Abstract
Zinc transporters play important roles in all eukaryotes by maintaining the rational zinc concentration in cells. However, the diversity of zinc transporter genes (ZTGs) remains poorly studied. Here, we investigated the genetic diversity of 24 human ZTGs based on the 1000 Genomes data. Some ZTGs show small population differences, such as SLC30A6 with a weighted-average FST (WA-FST = 0.015), while other ZTGs exhibit considerably large population differences, such as SLC30A9 (WA-FST = 0.284). Overall, ZTGs harbor many more highly population-differentiated variants compared with random genes. Intriguingly, we found that SLC30A9 was underlying natural selection in both East Asians (EAS) and Africans (AFR) but in different directions. Notably, a non-synonymous variant (rs1047626) in SLC30A9 is almost fixed with 96.4% A in EAS and 92% G in AFR, respectively. Consequently, there are two different functional haplotypes exhibiting dominant abundance in AFR and EAS, respectively. Furthermore, a strong correlation was observed between the haplotype frequencies of SLC30A9 and distributions of zinc contents in soils or crops. We speculate that the genetic differentiation of ZTGs could directly contribute to population heterogeneity in zinc transporting capabilities and local adaptations of human populations in regard to the local zinc state or diets, which have both evolutionary and medical implications.
Zinc (Zn), an essential trace mineral, is required for the structures and functions of many proteins, including enzymes and transcription factors, and is critical for their biological activities, such as cellular metabolism and gene expression1,2. Previous studies have estimated that >3% or even as much as 10% of human proteins are zinc-binding proteins3, indicating that the maintainability of cellular zinc concentrations and distribution in human cell compartments is of extreme importance. Generally, there are 24 zinc transporter genes (ZTGs) involved in cellular zinc homeostasis in the human genome4,5 among which 10 genes of SLC30A family lower intracellular cytoplasmic zinc by mediating zinc efflux from cells or influx into intracellular vesicles6, while 14 genes of SLC39A family transporters mobilize zinc in the opposite direction7.
Because of the fundamental roles in biological processes, mutations in ZTGs are likely to break the balance of zinc in cell compartments, resulting in improper biological functions of zinc-dependent proteins and subsequent severe diseases or impaired development. For instance, studies have shown that acrodermatitis enteropathica (OMIM 201100) was caused by the loss of function of one of or both SLC39A4 alleles, leading to a diminished uptake of dietary zinc, increased sensitivity to zinc deficiency, and severe growth retardation8. Some reports suggested that changes in intercellular zinc level are associated with cancer progression as the expression of zinc transporters in many cancers altered. For example, increased expression of ZIP6, ZIP7 and ZIP10 have been observed to contribute to zinc hyper-accumulation in breast tissue and breast cancer9. Other investigations also indicated that ZIP6 and ZIP10 play critical roles in breast cancer progression10,11. Similarly, elevated ZIP4 expression level in pancreatic cell is associated with pancreatic cancer12,13,14,15. Besides cancers, zinc transporters are involved in some complex diseases. Sladek et al. found that rs13266634, a non-synonymous SNP (R325W) in SLC30A8 is related to type 2 diabetes16. SLC30A8 encodes a zinc transporter which is expressed solely in secretory vesicles of β-cells and the overexpression of SLC30A8 in insulinoma cells increases glucose-simulated insulin secretion17. Another example is that SLC39A8 was observed to have relationship with body mass index18. Such disruptive mutations that reduce fitness would be removed from population by purified selection.
On the other hand, functionally important ZTGs may also be subjected to positive selection for acquired changes that increase fitness. Recently, Engelken and Carnero et al. reported that the human intestinal zinc uptake transporter, SLC39A4, has undergone positive selection in Sub-Saharan African populations19. By carrying out cell transfection assays of putative functional variants, they validated an amino acid change (L372V) in SLC39A4 which was shown to lead to reduced zinc uptake in the African isoform. According to previous studies, zinc contents in soils or crops are extremely diverse across continents or countries and it is estimated that African populations have undergone severe zinc deficiency, according to various kinds of zinc deficiency indicators20. We hypothesize that, due to the uneven global distribution of absorbable zinc in soils, crops and different diet habits, some ZTGs with adaptable variations in different populations might be underlying natural selection as their transporting capability changed, thus maintaining the balance of intercellular or serous zinc in the human body.
We, therefore, systematically analyzed the patterns of genetic diversity and signals of natural selection for 24 14ZTGs in 14 worldwide populations. In this study, we showed that ZTGs harbor many more highly population-differentiated variants compared with random genes and discussed the potential underlying forces shaping the genetic diversity of ZTGs. Further, we reported that SLC30A9 was underlying natural selection in both East Asians (EAS) and Africans (AFR) but in different directions. By performing a correlation, we found that the evolutionary force underlying the selective sweep of SLC30A9 may be the uneven worldwide zinc distribution in soils or corps. Moreover, we predicted 17 potentially functional SNPs, which may guide the study of molecular mechanism of ZTGs. Our results may subsequently increase our understanding of the evolutionary forces that affect ZTGs, as well as augmenting our knowledge of gene function on zinc homeostasis in different populations and the mechanisms of zinc-related diseases.
Results
Genetic differentiation of ZTGs among populations
We first performed an analysis of molecular variance (AMOVA) to examine whether genetic variance among four continental regions is significantly different from populations within each region (Table S1 and Figure S1). Our analysis showed that SLC30A9 and SLC30A3 had a greater proportion of variance among continental groups than within group (empirical P < 0.05), indicating that the two genes were genetically differentiated among continental populations. On the contrary, SLC30A6 showed a much lower proportion of variance between groups compared with most of the other genes genome-wide (empirical P < 0.05), suggesting that SLC30A6 may be functionally conserved.
We next calculated a weighted-average FST (referred to as WA-FST) for each gene. WA-FST employs a weighted average of the FST values calculated from variants in a gene or a genomic region. Among the 24 14ZTGs, SLC30A9 and SLC30A3 showed high WA-FST values (> = 0.19, top 5% cutoff of the whole genome), indicating that the variation composition is substantially different between populations. Therefore, the functional variants associated with specific haplotypes are also distributed heterogeneously across populations. On the contrary, SLC30A6 with a low WA-FST (< = 0.128, empirical P < 0.05) may have limited genetic heterogeneity across populations (Table S1 and Figure S2).
