Abstract
Antiretroviral treatment history and past HIV-1 genotypes have been shown to be useful predictors for the success of antiretroviral therapy. However, this information may be unavailable or inaccurate, particularly for patients with multiple treatment lines often attending different clinics. We trained statistical models for predicting drug exposure from current HIV-1 genotype. These models were trained on 63,742 HIV-1 nucleotide sequences derived from patients with known therapeutic history, and on 6,836 genotype-phenotype pairs (GPPs). The mean performance regarding prediction of drug exposure on two test sets was 0.78 and 0.76 (ROC-AUC), respectively. The mean correlation to phenotypic resistance in GPPs was 0.51 (PhenoSense) and 0.46 (Antivirogram). Performance on prediction of therapy-success on two test sets based on genetic susceptibility scores was 0.71 and 0.63 (ROC-AUC), respectively. Compared to geno2pheno[resistance], our novel models display a similar or superior performance. Our models are freely available on the internet via www.geno2pheno.org. They can be used for inferring which drug compounds have previously been used by an HIV-1-infected patient, for predicting drug resistance, and for selecting an optimal antiretroviral therapy. Our data-driven models can be periodically retrained without expert intervention as clinical HIV-1 databases are updated and therefore reduce our dependency on hard-to-obtain GPPs.
1. Introduction
Prolonged chemotherapy against the human immunodeficiency virus type 1 (HIV-1) bears the risk of selection of resistant viral strains, ultimately leading to therapy failure [1–6]. Once a drug-resistant HIV-1 variant has been selected in a host, it can be transmitted to another host [6,7]. Furthermore, drug-resistant viral variants are permanently archived in the body of the host and can promptly reemerge if drug pressure conveys them a competitive advantage to other viral variants [8]. In order to prevent premature therapy failure, the susceptibility of an HIV-1 variant to available antiretroviral drugs can be measured phenotypically or genotypically [4,9–12]. Due to the high cost, limited accessibility and high turnaround time of phenotypic resistance assays, genotypic resistance determination has become the standard of care [4,9]. Phenotypic resistance assays afford direct, quantitative resistance assessments that take into account resensitizing mutations [13], as well as complex mutational patterns [14]. However, certain drugs show significantly decreased in-vivo efficacy at very low in-vitro susceptibility changes which are close to the inherent variability of the phenotypic assay [15]. Furthermore, viral strains with mutations that do not directly cause resistance, but are strongly associated with the emergence of drug resistance, may be deemed susceptible by in-vitro phenotypic drug-resistance assays. If the respective drugs are taken by patients harboring these strains, resistant variants will promptly emerge and compromise virologic response to therapy [16].
Determination of genotypic resistance is performed by sequencing the viral genes coding for the targets of antiretroviral drugs, and subsequently interpreting the resulting nucleotide sequence [12]. A handful of tools exist for interpreting HIV-1 genotypes with respect to drug resistance. Drug-resistance mutation tables list amino acid mutations that confer resistance to antiretroviral drugs [14,17]. Rules-based genotypic interpretation systems score an HIV-1 genotype according to a set of rules defined by experts. The score for each drug is subsequently discretized into two to five categories indicating increasing levels of resistance [18,19]. Data-driven genotypic interpretation systems rely on statistical models of drug resistance for interpreting an HIV-1 genotype. These models are trained on sets usually containing genotype-phenotype pairs (GPP) [20,21] generated with in vitro phenotypic assays, and can thus potentially inherit their advantages and disadvantages.
HIV-1 substitutions resulting from chemotherapy are frequently divided in two groups: major drug-resistance mutations and minor drug resistance mutations, which can also occur as natural polymorphisms [14,17,22–26]. While there is no consensus on the definition of these two groups of mutations, in the following, we list the defining criteria that tend to be used. Major drug resistance mutations are frequently present in viral genotypes from patients failing antiretroviral therapy, and appear very rarely in HIV-1 genotypes from therapy-naïve patients. In fact, detection of such mutations in drug-naïve patients is currently interpreted as transmission of a resistant variant from patients who have failed therapy. By themselves, major drug-resistance mutations can either be directly responsible for drug resistance, or be informative markers for drug resistance. The implications of a mutation with respect to drug resistance can be investigated through site-directed mutagenesis with subsequent phenotypic resistance testing of the produced viral variant [27]. Minor drug resistance mutations tend to be polymorphic, and do not cause drug resistance by themselves, although they may further decrease susceptibility to a drug in combination with major drug resistance mutations and / or compensate for decreased replicative capacity resulting from selection of major mutations. In population genetics, a polymorphism is defined as a substitution that is present in more than one percent of the population [26,28]. The role HIV-1 polymorphisms play in chemotherapeutic success remains controversial [23,24,29–31]. Certain polymorphisms may tend to accumulate during chemotherapy while also being present in drug-naïve patients, albeit with a reduced frequency [22,23]. Polymorphisms present at baseline may influence the drug susceptibility of an HIV-1 variant [26,30–32]. Differential polymorphism distribution among HIV-1 subtypes has been observed, however, significant implications for drug susceptibility only seem to originate from intra-subtype variability as opposed to inter-subtype variability [24,25,32]. The most convincing explanation for the subtype-specific distribution of natural polymorphisms seems to be the existence of subtype-specific resistance pathways rather than subtype-specific propensity for selecting drug resistance [32].
Before drug resistance assays became available, treatment history was frequently used for the selection of new drug regimens [33]. Nowadays, new drug regimens are sometimes selected on the basis of treatment history when no drug resistance test is available. Indeed, statistical models that use treatment history in place of the genotype for predicting the success of antiretroviral therapy have been reported to be comparable to those of models that use the genotype (and do not use therapy history) [34–37]. However, to our knowledge, these methods have not yet found their way into clinical practice. In our experience, in today’s settings using therapy history as a proxy for genotype incurs substantial loss of predictive power. At the same time, a statistically significant increase in performance can be achieved by simultaneously using treatment history and the genotype for predicting the success of antiretroviral therapy [34,38–43].
Drug exposure can be predicted from genotype since the virus acquires mutations as a result of being exposed to a drug. These mutations encompass but are not limited to drug-resistance mutations. Thus, while some of these mutations may indicate clinically relevant drug resistance, others may also solely indicate that the virus has changed as a result of drug exposure. As drug susceptibility is a prerequisite for the success of antiretroviral therapy, the detection of drug exposure may pose a risk for therapeutic success (Fig 1). Reporting drug exposure from genotype is relevant if either no established resistance-associated mutations are detectable and / or in cases in which no treatment-history information is available. In this work, we present statistical models that use HIV-1 genotypes to produce predictions of drug-exposure that are correlated with both therapeutic history and drug resistance. We have developed our method in close contact with prospective users. From the resulting experience, we expect the method to provide a significant clinical advance in bioinformatics-based therapy-success prediction.
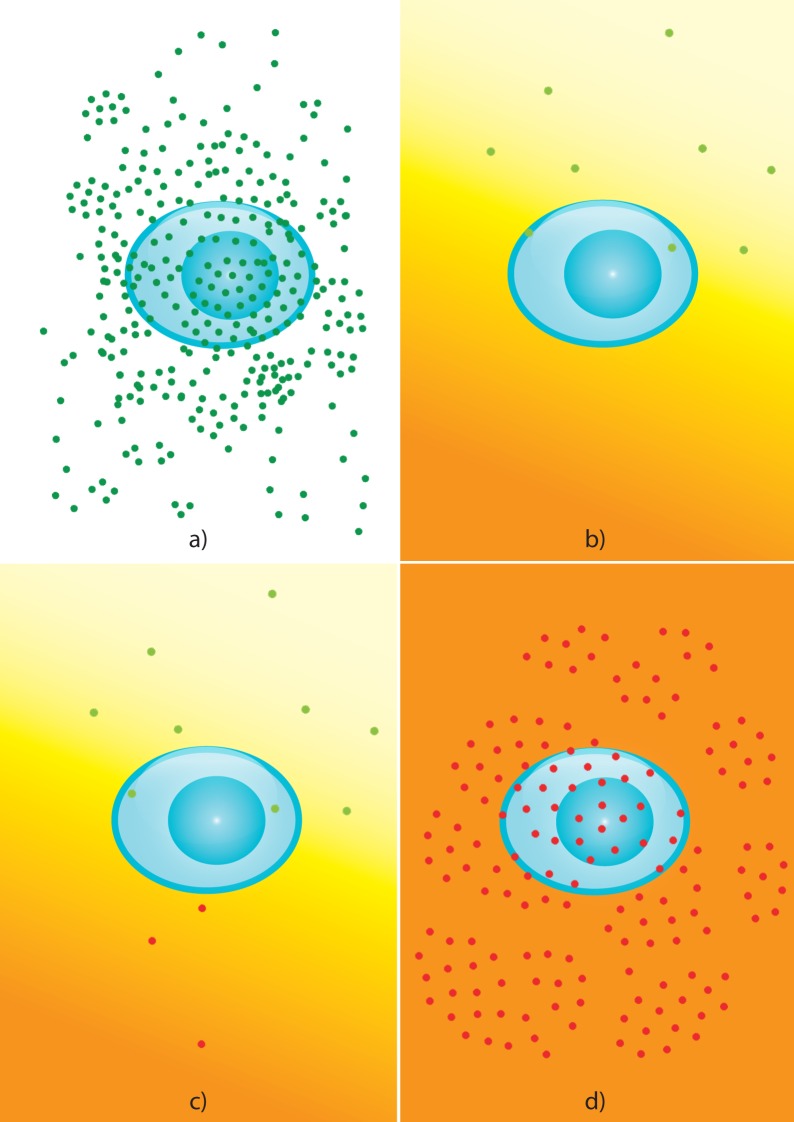
Fig 1. Relationship between drug exposure, drug resistance, and therapeutic success.
a) Prior to drug exposure, the virus typically does not carry drug-resistance mutations. In the absence of drug pressure, drug-susceptible virus can replicate at high titers (dark-green viral particles). b) If drug susceptibility is given, antiretroviral therapy frequently leads to the suppression of viral replication, which is a prerequisite for therapeutic success. While antiretroviral therapy is administered, however, drug concentrations fluctuate over the dosing interval and may vary within the different body compartments (orange-yellow gradient). This can give rise to sub-inhibitory concentrations in some compartments (light-yellow area in gradient), resulting in the selection of mutations that confer to the virus a selective advantage in the presence of the drug (light-green viral particles). These mutations need not result in virological therapy failure, since they may not enable the virus to replicate at high drug concentrations. c) Recurrence of sub-inhibitory drug concentrations can ultimately select for mutations that enable the virus to replicate even at the highest drug concentrations (red viral particles). d) The selection of drug-resistant virus leads to virological therapy failure: the virus replicates at high titers in spite of antiretroviral therapy.
Note that this article is largely based on another publication from which we amply quote [44].
2. Results
We trained models for predicting whether an HIV-1 variant had been previously exposed to a certain drug. One or two models were trained for each of the drugs considered in this study (Methods). Specifically, Exposure models were trained with HIV-1 sequences and information on drug exposure. The development sets of ExposurePheno models included genotype-phenotype pairs (GPPs) in addition to the data included in Exposure models. Since a sufficient number of HIV-1 sequences with information on drug-exposure was not available for all drugs, Exposure models could not be trained for all drugs. Additionally, we trained a model for discriminating between HIV-1 sequences from treatment-naïve patients and HIV-1 sequences from treatment-experienced patients. In the following, we refer to a number of datasets that we used for training and validating our models. For the comfort of the reader, we summarize the contents of each of these datasets in Table 1. Furthermore, we depict the relationships of each of the datasets in Figure A in S1 File.
