they may no longer function as a protease inhibitors and have potentially evolved a new function.
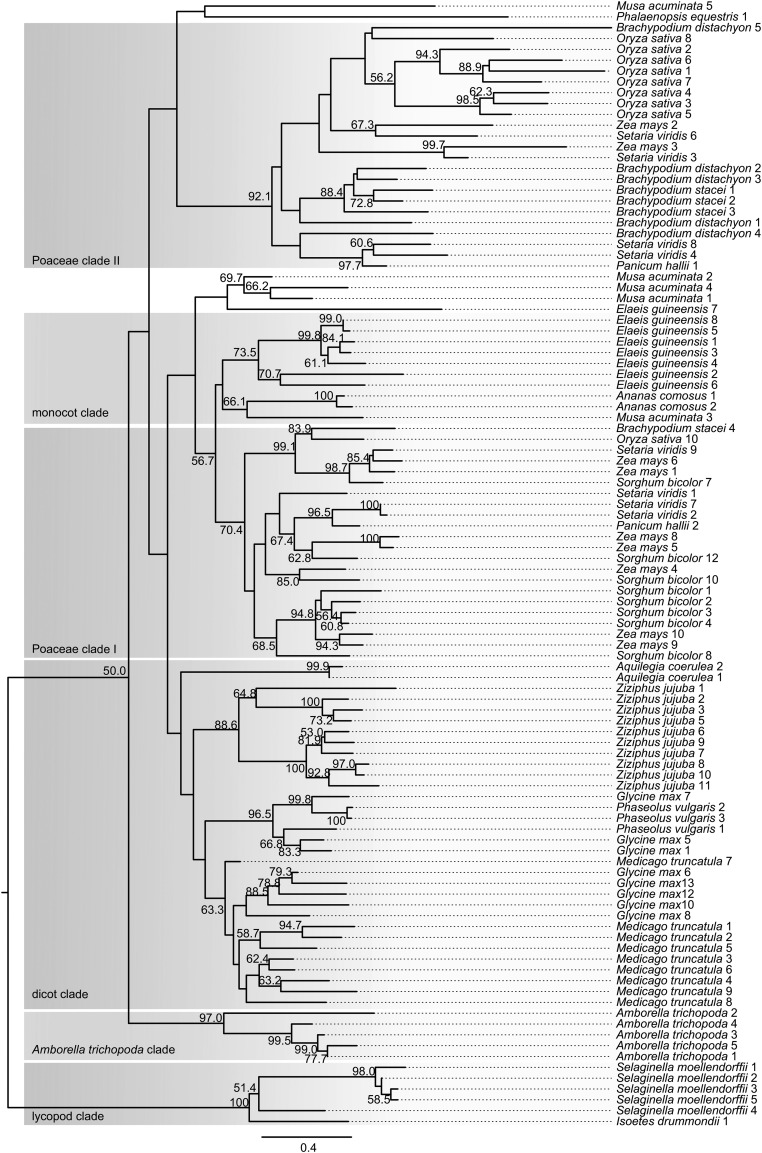
BBI-like sequences identified in angiosperms as well as those found in lycopods were used in phylogenetic analysis (Figure 6). The phylogenetic tree of full-length sequences was divided into six clades based on the branching pattern of the tree. The lycopod clade forms a species-specific clade with 100% bootstrap support, strongly suggesting that they share a single common ancestor. All angiosperm BBIs are predicted to have evolved from a single ancestral BBI sequence as they together form a single clade separate from lycopod BBIs. The dicot sequences all fall within a single clade. A. coerulea and Z. jujuba each form species-specific nodes with strong (≥88.6%) bootstrap support, suggesting the sequences belonging to these nodes probably duplicated following speciation. A duplication event occurring prior to the divergence of G. max and Phaseolus vulgaris likely resulted in two paralagous sequences, with one paralog being lost in P. vulgaris, as all Fabaceae sequences fall into two distinct bootstrap supported (≥63.3%) nodes.
Figure 6.
Phylogeny of BBI-Like Sequences in Plants.
Neighbor-joining tree from a protein distance matrix of 104 BBI-like sequences. Sequences were identified by searches within plant genome assemblies. Major clades are indicated and defined by branching patterns. The five S. moellendorffii sequences and the I. drummondii sequence were used to root the tree with the assumption they were evolutionarily most distant from all other sequences. Percentage bootstrap values from 1000 replicates of a distance neighbor-joining analysis are indicated at each node (only nodes with >50% bootstrap support are labeled). Identifiers and accession numbers are listed in Supplemental Data Set 1. The scale bar represents number of substitutions per site.
Nearly all monocot sequences fall within three clades designated the monocot clade, Poaceae clade I, and Poaceae clade II. The nodes leading to these three clades have weak bootstrap support; therefore, conclusions on the relationships between these groups cannot be made. Both Poaceae clades contain nodes with bootstrap support that contain sequences from more than one species, suggesting potential independent duplication events resulting in multiple paralogous sequences within Poaceae. The African oil palm (Elaeis guineensis) and pineapple (Ananas comosus), both sister to Poaceae, each form species-specific nodes with bootstrap support (≥73.5%) within the monocot clade. This suggests that the BBI sequences belonging to each species evolved from a single common ancestral sequence, so all monocot sequences share a common ancestral sequence.
