Abstract
Importance
Low-density lipoprotein (LDL) cholesterol-lowering alleles in or near NPC1L1 or HMGCR, encoding the respective molecular targets of ezetimibe and statins, have previously been used as proxies to study the efficacy of these lipid-lowering drugs. Alleles near HMGCR are associated with a higher risk of type 2 diabetes, mimicking the increased incidence of new-onset diabetes associated with statin treatment in randomized clinical trials. It is unknown whether alleles near NPC1L1 are also associated with the risk of type 2 diabetes.
Objective
To investigate whether LDL-lowering alleles in or near NPC1L1 and other genes encoding current or prospective molecular targets of lipid-lowering therapy (i.e. HMGCR, PCSK9, ABCG5/G8, LDLR) are associated with the risk of type 2 diabetes.
Design, Setting and Participants
The associations with type 2 diabetes and coronary artery disease of LDL-lowering genetic variants were investigated in meta-analyses of genetic association studies. Meta-analyses included 50,775 individuals with type 2 diabetes and 270,269 controls including three studies and 60,801 individuals with coronary artery disease and 123,504 controls from a published meta-analysis. Data collection took place in Europe and the United States between 1991 and 2016.
Exposure
LDL-lowering alleles in or near NPC1L1, HMGCR, PCSK9, ABCG5/G8, LDLR.
Main Outcomes and Measures
Odds ratio of type 2 diabetes and coronary artery disease.
Results
LDL-lowering genetic variants at NPC1L1 were inversely associated with coronary artery disease (odds ratio for a genetically-predicted reduction of 1 mmol/L in LDL cholesterol, 0.61; 95% confidence interval, 0.42-0.88; p=0.008) and directly associated with type 2 diabetes (2.42, 1.70-3.43; p<0.001). The odds ratio of type 2 diabetes for PCSK9 genetic variants was 1.19 (95% confidence interval, 1.02-1.38, p=0.03). For a given reduction in LDL cholesterol, genetic variants were associated with a similar reduction in coronary artery disease risk (I-squared for heterogeneity in genetic associations=0.0%; p=0.93). However, associations with type 2 diabetes were heterogeneous (I-squared=77.2%; p=0.002), indicating gene-specific associations with metabolic risk for LDL-lowering alleles.
Conclusions and Relevance
In this meta-analysis, exposure to LDL-cholesterol lowering genetic variants in or near NPC1L1 and other genes was associated with a higher risk of type 2 diabetes. These data provide insights into potential adverse effects of LDL cholesterol-lowering therapy.
Introduction
Treatment with statins, the pharmacological agents of choice for low-density lipoprotein (LDL) cholesterol-lowering therapy in cardiovascular prevention,1,2 is associated with weight gain and a higher incidence of new-onset type 2 diabetes.3–5 Ezetimibe, an inhibitor of the LDL cholesterol transporter Niemann-Pick C1-like 1 (NPC1L1),6,7 has been approved as a lipid-lowering agent, but it is unclear whether its use will also be associated with an adverse metabolic risk profile.
There is considerable interest in predicting the efficacy and safety of therapeutic targets early in the drug development process. Drug targets with supporting human genetic evidence have been shown to have lower attrition rates during drug development,8 while variation in genes encoding drug targets has been used to predict both the efficacy and safety of pharmacological perturbation of those targets.9,10 In particular, LDL-lowering alleles in HMGCR5,11 encoding the molecular target of statins, have been successfully used as genetic proxies to study the effects of these drugs.5,11 Furthermore, LDL-lowering alleles at HMGCR are associated with higher risk of type 2 diabetes and higher body mass index in genetic studies,5 mimicking the safety profile of statins in meta-analyses of randomized clinical trials.3–5
The efficacy of adding ezetimibe to simvastatin in secondary cardiovascular prevention was supported by the IMPROVE-IT trial.6,7 Immediately before and after the publication of the trial results, studies were reported describing the use of genetic variants at NPC1L1 to predict the efficacy of NPC1L1 inhibition in the prevention of coronary events.11,12 The purpose of this study was to use naturally-occurring LDL-lowering alleles at NPC1L1 to investigate the potential associations between NPC1L1 inhibition and the risk of type 2 diabetes. LDL-lowering alleles in or near genes encoding other current or prospective molecular targets of LDL-cholesterol lowering therapy were also studied.
Methods
Study design
The association of LDL-cholesterol lowering polymorphisms near NPC1L1 with the risk of type 2 diabetes was investigated in meta-analyses of genetic association studies. In addition to NPC1L1 polymorphisms, the association of LDL-lowering alleles in or near genes encoding other current or prospective molecular targets of LDL-cholesterol lowering therapy11 (i.e. HMGCR, PCSK9, ABCG5/G8, LDLR) with type 2 diabetes, coronary artery disease and continuous cardiometabolic traits was also studied. A summary of the studies participating in each analysis is presented in eTable 1.
Participants
The association of LDL-cholesterol lowering alleles with type 2 diabetes was estimated in a meta-analysis of 50,775 individuals with type 2 diabetes and 270,269 controls from the EPIC-InterAct study13, the UK Biobank study14 and the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM).15 An additional eleven studies (4,496 cases and 50,677 controls) previously reported by Swerdlow and colleagues5 were included in analyses of the association with type 2 diabetes of rs12916 in HMGCR (eFigure 1). The combined association of NPC1L1 genetic variants in subgroups of age, sex, and body mass index was analyzed in 14,657 unrelated cases of type 2 diabetes and 118,854 controls from EPIC-InterAct and UK Biobank with available individual-level genotyping data.