Table 1. Dataset cheat sheet.
| Dataset | Description | Input Variables | Target Variables |
|---|---|---|---|
| PRRT | Protease and reverse-transcriptase sequences from the EIDB and the LANLSD, along with the drug compounds previously used by the patient at the time of sequencing. | Sequence of protease and reverse transcriptase | Binary drug-exposure label for each protease inhibitor or reverse-transcriptase inhibitor |
| IN | Integrase sequences from the EIDB and the LANLSD, along with the drug compounds previously used by the patient at the time of sequencing. | Sequence of integrase | Binary drug-exposure label for each integrase inhibitor |
| TP | Past drug compounds and sequences in PRRT and in IN that were obtained during therapy pause. | Sequence of protease and reverse-transcriptase or integrase | Binary drug-exposure label for each drug |
| TPRRT | Test set of protease and reverse-transcriptase sequences and drug-exposure information. | Sequence of protease and reverse transcriptase | Binary drug-exposure label for each protease inhibitor or reverse-transcriptase inhibitor |
| TIN | Test set of integrase sequences and drug-exposure information. | Sequence of integrase | Binary drug-exposure label for each integrase inhibitor |
| DPRRT | Development set of protease and reverse-transcriptase sequences and drug-exposure information. | Sequence of protease and reverse transcriptase | Binary drug-exposure label for each protease inhibitor or reverse-transcriptase inhibitor |
| DIN | Development set of integrase sequences and drug-exposure information. | Sequence of integrase | Binary drug-exposure label for each integrase inhibitor |
| EuResistTCE | Test set of TCEs. Each TCE contains a protease and reverse-transcriptase baseline sequence, the drug compounds that were used in the therapy, and a label indicating therapeutic success or failure. | Baseline protease and reverse-transcriptase sequence for therapy | Binary therapy-success label |
| EuResistTCETP | Test set of TCEs whose baseline sequences were obtained during a therapy pause. Each TCE contains a protease and reverse-transcriptase baseline sequence, the drug compounds that were used in the therapy, and a label indicating therapeutic success or failure. | Baseline protease and reverse-transcriptase sequence for therapy | Binary therapy-success label |
| HIVdbExposure | Test set of protease and reverse-transcriptase sequences and drug-exposure information. | Sequence of protease and reverse transcriptase | Binary drug-exposure label for each protease inhibitor or reverse-transcriptase inhibitor |
| HIVdbTCE | Test set of TCEs. Each TCE contains a protease and reverse-transcriptase baseline sequence, the drug compounds that were used in the therapy, and a label indicating therapeutic success or failure. | Baseline protease and reverse-transcriptase sequence for therapy | Binary therapy-success label |
| Pheno | Dataset of GPPs. | Protease, reverse-transcriptase or integrase sequence | Resistance factors for different drugs |
| TPheno | Test set of GPPs. | Protease, reverse-transcriptase or integrase sequence | Resistance factors for different drugs or resistance categories |
| DPheno | Development set of genotype-phenotype pairs. | Protease, reverse-transcriptase or integrase sequence | Resistance factors for different drugs or resistance categories |
| NaïvePRRT | Dataset of protease and reverse-transcriptase sequences from treatment-naïve patients without TDR mutations. | Sequence of protease and reverse transcriptase | None |
| NaïveIN | Dataset of integrase sequences from treatment-naïve patients without TDR mutations. | Sequence of integrase | None |
| Exposuredrug | Cross-validation / development set for the compound drug. These datasets include sequences with drug exposure information but not GPPs. | Protease, reverse-transcriptase or integrase sequence | Binary drug-exposure label for drug |
| ExposurenaïvePRRT | Cross-validation /development set for models discriminating sequences from treatment-exposed and treatment-naïve patients. | Protease and reverse-transcriptase sequence | Binary label indicating whether sequence was obtained from therapy-naïve patient |
| ExposurePhenodrug | Cross-validation / development set for the compound drug. These datasets include sequences with drug exposure information and GPPs. | Protease, reverse-transcriptase or integrase sequence | Binary label indicating exposure or resistance to drug. Note that this label does not distinguish between drug exposure and drug resistance |
In the table above, the names of the datasets used in this study are tabulated along with a short description of their contents. The datasets are shown in order of appearance in Methods. Above, the term sequences refers to HIV-1 nucleotide sequences.
EIDB: EuResist Integrated Database; GPP: genotype-phenotype pair; LANLSD: Los Alamos National Laboratory Sequence Database; TCE: Therapy-Change Episode; TDR: transmitted drug resistance.
2.1. Dataset preparation
Prior to alignment, 48,666 nucleotide sequences with information on drug exposure were extracted from the EuResist Integrated Database (EIDB; http://www.euresist.org) [45] and aligned to reference sequences for the viral protease, reverse-transcriptase, and integrase. The alignment procedure yielded 38,754 sequences for protease and reverse-transcriptase (assigned to the PRRT dataset) and 6,214 integrase sequences (assigned to the IN dataset). PRRT and IN were further complemented with 36,774 and 5,262 sequences from therapy-naïve patients (short: therapy-naïve sequences), respectively, from the Los Alamos National Laboratory Sequence Database (LANLSD; http://www.hiv.lanl.gov/). The number of sequences in PRRT was reduced to 75,239 sequences after excluding sequences with more than 10% undetermined residues. After removal of duplicate sequences, PRRT included a total of 70,304 sequences (approximately 93% of the initially included sequences). The number of sequences in IN was reduced to 7,076 after excluding sequences with more than 10% undetermined residues. After duplicate removal, 5,523 sequences (approximately 48%) were left in IN. The number of sequences per subtype for PRRT and IN can be seen in Table 2. Sequences in PRRT and IN were randomly assigned either to the development sets DPRRT and DIN, respectively, or to the test sets TPRRT and TIN, respectively (Methods). Two additional test sets were created, TP and HIVdbExposure. TP contains sequences from TPRRT and TIN which were obtained during therapy pauses. HIVdbExposure was created from the treatment-change episode (TCE) repository in the HIV Drug Resistance Database (HIVdb) [14,46]. It contains nucleotide sequences and lists the sets of drug compounds that had been used by the patient before the sequence was obtained. The distribution of subtypes per sequence in HIVdbExposure can be seen in Table 2. Table 3 shows the number of sequences in datasets DPRRT, DIN, TPRRT, TIN, TP, and HIVdbExposure by drug exposure. In DPRRT, 37,557 sequences were therapy-naïve, of which 3,757 (10.0%) present transmitted drug resistance (TDR) [7]. A total of 1,917 sequences in DIN are therapy-naïve, of which 48 (approximately 2%) present TDR. Note that the duplicate removal procedure eliminated substantially more sequences from IN than from PRRT. TPRRT contains 2,056 therapy-naïve sequences, among which 219 (approximately 11%) present TDR, while TIN contains 154 therapy-naïve sequences with 3 (approximately 2%) presenting TDR. We applied the definition of the EuResist Standard Datum [40] to clinical HIV data in the EIDB and in the HIVdb TCE repository. This yielded two datasets of TCEs with binary labels for therapeutic success, the EuResistTCE (n = 1,650) and the HIVdbTCE (n = 1,000) datasets. Fig 2a) depicts the most frequent therapies in the EuResistTCE, while Fig 2b) does so for the TCEs in HIVdbTCE. The baseline sequences in EuResistTCE overlap with the sequences in PRRT partially; the baseline sequences of 619 TCEs are not included in PRRT. TCEs in EuResistTCE whose baseline sequences were obtained during a therapy pause were assigned to the EuResistTCETP test set. EuResistTCE contains 313 first-line therapies (19.0%) among which 44 (14.1%) present TDR in their baseline sequences. No therapy in HIVdbTCE is a first-line therapy. The Pheno dataset contains GPPs which were labeled susceptible or resistant using the resistance-factor (RF) cutoffs one and ten. Pheno was randomly split into the development and training sets DPheno and TPheno, respectively (Methods). The compositions of DPheno and TPheno are displayed in Table 4.
Table 2. Number of nucleotide sequences by subtype and dataset.
| Subtype | PRRT | IN | HIVdbExposure |
|---|---|---|---|
| B | 42,634 (61%) | 2,721 (49%) | 1,377 (1%) |
| C | 6,243 (9%) | 1,293 (23%) | 1 (< 1%) |
| A1 | 3,704 (5%) | 166 (3%) | 1 (< 1%) |
| G | 3,223 (5%) | 270 (5%) | 0 (0%) |
| 02_AG | 3,010 (4%) | 66 (1%) | 1 (< 1%) |
| 01_AE | 4,275 (6%) | 596 (11%) | 1 (< 1%) |
| D | 1,169 (2%) | 53 (1%) | 1 (< 1%) |
| F1 | 971 (1%) | 69 (1%) | 0 (0%) |
| 06_cpx | 312 (< 1%) | 89 (2%) | 0 (0%) |
| 07_BC | 651 (1%) | 4 (< 1%) | 0 (0%) |
| Other | 4,112 (6%) | 196 (4%) | 2 (< 1%) |
| Total | 70,304 | 5,523 | 1,381 |
Nucleotide sequences in the PRRT, IN, and HIVdbExposure datasets were subtyped with the Comet subtyping tool. Sequence counts for the ten most frequent subtypes are tabulated above. For each dataset, the percentage of nucleotide sequences with a particular subtype are stated in parenthesis.
Table 3. Number of sequences by dataset and drug exposure.
| DPRRT | DPRRT Comp. | DIN | DIN Comp. | TPRRT | TPRRT Comp. | TIN | TIN Comp. | TP | TP Comp. | HIVdbExposure | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ABC | 7,482 | 4,560 | 295 | 229 | 1,839 | 1,028 | 164 | 103 | 163 | 30 | 301 |
| AZT | 18,542 | 12,184 | 441 | 336 | 3,895 | 2,405 | 222 | 135 | 372 | 68 | 1,075 |
| d4T | 13,335 | 8,079 | 259 | 197 | 2,956 | 1,764 | 141 | 80 | 250 | 31 | 998 |
| ddC | 4,007 | 2,341 | 57 | 45 | 1,114 | 750 | 52 | 40 | 71 | 6 | 297 |
| ddI | 12,113 | 7,398 | 227 | 173 | 2,725 | 1,657 | 123 | 73 | 197 | 23 | 722 |
| FTC | 4,580 | 3,258 | 359 | 266 | 900 | 595 | 162 | 112 | 52 | 8 | 59 |
| 3TC | 20,730 | 13,416 | 525 | 394 | 4,191 | 2,543 | 262 | 151 | 390 | 70 | 0 |
| TDF | 9,546 | 6058 | 479 | 356 | 1,933 | 1,192 | 211 | 130 | 119 | 13 | 219 |
| DLV | 118 | 56 | 5 | 2 | 96 | 58 | 26 | 18 | 6 | 1 | 73 |
| EFV | 9,673 | 6,228 | 301 | 238 | 2,168 | 1,310 | 168 | 110 | 194 | 35 | 400 |
| ETR | 255 | 169 | 62 | 51 | 145 | 94 | 70 | 48 | 4 | 3 | 1 |
| NVP | 8,405 | 5,044 | 232 | 178 | 1,836 | 1,054 | 123 | 74 | 179 | 31 | 508 |
| RPV | 5 | 4 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| APV | 1,240 | 615 | 48 | 37 | 463 | 216 | 41 | 31 | 38 | 3 | 192 |
| ATV | 3,444 | 2,293 | 230 | 166 | 833 | 510 | 131 | 79 | 39 | 6 | 52 |
| DRV | 916 | 587 | 152 | 95 | 328 | 200 | 111 | 68 | 16 | 6 | 4 |
| FPV | 1,028 | 621 | 79 | 58 | 381 | 211 | 52 | 26 | 28 | 5 | 20 |
| IDV | 9,466 | 5,965 | 184 | 150 | 2,134 | 1,433 | 112 | 84 | 144 | 20 | 737 |
| LPV | 8,516 | 5,293 | 332 | 244 | 2,156 | 1,315 | 180 | 104 | 142 | 22 | 147 |
| NFV | 7,540 | 4,669 | 137 | 104 | 1,698 | 1,018 | 101 | 68 | 113 | 17 | 706 |
| SQV | 6,187 | 3,646 | 166 | 125 | 1,638 | 951 | 91 | 41 | 136 | 21 | 428 |
| TPV | 643 | 345 | 71 | 58 | 246 | 153 | 62 | 39 | 9 | 0 | 5 |
| EVG | 10 | 1 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RAL | 650 | 448 | 223 | 171 | 251 | 156 | 116 | 80 | 7 | 3 | 0 |
| Naïve | 37,577 | 37,577 | 1,917 | 1,917 | 2,056 | 2,056 | 154 | 154 | 3 | 3 | 0 |
| Total | 61,163 | 53,098 | 2,579 | 2,408 | 6,641 | 4,862 | 444 | 326 | 441 | 84 | 1,384 |
The numbers of sequences by drug exposure for the development and test datasets are tabulated above. Columns including the abbreviation Comp. in their headers indicate the numbers of sequences from a certain dataset and with a certain drug exposure whose complete drug exposure history is known. The complete drug exposure history for all sequences from the HIVdbExposure dataset is available.