M. acuminata BBI-like sequences showed the highest sequence divergence from other BBIs, with the five sequences analyzed falling into three distinct nodes. Potentially, species-specific duplication events occurred followed by sequence divergence. Only one M. acuminata sequence falls into the monocot clade and the other sequences form two distinct nodes from other monocot sequences. Sequences from other species do fall within these nodes, but with very low bootstrap support. Possibly, duplication events leading to the divergent sequences are independent of other monocots and could therefore be a result of the three whole genome duplication events in the Musa lineage (D’Hont et al., 2012). Alternatively, these divergent M. acuminata sequences could be the result of a duplication event in the ancestor of monocots and therefore could be paralogs of sequences found in the monocot clade and Poaceae clade II. Any definitive conclusions cannot be made due to the lack of bootstrap support for the branches leading to these sequences.
DISCUSSION
BBIs were first studied 70 years ago and since then their structure and function have been well defined (Bowman, 1946). Here, we report five BBI genes from the seedless, vascular plant S. moellendorffii. The genes identified are highly divergent from those in legumes and cereals but possess the conserved trypsin inhibitory motif. They also possess a conserved primary protein architecture, sharing a conserved pattern of Cys residues around the inhibitory loop motifs, and all but one S. moellendorffii BBI possesses a predicted ER signal. A predicted structure of a S. moellendorffii BBI modeled against other known BBIs suggests a similar double-headed structure. Together, these results support the notion that the BBI-like genes identified here are related to BBIs in angiosperms. Selaginella represents the most ancient extant lineage of vascular plants; therefore, the identification of these BBIs suggests an ancient ancestral origin of BBIs in angiosperms.
We determined that at least one S. moellendorffii BBI protein, BBI3, is a functional trypsin inhibitor. The structure of BBI3 is predicted to be double-headed according to homology-based modeling. However, due to high sequence divergence outside of the predicted inhibitory sequence motifs and the presence of a third potential inhibitory Lys residue, we further tested the predicted double-headed structure by analyzing modified proteins. By mutagenesis, we demonstrated that Ala substitutions at both Lys-67 and Lys-105 were required to abolish BBI3 inhibitor activity, suggesting that there are two independent sites that bind to and inhibit trypsin. Consistent with this, single Ala substitutions at Lys-67 and Lys-105 did not affect BBI3 inhibition. The residues Lys-67 and Lys-105 reside within the short BBI3 sequences that displayed high similarity to the angiosperm BBI inhibitory motifs and are predicted to share a similar loop structure from our protein model. Mutation of a third Lys residue that also fell within a short sequence that shared similarity to BBI inhibitory motifs, but was not predicted to be part of an inhibitory loop from our protein model, had no effect on BBI3 inhibition. Studies of short peptide mimics of the BBI inhibitory loop showed similar results when the P1 residue was mutated (Terada et al., 1978; Domingo et al., 1995).
Since BBIs had thus far only been described in legumes and cereals (Mello et al., 2003; Qi et al., 2005), and these two plant families are distantly related, we reasoned that if the BBIs in these two families share an ancient ancestral origin, they are likely more widely distributed in angiosperms. We demonstrated that BBIs are indeed found in angiosperm lineages outside the legumes and grasses. Supporting a hypothesis for a common ancestral origin for angiosperm BBIs, we identified BBI-like sequences in what is considered to be the most basal living representative of angiosperms, A. trichopoda (Soltis et al., 2008).
BBIs appear to be widespread throughout the monocot lineage, being absent from only one of the genomes searched. In the evolutionary scheme proposed by Mello et al. (2003), the loss of two Cys residues in the monocot lineage results in the loss of functionality of the second inhibitory loop and the double-headed structure characteristic of dicot BBIs. Our findings support this model, as we observed a loss of Cys residues in monocot BBI-like sequences; however, we also demonstrated that the loss of the two inhibitory loop-forming Cys residues potentially occurred following the divergence of monocots from dicots, as we found double-headed BBI-like sequences in the monocot M. acuminata.
Given our identification of the Selaginella BBIs and the now apparent widespread distribution in angiosperms, we expected to find BBI-like sequences in gymnosperms, which diverged after lycopods, but prior to the evolution of angiosperms. However, searches failed to identify any BBI-like sequences in P. taeda. This could be due to the lack of genetic data for other gymnosperm species, or potentially the BBIs might have been lost during evolution. Alternatively, BBI-like proteins might have evolved independently in Selaginella and are not related to those found in angiosperms. However, this is unlikely given the double-headed structure of the inhibitory motifs and shared inhibitory function.
