Abstract
Although chimeric repressors such as the Arabidopsis TCP3 repressor are known to have significant effects on flower morphology and color, their cellular-level effects on flower petals are not understood. The promoter sequences of the genes expressed in the flowers of cyclamen, a representative potted flower grown during the winter season, are also unknown. Here, we isolated eight promoters from cyclamen genes that are reportedly expressed in the petals. These promoters were then fused to four chimeric repressors and introduced into the model flower torenia to screen for effective combinations of promoters and repressors for flower breeding. As expected, some of the constructs altered flower phenotypes upon transformation. We further analyzed the effects of chimeric repressors at the cellular level. We observed that complicated petal and leaf serrations were accompanied by excessive vascular branching. Dichromatism in purple anthocyanin was inferred to result in bluish flowers, and imbalanced cell proliferation appeared to result in epinastic flowers. Thus, the genetic constructs and phenotypic changes described in this report will benefit the future breeding and characterization of ornamental flowers.
Introduction
The breeding of new flower cultivars with various phenotypes, such as different petal colors and shapes, is key to promoting flower use. Straightforward destruction of endogenous genes, introduction of new metabolic pathways from different species through transformation, and random mutagenesis using ion beam irradiation have been effective in generating double flowers as well as altering flower color, pigmentation patterns, and shapes (for example, refs 1–5). In addition to these approaches, the use of chimeric repressors to confer different characteristics upon flowers in a dominant genetic manner has also been tested.
Chimeric repressors are artificially generated by combining transcription activators with peptides known as repression domains. In plants, class II ethylene-responsive element-binding factor and TFIIIA-type zinc finger repressors, including SUPERMAN, share the ‘EAR motif’.6 Short peptides that include this motif function as repression domains, changing transcription activators into transcription suppressors. The most frequently used and strongest repression domain is ‘SRDX,’ which is a leucine-rich peptide consisting of 12 amino acid residues (LDLDLELRLGFA). This peptide was obtained by altering the native sequence of the repression domain of SUPERMAN.7,8 The wide range of flower phenotypes generated to date using chimeric repressors is listed in the FioreDB database (http://www.cres-t.org/fiore/public_db/index.shtml; refs 9,10).
Due to the ease of transformation compared with other ornamental crops, torenia (Torenia fournieri Lind.) is used as a model system for transgenic flower breeding. Various chimeric repressors of Arabidopsis (Arabidopsis thaliana (L.) Heynh.) transcription factors have been surveyed in torenia.11 According to the comparison of all such chimeric repressors examined in torenia thus far, repressors of TCP (Teosinte branched1, Cycloidea and Pcf) and SEP3 (SEPallata3) are among those causing the clearest phenotypic changes.12–15 Endogenous promoters of torenia have been used to drive the Arabidopsis TCP3 repressor to change torenia flower phenotypes specifically and in various ways.16 Thus, variation in promoters and chimeric repressors promotes the breeding of a variety of transgenic flowers. However, despite the publication of several relevant reports, the physiological effects of chimeric repressors linked to phenotypic changes in transgenic flowers, such as petal serration, have not been examined in ornamental flowers.
In the present study, we first isolated promoters from cyclamen (Cyclamen persicum Mill.), which is a representative potted winter-flowering plant. We identified eight promoter sequences of genes known to be expressed in cyclamen petals. We then cloned these promoters and used them to drive four different chimeric repressors in torenia. Examination of the transgenic torenia flowers harboring these genetic constructs allowed us to characterize the effects of these cyclamen promoters and chimeric repressors. Further analyses of the transgenic lines suggested morphological or biochemical effects linked to the phenotypic changes in the flowers.
Materials and methods
Isolation of cyclamen promoters
The full-length cDNA sequences of CpAP1, CpPI, CpAP3A, CpAP3B, CpSEP2, CpSEP3, CpFLC, CpCHS, CpDFR and CpOMT were previously reported (Table 1). Three gene-specific primers (GSP1, GSP2 and GSP3; Supplementary Table S1) were designed in reverse orientation corresponding to the 5′-untranslated region (UTR) or the first exon of each cDNA sequence, and additional gene-specific primers were designed for some of the genes to extend the promoter sequences. Genomic DNA was extracted from cyclamen (Cyclamen persicum Mill. cultivar ‘Mini Loyal Purple’) leaves using ‘PVPP buffer’ containing 4 m NaCl and was purified via ultracentrifugation.17,18 The promoter sequences were amplified by adaptor PCR with a ‘mismatched DS4 adaptor’ (ref. 19; Supplementary Table S1), which potentially has a better capability to amplify target sequences. Briefly, genomic DNA was digested with eight different restriction enzymes (DraI, EcoRV, ScaI, SspI, HindIII, KpnI, SacI and SpeI) and then ligated to adaptors and amplified by nested PCR using gene-specific primers and adaptor primers (DS4-AP1, DS4-AP2 or DS4-AP3; Supplementary Table S1). PCR was performed with KOD Plus Neo polymerase (Toyobo, Osaka, Japan) for 35 cycles, with an annealing temperature of 68 °C and an extension time of 5 min. The amplified fragments were recovered from the agarose gels, TA-cloned into plasmids, and sequenced. The promoters were then amplified from the genomic DNA using cloning primers (CP-F and CP-R; Supplementary Table S1) in which HindIII (5′- AAGCTT-3′) or BamHI (5′- GGATCC-3′) sites were attached at either end for subcloning (underlined in Supplementary Table S1). Amplification was performed with KOD Plus Neo polymerase for 45 cycles, with an annealing temperature of 68 °C and an extension time of 5 min. The accession numbers and lengths of the isolated promoters are shown in Table 1.
Table 1. Properties of cyclamen petal genes (CPGs) and their promoters.
| Gene | MADS class |
INSDCa
accession number |
Promoter lengthb | |
|---|---|---|---|---|
| Gene | Promoter | (bp) | ||
| CpAP1 | A | AB600230.1 | LC176494 | 1596 |
| CpPI | B | AB600234.2 | LC176500 | 2030 |
| CpAP3A | B | AB600231.1 | LC176495 | 1179 |
| CpAP3B | B | AB600232.1 | LC176496 | 1554 |
| CpSEP2 (CpMADS1)c | E | AB600235.1 | LC176501 | 1863 |
| CpSEP3 (CpMADS2)c | E | AB600236.1 | — | — |
| CpFLC (CpMADS3)c | — | AB600233 | — | — |
| CpCHSd | — | DD354767 | LC176497 | 1555 |
| CpDFRd | — | DD354768 | LC176498 | 1528 |
| CpOMTd | — | DD248507 | LC176499 | 1655 |
International Nucleotide Sequence Database Collaboration (collaboration between DDBJ, EMBL-EBI, and NCBI; http://www.insdc.org/).
Lengths of the isolated sequences, upstream of the start codon (ATG).
The original names for CpSEP2, CpSEP3, and CpFLC were CpMADS1, CpMADS2 and CpMADS3, respectively.25
These genes are expressed in cyclamen petals, as their cDNA sequences were isolated from cyclamen petals (Japan patents 2006-115861 and 2005-312388; https://www.j-platpat.inpit.go.jp/web/tokujitsu/tkbs/TKBS_GM101_Top.action, Japanese).
Subcloning
The protein-coding regions of AtTCP3, CpTCP1B, AtSEP3, and CpSEP3 (International Nucleotide Sequence Database Collaboration (INSDC) accession numbers NM_104201.3, LC186052, NM_180622.2 and AB600236.1, respectively) were cloned into the BamHI–SmaI site of the p35SSRDXG vector20 to produce 35Sp:CR vectors. Single-nucleotide (silent) mutations were introduced into the AtSEP3 and CpSEP3 sequences to eliminate restriction sites without changing the encoded protein sequences. Cyclamen promoter fragments were introduced into the HindIII-BamHI sites of 35Sp:CR vectors or into the pG2-35S-GUS vector to produce 32 CPGp:CR vectors and four CPGp:GUS vectors, respectively. The regions corresponding to each transgene were transferred into the binary vector pBCKH (for hygromycin selection of transgenic plants) or pBCKK (for kanamycin selection of transgenic plants; ref. 20) using the Gateway system (Thermo Fisher Scientific, Waltham, MA, USA).