EPIC-InterAct is a case-cohort study nested within the European Prospective Investigation into Cancer and Nutrition (EPIC) study, a cohort study of 500,000 European participants followed-up for an average of 8 years.13 Eight of the ten constituent EPIC cohorts agreed to take part in EPIC-InterAct leaving 455,680 participants for screening. Individuals were excluded from EPIC-InterAct if they did not have stored blood (n=109,625) or information on diabetes status (n=5,821; 1.3% of participants screened for inclusion). From the remaining 340,234 participants, 12,403 individuals who developed type 2 diabetes during follow-up constituted the incident case group of EPIC-InterAct and a random group of 16,154 individuals free of diabetes at baseline constituted the subcohort group of EPIC-InterAct.13 Subcohort participants were previously shown to be representative of eligible EPIC participants within each country.13 Data on a total of 20,831 participants with available genotyping (with no overlap with DIAGRAM15) were included in the main analysis, while data on all the 22,494 participants with available genotyping were included in subgroup analyses. Type 2 diabetes status was available in all participants. Individuals without genotype data were excluded from the study. Data collection took place between 1991 and 2016. Participant characteristics and genotyping methods have been previously reported in detail13 and are summarized in Table 1 and eTable 2.
Table 1. Participants of EPIC-InterAct, UK Biobank and DIAGRAM.
| Variable | EPIC-InterAct | UK Biobank | DIAGRAM | |||
|---|---|---|---|---|---|---|
| Type 2 diabetes | Subcohort (non-cases) | Type 2 diabetes | Controls | Type 2 diabetes | Controls | |
| Country | Multiple European countries | United Kingdom | Europe and United Statesc | |||
| Genotyping chip | Illumina 660w quad and Illumina CoreExome chip | Affymetrix UK Biobank Axiom Array | Multipled | |||
| Imputation panel | Haplotype Reference Consortium | 1000 Genomes Phase 3 plus UK10K | HapMap | |||
| Number | 10,071a | 12,423a | 6,627 | 143,765 | 34,840 | 114,981 |
| Age, mean years (SD) | 56 (8) | 52 (9) | 60 (7) | 56 (8) | 59 (10) | 54 (14) |
| Female sex, N (%) | 5,037 (50) | 7,713 (62) | 2,349 (35) | 77,397 (54) | 14,621 (42) | 60,377 (53) |
| Smoking status, current smokers N (%) | 2,830 (28) | 3,240 (26) | 811 (12) | 18,149 (13) | NA | NA |
| BMI in kg/m2, mean (SD) | 29.7 (4.8) | 25.8 (4.1) | 31.9 (5.9) | 27.3 (4.7) | 29.7 (5.9) | 26.5 (4.5) |
| Waist-to-hip ratio | 0.92 (0.09) | 0.85 (0.09) | 0.95 (0.08) | 0.87 (0.09) | NA | NA |
| Systolic blood pressure in mmHg, mean (SD) | 144 (20) | 132 (19) | 141 (17) | 138 (19) | NA | NA |
| Diastolic blood pressure in mmHg, mean (SD) | 87 (11) | 82 (11) | 82 (10) | 82 (10) | NA | NA |
| LDL cholesterol in mmol/L, mean (SD) | 4.0 (1) | 3.8 (1) | NAb | NAb | NA | NA |
| HDL cholesterol in mmol/L, mean (SD) | 1.2 (0.4) | 1.5 (0.4) | NAb | NAb | NA | NA |
| Triglycerides in mmol/L, median (IQR) | 1.7 (1.2-2.5) | 1.1 (0.8-1.6) | NAb | NAb | NA | NA |
Abbreviations: N, number of participants; BMI, body mass index; SD, standard deviation; LDL, low-density lipoprotein cholesterol; HDL, high-density lipoprotein cholesterol; IQR, interquartile range; NA, not available.
A total of 9,308 type 2 diabetes cases and 11,523 non-cases were included in the main analysis of the association of genetic variants with type 2 diabetes after the exclusion of participants overlapping with DIAGRAM.
Blood lipids concentrations are being measured in the UK Biobank study, with data release currently planned for the end of 2016.
DIAGRAM had a small South Asian component accounting for 2.44% of participants.
Affymetrix Human SNP Array 6.0; Illumina HumanHap 300, 300/370 and 550; Affymetrix Genechip 500K & MIPS 50K; Cardio-Metabolchip.
UK Biobank is a population-based cohort of 500,000 people aged between 40-69 years who were recruited in 2006-2010 from several centers across the United Kingdom.14 The association of genetic variants with prevalent type 2 diabetes was estimated in 6,627 cases and 143,765 controls of the UK Biobank dataset who had available genotype data. Genotyping was attempted in 152,770 individuals and failed in only 480 instances (0.3%). Among a total of 152,290 participants with available genotype data, type 2 diabetes status was adjudicated in 150,392 (98.8%) participants. Type 2 diabetes was defined on the basis of self-reported physician diagnosis at nurse interview or digital questionnaire, age at diagnosis > 36 years, and use of oral anti-diabetic medications. Data collection took place between 2006 and 2016. Participant characteristics and genotyping information are reported in Table 1 and eTable 2.
DIAGRAM is a research consortium that published the largest meta-analysis of genome-wide association studies for type 2 diabetes in individuals of European descent.15 Type 2 diabetes association results were made publicly available for up to 34,840 cases and 114,981 controls from 38 genetic association studies with a case-control or cohort design.15 Fifty percent of the participants were women and the average age was 55 years.15 Imputation was performed using the HapMap reference panel.15 Participant exclusion criteria encompassed duplicate samples, relatedness, mismatch between self-reported and genotype-determined sex, outlying heterozygosity and non-European descent. Type 2 diabetes status was available in all participants. Data collection took place between 2002 and 2012. Participant characteristics are reported in Table 1 and further characteristics of studies included in the DIAGRAM meta-analysis were reported previously in detail.15
The likelihood of bias for studies participating in this meta-analysis was deemed low on the basis of: (a) the low proportion of participants with missing data on exposure or outcome, (b) the high-quality genotyping or imputation of genetic variants included in the study (eTable 2), (c) the low likelihood of bias by case-status in genotyping errors or genotype misclassification, (d) the consideration that if any non-differential misclassification of exposure or outcome occurred, that would result in a bias towards the null and (e) the consideration that genetic variants are less likely to be affected by confounding or reverse causality.16,17 On this basis, studies were deemed suitable for pooling by meta-analysis.