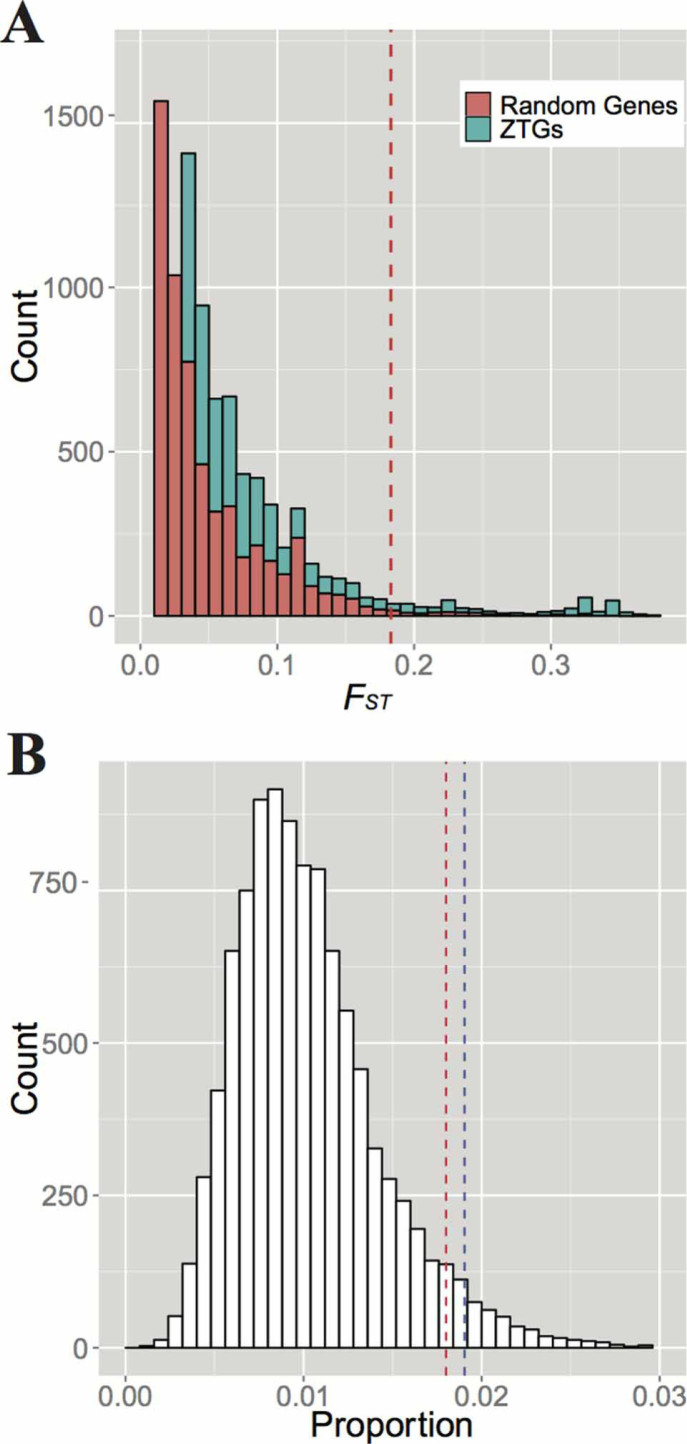
We then calculated the unbiased locus-specific FST, following Weir and Hill21 in determining the highly differentiated loci in ZTGs (see Materials and Methods). A locus-specific FST measures the apportionment of genetic variation of one specific SNP between populations. A high locus-specific FST value indicates that the corresponding allele is substantially different between populations, while SNPs with low locus-specific FST have small genetic difference and little functional heterogeneity across populations. The locus-specific FST values varied widely among SNPs in ZTGs, with the maximum FST for SNP rs1871534 in SLC39A4 at 0.763 and the average value at 0.018. We used the highest 1% of the genome-wide locus-specific FST (0.183) as the cutoff for a significant signal of population differentiation. As a result, compared to the FST distributions of 24 random genes, ZTGs showed significantly higher percentages of variants with high FST and average FST values (the mean FST value for random genes is 0.015). In total, there were 361 (of 19888) (1.82%) SNPs in ZTGs with FST > = 0.183, while the percentage was only 0.56% (131 of 23,237) in random genes (Figure 1A). In particular, 17% SNPs in SLC30A9, 8% SNPs in SLC30A3 and 6% SNPs in SLC30A4 harbored genetic variants with significantly high FST. The proportions in SLC30A1, SLC30A10, SLC30A5 and SLC30A6 were relatively lower than the other genes (Figure S3). Finally, a permutation test (see Materials and Methods) (P = 0.048) was applied to exclude the possibility that the population differentiations in ZTGs resulted from sampling effect (Figure 1B). When we used the top 5% of the genome-wide locus-specific FST as the cutoff for a significant signal of population differentiations, the P-value (<0.01) was also significant (see next section for more details). All these results suggested that the differentiations of ZTGs among populations were unlikely to be caused by any stochastic factors.
Figure 1. ZTGs show a higher proportion of locus-specific FST when compared to random genes.
(A) The locus-specific FST distribution of all sites located in 24 14ZTGs and 24 random genes. In the figure, the dashed line represents the cutoff with the empirical P-value of 0.01, i.e. FST = 0.183, the highest 1% of the genome-wide locus-specific FST. (B) Distribution of a 10,000 times permutation for the proportion of high FST in 24 random genes. The red dashed line represents the cutoff with the empirical P-value of 0.05, i.e., proportion = 0.018, while the blue line represents the proportion of loci with high FST in ZTGs (0.0182).
Population differentiations and potential effects of functional SNPs
The above analyses focused on general patterns of FST distribution of all loci in ZTGs. However, the genetic variants that directly affect protein functions and gene expressions are functional SNPs, which are more likely to have true associations with zinc transporting or diseases. In our study, SNPs that (i) change the amino acid sequence of protein (non-synonymous SNPs, nSNPs), (ii) correlate with gene expression (eQTL (expression quantitative trait loci) SNPs), (iii) alter splicing (splicing SNPs) and (iv) are associated with diseases (in the GWAS catalog or other databases) were investigated (see Materials and Methods).
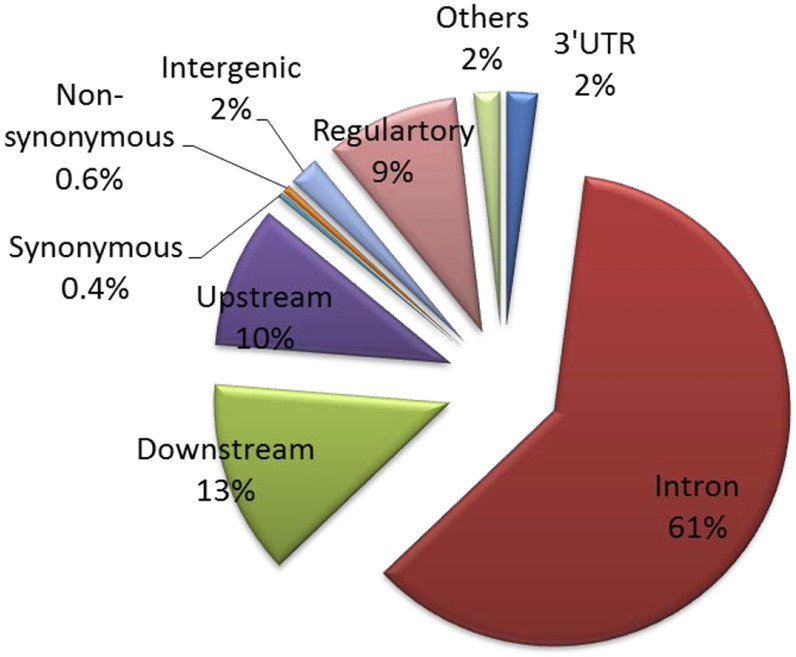
Compared to the FST distributions of random genes, the ZTGs showed significantly higher percentages of variants with high FST; i.e. there were 932 out of a total 19,888 14SNPs with FST > = 0.092 (the top 5% FST cutoff) (P < 0.01, 10,000 permutations). We first predicted the functional types of all 19,888 14SNPs in ZTGs and 5,728 14SNPs located within the upstream and downstream 10 14Kb of ZTGs by consulting the variance effect prediction tools from the Ensembl website (Figure 2). As shown in Figure 2, 61% SNPs were located in the intronic region, 2% SNPs in the intergenic region, 13% SNPs in the downstream region and 10% SNPs in the upstream region, respectively. In particular, 195 14nSNPs and 47 splicing SNPs were observed. Furthermore, we found an additional 25 14eQTL SNPs in RegulomeDB, 16 14SNPs in the GWAS catalog, and 3 14SNPs in the GKB database. From those 25,616 14SNPs, we screened 17 functional SNPs with FST > = 0.092, including 7 14nSNPs, 1 splicing SNP, 6 14eQTL SNPs and 3 disease-related SNPs (2 in the GWAS catalog and 1 in GKB) (Table 1).