Plant materials, plant growth conditions and transformation
In vitro cloned seedlings of torenia (Torenia fournieri Lind. cultivar ‘Crown Violet’ and the hybrid torenia cultivar ‘Summerwave Blue’) were maintained on sterilized media in transparent plastic boxes at 25 °C under white fluorescent light (photosynthetic photon flux density (PPFD)=150 μmol m−2 s−1) with a 16-h/8-h light/dark photoperiod. For transformation, bulk (mixture) or individual plant expression vectors carrying the appropriate constructs were introduced into Agrobacterium tumefaciens strain EHA105 via electroporation, and the Agrobacterium were subsequently infiltrated into leaf discs as previously described.21 Briefly, thousands of 5-mm leaf discs were prepared from in vitro cultures, inoculated with Agrobacterium, then co-cultured on media without antibiotics, and transformed calli were generated on selection media containing antibiotics. One or two transgenic lines were typically obtained from 100 leaf discs. The number of total leaf discs was ~8000. Regenerated plant cultures were maintained on media containing trehalose, and the media were renewed every 2 months.22 Transformants were then transferred to pots and grown in a sunlit greenhouse.
The transformants generated with bulk plasmids were examined via nested PCR to determine the inserted constructs (Supplementary Figure S4). The first round of PCR was performed with ‘Whole-F1’ and ‘Whole-R1’ primers. The first PCR product (diluted 1/1000 in solution) was then amplified using a combination of the ‘Whole-F2’ primer, individual promoter-specific primers (PSP; Supplementary Table S1), and the mixture of all repressor-specific primers (RSP; Supplementary Table S1). Both the first and second rounds of PCR were performed with KOD Plus Neo polymerase for 40 cycles, with an annealing temperature of 55 °C and an extension time of 2 min. The repressor genes (AtTCP3-RD, CpTCP1B-RD, AtSEP3-RD or CpSEP3-RD) introduced into the transformants were finally determined based on the PCR product sizes (approximately 650 bp for CpTCP1B-RD, 500 bp for CpSEP3-RD, 400 bp for AtSEP3-RD, and 200 bp for AtTCP3-RD). The whole-F2 primer generates large DNA fragments when there is no construct corresponding to the target promoter sequence. Inclusion of the whole-F2 primer in the second PCR is necessary to reduce non-specific amplification. This scheme was repeated for all CPG promoters that may have been introduced into the target line. Tetraploid transformants and those transformants harboring multiple constructs were not used for phenotypic analyses. As described in the ‘Results’ section, 95 transgenic crown violet lines possessing single constructs were generated via transformation with individual constructs (59 lines) or bulk constructs (36 lines); 52 transgenic crown violet lines were generated with a bulk plasmid of 8 constructs (CpOMTp and CpSEP2p constructs, inserts not determined); and 12 transgenic summerwave blue lines were generated with the CpAP3Bp-CpTCP1B-RD construct.
GUS staining
GUS staining was performed with T1 (first generation) transgenic plants (Figure 2). Plant tissues were pretreated with 90% acetone and placed under vacuum for 30 min in 100 mm sodium phosphate buffer (pH 7.0) containing 0.1% Triton X-100, 0.5 mg 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc), 10 mm ethylenediaminetetraacetic acid (EDTA), 5 mm potassium ferricyanide and 5 mm potassium ferrocyanide. The tissues were then incubated in this solution at 37 °C in the dark overnight. The stained tissues were washed with 70% ethanol, treated with ethanol-acetic acid (6:1, v/v) overnight, washed again with 70% ethanol, and treated with chloral hydrate solution (chloral hydrate:glycerol:sterile water=8 g:1 mL:2 mL) overnight. Images were obtained using a single-lens reflex digital camera with a white backlight to observe whole staining or with a stereoscopic microscope (MZ16 FA, Leica, Wetzlar, Germany) to observe staining in small areas.
Figure 2.
CPGp-GUS lines. (a) The genetic constructs introduced. Four cyclamen petal gene (CPG) promoters (CpPIp, CpDFRp, CpOMTp, and CpAP3Bp) were used to drive GUS gene expression. ‘NOS ter’ represents the NOPALINE SYNTHASE terminator. (b) GUS staining of the CpPIp-GUS line (left to right: flower, floral bud, leaf, and blotch), the CpDFRp-GUS line (flower, floral bud and leaf), the CpOMTp-GUS line (flower, floral bud and leaf), and the CpAP3Bp-GUS line (inset shows the anther excised from floral bud).
Vascular structure
Leaves were imaged using a single-lens reflex digital camera with a white backlight. The resulting digital images were retouched using Adobe Photoshop software (Adobe Systems Inc., San Jose, CA, USA) by converting the color image to a black and white image and then adjusting the brightness and contrast to make the vasculature clear (Figure 1f). Petals were treated with ethanol-acetic acid overnight and then placed in chloral hydrate solution overnight. The vasculature was observed under dark-field conditions with a stereoscopic microscope (MZ16 FA, Leica; Figure 6b).
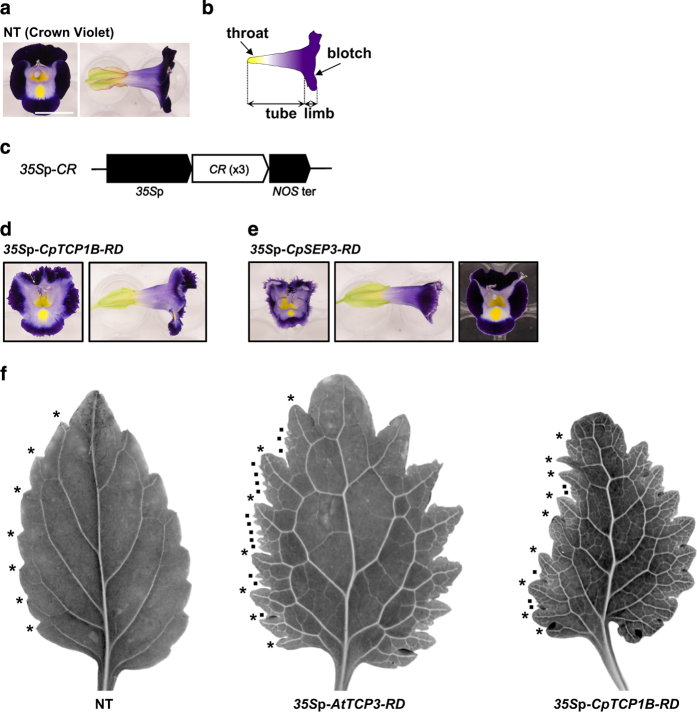
Figure 1.
Structure of torenia flowers and the phenotypes of 35Sp-CR lines. (a) A non-transgenic (NT) Crown Violet flower. Bar=1 cm. (b) Schematic representation of the flower structure. (c) The genetic constructs introduced. 35Sp was combined with three chimeric repressor (CR) genes (CpTCP1B-RD, CpSEP3-RD or AtTCP3-RD). (d) A transgenic line expressing 35Sp-CpTCP1B-RD. (e) Two transgenic lines expressing 35Sp-CpSEP3-RD. (f) Leaf silhouette and vascular structure of the non-transgenic line, the 35Sp-AtTCP3-RD line, and the 35Sp-CpTCP1B-RD line. The primary serrations are indicated with asterisks, and the secondary serrations are indicated with dots only on the left side of the leaves.
