Abstract
Age-related macular degeneration (AMD) is a devastating neurodegenerative disease and a major cause of blindness in the developed world. Owing to its complexity and the lack of an adequate human model that recapitulates key aspects of the disease, the molecular mechanisms of AMD pathogenesis remain poorly understood. Here we show that cultured human retinal pigment epithelium (RPE) from AMD donors (AMD RPE) are functionally impaired and exhibit distinct phenotypes compared with RPE cultured from normal donors (normal RPE). Accumulation of lipid droplets and glycogen granules, disintegration of mitochondria, and an increase in autophagosomes were observed in AMD RPE cultures. Compared with normal RPE, AMD RPE exhibit increased susceptibility to oxidative stress, produce higher levels of reactive oxygen species (ROS) under stress conditions, and showed reduced mitochondrial activity. Measurement of the ratio of LC3-II/ LC3-I, revealed impaired autophagy in AMD RPE as compared with normal RPE. Autophagic flux was also reduced in AMD RPE as compared with normal RPE, as shown by inability of AMD RPE to downregulate p62 levels during starvation. Impaired autophagic pathways were further shown by analyzing late autophagic vesicles; immunostaining with lysosome-associated membrane protein 1 (LAMP-1) antibody revealed enlarged and annular LAMP-1-positive organelles in AMD RPE as opposed to smaller discrete puncta observed in normal RPE. Our study provides insights into AMD cellular and molecular mechanisms, proposes dysfunctional autophagy as an underlying mechanism contributing to the pathophysiology of the disease, and opens up new avenues for development of novel treatment strategies.
Age-related macular degeneration (AMD) is a major cause of vision loss among elderly in developed countries worldwide. AMD affects approximately 30–50 million people, 1, 2 and its prevalence is expected to double by 2050.3
AMD initially affects the retinal pigment epithelium (RPE), a monolayer of pigmented and polarized central nervous system (CNS) tissue, and over time, leads to secondary loss of photoreceptor cells.2, 4
AMD is a multifactorial disease3 and its pathogenesis remains largely elusive. Mounting evidence suggests a complex interaction of genetic, environmental and metabolic factors contributing to the pathology of AMD.5 Impaired RPE function in dry AMD causes the formation of extracellular deposits called drusen,6 which accumulate between the RPE and Bruch's membrane (BM). The wet form of AMD involves choroidal neovascularization followed by formation of a disciform scar.7 Dry AMD could transform to the wet form at the latest stage of the disease.2 However, a patient could also present both forms at different stages of AMD.
The RPE has important roles in retinal homeostasis. It functions as a retinal blood barrier, a source of polarized growth factor release and transporter of ions, water and metabolic products from the subretinal space to the blood 8, 9 The RPE delivers blood-derived nutrients to photoreceptors, absorbs light and performs phagocytosis of the outer segments of photoreceptors.10
Numerous studies have reported on age-related physiological changes in RPE,11 including mitochondrial DNA damage,12 accumulation of lipofuscin,13 elevated amyloid β production,14 enhanced tissue factor (TF) expression,15 increased acidic beta-galactosidase activity suggestive of lysosomal dysfunction16 and altered expression of RPE structural proteins.17
Autophagy is a lysosome-mediated degradation process for non-essential or damaged cellular constituents to supply the cell with energy and to maintain homeostasis.18, 19 Recently, much interest has focused on the role of autophagy in health and disease.20, 21 Moreover, lipid droplets have been identified as an autophagy substrate22 and the impact of autophagy on mitochondrial function has been discussed.23, 24 The latter is particularly relevant to degenerative diseases, such as AMD, in which oxidative stress occurs over time. Recently, dysregulated autophagy in RPE was shown to increase susceptibility to oxidative stress and AMD.25 Another study reported that activation of autophagy clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human RPE cells.26
Several reviews also propose a role for autophagy in AMD,27, 28, 29 however, direct evidence for altered autophagic function in the pathophysiology of AMD has yet to be documented in an in vitro cellular model.
Here, we analyzed the phenotype and function of RPE cultures established from human donor eyes. In this novel system for studying AMD, we show that autophagy is selectively dysregulated in AMD RPE contributing to disease phenotypes.
Results
Human RPE from AMD donors exhibit distinct disease phenotypes
We developed AMD RPE cultures by isolating RPE from macula region of five AMD donors (AMD RPE) and five normal donors (normal RPE) according to established protocols.30 We purified RPE cells by magnetic cell sorting, using specific antibodies as described below and cultured them at 5% oxygen concentration to avoid oxidative stress. We performed the genetic study of single-nucleotide polymorphisms (SNPs) for AMD susceptibility loci (Table 1). Interestingly, AMD donor# 9, which had a protective allele, nevertheless developed AMD, possibly due to heavy smoking (40 years of two packs of cigarettes per day), whereas control donors #6 and #10 despite presenting the AMD susceptibility loci did not develop disease, providing further evidence for the multifactorial origin of AMD.
Table 1. Genotyping and clinical information of AMD and control RPE.
| Donor Id# | Donor age (gender) | Clinical diagnosis | CFH (C: risk) | HTRA1 (A: risk) | LOC (T: risk) | Factor B (T: protective) | C2 (C: protective) | Smoking | Cause of death | Enucleation (h) |
|---|---|---|---|---|---|---|---|---|---|---|
| 006 | 72 (M) | CONTROL | CT | AG | GT | CC | GG | Quit in 1993 | Chronic obstructive pulmonary disease | 12 |
| 010 | 80 (M) | CONTROL | CC | AG | GT | CC | GG | Quit in 1984 | Acute myocardial infraction | 9.4 |
| 015 | 11 (M) | CONTROL | TT | AG | GT | CC | GG | No | Multiple trauma | 9.5 |
| 023 | 17 (M) | CONTROL | CT | GG | GG | CT | GG | N/D | Motor vehicle accident | 9 |
| 025 | 50 (M) | CONTROL | TT | GG | GG | CC | GG | No | Myocardial infraction | 17 |
| 009 | 68 (F) | AMD | TT | GG | GG | TT | GG | 2 ppd for 40 years | Stroke | 3 |
| 014 | 82 (M) | AMD | CT | AG | GT | CC | GG | 1 ppd for 40 years | Cardial related | 12 |
| 017 | 81 (M) | AMD | CC | AA | TT | CC | GG | N/D | Cardial arrest | 10.5 |
| 019 | 80 (F) | AMD | CT | GG | GG | CC | GG | No | GI bleed | 9 |
| 032 | 75 (F) | AMD | CT | AA | TT | CC | GG | No | Pancreatic cancer | 7 |
Abbreviations: F, female; M, male
The five clinically diagnosed AMD donors and five clinically normal donors (control) from which primary RPE cultures were established. The cause of death and time of enucleation are indicated. Genotyping data for known AMD-associated Single Nuclear Polymorphisms showing the haplotypes of each donor, carrying risk or protective alleles as marked in bold entries
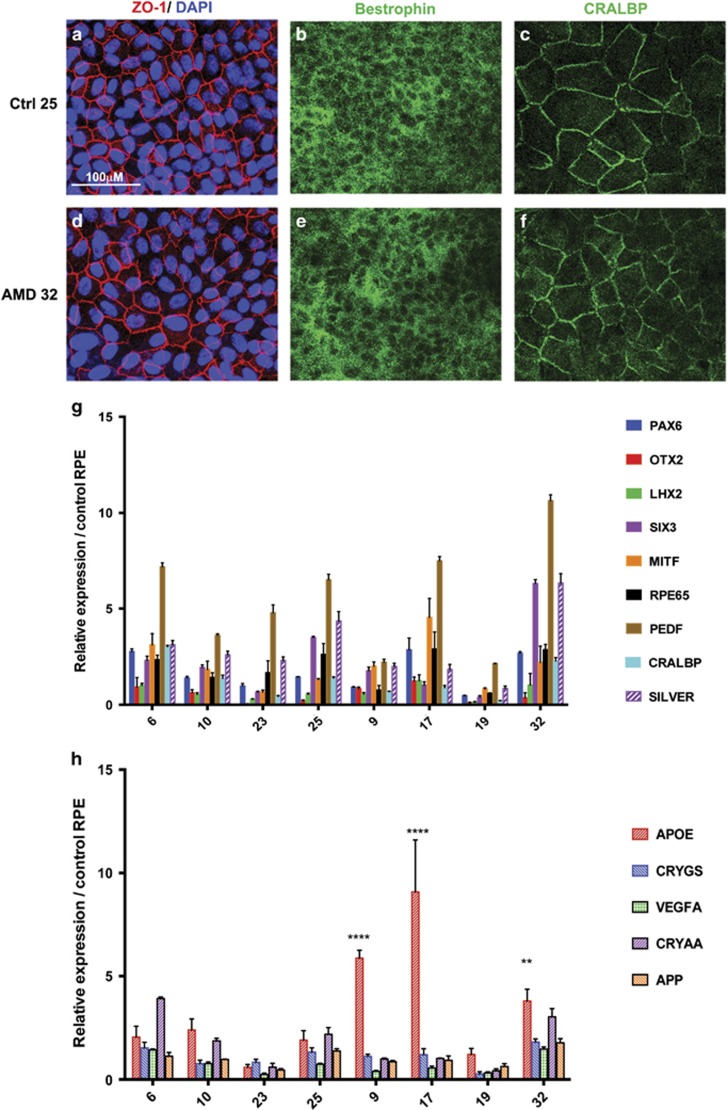
SNPs analysis was performed to obtain genetic information for AMD-associated loci for our samples, and not to make any association of genetics with cellular phenotypes because of the small sample size in our study. Table 1 summarizes the age, gender and genetic characteristics of the cultured RPE. We also analyzed the cells for expression of specific RPE proteins and genes (Figure 1). Our stained RPE were positive for ZO-1, Bestrophin and the retinaldehyde-binding protein (CRALB), which binds to 11-cis-retinal in the visual cycle, and whose function is associated with normal dark adaptation.31
Figure 1.