BBIs appear to have been lost in several angiosperm lineages. The main mechanisms for gene loss, as reviewed by Albalat and Cañestro (2016), are through unequal crossing-over during meiosis, the mobilization of a transposable element leading to physical loss of the gene from the genome, or through the introduction of iterative mutations resulting in pseudogenization or new functionality. Examples of BBI gene loss have been observed in pea (Pisum sativum) germplasm collections, with evidence of partial BBI sequences resulting from premature stop codons (Clemente et al., 2015). However, we failed to identify any sequences, including partial sequences, showing similarity to BBIs in whole plant lineages including rosid class II and the asterids, suggesting gene loss occurred by complete gene loss or through the introduction of mutations resulting in the sequence becoming unrecognizable as a BBI. The loss of BBIs is likely prevalent through angiosperm evolution as a result of functional redundancy, as there are several families of protease inhibitors in plants that could compensate for the loss of BBIs (Habib and Fazili, 2007). For example, the serpin family of protease inhibitors is widespread throughout angiosperms, including species lacking BBIs such as Arabidopsis and cucumber (Cucumis sativus; Roberts and Hejgaard, 2008). Species-specific gene loss has been observed for other protein-based plant defenses such as polyphenol oxidases (Tran et al., 2012). Gene loss is common and occurs more frequently for genes coding for nonessential proteins that have minor influences on plant fitness (Hirsh and Fraser, 2001; Krylov et al., 2003).
The evolutionary split between Isoetes and Selaginella is predicted to have occurred 370 million years ago (Arrigo et al., 2013). Therefore, the common ancestor of the BBI-like sequences we have found in Isoetes and Selaginella is at least this old and predates the evolution of angiosperms, which are predicted to have evolved between 167 to 199 million years ago (Bell et al., 2010). Both the Isoetes and Selaginella BBI-like sequences have a double-headed motif that is observed in legume BBIs; therefore, it can be predicted that the ancestor of both lycopod and angiosperm BBIs had a double-headed motif. This is supported by the identification of BBI-like sequences in the basal angiosperm species A. trichopoda. Although the double-headed motif likely arose from an internal gene duplication of a single inhibitory loop, we found no current-day examples of a single-headed BBI other than those found in cereals, whose sequences suggest they lost two Cys residues resulting in the second loop becoming nonfunctional.
In conclusion, we have shown that the highly conserved BBI inhibitory motif has been maintained through vascular plant evolution since at least prior to the divergence of Isoetes and Selaginella. This supports the many studies on synthetic peptide mimics of this inhibitory loop showing that the CT[K/R]SIPPXC motif is optimal for trypsin inhibition. Having identified BBIs throughout the angiosperm lineage, it is clear that BBIs in angiosperms share a common ancestor. We propose that the lycopod BBIs identified here share an ancient ancestral origin with the well-characterized BBIs in angiosperms given the sequence similarity at the conserved loops, a shared protein architecture, and the common mode of action.
METHODS
BLAST Searches Using the CTKSIPPIC Motif as Bait
To determine whether the sequence CTKSIPPIC existed in different protein contexts other than SFTI-1 and the legume and cereal BBIs, we performed a BLASTp analysis in the BLAST portal of Phytozome v11.0 excluding the genus Helianthus (sunflower SFTI-1) and Fabaceae or Poaceae families (well-known BBI-containing families). The top matches in this analysis were two unknown proteins from Selaginella moellendorffii (BBI1 GenBank: XP_002980999; BBI2 GenBank: XP_002981001). To assess the similarity to other BBIs, the BBI-like protein sequences were used as bait sequences in a BLASTp search in the NCBI nonredundant protein database restricted to Fabaceae and Poaceae sequences. The sequence similarity between the S. moellendorffii BBI-like genes and their corresponding BLASTp hits were calculated in a global alignment using the default settings of the ExPASy LALIGN program.
Plant Tissue
Cuttings were taken from S. kraussiana and S. martensii growing at Hilltop Nursery (Menzies Creek, Victoria, Australia), frozen on dry ice, and shipped to Western Australia (entry into WA under Quarantine Inspector’s Direction No. KH090713). Cuttings were taken from S. moellendorffii (catalog no. 3228; Plant Delights Nursery) growing at the Joint BioEnergy Institute. Plant Delights Nursery was the source of the S. moellendorffii used to generate the published genome sequence (Banks et al., 2011). The S. moellendorffii cuttings were preserved in RNAlater RNA Stabilization Reagent (Ambion) and shipped on ice packs to Western Australia.
RNA Extraction from S. moellendorffii, S. kraussiana, and S. martensii
Tissue from each species was ground with glass beads to a fine powder under liquid nitrogen. Some of the thicker S. martensii stems were removed during the grinding. Total RNA from ∼0.3 mL of frozen tissue powder was extracted as previously described (Mylne et al., 2012) using phenol:chloroform and a 2 M lithium chloride RNA-selective precipitation. Contaminating genomic DNA was removed by digesting total RNA with DNase and subsequent purification with a NucleoSpin RNA Clean-up kit (Macherey-Nagel). The resulting total RNA was analyzed on a NanoDrop Spectrophotometer for its A260/A280 and A260/A230 ratios, and all three samples displayed distinct banding when run on a 1% agarose gel, implying the mRNA was intact.