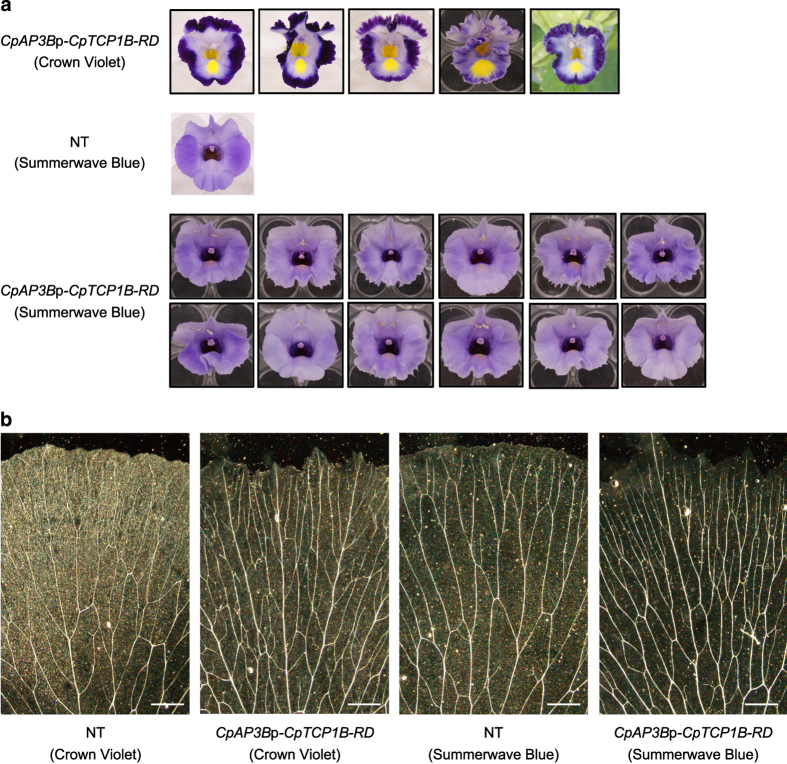
Figure 6.
Phenotypes of CpAP3Bp-CpTCP1B-RD lines. (a) Five CpAP3Bp-CpTCP1B-RD lines of ‘Crown Violet,’ the non-transgenic cultivar ‘Summerwave Blue,’ and twelve CpAP3Bp-CpTCP1B-RD lines of ‘Summerwave Blue.’ (b) Vascular structures in the petals of non-transgenic ‘Crown Violet,’ a CpAP3Bp-CpTCP1B-RD line of ‘Crown Violet,’ non-transgenic ‘Summerwave Blue,’ and a CpAP3Bp-CpTCP1B-RD line of ‘Summerwave Blue.’ Bars=1 mm.
Petal cross-sections and scanning electron microscopy
To prepare petal cross-sections, agar with an isotonic solute was produced (2% agar, 500 mm trehalose, 5 mm CaCl2, 0.2% Tween-20). Side lobes excised from open flowers were sliced at a thickness of 55 μm with a vibrating microslicer (DTK-1000, Dosaka EM, Kyoto, Japan) and embedded in the agar. The slices were subsequently observed with a light microscope (AX70, Olympus, Tokyo, Japan; Figure 8e). The adaxial and abaxial surfaces of side lobes excised from open flowers were also observed with a scanning electron microscope (VE-7800, Keyence, Osaka, Japan; Figure 8b).
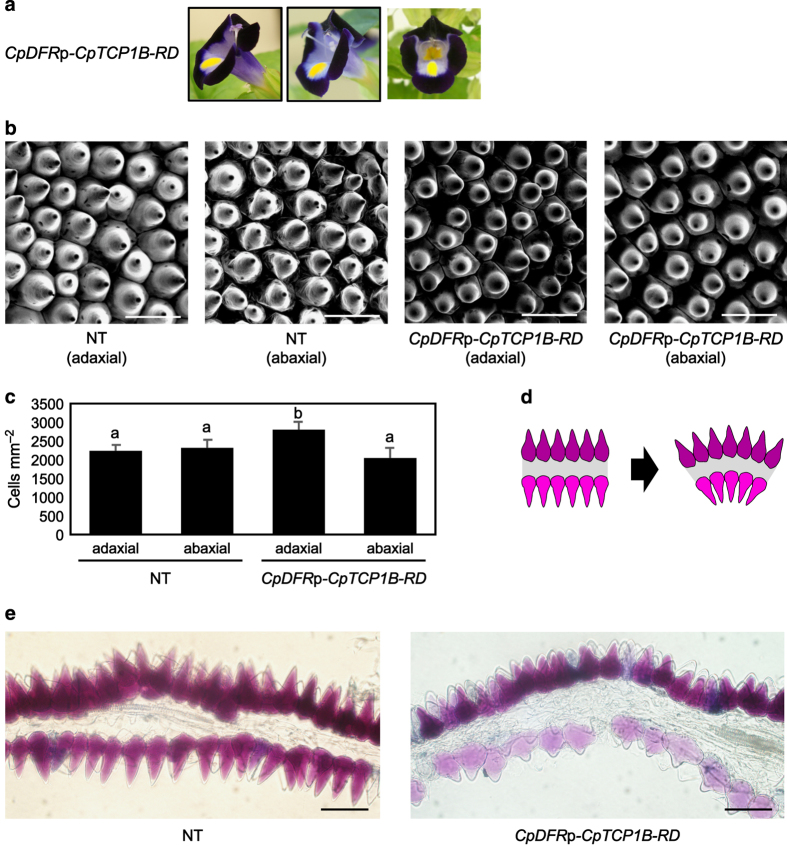
Figure 8.
Microscopy analysis of CpDFRp-CpTCP1B-RD lines. (a) Phenotypes of three CpDFRp-CpTCP1B-RD lines. (b) Conical cells of the petals imaged via scanning electron microscopy on the adaxial/abaxial sides of the non-transgenic line and curled CpDFRp-CpTCP1B-RD line. (c) Conical cell density on petal surfaces. The data represent the mean and standard deviation (n=4). The values labeled with different letters (a or b) are significantly different from each other by Student’s t-test (P<0.05). (d) Schematic representation of the effect of differential cell densities. (e) Cross-section of the lobe of the non-transgenic line and curled CpDFRp-CpTCP1B-RD line. Bars=50 μm in all images.
Colorimetry
The brightness of eight different parts of the same flowers was measured in images of flowers using Adobe Photoshop software (Figures 5b and 7b). The surface reflectance of the adaxial side of the side lobes was measured with an ultraviolet/visible (UV/VIS) spectrophotometer (UV-2450, Shimadzu, Kyoto, Japan; Figure 7c). RGB color under flat white light was calculated using a standardized CIE 1931 RGB color matching function. Hue, lightness, saturation, and coordinates in a round diagram were also calculated following previous reports (refs 23,24; Figure 7d). The recently developed round diagram is based on the RGB color system, which distributes hue angles and saturation values equally around a circle, consistent with our perception of color.24
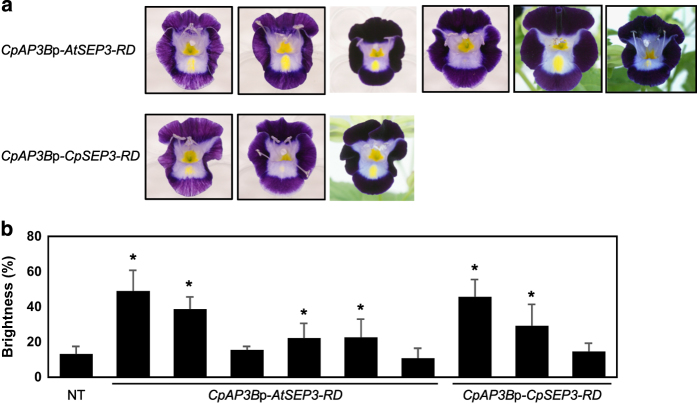
Figure 5.
Comparison between the effects of AtSEP3-RD and CpSEP3-RD. (a) Phenotypes of six CpAP3Bp-AtSEP3-RD lines and three CpAP3Bp-CpSEP3-RD lines. (b) Brightness of the lobes measured in the non-transgenic line, CpAP3Bp-AtSEP3-RD lines, and CpAP3Bp-CpSEP3-RD lines. The data represent the mean and standard deviation (n=8). Asterisks indicate significant differences from the non-transgenic line by Student’s t-test (P<0.05). The data for transgenic lines correspond to flowers shown in (a) in the same order.
