Abstract
In the search for genes that define critical steps of relapse in pediatric T-cell acute lymphoblastic leukemia (T-ALL) and can serve as prognostic markers, we performed targeted sequencing of 313 leukemia-related genes in 214 patients: 67 samples collected at the time of relapse and 147 at initial diagnosis. As relapse-specific genetic events, we identified activating mutations in NT5C2 (P=0.0001, Fisher's exact test), inactivation of TP53 (P=0.0007, Fisher's exact test) and duplication of chr17:q11.2-24.3 (P=0.0068, Fisher's exact test) in 32/67 of T-ALL relapse samples. Alterations of TP53 were frequently homozygous events, which significantly correlated with higher rates of copy number alterations in other genes compared with wild-type TP53 (P=0.0004, Mann–Whitney's test). We subsequently focused on mutations with prognostic impact and identified genes governing DNA integrity (TP53, n=8; USP7, n=4; MSH6, n=4), having key roles in the RAS signaling pathway (KRAS, NRAS, n=8), as well as IL7R (n=4) and CNOT3 (n=4) to be exclusively mutated in fatal relapses. These markers recognize 24/49 patients with a second event. In 17 of these patients with mostly refractory relapse and dire need for efficient treatment, we identified candidate targets for personalized therapy with p53 reactivating compounds, MEK inhibitors or JAK/STAT-inhibitors that may be incorporated in future treatment strategies.
Introduction
Approximately 20% of children with precursor T-cell acute lymphoblastic leukemia (T-ALL) experience a relapse1 and only 20% of relapsed patients can be cured with current salvage protocols.2 Treatment after first relapse of T-ALL is uniform, using very intensive chemotherapy to induce second remission followed by allogeneic stem cell transplantation.3, 4 This treatment fails in the majority of patients,2, 5 indicating the need to identify patients who may benefit from experimental therapies. With the intensity of both frontline and relapse therapies, treatment-related mortality is a significant cause of treatment failure. Therefore, further intensification of chemotherapy is not an option resulting in the need for accurate prognostic markers and for identification of druggable targets.
Relapse evolves via selection of clones and acquisition of mutations.6, 7 Nevertheless, as no common biological determinants were identified to explain these events, our understanding of the mechanisms underlying relapse and treatment resistance is still limited. Activating mutations of the nucleotidase NT5C2 were identified in approximately 20% of relapsed, but not primary T-ALL patients and confer resistance to chemotherapy in vitro.6, 8 TP53 mutations have previously been reported to be acquired in relapse in up to 24% of patients and correlated with poor prognosis.9, 10 Although intensively investigated,10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 clinically meaningful genetic markers for risk stratification or for targeted treatment could neither be established in primary nor in relapsed T-ALL.
To identify potential prognostic biomarkers in relapsed pediatric T-ALL and to define critical steps in disease progression and in resistance to treatment, we subjected a large cohort of 214 pediatric T-ALLs to targeted sequencing. We used the Haloplex target capture technique to analyze 313 leukemia-related genes in 147 samples collected at initial diagnosis and in 67 samples at the time of relapse. In addition to single-nucleotide variants (SNVs) and small insertions/deletions (InDels), we identified copy number alterations (CNAs) affecting target genes by analyzing coverage data.
Materials and methods
Patients' clinical characteristics
Altogether leukemic samples of 214 patients were analyzed: 67 relapse samples (REL) and 147 samples collected at initial diagnosis (INI). No matched primary and relapse samples were included in our study. Of the initial diagnosis patients, 31 were treated according to ALL-BFM 2000 and 116 patients according to AIEOP-BFM ALL 2009 protocol. All relapse patients were recruited from the ALL-REZ BFM 2002 trial. Clinical characteristics of the analyzed patients were compared with the remaining patients from the cohort (Supplementary Table 1). Except for white blood cell count, the distribution of patients' features was representative for the entire cohort. Enrichment for patients with high white blood cell counts is a likely consequence of selection for the samples with sufficient DNA amounts for the analyses performed here. Bone marrow or blood samples were enriched for mononuclear cells by Ficoll density gradient centrifugation. DNA was purified from mononuclear cells using the Gentra Puregene Cell Kit (Qiagen, Hilden, Germany). From one patient (PATNR: 82) with an isolated extramedullary relapse DNA was extracted from a lymph node. MRD (minimal residual disease) response was assessed as described before.1, 17 The study was approved by the institutional review boards of the Charité Universitätsmedizin Berlin and the Medical Faculty Heidelberg. Informed consent was obtained in accordance with the Declaration of Helsinki.
Targeted deep sequencing
The Haloplex Target Enrichment Kit (Agilent, Santa Clara, CA, USA) covered 324 genes comprising 5964 regions (Supplementary Table 2). In all, 58 348 amplicons covered a total of 3.04 Mbp. Target genes were selected based on previously published studies.6, 8, 20, 21, 22, 23, 24, 25
A pilot study confirmed that reducing the reaction volume during library preparation resulted in a complexity of libraries equivalent to the standard reaction volume (Supplementary Results, Supplementary Figure 1). To save on input sample DNA and on costs, all subsequent reactions were performed in half a standard reaction volume. DNA was quantified using Qubit dsDNA BR Assay kit (Life Technologies, Darmstadt, Germany). Starting material was 112.5 ng of genomic DNA. The volume of all the reagents described in the manufacturer's instructions (Version D.5, May 2013) was reduced by half. Libraries were pooled in batches of 43 (1) or 44 (5) samples. Each batch was sequenced as 100 bp paired reads on one lane using an Illumina HiSeq 2000 instrument (Illumina, San Diego, CA, USA). VarScan26 was used to detect both SNVs and small insertions and deletions. Coverage profiles were used to identify copy number variations (for details see Supplementary Methods).
Multiplex ligation-dependent probe amplification
The commercially available SALSA MLPA P383 T-ALL probe mix (MLPA (multiplex ligation-dependent probe amplification); MRC-Holland, Amsterdam, The Netherlands) and a custom-made probe set based on the SALSA MLPA P200-A1 probe mix (MRC-Holland; Supplementary Table 3) were used for the detection of specific copy number variations (Supplementary Methods).
