Abstract
Inactivating mutations of the CREBBP acetyltransferase are highly frequent in diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL), the two most common germinal-center (GC) derived cancers. However, the role of CREBBP inactivation in lymphomagenesis remains unclear. Here we show that CREBBP regulates enhancer/super-enhancer networks with central roles in GC/post-GC cell fate decisions, including genes involved in signal transduction by the B-cell receptor and CD40 receptor, transcriptional control of GC and plasma cell development, and antigen presentation. Consistently, Crebbp-deficient B-cells exhibit enhanced response to mitogenic stimuli and perturbed plasma cell differentiation. While GC-specific loss of Crebbp was insufficient to initiate malignant transformation, compound Crebbp-haploinsufficient/BCL2-transgenic mice, mimicking the genetics of FL and DLBCL, develop clonal lymphomas recapitulating the features of the human diseases. These findings establish CREBBP as a haploinsufficient tumor suppressor gene in GC B-cells and provide insights into the mechanisms by which its loss contributes to lymphomagenesis.
Keywords: CREBBP, germinal center, lymphoma, acetylation, enhancer
INTRODUCTION
Modulation of chromatin accessibility through histone modifications is a key step in the regulation of gene transcription, and disruption of histone modifying enzymes by genetic lesions has emerged as a common feature of cancer, including lymphoma (1,2). Among these lesions, inactivating mutations and deletions of the gene encoding for the CREBBP acetyltransferase (or, less frequently, its paralogue EP300) are highly recurrent in diffuse large B cell lymphoma (DLBCL, 30% of cases) and follicular lymphoma (FL, 60% of cases), the two most common types of germinal center (GC) derived B cell non-Hodgkin lymphoma, together accounting for over 70% of all diagnoses (3–8). CREBBP mutations have also been observed in a third malignancy originating from the GC, i.e. Burkitt lymphoma (9), while they are generally absent in non-GC derived mature B cell malignancies such as mantle cell lymphoma, suggesting a prominent role for the perturbation of epigenetic mechanisms in the malignant transformation of GC B cells.
Genetic-driven losses of CREBBP and EP300 are predominantly monoallelic, mutually exclusive, and are accompanied by expression of the residual wild type allele, a pattern consistent with a haploinsufficient tumor suppressor role (6). Indeed, a pathogenic effect for dose reduction of CREBBP/EP300 is demonstrated by the fact that germline loss of a single CREBBP allele by mutation or deletion is the causative genetic event in Rubinstein-Taybi syndrome, a rare autosomal congenital disorder that is also associated with tumor predisposition (10). Interestingly, phylogenetic analysis of tumor evolution during FL progression and transformation to DLBCL indicates that genetic lesions in epigenetic modifiers, including CREBBP and the methyltransferase KMT2D, are already present in a common precursor clone before divergent evolution to FL or DLBCL, suggesting a role early in the history of tumor clonal expansion (5,8,11).
CREBBP and EP300 are highly conserved, ubiquitously expressed enzymes that belong to the KAT3 family of acetyltransferases. They interact with over 400 proteins (12) and function as global transcriptional coactivators through the modification of lysines on both histone and non-histone nuclear proteins, also including well known proto-oncogenes (e.g. the BCL6 transcriptional repressor) (13) and tumor suppressor genes (e.g. TP53) (14–16). In accord with their involvement in multiple cellular processes, constitutional homozygous null mice for either Crebbp or Ep300 are early embryonic lethal, and the same is true for the compound Crebbp/Ep300 double heterozygous mice (17), consistent with the notion that the combined amount of these two proteins is limiting in the cell. Furthermore, while a partially redundant function has been invoked for CREBBP and EP300 during development, studies using conditional knock-out mice indicate that, in certain cellular contexts, they can exert distinct roles (18–21). Nonetheless, a comprehensive investigation of the tissue-specific requirement for CREBBP is lacking.
In DLBCL and FL, CREBBP mutations (both truncating and missense in the HAT domain) impair its ability to catalyze acetylation of TP53 as well as to acetylate and inactivate the function of BCL6, providing one mechanism by which loss of its activity may favor the malignant transformation of GC B cells (6). However, it is conceivable that reduced expression of CREBBP will have broad repercussions on gene transcription. While several studies have examined its role during hematopoiesis, including early B and T cell development (18–21), the transcriptional network regulated by CREBBP in the unique environment of the GC, and the mechanism by which genetic-driven inactivation of its function contributes to their malignant transformation remain unknown. The aim of this study was to explore the role of CREBBP loss in the biology of normal and transformed GC B cells, by integrating functional epigenomics in human cells and mouse genetics approaches.