Figure 7.
Colorimetric analysis of CpCHSp-AtSEP3-RD lines. (a) Phenotypes of five CpCHSp-AtSEP3-RD lines. (b) Brightness of the lobes measured in the non-transgenic line and CpCHSp-AtSEP3-RD lines. The data represent the mean and standard deviation (n=8). Asterisks indicate significant differences from the non-transgenic line by Student’s t-test (P<0.05). The data for the transgenic lines correspond to the flowers shown in (a) in the same order. (c) Reflectance spectra of visible light on the lobes of the non-transgenic line (black and gray lines) and of a light-colored CpCHSp-AtSEP3-RD line (violet and purple lines). The measurements were performed in two replications with different flowers; each replication produced similar results, and the lines (black and gray, or violet and purple) overlap with each other. The blue, green, and red bars at the bottom of the graph indicate the approximate color of light at each wavelength. (d) Colors (hue and saturation) of the non-transgenic line and a CpCHSp-AtSEP3-RD line plotted on a round color diagram.
Results
Isolation of cyclamen petal gene promoters
To express the chimeric repressors under the control of a variety of cyclamen promoters, we isolated 1.5-kb or longer upstream sequences (promoter sequences) of cyclamen petal genes (referred to as CPGs hereafter). Eleven homeotic MADS-box genes have been isolated from cyclamen, including CpAP1, CpPI, CpAP3A, CpAP3B, CpSEP2, CpSEP3, and CpFLC, which are all expressed in the petals (ref. 25; Table 1). In addition to these seven CPGs, we targeted three genes involved in anthocyanin pigmentation (CpCHS, CpDFR and CpOMT), which are also expressed in the petals (see footnote of Table 1). Here, the abbreviations AP, PI, FLC, CHS, DFR and OMT represent apetala, pistillata, flowering locus c, chalcone synthase, dihydroflavonol 4-reductase, and O-methyltransferase, respectively.
After adaptor PCR analysis, we successfully isolated the promoter sequences of 8 of the 10 target CPGs (CpAP1, CpPI, CpAP3A, CpAP3B, CpSEP2, CpCHS, CpDFR, and CpOMT). These promoter sequences are abbreviated as CpAP1p, CpPIp, CpAP3Ap, CpAP3Bp, CpSEP2p, CpCHSp, CpDFRp, and CpOMTp, respectively. All isolated promoter sequences (CPGp sequences), except for CpAP3Ap (1179 bp), were longer than 1.5 kb, ranging from 1528 to 2030 bp (Table 1).
In this study, the transformed material was the torenia cultivar ‘Crown Violet’ (Figure 1a,b) unless otherwise indicated. Torenia presents flowers with compound petals (gamopetalous corolla). The basal part of the corolla (referred to as the ‘throat’ hereafter) is yellow, while the middle part is light purple. The basal and middle parts of the corolla are collectively referred to as the ‘tube.’ The top part of the corolla (the ‘limb’) is dark purple and is divided into four parts (‘lobes’). The lobe margins are rounded, and there is a yellow spot on the basal lobe of the corolla, which is referred to as the ‘blotch.’ The dark purple color of the limbs is caused by anthocyanin pigments.26
To examine promoter activity, we generated one transgenic torenia line expressing the GUS (β-glucuronidase) gene under four representative CPG promoters (CpPIp, CpDFRp, CpOMTp and CpAP3Bp; Figure 2a). These transgenic plants were stained with a buffer containing 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-Gluc), which is catabolized by the GUS enzyme to form a blue pigment. Blue pigmentation was observed only in the blotch in the CpPIp-GUS line (Figure 2b), indicating that CpPIp primarily drives gene expression in blotch cells. Weak staining was observed in the limbs of open flowers of the CpDFRp-GUS line (Figure 2b), while no staining was observed in the CpOMTp-GUS line (Figure 2b). The CpAP3Bp-GUS line exhibited the greatest density of blue pigmentation among the four promoters examined (Figure 2b). Thus, strong staining was observed in the limbs of open flowers and in the anthers of floral buds, with weaker staining observed in the tubes of open flowers and in the basal part of the stem.
Preparation of chimeric repressors
In this study, we used four chimeric repressors to modify torenia flowers. In addition to AtTCP3-RD and AtSEP3-RD, whose effects have been previously reported in torenia,13,14 a gene homologous to AtSEP3 and a putative allele of CpTCP1 from cyclamen were isolated, and corresponding chimeric repressors (CpSEP3-RD and CpTCP1B-RD) were prepared. Expression vectors in which CpTCP1B-RD or CpSEP3-RD was driven by the Cauliflower mosaic virus 35S promoter (35Sp) were used for functional characterization of these repressors, as was the expression vector of AtTCP3-RD for comparison (Figure 1c).
We generated one transgenic torenia line harboring the 35Sp-CpTCP1B-RD construct (Figure 1d) and two transgenic torenia lines harboring the 35Sp-CpSEP3-RD construct (Figure 1e). The line expressing 35Sp-CpTCP1B-RD possessed serrated petals. One of the lines expressing 35Sp-CpSEP3-RD exhibited jaggy petals, which were more pointed than the serrated petals. Compared with previous reports on 35Sp-AtTCP3-RD and 35Sp-AtSEP3-RD, 35Sp-CpTCP1B-RD showed similar effects to 35Sp-AtTCP3-RD, while 35Sp-CpSEP3-RD showed similar effects to 35Sp-AtSEP3-RD in transgenic torenia petals.
Torenia lines expressing 35Sp-AtTCP3-RD or 35Sp-CpTCP1B-RD produce petals and leaves with deeper and more complicated serrations. That is, non-transgenic leaves exhibit only ‘primary’ serrations, but the transgenic lines exhibit small ‘secondary’ serrations formed on the primary serrations as well as deeper primary serrations (Figure 1f). In our observations, we noted that the transgenic lines with more complicated serrations displayed more vascular branching in the leaves compared with the leaves of the non-transgenic line (Figure 1f). Vascular branching was particularly promoted at the leaf margins of these transgenic lines.
Expression of chimeric repressors under the control of cyclamen petal gene promoters
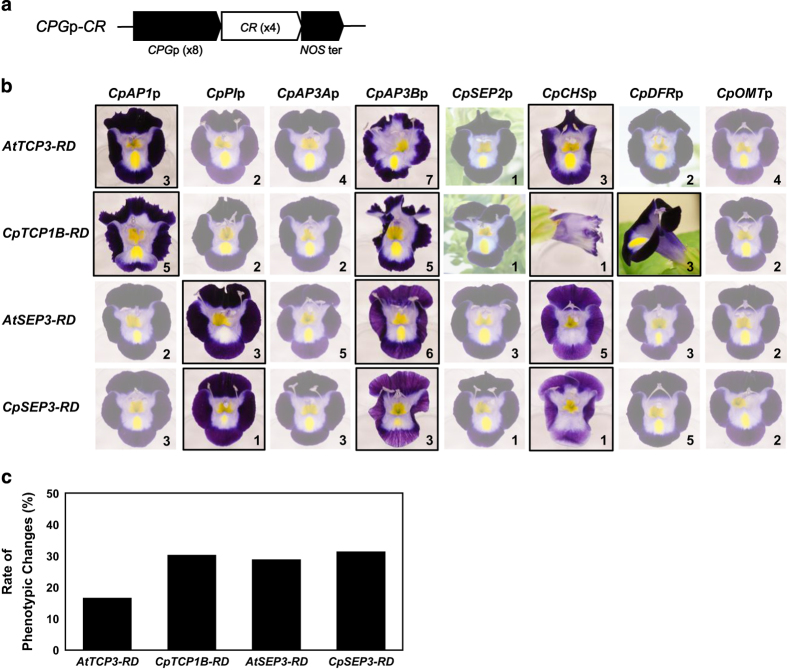
We prepared 32 different expression vectors using different combinations of the eight CPGp sequences and the four chimeric repressors (CPGp-CR vectors; Figure 3a). Torenia plants were transformed with each vector separately, and 59 lines were generated from these transformations. An additional 36 lines were generated through bulk transformation, whose T-DNA inserts were determined via PCR. Figure 3b shows representative lines with the most clearly modified flower phenotypes associated with each construct. Thirteen constructs resulted in phenotypes that differed from those of the non-transgenic line. CpAP1p generated flowers with jaggy margins when driving AtTCP3-RD or CpTCP1B-RD. CpPIp generated flowers without a blotch or with a smaller blotch (blotchless flowers) when driving AtSEP3-RD or CpSEP3-RD. CpAP3Bp generated serrated and wavy flowers when driving AtTCP3-RD or CpTCP1B-RD. CpAP3Bp also generated blotchless and striped flowers when driving AtSEP3-RD or CpSEP3-RD. CpCHSp generated ‘trumpet-shaped’ flowers with outer-curling limbs when driving AtTCP3-RD and ‘tube’ flowers without limbs when driving CpTCP1B-RD. CpCHSp generated blotchless and light-colored flowers when driving AtSEP3-RD or CpSEP3-RD. CpDFRp generated trumpet-shaped flowers when driving CpTCP1B-RD. CpAP3Ap, CpSEP2p and CpOMTp combined with four repressors did not generate any phenotypic changes.