Figure 2. The variant effect prediction of loci in ZTGs, including up- and down-stream 10-kb regions.
Table 1. Summary information of highly differentiated functional SNPs at ZTGS. It is noted that in the column “allele”, the first allele is the ancestral state, while the second one with highlight is the derived state. PRD, probably disease-causing; BNG, benign; iTMD, in transmembrane domain; NT, N-terminus; nTMD, near transmembrane domain; IV, intro variant; UGV, upstream gene variant; RRV, regulatory region variant; LS- FST, Locus-specific FST.
| rsID | Gene | Position | Allele | FEUR | FASN | FAFR | FAMR | LS-FST | Type | Database | Effect or Related | PolyPhen-2 | Topology and location | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1871534 | SLC39A4 | 8:145639681 | G/C | 0.002 | 0.000 | 0.873 | 0.081 | 0.763 | Missense | RegulomeDB | Leu/Val | PRD: 0.991 | iTMD (Fig. S4A) | |
| rs1047626 | SLC30A9 | 4:42003671 | A/G | 0.750 | 0.953 | 0.126 | 0.737 | 0.387 | Missense | - | Met/Val | BNG: 0.000 | NT (Fig. 6C) | |
| rs2466517 | SLC39A11 | 17:70943990 | T/C | 0.001 | 0.002 | 0.343 | 0.055 | 0.212 | Missense | RegulomeDB | Thr/Ala | BNG: 0.000 | nTMD (Fig. S4B) | |
| rs11011935 | SLC39A12 | 10:18280113 | T/C | 0.000 | 0.000 | 0.238 | 0.022 | 0.158 | Missense | RegulomeDB | Phe/Leu | BNG: 0.000 | nTMD (Fig. S4C) | |
| rs2010519 | SLC39A13 | 11:47431728 | G/A | 0.001 | 0.012 | 0.221 | 0.000 | 0.135 | Missense | - | Glu/Gly | BNG: 0.000 | NT (Fig. S4D) | |
| rs2272662 | SLC39A4 | 8:145639726 | T/C | 0.541 | 0.496 | 0.087 | 0.416 | 0.130 | Missense | RegulomeDB | Thr/Ala | BNG: 0.012 | iTMD (Fig. S4A) | |
| rs75920625 | SLC39A4 | 8:145639654 | T/C | 0.001 | 0.000 | 0.135 | 0.018 | 0.092 | Missense | RegulomeDB | Thr/Ala | BNG: 0.012 | nTMD (Fig. S4A) | |
| rs61756712 | SLC30A4 | 15:45814306 | A/G | 0.001 | 0.008 | 0.342 | 0.045 | 0.224 | Splice | RegulomeDB | - | - | ||
| rs759071 | SLC39A3 | 19:2728577 | G/A | 0.327 | 0.632 | 0.817 | 0.283 | 0.168 | eQTL | RegulomeDB | - | - | ||
| rs6832846 | SLC39A8 | 4:103181749 | A/G | 0.787 | 0.526 | 0.986 | 0.705 | 0.112 | eQTL | RegulomeDB | - | - | ||
| rs151368 | SLC39A8 | 4:103181113 | A/T | 0.787 | 0.526 | 0.986 | 0.705 | 0.112 | eQTL | RegulomeDB | - | - | ||
| rs11889699 | SLC39A10 | 2:196528101 | A/G | 0.588 | 0.366 | 0.127 | 0.377 | 0.108 | eQTL | RegulomeDB | - | - | ||
| rs17278473 | SLC39A8 | 4:103243101 | C/T | 0.400 | 0.024 | 0.179 | 0.324 | 0.097 | eQTL | RegulomeDB | - | - | ||
| rs151372 | SLC39A8 | 4:103174196 | C/T | 0.787 | 0.528 | 0.951 | 0.700 | 0.093 | eQTL | RegulomeDB | - | - | ||
| rs950027 | SLC30A4 | 15:45801035 | T/C | 0.452 | 0.947 | 0.953 | 0.652 | 0.210 | IV | GWAS | Response to fenofibrate | - | ||
| rs11264736 | SLC39A1 | 1:153939130 | T/C | 0.494 | 0.949 | 0.730 | 0.666 | 0.123 | UGV | GWAS | Lentiform nucleus volume | - | ||
| rs17060812 | SLC39A14 | 8:22228828 | C/T | 0.039 | 0.066 | 0.341 | 0.070 | 0.111 | RRV | PharmGKB | Nortriptyline | - | ||
All of the 17 functional SNPs showed high FST values across populations and therefore were assumed to have a higher potential functional impact on zinc transporting or were associated with diseases. Non-synonymous SNPs can cause a structural change in the protein product, which potentially leads to a minor or major phenotypic change. We applied Polyphen-222 to predict how amino acid variants might change the function of ZTGs peptides (Table 1). Interestingly, rs1871534 was predicted to be potentially damaging, with a score of 0.991; it was located in the second transmembrane domain (TMD) in SLC39A4, predicted by HMMTOP (Table 1 and Figure S4) and Engelkin et al. have proved that it affects zinc transporting ability of SLC39A4 in in vitro experiments23. Besides nSNPs, other types of SNPs may also be important. One splicing SNP (rs61756712), 6 eQTL SNPs (rs759071, rs6832846, rs151368, rs11889699, rs17278473 and rs151372) and 1 regulatory region variant (rs17060812) may both contribute to gene expression and thus alter intercellular zinc levels. Moreover, two SNPs included in GWAS catalog were identified. For example, rs950027 in SLC30A4 was reported to be related with response to fenofibrate24, and rs11264736 in SLC39A1 was associated with lentiform nucleus volume, which can lead to disorders, including Parkinson's disease25. Therefore, the 17 highly differential functional SNPs could contribute to population heterogeneity in zinc transporting capability, providing guidance for studying the transporting mechanism of ZTGs at a molecular level.
Frequency distributions of functional SNPs among different populations
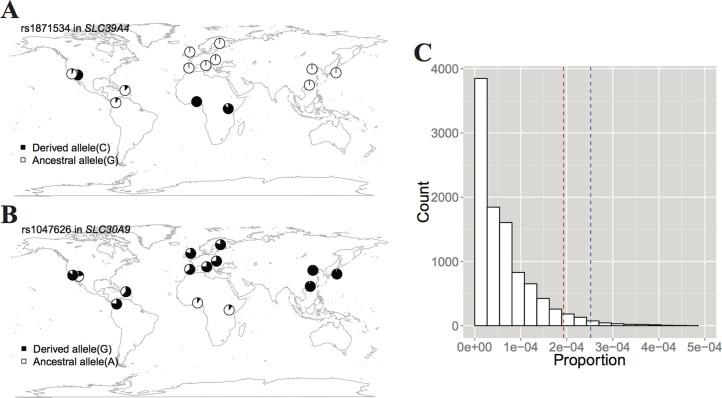
We further analyzed the distribution of allele frequencies of candidate SNPs in different populations worldwide (Table 1). Interestingly, the 7 14nSNPs in the 17 candidate SNPs had two distinct patterns. One is that the derived allele frequencies (DAF) of 5 14nSNPs (rs1871534, rs2466517, rs11011935, rs2010519 and rs75920625) were much higher in Africans compared with non-Africans in whom the DAF were close to 0 (Figures 3A and S5A–D). For instance, the DAF of rs1871534 was 0.006, 0.000 and 0.983 in CEU, CHB and YRI, respectively. The other pattern showing completely opposite trends was that rs1047626 and rs2272662 had high DAF in Europeans and East Asians but the derived alleles of the two SNPs were nearly absent in Africans (Figures 3B and S5E). In particular, the DAF of rs1047626 in SLC30A9 was amost 1 in East Asians (CHB: 0.964) but was 0.080 in YRI (Table 1). In spite of the different patterns, they showed the same trends indicating Africans and non-Africans were different in these nSNPs. A 10,000 permutations was implemented to validate that nSNPs with high FST (> = 0.092, the top 5% FST cutoff), and high DAF in YRI but low DAF (<0.05) in CEU and CHB were significantly enriched in ZTGs (P = 0.019) (Figure 3C). Therefore, it is unlikely the specific pattern of these candidate SNPs have been caused by genetic drift, possibly resulting from natural selection instead.