RESULTS
CREBBP is a major regulator of enhancer networks in the germinal center
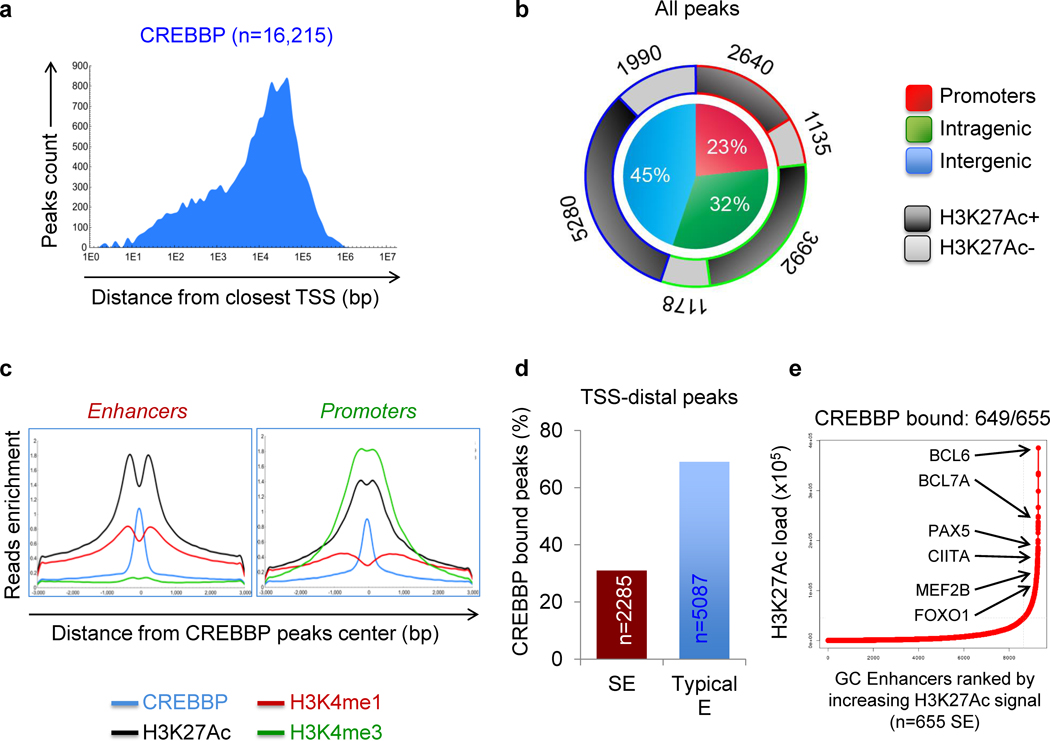
In order to define the genome-wide binding pattern of CREBBP in the GC, we performed chromatin immunoprecipitation and massively parallel sequencing (ChIP-Seq) in two independent pools of purified human GC B cells (n=3–5 donors/pool) with antibodies directed against CREBBP and, in parallel, against specific histone modifications (H3K4me1, H3K4me3, H3K27me3 and H3K27Ac) denoting well-characterized functional states of the bound chromatin. CREBBP-mediated histone acetylation is expected to be genome-wide (22) and, consistently, we identified 16,215 genomic regions (6,494 unique genes) that were significantly and reproducibly enriched in CREBBP binding in both biological replicates (P < 10−12) (Fig. 1A). The vast majority of these regions (n = 12,440, 76.7%) were localized distal from the transcription start site (TSS) of the closest gene (5,170 intragenic, 32.0%; and 7,270 intergenic, 44.7%), suggesting possible association with enhancers, while only 3,775 (23.3%) were represented by proximal promoter regions (–2/+1 kb from TSS) (Fig. 1A,B). CREBBP-bound regions were enriched in epigenetic marks of transcriptionally active chromatin, consistent with the notion that CREBBP acts to promote transcription via its acetyltransferase activity (Fig. 1B) (17). In particular, we found significant overlap between CREBBP occupancy and H3K27Ac (73% of all chromatin-bound CREBBP), along with either H3K4me3 at TSSs, indicative of active promoters (n = 3,135 peaks, 19%), or H3K4me1 in the absence of H3K4me3 at TSS-distal sites, a feature of active enhancers (n = 7,372 peaks, 45%)(23,24) (Fig. 1C). This chromatin profile suggests that CREBBP accesses DNA in nucleosome-free promoter/enhancer regions, possibly via binding to specific transcription factors.
Figure 1. CREBBP predominantly occupies enhancer regions in human GC B cells.
A, Genomic distribution of CREBBP-bound regions relative to the closest TSS. Only peaks identified in two independent ChIP-seq experiments performed on different pools of centroblasts (CB) were considered in the analysis. B, Distribution pattern of CREBBP peaks relative to UCSC annotated genes. Pie chart shows the percentage of CREBBP peaks that overlap with gene promoters (–2/+1 kb from the closest TSS; red), reside within intragenic regions (i.e., exon or intron; green) or are located in intergenic regions (i.e., upstream or downstream of the nearest annotated gene; blue). The overlap with H3K27Ac marks at these regions is indicated in the doughnut chart as different shades of grey. C, H3K4me1, H3K4me3 and H3K27Ac ChIP-seq density profiles observed in GC B cells at CREBBP-bound promoters and enhancer regions, defined as described in Methods. The plots indicate normalized mean ChIP-seq density, relative to the CREBBP peak summit, set as 0 (in blue). D, Percentage of CREBBP-bound, H3K27Ac+ regions at TSS distal sites identified as super-enhancers (SE) and typical enhancers (E). The number of peaks is given inside the bar. E, Rank order of increasing H3K27Ac fold enrichment at enhancer loci in normal CBs.