3TC: lamivudine, ABC: abacavir, AZT: zidovudine, d4T: stavudine, ddC: zalcitabine, ddI: didanosine, FTC: emtricitabine, TDF: tenofovir, DLV: delavirdine, EFV: efavirenz, ETR: etravirine, NVP: nevirapine, RPV: rilpivirine, APV: amprenavir, ATV: atazanavir, DRV: darunavir, FPV: fosamprenavir, IDV: indinavir, LPV: lopinavir, NFV: nelfinavir, SQV: saquinavir, TPV: tipranavir, EVG: elvitegravir, RAL: raltegravir
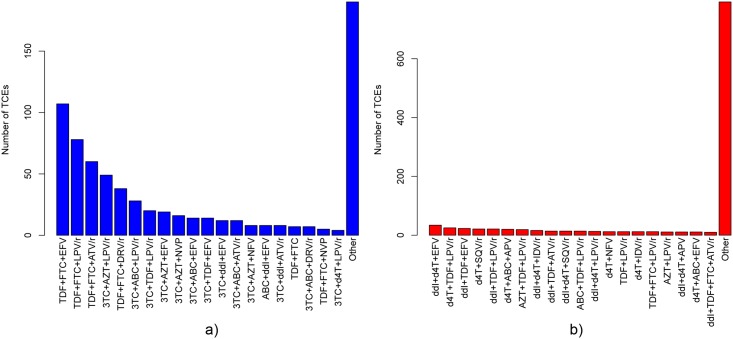
Fig 2. Drug-combination counts for therapies in EuResistTCE and HIVdbTCE.
The frequencies of the 20 most-frequent drug combinations in EuResistTCE (a) and HIVdbTCE (b) datasets are displayed above. 3TC: lamivudine, ABC: abacavir, AZT: zidovudine, d4T: stavudine, ddI: didanosine, FTC: emtricitabine, TDF: tenofovir, EFV: efavirenz, NVP: nevirapine, APV: amprenavir, ATV: atazanavir, DRV: darunavir, IDV: indinavir, LPV: lopinavir, NFV: nelfinavir, SQV: saquinavir.
Table 4. Number of phenotypes by drug in the pheno datasets.
| Antivirogram | PhenoSense | Susceptible | Resistant | Total | |
|---|---|---|---|---|---|
| DPheno | |||||
| 3TC | 905 | 1546 | 346 | 1362 | 2451 |
| ABC | 840 | 1473 | 531 | 186 | 2313 |
| AZT | 855 | 1567 | 801 | 773 | 2422 |
| d4T | 889 | 1573 | 1031 | 60 | 2462 |
| ddC | 821 | 451 | 371 | 47 | 1272 |
| ddI | 891 | 1575 | 654 | 59 | 2466 |
| TDF | 633 | 1234 | 850 | 33 | 1867 |
| DLV | 1016 | 1638 | 794 | 1091 | 2654 |
| EFV | 1106 | 1652 | 924 | 1127 | 2758 |
| ETR | 363 | 476 | 304 | 156 | 839 |
| NVP | 1170 | 1653 | 772 | 1447 | 2823 |
| RPV | 91 | 176 | 62 | 75 | 267 |
| ATV | 774 | 1134 | 401 | 978 | 1908 |
| DRV | 282 | 629 | 400 | 178 | 911 |
| FPV | 1088 | 1695 | 917 | 859 | 2783 |
| IDV | 1151 | 1734 | 782 | 1229 | 2885 |
| LPV | 1040 | 1468 | 665 | 1279 | 2508 |
| NFV | 1185 | 1780 | 483 | 1584 | 2965 |
| SQV | 1181 | 1741 | 985 | 1039 | 2922 |
| TPV | 742 | 854 | 584 | 191 | 1596 |
| EVG | 97 | 598 | 112 | 137 | 695 |
| RAL | 97 | 630 | 336 | 148 | 727 |
| TPheno | |||||
| 3TC | 115 | 166 | 37 | 158 | 281 |
| ABC | 107 | 166 | 60 | 25 | 273 |
| AZT | 107 | 165 | 92 | 88 | 272 |
| d4T | 110 | 168 | 122 | 6 | 278 |
| ddC | 105 | 46 | 38 | 5 | 151 |
| ddI | 111 | 168 | 72 | 7 | 279 |
| TDF | 87 | 132 | 87 | 4 | 219 |
| DLV | 126 | 169 | 81 | 125 | 295 |
| EFV | 141 | 171 | 105 | 136 | 312 |
| ETR | 43 | 52 | 36 | 15 | 95 |
| NVP | 146 | 175 | 82 | 170 | 321 |
| RPV | 14 | 21 | 13 | 10 | 35 |
| ATV | 85 | 131 | 42 | 115 | 216 |
| DRV | 22 | 79 | 50 | 20 | 101 |
| FPV | 110 | 193 | 88 | 105 | 303 |
| IDV | 125 | 194 | 76 | 142 | 319 |
| LPV | 113 | 172 | 76 | 151 | 285 |
| NFV | 127 | 199 | 48 | 189 | 326 |
| SQV | 129 | 195 | 105 | 119 | 324 |
| TPV | 80 | 106 | 56 | 22 | 186 |
| EVG | 17 | 61 | 9 | 11 | 78 |
| RAL | 17 | 65 | 36 | 21 | 82 |
The numbers of phenotypes by drug in the DPheno and TPheno datasets are tabulated above. Phenotypes were measured with the Antivirogram™ or PhenoSense™ assays. Resistance-factor cutoffs one and ten were used for dichotomizing phenotypes into susceptible and resistant.
3TC: lamivudine, ABC: abacavir, AZT: zidovudine, d4T: stavudine, ddC: zalcitabine, ddI: didanosine, FTC: emtricitabine, TDF: tenofovir, DLV: delavirdine, EFV: efavirenz, ETR: etravirine, NVP: nevirapine, RPV: rilpivirine, APV: amprenavir, ATV: atazanavir, DRV: darunavir, FPV: fosamprenavir, IDV: indinavir, LPV: lopinavir, NFV: nelfinavir, SQV: saquinavir, TPV: tipranavir, EVG: elvitegravir, RAL: raltegravir
2.2. Training of models for predicting drug exposure
We trained linear Support Vector classifiers (SVC) [47,48] for discriminating between sequences from viruses with and without previous exposure to a certain drug. We trained SVCs on two kinds of development sets, Exposure or ExposurePheno. Specifically, each sequence in the development sets DPRRT and DIN included binary labels indicating whether exposure to a particular drug had occurred or not. We used DPRRT and DIN for creating one Exposuredrug development set for each drug and subsequently trained one SVC on each of these development sets. We additionally created the development set ExposurenaïvePRRT in which labels indicate whether viral sequences were derived from therapy-naïve or therapy-experienced patients. Subsequently, we trained one SVC on ExposurenaïvePRRT. For creating the ExposurePhenodrug development sets, we extended the data in the Exposuredrug development sets with GPPs from DPheno. In the ExposurePhenodrug development sets, viral sequences from GPPs labeled as susceptible to the drug in question are treated as not having being exposed to the drug. Conversely, GPPs labeled as resistant to the drug in question are treated as having been exposed to the drug. We trained an SVC with each ExposurePhenodrug development set. We do not consider the binary output of the SVC classifier but rather the reported signed distance from the decision boundary, a real number. We call this number drug-exposure score (DES).
2.3. Assessment and comparison of performance
We constructed DES models with 10-fold cross validation on the respective development sets. Then we used the DES reported by the resulting models and the predicted resistance factor for geno2pheno[resistance], respectively to calculate and compare AUC performance of both models. This was done for the test sets TPRRT, TIN, TP, HIVdbExposure, TPheno, EuResistTCE, EuResistTCETP, and HIVdbTCE. Among the 7,275 protease and reverse-transcriptase nucleotide sequences contained in TPRRT and EuResistTCE, 23 (<0.01%) were not processed by geno2pheno[resistance] due to low sequence similarity. For the sake of performance comparison, these sequences were excluded. In the following, mean performances for the tested models are stated. In order to be able to compare the different models, these means were calculated only with the performances of the drugs that are common to Exposure and ExposurePheno models, as well as to geno2pheno[resistance]. p-values were calculated with a two-sided Wilcoxon signed-rank test [49].
2.3.1. Assessment of performance for predicting drug exposure via cross validation
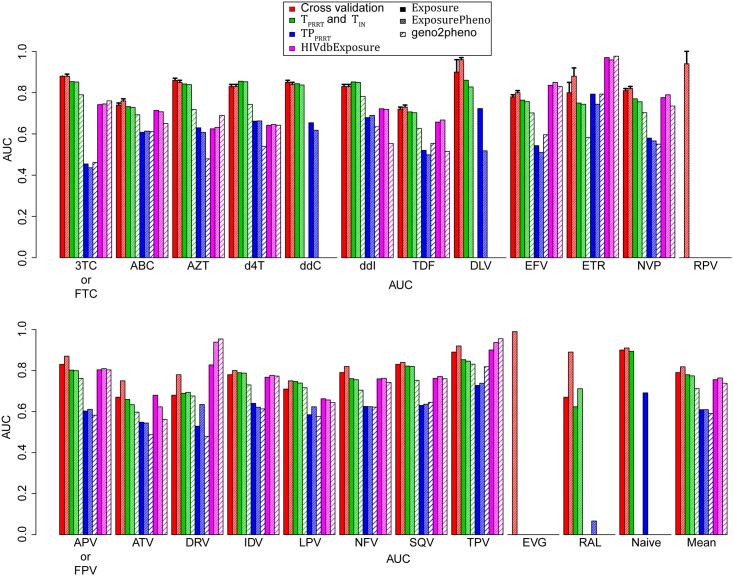
We cross validated the SVC on each Exposuredrug and each ExposurePhenodrug development set, as well as on the ExposurenaïvePRRT development set. Specifically, we performed ten repetitions of a five-fold cross validation on each development set while testing a series of values for the SVC c parameter (see Methods). One value of the c parameter was chosen for each development set. The mean drug-wise cross-validation performances (area under the receiver operating characteristic curve; AUC) for the chosen values of c ranged between 0.67 and 0.99. Models trained on ExposurePheno cross-validation sets had a higher mean cross-validation performance (μ = 0.82; σ = 0.05) than those trained on Exposure cross-validation sets (μ = 0.79; σ = 0.07; p < 0.003). The p-value quantifies the difference in the AUC distributions between Exposure and ExposurePheno models. Individual performances are depicted in Fig 3.
Fig 3. Performance of drug-exposure prediction.