For the genetic variants included in these analyses, LDL cholesterol association estimates were obtained from genetic association results in up to 188,577 participants of the Global Lipids Genetics Consortium.18 In addition to type 2 diabetes, the association of these LDL-lowering alleles with coronary artery disease and continuous cardiometabolic traits was also estimated in large meta-analyses of genome-wide association studies. For coronary artery disease, data were from the CARDIoGRAMplusC4D Consortium meta-analysis (60,801 cases and 123,504 controls).19 For glycaemic traits, including fasting glucose20,21 (N=133,010), glucose two hours after an oral glucose challenge20,22 (N=42,854) and fasting insulin levels20,21 (natural-logarithm transformed; N=108,557), data were from the MAGIC Consortium.20–22 For anthropometric traits, including body mass index (N=333,495) and waist-to-hip ratio (N=224,047), data were from the GIANT consortium.23,24 For details, see eTable 1.
In exploratory analyses, the burden of protein-truncating and “probably deleterious” missense variants in NPC1L1, HMGCR, PCSK9, ABCG5, ABCG8 and LDLR was estimated from exome sequencing studies of 8,373 type 2 diabetes cases and 8,466 controls (AMP-T2D Program; T2D-GENES Consortium, SIGMA T2D Consortium. 2016 May 26; http://www.type2diabetesgenetics.org/).
Selection of genetic variants
The combined association of two LDL cholesterol lowering genetic variants near NPC1L1 with type 2 diabetes constituted the primary analysis of the study (Table 2). These variants were identified as having distinct effects on LDL cholesterol levels in approximate conditional analyses using the GCTA software25,26 (see methodology description below; eFigure 2). In sensitivity analyses, the combined association of five LDL-lowering alleles near NPC1L1, previously used to predict the efficacy of ezetimibe,11 was also investigated (eTable 3).
Table 2. LDL-cholesterol lowering polymorphisms at NPC1L1 and other genes and their association with type 2 diabetes and coronary artery disease.
| Gene | dbSNP rsID | Genomic coordinate, chromosome and position | Effect / other allele | Effect allele frequency, mean (range) | LDL, N | Beta (95% CI) per allele in mmol/La | p-value | OR of coronary artery disease (95% CI) per alleleb | p-value | OR of type 2 diabetes (95% CI) per allelec | p-value | I-squared | Heterogeneity p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NPC1L1 | rs2073547 | chr7:44582331 | A / G | 0.81 (0.81, 0.82) | 169,889 | -0.049 (-0.058, -0.039) | 2 x 10-21 | 0.980 (0.957, 1.003) | 0.09 | 1.051 (1.027, 1.075) | 2 x 10-05 | 0.0% | 0.48 |
| rs217386 | chr7:44600695 | A / G | 0.42 (0.41, 0.44) | 173,021 | -0.036 (-0.044, -0.029) | 1 x 10-19 | 0.979 (0.960, 0.998) | 0.03 | 1.027 (1.009, 1.045) | 0.003 | 0.0% | 0.68 | |
| HMGCR | rs12916 d | chr5:74656539 | T / C | 0.58 (0.57, 0.60) | 168,357 | -0.073 (-0.081, -0.066) | 8 x 10-78 | 0.965 (0.947, 0.983) | 0.0002 | 1.029 (1.012, 1.046) | 0.0007 | 46.4% | 0.03 |
| rs5744707 | chr5:74890618 | A / G | 0.90 (0.90, 0.91) | 172,928 | -0.055 (-0.067, -0.043) | 6 x 10-19 | 0.970 (0.941, 0.999) | 0.04 | 0.983 (0.956, 1.011) | 0.24 | 2.8% | 0.38 | |
| rs16872526 | chr5:74675717 | T / G | 0.91 (0.90, 0.92) | 173,009 | -0.041 (-0.054, -0.027) | 2 x 10-08 | 0.988 (0.959, 1.018) | 0.44 | 1.016 (0.985, 1.047) | 0.32 | 50.1% | 0.11 | |
| PCSK9 | rs11591147 | chr1:55505647 | T / G | 0.02 (0.01, 0.02) | 77,417 | -0.497 (-0.532, -0.462) | 9 x 10-143 | 0.774 (0.692, 0.866) | 7 x 10-06 | 1.089 (1.010, 1.174) | 0.03 | 0.0% | 0.39 |
| ABCG5/G8 | rs4299376 | chr2:44072576 | T / G | 0.69 (0.68, 0.70) | 144,861 | -0.081 (-0.090, -0.072) | 4 x 10-72 | 0.950 (0.931, 0.970) | 1 x 10-06 | 1.011 (0.990, 1.032) | 0.29 | 2.2% | 0.38 |
| LDLR | rs6511720 | chr19:11202306 | T / G | 0.11 (0.10, 0.12) | 170,608 | -0.221 (-0.233, -0.209) | 4 x 10-262 | 0.882 (0.853, 0.912) | 1 x 10-13 | 1.028 (0.999, 1.057) | 0.05 | 0.0% | 0.93 |
Polymorphism names reported in the table are rsID entries from dbSNP release 147.
Genomic coordinates represent chromosome and physical position of genetic variants according to the Human Reference Genome Build 37.
Abbreviations: LDL, low-density lipoprotein cholesterol; N, number of participants; CI, confidence interval; CAD, coronary artery disease; T2D, type 2 diabetes; OR, odds ratio.