Low-coverage whole genome sequencing
Libraries for low-coverage WGS (whole genome sequencing) were prepared using NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs, Frankfurt am Main, Germany) from 100 ng of genomic DNA. Ten samples were pooled and sequenced on one Illumina HiSeq 2000 lane. Mean DNA sequence coverage was 3-fold (range 2–5-fold).
Sanger sequencing
Sanger sequencing of PTEN exon 7 and of NOTCH1 PEST, TAD, HD-C and HD-N domains was done as described before.16, 18
Statistical analyses
Statistical analyses were performed using GraphPad Prism 6.0. Prognostic factors analyses were conducted using either the SAS program (SAS-PC, v. 9.1, SAS Institute Inc., Cary, NC, USA) or R27(Package: Survival;28 for details, see Supplementary Methods).
Results
Targeted NGS identifies SNVs/InDels and CNAs with high sensitivity
We designed a Haloplex Panel targeting leukemia-related genes to cover 5964 exons of 324 genes that we had compiled on the basis of previously published data sets.6, 8, 20, 21, 22, 23, 24, 25 Of the 324 genes, 313 were sufficiently covered to be analyzed in all 214 pediatric T-ALL patients. Average coverage of all the exons in 214 samples was 424 (median=417). Ninety-seven percent of the exons were covered more than 30-fold. To determine the accuracy of the Haloplex panel and our analytic setup for the detection of SNVs and InDels, we compared next-generation sequencing (NGS) data with conventional sequencing of the NOTCH1 and PTEN genes in 144 patients. The sensitivity of the targeted NGS approach was 93% (67/72) and 94% (17/18) for NOTCH1 and PTEN mutations, respectively (for details, see Supplementary Results).
In addition to the DNA sequence, targeted sequencing also delivers the read count distribution that can be used to estimate the copy number of a specific DNA fragment.29, 30 For regions covered by the panel, CNA detection by coverage data resulted in a sensitivity of 99% as validated by two independent methods: MLPA and low-coverage WGS (see Supplementary Results).
SNVs and CNAs in leukemia-related genes are similarly common at the time of initial disease and at the time of relapse
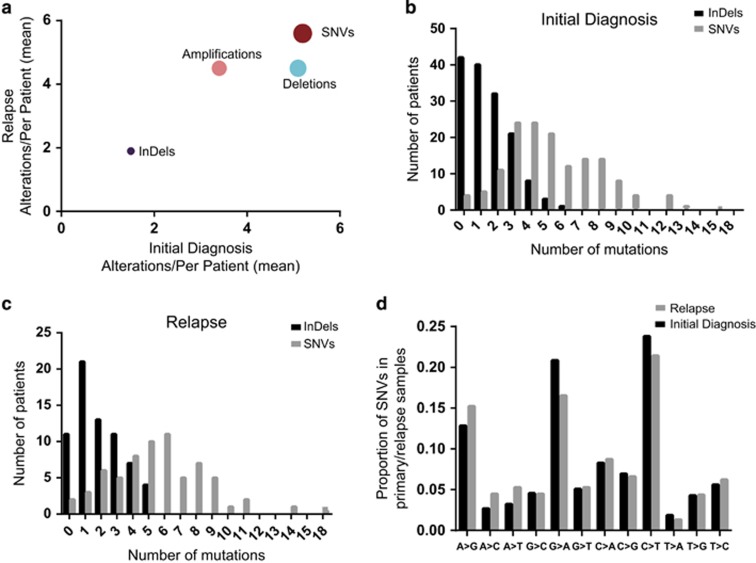
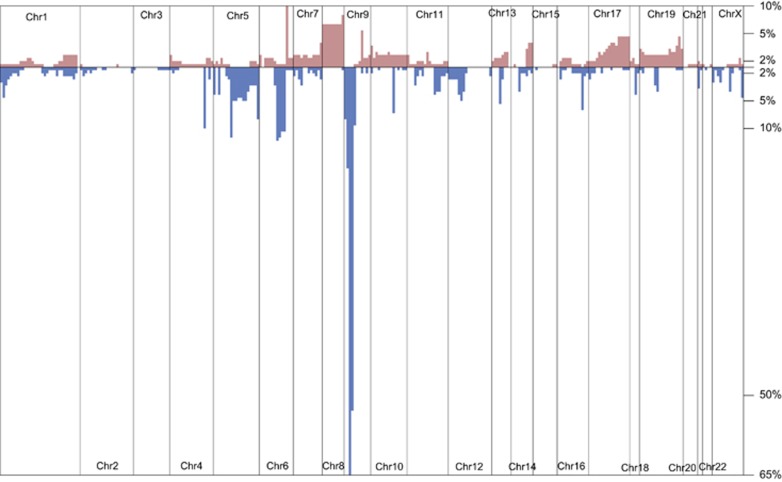
On average, gene panel sequencing detected seven SNVs or InDels per sample (in total 1496; Supplementary Table 5). We observed similar frequencies of SNVs in leukemia-related genes in the group of patients with initial disease and in the group of relapse patients (Figure 1a). InDels tended to be more frequent in relapse compared with primary leukemia (P=0.0618, Fisher's exact test; Figures 1b and c) suggesting that InDels may be a part of the mutational pattern induced by chemotherapy. Transitions constituted 61% of missense mutations with exchanges of C>T (22%) being the most common nonsynonymous mutations (Figure 1d), indicating that deamination of cytosine is a common mechanism of mutagenesis.31 Most of the mutations were found at allele frequencies of 35 to 65% (Supplementary Figure 2), indicating heterozygosity. Only few mutations were predicted to be homozygous, but the unknown blast content and subclonality of the samples do not allow for an unequivocal discrimination between homozygous and heterozygous events. We were able to perform CNA analyses in 202 of 214 samples (144 primary and 58 relapsed T-ALLs; Figure 2) and found that the average number of genes affected by CNAs in the leukemia-related genes of our panel does not differ between initial disease and relapse in pediatric T-ALL (INI: 8.6 vs REL: 9). As expected, CNAs most commonly affected chromosome 9 with CDKN2A and CDKN2B deletions detected in 66% and 55% of all patients, respectively.32 We observed recurrent deletions of regions on chr6:q14-15 (10%) and on chr5:q22-35 (up to 11% for APC INI: 20; REL: 3). Duplications of chromosome 8, including MYC, were the most common amplifications found in 7% of all patients (INI: 11; REL: 6; Figure 2, for details see Supplementary Results).