Figure 3. Worldwide derived allele frequency (DAF) distributions of highly differential nSNPs.
(A) DAF of rs1871534 in SLC39A4, which is nearly fixed in Africans but almost absent in non-Africans. (B) DAF of rs1047626 in SLC30A9. This SNP shows a pattern opposite of rs1871534. (C) 10,000 times permutation for the proportion of nSNPs with a high FST (> = 0.092) and a low DAF (<0.05) in CEU and CHB in random genes. The red and blue dashed lines represent the cutoff with the empirical p value 0.05 and a proportion in ZTGs, respectively. nSNPs with a high FST and a low DAF in CEU and CHB are significantly enriched in ZTGs (P = 0.019), which is possibly caused by natural selection. World maps here were created by R packages (http://www.r-project.org/).
Detecting regional natural selection in ZTGs
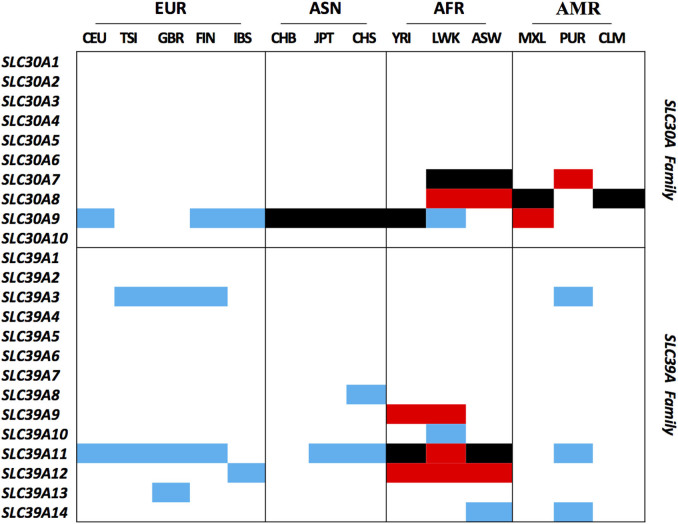
The AMOVA and FST analyses revealed overall patterns of ZTGs genetic differentiations among worldwide populations. We then applied two commonly used statistics, the integrated haplotype score (iHS) and the composite likelihood ratio (CLR) (see Materials and Methods), to detect the signals of selection in the ZTGs. We found ZTGs exhibiting evidence of recent positive selection. There was a certain consensus between the results of the iHS and CLR tests, which supported the reliability of the analysis (Figure 4). Figure 4 illustrates the signals of positive selections for ZTGs in different populations. The iHS test supported the indication of selection on several genes. SLC30A7 exhibited signals in two African populations (LWK and ASW) (Figures 4 and S6). SLC30A8 appeared to have been selected in primarily American populations (MXL and CLM) (Figures 4 and S7). SLC30A9 underwent selection, as indicated by iHS, in all of the Asian populations, three European populations (CEU, FIN and IBS) and two African populations (YRI and LWK) (Figures 4 and S8). All populations shared the iHS signals in SLC39A11 except IBS, CHB, LWK, MXL and CLM (Figures 4 and S9). There were 6 genes (SLC30A7, SLC30A8, SLC30A9, SLC39A9, SLC39A11 and SLC39A12) exhibiting significant signals in the CLR test. For instance, SLC39A9 shows strong selection signals in YRI and LWK. Four genes (SLC30A7, SLC30A8, SLC30A9 and SLC39A11) showed overlapping signals in both methods in some populations. For example, SLC30A7 harbored both iHS and CLR signals in LWK and ASW with SLC30A8 in MXL and CLM. SLC39A11 had overlapping signals for both methods in YRI and ASW. Specifically, a previous study reported that SLC30A9 was strongly selected in East Asian populations, and it is among the top 10 selection signals in the genome-wide detection of positive selection in human populations26. However, we found that SLC30A9 underwent selection in different continental groups, including Africans and East Asians using both iHS and CLR tests (Figures 4 and S8), suggesting that the selective signals in SLC30A9 were very strong.
Figure 4. Global natural selection pattern for ZTGs.
The blue blocks indicate the significant signals found by the iHS test; the red color indicates the significant signals found by Sweep Finder, and black indicates the overlapping signals for both methods.
Directional selections acted on SLC30A9 differently in Africans and East Asians
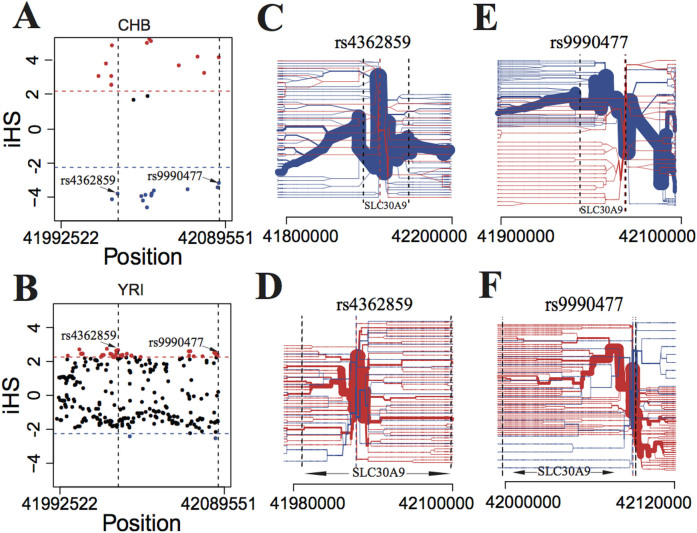
We further analyzed the selected haplotypes of SLC30A9 in each population and found that the haplotype favored in the African population was different from that found in East Asians and Europeans (Figure 5). The iHS test generally takes SNPs with minor allele frequency (MAF) large than 5% into consideration. Therefore, there were only 28 and 294 (of 1294) loci with iHS values in CHB and YRI, respectively, leaving numerous numbers of alleles in SLC30A9 which were nearly fixed and thus could not be estimated. The SNP with a large positive iHS value indicates that the ancestral allele of this respective site hitchhiked with the selected locus, while the SNP with a large negative iHS value indicates the derived allele was favored by selection. According to the iHS distribution of SLC30A9 in CHB, there was a high proportion of selected SNPs (SNPs with a significant iHS value, P < 0.05) with large negative values (13 of 26) (Figure 5A), while in YRI most of the selected SNPs harbored large positive iHS values (53 of 55) (Figure 5B). That is, the selected haplotype in CHB included approximately 50% of the derived alleles, while merely 3.6% (2 of 55) of selected SNPs hitchhiked with the extended haplotype in YRI, indicating that the haplotypes selected in these two populations were probably different. Moreover, not only were there differences in the ratios of the derived and ancestral alleles selected in CHB and YRI, the SNPs with significant iHS values were unmatched in each population. There were only two selected SNPs (rs4362859 and rs9990477) that were shared by the extended haplotypes in CHB and YRI (Figures 5A–B and S10A–B). For these two SNPs in CHB and YRI, haplotype bifurcation diagrams were drawn27, which allow a better understanding of the origin of an observed footprint of selective sweeps (Figure 5C–E). As shown in Figure 5C and E, when the haplotype of SLC30A9 harbored the derived alleles of rs4362859 (T) and rs28620429 (C) (blue color), it had long-range haplotype homozygosity, which was unusual given their frequency in CHB. However, if the haplotype included ancestral alleles (red color), it extended less compared with the comprised derived ones in CHB. However, the trend in YRI was completely the opposite. That is, the extended haplotype in YRI comprised the ancestral allele of rs4362859 (C) and rs28620429 (T), while the haplotype that included derived alleles had short-range homozygosity (Figure 5D and F). Therefore, according to the differences in the ratios of derived and ancestral alleles selected in CHB and YRI, as well as the unmatched selected SNPs in each population, we inferred that the selected haplotypes in YRI and CHB were different.