(a-f) Isolation and characterization of RPE from donors. The isolated RPE from AMD and control donors express RPE-specific proteins ZO-1, Bestrophin and CRALBP. A representative image of immunostaining is shown for each group. Bar represents 100 μM. (g) Gene expression analysis by qRT-PCR confirming the expression of RPE marker genes in the RPE isolated from donors (controls 6, 10, 23 and 25; AMD 9, 17, 19 and 32). (h) Gene expression analysis of AMD-associated genes in AMD and normal RPE. **P<0.01, ****P<0.0001
Gene expression analysis was performed by qPCR and showed expression of all RPE-specific genes in both the AMD and normal RPE cultures. We also analyzed the expression of AMD-associated genes and interestingly found that the expression of apolipoprotein E (APOE) was increased in three out of four tested AMD RPE as compared with four normal RPE. APOE is a prominent component of drusen, has a central role in lipid metabolism and is believed to be involved in lipid trafficking in AMD.32
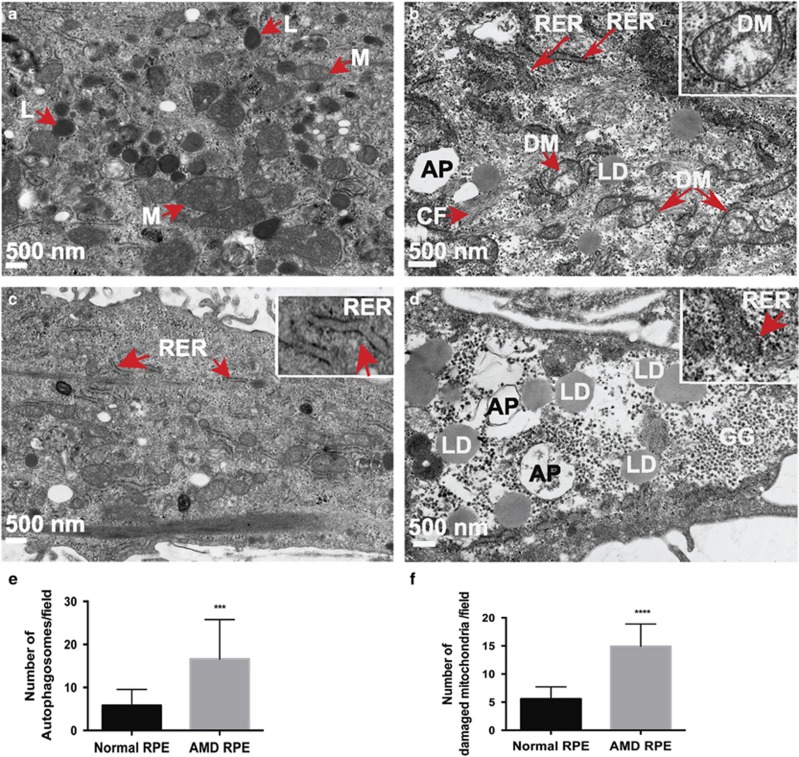
Electron microscopy (EM) imaging (Figure 2) revealed that AMD RPE cultures exhibited distinct disease phenotypes compared with normal RPE. AMD RPE (Figures 2b and d) contained less dense cytoplasm, a higher content of lipid droplets, increased number of glycogen granules and enlarged autophagosomes (APs). The space between the two membranes of the rough endoplasmic reticulum (RER) appeared to be wider and irregular in AMD RPE (Figure 2b and inset 2d), whereas it appeared rather uniform in normal RPE (Figure 2c and inset 2c). Cytoskeletal filaments formed fascicles and appeared condensed (Figure 2b) and mitochondria appeared disintegrated in AMD RPE compared with normal RPE (Figure 2b). We also counted the number of APs and damaged mitochondria and performed a blind comparison of the EM images of normal and AMD RPE (n=3). For each sample, we counted four random regions per cell. The images used for counting were all the same size and same magnification. The mean±S.D. and the P-values were calculated for statistical significance. We observed significant difference between the numbers of APs and disintegrated mitochondria (DM) in AMD RPE as compared with normal RPE (Figures 2e and f).
Figure 2.
AMD RPE exhibit disease phenotypes. (a-d) EM images of normal 10 (a and c) and AMD 9 (b and d) RPE showing disease phenotypes. Red arrows indicate the observed morphological differences. Higher magnification insets show the observed phenotypes (in b and d), in comparison with the normal cellular structures (in c). L, lysosomes; M, mitochondria; RER, rough ER; LD, lipid droplets; F, cytoskeletal fascicles. (e and f) Number of APs and damaged mitochondria were counted (n=3) in four random regions per cell. The images used for counting were all the same size and same magnification. The mean±S.D. and the P-values were calculated for statistical significance. P-value equals 0.0002 (***) and 0.0001 (****) for (e and f), respectively
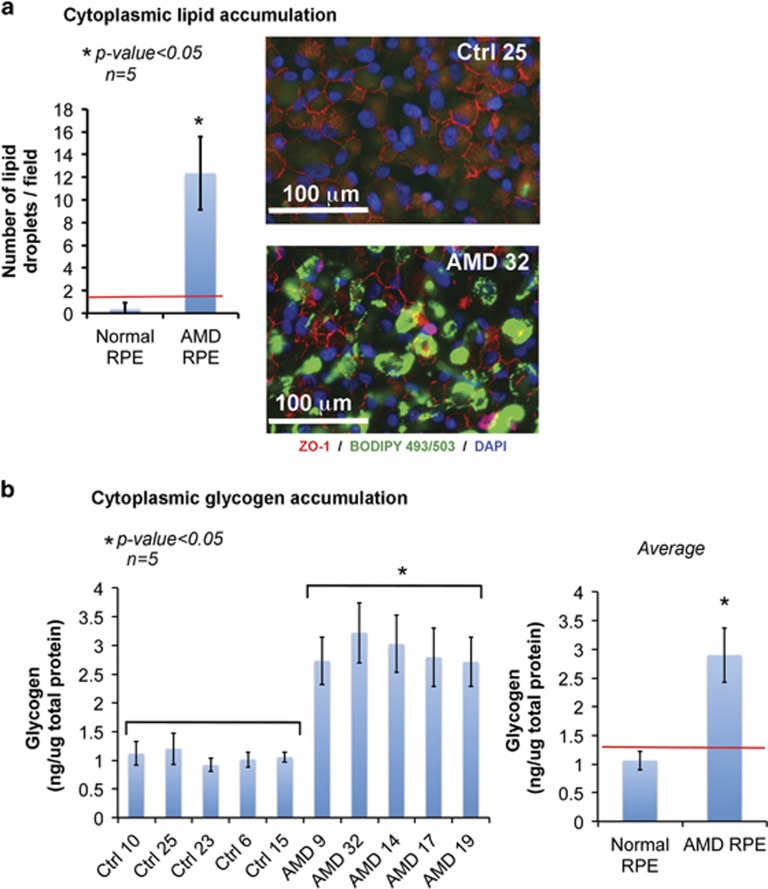
Quantification of lipid droplets and glycogen granules by fluorescence staining and colorimetric assay, respectively, revealed higher levels of lipid droplets and glycogen granules in AMD RPE as compared with normal RPE, further confirming the EM phenotypes (Figures 3a and b). The distinct phenotypes identified in AMD RPE support the use of these cells as an in vitro model to study the underlying mechanisms responsible for the observed phenotypes.