Performance of drug-exposure prediction was assessed with 10-fold cross validation on the development set and four test sets. Test sets TPRRT and TIN were obtained from the EuResist database and contain protease and reverse-transcriptase and integrase sequences, respectively. TP is a subset of TPRRT ∪ TIN and contains nucleotide sequences that were measured during therapy pauses. HIVdbExposure was obtained from the HIVdb TCE repository and contains protease and reverse-transcriptase sequences. Performance on the test sets was compared to that of geno2pheno[resistance]. Bars depicting mean performances were calculated only using drugs that are common to Exposure and ExposurePheno models, as well as to geno2pheno[resistance]. Error bars indicate the standard deviation. 3TC: lamivudine, ABC: abacavir, AZT: zidovudine, d4T: stavudine, ddC: zalcitabine, ddI: didanosine, FTC: emtricitabine, TDF: tenofovir, DLV: delavirdine, EFV: efavirenz, ETR: etravirine, NVP: nevirapine, RPV: rilpivirine, APV: amprenavir, ATV: atazanavir, DRV: darunavir, FPV: fosamprenavir, IDV: indinavir, LPV: lopinavir, NFV: nelfinavir, SQV: saquinavir, TPV: tipranavir, EVG: elvitegravir, RAL: raltegravir.
2.3.2. Assessment of performance for predicting drug exposure on test sets
The performances of DES for predicting drug exposure on the TPRRT, TIN, TP, and HIVdbExposure test sets are depicted in Fig 3. The performance of geno2pheno[resistance] on TPRRT, TP, and HIVdbExposure sets can be seen in Fig 3 as well. In the following, p-values quantify the difference in the AUC distributions between Exposure models, ExposurePheno models or geno2pheno[resistance]. The best mean performance on the TPRRT dataset could be attained by Exposure models (μ = 0.78; σ = 0.06), while the mean performance of geno2pheno[resistance] was lower (μ = 0.71; σ = 0.07; p < 10−4). On the TIN dataset, DES performance for RAL was comparable for models trained on ExposurePheno cross-validation sets (AUC = 0.71), but not for those trained on Exposure cross-validation sets (AUC = 0.62). On the HIVdbExposure dataset, the best mean performance with lowest standard deviation was achieved with Exposure models (μ = 0.76; σ = 0.09), while geno2pheno[resistance] achieved a lower mean performance (μ = 0.74; σ = 0.14; p = 0.43). The best mean performance on TP could be attained with Exposure and ExposurePheno models (μ = 0.61; σ = 0.08), while geno2pheno[resistance] displayed a lower mean performance (μ = 0.59; σ = 0.10; p = 0.3778).
2.3.3. Assessment of performance for predicting drug resistance
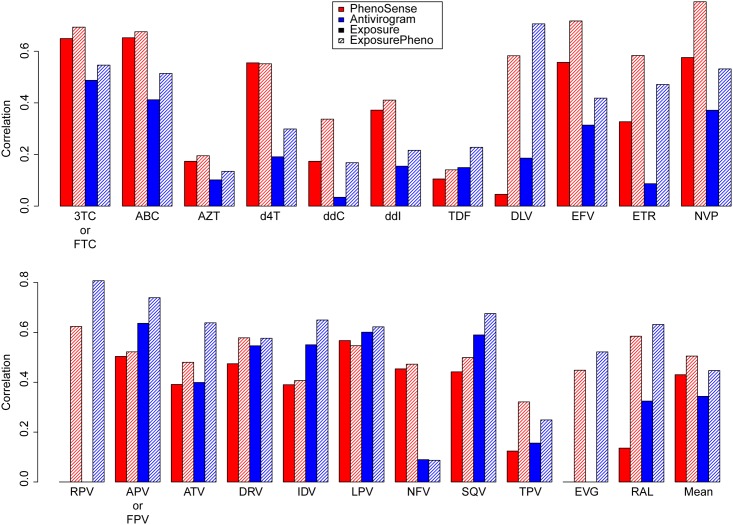
Fig 4 shows the correlation of DES with the logarithmized resistance factors from the TPheno dataset. DES models trained on ExposurePheno cross-validation sets could attain substantially higher mean correlations (μAntivirogram = 0.46; μPhenoSense = 0.51; σAntivirogram = 0.2; σPhenoSense = 0.17) than those trained on Exposure cross-validation sets (μAntivirogram = 0.34; μPhenoSense = 0.41; σAntivirogram = 0.2; σPhenoSense = 0.18). We consider this sufficient correlation for the intended use of our software. Furthermore, it should be noted that the between-test correlation of PhenoSense and Antivirogram is weak (r = 0.36) [11].
Fig 4. Correlation of drug-exposure scores with logarithmized resistance factors.
Genotypes in TPheno were interpreted with drug-exposure models. The correlation of the resulting drug-exposure scores with the corresponding logarithmized resistance factors is displayed above. Note that drug-resistance assays (either Antivirogram™ or PhenoSense™) are denoted by the colors of the bars, while the drug-exposure model types (Exposure or ExposurePheno) are denoted by the shading of the bars. Bars depicting the mean performances were calculated with the drugs for which Exposure and ExposurePheno models are available. Error bars indicate the standard deviation. 3TC: lamivudine, ABC: abacavir, AZT: zidovudine, d4T: stavudine, ddC: zalcitabine, ddI: didanosine, FTC: emtricitabine, TDF: tenofovir, DLV: delavirdine, EFV: efavirenz, ETR: etravirine, NVP: nevirapine, RPV: rilpivirine, APV: amprenavir, ATV: atazanavir, DRV: darunavir, FPV: fosamprenavir, IDV: indinavir, LPV: lopinavir, NFV: nelfinavir, SQV: saquinavir, TPV: tipranavir, EVG: elvitegravir, RAL: raltegravir.
2.3.4. Assessment of performance for predicting therapy success
We tested the performance of DES models and of geno2pheno[resistance] in predicting the binary therapy-success labels of the TCEs in EuResistTCE, EuResistTCETP, and HIVdbTCE. For this purpose, we used DES and geno2pheno[resistance], respectively, for calculating a genetic susceptibility score (GSS) for each TCE, an additive score that rates the susceptibility of the virus to the used drugs. For GSS calculation, the predictions of DES models and of geno2pheno[resistance] were translated into probability scores (Methods). The GSS for a TCE is the sum of the probability scores for its constituent drug compounds. The performances of GSS calculated with Exposure models, ExposurePheno models, and geno2pheno[resistance], respectively, when predicting binary labels for therapeutic success are displayed in Table 5. On the EuResistTCE dataset, the best performance could be attained with Exposure and ExposurePheno models (AUC = 0.71), while the performance of geno2pheno[resistance] was lower (AUC = 0.68). On therapies with baseline sequences measured during a therapy pause (EuResistTCETP), ExposurePheno models displayed the best performance (AUC = 0.73), while the performance of geno2pheno[resistance] was lower (AUC = 0.66). The best performance on HIVdbTCE is displayed by geno2pheno[resistance] (AUC = 0.64), while the performance of drug-exposure models was lower (AUC ≤ 0.63). On average, ExposurePheno models displayed the highest performance with lowest standard deviation in predicting therapeutic success (μ = 0.69; σ = 0.05). The average performance of geno2pheno[resistance] when predicting therapeutic success was lower (μ = 0.66; σ = 0.02; p = 0.5).
Table 5. Performance of prediction of therapy-success for therapies in EuResistTCE, EuResistTCETP, and HIVdbTCE.
| Exposure | ExposurePheno | geno2pheno[resistance] | |
|---|---|---|---|
| EuResistTCE | 0.71 | 0.71 | 0.68 |
| EuResistTCETP | 0.72 | 0.73 | 0.66 |
| HIVdbTCE | 0.62 | 0.63 | 0.64 |
Therapy success was predicted for therapies in the EuResistTCE, EuResistTCETP, and HIVdbTCE test using three different genetic susceptibility scores (GSS) for each therapy. The first GSS was obtained with Exposure models, the second GSS with ExposurePheno models and the third GSS with geno2pheno[resistance]. Above, the performances of the three different GSS are tabulated for each dataset. Performances were quantified with the area under the receiver operating characteristic curve.
2.4. Assessment of performance of drug-exposure models with cutoff-based categorization
HIV-1 nucleotide sequences can be submitted to our web service for interpretation with ExposurePheno models (see Discussion). For the purpose of facilitating the use of DES by human experts, we estimated cutoffs for translating DES into clinically meaningful categories. We estimated two sets of cutoffs (see Methods for details). DEMax cutoffs translate DES into categories describing degrees of drug exposure (Table A in S1 File), while pheno cutoffs translate DES into categories describing degrees of drug resistance (Table B in S1 File). For determining and testing pheno cutoffs, we applied clinically relevant RF cutoffs to PhenoSense GPPs in Pheno (Table C in S1 File). After discretization of DES with DEMax cutoffs, we calculated their performance when predicting drug exposure on TPRRT, TIN, TP, and HIVdbExposure in terms of AUC (Table D in S1 File). Furthermore, we discretized DES with pheno cutoffs and calculated their misclassification rates when predicting drug resistance in TPheno (Table E in S1 File). The application of cutoffs to DES can be associated with a mild loss in predictive performance. When predicting drug exposure without application of cutoffs, average performances (AUC; mean calculated with all drugs for which an ExposurePheno model is available) on the TPRRT and TIN, TP, and HIVdbExposure datasets are 0.77 (0.06), and 0.57 (0.1), and 0.76 (0.11), respectively. After application of cutoffs, mean performance in predicting drug exposure is 0.76 (0.06), 0.58 (0.14), and 0.76 (0.11), respectively. When predicting phenotypic drug resistance, performance with and without application of cutoffs is difficult to compare since different performance measures are required for each case. Specifically, without application of cutoffs, performance was measured with the Pearson correlation coefficient, while with application of cutoffs, performance was measured with the misclassification rate. With application of cutoffs, the mean misclassification rate is 0.27 (0.1) for all GPPs, 0.11 (0.06) for susceptible-labeled GPPs, 0.1 (0.06) for intermediate-labeled GPPs, and 0.05 (0.03) for resistant-labeled GPPs.
3. Discussion
DES models constitute data-driven interpretation systems for HIV-1 protease, reverse-transcriptase, and integrase sequences. Two versions of DES models were trained and tested. Specifically, one version of the models is solely trained on genotypes and drug exposure information (Exposure models), while the other version additionally includes GPPs (ExposurePheno models). When compared to ExposurePheno models, Exposure models show a high performance when predicting drug exposure, but their correlation with RFs and their performance when predicting antiretroviral therapy success are lower. We chose to include GPPs in the training sets of ExposurePheno models for the following reasons. Both drug exposure and drug resistance are predictive of success of antiretroviral therapy [34,38–40]. The major factor leading to viral drug resistance is exposure to antiretroviral drugs. Specifically, drug resistance arises through the selection of HIV-1 strains with mutations that confer a replicative advantage in the presence of the drug. Thus, drug exposure indirectly causes drug resistance and therefore, both drug exposure and drug resistance are correlated with certain mutations in the genome of HIV-1. Nevertheless, drug exposure and drug resistance are not redundant, but can complement each other. For this reason, simultaneous interpretation of HIV-1 genotypes with respect to drug exposure and to drug resistance is useful for the prediction of the success of antiretroviral therapy. ExposurePheno models consider drug exposure and drug resistance jointly. For the purpose of including GPPs in the training set of classification models, RFs required categorization. Thus, we replaced the RFs in the GPPs with the labels susceptible and resistant. For the purpose of labeling, the RF cutoffs one and ten were applied to all GPPs, regardless of the drug-resistance test (Antivirogram or PhenoSense) and of the tested drug. GPPs with RFs between one and ten were not used for training the models. When clinically relevant categorization of GPPs is intended, different cutoffs for each drug and drug resistance test must be used [15]. However, rather than producing clinically relevant labels for training, we aimed at discriminating fully susceptible GPPs from those that have developed resistance to an extent well beyond the variability arising from the drug resistance test itself, for the following reasons. First, drug resistance is a continuum, and the creation of training instances with a clear separation in this continuum is adequate for the training of binary classification models. Second, clinically relevant cutoffs are selected under the (implicit) consideration of the pharmacokinetic properties of a drug. For example, the use of ritonavir as a booster for protease inhibitors (PIs) leads to an increased and sustained concentration of PIs in the body [50]. For this reason, clinically relevant cutoffs for boosted PIs are shifted upwards with respect to their unboosted counterparts [15]. However, we aim at discriminating viral sequences that display mutations as a consequence of drug exposure (or as the cause of resistance), without regard for drug concentrations in the blood of patient. The cutoffs one and ten are adequate for combining GPPs produced with the Antivirogram and PhenoSense assays; if other assays are used, other cutoffs might need to be selected. One advantage of ExposurePheno models over Exposure models is their higher performance. Another advantage is that they can make use of an additional data source, the GPPs. The use of GPPs allowed for the training of models for two additional drugs (EVG and RPV).