Figure 5. SLC30A9 shows selective signatures in both Africans and East Asians.
(A), (B) Plots of iHS values for SNPs of SLC30A9 in CHB (A) and YRI (B). The values of standardized iHS for sNPS with minor allele frequency >5% are plotted against the the genomic positions (hg19). The 95% cutoff value of the empirical distribution of standardized iHS is given in each plot (dashed line). Red and blue dots representing ancestral and derived alleles of SNPs were selected, respectively. Only rs4362859 and rs9990477 harbor significant but opposite iHS values in both CHB and YRI. (C), (D) Haplotype bifurcation diagrams for the core haplotype carrying rs4362859 at SLC30A9 in CHB (C) and YRI (D). In CHB, the haplotype carrying the derived allele of rs4362859 (blue color) has long-range homozygosity that is unusual given their frequency, while the haplotype carrying the ancestral allele of rs4362859 (red color) is favored. (E), (F) Haplotype bifurcation diagrams for core haplotype carrying rs9990477 at SLC30A9 in CHB (E) and YRI (F). The pattern of rs9990477 is similar with that of rs4362859.
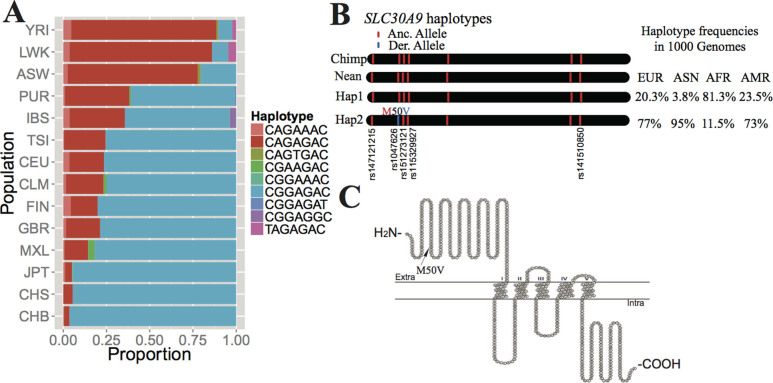
Next, we investigated the haplotype abundance distribution for SLC30A9 in each population. Non-synonymous SNPs that may cause a structural change in protein can potentially lead to a minor or major phenotypic change. We observed that 9 different kinds of haplotypes of SLC30A9 were composed of 7 14nSNPs (Table S2): rs147121215, rs1047626, rs151273121, rs115329927, rs2581423, rs181235146 and rs141510850. There were two main haplotypes that constituted more than 90% of the haplotypes for every population (Figure 6A and Table S2). Strikingly, the haplotype particularly enriched in CHB was CGGAGAC (Haplotype 2, H2) (96.4%), which was nearly fixed. The frequency of H2 in CEU and YRI was 76.5% and 0.08%, respectively (Figure 6A and Table S2), while CAGAGAC (Haplotype 1, H1) was the most common haplotype in African populations (84.1% in YRI). Comparing H1 with H2, we found that the only difference was that H2 contained the derived allele in rs1047626, while H1 had the ancestral one (Figures 6B and S11). The rs1047626 is an nSNP with a high FST value (0.387) (Table 1), changing the 50th amino acid in SLC30A9 from methionine acid to valine (M50V) (Figure 6C). Previous studies also screened a selection signal at this locus26,28, indicating that it has a high probability to be the causal SNP for the selection signal in East Asia.
Figure 6. Analysis of putative selected haplotypes carrying 7 14nSNPs in SLC30A9.
(A) Haplotypes shared among different continental groups. African populations are more likely to harbor haplotype CAGAGAC, while in non-African populations, haplotype CGGAGAC is much more pervasive, especially in East Asians in whom the haplotype frequency is nearly fixed. (B) Graphical depictions of SLC30A9 haplotypes constructed from 7 14nSNPs with haplotype frequencies derived from the 1000 Genomes Project. Hap1 (CAGAGAC) is the same as the haplotype in a chimpanzee (Chimp) and a Neanderthal (Nean). Hap2 (CGGAGAC) is different from Hap1 at rs1047626 of which the derived allele can alter an amino acid change from methionine acid to valine (M50V). (C) Predicted membrane topology of human SLC30A9 generated using HMMTOP and visualized with TeXtopo. Location of rs1047626 carried by the possible selected haplotype is indicated.
The correlation between haplotype frequencies and zinc deficiency
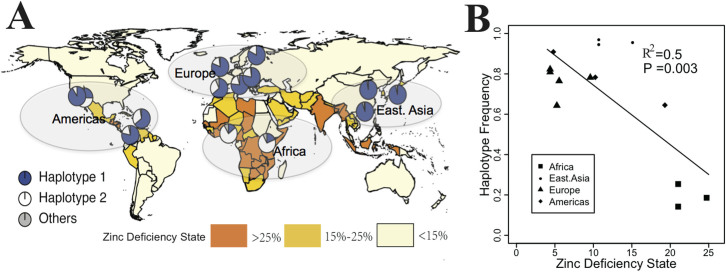
Finally, to explore the possible reasons underlying this interesting distribution pattern of haplotypes in SLC30A9, we performed a correlation between the haplotype frequency of H2 in each population and the corresponding zinc deficiency status. Previous studies have estimated the global prevalence of zinc deficiencies based on zinc availability in national food supplies and the prevalence of stunting20. Their preceding results have indicated that inadequate dietary zinc intake may particularly occur in Sub-Saharan Africans and south Asians. For example, about 20%, 11% and 10% of the population have inadequate zinc intake in YRI, CHB and CEU, respectively20. Most of the countries in Africa were marked as being in the “HIGH” risk category of zinc deficiency. Here we directly adopted the zinc related data previous researchers used to analyze the relationship between zinc status and populational haplotype distributions (Figure 7A and Table S3). As shown in Figure 7A, populations in those areas with high prevalence of zinc deficiency (YRI and LWK) are more likely to harbor H1 of SLC30A9. For instance, in YRI the frequency of H1 was 84.1%, but the zinc deficiency state was 21%, which is the highest among all populations. However, H2 was dominant in the populations with a low prevalence of zinc deficiency. For example, the frequencies of H2 in CEU (76.5%) and CHB (96.4%) were much higher than in YRI (8%), but the zinc deficiency states in CEU (9.6%) and CHB (11%) were lower than in YRI (21%). We then demonstrated a strong correlation between the haplotype frequency of H2 and the zinc deficiency state (R2 = 0.5, P = 0.003, Figure 7B). We further included additional 51 populations from Human Genome Diversity Project (HGDP)29 in our analysis and also observed strong correlation between H2 and zinc deficiency (R2 = 0.38, P = 3.4 × 10−8, Figure S12), giving further support to natural selection acting on SLC30A9.