LDL cholesterol data were from the Global Lipids Genetics Consortium.18
Coronary artery disease data were from 60,801 coronary artery disease cases and 123,504 controls from the CARDIoGRAMplusC4D Consortium.19
Type 2 diabetes data were from 50,775 cases of type 2 diabetes and 270,269 controls from EPIC-InterAct,13 UK Biobank14 and DIAGRAM.15
In addition to EPIC-InterAct,13 UK Biobank14 and DIAGRAM.15, type 2 diabetes association analyses of rs12916 included eleven studies (4,496 cases and 50,677 controls) previously reported by Swerdlow and colleagues.5 The total sample size of this analysis was of 55,271 cases of type 2 diabetes and 320,946 controls.
For comparison with NPC1L1, other LDL-lowering alleles in or near genes encoding other current or prospective molecular targets of LDL-cholesterol lowering therapy (i.e. HMGCR, PCSK9, ABCG5/G8, LDLR) were studied.11 Three LDL-lowering polymorphisms in or near HMGCR, previously demonstrated to mimic the efficacy and metabolic effects of statins,5,11 were analyzed (Table 2). At the ABCG5/G8 and LDLR loci, polymorphisms previously used to investigate genetic relationships between LDL cholesterol and coronary artery disease11 were studied (Table 2). At the PCSK9 locus, in addition to the rs11591147 (p.R46L) variant (Table 2), the combined association of up to an additional eight likely-independent LDL-lowering polymorphisms was investigated (eFigure 3). Genetic variants included in the analyses were strongly and specifically associated with LDL cholesterol (eFigure 4).
Approximate conditional analyses on large-scale LDL-cholesterol association data from the Global Lipids Genetics Consortium18 using the GCTA software25,26 were performed in order to identify distinct association signals for LDL cholesterol at the NPC1L1 and PCSK9 loci. This approach uses genetic association results in addition to the linkage disequilibrium pattern in a reference population to estimate the association of genetic variants in a region after accounting for one or more index genetic variants. In so doing, the software allows for the identification of likely-independent association signals in a given region using result-level data. At the PCSK9 locus, in a smaller sample of individuals with individual-level genotypes, formal conditional analyses of the association with LDL cholesterol of polymorphisms after adjusting for rs11591147 genotype status were also conducted (eFigure 3).
Genetic reference information
HUGO Gene Nomenclature Committee27 (URL: www.genenames.org) gene names for the investigated genes were: HGNC:7898 (NPC1L1), HGNC:5006 (HMGCR), HGNC:20001 (PCSK9), HGNC:13886 (ABCG5), HGNC:13887 (ABCG8), HGNC:6547 (LDLR). Genomic coordinates reported in the manuscript represent the chromosome and physical position of genetic variants according to the Human Reference Genome Build 37 (URL: http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/). Polymorphism names reported in the manuscript represent rsID entries from dbSNP release 147 (URL: http://www.ncbi.nlm.nih.gov/SNP/).
Statistical analysis
Genetic association data for the meta-analyses were either generated or gathered from available sources at the MRC Epidemiology Unit, University of Cambridge (United Kingdom). For each genetic variant and outcome, inverse variance weighted meta-analyses using fixed-effect models was used to obtain pooled estimates. The I-squared statistic was used to quantify heterogeneity. For each gene, associations of LDL-lowering genetic variants and outcomes was estimated using Mendelian randomization statistical methodology.17 Estimates of “genetic variant to LDL-cholesterol” and “genetic variant to outcome” associations were used to calculate estimates of “LDL-cholesterol reduction to outcome” association at each gene.17 When multiple genetic variants at a given gene were included in the model, estimates were pooled with a weighted generalized linear regression method that accounts for the correlation between genetic variants.17 The correlation values were obtained from the SNAP software28 or from the 1000 Genomes Project data on individuals of European ancestry (URL:http://browser.1000genomes.org/; eTable 4). Results were scaled to represent the odds ratio per 1 mmol/L genetically-predicted reduction in LDL cholesterol. Absolute risk differences were estimated assuming that the incidence rate of type 2 diabetes in the InterAct study subcohort would be the baseline incidence rate in “non-exposed” individuals (i.e. 3.76 incident cases per 1000 person-years of follow-up).13 This baseline rate was then multiplied by the odds ratio estimated from genetic analyses to obtain the “at risk” incidence rate. The absolute risk difference estimate was the “at risk” incidence rate minus the baseline incidence rate. Absolute risk differences were expressed in incident events per 1000 person-years for a 1 mmol/L genetically-predicted reduction in LDL cholesterol. Statistical analyses were conducted using STATA v14.1 (StataCorp, College Station, Texas 77845 USA), R v3.2.2 (The R Foundation for Statistical Computing), and METAL.29 A two-tailed p-value of p<0.05 was considered statistically significant.
Results
LDL cholesterol lowering alleles at NPC1L1 and risk of type 2 diabetes
LDL cholesterol lowering alleles at the NPC1L1 locus were inversely associated with coronary artery disease and directly associated with type 2 diabetes, both individually (Table 2) and collectively (odds ratio of type 2 diabetes per 1 mmol/L genetically-predicted LDL cholesterol reduction, 2.42; 95% confidence interval, 1.70-3.43, p<0.001; estimated absolute risk difference, 5.3 incident cases per 1000 person-years for a 1 mmol/L genetically-predicted reduction in LDL cholesterol; Figure 1). For both polymorphisms, estimates of the association with type 2 diabetes were consistent across the studies included in the meta-analysis (eFigure 1). In the periphery of the NPC1L1 locus, approximately 400 kilobases from the lead rs2073547 polymorphism, there was a known association signal for type 2 diabetes and glycemic traits near the GCK gene.15,20,21 After accounting for variation in GCK, the association of with type 2 diabetes at NPC1L1 did not change (eTable 3). Association estimates also remained unchanged when modeling the association of five polymorphisms previously used by Ference et al.11 as a proxy for NPC1L1 inhibition (eTable 3). In 14,657 cases of type 2 diabetes and 118,854 controls for whom we had access to individual-level genotyping data, there was no evidence of heterogeneity in the association between NPC1L1 alleles and type 2 diabetes in analyses stratified by age, sex or body mass index (eFigure 5). In exome sequencing association results, there was no evidence of enrichment of NPC1L1 protein truncating alleles in type 2 diabetes cases compared with controls (odds ratio of type 2 diabetes for individuals carrying a truncating allele, 1.12; 95% confidence interval, 0.88-1.43; p=0.34), but missense variants in NPC1L1 predicted to be “probably deleterious” were overrepresented in individuals with type 2 diabetes compared with controls (1.26, 1.07-1.47; p=0.005).