Notably, 2,285 (31%) of these “active” enhancer peaks were located within predicted GC specific super-enhancers (SE), as determined by the ROSE algorithm based on ranked ordering of H3K27Ac enrichment (see Methods) (25) (Fig. 1D). Indeed, virtually all GC SE (n = 649/655, 99%) were found to be loaded by CREBBP (Supplementary Table S1), including many that could be assigned to master regulators of the GC (namely BCL6, MEF2B and MEF2C, among others) (26,27) or to molecules (e.g. CIITA, CD74) involved in MHC class II antigen presentation, a program recently found to be disturbed in tumors lacking functional CREBBP alleles (11)(Fig. 1E and Supplementary Fig. S1). Collectively, these data indicate that CREBBP is preferentially associated with enhancer regions and suggest an important function for this acetyltransferase as a modulator of enhancer networks in GC B cells.
The CREBBP-regulated program is enriched in pathways activated in the GC light zone
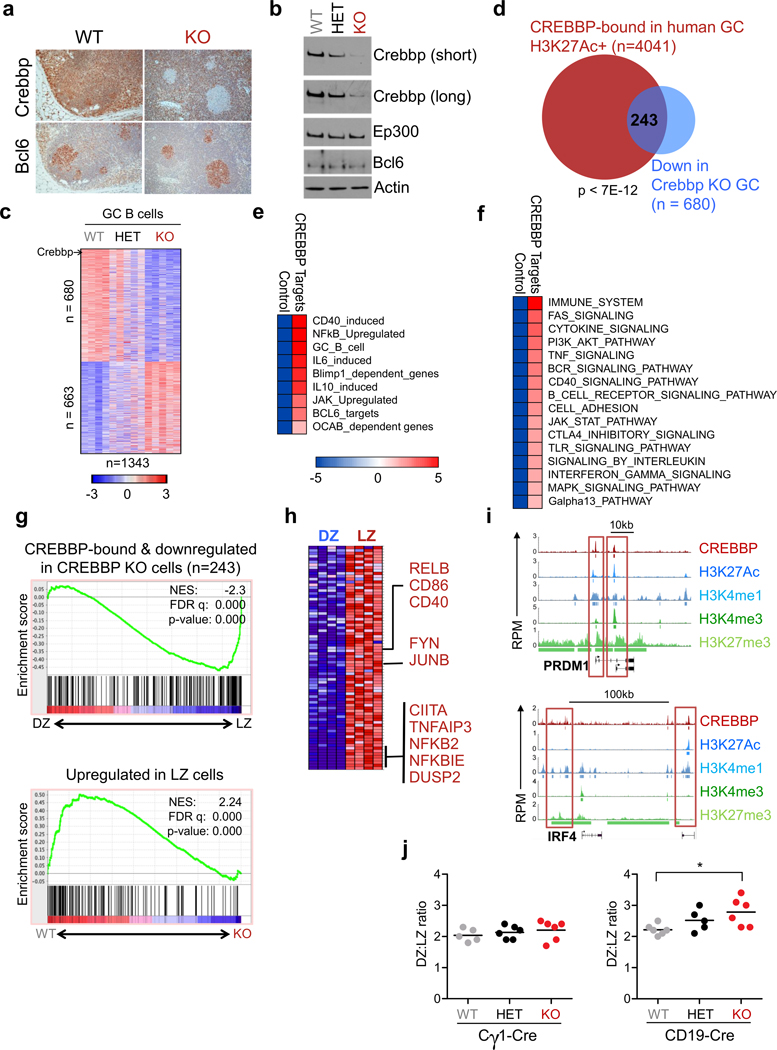
CREBBP and EP300 are ubiquitously expressed during B cell development (Supplementary Fig. S2a–d), and presumably regulate distinct sets of genes in a context specific manner. In order to determine functionally relevant targets regulated by CREBBP in the GC, we integrated the human ChIP-seq data with gene expression profile analysis of GC B cells purified from mice (Crebbpfl/fl, Crebbpfl/+ and Crebbp+/+)(28) in which the Crebbp gene can be conditionally deleted as early as 2 days after the initiation of the GC reaction via the Cγ1-Cre recombinase (29), following immunization with the T cell dependent antigen sheep red blood cells (SRBC). We did not detect apparent differences in the fraction of GC B cells (B220+CD95+PNAhi) across the three genotypes, even though efficient ablation of Crebbp expression was documented at both RNA (not shown) and protein level (Fig. 2A,B). However, supervised analysis of gene expression data identified 1343 transcripts that were differentially expressed in CREBBP-proficient vs CREBBP-deficient cells, of which 680 were down-regulated and 663 were upregulated in the knock-out (Student’s t-test, p<0.05, FC≥1.2, FDR<0.5) (Fig. 2C and Supplementary Table S2). Since CREBBP-mediated acetylation facilitates active transcription, we focused on genes whose mRNA levels were reduced in the absence of Crebbp, and intersected these data with the list of genes that are bound by CREBBP, decorated by H3K27Ac, and expressed in human GC B cells, as determined by RNA-seq analysis of bulk CD77+ cells or by GEP of cells purified from the dark zone (DZ) and light zone (LZ) compartment. This approach revealed a restricted group of 243 genes that presumably represent bona-fide direct targets of CREBBP activity, at least in steady state conditions (Fig. 2D and Supplementary Table S3).