Transcriptome Sequencing, Assembly, and Mining Data for BBI-Like Sequences
Sequencing libraries were generated using the TruSeq Stranded Total RNA LT with Ribo-Zero Plant kit (Illumina) with 300 to 1000 ng of purified total RNA according to the manufacturer’s instructions. Sequencing was then performed on an Illumina HiSeq 1500 instrument as 101-bp single read runs or 2 × 101-bp paired-end read runs.
The de novo transcriptome assemblies were done as described (Jayasena et al., 2014). Raw reads were inspected for quality using FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Quality trimming and filtering were done using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Raw reads were trimmed to maintain a phred score of 30, which sets the base call accuracy to be 99.9%, and the minimum length after trimming was set at 50. Trimmed reads were filtered with a quality threshold of 22, and the percentage of bases that match the quality threshold was set to 90.
De novo transcriptomes were assembled using CLC Genomics Workbench 7.0 (CLCBio). RNA from S. moellendorffii was sequenced only as single reads. S. martensii and S. kraussiana were sequenced as paired-end reads. Each data set was assembled initially with the default CLC settings (i.e., word size: 23; bubble size: 50) and subsequently with three different word sizes (i.e., 30, 40, and 60), which determines the size the reads are fragmented into before being assembled, keeping all other parameters at default settings. In each case, reads were mapped back to contigs keeping the mapping parameters as follows: mismatch cost, 2; insertion cost, 3; deletion cost, 3; length fraction, 0.8; similarity fraction, 0.8; update contigs, yes. Relevant statistics are presented in Table 2. In this way, four transcriptomes for each of the three species were assembled.
Table 2. De Novo Transcriptome Assembly Statistics.
| Species | Raw Reads | Clean Reads | Word Size | Contigs | N50 |
|---|---|---|---|---|---|
| S. moellendorffii | 63,151,678 | 56,711,459 | 40 (Single) | 44,564 | 584 |
| S. martensii | 90,146,302 | 82,241,287 | 40 (Paired) | 51,848 | 557 |
| S. kraussiana | 145,585,252 | 135,198,965 | 60 (Paired) | 44,996 | 780 |
| I. drummondii | 103,198,884 | 68,341,138 | 60 (Paired) | 264,900 | 427 |
The number of contigs, number of raw and clean reads, the word size used to generate each assembly, and the N50 statistic (summary of the lengths of the longest contigs until 50% of the total contig length is reached) are shown.
Each assembly was queried using tBLASTn for the two BBI-like genes identified from the S. moellendorffii genome.
The sequence referred to from S. stauntoniana was found within the NCBI short read archive (run ERR364347) with the specific sequence ID within the data set being ERR364347.14864137.1. The pertinent region of the translated sequence encoded TVCTMSYPPSCFCTKAPPNCGVGSSCCSD, in which the CTKSIPPIC-like motifs are underlined.
Cloning of S. moellendorffii BBI-Like Sequences
The 5′ and 3′ RACE-ready cDNA was generated using the SMARTer RACE cDNA amplification kit (Clontech). Primers for 3′ and 5′ RACE were designed against sequences from the assembled transcriptome (for BBI3 and BBI4) and from the published genome (for BBI1 and BBI2). All primers are included in Supplemental Table 1. Sequences were amplified using Taq DNA polymerase. Purified 3′ and 5′ RACE PCR products were cloned into pGEM-T Easy vector (Promega) and sequenced. Primers designed based on the RACE results were used to clone full-length sequences with Taq DNA polymerase. For each PCR-amplified product, at least three independent clones were sequenced to account for errors induced by Taq DNA polymerase. Because of the high sequence similarity between BBI2 and BBI3, a single primer was designed directly upstream of the poly(A) tail that matched both the BBI2 and BBI3 3′ untranslated region sequences. BBI2 and BBI3 full-length sequences were obtained with the same primer pair.
Sequencing and Validation of BBI Genes
Searches of the S. moellendorffii transcriptome identified two BBI-like sequences (BBI3 and BBI4) (Figure 2). These sequences were not initially found in the published genome using BLASTp searches. Using the RNA sequences in BLASTn searches, BBI3 and BBI4 were found in the genome v1.0 (BBI3, gene locus: scaffold 54: 291,062…291,531; BBI4, gene locus: scaffold 53: 389,315. . .389,727). To verify the sequences identified in both published genome and our transcriptome assembly, the sequences were amplified by 5′ and 3′ RACE, cloned, and sequenced. All sequences were further verified by amplification of an identical sequence from genomic DNA, which also demonstrated the genes for these transcripts are intronless.