Figure 3.
Lipid and glycogen accumulation in AMD RPE. (a) (Left): graph showing the quantification of lipid droplets in normal and AMD RPE, as determined by counting the lipid droplets in three random fields of stained images (shown in right panel), in the five normal and five AMD RPE (n=5). Asterisk (*) indicates statistically significant difference in number of lipid droplets between control and AMD RPE. (a) (Right): analysis of cytoplasmic lipid by fluorescence staining in control and AMD RPE, showing higher intensity of lipid staining in the AMD RPE. (b) (Left): measurement of glycogen accumulation by colorimetric assay showing statistically significant higher concentration in the AMD RPE compared with control RPE (n=5). (b) (Right): graph of the average glycogen concentrations calculated from five normal and five AMD RPE shown in the left graph. Red line indicates the threshold that separates AMD and normal RPE. Asterisk (*) indicates statistically significant differences in glycogen concentrations between the AMD and control RPE. P-values in (a and b) are determined by ANOVA followed by Tukey's test, with P<0.05
AMD RPE show increased susceptibility to oxidative stress, produce higher levels of reactive oxygen species (ROS) and exhibit lower mitochondrial activity
To assess the functional consequences of the observed phenotypes, we first established chronic oxidative stress conditions using a series of increasing H2O2 concentrations. Under these conditions, RPE cells can be cultured up to 48 h with increasing doses of H2O2 to study their susceptibility to oxidative stress.
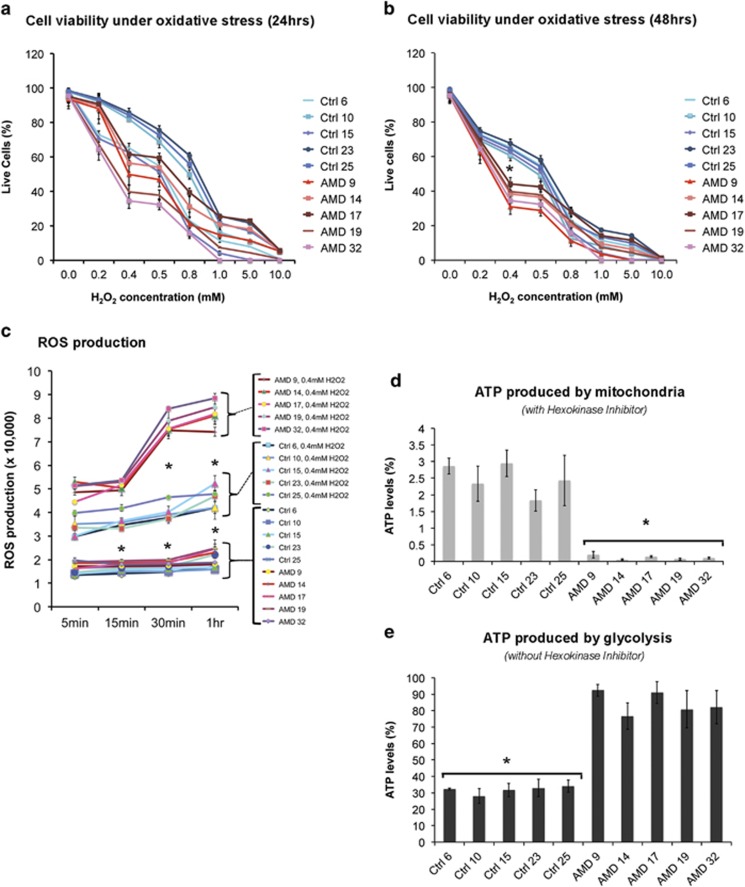
Cell viability assays using 0.2–10 mM of H2O2 for 24 and 48 h revealed that AMD RPE and normal RPE are affected differently by oxidative stress. Our data demonstrated that while H2O2 treatment for 24 h only partially separated the AMD RPE from normal RPE (Figure 4a), 48 h of H2O2 treatment revealed significantly higher vulnerability for AMD RPE (P-value: 0.01). The AMD RPE died faster and at a lower concentration of H2O2 compared with normal RPE (Figure 4b).
Figure 4.
AMD RPE are more susceptible to oxidative stress and show lower mitochondrial activity. (a and b) Cell viability assays of AMD and control RPE treated with increasing concentrations of H2O2 for 24 h (a) and 48 h (b). Higher susceptibility of the AMD RPE under oxidative stress conditions is observed in 48 h. (c) ROS production under stress is significantly higher in AMD RPE. (d and e) AMD RPE have significantly lower mitochondrial activity, as indicated by their ATP levels measured by a luminescence assay in the presence of hexokinase inhibitor. (d) ATP levels produced by mitochondria are significantly lower in AMD RPE as measured in the presence of hexokinase inhibitor. (e) ATP levels produced by glycolysis are higher in AMD RPE as measured in the absence of hexokinase inhibitor. The difference in ATP levels measured in the presence (d) and absence (e) of hexokinase inhibitor show glycolysis as the major source of ATP production in AMD RPE. Asterisks (*) indicate statistical significance, determined by the ANOVA analysis followed by Tukey's test (P-value<0.05, n=5 for each sample)
It has been proposed that mitochondrial dysfunction leads to elevated ROS production.33 To examine whether structural defects observed in mitochondria (Figure 2) correlate with elevated oxidative stress, we measured ROS production in the presence of 0.4 mM H2O2 for 5 min to 1 h in AMD RPE and normal RPE. Our data showed that AMD RPE produce significantly higher ROS levels than normal RPE under oxidative stress (Figure 4c).
A recent report showed that mtDNA damage positively correlated with the severity of AMD, whereas repair capacity was negatively correlated.12 To analyze mitochondrial activity in AMD and normal RPE, we measured ATP levels. When we treated the cells with hexokinase inhibitor to block the glycolytic ATP production, the ATP produced by mitochondria was significantly lower in AMD RPE (Figure 4d). Conversely, ATP levels measured in the absence of hexokinase inhibitor revealed that glycolytic ATP production is significantly higher in AMD compared with normal RPE (Figure 4e), suggesting that in AMD RPE, ATP is produced primarily through glycolysis. These deficits in mitochondrial function correlate well with the altered mitochondrial morphology observed by EM (Figure 2b).