Figure 1.
(a) Mean of SNVs, InDels, amplifications and deletions in initial diagnosis (x) plotted against relapse (y). Distribution of the number of SNVs (gray) and InDels (black) detected in the analyzed genes in the (b) initial diagnosis patients and in (c) relapse patients. (d) Frequencies of different types of missense mutations in primary and relapse samples.
Figure 2.
Frequencies of copy number alterations (red—mplifications; blue—deletions) detected in 202 T-ALL patients for whom coverage data from targeted sequencing were available. CNAs were plotted against their chromosomal position.
Leukemogenic driver genes are characterized by high mutation densities
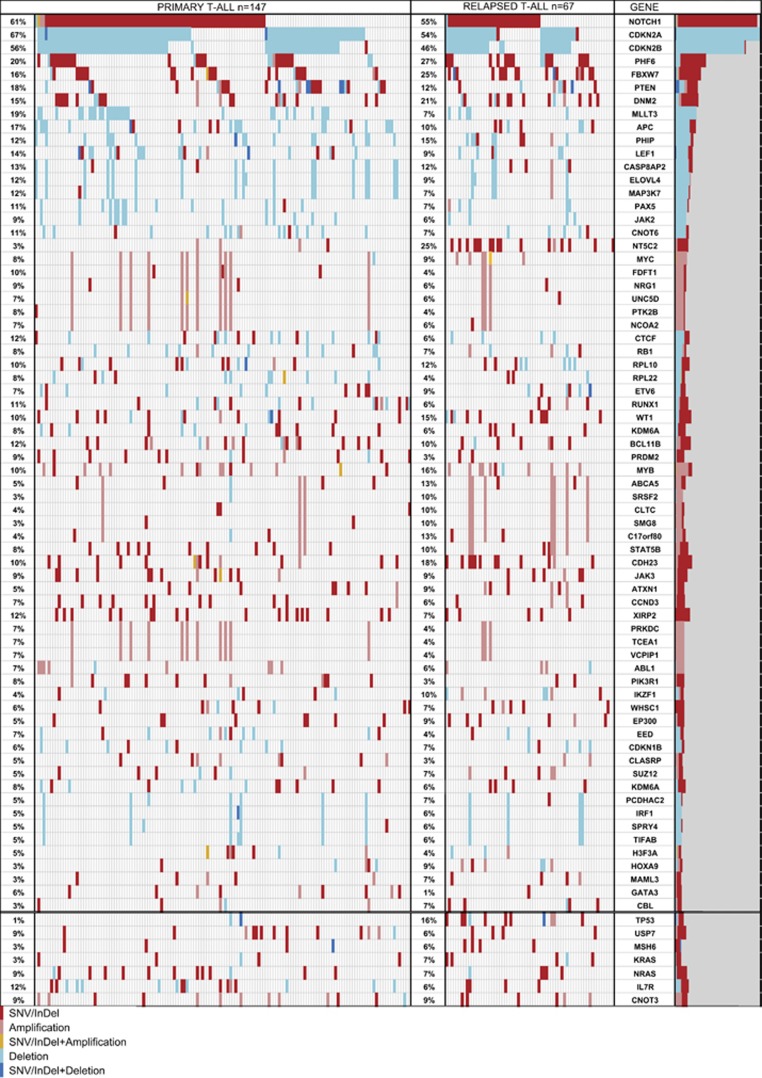
We found known leukemia drivers to be mutated at the expected frequency21, 33, 34 (Figure 3). This includes among others NOTCH1, PHF6, FBXW7 and PTEN, that had previously been found to be mutated at the frequency of approximately 50%, 16%, 20% and 17%, respectively. Other genes that were mutated in more than 10% of all patients were DNM2, XIRP2 and CDH23. One hundred and eighty-two additional genes recurrently carried SNVs and InDels in less than 10% of patients.
Figure 3.
Comparison of the pattern of alterations in initial diagnosis patients (n=147) and in relapse (n=67) in the genes with high mutation density of SNVs/InDels (>1.9/Mbp; Supplementary Table 7) or/and high frequency of CNA (>5% of patients; data available for 202 patients: 59 REL and 144 INI). Frequencies refer to all mutations (SNVs/InDels+CNAs), alterations were sorted according to their frequency and position in the genome. Red—SNV/InDel; pink—amplification; orange—SNV/InDel+amplification; pale blue—deletion; dark blue—SNV/InDel+deletion.