The interpretations provided by DES models can be used to addresses three questions: (1) Was an HIV-1 variant exposed to a certain drug? (2) Is an HIV-1 variant resistant to a certain drug? and (3) How does the effect of a drug combination therapy decompose into effects of its constituent drugs? In the following, we propose how DES models can be used for addressing the three questions mentioned above.
Ad question (1): When the prediction of drug exposure is required, we propose two ways in which DES can be used. For quantification and comparison of the degree of drug exposure between at least two groups of patients, we do not recommend translating DES into categories (by using cutoffs), since this leads to loss of precision. Instead, DES should be directly used for detecting differences between groups. Note that comparison of DES for different drugs requires normalization, e.g. via the calculation of z-scores (this is provided by our web service). If the prediction of the drug exposures of individual patients is required, cutoffs can be used in order to translate DES into clinically meaningful categories (this is also provided by our web service).
Ad question (2): When predicting drug resistance with DES, one should bear in mind that the correlation of DES with log RFs is weak to strong, depending on the drug in question (Fig 4). Correlation with drug resistance to the nucleotide and nucleoside reverse-transcriptase inhibitors (NRTIs) AZT, d4T, ddC, ddI, and TDF is lower than with resistance to other drugs. We interpret this to be the result of the high similarity of the resistance profiles among these NRTIs [17]. Mutations conferring resistance to one of these NRTIs confer resistance to the other NRTIs (cross-resistance) and are thus less discriminative of exposure and resistance to any specific drug among the NRTIs we mention above. Nonetheless, the correlation is sufficient for predicting the susceptible-intermediate-resistant (SIR) label of GPPs discretized with clinically relevant cutoffs (Table E in S1 File). While a mean misclassification rate of 0.27 (0.1) seems high, most of errors arise from misclassifying intermediate-labeled GPPs (μ = 0.1; σ = 0.06), for which the clinical relevance is uncertain [12], and from misclassification of susceptible-labeled GPPs that are predicted to be intermediate or resistant (μ = 0.1; σ = 0.06). For the drugs AZT, d4T, ddI, and TDF, misclassification of susceptible- and intermediate-labeled GPPs as resistant is especially high, which we also attribute to the high similarity of their resistance profiles. Misclassifying susceptible-labeled GPPs as intermediate or resistant can be clinically adequate, for the following reason. Phenotypic resistance measurements do not account for mutations that do not cause drug resistance at the time of resistance testing, but are indicative that drug resistance can be easily developed in the future [16]. Therefore, when such resistance mutations are present in the viral baseline genotype of a patient, and even if phenotypic resistance measurements indicate full drug susceptibility, classification of the genotype as non-susceptible will prevent selecting a combination of drug compounds that could quickly fail due to the emergence of drug resistant viral variants. Nonetheless, misclassifying susceptible-labeled GPPs as intermediate or resistant could also lead to rejection of a drug for treating a patient although the drug could have been a good choice. When the prediction of the results of phenotypic resistance tests is required, we recommend the use of interpretation systems that have been specifically designed and validated for this purpose, e.g. geno2pheno[resistance]. DES predictions are especially useful in two situations: first, when (imminent) drug resistance is not detected by other methods because the process of selection of drug-resistant variants has led to the selection of certain mutations that have not (yet) resulted in clinically relevant drug-resistance (Fig 3; ExposurePheno models have a higher performance in predicting drug exposure than geno2pheno[resistance]). Second, when drug-resistant HIV-1 variants are in the process of reverting to the wild type after the removal of drug pressure (Fig 3 and Table 5; ExposurePheno models have a higher performance in predicting drug exposure and therapeutic success than geno2pheno[resistance] when genotypes were obtained during therapy pauses). Decision support for the choice of the use of the tool will be given in our follow-up paper.
Ad question (3): DES are predictive of therapeutic success (Table 5). In order to facilitate the use of DES for deciding which drug could be useful components of combination antiretroviral therapy, as well as for interpretation of DES by human experts, our online web service offers the following features. (i) Calculation of DES. (ii) Calculation of z-scores, which normalize DES with respect to their distribution in therapy patients. These z-scores can be useful when DES for different drugs need to be compared or merged for the analysis of clinical data. (iii) Translation of DES into categories related to drug exposure and drug resistance via cutoffs. HIV-1 can mutate as a result of exposure to antiretroviral drugs, which does not necessarily entail clinically relevant drug resistance. Drug-exposure categories help the user to determine whether a viral variant has changed as a result of drug exposure. If a viral variant is not rated unexposed for a certain drug, drug resistance to that drug should be at least suspected. Drug-resistance categories indicate whether viral mutations are not only indicative of exposure to a particular drug, but also indicative of clinically relevant drug resistance. If a viral variant is not rated susceptible, drug resistance is highly likely. Drug-exposure and drug-resistance categories are useful when selecting the drug components of antiretroviral therapy. However, predictions are provided for each drug individually (as in most drug-resistance interpretation tools). Thus, the selection of an adequate drug combination under consideration of DES is still left to the expert. In a follow-up paper we will report on a DES-based model that does not require expert selection of drug combinations. Specifically, we are currently testing DES as input features for a model for predicting the success of combinations of antiretroviral drugs. This model will exploit DES for selecting the compounds of antiretroviral therapy. (iv) Presentation of the basis of the predictions by displaying the residues with the largest influence on the prediction.
In summary, in this study, we present a novel approach for training a data-driven interpretation system for drug exposure and drug resistance. We show that models trained on HIV-1 sequences from patients with known drug history can be used for predicting drug exposure, drug resistance, and therapeutic success, even if no GPPs are used. The inclusion of GPPs in the training sets of the models boosted the performance of the models when predicting in-vitro phenotypic drug resistance measurements and therapeutic success, but not when predicting drug exposure. Compared to geno2pheno[resistance], the method could attain higher mean performances when predicting drug exposure and therapeutic success. The difference in performance was only statistically significant at the 5% level when predicting drug exposure on TPRRT. Note that many of the drugs in HIVdbTCE are not used any more due to their toxicity profiles or their comparatively low potency. A large advantage of DES models is that they are trained on clinical HIV data and freely available GPPs. In conjunction with a frequently updated database with HIV-1 data from routine clinical practice, such as the EIDB, DES models can be automatically updated on a regular basis. Thus, these models allow us to reduce our dependency on hard-to-obtain GPPs for offering a publicly available data-driven genotypic drug-exposure and drug-resistance interpretation system that is kept up to date. While regularly updatable interpretation systems are clearly the appropriate method for accounting for the growing richness of clinical data, innovative procedures may have to be put in place for adequate certification of such systems. DES models for protease and reverse transcriptase inhibitors have been integrated into the geno2pheno[resistance] server http://www.geno2pheno.org. Support for integrase inhibitors will follow. After a sequence has been submitted for prediction, the tab labeled Drug Exposure must be selected in order to view DES predictions. Note that in the input tab, sample nucleotide sequences can be loaded by selecting the appropriate action. On the website, mutations with the highest influence on the prediction are displayed. These are ordered by the magnitude of their influence. Mutations colored in red increase DES, while those colored in green decrease it.
4. Materials and methods
4.1. Ethics statement
All data considered in this study had been previously de-identified. For this reason, consent was neither required nor given by human subjects.
4.2. Drugs considered in this study
In this study, the following antiretroviral drugs are considered: lamivudine (3TC), abacavir (ABC), zidovudine (AZT), stavudine (d4T), zalcitabine (ddC), didanosine (ddI), emtricitabine (FTC), tenofovir (TDF), delavirdine (DLV), efavirenz (EFV), etravirine (ETR), nevirapine (NVP), rilpivirine (RPV), amprenavir (APV), atazanavir (ATV), darunavir (DRV), fosamprenavir (FPV), indinavir (IDV), lopinavir (LPV), nelfinavir (NFV), saquinavir (SQV), tipranavir (TPV), raltegravir (RAL) and elvitegravir (EVG). Other antiretroviral drugs were excluded due to insufficient data.
4.3. Dataset construction
In the following, we describe a number of datasets that we used for training and validating our models. For the comfort of the reader, we summarize the contents of each of these datasets in Table 1. Furthermore, we depict the relationships of each of the datasets in Figure A in S1 File.
4.3.1. Datasets of genotypes and therapeutic history
The PRRT and IN datasets contain HIV-1 nucleotide sequences and information on the antiretroviral compounds that were used before each sequence was obtained. When constructing these datasets, we disregarded episodes of treatment with a drug that lasted less than 30 days. The PRRT dataset was constructed by pooling 70,304 HIV-1 protease (PR) and reverse-transcriptase (RT) nucleotide sequences from two sources: 37,799 sequences from the EuResist Integrated Database (EIDB; http://www.euresist.org, downloaded April 11th, 2014) [45], 9,627 of which were derived from drug-naïve patients (short: drug-naïve sequences), and 32,506 drug-naïve sequences from the Los Alamos National Laboratory Sequence Database (LANLSD; http://www.hiv.lanl.gov/; downloaded on March 31st, 2015). Among the sequences in the PRRT dataset that were derived from therapy-experienced patients (short: drug-exposed sequences), 18,328 sequences were derived from patients whose complete drug history was available at the time of sequencing. The IN dataset includes a total of 5,523 integrase (IN) nucleotide sequences with the following characteristics: 3,382 sequences were extracted from the EuResist database, 1,240 of which are drug-naïve, 397 have been exposed to an integrase inhibitor (INI) and possibly other drugs, and 1,745 have been exposed only to drugs whose target is different from integrase. The complete drug history was available for 1,432 of the drug-exposed integrase sequences. Additionally, 3,782 drug-naïve integrase sequences from the LANLSD (downloaded on March 31st, 2015) were added to the IN dataset. Inclusion criteria for the sequences were as follows. (1) Alignment with the MutExt software (http://www.schuelter-gm.de/mutext.html) must not have produced an error due to low sequence similarity to the reference sequence (2) at most 10% of the residues of the considered protein regions could not be determined by the sequencing procedure (considered protein regions are listed in Section Subtype Determination, Sequence Alignment and Encoding), (3) the amino-acid sequence resulting from nucleotide translation must be unique within the dataset, unless drug exposure differed between duplicates. The order of appearance of the sequences in the dataset determined which duplicate sequence was excluded, with sequences appearing first preempting inclusion of sequences appearing later. Older reverse-transcriptase sequences not covering amino-acid positions 221–230 were excluded as well.
PRRT and IN were split into development and test sets, as follows. For the purpose of rigorous validation, HIV-1 nucleotide sequences derived from the same patient were not allowed to be simultaneously present in the development and test sets. In the following, dataset nomenclature consists of an abbreviation describing a characteristic of the dataset and optionally PRRT or IN in subscript. The letters in subscript indicate whether the dataset is a subset of the PRRT or of the IN dataset. All HIV-1 nucleotide sequences in PRRT and IN that were obtained during a therapy pause were assigned to TP (n = 441). This dataset is only used for testing purposes, since we deem therapy-pause sequences valuable for testing and an insignificant minority in the much larger training set. In order to make sure that the test sets are patient-wise disjoint with respect to the development set, a set of test patients P was created iteratively. Initially, P included all patients with sequences in TP. Further patients from PRRT and IN were subsequently added to P by random selection until the number of available sequences for the patients in P was approximately 10% of the number of sequences in PRRT and IN. The test sets TPRRT and TIN contain the protease and reverse-transcriptase sequences available for the patients in P, respectively. The development sets DPRRT and DIN contain the sequences in PRRT and IN, respectively, that are not included in TPRRT, and TIN.