Figure 3.
CPGp-CR lines. (a) The genetic constructs introduced. Eight CPG promoters (CpAP1p, CpPIp, CpAP3Ap, CpAP3Bp, CpSEP2p, CpCHSp, CpDFRp and CpOMTp) were combined with four chimeric repressors (AtTCP3-RD, CpTCP1B-RD, AtSEP3-RD, or CpSEP3-RD) in independent vectors. (b) Representative lines (those that were most clearly modified) transformed with each construct. The images are aligned by the introduced promoters and chimeric repressors. Phenotypically modified flowers are highlighted and surrounded with black lines. Images of phenotypically modified flowers are also surrounded with black lines in the other figures. The numbers at the bottom-right corner in each image indicate the numbers of transgenic lines generated. (c) The average rates of phenotypic changes for every chimeric repressor.
The absence of phenotypic changes associated with the constructs driven by CpSEP2p or CpOMTp was supported by the finding that a population of an additional 52 lines transformed with a bulk plasmid consisting of eight constructs driven by these promoters did not show any phenotypic changes (Supplementary Figure S1). Although it was not determined which construct was introduced into each line, eight constructs containing CpSEP2p or CpOMTp were expected to be randomly introduced into these lines. Excluding this population, one to seven transgenic lines per 32 constructs (approximately three lines, on average) were analyzed (Supplementary Figure S2). Approximately half of the lines were modified from the non-transgenic line when the 13 constructs were introduced, and the phenotypes were similar to each other when the transgenic lines possessed the same construct. It is possible that the remaining 19 constructs may produce phenotypic changes if larger populations of transgenic plants are generated. The average rate of phenotypically modified transgenic lines in the CPGp-CR population was ~30% for CpTCP1B-RD, AtSEP3-RD and CpSEP3-RD, while the rate for AtTCP3-RD was less than 20% (Figure 3c).
Comparison between the chimeric repressors derived from Arabidopsis and cyclamen
AtTCP3-RD and CpTCP1B-RD caused similar phenotypic changes in transgenic torenia flowers, although both the degree and the rate of phenotypic changes appeared to be greater in association with CpTCP1B-RD. However, AtSEP3-RD and CpSEP3-RD caused similar phenotypic changes and showed a similar degree and rate of phenotypic changes (Figures 3b and Supplementary Figure S2). These trends were reconfirmed through an analysis of representative constructs.
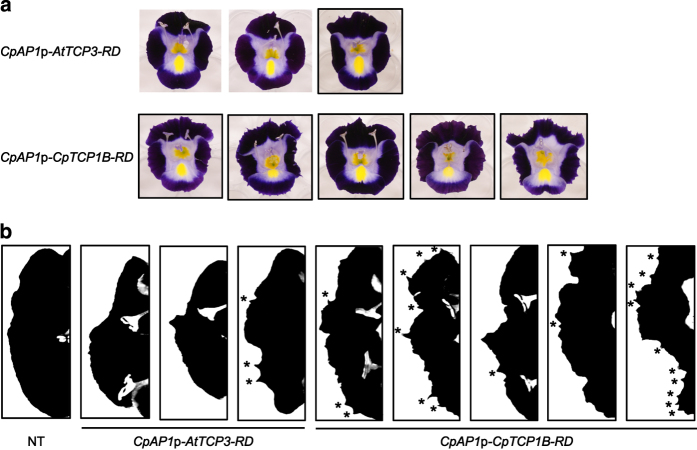
The CpAP1p-AtTCP3-RD and CpAP1p-CpTCP1B-RD constructs were chosen for comparison of the effects of AtTCP3-RD and CpTCP1B-RD. Both constructs generated serrations at the lobe margins, but the degree of serration tended to be greater in CpAP1p-CpTCP1B-RD lines (Figure 4). The CpAP3Bp-AtSEP3-RD and CpAP3Bp-CpSEP3-RD constructs were chosen for a comparison of the effects of AtSEP3-RD and CpSEP3-RD. Both constructs decreased the purple coloration of the lobes and the yellow coloration of the blotch (Figure 5a). The degree of phenotypic changes was quantified based on the brightness of the lobes (Figure 5b); when the purple coloration is reduced by the chimeric repressors to a greater extent, the lobes become brighter. The distribution of lobe brightness in the transgenic lines was similar between AtSEP3-RD and CpSEP3-RD, suggesting that AtSEP3-RD and CpSEP3-RD have similar effects in torenia.
Figure 4.
Comparison between the effects of AtTCP3-RD and CpTCP1B-RD. (a) Phenotypes of three CpAP1p-AtTCP3-RD lines and five CpAP1p-CpTCP1B-RD lines. (b) Silhouettes of the top lobes of the non-transgenic line, CpAP1p-AtTCP3-RD lines, and CpAP1p-CpTCP1B-RD lines. Serrations are indicated by asterisks. The silhouettes of the transgenic lines correspond to the flowers shown in (a) in the same order.
Microscopic analysis of serrated flowers
Although the beauty of flowers will always be subjective depending on personal preferences, the flowers among these lines showing the greatest degree of modification are the serrated, wavy (and light-colored) flowers associated with CpAP3Bp-CpTCP1B-RD, the blotchless, light-colored flowers associated with CpCHSp-AtSEP3-RD, and the trumpet-shaped flowers associated with CpDFRp-CpTCP1B-RD. In addition, all flowers on individual plants showed similar, consistent phenotypes. Examples of plants producing these three types of flowers are shown in Supplementary Figure S3. The flowers generated by these three constructs were further analyzed.
Phenotypic changes were observed to varying degrees in all five CpAP3Bp-CpTCP1B-RD lines of the ‘Crown Violet’ cultivar (Figure 6a). We also generated 12 CpAP3Bp-CpTCP1B-RD lines of the torenia cultivar ‘Summerwave Blue,’ all of which exhibited serrations and/or slight reductions in purple coloration (Figure 6a). We observed vascular structures on the serrated side lobes of CpAP3Bp-CpTCP1B-RD lines (Figure 6b). Vascular branching was enhanced by the introduction of the CpAP3Bp-CpTCP1B-RD construct in both the crown violet and summerwave blue genetic backgrounds.