Figure 7. Correlation between zinc deficiency state and frequency of putative selected haplotype.
(A) Estimated country-specific prevalence of zinc deficiency and worldwide haplotype frequency distribution for SLC30A9. Population zinc deficiency data are based on the composite nutrient composition database, IZINCG physiological requirements, the Miller Equation to estimate zinc absorption and an assumed 25% inter-individual variation in zinc intake (Ref. 20, see Table S3). (B) Correlation test for the zinc deficiency and frequency of Hap2. World map was created by R packages (http://www.r-project.org/).
Discussion
With recent developments of high-through DNA genotyping and sequencing technologies, genome-wide scans for genes that have been targeted by selection have become feasible26,28,30,31,32. These studies screened out several categories of genes that have undergone natural selection, for example, electron transporter genes and peroxisome transporter genes. But it was still necessary to unfold which changes in environment or habit that one category of genes have adapted to. Our work provides such an example that may advance our understanding of human evolution and molecular evolution. In this study, we found that ZTGs have very different population differentiation patterns, both globally and regionally. Some genes have higher global population differentiation levels, such as SLC30A9 and SLC30A3, while SLC30A6 is more conserved (Figure S1). Moreover, our results showed that ZTGs exhibit significantly larger percentages of genetic variants with high FST than random genes do, indicating there are great genetic differentiations in ZTGs among populations (Figure 1A). Generally, several factors can influence population differentiation of a certain gene, such as natural selection, genetic drift and migration33,34. Under neutral evolution, population differentiation is influenced solely by genetic drift, which increases differentiation versus migration, which decreases differentiation. These two factors, drift and migration, are expected to have the same average effect across the genome. However, natural selection impacts population differentiation only in specific regions33,34. We ruled out the possibility that drift could cause this differentiation landscape in ZTGs by comparing 24 14ZTGs with random genes (Figure 1B). Despite the marginally significant P-value (0.048) of permutation, we suggested that natural selection had acted on ZTGs. Furthermore, another analysis that indicating nSNPs with high FST and low DAF (<0.05) in CEU and CHB are significantly enriched in ZTGs (P = 0.019) (Figure 3C), which has provided more evidence that natural selection events may have occurred on ZTGs.
The analysis above was based on locus-specific FST, which revealed patterns of genetic differentiations that are incompatible with neutral expectations in ZTGs among worldwide populations. We then applied two commonly used approaches, iHS and CLR, to detect natural selection in ZTGs; several genes exhibited significant signals (Figure 4). In general, there were selective sweeps in three genes (SLC30A8, SLC30A9 and SLC39A11) that are shared among different continental populations. In principle, haplotypes inherited from ancestral populations might also lead to sharing selective signals between populations. However, this is likely a small effect because recombination for >1000 generations will break down such unusually long haplotypes30. Therefore, this might be due to selective events these populations have experienced.
Among these 3 genes, SLC30A9 was reported to be strongly selected in East Asian populations; it was among the top 10 selective signals in the genome-wide detection of positive selection in human populations26,28. However, we detected the selective signal in this gene not only in East Asians, but also in Europeans and Africans (Figures 4–5 and S8). This is probably because the footprints that selection left in European populations and African populations were not strong enough to be screened by the genome-wide selection scan in previous studies. Surprisingly, the selective pressure directions in East Asians and Africans were the opposite. It is notable that two functional haplotypes, H1 and H2, are dominant in Africans and East Asians, respectively (Figures 5 and 6). A linear regression was performed to reveal the relationship between the zinc deficiency state and H2 of SLC30A9 in corresponding populations (Figure 7 and S12). Although the zinc deficiency data used here may not be similar with those of the past and could also be biased due to economic states and dietary habits, these are the best data available so far to investigate the underlying selective force of SLC30A9. The significant result of correlation suggests that individuals in continents with low deficiency states are more likely to harbor H2 to perform zinc transporting than those individuals in continents with high deficiency states. We speculated that the zinc transporting ability of SLC30A9 with H1 and H2 is different and the local zinc concentration affected frequency of the allele which was favored by natural selection. That is, ancient Africans were forced to adapt to low zinc concentrations, while ancient East Asians and Europeans adapted to the high zinc concentrations so the serum or cell concentration could be maintained properly. Further, preceding investigations proved that zinc concentrations or deficiencies can regulate the transcriptional expression of SLC30A9 and other ZTGs in many model organisms, such as zebra fish35 and rats36, supporting our inference that local zinc concentration could be a putative selective force shaping the asymmetric distribution pattern of haplotypes in SLC30A9. It is possible that the N-terminal of a transporter protein could affect its transporting ability37. A non-synonymous SNP, rs1047626, located in the N-terminal (50th amino acid) of SLC30A9 (Figure 6C) shows a high FST value (0.387) (Table 1). We have speculated that it is the putative causal SNP, resulting in different transporting abilities between H1 and H2.
The correlation (Figure 7) between zinc deficiency and the frequency of H2 in SLC30A9, which exhibited a great proportion of high FST (Figure S3) and significant selective signals (Figure 4), provides us a good example for investigating the underlying force that results in high differentiations of the ZTGs across different ethnic groups. Many other genes in ZTGs, such as SLC30A3 and SLC30A4, which harbored a great proportion of high FST and SLC30A7, SLC30A8 and SLC39A11, which showed significant selective signals probably exhibited a similar pattern with SLC30A9, because the regulation of serum or cellular zinc concentrations is a complex biological process in which many ZTGs are definitely involved. Therefore, we speculate that some genes in ZTGs were also selected by the uneven continental distribution of absorbable zinc in soils and crops or otherwise in the food chain, since humans spread from Africa and colonized most of the globe. Local adaptation consequently resulted in high differentiations of ZTGs across different ethnic groups. Other evolutionary scenarios cannot be entirely ruled out and should be investigated in detail. One possible scenario is that, in addition to transporting zinc, ZTGs transport other microelements, such as cadmium and manganese38, which may also make ZTGs adapt to the environment, leading to high differentiation. What's more, not only does it act as a transporter, SLC30A9 is located in nucleus and performs as a nuclear receptor co-activator to regulate gene expression39. “Nutritional immunity”40,41 may also explain the evolutionary force. According to the nutritional immunity hypothesis, the human host restricts access to certain micronutrients so that pathogens become less virulent. One recent study used this hypothesis to interpret the selective force of SLC39A4 in Sub-Saharan Africa19. The findings have indicated that the underlying evolutionary force that led to population differentiations of ZTGs still needs to be investigated further.
In summary, proteins coded by Zinc Transporter Genes (ZTGs) play pivotal roles in decreasing or increasing zinc concentrations in cells and thus keep homeostasis of zinc in human body. Genetic variations in ZTGs could directly contribute to population heterogeneity in zinc transporting capabilities. In this study, we outline for the first time the genetic differentiation of worldwide populations and footprints of natural selection upon a comprehensive list of ZTGs. We demonstrated that high differentiations exist in ZTGs among populations. Besides, we identified 17 potentially functional SNPs with allele frequency highly differentiated among populations, which may affect either the protein structures or the expression levels of ZTGs. These results may enhance our understanding of the importance of zinc levels in human evolutionary history and facilitate further functional studies of ZTGs and medical studies on worldwide nutrient problem as well as zinc-related diseases.