Autophagy is dysfunctional in AMD RPE
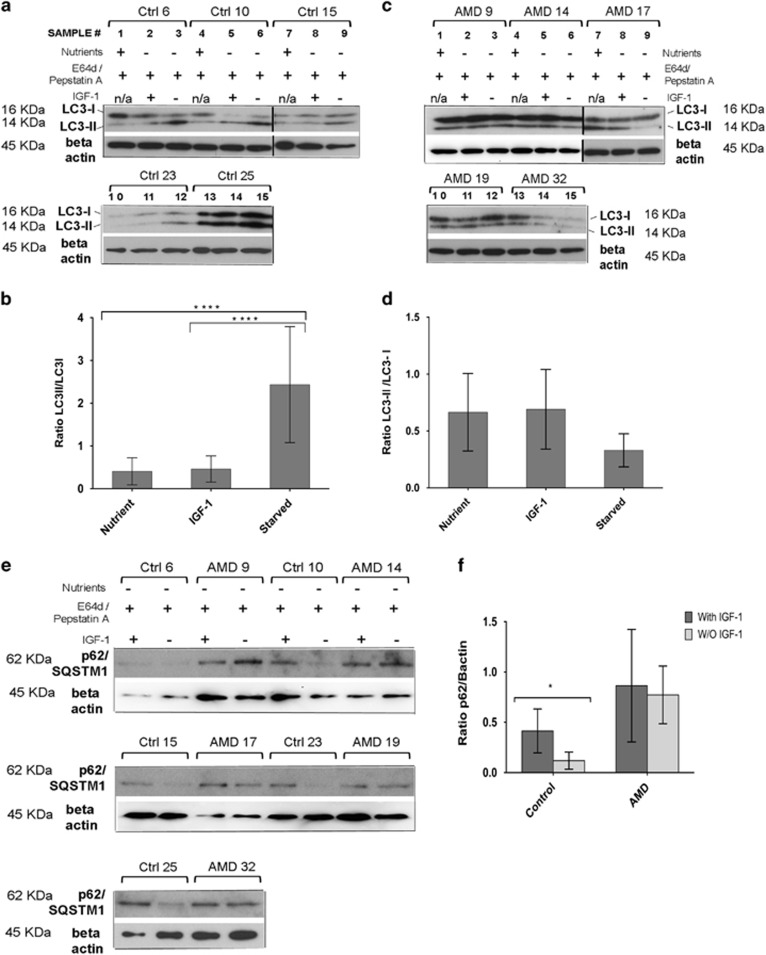
To investigate the biological relevance of AP accumulation observed in our EM data (Figures 2b and d) and to directly study the role of autophagy in the pathophysiology of AMD, we measured autophagy by inducing nutrient starvation in the presence of lysosomal inhibitors and in the presence or absence of the insulin growth factor (IGF-1). A commonly accepted method to monitor autophagic flux is to assay processing of ubiquitin-like microtubule-associated protein 1 light chain LC3-I, which after lipidation becomes LC3-II, is inserted into the inner and outer membranes of the AP and is eventually degraded.34 Therefore, to obtain an accurate measurement of the endogenous levels of LC3-II, we pre-treated the AMD and normal RPE with E64d and pepstatin A35 to inhibit lysosomal degradation. Under these conditions, the levels of the lipidated form of LC3-II relative to unprocessed LC3-I are an accurate measurement of autophagic flux. Our results revealed that normal RPE rapidly induced autophagy after starvation in the absence of IGF-1 (Figures 5a and b), whereas AMD RPE failed to increase the ratio of LC3-II/LC3-I, under the same conditions (Figures 5c and d). Therefore, the addition of IGF-1, expected to suppress autophagy through activation of the AKT/mTOR signaling in normal cells, did not seem to decrease the ratio of LC3-II/LC3-I in AMD RPE (Figures 5c and d), as shown by the densitometry of western blots of three independent experiments (Figures 5b–d). These observations suggest that induction of autophagy in response to starvation is impaired or limited in AMD RPE. Although autophagy dynamics could vary from sample to sample and based on the severity of the disease phenotypes and stage of the disease in each sample, we observed a general trend of lower autophagy dynamics in AMD as compared with normal RPE.
Figure 5.
Autophagy is dysfunctional in AMD RPE. (a and c) Analysis of autophagy dynamics in normal RPE (a), and AMD RPE (c), n=5. LC3 immunoblots of control and AMD RPE under three different conditions, nutrient, starvation in the presence of IGF-1, and starvation in the absence of IGF-1 are shown. Beta actin is used as a normalization control. Spliced membranes are indicated by the vertical lines. (b and d) The ratios of the LC3-II/LC3-I levels as determined by densitometry are illustrated in the graphs, showing that an increase in autophagy dynamics in the absence of IGF-1 under starvation conditions is observed only in normal, but not in AMD RPE. Densitometry was performed on three repeats of the experiment for each sample in five normal and five AMD RPE (b and d). Asterisks in (b) represent P-value <0.0001 of LC3-II/LC3-I ratios as determined by one-way Anova followed by Tukey's test. (e and f) Autophagic flux is lower in AMD RPE as compared with normal RPE. (e) Immunoblot of p62, demonstrating lower autophagic flux in AMD RPE as shown by inability of AMD RPE to downregulate p62 levels during starvation in the absence of IGF-1. Beta actin is used as a normalization control. (f) Relative expression of p62 in control and AMD RPE in the presence and absence of IGF-1, as determined by densitometry analysis of the immunoblot in (e), n=5. The asterisks (*) indicate statistical significance determined by ANOVA followed by Tukey's test (P<0.05)
p62, also called sequestosome 1 (SQSTM1), possesses a short LC3 interaction region that facilitates direct interaction with LC3 and causes p62 degradation by autophagy.36, 37, 38 p62 is known to accumulate in cytoplasmic and nuclear ubiquitinated protein aggregates in various neurodegenerative diseases.39 As p62 degradation depends on autophagy, the level of p62 protein is another indicator of autophagic flux.40
To investigate autophagic flux in AMD RPE, we compared p62 levels in AMD RPE and normal RPE under the same experimental conditions as described above. As shown in Figures 5e and f, under starvation and in the absence of IGF-1 the levels of p62 were decreased in normal RPE as compared with p62 levels under starvation and in the presence of IGF-1. However, under the same conditions, AMD RPE were unable to downregulate p62 levels (Figure 5f, n=5).
Therefore, despite the observed accumulation of APs by EM (Figures 2b and d), AMD RPE cultures failed to induce autophagy during starvation suggesting dysfunctional autophagy and significantly lower autophagic flux in AMD RPE.
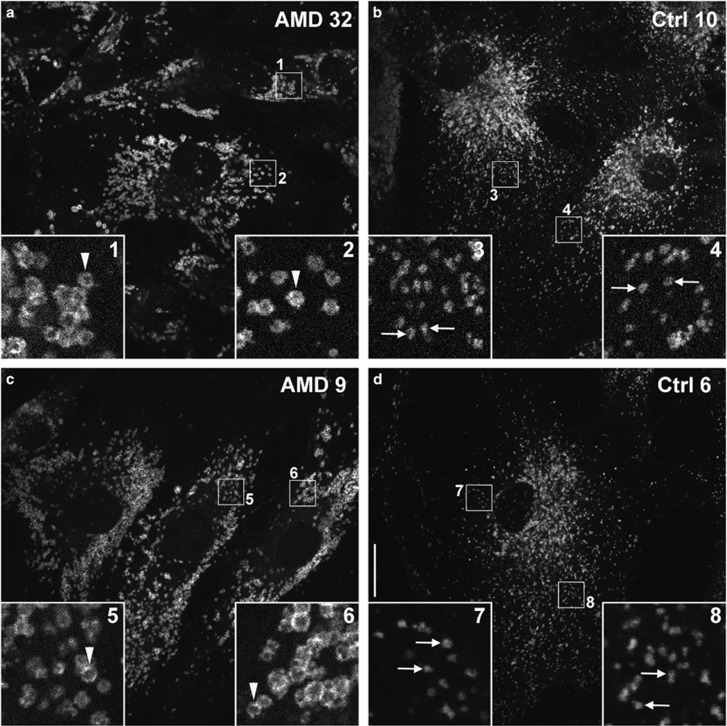
We further analyzed the late autophagic vesicles, autolysosomes by immunostaining with lysosomal-associated membrane protein 1 (LAMP-1) antibody in AMD and normal RPE cultures. Our data showed that LAMP-1-positive organelles in AMD RPE are enlarged and annular, as opposed to the smaller discrete puncta observed in normal RPE (compare arrowed structures with arrowheads in magnified insets within Figure 6). This enlarged morphology suggests inefficient degradation of cellular debris or inefficient recycling of membranes within the lysosomes of AMD RPE. Together, these observations strongly suggest dysfunctional autophagy in AMD RPE that translates into lower autophagic flux and accumulation of cytoplasmic debris.
Figure 6.