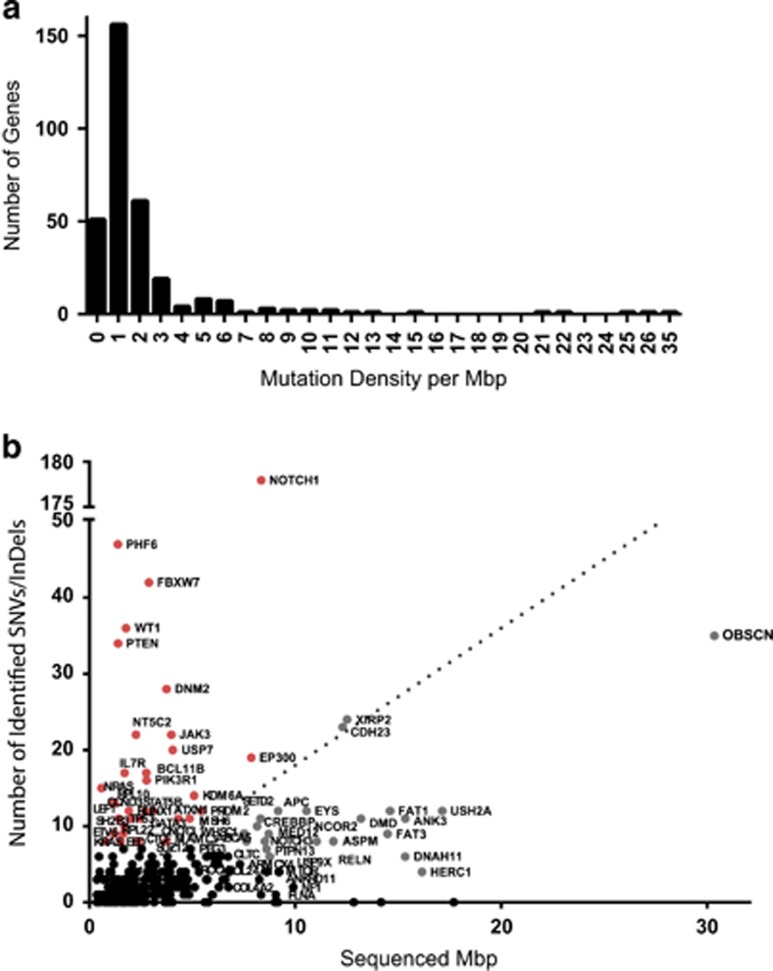
We next assessed which of the recurrently mutated genes likely represent drivers of leukemogenesis. The best way to distinguish driver genes from genes with randomly acquired (passenger) mutations would be through their pattern of mutations.35 However the low frequency of mutations in most of the analyzed genes precludes such an analysis. Therefore, we have calculated the mutation density for each gene by dividing the number of detected nonsynonymous SNVs and InDels by the length of the targeted exons per gene (mutations per Mbp; Supplementary Table 6). The mean mutation density for the regions covered by the panel was 1.8/Mbp. Most of the genes (77%, Figure 4) showed a mutation density below the mean. Genes with low mutation densities are likely to be randomly mutated and probably do not have a specific role in leukemogenesis, although some large genes in this category such as OBSCN, EYS, DMD, FAT1, USH2A and ANK3 with a low mutation density between 0.7 and 1.2 were found to be mutated in a notable proportion of patients. This analysis confirmed the known leukemogenic role of genes with high mutation densities (>20/Mbp: PHF6, NRAS, PTEN, NOTCH1, WT1; >10/Mbp: FBXW7, KRAS, IL7R, NT5C2, CCND3, RPL10, RPL22). XIRP2 and CDH23, both mutated in more than 20 patients but without an obvious link to leukemogenesis, had a much lower mutation density of 1.9/Mbp.
Figure 4.
(a) Histogram of the distribution of mutation density in the analyzed genes (b) Mutation density shown as length of the targeted exons (Mpb) plotted against the absolute number of detected SNVs/InDels; red—known leukemia drivers or cancer-related genes; gray—genes that show low mutation density (<2), regardless of the high mutation frequency observed in some of them (for example, OBSCN, XIRP2, CDH23); black—genes that carry a low absolute number of mutations (n<10).
T-ALL relapses are characterized by a limited number of specific alterations in leukemia-related target genes
As shown previously, NT5C2 was the gene that most frequently carried relapse-specific mutations.8, 36 Twenty NT5C2 SNVs were found in 16 (24%) of the 67 relapse samples and one NT5C2 SNV was carried by leukemic cells of a patient at initial diagnosis (P=0.0001, Fisher's exact test). Thirteen out of 21 mutations detected in NT5C2 were found in the hotspot positions R238, R367 and D407. These mutations cause a gain of function of this nucleotidase thus promoting the inactivation of nucleoside analogs that are used in the chemotherapy of T-ALL 8. Out of the 21 mutations detected in NT5C2, 15 (71%) were, as reported before,6 subclonal with an allele frequency (AF) of below 0.7 in proportion to the average of the AF in the major clone represented by mutations with the highest AF in the respective sample (Supplementary Table 5). This subclonality of NT5C2 mutations suggests that these have been acquired later than the presumably clonal relapse-initiating event. Therefore, NT5C2 mutations in T-ALL are likely not the relapse-initiating events. This observation is consistent with the lack of prognostic relevance of NT5C2 mutations in relapsed T-ALL (see below).
TP53 was the second most common gene to be mutated in a relapse-specific fashion. Only 2/147 samples at initial disease carried alterations of TP53, whereas we detected TP53 SNVs (7 missense, 1 stopgain, 1 frameshift deletion) or large deletions (2) in 9/67 samples of relapse patients (P=0.0007, Fisher's exact test). Mutations of TP53 detected at relapse were individually non-recurrent and affected either the transactivation or DNA-binding domains. SIFT,37 MutationTaster38 and PolyPhen-239 concordantly predicted these mutations to be damaging for p53 function (Supplementary Table 5). None of the TP53 mutations were detected at the known activating hotspots.40 Patients who carried SNVs or deletions of TP53 showed higher mutation rates in other genes compared with those with wild-type TP53 (31 vs 14 alterations per patient, P=0.0002, Mann–Whitney's test). This difference was particularly notable in the frequencies of CNAs (mutTP53: 24 vs wtTP53: 8 per patient, P=0.0004, Mann–Whitney's test), whereas the difference in SNVs and InDels was less pronounced (mutTP53: 8 vs wtTP53: 6 per patient, P=0.0036, Mann–Whitney's test). These findings suggest that disruption of TP53 function in T-ALL results in an accumulation of somatic duplications and deletions.
In addition, we identified amplifications of a region on chromosome 17 q11.2-24.3, represented in our panel by STAT5B, STAT5A, STAT3, DHX8, SMG8, CLTC, ABCA5, C17orf80, to be significantly enriched in relapse (REL 7/58 vs INI 3/144, P=0.0068, Fisher's exact test). Because targeted sequencing does not allow to define exact borders of CNAs, three of the samples that carry amplifications of large regions of chromosome 17 were analyzed by low-coverage WGS. We thus resolved the amplified region to chr17: q21.23-q25.3 in two samples (PATNR: 20; 22) and to chr17: p11.2-q25.3 in the third sample (PATNR: 50). There was no significant association of amplifications of chromosome 17 with prognosis in relapse. However, this region is enriched in genes of the JAK/STAT pathway that has been implicated in leukemogenesis25 and in the ABCA gene family that has been implicated in drug resistance.41 As these two groups of genes may offer options for targeted treatment,41, 42, 43 it will be important to identify this amplification in future studies.