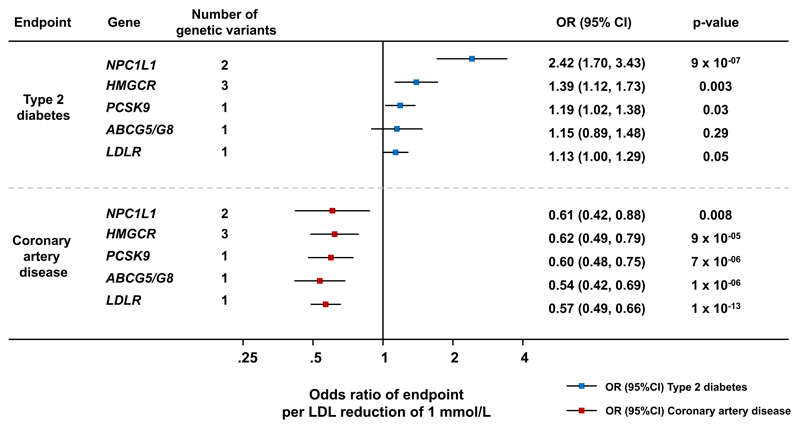
Figure 1. Odds ratio of coronary artery disease and type 2 diabetes associated with LDL-lowering genetic variants in or near investigated genes.
Coronary artery disease data were from 60,801 coronary artery disease cases and 123,504 controls from the CARDIoGRAMplusC4D Consortium.19 Type 2 diabetes data were from 50,775 cases of type 2 diabetes and 270,269 controls from EPIC-InterAct,13 UK Biobank14 and DIAGRAM.15 In addition to EPIC-InterAct,13 UK Biobank14 and DIAGRAM.15, type 2 diabetes association analyses of rs12916 at HMGCR included eleven studies (4,496 cases and 50,677 controls) previously reported by Swerdlow and colleagues.5 Therefore the sample size of HMGCR genetic variants association with type 2 diabetes was of up to 55,271 cases of type 2 diabetes and 320,946 controls. All results are scaled to represent the odds ratio per 1 mmol/L genetically-predicted reduction in LDL cholesterol. Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio; CAD, coronary artery disease; T2D, type 2 diabetes; LDL, low-density lipoprotein cholesterol.
Associations with type 2 diabetes at other genes
As previously reported,5,11 LDL cholesterol lowering alleles at HMGCR were also associated with type 2 diabetes and coronary artery disease in opposite directions (Table 2 and Figure 1). An association of the loss-of-function p.R46L (rs11591147) variant in PCSK9 with higher risk of type 2 diabetes was also found (odds ratio of type 2 diabetes per 1 mmol/L genetically-predicted LDL cholesterol reduction, 1.19; 95% confidence interval, 1.02-1.38, p=0.03; estimated absolute risk difference, 0.7 incident cases per 1000 person-years for a 1 mmol/L genetically-predicted reduction in LDL cholesterol; Table 2 and Figure 1). At PCSK9, analyses of the LDL cholesterol association data suggested the presence of distinct association signals. In formal conditional analyses, there was evidence of at least two distinct association signals (rs11591147 and rs471705; eFigure 3). Using the GCTA software,25,26 approximate conditional analyses suggested the presence of nine distinct association signals (rs11591147 plus eight additional genetic variants; eFigure 3). Inclusion of these additional signals gave similar associations with type 2 diabetes as the p.R46L variant alone (odds ratio of type 2 diabetes per 1 mmol/L genetically-predicted reduction in LDL cholesterol using rs11591147 plus rs471705, 1.21, 95% confidence interval, 1.04-1.41, p=0.01; and 1.16, 1.03-1.31, p=0.02, using rs11591147 plus the eight additional polymorphisms; eTable 3). The association with type 2 diabetes of LDL-lowering alleles at the ABCG5/G8 and LDLR loci did not reach statistical significance. There was no evidence of association with type 2 diabetes for missense variants predicted to be “probably deleterious” or protein truncating alleles in the HMGCR, PCSK9, ABCG5, ABCG8 and LDLR genes (eTable 5), but the confidence intervals around risk estimates were generally wide, reflecting the low prevalence of these genetic variants and the relatively small sample size of this analysis.
Evidence of gene-specific associations with type 2 diabetes risk
In analyses of the association with disease risk for a given genetically-predicted reduction in LDL cholesterol, there was a similar reduction in coronary artery disease risk across genes (I-squared for heterogeneity in genetic associations = 0.0%; p=0.93, Figure 1). However, for the same reduction in LDL cholesterol, the association with type 2 diabetes risk differed by gene (I-squared = 77.2%; p=0.002, Figure 1). The different magnitudes and directions of association of LDL-lowering alleles with continuous glycemic and anthropometric traits suggested gene-specific mechanisms underlying the altered risk of type 2 diabetes (eFigure 6). For example, at the HMGCR locus there were associations with body mass index and waist-to-hip ratio, while at the PCSK9 locus there were associations with higher fasting glucose and two hour glucose (eFigure 6).