Autophagolysosomes are swollen in AMD RPE. Swollen LAMP-1-positive organelles, commonly indicative of defective lysosomal function, are consistently observed in AMD RPE (white arrowheads, AMD 32 (a) and AMD 9 (c)), but not in healthy RPE (white arrows, Ctrl 10 (b) and Ctrl 6 (d)). Insets (1–8) are × 5 magnifications of indicated boxed regions, scale bar represents 20 μm
Discussion
Our study identifies morphological and functional differences between RPE cell cultures established from healthy human donors and donors with AMD. Hallmark morphological changes associated with AMD RPE included accumulation of APs, glycogen granules and lipid droplets, as well as DM. Functional analysis revealed increased vulnerability to oxidative stress, higher ROS production under oxidative stress and lower mitochondrial activity in AMD compared with normal RPE. Analysis of autophagic efficiency showed dysfunctional autophagic activity and inability to induce autophagy in response to starvation in AMD RPE. Collectively, our study suggests that impaired autophagy dynamics in RPE contributes to the pathophysiology of AMD. These observations provide insights into biochemical pathways that are affected in AMD and may have important implications for development of potential therapeutic interventions for this devastating neurodegenerative disease.
Distinct AMD disease phenotypes
Despite the high prevalence of AMD, to date, there exists no disease altering treatment for dry AMD and millions of people worldwide continue to suffer from this debilitating disease.1, 41
A major limitation in understanding the pathophysiology of AMD is its complexity and the lack of a single animal model42 and absence of an in vitro model that could fully replicate key features of the human disease. This is primarily due to the multifactorial origin of AMD, such as human genetic polymorphisms and long-term exposure to environmental factors43 that induce epigenetic changes.
Here, we characterized cultured RPE from AMD donors and controls and showed that AMD RPE exhibit distinct pathological alterations and functional deficits compared with normal RPE.
In concordance with our EM observations showing AP accumulation, Wang et al.44 reported that drusen in AMD eye sections contained increased levels of autophagic markers. Similarly, increased flavoprotein fluorescence, suggesting elevated mitochondrial dysfunction, has been observed in nonexudative eyes with AMD.45 A recent study showed that a marine n-3 polyunsaturated fatty acid that induces a transient increase in cellular ROS levels in RPE activates the oxidative stress response regulator NFE2L2/NRF2 and simultaneously increases intracellular protein aggregates containing p62 and autophagy.46
We also observed that the space between the two membranes of the RER to be wider and irregular in AMD RPE (Figure 2b and inset 2d), whereas it appeared rather uniform in normal RPE (Figure 2c and inset 2c). The double membranes of APs are derived from ribosome-free portions of the endoplasmic reticulum (ER)47 and a number of pathological conditions can cause ER stress,48 which has a significant role in several pathologies including neurodegenration, diabetes and cancer.49, 50 The effect of ER stress in AMD has also been discussed;51 VEGF secretion is readily induced by such ER stress, and could also be implicated in lysosomal accumulation of lipofuscin within RPE.51
Our gene expression analysis revealed increased APOE expression in AMD RPE relative to normal RPE. APOE is synthetized by RPE and is one component of drusen.52 It has been shown that human fetal RPE cultured on porous membranes can secrete deposits that are rich in APOE and that exposure to serum could further result in accumulation of drusen components.53 This cell culture system mimics drusen formation and could be used for investigating AMD molecular mechanisms. Our observation on APOE gene expression correlates with accumulation of lipid droplets that we observed in AMD RPE (Figure 3a and Figures 2b and d), and is directly related to AMD disease mechanisms. This increased APOE expression in AMD RPE is an interesting observation and could be analyzed further in a larger cohort of AMD and normal RPE.
Susceptibility to oxidative stress, increased ROS production and dysfunctional mitochondria in AMD
RPE cells are constantly exposed to light-induced oxidative stress,54 that over time may damage RPE tight junctions and disrupt the retinal blood barrier.55 Oxidative stress has long been hypothesized as a key factor in the development of AMD.56 However, the mechanisms underlying the increased susceptibility to oxidative stress in AMD remain unclear. Here, we have established a chronic oxidative stress condition that allowed us to study cell viability of AMD and normal RPE up to 48 h in the presence of a wide range of H2O2 concentrations. Our data clearly show that AMD RPE exhibit increased susceptibility to chronic oxidative stress. We further demonstrate that AMD RPE produce increased levels of ROS when exposed to oxidative stress as compared with normal RPE.
It is well established that mitochondria are the major source of ROS production and that excess of ROS also induces mitochondrial damage, which can lead to pathologies.57 Recent studies show that dynamic increase in extracellular ATP accelerates photoreceptor cell apoptosis and that ATP induces photoreceptor death and retinal remodeling in rats.58, 59 Our measurement of mitochondrial activity in AMD and normal RPE confirmed decreased ATP production by mitochondria, increased ATP production by glycolysis in AMD as compared with normal RPE and identified glycolysis as the main source of ATP production in AMD RPE. Consistent with our observations, it has recently been shown that in an early phase of hepatocyte failure, an adaptive metabolic shift, from generating energy predominantly from oxidative phosphorylation to glycolysis occurs. This allows maintenance of energy homeostasis during early stages of liver injury, however, leads to hepatocyte dysfunction during terminal stages of chronic liver disease as hepatocytes are unable to sustain high levels of energy production from glycolysis.60
Dysfunctional autophagy in AMD
Defects in autophagic pathways have been associated with neurodegenerative diseases,61, 62 and much interest has focused on the role of autophagy in AMD.25, 63
The impact of autophagy on mitochondrial function and accumulation of reactive species has been reported,24 and is particularly relevant to degenerative diseases caused by accumulation of oxidative stress over time, where dysfunction in both mitochondrial and autophagic pathways are involved.24 APs and mitochondria are suggested to be interrelated homeostatic elements; defects in one element likely impair the other, resulting in increased risk for various human diseases.64 How these organelles interact and contribute to the pathophysiology of AMD remains unknown.
Recent evidence links the cellular quality control system, in particular autophagy/mitophagy and mitochondrial dynamics with age-related diseases.65 Autophagy loss results in mitochondrial dysfunction and accumulation of organelles, proteins and ROS,66 and affects cell viability and induces cell death.67
In AMD, RPE dysfunctional autophagy/mitophagy could lead to mitochondrial disintegration by affecting the mitochondrial fission/fusion ratio that is crucial for maintaining functional mitochondria.68 Dysfunctional mitochondria in AMD RPE can further translate into an increased oxidative stress response and a shift from oxidative phosphorylation to glycolytic ATP production. Furthermore, dysfunctional mitochondria can affect lipid and glucose metabolism resulting in cytoplasmic lipid and glycogen accumulation, two cellular phenotypes that we observed in AMD RPE. A critical role for autophagy in lipid metabolism is reported, with inhibition of autophagy leading to increased triglyceride storage in lipid droplets.22 These observations could also explain the accumulation of lipid droplets in AMD RPE because of dysfunctional autophagy.
Another possible explanation for difference in autophagic flux, stress response and glycolytic ATP production, could be due to an overactive mTOR pathway in AMD RPE. Studies have shown direct control of mitochondrial function by mTOR69 with inhibition of mTOR enhancing aerobic glycolysis.69 In addition, p62 is suggested to be a key regulator of nutrient sensing in this pathway 70 with overexpression favoring more efficient activation of the mTOR pathway.70 Our observed increased p62 levels during starvation in AMD RPE could impact mTOR activity. Although the mTOR pathway is important for mitochondrial activity and biogenesis,71 overactive mTOR could inhibit autophagy/mitophagy72 and subsequently induce accumulation of damaged mitochondria in AMD RPE. Further studies need to be performed to elucidate these interactions in AMD RPE.
Our observation of glycogen accumulation and autophagic dysfunction in AMD RPE is in accordance with recent studies revealing that glycogen accumulation accounts for the neurodegeneration, as well as impaired autophagy.73
A recent study reported on increased markers of autophagy and exosomes in the drusen of AMD donor eyes and speculated that increased autophagy and release of intracellular proteins via exosomes by the aged RPE may contribute to drusen formation and AMD.44 Consistent with these morphological observations, we found an increased number of APs in AMD compared with normal RPE. However, mechanistic studies analyzing the ratio of LC3-II/LC3-I under starvation revealed that despite AP accumulation, the efficiency of autophagy is reduced in AMD RPE, and that these cells failed to induce autophagy in response to starvation, contrary to what was speculated by Wang et al. In addition, analysis of p62 levels further revealed lower autophagic flux in AMD compared with normal RPE. This failure to induce autophagy in response to starvation stress could be due to the difference in basal autophagic flux in AMD as compared with normal RPE. Furthermore, LAMP-1 immunostaining clearly demonstrated swollen vesicles likely indicative of dysfunctional autophagolysosomes in AMD RPE.