Fatal outcome in relapsed T-ALL is heralded by the mutational profile
Clinical follow-up data were available for 66 of the 67 relapse patients. Forty-nine of these patients (74%) suffered an event (second relapse, death or secondary malignancy), whereas 17 (26%) could be salvaged by relapse therapy.
TP53 mutations were most highly predictive of a second event. For one out of nine patients with alterations in TP53 in leukemic cells, clinical data were not available. All other patients who carried a total of nine TP53 mutations and/or deletions died within 9 months after first relapse, whereas 17 of 58 patients without TP53 mutation (29%) survived (P=0.001, log-rank test). Five of ten TP53 mutations detected in our entire cohort had an AF equal or above 74%. Such high allele frequencies were found in only 106/1507 of all SNVs and InDels in our analysis (Supplementary Figure 2), suggesting that TP53 mutations were more frequently homozygous events than mutations in the other analyzed genes. Two were a consequence of loss of heterozygosity through a deletion of the second allele (patients GW15, PATNR: 14). In three more samples, no CNAs were detected but all SNVs and single-nucleotide polymorphisms in TP53 were homozygous, indicating that loss of heterozygosity had occurred via uniparental disomy. Therefore, there is a strong selective pressure that results in the acquisition of homozygous TP53 inactivation in relapsed pediatric T-ALL.
By contrast, NT5C2 mutations, highly enriched in relapse, had no prognostic impact in our series. Although patients who harbored NT5C2 mutations had relapsed earlier (P=0.01, Fisher's exact test, Supplementary Table 7), the frequency of NT5C2 mutations did not differ between patients who suffered a second event (12/49) and patients who were salvaged by second line treatment (4/17; P=1, Fisher's exact test).
We next focused on mutations in those genes that were not significantly enriched in relapse, but when found at relapse occurred only in samples from patients who suffered a second event and death. These were USP7, MSH6, KRAS, NRAS, CNOT3 and IL7R.
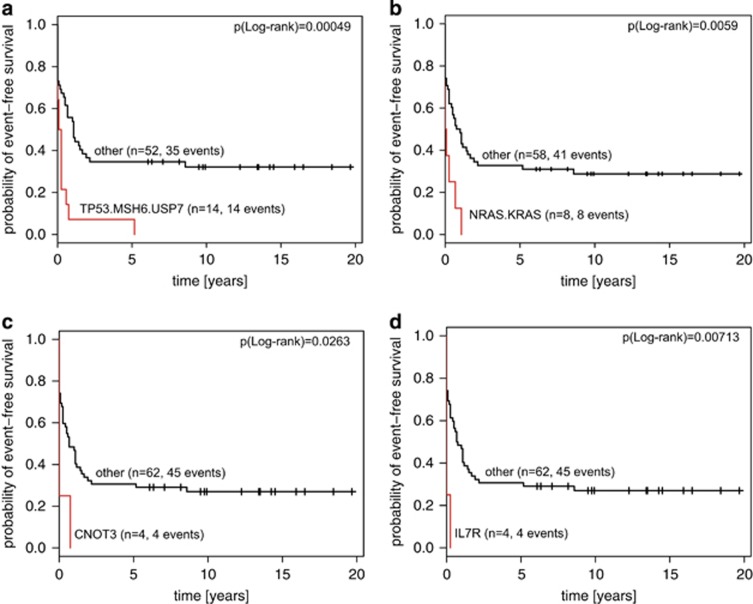
Mutations in USP7, that deubiquitinates p53 preventing its degradation and enhancing p53-dependent transcription regulation, cell growth repression and apoptosis,44 were found in four relapse samples. Three patients carried a frameshift insertion of USP7 and one carried a missense variant predicted to be damaging for the structure of the protein (Supplementary Table 5). All except one were located in the catalytic domain of USP7. Another gene that is responsible for maintenance of genetic stability and tumor suppression and which was mutated exclusively in relapse patients with fatal outcome was the DNA mismatch repair gene MSH6. It recognizes mismatched nucleotides before their repair. Six MSH6 mutations were found in four relapse samples. All samples carried at least one mutation in the sequence coding for the conserved MutS domain. We detected three MSH6 mutations leading to a stop codon, two InDels and one missense mutation concordantly predicted to be deleterious by SIFT,37 Polyphen39 and MutationTaster.38 Together, mutations in TP53, USP7 and MSH6 define a group of fatal relapsed leukemias predicted to be defective in surveillance of DNA integrity (P=0.00049, log-rank test, Figure 5a).
Figure 5.
Kaplan–Meier plots of event-free survival in the group of 66 relapse patients. (a) Patients who carry SNV/InDel or deletions in TP53/MSH6/USP7 vs other. (b) Patients who carry mutations in KRAS or NRAS vs other. (c) Patients who carry SNVs in CNOT3 vs other. (d) Patients who carry mutations in IL7R vs other.
The Ras/Raf/MEK/ERK pathway has a crucial role in the transmission of proliferative signals from membrane-bound receptors. Ras mutations were found in eight relapsed patients, while no mutation of the downstream target of Ras signaling, BRAF, was detected in our cohort. Seven out of eight NRAS and KRAS mutations were detected at known activating hotspots (G12, A59, Q61 and A146) or in their close proximity (V14). All relapsed patients with KRAS or NRAS mutations in leukemic samples died, six out of eight within 3 months after relapse, indicating that these mutations are associated with treatment resistance (P=0.0059, log-rank test, Figure 5b).