We cloned a fifth BBI-like sequence (BBI5) using primers originally designed for BBI3 (Figure 2). We found BBI5 through searches in the genome v1.0 (scaffold 47: 346,127…346,596). Following identification of five BBI-like sequences, the RNA-seq reads were mapped to all five BBI-like sequences (Table 1; Supplemental Figure 1). Reads supporting each sequence indicated all were expressed. In the current model for BBI2 (GenBank: XM_002980955.1), an intron is predicted. However, upon cloning the full-length sequence from cDNA, we found no intron in the sequence. We verified this by mapping the RNA-seq reads onto the genomic sequence with and without the predicted intron present (Supplemental Figure 4). RNA-seq reads mapped to the predicted intron sequence, but not across the exon-exon junction with the predicted intron manually removed, confirming that BBI2 is actually intronless.
S. moellendorffii BBI mRNA Abundance
To estimate the relative abundance of each BBI gene at the mRNA level, clean RNA-seq reads from S. moellendorffii mRNA (NCBI SRA BioSample Accession: SAMN05958544) were mapped onto the ORF of each gene using the CLC Genomics Workbench 6.5.1 (CLC bio). To avoid nonspecific reads mapping due to the high sequence similarity between the BBI-like genes, stringent parameters were used. Mapping parameters were set to a length fraction of 1.0 and similarity fraction of 1.0 to allow only perfect matches to map to each gene.
To determine if BBI2 was intronless as we predicted, reads were mapped to genomic DNA with and without the currently annotated (GenBank: XP_002981001) intron using the same parameters above (Supplemental Figure 4).
Homology Modeling of S. moellendorffii BBI3
A protein model of BBI3 was generated using EasyModeller 4.0, a graphical user interface for MODELLER. Only the BBI-like domain (BBI346-116) was used for the protein model. The model was built against the following templates: soybean (Glycine max) BBI (1BBI), the snail medick (Medicago scutellata) BBI (1MVZ), and cowpea (Vigna unguiculata) BBI (2R33), which show 28, 31, and 28% sequence similarity with the BBI3 BBI domain, respectively.
Recombinant Protein Expression, Purification, and Mutagenesis
The protein encoded by BBI3 was chosen for expression in Escherichia coli as it had the highest number of reads map to its sequence, indicating that it is the most abundant BBI (Table 2; Supplemental Figure 1). A synthetic BBI3 ORF that included an N-terminal six-His tag and a TEV protease cleavage site in lieu of its ER signal was designed with optimal codon usage for E. coli (GENEART) (Supplemental Figure 2). The sequence was subcloned into the BamHI and SalI sites of pQE30 (Qiagen). The pQE30-BBI3 construct and the suppressor plasmid pREP4 (Qiagen) were cotransformed into the E. coli strain Shuffle Express (New England Biolabs; catalog no. C3028H) for protein production.
To test the predicted P1 inhibitory residues of BBI3, a series of mutant sequences were generated by site-directed mutagenesis (Zheng et al., 2004). Three BBI3 motifs are similar to the conserved BBI inhibitory motif. In all three cases, the putative P1 Lys was mutated to Ala by site-directed PCR mutagenesis using primers listed in Supplemental Table 1.
E. coli expressing BBI3 or BBI3 mutants were grown in 750 mL Luria-Bertani medium in 2-liter flasks at 30°C to an OD600 of 0.8, induced by isopropyl β-d-1-thiogalactopyranoside addition to 1 mM, and incubated overnight at 16°C. Bacterial pellets (∼5 mL) were resuspended in 25 mL lysis buffer (50 mM Tris-HCl, pH 8.0, 1 M sodium chloride, and 30 mM imidazole) and lysed by sonication. After centrifugation, the cleared lysate was incubated with Ni-NTA resin (Bio-Rad) at 4°C overnight with mild agitation. The resin was washed with lysis buffer and eluted in elution buffer (50 mM Tris-HCl, pH 8.0, 100 mM sodium chloride, and 300 mM imidazole). The protein was further purified by gel filtration chromatography (BioLogic DuoFlow; Bio-Rad) on a 10/300 Superdex 200 Increase column (GE Healthcare) or 10/300 Superdex 75 GL column (GE Healthcare) in gel filtration buffer (20 mM Tris-HCl, pH 8.0, and 50 mM sodium chloride).
Trypsin Inhibition Assay
Inhibition was determined as described (Prasad et al., 2010). Protein concentration was determined with a Pierce BCA Protein Assay Kit (Thermo Scientific) using BSA as a standard. BBI from soybean was used as a positive control (Sigma-Aldrich; catalog no. T9777). The inhibitors were diluted to 100 µg/mL in gel filtration buffer and 5 μL was added to 20 μL of 25 µg/mL trypsin from bovine pancreas (Sigma-Aldrich) dissolved in 50 mM Tris-Cl and 20 mM calcium chloride, pH 8.0, and incubated for 15 min at 37°C. Residual trypsin activity was determined by the addition of 125 μL of 1 mM N-α-benzoyl-dl-arginine-p-nitroanilide (BAPNA) substrate (Sigma-Aldrich) dissolved in 99% 50 mM Tris-HCl, 20 mM calcium chloride, pH 8.0, and 1% (v/v) dimethyl sulfoxide and incubation for 45 min at 37°C. The reaction was stopped with the addition of 25 μL of 30% (v/v) acetic acid. The absorbance was measured at 410 nm.