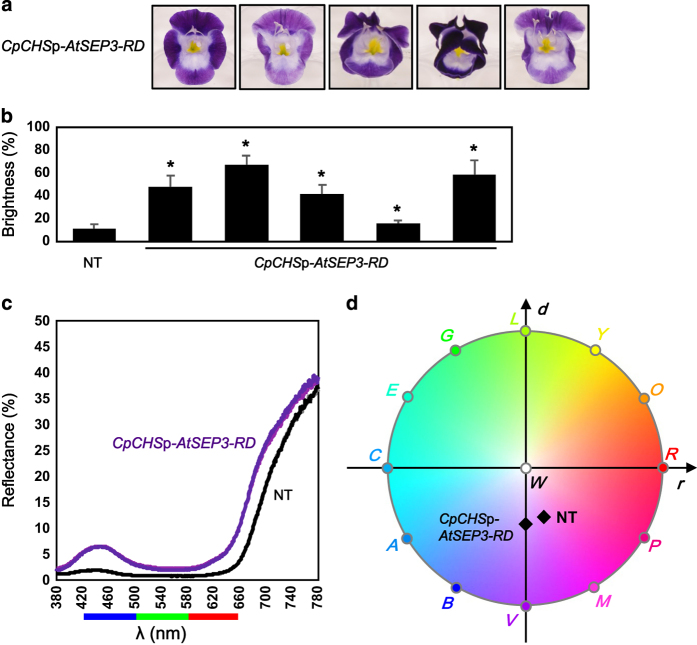
Colorimetric analysis of light-colored flowers
Transgenic lines harboring the CpCHSp-AtSEP3-RD construct produced blotchless and light-colored flowers (Figure 7a). The lobe colors of all five lines were brighter than those of the non-transgenic line (Figure 7b). Although it may not be clear from the digital images (due to a problem with color regeneration from the digital camera), the light-colored transgenic flowers tended to be slightly more bluish than the non-transgenic flowers. To examine this phenomenon more precisely, we analyzed the side lobes of one of the CpCHSp-AtSEP3-RD lines following rigorous colorimetric procedures.
First, we measured the reflectance spectra of visible wavelengths on the lobes with a spectrophotometer (Figure 7c). Wavelengths in the ranges of 420–500, 500–580 and 580–660 nm roughly correspond to blue, green and red coloration, respectively.24 The reflectance recorded in the non-transgenic line consisted of a medium peak in the blue range (~440 nm), low values in the green range, and a large slope in the red range. Reflectance was enhanced for all wavelengths in the CpCHSp-AtSEP3-RD line, but the positions of the blue peak and the red slope were similar to those in the non-transgenic line.
We transformed the visible light spectra of the reflectance of the lobes to RGB color using an ‘RGB color matching function.’ In general, color is composed of three factors: hue (for example, red, yellow, green and purple), lightness (brightness), and saturation (vividness). In colorimetry, hue is expressed as the hue angle (0–360°), while lightness and saturation are expressed as ratios (0–100%). The non-transgenic line and the CpCHSp-AtSEP3-RD line exhibited hue angles (HRGB value) of 290° and 269°, lightness values (LRGB) of 4% and 12%, and saturation values (SRGB2) of 37% and 40%, respectively. The lobe colors were then plotted on a ‘round diagram’ (Figure 7d).
Microscopic observation of trumpet-shaped flowers
Trumpet-shaped flowers were generated in two of the three CpDFRp-CpTCP1B-RD lines (Figure 8a). We examined the effect of this construct on the generation of trumpet-shaped flowers by performing microscopic observations of the side lobes of both the non-transgenic line and a trumpet-shaped CpDFRp-CpTCP1B-RD line.
We directly observed the density of conical cells on the adaxial (upper) and abaxial (lower) surfaces of the side lobes in the non-transgenic line and the CpDFRp-CpTCP1B-RD line through scanning electron microscopy (Figure 8b). The conical cell density was significantly greater on the adaxial surface than on the abaxial surface of the CpDFRp-CpTCP1B-RD line (Figure 8c). Figure 8d illustrates a possible geometric effect of increased conical cell density on the adaxial surface of the lobes, while Figure 8e shows cross-sections of fresh lobes prepared from both the non-transgenic and CpDFRp-CpTCP1B-RD lines.
Discussion
Activities of CPG promoters in torenia and their relationship with flower phenotypes
The sizes of the isolated promoter sequences (all longer than 1.5 kb, except for CpAP3Ap) are expected to be sufficient for the expression of downstream genes because conserved domains are found within 1.0 kb of promoter sequences for dicot MADS-box genes.27 In addition, floral gene promoters ranging from 1394 bp to 2083 bp derived from Arabidopsis or torenia have been successfully used previously for gene expression in torenia petals.16 Different combinations of CPG promoters and chimeric repressors generated different types of flowers in torenia, although some specific combinations did not result in any phenotypic changes (Figure 3). The observed phenotypic changes in the petals fell into three categories: changes in lobe morphology (e.g., serrated lobes), changes in the yellow coloration of the blotch, and changes in the purple coloration of the limbs. Figure 9 shows the phenotypic changes in these categories caused by different combinations of CPG promoters and chimeric repressors (TCP-RD or SEP3-RD). For example, because transgenic lines harboring CpPIp-SEP3-RD constructs exhibited a blotchless phenotype, they were considered modified in category ‘Y’ (yellow) but not in categories ‘L’ (lobes) and ‘P’ (purple). In contrast, transgenic lines harboring CpAP3Bp-TCP-RD constructs were modified in categories ‘L’ and ‘P,’ but these lines always exhibited normal blotch colors.
Figure 9.
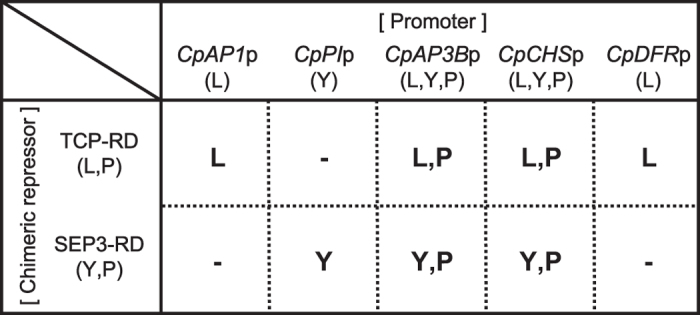
Relationship between CPG promoters, chimeric repressors, and the categories of flower phenotypes. The phenotypes of transgenic torenia flowers generated by a combination of five CPG promoters (CpAP1p, CpPIp, CpAP3Bp, CpCHSp or CpDFRp) and two types of chimeric repressors (TCP-RD or SEP3-RD) were categorized as ‘L’ (lobe morphology), ‘Y’ (yellow color of the blotch), or ‘P’ (purple color of the petal). The potential effects of the CPG promoters and chimeric repressors, as deduced from the flower phenotypes, are also indicated in parentheses.
This categorization of the observed phenotypic changes in the transgenic lines allowed us to deduce the capacity of CPG promoters and chimeric repressors to modify these respective categories of flower phenotypes. Thus, TCP repressors can modify L-type and P-type phenotypes, while SEP3 repressors can modify Y-type and P-type phenotypes. CpAP1p and CpDFRp can modify L-type phenotypes; CpPIp can modify Y-type phenotypes; and CpAP3Bp and CpCHSp can modify all three categories. Each phenotype category is modified only when both the promoter and repressor have the ability to modify it. This model, based on the differential activities of both CPG promoters and chimeric repressors, would reasonably explain the variable flower phenotypes generated by different constructs.
The tissue-specific activities of the four promoters (Figure 2) are also consistent with this model. CpPIp activity was specifically detected in the blotch, which explains why CpPIp can modify only the Y-type phenotype and why the flower throat remained yellow in the lines possessing the CpPIp-SEP3-RD construct. In contrast, CpAP3Bp activity was observed in the whole petal, explaining why this promoter can modify all three phenotypic categories. CpDFRp was only weakly active in the limbs, and this promoter specifically modified L-type phenotypes. L-type phenotypes may be modified by TCP repressors more easily compared with P-type phenotypes. No activity of CpOMTp was detected in our experiments, and this promoter accordingly did not cause any changes in flower phenotypes. As CpAP3Ap or CpSEP2p also did not cause phenotypic changes, these two promoters may not be active in torenia flowers.