Figure 2. The CREBBP-modulated network is enriched in signaling pathways upregulated in the LZ.
A, Immunohistochemical analysis of Crebbp (top) and Bcl6 (bottom) in representative serial sections from Crebbpfl/fl Cγ1-Cre (KO) and Crebbp+/+ Cγ1-Cre (WT) mouse spleens, analyzed 10 days after SRBC immunization. B, Western blot analysis of Crebbp and Ep300 in splenic GC B cells (B220+CD95+PNAhi) sorted from SRBC-immunized mice (n=2 mice/genotype, pooled together). Bcl6 and actin control for the identity of the fractions and for loading, respectively. C, Heat map of differentially expressed genes in sorted Crebbpfl/fl vs Crebbp+/+ GC B cells from the Cγ1-Cre cohort (Crebbpfl/+ samples shown by default) (Student’s t-test, corrected P<0.05, FC≥1.2). Colors visualize differences in the Z score (see scale bar). D, Overlap between CREBBP target genes displaying epigenetic marks of activation and genes down-regulated in Crebbp-deficient mouse GC B cells. The complete list of these genes is provided in Supplementary Table S3. The significance of the overlap was calculated by the hypergeometric distribution. E–F, Pathway enrichment analysis of CREBBP core targets (i.e. genes bound by CREBBP in human GC B cells, enriched in active histone marks, and down-regulated in Crebbpfl/flCγ1-Cre GC B cells) using the Lymphochip database and MSigDB. Only pathways relevant to B cell biology are shown in the figure, and the complete lists are available in Supplementary Table S4. Statistical significance was calculated by the hypergeometric distribution and is shown in the color key. G, GSEA analysis of CREBBP core targets along the T-score rank of transcripts expressed in human DZ vs LZ GC B cells (top). The reverse analysis (LZ-upregulated genes in Crebbp wild type vs knock-out GC B cells) is shown at the bottom. H, Leading edge associated with the GSEA shown in G. Selected CREBBP bound genes with relevant roles in the LZ physiology are indicated. I, ChIP-seq plots of CREBBP and H3K27 binding at PRDM1 and IRF4. Read density tracks of H3K4me1, H3K4me3 and H3K27me3 ChIP-Seq enrichment are also indicated. Boxed areas highlight regions showing significant CREBBP binding. J, Quantification of DZ (Cxcr4hiCd86lo) vs LZ (Cxcr4loCd86hi) cells ratio in the GC B cell (B220+CD95+PNAhi) population of Cγ1-Cre and CD19-Cre mice (see Supplementary Fig. S4D,H for the gating strategy). In the graph, each dot corresponds to one mouse, and the horizontal bar indicates the mean. WT, Crebbp+/+; HET, Crebbpfl/+; KO, Crebbpfl/fl. Only statistically significant p values are indicated. * p<0.05, one way ANOVA.
When interrogated for signatures from the Lymphochip database (30) and MSigDB (31), these genes were significantly enriched for functional annotations related to signal transduction programs that are central to GC B cell physiology and pathology, including positive regulators of the B cell receptor (FYN) and CD40 receptor (CD40), as well as downstream targets involved in NF-κB activity (NFKBIE, TNFAIP3, NFKB2), chemokine/cytokine responses (IL6, IL4RA, JUNB), lymphocyte migration, and previously identified targets of CREBBP in mature B cells, such as CIITA and Bcl2a1 (11,21)(Fig. 2E,F, Supplementary Fig. S3A and Supplementary Table S4). Thus, despite its genome-wide occupancy, Crebbp deficiency alone appears to modulate specific functions that are key to GC biology.
In particular, the signaling pathways uncovered above are known to be engaged in the LZ of the GC, where B cells receive specific cues to differentiate into plasma cells or memory B cells, or to recirculate into the DZ for additional rounds of mutations and selection (27,32). We thus used the more sensitive GSEA to ask whether the CREBBP targets are preferentially upregulated during the DZ to LZ transition. Analysis of both human and mouse transcriptional signatures using the top 500 most significantly bound genes or the 243 core targets confirmed the significant enrichment of the CREBBP-cistrome along the ranked gene expression changes associated with the GC LZ, and the same was true in the reverse analysis, using the list of LZ upregulated genes (33) (Fig. 2G,H, Supplementary Fig. S3B and Supplementary Table S5). These data suggest that CREBBP is implicated in LZ-related functions including recirculation, differentiation and exit from the GC. Indeed, CREBBP binding was also found at the regulatory sequences of known master regulators of plasma cell differentiation, including two well-characterized elements located upstream to the TSS of both PRDM1 isoforms alpha and beta (Fig. 2I), and two putative regulatory regions located upstream and downstream of the IRF4 promoter (27), supporting the involvement of CREBBP in the decision of LZ B cells to enter terminal differentiation.