Methods
Genetic variation data
We analyzed the latest release of the data (version 3 of phase 1, March 2012 release) from the 1000 Genomes Project with autosomal SNPs of 1,092 individuals representing 14 populations worldwide42. According to the 1000 Genomes Project Steering Committee, the 14 populations were derived from four ancestries: East Asian ancestry (ASN: CHB, CHS, JPT), African ancestry (AFR: ASW, LWK, YRI), European ancestry (EUR: GBR, FIN, IBS, TSI, CEU) and American ancestry (AMR: CLM, MXL, PUR). The ancestry information of SNPs was obtained from the 1000 Genomes Project (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/analysis_results/supporting/ancestral_alignments/).
Obtaining gene information for zinc transporter genes
Mammalian zinc transporters come from two major families, the SLC30 (ZnT) family and the SLC39 (Zip) family43. There are 10 14ZnT transporters and 14 14Zip transporters encoded in the human genome. Their genes are designated as SLC30A1-10 and SLC39A1-14, respectively. Gene coordinate information was obtained from the UCSC Table Browser (http://genome.ucsc.edu/cgi-bin/hgTables) to infer the start and end position (hg19) for each gene (Table S4 and Figure S13).
Population differentiation and FST estimation
To evaluate whether the genetic variance between the four continental regions is significantly different from the genetic variance among populations within each region, AMOVA was carried out using Arlequin44. To investigate global differentiation among ZTGs, we calculated the weighted average FST (WA - FST) of multiple sites from haplotypes of each gene across all 14 populations. The genetic differentiation between populations of each locus was measured using the unbiased estimates of FST, following Weir and Hill21 with a python script. We determined empirical cutoffs for the top 1% and 5% of signals genome-wide. Thus, loci or genes with an FST value greater than the cutoffs were considered as highly differential SNPs (selected SNPs) or genes. As a result, the highest 1% and 5% of the genome-wide locus-specific FST is 0.183 and 0.092 with the average being 0.017.
Functional SNPs prediction
The functional effects of each SNP from each ZTGs gene were obtained based on the variance effect prediction tools from the Ensembl database45. The SNPs that affect gene expression were studied based on the RegulomeDB database46. In addition, we studied the SNPs with clear clinical effects and disease-related effects, which were then collected and annotated in the PharmGKB database and the GWAS catalog, respectively47,48.
Permutation test
Two kinds of permutation were used in our study. One was used to discern that the FST distribution of ZTGs was different from random genes of the whole genome. We randomly sampled 24 unrepeated genes from the whole genome gene list 10,000 times. In every instance, we calculated the proportion of SNPs with FST values higher than 0.183. Finally, we obtained 10,000 percentage values representing the whole genome and 1 value (0.0182) for ZTGs; we then sorted these values in descending order. Thus, the index (478 of 10,000) of value for ZTGs represents the P-value of this permutation, which is 0.0478. The distribution of percentage values shows that the ratio (0.0182) of ZTGs is larger than the highest 5% cutoff value (0.0180) of the whole genome (Figure 1B).
The other permutation attempts to examine whether nSNPs with high FST (> = 0.092, top 5 percentile of FST values) in Africans are enriched in ZTGs, and to exclude the possibility that drift leads to this pattern. We randomly sampled 24 genes from the UCSC database without repetition 10,000 times. Every time, for random genes we calculated the proportion that the number of nSNPs with high DAF in YRI but low DAF (<0.05) in CEU and CHB accounted for the number of SNPs with high FST.
Detecting signals of selection
Two approaches, the integrated haplotype score (iHS) and the composite likelihood ratio (CLR) test, were used to detect the signals of recent positive selection. Because of the hitchhiking effect, positive selection might bring a selected allele into high frequency rapidly enough that recombination does not have time to break down this haplotype, resulting in a long haplotype in high frequency49. The iHS test is based on the long haplotype, which is a distinctive signature that could not be expected under neutral drift30. It has been shown to possess power enough to identify recent, incomplete sweeps. The standardized iHS scores were calculated for every SNP with minor allele frequency >5% by an R package, rehh50. For every gene in each population, we screened the iHS value of each locus and inferred a positive selection signal if there are 7 or more loci with |iHS| equal to or more than the top 5% of genome-wide signals in any continuous 50-SNPs bin of this gene region.
The CLR test, a model-based method, is a statistic to compute the likelihood ratio of selective sweeps by comparing the spatial distribution of allele frequencies in a given window, compared to the frequency spectrum of null distribution, such as all the autosomal regions51. In this study, the SweepFinder52 program was used to carry out the calculation. For the CLR test, we calculated the standardized CLR score of each population for the entire autosomal regions and took the values with an empirical P-value of 0.05 as the cutoff to detect a natural selection signal at given ZTGs genes.
Haplotype analysis
To visualize the long haplotype of SLC30A9, EHH plot and bifurcation diagrams were drawn using an R package rehh50. SLC30A9 spans about 97 14Kb in the chromosome region 4p13. We defined that SLC30A9 haplotypes were composed of 7 non-synonymous SNPs (Table S2): rs147121215, rs1047626, rs151273121, rs115329927, rs2581423, rs181235146 and rs141510850. Then, we counted the number of haplotypes composed of these chosen SNPs and computed the corresponding proportion in each population using python script.
Correlation test
Three indicators of zinc status at the population level has been recommended: (1) the percentage of the population with plasma (serum) zinc concentrations below an appropriate cut-off, (2) the prevalence of usual dietary zinc intakes below the Estimated Average Requirement (EAR), and (3) stunting prevalence53,54,55,56. The zinc status data for specific regions or countries was directly downloaded from a published paper that estimated the global prevalence of zinc deficiency20. We used a proportion of the population with inadequate zinc intake as an indicator for zinc deficiency and investigated its correlation with H2 (CGGAGAC) of SLC30A9, which is strikingly common in East Asians. All significance tests were performed using R packages (http://www.r-project.org/).
Membrane protein topology and phenotype variation prediction
The transmembrane helices and topology of ZTGs were predicted using HMMTOP23 and visualized with TeXtopo57. To predict how amino acid variants might change the function of the peptides of ZTGs, Polyphen-222 was used.
Supplementary Material
Supplementary Information
Acknowledgments
These studies were supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (XDB13040100), by the National Science Foundation of China (NSFC) grants 91331204, 31370505, 31301083, and 31171218, by the Knowledge Innovation Program of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (2013KIP108). S.X. is Max-Planck Independent Research Group Leader and member of CAS Youth Innovation Promotion Association. S.X. also gratefully acknowledges the support of the National Program for Top-notch Young Innovative Talents of The "Wanren Jihua". We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript. All funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no competing financial interests.
Author Contributions S.X. and J.L. conceived and designed the study. C.Z. and J.L. analyzed data, with contribution from L.T., D.L., K.Y. and Y.Y. C.Z., J.L. and S.X. wrote the paper. All authors contributed to revision and review of the manuscript.