4.3.2. EuResistTCE dataset and standard datum definition
In order to test the performance of our models in predicting therapeutic success, we created the EuResistTCE test set, as follows. We extracted a total of 9,201 therapy-change episodes (TCEs) from the EIDB [45]. These TCEs were constructed according to the definition of the EuResist Standard Datum [40]. In contrast to the EuResist Standard Datum, however, viral-load (VL) measurements were constrained to those not reaching a lower limit of quantification greater or equal than 50 HIV-1 RNA copies per milliliter of blood plasma. In summary, each TCE includes a protease and reverse transcriptase baseline sequence, the compounds that were prescribed to the patient, a baseline and a follow-up viral load (VL), and a binary label indicating whether the therapy was successful or not. The follow-up VL must have been measured within 4–12 weeks after therapy start, preferring the VL closest to week 8 after therapy start. Therapy success at follow up is defined as an at least 100-fold reduction in the VL or a VL of less than 400 HIV-1 RNA copies per ml of blood plasma. This definition of therapy success was used for producing binary labels for the TCEs. To allow performance comparison, only therapies including the following antiretroviral drugs were considered: 3TC, ABC, AZT, d4T, ddI, FTC, TDF, EFV, ETR, NVP, APV, ATV, DRV, FPV, IDV, LPV, NFV, SQV, TPV and ritonavir as a boosting agent (/r). Therapies including unboosted protease inhibitors (except for NFV, since the drug cannot be boosted) were excluded due to their comparatively inferior potency.
The baseline sequences of the EuResist TCEs partially overlap with the sequences in the datasets described above. A minority of baseline sequences were not included in any of the datasets described above because we could not ascertain whether drug exposure had occurred or the patient was therapy-naïve at the time of sequencing. We created a set of test TCEs, EuResistTCE, with a fraction of the initially extracted EuResist TCEs. Specifically, EuResistTCE only contained TCEs with baseline sequences that had not been derived from a patient with an HIV-1 nucleotide sequence in DPRRT or in DIN. A subset of the TCEs in EuResistTCE includes baseline sequences which were obtained during a therapy pause. We refer to these TCEs as EuResistTCETP.
4.3.3. HIVdbExposure and HIVdbTCE datasets
For testing the performance of our models in predicting drug exposure and therapeutic success, we created the HIVdbExposure and the HIVdbTCE test sets, respectively. The TCE repository in the HIV Drug Resistance Database was downloaded in its entirety on November 21st, 2013 [14,46]. The TCE repository contains 58 TCEs from the EuResist database, which were discarded. A total of 1,384 sequences with drug-exposure information could be extracted from the repository. We assigned these sequences to the HIVdbExposure test set. For creating the HIVdbTCE test set, the EuResist Standard Datum definition was applied to therapies in HIVdbTCE whose drug compounds are investigated in this study (with the exception of ddC and raltegravir (RAL) for the sake of performance comparison).
4.3.4. Datasets of genotype-phenotype pairs
A total of 7,597 GPPs were downloaded from the HIV Drug Resistance Database [14] on April 15th, 2015 (Pheno dataset). The phenotypic drug-resistance assays used for producing the phenotypes were constrained to Antivirogram® [51] and PhenoSense®[52]. The genotypes are provided in the form of substitutions with respect to the reference sequence consensus B [14]. 3,323 GPP quantify protease-inhibitor (PI) resistance, 3,477 reverse-transcriptase-inhibitor (RTI) resistance, and 797 INI resistance. The TPheno test set was created from the Pheno dataset by randomly sampling approximately 10% of the GPP. The rest of the GPPs in Pheno were assigned to the DPheno development set. For training our models with the GPPs, we categorized the resistance factors (RFs) in DPheno as susceptible or resistant. Specifically, GPPs with RFs lower or equal to one were categorized as susceptible, while GPPs with RFs greater or equal to ten were categorized as resistant. GPPs with RFs between one and ten were not used for training our models.
4.3.5. NaïvePRRT and NaïveIN datasets
Transmitted drug resistance (TDR) in PR- and RT-naïve sequences was defined as the presence of at least one mutation in the list of drug resistance mutations for surveillance of transmitted HIV-1 drug resistance (DRMT) [7]. Since the list of DRMT only contains PR and RT mutations, TDR in IN sequences was defined as the presence of an INI drug-resistance mutation in the 2013 IAS list [17]. Following the methodology used for establishing the list of DRMT, INI drug-resistance mutations with a prevalence greater than 0.5% among sequences from the LANLSD in IN were not regarded as indicative of TDR [28]. The NaïvePRRT and NaïveIN were created by randomly sampling 2,500 LANLSD sequences without TDR from the PRRT and IN datasets, respectively. These sequences are not included in TPRRT, TIN, DPRRT or DIN. NaïvePRRT and NaïveIN are used by our web service for z-score calculation.
4.4. Subtype determination, sequence alignment and encoding
The subtype distribution in the PRRT and IN datasets was determined with the COMET subtyping tool [53]. Nucleotide sequences in PRRT and IN were aligned against HXB2 and translated, using MutExt (http://www.schuelter-gm.de). The resulting amino-acid sequences, along with amino-acid sequences in the Pheno dataset, were represented vectorially with a binary encoding. The vectorial representation considers substitutions, deletions and the presence of insertions within the following HXB2 amino-acid positions: protease 3–99, reverse transcriptase 40–230, and integrase 30–260. The presence of deletions and insertions was encoded for each amino-acid position, while the amino acids of which a specific insertion consists were not encoded.
4.5. Creation of exposure and ExposurePheno development sets
DPRRT and DIN were used for constructing the development sets Exposuredrug for drug ∈ {ABC, AZT, d4T, ddC, ddI, 3TC/FTC, TDF, EFV, ETR, NVP, RPV, ATV, DRV, APV/FPV, IDV, LPV, NFV, SQV, TPV, RAL, EVG} which contain an equal number of sequences exposed and not exposed to a certain drug, along with binary labels indicating exposure to the drug. Sequences not exposed to the drug were randomly selected from DPRRT or DIN, as they were in excess; these sequences were required to have been derived from patients whose complete drug exposure history is recorded. Where possible, half of the sequences not exposed to the drug were drug-naïve, and half of them were exposed to some other drug. A development set ExposurenaïvePRRT containing an equal number of drug-naïve and drug-experienced PRRT sequences was constructed as well. An ExposurenaïveIN development set was not created due to the fact that only a sufficient number of RAL-exposed integrase sequences was available.
The ExposurePhenodrug development sets were created from the Exposuredrug sets with additional supplementation of some GPPs from the DPheno dataset. Specifically, genotypes with corresponding RFs classified as resistant were treated as drug-exposed sequences while those with corresponding RFs classified as susceptible were treated as sequences not exposed to the drug in question. Genotypes with corresponding RFs between the two cutoffs were not used for training (see Phenotypic Resistance Cutoffs). This procedure incremented the number of available drug-exposed sequences and allowed for the creation of the development sets ExposurePhenoRPV and ExposurePhenoEVG, as the number of available drug-exposed sequences for RPV and EVG was very low. Development sets for dolutegravir could not be created, as neither a sufficient number of resistant phenotypes nor a sufficient number of drug-exposed sequences were available.
4.6. Training and selection of models for predicting drug exposure
For performing five repetitions of a 10-fold cross validation, each Exposuredrug and each ExposurePhenodrug set was randomly partitioned five times into ten folds. Each fold contained an equal proportion of sequences with and without exposure to the drug in question. The partitions were used to cross validate linear support-vector classifiers (SVCs) [47,48] discriminating between sequences with and without exposure to a certain drug. The vectorial representation used to train each drug-specific model was constrained to the vector elements describing the drug’s target protein (protease, reverse transcriptase or integrase). Each cross validation was performed with a certain value for the regularization parameter c for the SVM, specifically, c ∈ {2−8, 2−7, …, 22}. Performance was measured in terms of the area under the receiving-operator-characteristic curve (AUC) [54,55]. The signed distance to the classification hyperplane (also called decision value) was used as a score for predicting drug exposure. Thus, we call such decision values drug-exposure scores (DES). For each cross-validation set and vectorial representation, the model with the lowest value of c whose average performance was not significantly lower than the best average performance was selected (Benjamini-Hochberg-corrected Wilcoxon signed-rank test [49] with a significance threshold of 0.05). Finally, each cross-validation set and vectorial representation was used without partitioning to train a final SVC with the selected value of c. We refer to these SVCs as the final DES models.
4.7. Assessment and comparison of performance
The performance of the drug-exposure models was compared to that of geno2pheno[resistance] 3.3 (http://www.geno2pheno.org) [20]. The output of geno2pheno[resistance] includes a prediction of the resistance-factor (RF). Since geno2pheno[resistance] uses its own alignment program, performance comparison was constrained to the set of sequences which could be aligned without errors by geno2pheno[resistance]. Furthermore, the drug ddC was also excluded from performance comparison, as it is not supported by geno2pheno[resistance] any more.
Assessment of performance
Sequences in TPRRT, TIN, TP, EuResistTCE, EuResistTCETP, HIVdbTCE, HIVdbExposure, and TPheno were interpreted with the final drug-exposure models and geno2pheno[resistance]. Performance was assessed in three respects. First, the performance of DES and of geno2pheno[resistance] when predicting drug exposure was assessed using TPRRT, TIN, TP, and HIVdbExposure. These datasets contain HIV-1 sequences and a binary matrix indicating the previous exposure of these sequences to each individual drug compound. The area under the receiver operating characteristic curve (AUC) was used as a performance measure. Second, the performance of DES when predicting drug resistance was quantified with the correlation between DES for the genotypes in TPheno and the corresponding log RFs. Unfortunately, performance in predicting drug resistance could not be compared to that of geno2pheno[resistance], since the genotypes in TPheno were only available as amino-acid sequences and geno2pheno[resistance] requires nucleotide sequences as an input. Third, the performance of DES and of geno2pheno[resistance] when predicting therapy success was assessed with EuResistTCE, EuResistTCETP, and HIVdbTCE. For this purpose, DES and RF predictions were converted to probability scores. Specifically, DES, which are SVM decision values, were converted to probability scores as described by Platt [56]. Predicted RFs were converted to probability scores by fitting a two-component Gaussian-mixture model. In the Gaussian mixture model, one Gaussian is fitted to RFs that belong to the susceptible population, while the other Gaussian is fitted to RFs that belong to the resistant population. Subsequently, a sigmoid function is used for estimating the probability of resistance [20]. We define the probability of susceptibility as one minus the probability of resistance. Probability scores were used for calculating a genetic susceptibility score (GSS) for each therapy. A GSS consisted of the sum of the individual probability scores for each drug in the regimen. For each therapy, three GSS were calculated. The first two GSS were calculated using the probability scores derived with DES from Exposure and ExposurePheno models, respectively, while the third GSS was calculated with the probability scores derived with geno2pheno[resistance]. Performance in predicting therapeutic success was quantified with the AUC. Significance values in the Results section were calculated with a two-sided Wilcoxon signed-rank test [49].
4.8. Determination of parameters for our web service
Our drug-exposure models are freely available online (http://www.geno2pheno.org; see Discussion). For the purpose of facilitating the use of DES by human experts, we calculated two sets of parameters. The first set of parameters is used for calculating z-scores of DES. It includes a mean and a standard-deviation value for each drug, calculated with the nucleotide sequences in NaïvePRRT and NaïveIN. The second set of parameters includes cutoffs which translate DES into clinically meaningful categories related to drug exposure and drug resistance. For the purpose of displaying the sequence features with the largest influence on the predictions of DES models, we translated the Support Vectors and corresponding Support-Vector coefficients of each SVC into a linear function. In the following, we detail on the procedures we used for determining the z-score parameters, the cutoffs, and for extracting the weights for the input features.