The observed activity of CpAP3Bp in the petals and stamens is in accord with its phylogenetic classification as a class-B MADS-box gene, although no phenotypic changes were observed in the stamens in this study. The blotch-specific activity of CpPIp was unexpected, considering its classification as a class-B MADS-box gene, and the GUS staining associated with CpDFRp in this study appeared weaker than the GUS staining associated with the torenia DFR promoter in a previous study.16 Torenia may be lacking in trans regulators that properly recognize and strongly drive the transgenic cis regulators CpPIp and CpDFRp. Due to their specific expression patterns, the activities of these promoters may result in a lack of phenotypic changes in transgenic torenia in some cases and unexpected flower phenotypes in others.
Role of TCP transcription factors in the regulation of cell proliferation
TCP transcription factors are known to regulate cell proliferation.28 Among the TCP family genes, AtTCP3 and CpTCP1 are close homologs of the CINCINNATA gene (AmCIN) of snapdragon (Antirrhinum majus; ref. 15). A mutation in the AmCIN gene causes crinkly (curly) leaves, particularly at the margins, and arrests the growth of the lobes in snapdragon,29 while a snapdragon mutant harboring mutations in the TCP genes DICHOTOMA and CYCLOIDEA exhibits a rounded corolla.30 Thus, TCP has various effects on flower morphogenesis.
The curly lobes observed in the CpDFRp-CpTCP1B-RD line (Figure 8a) were consistent with the mutant phenotype of the AmCIN gene in snapdragon. Outer curling of the lobes (that is, epinasty) is caused by an imbalance in cell proliferation and/or cell expansion between the adaxial (inner) and abaxial (outer) surfaces of the lobes. Microscopic analysis revealed an increased density of conical cells only on the adaxial surface of the CpDFRp-CpTCP1B-RD line (Figure 8c), indicating that CpTCP1B-RD accelerated cell proliferation on the adaxial surface of transgenic plants. The conical cells of the CpDFRp-CpTCP1B-RD line also tended to be flatter than those of the non-transgenic line (Figure 8e), similar to what was observed in a snapdragon AmCIN gene mutant.31
In addition to the petal curvature caused by CpDFRp-CpTCP1B-RD, the TCP repressors also generated complicated serrations in leaves and petals (Figures 1 and 6). The effects of TCP repressors on serrations can be divided into two categories. First, TCP repressors cause deeper primary serrations. This effect is consistent with the CUC2 (CUp-shaped Cotyledon 2)-mediated formation and regulation of leaf serrations.32 Indeed, CUC2 expression is elevated in Arabidopsis plants with mutations in TCP genes.33 Second, TCP repressors generate small secondary serrations in leaves (Figure 1f) and possibly in petals (Figure 6b). The positions of the secondary serrations appear to overlap with the positions at which additional vasculature is formed in transgenic lines. This positive and possibly simultaneous regulation of the formation of secondary serrations and vascular branching should be independent of the CUC2 pathway because CUC2 is known to neither induce secondary serrations nor regulate vascular branching.32
A mutation in the FRILL1 gene also causes frilled flowers in Arabidopsis,34 and the vascular pattern is altered in such flowers.35 The frilled phenotype of this mutant is attributed to the differential regulation of endoreduplication,36 a phenomenon in which some diploid (2C) cells are converted to tetraploid (4C), octaploid (8C) or hexadecaploid (16C) cells with an enlarged cell size. Cell ploidy analyses have shown that endoreduplication frequently occurs in vegetative cells of Arabidopsis (a diploid) (for example see refs. 37,38). However, there is no sign of endoreduplication in diploid torenia leaves; i.e., there is only a single peak corresponding to diploid (2C) cells (Kasajima et al., unpublished). Although we do not have sufficient data to reach a definite conclusion, vascular branching, rather than endoreduplication, may be linked to the serrations observed in torenia.
Phenotypic changes caused by SEP3 repressors are not necessarily strong, but the reduction of purple coloration may be a sign of the conversion of petals to sepals. This possibility could be examined using gene expression analysis in future studies.
Why do transgenic plants with CpCHSp-SEP3-RD constructs produce bluish flowers?
CpCHSp-SEP3-RD constructs generated blotchless, light-colored flowers (Figure 3). The characteristics of these flowers lie partly in the simple balance of colors, in the absence of a yellow blotch, and in lighter lobe colors. The lobe coloration of these lines also appeared somewhat bluish when directly observed with the naked eye, which was confirmed by colorimetric analysis (Figure 7). Judging from the obtained colorimetric values, the non-transgenic flowers were magenta (red-purple), while the CpCHSp-AtSEP3-RD flowers were violet (blue-purple) and three times lighter. The purple petal color of torenia is caused by anthocyanins.26
The color of purple flowers is affected by changes in the spectra of incident light. Therefore, the abovementioned flower colors (magenta and violet) were calculated under a hypothetical ‘flat’ white light, which mimics the solar spectrum, with an even distribution of spectral power across all visible wavelengths (380–780 nm). The color of purple flowers, including that of purple torenia flowers, becomes bluish under white light-emitting diode (LED) light and reddish under incandescent light due to the colorimetric phenomenon known as the ‘alexandrite effect’ or ‘color change’.24 This may be the principal reason that the color (hue) of a given purple flower appears different between images.
The observed difference in the hues between the non-transgenic line and the CpCHSp-AtSEP3-RD line may have been caused by a colorimetric phenomenon other than changes in the chemical structure of anthocyanins. In a previous report, a bluish torenia line (cultivar ‘Crown Violet’) was generated by suppressing the DFR gene.26,39 Suppression of the DFR gene modified co-pigment accumulation and shifted the spectral pattern of light absorbance to the right (that is, to longer wavelengths). However, there was no obvious shift or changes in the shape of the reflectance spectrum in the CpCHSp-AtSEP3-RD line (Figure 7c), except that the entire spectrum was magnified to approximately three times that of the non-transgenic line. Although we did not measure anthocyanin concentrations in this study, we deduced that ‘dichromatism’ causes the hue to change in the presence of different concentrations of purple anthocyanins. Strong red reflectance will cause the flower color to be reddish at high anthocyanin concentrations in the non-transgenic line, whereas intermediate blue reflectance will be enhanced in the presence of lower concentrations of purple anthocyanins in the CpCHSp-AtSEP3-RD line, causing the flower to become bluish. The exponential relationship between light transmittance and pigment concentrations, as described by the Beer-Lambert law, causes this kind of alteration in the balance between the intensities of different wavelengths at different pigment concentrations.23 The concentration and chemical structure of anthocyanins in the petals of CpCHSp-AtSEP3-RD lines should be examined in the future to obtain further insight into the color of these flowers.
Conclusion
In this study, we newly isolated 8 promoters of genes expressed in cyclamen petals and prepared 32 constructs. We then identified 13 genetic constructs that modify flower phenotypes, including CpCHSp-AtSEP3-RD, CpDFRp-CpTCP1B-RD, and CpAP3Bp-CpTCP1B-RD, by ‘screening’ the constructs in the model flower torenia. These three constructs generated pale bluish petals, trumpet-like petals, and serrated petals, respectively. We also examined the cellular effects underlying these phenotypic changes and deduced that the changes are linked to dichromatism, imbalanced cell proliferation, and excessive vascular branching, respectively. Taken together, the information and genetic resources generated in this study will benefit future molecular flower breeding efforts and further characterization of these valuable transgenic plants.
Acknowledgments
We thank Satoko Ohtawa for assistance in the generation of transgenic plants, Ms. Yoshiko Kashiwagi for assistance in the maintenance of potted plants, Hiroyasu Yamaguchi for assistance in the cell ploidy analysis, and Takaaki Nishijima and Tomoya Niki for their advice regarding microscopy. We also appreciate the kind cooperation of researchers at Hokko Chemical Industry Co., Ltd. in the analysis of cyclamen genes. The CpSEP3 and CpTCP1B genes were kindly provided by Hokko Chemical Industry. This work was supported by the Scientific Technique Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (Japan).
Footnotes
Supplementary Information for this article can be found on the Horticulture Research website (http://www.nature.com/hortres)
The authors declare no conflict of interest.