CNOT3 and IL7R are similarly associated with early treatment failure after relapse (Figures 5c and d). CNOT3 was previously identified as a tumor suppressor in 8% of adult T-ALLs.23 Out of four CNOT3 mutations, three were found in a region coding for the N-terminal coiled-coil domain and one directly on the border with a linker region (CNOT3-M). One of the mutations was an exchange of Arginine 57 in a hotspot position previously reported to interfere with the tumor suppressor function of CNOT3.23 One of the mutations is a frameshift insertion and the effect of the two remaining mutations is damaging for the protein structure based on predictive algorithms. All four patients harboring CNOT3 mutations at relapse died, three of them within 1 month after diagnosis. The same applies to patients with IL7R mutations. All IL7R mutations were found at the hotspot positions coding for Leucine 242 and 243, three out of four being nonframeshift insertions introducing an unpaired cysteine and inducing ligand-independent constitutive hyperactivation of IL7R-mediated signal transduction.42, 45
Mutations of genes governing DNA integrity (TP53, USP7, MSH6; n=15 patients) and in the key players of the RAS signaling pathway (KRAS, NRAS; n=8 patients) combined with alterations of IL7R (n=4) and CNOT3 (n=4) represent a prognostic signature that recognizes 24/49 (49%) relapse patients with fatal outcome. Notably, out of those 23 patients for whom clinical data are available, 13 were resistant to treatment and did not reach second remission. Six more patients suffered from an early second relapse and died before proceeding to stem cell transplantation. Of the four patients who proceeded to stem cell transplantation, three died after relapse and one experienced a secondary malignancy. In comparison, only nine of 43 patients without high-risk mutations failed to reach second remission (P=0.0058, Fisher's exact test).
Discussion
Recurrent SNVs and InDels
We compared mutational patterns of point mutations and CNAs in a large group of 147 primary and 67 relapsed T-ALL patients by targeted sequencing of more than 300 genes. The distribution of the most common SNVs/InDels detected in our cohort is in accordance with published data, reproducing the frequent association of T-ALL with mutations in NOTCH1, FBXW7, PHF6, PTEN, IL7R, WT1 and others34, 46 (Figure 3). Although we did not identify novel common driver genes that were mutated in more than 10% of patients, 140 of 147 primary T-ALL samples carried at least one mutation in a gene that was mutated in 2 to 10% of all patients. We hypothesize that at least some of those genes that are recurrently mutated at low frequency may contribute to leukemogenesis and that these rare genetic alterations contribute considerably to the genetic diversity of T-ALL.
In our previous whole-exome sequencing analysis of matched samples from primary and relapsed T-ALL, we observed that the number of detected SNVs and InDels between initial diagnosis and relapse doubled,6 while the numbers detected by targeted sequencing as performed here were not significantly higher in relapse. This is likely explained by exome sequencing identifying both, the accumulation of driver and passenger mutations. By contrast, leukemia-related mutations detected by targeted sequencing do not show a major change in number during the evolution to relapse.
Panel sequencing allows for robust detection of CNAs
The feasibility of targeted NGS for the detection of CNAs was shown by validation by two independent methods: MLPA and WGS. Moreover, genome-wide copy number analyses using Affymetrix single-nucleotide polymorphism arrays in 73 (ref. 32) and 50 (ref. 47) T-ALL patients show a high concordance in terms of frequencies of deletions and amplifications with those found in genes covered by our analyses. Within the group of selected regions contained in the Haloplex panel, genes were more frequently affected by duplications or deletions than by point mutations and InDels (mean number of SNVs/InDels per gene: 7, median: 7; CNA: mean: 8.6, median: 5). In 31 of 202 (15%) samples for which CNA data were available we observed large regions to be affected, possibly affecting entire chromosomes. Assuming that the proportion of genes affected by CNA within our panel is representative for the entire genome, we estimate that in average between two and three percent of the genome is altered by copy number changes in pediatric T-ALL.
TP53 and Ras genes confer a dismal prognosis in relapsed T-ALL
Mutations detected in NT5C2 and in TP53 were significantly enriched in relapse. Activating NT5C2 mutations were previously reported to be relapse-specific36 and to drive resistance against 6-mercaptopurine and 6-thioguanine in relapsed ALLs.8 Twelve out of 16 NT5C2-positive relapse patients carried at least one mutation of the known activating hotspot. In the four remaining leukemic samples, missense exchanges at positions R34Q, R195Q and T489M and nonframeshift deletion of four amino acids at positions 396-400 were detected.
NT5C2 mutations are associated with early first relapse, indicating that the acquisition of NT5C2 mutations is driven by selective pressure. Induction failure after relapse is not predicted by NT5C2 mutations, most likely because derivatives of 6-mercaptopurine are not a component of induction treatment after relapse.
Alterations of TP53 were reported to be gained at relapse in 54% of 23 ALL samples and found to predict chemotherapy resistance and poor outcome in first relapse of childhood B-cell-precursor ALL.10 In T-ALL, TP53 mutations were previously detected in first relapse in 12 of 51 patients (24%).9 Patients with TP53 mutations in leukemic samples experienced a shorter duration of survival and were significantly less likely to have achieved a complete second remission than patients without TP53 mutations.9 Our results confirm and extend these earlier findings. We find TP53 to be mutated in a relapse-specific manner with high allele frequencies, indicating homozygous events. Moreover, TP53 mutants were characterized by a higher overall mutation rate in comparison with TP53 wild-type patients, which was particularly evident by the accumulation of CNAs. It has been demonstrated that TP53 mutations result in the accumulation of genomic rearrangements in several cancer entities.48, 49, 50 However, we cannot formally exclude that mutations in TP53 are not a cause but the consequence of genomic instability.
T-ALLs that showed such genetic instability as a result of TP53 mutation failed treatment within 9 months after relapse. When we combine TP53 alterations with recurrent mutations in MSH6 and USP7, which also interfere with the regulation of DNA surveillance, we can define a group of approximately 20% of patients with invariably fatal outcome in first relapse (15/67).
Mutations in NRAS and KRAS recognize eight relapsed patients with fatal outcome, although neither of these mutations had significant prognostic impact in primary disease (P=0.15 and 0.49, respectively, Gray's test). Deregulation of the Ras/Raf/MEK pathway has repeatedly been implicated in resistance to chemotherapy.19, 51 This may be a result of constitutive activation of its direct components (NRAS, KRAS and BRAF), upstream receptors (EGFR, FLT3) or chimeric chromosomal translocations (BCR–ABL, TEL–PDGFR). Targeting this pathway may be a novel therapeutic approach for drug resistant leukemia, which is often cross-resistant to multiple chemotherapeutic drugs.52 With relapsed T-ALL being a rare entity, it is fortunate that numerous compounds targeting the Ras/Raf/MEK pathway, such as the small molecules trametinib or selumetinib, have been tested in other conditions and could potentially be repurposed for the treatment of T-ALL.