Comprehensive BLAST Searches and Phylogenetic Analysis
BLAST searches to identify BBI-like sequences from other species were performed against the NCBI and phytozome genome databases using BBI sequences from legumes and cereals as well as the S. moellendorffii BBI sequences as baits. A list of genome version numbers used for tBLASTn searches is included in Supplemental Data Set 1. Searches were completed by tBLASTn using an E-value threshold of 50. Sequences that shared similarity around the conserved motif were collected and pooled into a single file. Only full-length sequences were used for analysis. Accession numbers for all sequences are included in Supplemental Data Set 1. These sequences were analyzed in a PFAM batch sequence search, and each sequence was found to have homology with the conserved BBI domain.
An alignment using the predicted amino acid sequences from all BBI-like sequences identified through tBLASTn searches, as well as those from S. moellendorffii and Isoetes drumondii, was generated with ClustalW (2.0.12) followed by manual editing with BioEdit Sequence Alignment Editor v7.2.5 (Hall, 1999). Other than manually adjusting the alignment, no deletions were made to avoid reducing the accuracy of the analysis. According to a comprehensive study of filtering methods by Tan et al. (2015), all current filtering methods reduce accuracy of the resulting phylogenetic analysis. A phylogenetic tree was generated from a protein distance matrix using neighbor joining methods using the Jones-Taylor-Thornton model in the Phylip package (v 3.67). Highly similar sequences were not included. Sequences that resulted in very long branch lengths were removed as potential gene model errors. Following the removal of these sequences, the alignment and phylogenetic analysis was repeated with 105 BBI-like sequences (Supplemental File 1). Bootstrap values were calculated from 1000 phylogenetic constructions using a distance neighbor-joining analysis and mapped back onto the original tree. The phylogenetic tree graphical representation was generated with FigTree (v 1.4).
Sequencing BBI-Like Genes from Banana and Quillwort
Leaf tissue was collected from a Cavendish banana (Musa acuminata) tree sourced from Bunnings Warehouse. Live I. drummondii plants were collected by Kingsley Dixon from Alison Baird Reserve, Kenwick, in Western Australia.
I. drummondii was sequenced by Illumina NextSeq500 as 2×151 pair-end read length runs. The transcriptome was assembled as described for Selaginella species. Relevant statistics are presented in Table 2.
To confirm the presence of BBI-like sequences in M. acuminata, genomic DNA was extracted using the DNeasy Plant Minikit (Qiagen). RNA was extracted from I. drummondii as described for Selaginella species. Reverse transcription of this RNA was performed using a ProtoScript II first-strand cDNA synthesis kit (NEB). Primers designed to amplify the complete ORF from cDNA or genomic DNA are included in Supplemental Table 1. Purified PCR products were cloned into pGEM-T Easy (Promega) and sequenced. At least two independent clones were used to account for errors introduced by Taq DNA polymerase.
Accession Numbers
Sequence data generated for this article can be found in the NCBI Short Read Archive and GenBank under accession numbers SRP092379 and KY069178-KY069186, respectively. Protein Data Bank codes for data used in figures are as follows: SFTI-1 1JBL; Gmaa 1BBI; HvuBBIa 2FJ8; MscBBI 1MVZ. GenBank accession numbers for sequences used in figures are Gmab (ACU13240) and HvuBBIb (BAJ91702).
Supplemental Data
Supplemental Figure 1. Relative BBI transcript abundance in S. moellendorffii tissue shown as the number of clean RNA-seq reads that mapped to each open reading frame.
Supplemental Figure 2. BBI3 synthetic protein sequence used for recombinant protein production and SDS-PAGE gel of expressed protein before and after TEV cleavage to remove the 6-His tag.
Supplemental Figure 3. Trypsin inhibition by S. moellendorffii BBI3 and Glycine max BBI.
Supplemental Figure 4. Mapping of RNA-seq reads to genomic DNA sequence shows that BBI2 is intronless.
Supplemental Table 1. All primers used in this study.
Supplemental Data Set 1. Accession numbers and sequence information for sequences indicated in Figure 4.
Supplemental File 1. Sequence alignment of full-length BBI sequences used to generate the BBI phylogeny shown in Figure 6.
Supplementary Material
Acknowledgments
We thank J. Heazlewood for S. moellendorffii cuttings and staff at Hilltop Nursery for cuttings from S. kraussiana and S. martensii. A.M.J. was supported by a University International Stipend and a Scholarship for International Research Fees from UWA. A.S.J. was supported by an International Postgraduate Research Scholarship and an Australian Postgraduate Award from UWA. J.Z. was supported by an International Postgraduate Research Scholarship and an Australian Postgraduate Award from UWA. G.J.K. was supported by a Hackett Postgraduate Scholarship from UWA. J.W. was supported by an Australian Research Council (ARC) Centre of Excellence Grant CE140100008. J.S.M. was supported by an ARC Future Fellowship (FT120100013). This work was supported by ARC Grant DP130101191.