References
- Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, Brugliera F, Holton TA, Karan M et al. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol 2007; 48: 1589–1600. [DOI] [PubMed] [Google Scholar]
- Klocko AL, Borejsza-Wysocka E, Brunner AM, Shevchenko O, Aldwinckle H, Strauss SH. Tarnsgenic suppression of AGAMOUS genes in apple reduces fertility and increases floral attractiveness. PLoS ONE 2016; 11: e0159421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N, Aida R, Kishimoto S, Ishiguro K, Fukuchi-Mizutani M, Tanaka Y et al. Genetic engineering of novel bluer-colored chrysanthemums produced by accumulation of delphinidin-based anthocyanins. Plant Cell Physiol 2013; 54: 1684–1695. [DOI] [PubMed] [Google Scholar]
- Ono E, Fukuchi-Mizutani M, Nakamura N, Fukui Y, Yonekura-Sakakibara K, Yamaguchi M et al. Yellow flowers generated by expression of the aurone biosynthetic pathway. Proc Natl Acad Sci USA 2006; 103: 11075–11080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Aida R, Niki T, Yamaguchi H, Narumi T, Nishijima T et al. High-efficiency improvement of transgenic torenia flowers by ion beam irradiation. Plant Biotechnol 2008; 25: 81–89. [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 2001; 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett 2002; 514: 351–354. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain in Arabidopsis. Plant J 2003; 34: 733–739. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Umemura Y, Ikeda M, Shikata M, Koyama T, Matsui K et al. FioreDB: a database of phenotypic information induced by the chimeric repressor silencing technology (CRES-T) in Arabidopsis and floricultural plants. Plant Biotechnol 2008; 25: 37–43. [Google Scholar]
- Mitsuda N, Takiguchi Y, Shikata M, Sage-Ono K, Ono M, Sasaki K et al. The new FioreDB database provides comprehensive information on plant transcription factors and phenotypes induced by CRES-T in ornamental and model plants. Plant Biotechnol 2011; 28: 123–130. [Google Scholar]
- Shikata M, Narumi T, Yamaguchi H, Sasaki K, Aida R, Oshima Y et al. Efficient production of novel floral traits in torenia by collective transformation with chimeric repressors of Arabidopsis transcription factors. Plant Biotechnol 2011; 28: 189–199. [Google Scholar]
- Gion K, Suzuri R, Shikata M, Mitsuda N, Oshima Y, Koyama T et al. Morphological changes of Rosa x hybrida by a chimeric repressor of Arabidopsis TCP3. Plant Biotechnol 2011; 28: 149–152. [Google Scholar]
- Narumi T, Aida R, Koyama T, Yamaguchi H, Sasaki K, Shikata M et al. Arabidopsis chimeric TCP3 repressor produces novel floral traits in Torenia fournieri and Chrysanthemum morifolium. Plant Biotechnol 2011; 28: 131–140. [Google Scholar]
- Shikata M, Ohme-Takagi M. The utility of transcription factors for manipulation of floral traits. Plant Biotechnol 2008; 25: 31–36. [Google Scholar]
- Tanaka Y, Yamamura T, Oshima Y, Mitsuda N, Koyama T, Ohme-Takagi M et al. Creating ruffled flower petals in Cyclamen persicum by expression of the chimeric cyclamen TCP repressor. Plant Biotechnol 2011a; 28: 141–147. [Google Scholar]
- Sasaki K, Yamaguchi H, Kasajima I, Narumi T, Ohtsubo N. Generation of novel floral traits using a combination of floral organ-specific promoters and a chimeric repressor in Torenia fournieri Lind. Plant Cell Physiol 2016; 57: 1319–1331. [DOI] [PubMed] [Google Scholar]
- Kasajima I, Sasaki K, Tanaka Y, Terakawa T, Ohtsubo N. Large-scale extraction of pure DNA from mature leaves of Cyclamen persicum Mill. and other recalcitrant plants with alkaline polyvinylpolypyrrolidone (PVPP). Sci Hortic 2013; 164: 65–72. [Google Scholar]
- Kasajima I. Protocol for DNA extraction from any plant species (alkaline PVPP method). Protocol Exchange 2017; doi:10.1038/protex.2017.004.
- Kasajima I, Ohtsubo N, Sasaki K. Faster, safer, and better DNA purification by ultracentrifugation using GelRed stain and development of mismatch oligo DNA for genome walking. Biosci Biotechnol Biochem 2014; 78: 1902–1905. [DOI] [PubMed] [Google Scholar]
- Oshima Y, Mitsuda N, Nakata M, Nakagawa T, Nagaya S, Kato K et al. Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T). Plant Biotechnol 2011; 28: 201–210. [Google Scholar]
- Aida R, Shibata M. Agrobacterium–mediated transformation of torenia (Torenia fournieri). Breed Sci 1995; 45: 71–74. [Google Scholar]
- Yamaguchi H, Sasaki K, Shikata M, Aida R, Ohtsubo N. Trehalose drastically extends the in vitro vegetative culture period and facilitates maintenance of Torenia fournieri plants. Plant Biotechnol 2011; 28: 263–266. [Google Scholar]
- Kasajima I, Sasaki K. Dichromatism causes color variations in leaves and spices. Color Res Appl 2015; 40: 605–611. [Google Scholar]
- Kasajima I. Alexandrite-like effect in purple flowers analyzed with newly devised round RGB diagram. Sci Rep 2016; 6: 29630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Yamamura T, Terakawa T. Identification and expression analysis of the Cyclamen persicum MADS-box gene family. Plant Biotechnol 2011b; 28: 167–172. [Google Scholar]
- Aida R, Yoshida K, Kondo T, Kishimoto S, Shibata M. Copigmentation gives bluer flowers on transgenic torenia plants with the antisense dihydroflavonol-4-reductase gene. Plant Sci 2000b; 160: 49–56. [DOI] [PubMed] [Google Scholar]
- Bodt SD, Theissen G, de Peer YV. Promoter analysis of MADS-box genes in eudicots through phylogenetic footprinting. Mol Biol Evol 2006; 23: 1293–1303. [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 1999; 18: 215–222. [DOI] [PubMed] [Google Scholar]
- Nath U, Crawford BCW, Carpenter R, Coen E. Genetic control of surface curvature. Science 2003; 299: 1404–1407. [DOI] [PubMed] [Google Scholar]
- Cronk QCB. Plant evolution and development in a post-genomic context. Nat Rev Genet 2001; 2: 607–619. [DOI] [PubMed] [Google Scholar]
- Crawford BCW, Nath U, Carpenter R, Coen ES. CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol 2004; 135: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C et al. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci USA 2011; 108: 3424–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Sato F, Ohme-Takagi M. A role of TCP1 in the longitudinal elongation of leaves in. Arabidopsis Biosci Biotech Biochem 2010; 74: 2145–2147. [DOI] [PubMed] [Google Scholar]
- Hase Y, Tanaka A, Baba T, Watanabe H. FRL1 is required for petal and sepal development in Arabidopsis. Plant J 2000; 24: 21–32. [DOI] [PubMed] [Google Scholar]
- Carland F, Fujioka S, Takatsuto S, Yoshida S, Nelson T. The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 2002; 14: 2045–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase Y, Fujioka S, Yoshida S, Sun G, Umeda M, Tanaka A. Ectopic endoreduplication caused by sterol alteration results in serrated petals in Arabidopsis. J Exp Bot 2005; 56: 1263–1268. [DOI] [PubMed] [Google Scholar]
- Kasajima I, Yoshizumi T, Ichikawa T, Matsui M, Fujiwara T. Possible involvement of ploidy in tolerance to boron deficiency in Arabidopsis thaliana. Plant Biotechnol 2010; 27: 435–445. [Google Scholar]
- Yoshizumi T, Tsumoto Y, Takiguchi T, Nagata N, Yamamoto YY, Kawashima M et al. Increased level of POLYPLOIDY1, a conserved repressor of CYCLINA2 transcription, controls endoreduplication in Arabidopsis. Plant Cell 2006; 18: 2452–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida R, Kishimoto S, Tanaka Y, Shibata M. Modification of flower color in torenia (Torenia fournieri Lind.) by genetic transformation. Plant Sci 2000a; 153: 33–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.