Discussion
In this meta-analysis, exposure to LDL-cholesterol lowering genetic variants in or near the NPC1L1 gene was associated with a higher risk of type 2 diabetes. This finding is consistent with the results of a small-scale open label randomized clinical trial, showing increased glycated hemoglobin in association with ezetimibe treatment.30 Blazing et al. reported that the addition of ezetimibe to simvastatin for secondary cardiovascular prevention in the IMPROVE-IT trial resulted in a small and not statistically significant increase in risk of new-onset diabetes (9% relative risk increase per 1 mmol/L reduction in LDL cholesterol).31 However, IMPROVE-IT results may not be sufficient to rule out an effect of inhibiting Niemann-Pick C1-like 1 on diabetes risk because: (1) some of the effects of NPC1L1 inhibition may be apparent only after several years of treatment; (2) the risk of type 2 diabetes in individuals with a history of acute coronary syndrome yet free from type 2 diabetes in IMPROVE-IT may not reflect that of the general population on which this genetic analysis is based; (3) limited compliance to drug treatment, as observed in IMPROVE-IT,7 may dilute etiologic effect estimates. By analogy, the association of statin treatment with higher diabetes risk was only demonstrable in a meta-analysis of several randomized clinical trials including more than 90,000 individuals.3 Therefore, these results warrant the continued monitoring of the glycemic effects of ezetimibe in randomized clinical trials and clinical practice particularly in a primary prevention setting.
The results of this study show that multiple LDL-lowering mechanisms, including those mediated by the molecular targets of available LDL-lowering drugs (i.e. statins, ezetimibe, and PCSK9 inhibitors), are associated with adverse metabolic consequences and increased type 2 diabetes risk. These findings are consistent with other studies of the association with type 2 diabetes of genetic scores aggregating multiple polymorphisms affecting LDL cholesterol and other lipid fractions.32 They are also consistent with the observation that patients with familial hypercholesterolemia are less likely to have type 2 diabetes.33 The genes which were associated both with lower LDL cholesterol levels and higher type 2 diabetes risk impact on LDL cholesterol by distinct pathways including cholesterol absorption (NPC1L1),34 endogenous cholesterol synthesis (HMGCR)35 and internalization of cholesterol-rich particles into the cell (PCSK9).36,37 For a similar reduction in LDL cholesterol, the association with type 2 diabetes differed by gene which would be consistent with the mediation of their associations by different mechanisms. Besseling et al. have proposed that an increased internalization of cholesterol into pancreatic beta-cells may result in impaired secretion of insulin,33 a hypothesis supported by murine experimental models.38 LDL cholesterol lowering alleles at HMGCR are associated with higher fasting insulin and body mass index, suggesting an insulin resistance-related mechanism.5 Finally, in contrast with early evidence showing metabolic benefits of NPC1L1 knock-out in mice,39 recent studies suggest that its over expression in the liver may suppress gluconeogenesis and, therefore, that its inhibition could perhaps enhance glucose production.40 Overall, these results indicate complex relationships between the mechanisms leading to lower LDL cholesterol and metabolic risk.
Contrary to previous, smaller-scale investigations,41 there were associations of the p.R46L variant in PCSK9 (rs11591147) with a higher risk of type 2 diabetes, and higher fasting and two hour glucose. These associations have to be interpreted with caution, given the level of statistical significance for the association and the context of multiple comparisons presented in this study. This finding nonetheless suggests that the effect of LDL-lowering drugs on increased diabetes risk might extend to the newly-developed PCSK9 inhibitors, encouraging further genetic and clinical trial investigations.
In general, unlike the association of LDL-lowering alleles with cardiovascular risk, the association of these alleles with metabolic risk appears to be gene-specific, which in turn might suggest that the adverse consequences of lipid-lowering agents on diabetes risk could be target-specific. This may have clinical implications for the future of lipid-lowering therapy in the context of the growing number of approved drugs acting on different molecular targets. The overall safety profile of these drugs, including the magnitude of risk of new-onset type 2 diabetes, may be relevant to the choice of specific agent for subsets of the patient population, for example those at high risk of type 2 diabetes who are also candidates for lipid-lowering therapy.
A number of assumptions and potential limitations of the genetic approach used in this study should be considered. “Mendelian randomization” generally assumes that genetic variants are associated with the endpoint exclusively via the risk factor of interest.16,17 The strong and specific association with LDL cholesterol, the well-known role of target genes in LDL cholesterol metabolism and the use of conditionally-distinct genetic variants at given loci strengthen the validity of the genetic models used in this study. Similar to previous examples,5,11,42 the aim of this study was to use genetic variants that “mimic” the action of pharmacological therapy and therefore “pleiotropy” (i.e. the association with variables other than LDL cholesterol) may be more informative than concerning. For instance, HMGCR genetic variants are associated with higher body mass index, consistent with the effects on body weight observed in randomized clinical trials of statins.5 However, the consequences of modest reductions in LDL cholesterol associated with LDL-lowering alleles over several decades, as assessed in this study, may differ from the short-term pharmacological inhibition of a molecular target in randomized clinical trials or clinical practice. Finally, several of the included studies were population-based and therefore association estimates from these studies may not be applicable to patient groups in whom a particular therapy is indicated.
Conclusions
In this meta-analysis, exposure to LDL-cholesterol lowering genetic variants in or near NPC1L1 and other genes was associated with a higher risk of type 2 diabetes. These data provide insights into potential adverse effects of LDL cholesterol-lowering therapy.
Supplementary Material
Key Points.
Question: Are LDL-cholesterol lowering alleles at NPC1L1 or other genes associated with the risk of type 2 diabetes?
Findings: In a meta-analysis of genetic association studies including 50,775 individuals with type 2 diabetes and 270,269 controls, LDL-lowering polymorphisms at NPC1L1 were associated with a statistically significant odds ratio for type 2 diabetes of 2.42 per genetically-predicted reduction of 1 mmol/L in LDL cholesterol. LDL-lowering polymorphisms at HMGCR and PCSK9 were also associated with a higher risk of diabetes.
Meaning: These data provide insights into potential adverse effects of LDL cholesterol-lowering therapy.