In addition, mutations in CNOT3 and IL7R are exclusively found in patients experiencing treatment failure after first relapse. Although a correlation of IL7R mutations with prognosis in primary T-ALL could not be established,43 IL7R mutations in relapse were correlated with very short survival (P=0.007, log-rank test). These findings have potential therapeutic implications as it was shown that T-ALL cells harboring IL7R mutations are sensitive to JAK–STAT pathway inhibitors.42
Although the low frequency of these mutations individually precludes a separate analysis as prognostic biomarkers, a combination of these functionally related genes identifies 49% (24 of 49) of all patients that fail relapse treatment, mainly as a result of nonresponse. Twelve of these 24 patients would have been candidates for targeted treatment with either JAK–STAT or MEK inhibitors. Six patients that carry missense mutations in TP53 could potentially benefit from agents that restore the active conformation of mutant p53 and induce p53-dependent suppression of tumor cell growth as currently being tested in other cancer indications.53, 54 We are currently developing xenograft models of genetically well-defined leukemias that will allow correlating the mutational profile with drug sensitivity and will thus enable us to directly test the potential to use the signature reported here to design new targeted treatment strategies.
In reverse, the absence of mutations in TP53, MSH6, USP7, NRAS, KRAS, CNOT3 and IL7R defines a group among T-ALL relapse patients with a 40% chance of survival with conventional relapse treatment (Supplementary Figure 3).
Acknowledgments
We thank the following institutions for grants: German Consortium for Translational Cancer Research (DKTK), ‘Tour der Hoffnung', Manfred Lautenschläger Stiftung, European Commission (FP7, ERA-NET on Translational Cancer Research, TRANSCALL to MUM). JBK received fellowship from the Heidelberg Research Center for Molecular Medicine. ORB received grants from the Excellence Initiative, Innovation Fund FRONTIER, University of Heidelberg. We thank the EMBL GeneCore sequencing team.
Author contributions
PR-P and JBK designed the experiments, analyzed the data, interpreted it and wrote the manuscript. JH provided statistical analyses for the relapsed cohort and edited the manuscript. MZ provided statistical analyses for the patients at initial diagnosis. TR performed WGS analysis and supported bioinformatic analyses. ORB and GS performed Sanger sequencing for validation cohort and their analysis. EO performed NGS library preparation. JCS was involved in design of the target capture panel. MSt, MSc, GC, RK-S and CE provided the patients samples and clinical data. VB advised on the methodology. JOK, MUM and AEK conceptualized and supervised the research and contributed to the writing of the manuscript. All the authors read and approved the manuscript.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
The authors declare no conflict of interest.
Supplementary Material
References
- Schrappe M, Valsecchi MG, Bartram CR, Schrauder A, Panzer-Grumayer R, Moricke A et al. Late MRD response determines relapse risk overall and in subsets of childhood T-cell ALL: results of the AIEOP-BFM-ALL 2000 study. Blood 2011; 118: 2077–2084. [DOI] [PubMed] [Google Scholar]
- von Stackelberg A. Charité Universitätsmedizin Berlin, 2015, with reference to the trials ALL-REZ BFM 83 - 2002.
- Locatelli F, Schrappe M, Bernardo ME, Rutella S. How I treat relapsed childhood acute lymphoblastic leukemia. Blood 2012; 120: 2807–2816. [DOI] [PubMed] [Google Scholar]
- Henze G, von Stackelberg A, Eckert C. ALL-REZ BFM—the consecutive trials for children with relapsed acute lymphoblastic leukemia. Klin Padiatr 2013; 225: S73–S78. [DOI] [PubMed] [Google Scholar]
- Tallen G, Ratei R, Mann G, Kaspers G, Niggli F, Karachunsky A et al. Long-term outcome in children with relapsed acute lymphoblastic leukemia after time-point and site-of-relapse stratification and intensified short-course multidrug chemotherapy: results of trial ALL-REZ BFM 90. J Clin Oncol 2010; 28: 2339–2347. [DOI] [PubMed] [Google Scholar]
- Kunz JB, Rausch T, Bandapalli OR, Eilers J, Pechanska P, Schuessele S et al. Pediatric T-lymphoblastic leukemia evolves into relapse by clonal selection, acquisition of mutations and promoter hypomethylation. Haematologica 2015; 100: 1442–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 2008; 322: 1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med 2013; 19: 368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diccianni MB, Yu J, Hsiao M, Mukherjee S, Shao LE, Yu AL. Clinical significance of p53 mutations in relapsed T-cell acute lymphoblastic leukemia. Blood 1994; 84: 3105–3112. [PubMed] [Google Scholar]
- Hof J, Krentz S, van Schewick C, Korner G, Shalapour S, Rhein P et al. Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J Clin Oncol 2011; 29: 3185–3193. [DOI] [PubMed] [Google Scholar]
- Clappier E, Collette S, Grardel N, Girard S, Suarez L, Brunie G et al. NOTCH1 and FBXW7 mutations have a favorable impact on early response to treatment, but not on outcome, in children with T-cell acute lymphoblastic leukemia (T-ALL) treated on EORTC trials 58881 and 58951. Leukemia 2010; 24: 2023–2031. [DOI] [PubMed] [Google Scholar]
- Ferrando A. NOTCH mutations as prognostic markers in T-ALL. Leukemia 2010; 24: 2003–2004. [DOI] [PubMed] [Google Scholar]
- Krieger D, Moericke A, Oschlies I, Zimmermann M, Schrappe M, Reiter A et al. Frequency and clinical relevance of DNA microsatellite alterations of the CDKN2A/B, ATM and p53 gene loci: a comparison between pediatric precursor T-cell lymphoblastic lymphoma and T-cell lymphoblastic leukemia. Haematologica 2010; 95: 158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel A, Schnittger S, Weissmann S, Kuznia S, Kern W, Kohlmann A et al. TP53 mutations occur in 15.7% of ALL and are associated with MYC-rearrangement, low hypodiploidy, and a poor prognosis. Blood 2014; 124: 251–258. [DOI] [PubMed] [Google Scholar]