To verify this hypothesis in vivo, we investigated the GC B cell response in Crebbp Cγ1-Cre mice analyzed upon SRBC immunization. No significant differences were detected between Crebbp-deficient and Crebbp-proficient littermates in the overall percentage of GC B cells as well as in the ratio between DZ and LZ B cells (Fig. 2J left and Supplementary Fig. S4A–D). Because epigenetic reprogramming may require multiple rounds of cell divisions to allow the replacement of modified histones by non-modified histones (34), and the developmental stage at which CREBBP mutations are introduced in the precursor tumor cell is not known, we then tested whether loss of Crebbp at earlier stages of B cell development had measurable effects on GC formation, by using CD19-Cre conditional knock-out mice (35). As reported (21), Crebbpfl/fl CD19-Cre animals showed significant reduction in overall percentage and number of splenic B220+ cells (not shown), with consequently reduced numbers of all mature B cell compartments (Supplementary Fig. S4E–H and Supplementary Fig. S5A–D for a complete characterization of the GC/post GC compartment). However, a mild but statistically significant increase could be appreciated in the DZ:LZ B cells ratio of the Crebbp knock-out mice, as compared to their wild type littermates (p<0.05, one-way ANOVA)(Fig. 2J right and Supplementary Fig. S4H), indicating a selective defect in their relative redistribution at the expense of LZ B cells. Taken together, these data are consistent with the involvement of CREBBP in the orderly activation of transcriptional programs that operate in the DZ to LZ transition during the GC reaction.
Opposing roles of BCL6 and CREBBP in the GC
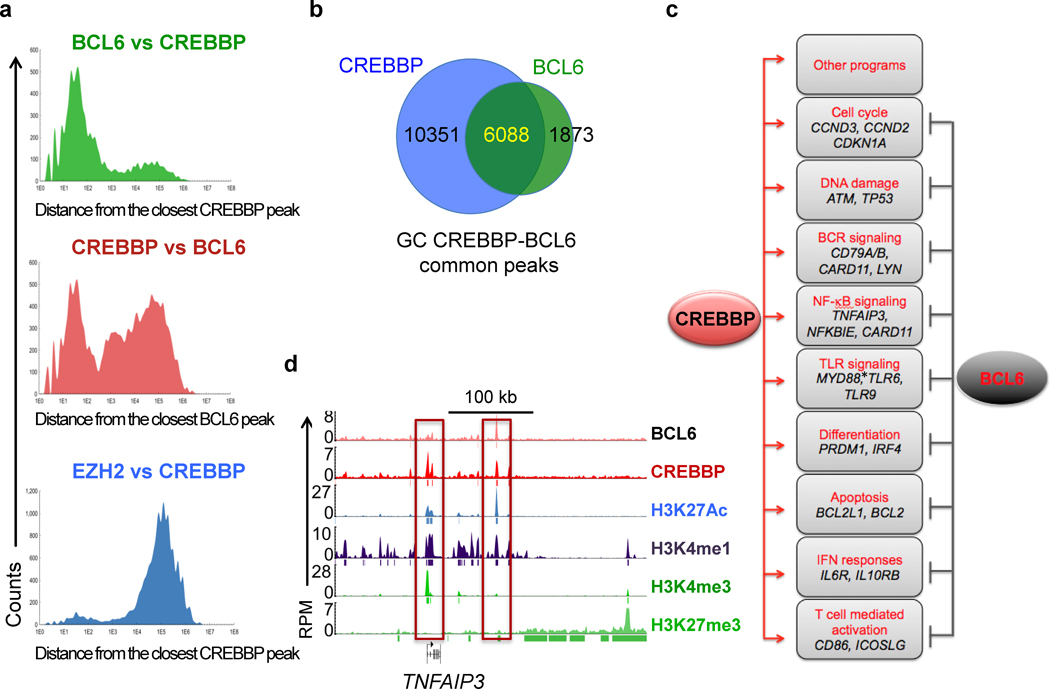
A relevant non-histone substrate of CREBBP mediated acetylation in GC B cells is the BCL6 proto-oncogene, where this post-translational modification acts as a negative modulator by disrupting its ability to recruit HDACs and to function as a transcriptional repressor (6,13). Yet, GSEA of the CREBBP-bound regions also identified a significant enrichment for previously reported BCL6 direct targets (Fig. 2E, Supplementary Fig. S3A and Supplementary Table S4) (26). Indeed, intersection of the CREBBP and BCL6 ChIP-seq data obtained from the same pools of GC B cells revealed that 87% (6088/7961) of the BCL6 peaks were consistently co-occupied by CREBBP (p << 0.0001), both at TSS proximal and at more distal sites that may represent cis-regulatory elements with enhancer features. In particular, most BCL6 bound peaks clustered in close proximity (100–1000 bp) to CREBBP bound chromatin, whereas CREBBP binding also occurred at a large number of additional loci, as expected (Fig. 3A,B). Integration of these data with transcriptomic analysis of human GC vs naïve and memory B cells pointed to a number of biological programs that are modulated by BCL6 and CREBBP in opposite fashion via binding to the same regulatory regions, including signal transduction from the BCR, TLR and CD40 receptor, NF-κB and Galpha13 signaling, T-cell-mediated B cell activation, and plasma cell differentiation (Fig. 3C,D and Supplementary Table S6). Additionally, genes implicated in DNA repair (ATM, TP53), protection from apoptosis (BCL2, BCL2L1) and cell proliferation (the cell cycle modulators CCND2 and CDKN1A) were also occupied by both proteins at promoter and/or enhancer sequences. Of note, many of the validated BCL6 target genes were decorated by both repressive and activating histone marks in a pattern consistent with a poised state, in agreement with their lack of expression in GC centroblasts at steady state conditions, and with their rapid induction upon specific signals.