Acknowledgement
Authors affiliated with the EPIC-InterAct Consortium: Claudia Langenberg, MD, PhD; Robert A. Scott, PhD; Stephen J. Sharp; Eva Ardanaz, MD PhD; Larraitz Arriola MD MSc; Beverley Balkau, PhD; Heiner Boeing, PhD; Panos Deloukas, PhD; Nita G Forouhi, FFPHM; Paul W Franks, PhD; Sara Grioni, BSc; Rudolf Kaaks, PhD; Timothy J Key, DPhil; Carmen Navarro, MD PhD MSc; Peter M Nilsson, PhD; Kim Overvad, PhD; Domenico Palli, MD; Salvatore Panico, MD; Jose-Ramón Quirós, MD; Elio Riboli, MD MPH, ScM; Olov Rolandsson, MD PhD; Carlotta Sacerdote, PhD; Elena C Salamanca, MSc; Nadia Slimani, PhD; Annemieke MW Spijkerman; Anne Tjonneland, DrMedSci; Rosario Tumino, MD MSc, DLSHTM; Daphne L van der A, PhD; Yvonne T van der Schouw, PhD; Mark I. McCarthy, MD, PhD; Inês Barroso, PhD; Nicholas J. Wareham, MB BS, PhD.
L.A.L had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. The authors gratefully acknowledge the help of the MRC Epidemiology Unit Support Teams, including Field Teams, Laboratory Team and Data Management Team. This research has been conducted using the UK Biobank resource.
Funding: MRC Epidemiology Unit funding: this study was funded by the United Kingdom’s Medical Research Council through grants MC_UU_12015/1, MC_PC_13046, MC_PC_13048 and MR/L00002/1. EPIC-InterAct Study funding: funding for the InterAct project was provided by the EU FP6 programme (grant number LSHM_CT_2006_037197). D. B. S. is supported by the Wellcome Trust grant n. 107064. M. McC. is a Wellcome Trust Senior Investigator and is supported by the following grants from the Wellcome Trust: 090532 and 098381. I. B. is supported by the Wellcome Trust grant WT098051. The funding bodies had no role in the design or conduct of the study; collection, management, analysis or interpretation of the data; preparation, review or approval of the manuscript or the decision to submit the manuscript for publication.
Footnotes
Disclosures: I. B. and spouse own stock in GlaxoSmithKline and Incyte. N.S. reports personal fees from Amgen, personal fees from Sanofi, personal fees from Merck, outside the submitted work. M. McC. reports grants from Eli Lilly, grants from Roche, grants from Astra Zeneca, grants from Merck, grants from Janssen, grants from Servier, grants and other from Novo Nordisk, grants from Sanofi Aventis, grants from Boehringer Ingelheim, grants and personal fees from Pfizer, grants from Takeda, outside the submitted work. S.O’R. reports personal fees from CVMED TASAP Pfizer Advisory Board, personal fees from AstraZeneca CVMD iMed External Scientific Panel, personal fees from MedImmune Cardiovascular & Metabolic Disease (CVMD) iMED Advisory Board, personal fees from ERX Pharmaceuticals Scientific Advisory Board, outside the submitted work. The other authors report no conflict of interest relative to this study.
References
- 1.Stone NJ, Robinson JG, Lichtenstein AH, et al. Treatment of blood cholesterol to reduce atherosclerotic cardiovascular disease risk in adults: synopsis of the 2013 American College of Cardiology/American Heart Association cholesterol guideline. Annals of internal medicine. 2014 Mar 4;160(5):339–343. doi: 10.7326/M14-0126. [DOI] [PubMed] [Google Scholar]
- 2.Cholesterol Treatment Trialists Consortium. Fulcher J, O'Connell R, et al. Efficacy and safety of LDL-lowering therapy among men and women: meta-analysis of individual data from 174,000 participants in 27 randomised trials. Lancet. 2015 Apr 11;385(9976):1397–1405. doi: 10.1016/S0140-6736(14)61368-4. [DOI] [PubMed] [Google Scholar]
- 3.Sattar N, Preiss D, Murray HM, et al. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials. Lancet. 2010 Feb 27;375(9716):735–742. doi: 10.1016/S0140-6736(09)61965-6. [DOI] [PubMed] [Google Scholar]
- 4.Preiss D, Seshasai SR, Welsh P, et al. Risk of incident diabetes with intensive-dose compared with moderate-dose statin therapy: a meta-analysis. JAMA : the journal of the American Medical Association. 2011 Jun 22;305(24):2556–2564. doi: 10.1001/jama.2011.860. [DOI] [PubMed] [Google Scholar]
- 5.Swerdlow DI, Preiss D, Kuchenbaecker KB, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2015 Jan 24;385(9965):351–361. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarcho JA, Keaney JF., Jr Proof That Lower Is Better--LDL Cholesterol and IMPROVE-IT. The New England journal of medicine. 2015 Jun 18;372(25):2448–2450. doi: 10.1056/NEJMe1507041. [DOI] [PubMed] [Google Scholar]
- 7.Cannon CP, Blazing MA, Giugliano RP, et al. Ezetimibe Added to Statin Therapy after Acute Coronary Syndromes. The New England journal of medicine. 2015 Jun 18;372(25):2387–2397. doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
- 8.Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nature genetics. 2015 Aug;47(8):856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 9.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nature reviews Drug discovery. 2013 Aug;12(8):581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 10.Scott RA, Freitag DF, Li L, et al. A genomic approach to therapeutic target validation identifies a glucose-lowering GLP1R variant protective for coronary heart disease. Science translational medicine. 2016 Jun 1;8(341):341ra376. doi: 10.1126/scitranslmed.aad3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ference BA, Majeed F, Penumetcha R, Flack JM, Brook RD. Effect of naturally random allocation to lower low-density lipoprotein cholesterol on the risk of coronary heart disease mediated by polymorphisms in NPC1L1, HMGCR, or both: a 2 x 2 factorial Mendelian randomization study. Journal of the American College of Cardiology. 2015 Apr 21;65(15):1552–1561. doi: 10.1016/j.jacc.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myocardial Infarction Genetics Consortium Investigators. Stitziel NO, Won HH, et al. Inactivating mutations in NPC1L1 and protection from coronary heart disease. The New England journal of medicine. 2014 Nov 27;371(22):2072–2082. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.InterAct Consortium. Langenberg C, Sharp S, et al. Design and cohort description of the InterAct Project: an examination of the interaction of genetic and lifestyle factors on the incidence of type 2 diabetes in the EPIC Study. Diabetologia. 2011 Sep;54(9):2272–2282. doi: 10.1007/s00125-011-2182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins R. What makes UK Biobank special? Lancet. 2012 Mar 31;379(9822):1173–1174. doi: 10.1016/S0140-6736(12)60404-8. [DOI] [PubMed] [Google Scholar]