Our studies identify specific disease phenotypes in a novel model for human AMD, and suggest that dysfunctional autophagy in RPE contributes to the pathophysiology of AMD.
Materials and methods
In vitro disease model
A total of 10 eyes from 5 organ donors (2 males and 3 females) clinically diagnosed with AMD and 10 eyes from 5 clinically normal male donors (5 male) were purchased from National Disease Research Interchange74(NDRI, Philadelphia, PA, USA). We excluded eyes of donors with other ocular diseases or diabetes from the study. The average time of postmortem enucleation was 9 h and eyes were received in <24 h. Samples with infectious diseases were excluded by serology tests performed by NDRI (Table 1). RPE from macula region were isolated according to established protocols30 and cultured in serum-free media75, 76 under controlled oxygen (5%) and CO2 (5%) conditions.77 Studies have shown that low (physiological) oxygen concentration promotes RPE growth77 and better protects from ROS-induced damage in vitro. RPE were then purified with magnetic-activated cell sorting by positive selection for epithelial cells using anti-BEST1 antibody (Abcam, Cambridge, MA, USA, 1/500) and anti-E-cadherin (Miltenyi Biotech, San Diego, CA, USA, 1/200); and by negative selection using a fibroblast-specific antibody (Miltenyi Biotech, 1/200) to remove fibroblasts. The purity of the sorted cells was confirmed by immunostaining with anti-ZO-1 (1/200) and anti-BEST1, and Real-Time PCR for RPE specific genes (Figures 1a and b). The RPE cells were grown in serum-free RPE media75 at 37 oC, with 5% O2 and 5% CO2.77
SNPs genotyping
RPE were tested for selected SNPs known to be associated with AMD by RT-PCR followed by sequencing analysis.78, 79
Immunostaining
RPE cells grown on transwells 6.5 mm inserts and 0.4 μm membrane (Costar, Corning, NY, USA) were fixed with 4% paraformaldehyde for 10–15 min followed by rinsing with PBS and incubation in blocking/permeabilizing buffer containing 0.5% BSA and 0.2% Tween 20 in PBS. Cells were stained using established protocols for the primary and secondary antibodies. Rabbit and mouse anti-ZO-1 (Invitrogen, Carlsbad, CA, USA) were used at 1/200 dilution. Mouse anti-CRALBP (Novus Biologicals, Littleton, CO, USA), and mouse anti-Bestrophin (Abcam) were used at 1/500. Anti-mouse and rabbit secondary antibodies (Molecular Probes, Carlsbad, CA, USA) were used at 1/1000. Stained cells were mounted with anti-fading medium (Invitrogen), and imaged using Olympus Fluoview microscope (Olympus, Center Valley, PA, USA).
Quantitative real-time polymerase chain reaction analyses
Total RNA was extracted with the RNeasy kit (Qiagen, Germantown, MD, USA), treated with RNase-free DNase I (Qiagen), and reverse transcribed with oligo-dT using the SuperScript III cDNA synthesis kit (Invitrogen). Quantitative PCR was performed with the QuantiTect SYBR Green PCR Kit (Qiagen). Specific primers (Supplementary Table 1) for each gene were designed with the PrimerQuest software (Integrated DNA Technologies, Coralville, IA, USA), and the cDNA sequences of each gene (GenBank, NCBI, NIH) were used to produce 100–250 bp PCR amplicons that span one or more exon/intron boundaries.
Electron microscopy
RPE cell cultures were rinsed with Dulbecco PBS (without Ca2+ and Mg2+), fixed twice in PBS-buffered glutaraldehyde (2.5% at pH 7.4) and PBS-buffered osmium tetroxide (0.5%), and embedded in epoxy resin. Thin sections (90nm) were collected on 200 μm mesh copper grids, dried for 24 h, and double-stained with uranyl acetate and lead citrate. Sections were viewed and photographed with JEOL JM-1010 EM.80
Lipid staining
Accumulation of lipids was assayed with the BODIPY 493/503 lipid-specific dye (Invitrogen) on RPE cultured on 24-well plates or 8-well slide chambers. Stained RPE were imaged using an EVOS FL microscope (Life Technologies, Carlsbad, CA, USA) and the number of green fluorescent lipid aggregates was counted in diseased versus normal RPE cells.
Glycogen concentration assay
Cytoplasmic accumulation of glycogen was assayed using the Glycogen Assay Kit (Sigma, St. Louis, MO, USA) on RPE monolayers grown in 96-well plates. Glycogen concentration was determined by a coupled enzyme assay, which produces a colorimetric (570 nm)/fluorometric (535/587 nm) product, proportional to the glycogen present.
Oxidative stress conditions
RPE were cultured at 80–90% confluency in 96-well plates, at 37 oC with 5% O2 and 5% CO2. Oxidative stress was induced with different concentrations of H2O2 ranging from 0 to 10 mM for 24 or 48 h, followed by cell viability measurements.
Cell viability assay
Cell viability of RPE monolayers on 96-well plates was measured under different concentrations of H2O2 (0–10 mM) following 30-min incubation with the PrestoBlue Reagent (Life Technologies); six wells used for each sample. Fluorescence measurements were performed using an Ultra 384 plate reader with 535nm excitation and 612 nm emission wavelengths. The difference between cell viability in normal RPE and AMD RPE was significant between 0.2 to 0.6 mM of H2O2. At 0.8 mM, most cells died and no significant difference was observed between the diseased and normal RPE. We found that the most significant difference was obtained at 0.4 mM of H2O2. However, the difference in cell viability was also significant at 0.3, 0.5 and 0.6 mM of H2O2. Therefore, for the experiments under stress conditions we used 0.4 mM of H2O2.
ROS measurement
Production of ROS under oxidative stress was measured using the OxiSelect™ Intracellular ROS Assay Kit (Cell Biolabs, San Diego, CA, USA). RPE monolayers were cultured in 96-well plates in serum-free RPE media76 for 20 h and then loaded with 1 mM of the cell-permeable fluorogenic probe 2'-7'-dichlorodihydrofluorescin diacetate for 1 h. Oxidative stress was induced with 2-h incubation in 0.4 mM H2O2. The fluorescence intensity of each sample, proportional to the ROS levels, was measured against the fluorescence of the provided standard at the indicated time-points using a Tecan (Morrisville, NC, USA) Ultra 384 plate reader.
Measurement of ATP levels
ATP levels of the RPE monolayers were measured with the Mitochondrial ToxGlo Assay (Promega, Madison, WI, USA). To assay the mitochondrial activity in AMD and normal RPE, the ATP measurement was performed following 2-h incubation with or without 10 μM of the bromopyruvate analog (3-BrPA), an inhibitor of the glycolytic enzyme hexokinase II (EMD Millipore, Billerica, MA, USA).
Autophagy dynamics assay
For measurement of LC3-II/LC3-I ratios, normal and AMD RPE cells were pre-treated with IGF-1 for 1 h and starved in HBSS for 4 h in the presence or absence of IGF-1 (50 nM). Lysosomal inhibitors E64d and Pepstatin A were added in the last 2 h of starvation at a concentration of 10 μg/ml and the cells were lysed and analyzed with immunoblot for LC3 and p62 based on the established protocol.35
Antibodies
(a) Primary antibodies
The primary antibodies used were as follows: rabbit anti-LC3, rabbit anti-p62/SQSTM1 and rabbit anti-beta actin from Cell Signaling Technology (Danvers, MA, USA) used at 1/1000, 1/1000 and 1/10 000, respectively; mouse anti-LAMP-1 from BD Biosciences (San Jose, CA, USA) (1/200); mouse anti-BEST1 from Abcam (1/500); mouse anti-CRALBP from Novus Biologicals (1/500); mouse anti-ZO-1 from Invitrogen (1/200); fibroblast-specific antibody (conjugated to magnetic beads, 1/200), mouse anti-E-cadherin (CD324, 1/200) from Miltenyi Biotech.