4.8.1. Calculation of z-scores from drug-exposure scores
We interpreted each sequence in NaïvePRRT with each DES model for predicting exposure to protease inhibitors (PIs) and reverse-transcriptase inhibitors (RTIs). Likewise, we interpreted each sequence in NaïveIN with each DES model for predicting exposure to INIs. For each drug, we calculated the mean and standard deviation of the resulting DES. Our web service calculates the z-score for a sequence s and compound drug according to the following formula
| (1) |
where zdrug(s) is the z-score for sequence s and compound drug, δdrug(s) is the DES for sequence s and compound drug, μdrug is the mean DES value calculated with the NaïvePRRT or NaïveIN datasets, and σdrug is the corresponding standard deviation.
4.8.2. Estimation of cutoffs of drug-exposure scores
Two sets of cutoffs were determined for each final DES model. The following goals are addressed by each set of cutoffs: (1) prediction of drug exposure and (2) prediction of drug resistance. Each set of cutoffs includes a lower and an upper cutoff for the corresponding DES models. The description of the methods used for determination of these cutoffs follows.
4.8.3. Cutoffs maximizing the performance of the prediction of drug exposure (DEMax cutoffs)
ExposurePhenodrug cross-validation sets were interpreted with the corresponding final DES models that were trained on them (this is also called calculation of reinsertion predictions). For each cross-validation set, an upper and a lower cutoff were estimated such that the AUC of the drug-exposure prediction is maximized. We call these cutoffs the DEMax cutoffs, and they allow for the discretization of a DES for a drug into the categories unexposed (U), possible exposure (PE) and probably exposed (E). A detailed description of the procedure with which DEMax cutoffs were determined follows.
Function (1) was defined for discretization of a value δs ∈ ℝ associated to a sequence s by using the lower and upper cutoffs cL, cU ∈ ℝ.
| (2) |
Let Δdrug ∈ ℝn be a vector of DES predicting the drug exposure of each of n sequences s to drug, and let Edrug ∈ {0,1}n be the corresponding vector of class labels, indicating whether each sequence s was exposed to the drug or not. Application of cutoffs cL, cU and Function (2) to a vector of DES Δdrug results in the discrete DES vector discretize(cL, cU, Δdrug). For each bootstrap replicate, an upper and a lower cutoff cL and cU were determined as
| (3) |
where AUC(discretize(cL, cU, Δdrug), Edrug) is the AUC quantifying the performance of discretize(cL, cU, Δdrug) in predicting exposure to drug for each sequence with a DES in Δdrug. AUC maximization was performed via grid search over cL and cU.
ExposurePhenodrug cross-validation sets were interpreted with the corresponding final models that were trained on them. Two thousand bootstrap replicates of the DES of each cross-validation set were created. For each bootstrap replicate and the corresponding class labels, an upper and a lower cutoff were determined by AUC maximization Eq (3). The resulting 2,000 upper and 2,000 lower cutoffs for each final drug-exposure model were averaged to yield the final set of cutoffs. We call these cutoffs the DEMax cutoffs. If a DES for a drug is less than both cutoffs for that drug, then we discretize that DES as unexposed (U). If a DES is greater or equal than the lower cutoff, but less or equal than the upper cutoff, we discretize that DES as intermediate exposure (IE). Finally, if a DES is greater than both cutoffs, then we discretize that DES as exposed (E).
4.8.4. Phenotypically-guided cutoffs for prediction of phenotypic in-vitro drug resistance (pheno cutoffs)
A set of clinically relevant cutoffs for PhenoSense GPPs was obtained from the HIVdb website [14] and is composed as follows. 3TC: 3 and 20; ABC: 3 and 6; AZT: 3 and 10; d4T: 1.5 and 2; ddI: 1.5 and 2; TDF 1.5 and 4; all non-nucleoside reverse-transcriptase inhibitors (NNRTIs): 3 and 10; and all INIs 4 and 20. The set of clinically-relevant cutoffs were used for discretizing PhenoSense GPPs in DPheno into the categories susceptible (S), intermediate (I) or resistant (R), henceforth called the true labels. The genotypes associated with these GPPs were interpreted with the final DES models. For each drug, an upper and a lower DES cutoff yield predicted GPP labels. These cutoffs, which we call pheno cutoffs, are determined such that the sum of the penalties quantifying the differences between the true labels and the predicted labels is minimized. An individual penalty equals one, if the true label was R and the predicted label was S. If the true label is I, and the predicted label S, the penalty equals 0.75. All other differences between true and predicted labels were penalized with the value 0.5, while the equality of true and predicted labels was not penalized. Pheno cutoffs allow for discretization of a DES for a drug as susceptible (S), intermediate (I) or resistant (R). Further details on the cutoff-determination procedure, including the rationale for choosing the penalty values follow.
The error matrix E ∈ ℝ3x3 Eq (4) was defined for penalizing the misclassification of a discretized value δs with label l ∈ {1,2,3} and predicted label
| (4) |
The rationale for choosing the values of the error matrix follows. Diagonal entries are zero, as correct classification incurs no penalty. From a clinical perspective, the worst kind of misclassification that can occur is the classification of a resistant viral strain (label 3) as susceptible (label 1), since the prescription of a therapy including a thus misclassified compound could compromise the susceptibility of all compounds in the therapy. Therefore, this kind of misclassification was assigned the maximum penalty, one. Misclassification of a resistant strain as intermediate (label 2) deserves a smaller penalty, as surpassing the lower cutoff indicates a clinically-relevant decrease in susceptibility, albeit implying that some susceptibility is given. Therefore, this kind of misclassification was assigned the penalty 0.75. All other types of misclassifications are considered equally undesirable, but less severe than the first two, and were assigned the penalty 0.5. Clinically-relevant cutoffs were used to discretize PhenoSense GPPs in DPheno with Function (2), yielding their labels. The genotypes s associated with these GPPs were interpreted with the DES models. For each drug involved in a GPP, 2,000 bootstrap replicates of the PhenoSense GPPs in DPheno were sampled. In order to assign to each of the three classes the same weight in this procedure, each bootstrap replicate was constructed using an equal number of GPPs with each label. For each drug, this number was equal to the maximum number of GPPs with a certain label. Each bootstrap replicate was used to determine a lower and an upper cutoff , which minimizes the sum of the penalties for each label l = discretize(cL, cU, RFs) with corresponding prediction for a resistance factor RF and a drug-exposure score DES associated with genotype s. The resulting 2,000 cutoff pairs for each drug and DES model were averaged, yielding the final phenotypically guided cutoffs. If a DES for a drug is less than both cutoffs for that drug, then we discretize that DES as susceptible (S). If a DES is greater or equal than the lower cutoff, but less or equal than the upper cutoff, we discretize that DES as intermediate (I). Finally, if a DES is greater than both cutoffs, then we discretize that DES as resistant (R).
4.8.5. Extraction of input-feature weights from drug-exposure models
For the purpose of displaying the input features (i.e. HIV-1 substitutions, insertions, and deletions) with the largest influence on a DES interpretation, we represented the SVCs that produce DES as linear functions. Let xi ∈ {0,1}p, i ∈ {1, …, n} be the Support Vectors for a given DES model, αi ∈ ℝ their corresponding Support-Vector coefficients, and ρ ∈ ℝ their intercept. The linear-function representation for a DES model is given by
| (5) |
where xs ∈ {0,1}p is the encoding for input sequence s. Given an encoded sequence xs, the linear Function (5) produces the same numerical output as the corresponding DES SVC. The linear function consists of an offset (also called y-axis intercept) and p coefficients that correspond to the components of the vectors that encode each sequence. The vector contains these coefficients (also called weights). Since the encoding of the sequence xs is binary, DES calculation can be performed by adding the offset to the coefficients that correspond to the input features that are present in sequence s. In our web service, we display for each drug a selection of features of the input sequence. These features have the largest absolute values of the coefficients in the linear-function representation of DES models. Features with positive coefficients increase DES and are displayed in red. Features with negative coefficients decrease DES and are displayed in green.
Supporting information
(DOC)
Acknowledgments
We acknowledge the contribution of the EuResist Network Study Group to acquiring the data for this work.
This work was supported by the German Health Ministry [MASTER/HIV-HEP].
Data Availability
Data used for this study were obtained from three sources: 1. The Los Alamos National Laboratoy Sequence Database (freely downloaded from https://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html); 2. The HIV Drug Resistance Database (freely downloaded from https://hivdb.stanford.edu/); 3. The EuResist Database (http://www.euresist.org/; obtained upon request). All of these three sources contain third-party data. The authors accessed the data for this study as described above. Access to the EuResist database is free of charge but requires the submission of a project proposal to the Scientific Board of the Project. Some of the authors of this work form part of the EuResist consortium. However, no special privilege was exercised in order to access EuResist data. The authors' membership in the EuResist consortium will not influence availability of the data to third parties. Interested researchers can obtain access to the same data that was available to the authors by submitting a project proposal to the Scientific Board of the EuResist consortium.