Figure 3. CREBBP is involved in the regulation of BCL6-repressed targets.
A, Distribution of CREBBP peaks relative to the closest BCL6 peak (top panel); the reciprocal analysis is shown in the middle panel, while the distribution of EZH2-bound regions relative to CREBBP is shown in the bottom panel as control. B, Venn diagram illuastrating the overlap between genes bound by CREBBP and genes bound by BCL6 in human GC B cells. C, Schematic representation of relevant pathways affected by BCL6 and CREBBP with opposing effects in GC B cells. The complete list of significantly enriched pathways among the list of genes co-bound by CREBBP and BCL6 is provided in Supplementary Table S6 (see also Methods). *, bound by CREBBP only. D, Genomic snapshots of CREBBP and histone marks peaks at the previously reported BCL6 target TNFAIP3.
Overall, the data described above suggest that CREBBP and BCL6 exert opposing functions in modulating a defined subset of transcripts with key roles in the GC reaction, and point to a dual mechanism by which CREBBP may restrain the potentially deleterious activity of BCL6 by both acetylating and inactivating its protein, and by directly facilitating the transcription of its transrepressed genes.
Loss of CREBBP interferes with terminal B cell differentiation
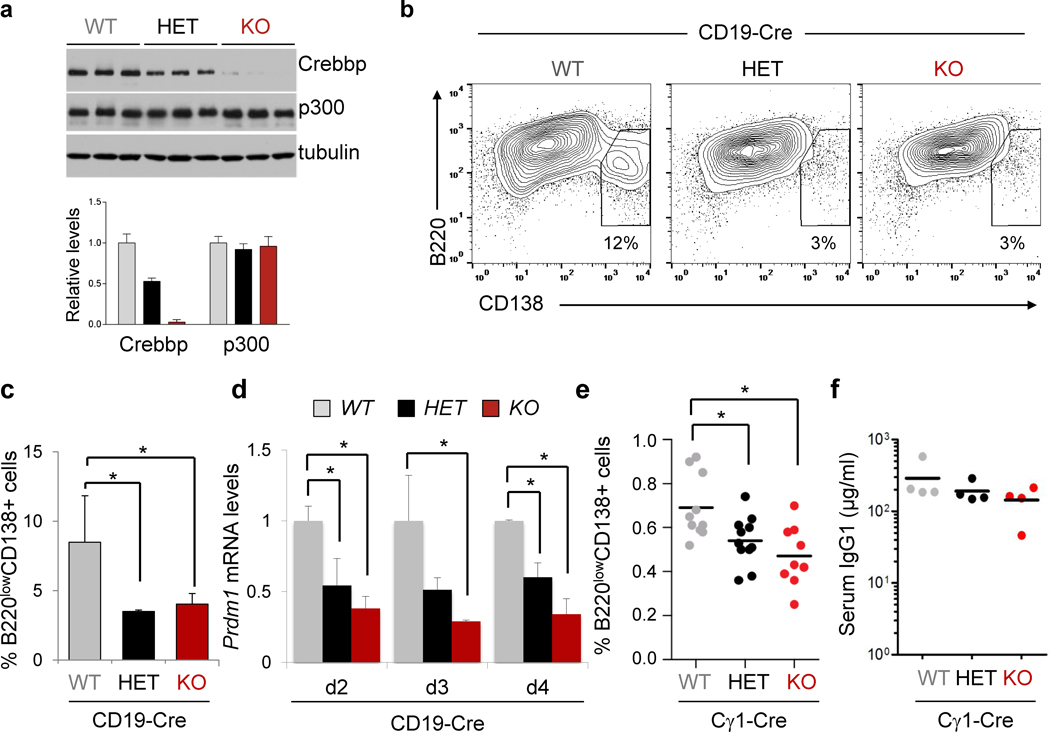
The finding of CREBBP occupancy in the regulatory sequences of PRDM1 and IRF4 prompted us to test whether reduced dosage of CREBBP affects the ability of GC B cells to differentiate into plasma cells. Because the GC reaction cannot be recapitulated in vitro, we first used an ex vivo system to assess the response of B220+ cells isolated from Crebbp+/+, Crebbpfl/+ and Crebbpfl/fl CD19-Cre mice to LPS and IL4, a mitogenic stimulus that induces proliferation as well as differentiation (Fig. 4A). We selected the CD19-Cre strain because the Cγ1-Cre mouse model requires activation of the Cre recombinase ex vivo, and the kinetics of the experiment may be too short to allow implementation of the epigenetic modifications that are necessary to induce phenotypic effects. In agreement with previous studies, flow cytometric analysis of CD138 expression performed at day 3 and 4 post-stimulation documented the appearance in the Crebbp+/+ population of cells expressing high levels of CD138 and low levels of B220, consistent with a plasmablastic phenotype (on average, 8.5% of cells). In contrast, only ∼3% of the Crebbpfl/+ and Crebbpfl/fl cells had upregulated CD138 (p<0.05; Student’s t-test) (Fig. 4B,C). This difference was not due to diminished proliferation, which is also required for plasma cell differentiation (36), as the percentage of cells that had undergone cell divisions in the Crebbpfl/fl population was even higher compared to wild type cells (see next paragraph). Interestingly, q-RT-PCR analysis of PRDM1 mRNA performed at day 2 through 4 post-stimulation revealed significantly reduced levels in both Crebbpfl/+ and Crebbpfl/fl B cells, consistent with a dose-dependent role of Crebbp in its transcriptional activation (Fig. 4D).