AUTHOR CONTRIBUTIONS
J.S.M. conceived the study. A.M.J. designed and performed all experimentation with the exception of the following: J.S.M. performed RNA extractions; A.S.J. and J.Z. assembled transcriptomes; D.S., O.B., and J.W. prepared libraries and performed sequencing. All authors analyzed the data. G.J.K. and C.B. optimized methods for protein purification. A.M.J. and J.S.M. wrote the manuscript with contributions from all authors.
Glossary
- BBI
Bowman-Birk Inhibitor
- ER
endoplasmic reticulum
- ORF
open reading frame
References
- Albalat R., Cañestro C. (2016). Evolution by gene loss. Nat. Rev. Genet. 17: 379–391. [DOI] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181: 1–20. [Google Scholar]
- Arrigo N., Therrien J., Anderson C.L., Windham M.D., Haufler C.H., Barker M.S. (2013). A total evidence approach to understanding phylogenetic relationships and ecological diversity in Selaginella subg. Tetragonostachys. Am. J. Bot. 100: 1672–1682. [DOI] [PubMed] [Google Scholar]
- Banks J.A. (2009). Selaginella and 400 million years of separation. Annu. Rev. Plant Biol. 60: 223–238. [DOI] [PubMed] [Google Scholar]
- Banks J.A., et al. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.D., Soltis D.E., Soltis P.S. (2010). The age and diversification of the angiosperms re-revisited. Am. J. Bot. 97: 1296–1303. [DOI] [PubMed] [Google Scholar]
- Birk Y. (1961). Purification and some properties of a highly active inhibitor of trypsin and α-chymotrypsin from soybeans. Biochim. Biophys. Acta 54: 378–381. [DOI] [PubMed] [Google Scholar]
- Birk Y., Gertler A., Khalef S. (1963). A pure trypsin inhibitor from soya beans. Biochem. J. 87: 281–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman D.E. (1946). Differentiation of soy bean antitryptic factors. Proc. Soc. Exp. Biol. Med. 63: 547–550. [DOI] [PubMed] [Google Scholar]
- Brauer A.B.E., Leatherbarrow R.J. (2003). The conserved P1′ Ser of Bowman-Birk-type proteinase inhibitors is not essential for the integrity of the reactive site loop. Biochem. Biophys. Res. Commun. 308: 300–305. [DOI] [PubMed] [Google Scholar]
- Brauer A.B.E., Domingo G.J., Cooke R.M., Matthews S.J., Leatherbarrow R.J. (2002). A conserved cis peptide bond is necessary for the activity of Bowman-Birk inhibitor protein. Biochemistry 41: 10608–10615. [DOI] [PubMed] [Google Scholar]
- Clemente A., Arques M.C., Dalmais M., Le Signor C., Chinoy C., Olias R., Rayner T., Isaac P.G., Lawson D.M., Bendahmane A., Domoney C. (2015). Eliminating anti-nutritional plant food proteins: the case of seed protease inhibitors in pea. PLoS One 10: e0134634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hont A., et al. (2012). The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488: 213–217. [DOI] [PubMed] [Google Scholar]
- Domingo G.J., Leatherbarrow R.J., Freeman N., Patel S., Weir M. (1995). Synthesis of a mixture of cyclic peptides based on the Bowman-Birk reactive site loop to screen for serine protease inhibitors. Int. J. Pept. Protein Res. 46: 79–87. [DOI] [PubMed] [Google Scholar]
- Elliott A.G., et al. (2014). Evolutionary origins of a bioactive peptide buried within Preproalbumin. Plant Cell 26: 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariani T., McBride J.D., Leatherbarrow R.J. (1999). The role of the P2′ position of Bowman-Birk proteinase inhibitor in the inhibition of trypsin. Studies on P2′ variation in cyclic peptides encompassing the reactive site loop. Biochim. Biophys. Acta 1431: 232–237. [DOI] [PubMed] [Google Scholar]
- Habib H., Fazili K.M. (2007). Plant protease inhibitors: a defense strategy in plants. Biotechnol. Mol. Biol. Rev. 2: 68–85. [Google Scholar]
- Hall T.A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Hirsh A.E., Fraser H.B. (2001). Protein dispensability and rate of evolution. Nature 411: 1046–1049. [DOI] [PubMed] [Google Scholar]
- Jayasena A.S., Secco D., Bernath-Levin K., Berkowitz O., Whelan J., Mylne J.S. (2014). Next generation sequencing and de novo transcriptomics to study gene evolution. Plant Methods 10: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov D.M., Wolf Y.I., Rogozin I.B., Koonin E.V. (2003). Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 13: 2229–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang C., Xu X., Wang J., Yu H., Lai R., Gong W. (2007). Trypsin inhibitory loop is an excellent lead structure to design serine protease inhibitors and antimicrobial peptides. FASEB J. 21: 2466–2473. [DOI] [PubMed] [Google Scholar]