- 15.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature genetics. 2012 Sep;44(9):981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? International journal of epidemiology. 2003 Feb;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 17.Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Statistics in medicine. 2016 May 20;35(11):1880–1906. doi: 10.1002/sim.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Global Lipids Genetics Consortium. Willer CJ, Schmidt EM, et al. Discovery and refinement of loci associated with lipid levels. Nature genetics. 2013 Nov;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nature genetics. 2015 Oct;47(10):1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott RA, Lagou V, Welch RP, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nature genetics. 2012 Sep;44(9):991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manning AK, Hivert MF, Scott RA, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nature genetics. 2012 Jun;44(6):659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saxena R, Hivert MF, Langenberg C, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nature genetics. 2010 Feb;42(2):142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015 Feb 12;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shungin D, Winkler TW, Croteau-Chonka DC, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015 Feb 12;518(7538):187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American journal of human genetics. 2011 Jan 7;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Ferreira T, Morris AP, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nature genetics. 2012 Apr;44(4):369–375. doi: 10.1038/ng.2213. S361–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic acids research. 2015 Jan;43(Database issue):D1079–1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008 Dec 15;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010 Sep 1;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeshita Y, Takamura T, Honda M, et al. The effects of ezetimibe on non-alcoholic fatty liver disease and glucose metabolism: a randomised controlled trial. Diabetologia. 2014 May;57(5):878–890. doi: 10.1007/s00125-013-3149-9. [DOI] [PubMed] [Google Scholar]
- 31.Blazing MA, on behalf of the IMPROVE-IT Investigators Incidence of new onset diabetes in the IMPROVE-IT trial: does adding ezetimibe to simvastatin increase risk compared to simvastatin alone? European Society of Cardiology Congress 2015. http://congress365.escardio.org/Search-Results?vgnextkeyword=C365PRESENTATION124439#.V0iX-FUrKUl. [Google Scholar]
- 32.Fall T, Xie W, Poon W, et al. Using Genetic Variants to Assess the Relationship Between Circulating Lipids and Type 2 Diabetes. Diabetes. 2015 Jul;64(7):2676–2684. doi: 10.2337/db14-1710. [DOI] [PubMed] [Google Scholar]
- 33.Besseling J, Kastelein JJ, Defesche JC, Hutten BA, Hovingh GK. Association between familial hypercholesterolemia and prevalence of type 2 diabetes mellitus. JAMA : the journal of the American Medical Association. 2015 Mar 10;313(10):1029–1036. doi: 10.1001/jama.2015.1206. [DOI] [PubMed] [Google Scholar]
- 34.Davis HR, Jr, Zhu LJ, Hoos LM, et al. Niemann-Pick C1 Like 1 (NPC1L1) is the intestinal phytosterol and cholesterol transporter and a key modulator of whole-body cholesterol homeostasis. The Journal of biological chemistry. 2004 Aug 6;279(32):33586–33592. doi: 10.1074/jbc.M405817200. [DOI] [PubMed] [Google Scholar]
- 35.Brown MS, Goldstein JL. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. Journal of lipid research. 1980 Jul;21(5):505–517. [PubMed] [Google Scholar]
- 36.Maxwell KN, Breslow JL. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proceedings of the National Academy of Sciences of the United States of America. 2004 May 4;101(18):7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjannet S, Rhainds D, Essalmani R, et al. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. The Journal of biological chemistry. 2004 Nov 19;279(47):48865–48875. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- 38.Brunham LR, Kruit JK, Pape TD, et al. Beta-cell ABCA1 influences insulin secretion, glucose homeostasis and response to thiazolidinedione treatment. Nature medicine. 2007 Mar;13(3):340–347. doi: 10.1038/nm1546. [DOI] [PubMed] [Google Scholar]
- 39.Labonte ED, Camarota LM, Rojas JC, et al. Reduced absorption of saturated fatty acids and resistance to diet-induced obesity and diabetes by ezetimibe-treated and Npc1l1-/- mice. American journal of physiology. Gastrointestinal and liver physiology. 2008 Oct;295(4):G776–783. doi: 10.1152/ajpgi.90275.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurano M, Hara M, Satoh H, Tsukamoto K. Hepatic NPC1L1 overexpression ameliorates glucose metabolism in diabetic mice via suppression of gluconeogenesis. Metabolism: clinical and experimental. 2015 May;64(5):588–596. doi: 10.1016/j.metabol.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 41.Bonnefond A, Yengo L, Le May C, et al. The loss-of-function PCSK9 p.R46L genetic variant does not alter glucose homeostasis. Diabetologia. 2015 Sep;58(9):2051–2055. doi: 10.1007/s00125-015-3659-8. [DOI] [PubMed] [Google Scholar]
- 42.Kathiresan S. Developing medicines that mimic the natural successes of the human genome: lessons from NPC1L1, HMGCR, PCSK9, APOC3, and CETP. Journal of the American College of Cardiology. 2015 Apr 21;65(15):1562–1566. doi: 10.1016/j.jacc.2015.02.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.