(b) Secondary antibodies
The secondary antibodies used were as follows: goat anti-rabbit HRP-linked from Cell Signaling Technology; donkey anti-mouse Alexa488-linked and donkey anti-mouse Alexa594-linked from Invitrogen; and anti-mouse IgG MicroBeads from Miltenyi Biotech.
Immunoblot analysis
Protein samples were extracted in radioimmunoprecipitation assay (RIPA) buffer (1% NP-40, 0.5% sodium deoxycholate, and 1% SDS in 1x PBS), containing freshly added protease and phosphatase inhibitor cocktail tablets (Roche Applied Science, Indianapolis, IN, USA), 1x Protease Inhibitor Cocktail Set I (EMD Millipore), 1 mM sodium vanadate, 50 mM sodium fluoride and 1 mM PMSF (Sigma-Aldrich, St. Louis, MO, USA). Protein concentrations were measured by Bradford assay (Bio-Rad, Hercules, CA, USA). Protein samples were analyzed using the NuPAGE electrophoresis and XCell western blot system (Invitrogen). Primary and secondary antibodies were used based on the manufacturer's instructions. Immunoreactive protein bands were visualized by the Clarity chemiluminescent substrate (Bio-Rad) followed by revealing with X-ray films. Densitometry was performed using the ImageJ software (https://ImageJ.nih.gov).
Statistical analysis
Quantitative functional and gene expression assays were performed three times, each sample represented by 3–6 replicas per experiment. Mean averages±S.D. were calculated. The statistical significance of the observed differences was tested by ANOVA analysis followed by Tukey's test in the experiments where comparisons were made between groups of samples (i.e., control versus AMD) or by t-test in the experiments where two samples were compared, using P-value<0.05.
Acknowledgments
We thank the donors of Macular Degeneration Research, a program of BrightFocus Foundation, and Prevention of Blindness Society of Metropolitan Washington (POB), for support of this research. The Georgetown-Lombardi Comprehensive Cancer Center Shared Resource facilities were used for qRT-PCR. We thank Dr. Yetrib Hathout, Children's National Health System, for sharing the normal and AMD RPE cells, Dr. Mones Abu-Asab, NEI/NIH for assistance in EM imaging.
Author contributions
NG: designed the experiments, performed experiments and wrote the manuscript; YC, YYX and GS: performed experiments; ACT: performed experiments and edited the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by GM Fimia
The authors declare no conflict of interest.
Supplementary Material
References
- Klein R, Chou CF, Klein BE, Zhang X, Meuer SM, Saaddine JB. Prevalence of age-related macular degeneration in the US population. Arch Ophthalmol 2011; 129: 75–80. [DOI] [PubMed] [Google Scholar]
- Gehrs KM, Anderson DH, Johnson LV, Hageman GS. Age-related macular degeneration—emerging pathogenetic and therapeutic concepts. Ann Med 2006; 38: 450–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein DB, Wittenborn JS, Zhang X, Honeycutt AA, Lesesne SB, Saaddine J. Forecasting age-related macular degeneration through the year 2050: the potential impact of new treatments. Arch Ophthalmol 2009; 127: 533–540. [DOI] [PubMed] [Google Scholar]
- Boulton M, Dayhaw-Barker P. The role of the retinal pigment epithelium: topographical variation and ageing changes. Eye (Lond) 2001; 15(Pt 3): 384–389. [DOI] [PubMed] [Google Scholar]
- Nowak JZ. Age-related macular degeneration (AMD): pathogenesis and therapy. Pharmacol Rep 2006; 58: 353–363. [PubMed] [Google Scholar]
- Abdelsalam A, Del Priore L, Zarbin MA. Drusen in age-related macular degeneration: pathogenesis, natural course, and laser photocoagulation-induced regression. Survey Ophthalmol 1999; 44: 1–29. [DOI] [PubMed] [Google Scholar]
- Ferris FL 3rd, Fine SL, Hyman L. Age-related macular degeneration and blindness due to neovascular maculopathy. Arch Ophthalmol 1984; 102: 1640–1642. [DOI] [PubMed] [Google Scholar]
- Dornonville de la Cour M. Ion transport in the retinal pigment epithelium. A study with double barrelled ion-selective microelectrodes. Acta Ophthalmol Suppl 1993; 209: 1–32. [PubMed] [Google Scholar]
- Hamann S. Molecular mechanisms of water transport in the eye. Int Rev Cytol 2002; 215: 395–431. [DOI] [PubMed] [Google Scholar]
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev 2005; 85: 845–881. [DOI] [PubMed] [Google Scholar]
- Kozlowski MR. RPE cell senescence: a key contributor to age-related macular degeneration. Med Hypotheses 2012; 78: 505–510. [DOI] [PubMed] [Google Scholar]
- Lin H, Xu H, Liang FQ, Liang H, Gupta P, Havey AN et al. Mitochondrial DNA damage and repair in RPE associated with aging and age-related macular degeneration. Invest Ophthalmol Vis Sci 2011; 52: 3521–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Valckenberg S, Fleckenstein M, Scholl HP, Holz FG. Fundus autofluorescence and progression of age-related macular degeneration. Survey Ophthalmol 2009; 54: 96–117. [DOI] [PubMed] [Google Scholar]
- Wang J, Ohno-Matsui K, Morita I. Elevated amyloid beta production in senescent retinal pigment epithelium, a possible mechanism of subretinal deposition of amyloid beta in age-related macular degeneration. Biochem Biophys Res Commun 2012; 423: 73–78. [DOI] [PubMed] [Google Scholar]
- Cho Y, Cao X, Shen D, Tuo J, Parver LM, Rickles FR et al. Evidence for enhanced tissue factor expression in age-related macular degeneration. Lab Invest 2011; 91: 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga H, Handa JT, Aotaki-Keen A, Sherwood SW, West MD, Hjelmeland LM. Beta-galactosidase histochemistry and telomere loss in senescent retinal pigment epithelial cells. Invest Ophthalmol Vis Sci 1999; 40: 197–202. [PubMed] [Google Scholar]
- Gu X, Neric NJ, Crabb JS, Crabb JW, Bhattacharya SK, Rayborn ME et al. Age-related changes in the retinal pigment epithelium (RPE). PLoS One 2012; 7: e38673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol 1966; 28: 435–492. [DOI] [PubMed] [Google Scholar]
- Finn PF, Dice JF. Proteolytic and lipolytic responses to starvation. Nutrition 2006; 22: 830–844. [DOI] [PubMed] [Google Scholar]
- Wang Y, Singh R, Xiang Y, Czaja MJ. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology 2010; 52: 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell 2011; 146: 682–695. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M et al. Autophagy regulates lipid metabolism. Nature 2009; 458: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy 2007; 3: 337–346. [DOI] [PubMed] [Google Scholar]
- Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J 2012; 441: 523–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter SK, Song C, Qi X, Mao H, Rao H, Akin D et al. Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy 2014; 10: 1989–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viiri J, Amadio M, Marchesi N, Hyttinen JM, Kivinen N, Sironen R et al. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One 2013; 8: e69563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter SK, Rao HV, Qi X, Cai J, Sugrue A, Dunn WA Jr. et al. Autophagy in the retina: a potential role in age-related macular degeneration. Adv Exp Med Biol 2012; 723: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen K, Petrovski G, Moe MC, Berta A, Kaarniranta K. Molecular mechanisms of retinal pigment epithelium damage and development of age-related macular degeneration. Acta Ophthalmologica 2012; 90: 299–309. [DOI] [PubMed] [Google Scholar]
- Ambati J, Fowler BJ. Mechanisms of age-related macular degeneration. Neuron 2012; 75: 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maminishkis A, Chen S, Jalickee S, Banzon T, Shi G, Wang FE et al. Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest Ophthalmol Vis Sci 2006; 47: 3612–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saari JC, Nawrot M, Kennedy BN, Garwin GG, Hurley JB, Huang J et al. Visual cycle impairment in cellular retinaldehyde binding protein (CRALBP) knockout mice results in delayed dark adaptation. Neuron 2001; 29: 739–748. [DOI] [PubMed] [Google Scholar]
- Ishida BY, Bailey KR, Duncan KG, Chalkley RJ, Burlingame AL, Kane JP et al. Regulated expression of apolipoprotein E by human retinal pigment epithelial cells. J Lipid Res 2004; 45: 263–271. [DOI] [PubMed] [Google Scholar]