Figure 4. Crebbp loss interferes with plasma cell differentiation.
A, Western blot analysis of Crebbp and Ep300 in splenic B cells isolated from the indicated mice and used for the ex vivo assay (top). The relative levels of Crebbp and Ep300 are quantified in the bottom panel. B, Dot plots of representative flow cytometric analysis of CD138 and B220, performed 72 hours post-stimulation. Numbers indicate the percentage of cells in the gate. C, Quantification of the data shown in B (mean ± SD; n=3 mice/genotype; * p<0.05, one way ANOVA). D, Q-RT-PCR analysis of Prdm1 mRNA expression in the same animals, performed at days 2–4 post-stimulation. Values correspond to the average of 3 mice, each analyzed in triplicate, ± SD (*p<0.05, one way ANOVA; one experiment of two that gave comparable results). E, Percentage of splenic plasma cells in Crebbpfl/fl, Crebbpfl/+ and Crebbp+/+ Cγ1-Cre mice, analyzed at day 10 post-SRBC immunization. Horizontal bars indicate the mean. Statistical significance was calculated by one way ANOVA (* p<0.05). F, IgG1 serum levels from Crebbpfl/fl , Crebbpfl/+ and Crebbp+/+ Cγ1-Cre mice, measured by ELISA 10 days after SRBC immunization. Horizontal bars indicate the mean. No statistically significant differences were observed (p = 0.18), although a trend towards lower levels can be appreciated in the Crebbp deficient animals. In the figure, WT denotes Crebbp+/+, HET is Crebbpfl/+, and KO indicates Crebbpfl/fl.
Consistent with these results, flow cytometric analysis of the plasma cell compartment in Crebbpfl/+ and Crebbpfl/fl Cγ1-Cre mice, compared to littermate controls, uncovered a mild yet significant decrease in the percentage of splenic CD138+B220− cells (p<0.05, One-way ANOVA), which was accompanied by a trend toward reduced levels of serum IgG1 (Fig. 4E,F; see also Supplementary Fig. S5C,D for the analysis of the CD19-Cre cohort). The inability to detect differences in bone marrow plasma cells indicates that the phenotypic changes imposed by Crebbp loss in vivo are subtle, and may be explained by the involvement of compensatory mechanisms (e.g. extra-GC plasma cells), along with the difficulty to quantitatively measure the GC-exiting plasma cell population (<1% of total lymphocytes) in the absence of a trackable marker, also considering the early time point (primary response) and the presence of residual cells that fail to delete Crebbp in both the Cγ1-Cre and CD19-Cre model (see Fig. 2B and Supplementary Fig. S4E). Nonetheless, these data suggest that a full complement of Crebbp may be necessary for optimal terminal differentiation of GC B cells, which is attained in part by remodeling the chromatin landscape of plasma cell master regulators and facilitating their accessibility to the transcriptional machinery.