- Luckett S., Garcia R.S., Barker J.J., Konarev A.V., Shewry P.R., Clarke A.R., Brady R.L. (1999). High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds. J. Mol. Biol. 290: 525–533. [DOI] [PubMed] [Google Scholar]
- McBride J.D., Brauer A.B.E., Nievo M., Leatherbarrow R.J. (1998). The role of threonine in the P2 position of Bowman-Birk proteinase inhibitors: studies on P2 variation in cyclic peptides encompassing the reactive site loop. J. Mol. Biol. 282: 447–458. [DOI] [PubMed] [Google Scholar]
- McBride J.D., Watson E.M., Brauer A.B., Jaulent A.M., Leatherbarrow R.J. (2002). Peptide mimics of the Bowman-Birk inhibitor reactive site loop. Biopolymers 66: 79–92. [DOI] [PubMed] [Google Scholar]
- Mello M.O., Tanaka A.S., Silva-Filho M.C. (2003). Molecular evolution of Bowman-Birk type proteinase inhibitors in flowering plants. Mol. Phylogenet. Evol. 27: 103–112. [DOI] [PubMed] [Google Scholar]
- Mylne J.S., Colgrave M.L., Daly N.L., Chanson A.H., Elliott A.G., McCallum E.J., Jones A., Craik D.J. (2011). Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat. Chem. Biol. 7: 257–259. [DOI] [PubMed] [Google Scholar]
- Mylne J.S., Chan L.Y., Chanson A.H., Daly N.L., Schaefer H., Bailey T.L., Nguyencong P., Cascales L., Craik D.J. (2012). Cyclic peptides arising by evolutionary parallelism via asparaginyl-endopeptidase-mediated biosynthesis. Plant Cell 24: 2765–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino N., Aoyagi H., Kato T., Izumiya N. (1977). Studies on the synthesis of proteinase inhibitors. I. Synthesis and activity of nonapeptide fragments of soybean Bowman-Birk inhibitor. J. Biochem. 82: 901–909. [DOI] [PubMed] [Google Scholar]
- Odani S., Ikenaka T. (1973). Studies on soybean trypsin inhibitors. 8. Disulfide bridges in soybean Bowman-Birk proteinase inhibitor. J. Biochem. 74: 697–715. [DOI] [PubMed] [Google Scholar]
- Park E.Y., Kim J.A., Kim H.W., Kim Y.S., Song H.K. (2004). Crystal structure of the Bowman-Birk inhibitor from barley seeds in ternary complex with porcine trypsin. J. Mol. Biol. 343: 173–186. [DOI] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Prakash B., Selvaraj S., Murthy M.R., Sreerama Y.N., Rao D.R., Gowda L.R. (1996). Analysis of the amino acid sequences of plant Bowman-Birk inhibitors. J. Mol. Evol. 42: 560–569. [DOI] [PubMed] [Google Scholar]
- Prasad E.R., Merzendorfer H., Madhurarekha C., Dutta-Gupta A., Padmasree K. (2010). Bowman-Birk proteinase inhibitor from Cajanus cajan seeds: purification, characterization, and insecticidal properties. J. Agric. Food Chem. 58: 2838–2847. [DOI] [PubMed] [Google Scholar]
- Qi R.F., Song Z.W., Chi C.W. (2005). Structural features and molecular evolution of Bowman-Birk protease inhibitors and their potential application. Acta Biochim. Biophys. Sin. (Shanghai) 37: 283–292. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D., Barrett A.J., Finn R. (2016). Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 44: D343–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T.H., Hejgaard J. (2008). Serpins in plants and green algae. Funct. Integr. Genomics 8: 1–27. [DOI] [PubMed] [Google Scholar]
- Schechter I., Berger A. (1967). On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27: 157–162. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., et al. (2008). The Amborella genome: an evolutionary reference for plant biology. Genome Biol. 9: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.K., Kim Y.S., Yang J.K., Moon J., Lee J.Y., Suh S.W. (1999). Crystal structure of a 16 kDa double-headed Bowman-Birk trypsin inhibitor from barley seeds at 1.9 A resolution. J. Mol. Biol. 293: 1133–1144. [DOI] [PubMed] [Google Scholar]
- Tan G., Muffato M., Ledergerber C., Herrero J., Goldman N., Gil M., Dessimoz C. (2015). Current methods for automated filtering of multiple sequence alignments frequently worsen single-gene phylogenetic inference. Syst. Biol. 64: 778–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada S., Sato K., Kato T., Izumiya N. (1978). Inhibitory properties of nonapeptide loop structures related to reactive sites of soybean Bowman-Birk inhibitor. FEBS Lett. 90: 89–92. [DOI] [PubMed] [Google Scholar]
- Tran L.T., Taylor J.S., Constabel C.P. (2012). The polyphenol oxidase gene family in land plants: Lineage-specific duplication and expansion. BMC Genomics 13: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Baumann U., Reymond J.L. (2004). An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.