- Murphy MP. Mitochondrial dysfunction indirectly elevates ROS production by the endoplasmic reticulum. Cell Metab 2013; 18: 145–146. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy 2007; 3: 542–545. [DOI] [PubMed] [Google Scholar]
- Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 2005; 1: 84–91. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007; 131: 1149–1163. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282: 24131–24145. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 2009; 452: 181–197. [DOI] [PubMed] [Google Scholar]
- Kuusisto E, Kauppinen T, Alafuzoff I. Use of p62/SQSTM1 antibodies for neuropathological diagnosis. Neuropathol Appl Neurobiol 2008; 34: 169–180. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on Huntingtin-induced cell death. J Cell Biol 2005; 171: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R, Klein BE, Lee KE, Cruickshanks KJ, Gangnon RE. Changes in visual acuity in a population over a 15-year period: the Beaver Dam Eye Study. Am J Ophthalmol 2006; 142: 539–549. [DOI] [PubMed] [Google Scholar]
- Fletcher EL, Jobling AI, Greferath U, Mills SA, Waugh M, Ho T et al. Studying age-related macular degeneration using animal models. Optom Vis Sci 2014; 91: 878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet 2012; 379: 1728–1738. [DOI] [PubMed] [Google Scholar]
- Wang AL, Lukas TJ, Yuan M, Du N, Tso MO, Neufeld AH. Autophagy and exosomes in the aged retinal pigment epithelium: possible relevance to drusen formation and age-related macular degeneration. PLoS One 2009; 4: e4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MG, Comer GM, Kawaji T, Petty HR, Elner VM. Noninvasive imaging of mitochondrial dysfunction in dry age-related macular degeneration. Ophthalmic Surg Lasers Imaging 2012; 43: 358–365. [DOI] [PubMed] [Google Scholar]
- Johansson I, Monsen VT, Pettersen K, Mildenberger J, Misund K, Kaarniranta K et al. The marine n-3 PUFA DHA evokes cytoprotection against oxidative stress and protein misfolding by inducing autophagy and NFE2L2 in human retinal pigment epithelial cells. Autophagy 2015; 11: 1636–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WA Jr.. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol 1990; 110: 1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonthal AH. Endoplasmic reticulum stress: its role in disease and novel prospects for therapy. Scientifica (Cairo) 2012; 2012: 857516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H. ER stress and diseases. FEBS J 2007; 274: 630–658. [DOI] [PubMed] [Google Scholar]
- Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol 2008; 3: 399–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Kauppinen A, Hyttinen JM, Toropainen E, Kaarniranta K. Endoplasmic reticulum stress in age-related macular degeneration: trigger for neovascularization. Mol Med 2010; 16: 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H et al. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci USA 2002; 99: 14682–14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LV, Forest DL, Banna CD, Radeke CM, Maloney MA, Hu J et al. Cell culture model that mimics drusen formation and triggers complement activation associated with age-related macular degeneration. Proc Natl Acad Sci USA 2011; 108: 18277–18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Nelson KC, Wu M, Sternberg P Jr., Jones DP. Oxidative damage and protection of the RPE. Prog Retin Eye Res 2000; 19: 205–221. [DOI] [PubMed] [Google Scholar]
- Negi A, Marmor MF. Experimental serous retinal detachment and focal pigment epithelial damage. Arch Ophthalmol 1984; 102: 445–449. [DOI] [PubMed] [Google Scholar]
- Beatty S, Koh H, Phil M, Henson D, Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Survey Ophthalmol 2000; 45: 115–134. [DOI] [PubMed] [Google Scholar]
- Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell 2012; 48: 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi S, Hisatomi T, Murakami Y, Terasaki H, Sonoda S, Asato R et al. Dynamic increase in extracellular ATP accelerates photoreceptor cell apoptosis via ligation of P2RX7 in subretinal hemorrhage. PLoS One 2013; 8: e53338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey KA, Greferath U, Aplin FP, Jobling AI, Phipps JA, Ho T et al. Adenosine triphosphate-induced photoreceptor death and retinal remodeling in rats. J Comp Neurol 2014; 522: 2928–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T, Bellance N, Damm A, Bing H, Zhu Z, Handa K et al. A switch in the source of ATP production and a loss in capacity to perform glycolysis are hallmarks of hepatocyte failure in advance liver disease. J Hepatol 2014; 60: 1203–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum Mol Genet 2009; 18: R169–R176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb RA, Carreira RS. Autophagy in health and disease. 5. Mitophagy as a way of life. Am J Physiol Cell Physiol 2010; 299: C203–C210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaarniranta K, Sinha D, Blasiak J, Kauppinen A, Vereb Z, Salminen A et al. Autophagy and heterophagy dysregulation leads to retinal pigment epithelium dysfunction and development of age-related macular degeneration. Autophagy 2013; 9: 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N. Mitochondria and autophagy: critical interplay between the two homeostats. Biochim Biophys Acta 2012; 1820: 595–600. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth SE, Calvani R, Marzetti E. The interplay between autophagy and mitochondrial dysfunction in oxidative stress-induced cardiac aging and pathology. J Mol Cell Cardiol 2014; 71: 62–70. [DOI] [PubMed] [Google Scholar]
- Garcia-Prat L, Martinez-Vicente M, Perdiguero E, Ortet L, Rodriguez-Ubreva J, Rebollo E et al. Autophagy maintains stemness by preventing senescence. Nature 2016; 529: 37–42. [DOI] [PubMed] [Google Scholar]
- Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest 2005; 115: 2679–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science 2012; 337: 1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A, Schreiber SL. Direct control of mitochondrial function by mTOR. Proc Natl Acad Sci USA 2009; 106: 22229–22232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell 2011; 44: 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 2013; 18: 698–711. [DOI] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15: 406–416. [DOI] [PubMed] [Google Scholar]
- Duran J, Gruart A, Garcia-Rocha M, Delgado-Garcia JM, Guinovart JJ. Glycogen accumulation underlies neurodegeneration and autophagy impairment in Lafora disease. Hum Mol Genet 2014; 23: 3147–3156. [DOI] [PubMed] [Google Scholar]
- An E, Sen S, Park SK, Gordish-Dressman H, Hathout Y. Identification of novel substrates for the serine protease HTRA1 in the human RPE secretome. Invest Ophthalmol Vis Sci 2010; 51: 3379–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronk AH, Vargis E. Methods for culturing retinal pigment epithelial cells: a review of current protocols and future recommendations. J Tissue Eng 2016; 7: 2041731416650838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamm DM, Melvan JN, Shearer RL, Pinilla I, Sabat G, Svendsen CN et al. A novel serum-free method for culturing human prenatal retinal pigment epithelial cells. Invest Ophthalmol Vis Sci 2008; 49: 788–799. [DOI] [PubMed] [Google Scholar]
- Knorr HL, Linde-Behringer M, Gossler B, Mayer UM. Human retinal pigment epithelium in vitro: influence of low oxygen tension, glucose and insulin. Ophthal Res 1993; 25: 226–234. [DOI] [PubMed] [Google Scholar]
- Sharma NK, Gupta A, Prabhakar S, Singh R, Sharma S, Anand A. Single nucleotide polymorphism and serum levels of VEGFR2 are associated with age related macular degeneration. Curr Neurovasc Res 2012; 9: 256–265. [DOI] [PubMed] [Google Scholar]
- Black JR, Clark SJ. Age-related macular degeneration: genome-wide association studies to translation. Genet Med 2016; 18: 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkinaki M, Abu-Asab M, Gunawardena N, Ahern G, Javidnia M, Young J et al. Klotho regulates retinal pigment epithelial functions and protects against oxidative stress. J Neurosci 2013; 33: 16346–16359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.