Funding Statement
This work was supported by Bundesministerium für Gesundheit, HIV-HEP-MASTER (IIA5-2013-2514AUK375). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Deeks SG. Determinants of virological response to antiretroviral therapy: implications for long-term strategies. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2000;30 Suppl 2:S177–184. [DOI] [PubMed] [Google Scholar]
- 2.Lohse N, Obel N, Kronborg G, Laursen A, Pedersen C, Larsen CS, et al. Declining risk of triple-class antiretroviral drug failure in Danish HIV-infected individuals. AIDS Lond. Engl. 2005;19:815–22. [DOI] [PubMed] [Google Scholar]
- 3.Lampe FC, Gatell JM, Staszewski S, Johnson MA, Pradier C, Gill MJ, et al. Changes over time in risk of initial virological failure of combination antiretroviral therapy: a multicohort analysis, 1996 to 2002. Arch. Intern. Med. 2006;166:521–8. 10.1001/archinte.166.5.521 [DOI] [PubMed] [Google Scholar]
- 4.Günthard HF, Aberg JA, Eron JJ, Hoy JF, Telenti A, Benson CA, et al. Antiretroviral Treatment of Adult HIV Infection: 2014 Recommendations of the International Antiviral Society—USA Panel. JAMA. 2014;312:410 10.1001/jama.2014.8722 [DOI] [PubMed] [Google Scholar]
- 5.Svicher V, D’Arrigo R, Alteri C, Andreoni M, Angarano G, Antinori A, et al. Performance of genotypic tropism testing in clinical practice using the enhanced sensitivity version of Trofile as reference assay: results from the OSCAR Study Group. New Microbiol. 2010;33:195–206. [PubMed] [Google Scholar]
- 6.Langford SE, Ananworanich J, Cooper DA. Predictors of disease progression in HIV infection: a review. AIDS Res. Ther. 2007;4:11 10.1186/1742-6405-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PloS One. 2009;4:e4724 10.1371/journal.pone.0004724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaccarelli M, Lorenzini P, Ceccherini-Silberstein F, Tozzi V, Forbici F, Gori C, et al. Historical resistance profile helps to predict salvage failure. Antivir. Ther. 2009;14:285–91. [PubMed] [Google Scholar]
- 9.Lundgren J, Gatell JM, Furrer H, Rockstroh J. Guidelines. Version 7.1. November 2014 [Internet]. 7.1. European Aids Clinical Society; [cited 2015 Apr 25]. http://www.eacsociety.org/files/guidelines-7.1-english.pdf
- 10.Mayer KH, Hanna GJ, D’Aquila RT. Clinical Use of Genotypic and Phenotypic Drug Resistance Testing to Monitor Antiretroviral Chemotherapy. Clin. Infect. Dis. 2001;32:774–82. 10.1086/319231 [DOI] [PubMed] [Google Scholar]
- 11.Wang K, Samudrala R, Mittler JE. Antivirogram or phenosense: a comparison of their reproducibility and an analysis of their correlation. Antivir. Ther. 2004;9:703–12. [PubMed] [Google Scholar]
- 12.Tang MW, Shafer RW. HIV-1 antiretroviral resistance: scientific principles and clinical applications. Drugs. 2012;72:e1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larder B, Kemp S, Harrigan P. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–9. [DOI] [PubMed] [Google Scholar]
- 14.Shafer RW. Rationale and uses of a public HIV drug-resistance database. J. Infect. Dis. 2006;194 Suppl 1:S51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winters B, Montaner J, Harrigan PR, Gazzard B, Pozniak A, Miller MD, et al. Determination of Clinically Relevant Cutoffs for HIV-1 Phenotypic Resistance Estimates Through a Combined Analysis of Clinical Trial and Cohort Data: JAIDS J. Acquir. Immune Defic. Syndr. 2008;48:26–34. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Lerma JG, Nidtha S, Blumoff K, Weinstock H, Heneine W. Increased ability for selection of zidovudine resistance in a distinct class of wild-type HIV-1 from drug-naive persons. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13907–12. 10.1073/pnas.241300698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer RW, et al. Update of the drug resistance mutations in HIV-1: March 2013. Top. Antivir. Med. 2013;21:6–14. [PMC free article] [PubMed] [Google Scholar]
- 18.Obermeier M, Pironti A, Berg T, Braun P, Däumer M, Eberle J, et al. HIV-GRADE: a publicly available, rules-based drug resistance interpretation algorithm integrating bioinformatic knowledge. Intervirology. 2012;55:102–7. 10.1159/000331999 [DOI] [PubMed] [Google Scholar]
- 19.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2006;42:1608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beerenwinkel N, Däumer M, Oette M, Korn K, Hoffmann D, Kaiser R, et al. Geno2pheno: Estimating phenotypic drug resistance from HIV-1 genotypes. Nucleic Acids Res. 2003;31:3850–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vermeiren H, Van Craenenbroeck E, Alen P, Bacheler L, Picchio G, Lecocq P. Prediction of HIV-1 drug susceptibility phenotype from the viral genotype using linear regression modeling. J. Virol. Methods. 2007;145:47–55. 10.1016/j.jviromet.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 22.Santos AF, Soares MA. HIV Genetic Diversity and Drug Resistance. Viruses. 2010;2:503–31. 10.3390/v2020503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theys K, Abecasis AB, Vandamme A-M. HIV-1 drug resistance: where do polymorphisms fit in? Future Microbiol. 2013;8:303–6. 10.2217/fmb.13.10 [DOI] [PubMed] [Google Scholar]
- 24.Langs-Barlow A, Paintsil E. Impact of Human Immunodeficiency Virus Type-1 Sequence Diversity on Antiretroviral Therapy Outcomes. Viruses. 2014;6:3855–72. 10.3390/v6103855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh K, Flores J, Kirby K, Neogi U, Sonnerborg A, Hachiya A, et al. Drug Resistance in Non-B Subtype HIV-1: Impact of HIV-1 Reverse Transcriptase Inhibitors. Viruses. 2014;6:3535–62. 10.3390/v6093535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantor R, Katzenstein D. Polymorphism in HIV-1 non-subtype B protease and reverse transcriptase and its potential impact on drug susceptibility and drug resistance evolution. AIDS Rev. 2003;5:25–35. [PubMed] [Google Scholar]
- 27.Garcia-Lerma JG, MacInnes H, Bennett D, Weinstock H, Heneine W. Transmitted Human Immunodeficiency Virus Type 1 Carrying the D67N or K219Q/E Mutation Evolves Rapidly to Zidovudine Resistance In Vitro and Shows a High Replicative Fitness in the Presence of Zidovudine. J. Virol. 2004;78:7545–52. 10.1128/JVI.78.14.7545-7552.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shafer RW, Rhee S-Y, Pillay D, Miller V, Sandstrom P, Schapiro JM, et al. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS Lond. Engl. 2007;21:215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scherrer AU, Ledergerber B, von Wyl V, Böni J, Yerly S, Klimkait T, et al. Minor Protease Inhibitor Mutations at Baseline Do Not Increase the Risk for a Virological Failure in HIV-1 Subtype B Infected Patients. Menéndez-Arias L, editor. PLoS ONE. 2012;7:e37983 10.1371/journal.pone.0037983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackie NE, Dunn DT, Dolling D, Garvey L, Harrison L, Fearnhill E, et al. The impact of HIV-1 reverse transcriptase polymorphisms on responses to first-line nonnucleoside reverse transcriptase inhibitor-based therapy in HIV-1-infected adults: AIDS. 2013;27:2245–53. 10.1097/QAD.0b013e3283636179 [DOI] [PubMed] [Google Scholar]
- 31.Hachiya A, Marchand B, Kirby KA, Michailidis E, Tu X, Palczewski K, et al. HIV-1 Reverse Transcriptase (RT) Polymorphism 172K Suppresses the Effect of Clinically Relevant Drug Resistance Mutations to Both Nucleoside and Non-nucleoside RT Inhibitors. J. Biol. Chem. 2012;287:29988–99. 10.1074/jbc.M112.351551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wainberg MA, Brenner BG. The Impact of HIV Genetic Polymorphisms and Subtype Differences on the Occurrence of Resistance to Antiretroviral Drugs. Mol. Biol. Int. 2012;2012:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carpenter CCJ. Antiretroviral Therapy for HIV Infection in 1996: Recommendations of an International Panel. JAMA. 1996;276:146 [PubMed] [Google Scholar]
- 34.Prosperi MCF, Rosen-Zvi M, Altmann A, Zazzi M, Di Giambenedetto S, Kaiser R, et al. Antiretroviral Therapy Optimisation without Genotype Resistance Testing: A Perspective on Treatment History Based Models. Poon AFY, editor. PLoS ONE. 2010;5:e13753 10.1371/journal.pone.0013753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Revell A, Boyd M, Wang D, Emery S, Gazzard B, Reiss P, et al. A comparison of computational models with and without genotyping for prediction of response to second-line HIV therapy: Modelling response to second-line HIV therapy. HIV Med. 2014;15:442–8. 10.1111/hiv.12156 [DOI] [PubMed] [Google Scholar]
- 36.Revell AD, Wang D, Wood R, Morrow C, Tempelman H, Hamers R, et al. An update to the HIV-TRePS system: the development of new computational models that do not require a genotype to predict HIV treatment outcomes. J. Antimicrob. Chemother. 2014;69:1104–10. 10.1093/jac/dkt447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prosperi MCF, De Luca A. Computational models for prediction of response to antiretroviral therapies. AIDS Rev. 2012;14:145–53. [PubMed] [Google Scholar]
- 38.Rosen-Zvi M, Altmann A, Prosperi M, Aharoni E, Neuvirth H, Sönnerborg A, et al. Selecting anti-HIV therapies based on a variety of genomic and clinical factors. Bioinforma. Oxf. Engl. 2008;24:i399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Revell AD, Wang D, Boyd MA, Emery S, Pozniak AL, De Wolf F, et al. The development of an expert system to predict virological response to HIV therapy as part of an online treatment support tool. AIDS Lond. Engl. 2011;25:1855–63. [DOI] [PubMed] [Google Scholar]
- 40.Zazzi M, Kaiser R, Sönnerborg A, Struck D, Altmann A, Prosperi M, et al. Prediction of response to antiretroviral therapy by human experts and by the EuResist data-driven expert system (the EVE study). HIV Med. 2011;12:211–8. 10.1111/j.1468-1293.2010.00871.x [DOI] [PubMed] [Google Scholar]
- 41.Saigo H, Altmann A, Bogojeska J, Mller F, Nowozin S, Lengauer T. Learning from past treatments and their outcome improves prediction of in Vivo response to anti-HIV therapy. Stat. Appl. Genet. Mol. Biol. 2011;10. [DOI] [PubMed] [Google Scholar]
- 42.Bogojeska J, Stöckel D, Zazzi M, Kaiser R, Incardona F, Rosen-Zvi M, et al. History-alignment Models for Bias-aware Prediction of Virological Response to HIV Combination Therapy. In: Lawrence N, Girolami M, editors. Proc. Fifteenth Int. Conf. Artif. Intell. Stat. AISTATS 2012. La Palma, Canary Islands, Spain: Journal of Machine Learning Research; 2012. p. 118–126.
- 43.Bickel S, Bogojeska J, Lengauer T, Scheffer T. Multi-task learning for HIV therapy screening. ACM Press; 2008. [cited 2016 Jul 22]. p. 56–63. http://portal.acm.org/citation.cfm?doid=1390156.1390164 [Google Scholar]
- 44.Pironti A. Improving and Validating Data-Driven Genotypic Interpretation Systems for the Selection of Antiretroviral Therapies [PhD Thesis]. [Saarbrücken, Germany]: University of Saarland; 2016.
- 45.Zazzi M, Incardona F, Rosen-Zvi M, Prosperi M, Lengauer T, Altmann A, et al. Predicting response to antiretroviral treatment by machine learning: the EuResist project. Intervirology. 2012;55:123–7. 10.1159/000332008 [DOI] [PubMed] [Google Scholar]
- 46.Rhee S-Y, Blanco J, Liu TF, Pere I, Kaiser R, Zazzi M, et al. Standardized representation, visualization and searchable repository of antiretroviral treatment-change episodes. AIDS Res. Ther. 2012;9:13 10.1186/1742-6405-9-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2:1–27. [Google Scholar]
- 48.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed New York: Springer; 2011. [Google Scholar]
- 49.Wilcoxon F. Individual Comparisons by Ranking Methods. Biom. Bull. 1945;1:80. [Google Scholar]
- 50.Kempf DJ, Marsh KC, Kumar G, Rodrigues AD, Denissen JF, McDonald E, et al. Pharmacokinetic enhancement of inhibitors of the human immunodeficiency virus protease by coadministration with ritonavir. Antimicrob. Agents Chemother. 1997;41:654–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hertogs K, de Béthune MP, Miller V, Ivens T, Schel P, Van Cauwenberge A, et al. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 1998;42:269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, et al. A Novel Phenotypic Drug Susceptibility Assay for Human Immunodeficiency Virus Type 1. Antimicrob. Agents Chemother. 2000;44:920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pineda-Peña A-C, Faria NR, Imbrechts S, Libin P, Abecasis AB, Deforche K, et al. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: Performance evaluation of the new REGA version 3 and seven other tools. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2013; [DOI] [PubMed] [Google Scholar]
- 54.R Core Team. R: A Language and Environment for Statistical Computing [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2014. http://www.R-project.org [Google Scholar]
- 55.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinforma. Oxf. Engl. 2005;21:3940–1. [DOI] [PubMed] [Google Scholar]
- 56.Platt J. Probabilistic Outputs for Support Vector Machines and Comparisons to Regularized Likelihood Methods Adv. Large Margin Classif. MIT Press; p. 61–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
Data used for this study were obtained from three sources: 1. The Los Alamos National Laboratoy Sequence Database (freely downloaded from https://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html); 2. The HIV Drug Resistance Database (freely downloaded from https://hivdb.stanford.edu/); 3. The EuResist Database (http://www.euresist.org/; obtained upon request). All of these three sources contain third-party data. The authors accessed the data for this study as described above. Access to the EuResist database is free of charge but requires the submission of a project proposal to the Scientific Board of the Project. Some of the authors of this work form part of the EuResist consortium. However, no special privilege was exercised in order to access EuResist data. The authors' membership in the EuResist consortium will not influence availability of the data to third parties. Interested researchers can obtain access to the same data that was available to the authors by submitting a project proposal to the Scientific Board of the EuResist consortium.