- Van Vlierberghe P, Ambesi-Impiombato A, De Keersmaecker K, Hadler M, Paietta E, Tallman MS et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood 2013; 122: 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandapalli OR, Zimmermann M, Kox C, Stanulla M, Schrappe M, Ludwig WD et al. NOTCH1 activation clinically antagonizes the unfavorable effect of PTEN inactivation in BFM-treated children with precursor T-cell acute lymphoblastic leukemia. Haematologica 2013; 98: 928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, Tolle G et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood 2006; 108: 1151–1157. [DOI] [PubMed] [Google Scholar]
- Kox C, Zimmermann M, Stanulla M, Leible S, Schrappe M, Ludwig WD et al. The favorable effect of activating NOTCH1 receptor mutations on long-term outcome in T-ALL patients treated on the ALL-BFM 2000 protocol can be separated from FBXW7 loss of function. Leukemia 2010; 24: 2005–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianfelici V, Chiaretti S, Demeyer S, Di Giacomo F, Messina M, La Starza R et al. RNA sequencing unravels the genetics of refractory/relapsed T-cell acute lymphoblastic leukemia. Prognostic and therapeutic implications. Haematologica 2016; 101: 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuurbier L, Petricoin EF 3rd, Vuerhard MJ, Calvert V, Kooi C, Buijs-Gladdines JG et al. The significance of PTEN and AKT aberrations in pediatric T-cell acute lymphoblastic leukemia. Haematologica 2012; 97: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest 2012; 122: 3398–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atak ZK, Gianfelici V, Hulselmans G, De Keersmaecker K, Devasia AG, Geerdens E et al. Comprehensive analysis of transcriptome variation uncovers known and novel driver events in T-cell acute lymphoblastic leukemia. PLoS Genet 2013; 9: e1003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet 2013; 45: 186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012; 481: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandapalli OR, Schuessele S, Kunz JB, Rausch T, Stutz AM, Tal N et al. The activating STAT5B N642H mutation is a common abnormality in pediatric T-cell acute lymphoblastic leukemia and confers a higher risk of relapse. Haematologica 2014; 99: e188–e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009; 25: 2283–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team RA Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Grambsch TMTaPMModeling Survival Data: Extending the Cox Model. Springer: New York, NY, USA, 2000. [Google Scholar]
- Bolli N, Manes N, McKerrell T, Chi J, Park N, Gundem G et al. Characterization of gene mutations and copy number changes in acute myeloid leukemia using a rapid target enrichment protocol. Haematologica 2015; 100: 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente C, Schwab C, Broux M, Geerdens E, Degryse S, Demeyer S et al. Targeted sequencing identifies associations between IL7R-JAK mutations and epigenetic modulators in T-cell acute lymphoblastic leukemia. Haematologica 2015; 100: 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Grigg GW. DNA methylation and mutation. Mutat Res 1993; 285: 61–67. [DOI] [PubMed] [Google Scholar]
- Remke M, Pfister S, Kox C, Toedt G, Becker N, Benner A et al. High-resolution genomic profiling of childhood T-ALL reveals frequent copy-number alterations affecting the TGF-beta and PI3K-AKT pathways and deletions at 6q15-16.1 as a genomic marker for unfavorable early treatment response. Blood 2009; 114: 1053–1062. [DOI] [PubMed] [Google Scholar]
- Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet 2010; 42: 338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer 2016; 16: 494–507. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz Jr LA, Kinzler KW. Cancer genome landscapes. Science 2013; 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet 2013; 45: 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9: 701–713. [DOI] [PubMed] [Google Scholar]
- Hedditch EL, Gao B, Russell AJ, Lu Y, Emmanuel C, Beesley J et al. ABCA transporter gene expression and poor outcome in epithelial ovarian cancer. J Natl Cancer Inst 2014; 106: dju149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, Paganin M et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet 2011; 43: 932–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, te Kronnie G et al. Gain-of-function mutations in interleukin-7 receptor-alpha (IL7R) in childhood acute lymphoblastic leukemias. J Exp Med 2011; 208: 901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cao M, Dong J, Li C, Xu W, Zhan Y et al. ABRO1 suppresses tumourigenesis and regulates the DNA damage response by stabilizing p53. Nat Commun 2014; 5: 5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shochat C, Tal N, Gryshkova V, Birger Y, Bandapalli OR, Cazzaniga G et al. Novel activating mutations lacking cysteine in type I cytokine receptors in acute lymphoblastic leukemia. Blood 2014; 124: 106–110. [DOI] [PubMed] [Google Scholar]
- Van Vlierberghe P, Pieters R, Beverloo HB, Meijerink JP. Molecular-genetic insights in paediatric T-cell acute lymphoblastic leukaemia. Br J Haematol 2008; 143: 153–168. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007; 446: 758–764. [DOI] [PubMed] [Google Scholar]
- Abaigar M, Robledo C, Benito R, Ramos F, Diez-Campelo M, Hermosin L et al. Chromothripsis Is a Recurrent Genomic Abnormality in High-Risk Myelodysplastic Syndromes. PLoS One 2016; 11: e0164370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Jhanwar SC, Testa JR. Chromothripsis in a case of TP53-deficient chronic lymphocytic leukemia. Leuk Res Rep 2012; 1: 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 2012; 148: 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta 2007; 1773: 1263–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving J, Matheson E, Minto L, Blair H, Case M, Halsey C et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood 2014; 124: 3420–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S, Bykov VJ, Ali D, Andren O, Cherif H, Tidefelt U et al. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol 2012; 30: 3633–3639. [DOI] [PubMed] [Google Scholar]
- Zawacka-Pankau J, Selivanova G. Pharmacological reactivation of p53 as a strategy to treat cancer. J Intern Med 2015; 277: 248–259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.