References
- Vallee B. L. & Auld D. S. Cocatalytic zinc motifs in enzyme catalysis. Proc Natl Acad Sci USA 90, 2715–2718 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee B. L. & Falchuk K. H. The biochemical basis of zinc physiology. Physiol Rev 73, 79–118 (1993). [DOI] [PubMed] [Google Scholar]
- Blasie C. A. & Berg J. M. Structure-based thermodynamic analysis of a coupled metal binding-protein folding reaction involving a zinc finger peptide. Biochemistry 41, 15068–15073 (2002). [DOI] [PubMed] [Google Scholar]
- Kambe T., Weaver B. P. & Andrews G. K. The genetics of essential metal homeostasis during development. Genesis 46, 214–228 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambe T., Yamaguchi-Iwai Y., Sasaki R. & Nagao M. Overview of mammalian zinc transporters. Cell Mol Life Sci 61, 49–68 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R. D. & Huang L. Efflux and compartmentalization of zinc by members of the SLC30 family of solute carriers. Pflügers Arch 447, 744–751 (2004). [DOI] [PubMed] [Google Scholar]
- Eide D. J. The SLC39 family of metal ion transporters. Pflügers Arch 447, 796–800 (2004). [DOI] [PubMed] [Google Scholar]
- Schmitt S. et al. An update on mutations of the SLC39A4 gene in acrodermatitis enteropathica. Hum Mutat 30, 926–933 (2009). [DOI] [PubMed] [Google Scholar]
- Taylor K. M. et al. The emerging role of the LIV-1 subfamily of zinc transporters in breast cancer. Mol Med 13, 396–406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagara N., Tanaka N., Noguchi S. & Hirano T. Zinc and its transporter ZIP10 are involved in invasive behavior of breast cancer cells. Cancer Sci 98, 692–697 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K. M., Morgan H. E., Johnson A., Hadley L. J. & Nicholson R. I. Structure-function analysis of LIV-1, the breast cancer-associated protein that belongs to a new subfamily of zinc transporters. Biochem J 375, 51–59 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. et al. Aberrant expression of zinc transporter ZIP4 (SLC39A4) significantly contributes to human pancreatic cancer pathogenesis and progression. Proc Natl Acad Sci USA 104, 18636–18641 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. et al. Down-regulation of ZIP4 by RNA interference inhibits pancreatic cancer growth and increases the survival of nude mice with pancreatic cancer xenografts. Clin Cancer Res 15, 5993–6001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. ZIP4 regulates pancreatic cancer cell growth by activating IL-6/STAT3 pathway through zinc finger transcription factor CREB. Clin Cancer Res 16, 1423–1430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen C., Yao Q. & Li M. ZIP4 Upregulates the expression of neuropilin-1, vascular endothelial growth factor, and matrix metalloproteases in pancreatic cancer cell lines and xenografts. Cancer Biol Ther 9, 235–241 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek R. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885 (2007). [DOI] [PubMed] [Google Scholar]
- Chimienti F. et al. In vivo expression and functional characterization of the zinc transporter ZnT8 in glucose-induced insulin secretion. J Cell Sci 119, 4199–4206 (2006). [DOI] [PubMed] [Google Scholar]
- Speliotes E. K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat genet 42, 937–948 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelken J. et al. Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa. PLoS Genet 10, e1004128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan Wessells K. & Brown K. H. Estimating the Global Prevalence of Zinc Deficiency: Results Based on Zinc Availability in National Food Supplies and the Prevalence of Stunting. PLoS One 7, e50568 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S. & Hill W. Estimating F-statistics. Annu Rev Genet 36, 721–750 (2002). [DOI] [PubMed] [Google Scholar]
- Adzhubei I. A. et al. A method and server for predicting damaging missense mutations. Nat methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady G. E. & Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850 (2001). [DOI] [PubMed] [Google Scholar]
- Aslibekyan S. et al. Preliminary evidence of genetic determinants of adiponectin response to fenofibrate in the Genetics of Lipid Lowering Drugs and Diet Network. Nutr Metab Cardiovasc Dis 23, 987–994 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar D. P. et al. Genome-wide association identifies genetic variants associated with lentiform nucleus volume in N = 1345 young and elderly subjects. Brain Imaging Behav 7, 102–115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C. et al. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C. et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 419, 832–837 (2002). [DOI] [PubMed] [Google Scholar]
- Grossman S. R. et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327, 883–886 (2010). [DOI] [PubMed] [Google Scholar]
- Cann H. M. et al. A Human Genome Diversity Cell Line Panel. Science 296, 261–262 (2002). [DOI] [PubMed] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X. & Pritchard J. K. A map of recent positive selection in the human genome. PLoS Biol 4, e72 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. T., Kodama G., Baldi P. & Moyzis R. K. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci U S A 103, 135–140 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman S. R. et al. Identifying recent adaptations in large-scale genomic data. Cell 152, 703–713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaresque P. L., Ballereau S. J. & Jobling M. A. Challenges in human genetic diversity: demographic history and adaptation. Hum Mol Genet 16, R134–139 (2007). [DOI] [PubMed] [Google Scholar]
- Nielsen R. et al. Darwinian and demographic forces affecting human protein coding genes. Genome Res 19, 838–849 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney G. P., Zheng D., Kille P. & Hogstrand C. The phylogeny of teleost ZIP and ZnT zinc transporters and their tissue specific expression and response to zinc in zebrafish. Biochim Biophys Acta 1732, 88–95 (2005). [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. & Windisch W. Influence of zinc deficiency on the mRNA expression of zinc transporters in adult rats. J Trace Elem Med Biol 17, 97–106 (2003). [DOI] [PubMed] [Google Scholar]
- Larson C. A. et al. The role of the N-terminus of mammalian copper transporter 1 in the cellular accumulation of cisplatin. Biochem Pharmacol 80, 448–454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno S., Yanagiya T. & Fujishiro H. The role of zinc transporters in cadmium and manganese transport in mammalian cells. Biochimie 91, 1218–1222 (2009). [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Yang C. K., Xia M., Ou C. Y. & Stallcup M. R. Role of GAC63 in transcriptional activation mediated by beta-catenin. Nucleic Acids Res 35, 2084–2092 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochan I. The role of iron in bacterial infections, with special consideration of host-tubercle bacillus interaction. Curr Top Microbiol Immunol 1–30 (1973). [DOI] [PubMed] [Google Scholar]
- Hood M. I. & Skaar E. P. Nutritional immunity: transition metals at the pathogen–host interface. Nat Rev Microbiol 10, 525–537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. . An integrated map of genetic variation from 1,092 human genomes. Nature 491, 1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten L. A. & Cousins R. J. Mammalian zinc transporters: nutritional and physiologic regulation. Annu Rev Nutr 29, 153–176 (2009). [DOI] [PubMed] [Google Scholar]
- Excoffier L., Laval G. & Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1, 47 (2005). [PMC free article] [PubMed] [Google Scholar]
- McLaren W. et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26, 2069–2070 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22, 1790–1797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whirl-Carrillo M. et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92, 414–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L. A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 106, 9362–9367 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. M. & Haigh J. The hitch-hiking effect of a favourable gene. Genet Res 23, 23–35 (1974). [PubMed] [Google Scholar]
- Gautier M. & Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics 28, 1176–1177 (2012). [DOI] [PubMed] [Google Scholar]
- Nielsen R. et al. Genomic scans for selective sweeps using SNP data. Genome Res 15, 1566–1575 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annu Rev Genet 39, 197–218 (2005). [DOI] [PubMed] [Google Scholar]
- Hess S. Y., Peerson J. M., King J. C. & Brown K. H. Use of serum zinc concentration as an indicator of population zinc status. Food Nutr Bull 28, 403S–429S (2007). [DOI] [PubMed] [Google Scholar]
- Hotz C. Dietary indicators for assessing the adequacy of population zinc intakes. Food Nutr Bull 28, 430S–453S (2007). [DOI] [PubMed] [Google Scholar]
- Fischer Walker C. L. & Black R. E. Functional indicators for assessing zinc deficiency. Food Nutr Bull 28, 454S–479S (2007). [DOI] [PubMed] [Google Scholar]
- Gibson R. S., Hess S. Y., Hotz C. & Brown K. H. Indicators of zinc status at the population level: a review of the evidence. Br J Nutr 99, S14–23 (2008). [DOI] [PubMed] [Google Scholar]
- Beitz E. TEXtopo: shaded membrane protein topology plots in LATEX2ε. Bioinformatics 16, 1050–1051 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information