Crebbp loss confers proliferative advantage
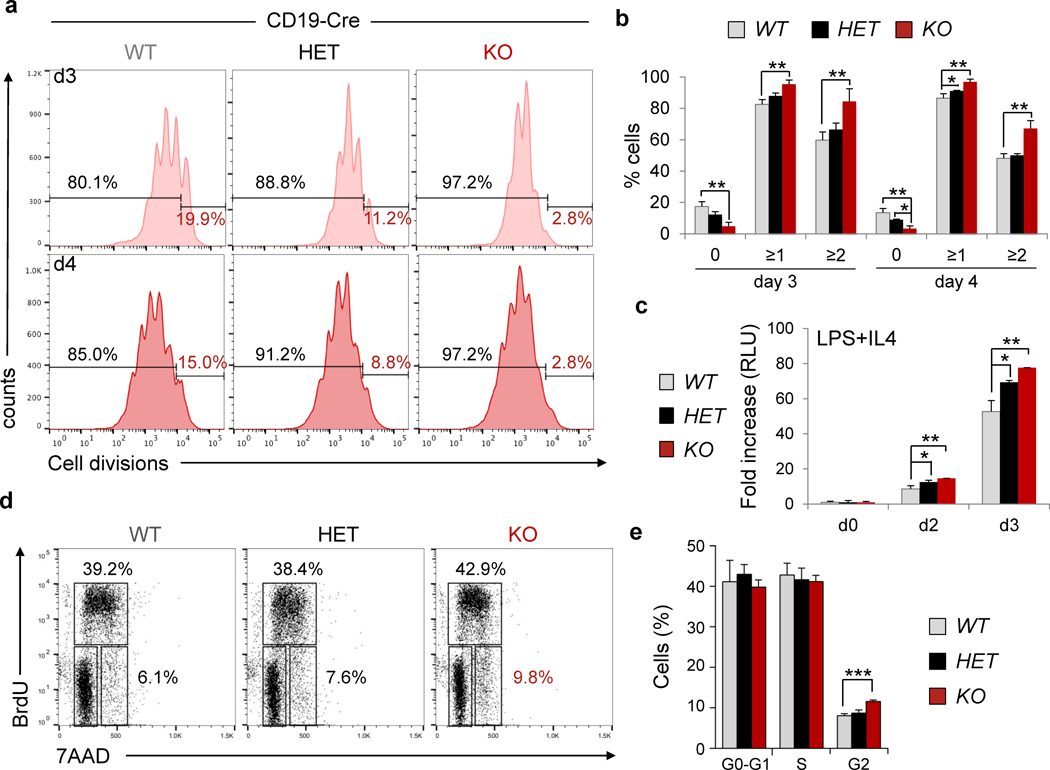
To further investigate the biological consequences of CREBBP haploinsufficiency in the mature B cell compartment, we measured the proliferative capacity of B220+ cells isolated from the spleen of CD19-Cre mice and stimulated with anti-CD40+IL4 (mimicking the signals delivered in vivo to induce T cell dependent B cell responses) or LPS+IL4. Flow cytometric analysis of CellTrace Violet labeled cells at day 3 and 4 post-stimulation revealed that a significantly higher number of cells had undergone cell division in both homozygous and, to a minor extent, heterozygous null Crebbp cultures, compared to wild-type (on average, 4.7% and 12.2% non-divided cells, respectively, vs 17.5% in the control mice)(p<0.05)(Fig. 5A,B). Consistently, viability was greater in the Crebbpfl/fl and Crebbp+/fl cultures as measured based on their metabolic activity (Fig. 5C and not shown). This finding seems to be due to an increased propensity of the Crebbp deficient population to initiate proliferation following mitogenic stimuli, as indicated by the almost complete absence of non-divided cells in this culture compared to the wild type control, while analysis of BrdU incorporation revealed a subtle yet measurable difference in the percentage of cells residing in the G2 phase of the cell cycle (Fig. 5D,E). Also consistent with enhanced proliferative activity, class switch recombination to IgG1 –a process known to require cell division– was significantly increased in Crebbp+/fl and Crebbpfl/fl cells overall, but was comparable to that of control cells when normalized by the number of cell divisions (not shown). Finally, while GC B cells from Crebbpfl/fl and Crebbp+/+ Cγ1-Cre mice were undistinguishable in vivo, a measurable increase in the fraction of BrdU+ GC B cells could be detected in the CD19-Cre cohort (not shown).
Figure 5. Crebbpfl/fl B cells exhibit proliferative advantage.
A, Representative cell proliferation profiles of B220+ splenocytes isolated from Crebbpfl/fl (KO), Crebbpfl/+ (HET), and Crebbp+/+ (WT), CD19-Cre mice, labeled with the CellTrace Violet dye and cultured in the presence of anti-CD40 and IL4 for 3 (top) and 4 (bottom) days. Individual peaks in the plot correspond to different numbers of cell divisions, measured by flow cytometric analysis. Numbers indicate the percentage of cells with one or more divisions (black) vs the undivided cells (red), and the quantification of three samples/genotype is shown in panel B, where values represent the mean and error bars represent the SD from the mean (* p<0.05, ** p<0.01, one way ANOVA). C, Relative growth of B220+ splenocytes treated with LPS and IL4 for 3 days, measured by the CellTiter Glo reagent. Data are indicated as fold changes in relative luciferase units (RLU), with the mean value of unstimulated cells, measured at day 0, arbitrarily set as 1 (mean ± SD; n=3 mice/genotype, in triplicate; * p<0.05, ** p<0.01, one way ANOVA). D, Representative cell cycle profiles of B220+ cells, labeled with BrdU and analyzed by flow cytometry at day 3 after stimulation with CD40 and IL4. Numbers indicate the percentage of cells in the gated population. E, Quantification of panel d (mean ± SD; n = 3 mice/genotype). *** p<0.001, one way ANOVA.
Together, these results indicate that loss of Crebbp provides proliferative advantage to B cells, reflecting in part an augmented ability to respond/start cell division in response to stimulation, and suggest an additional mechanism by which tumor-associated inactivating mutations might contribute to tumor development in the mature B cell lineage. Intriguingly, none of these effects were observed in the Cγ1-Cre background, despite deletion of the Crebbp protein was efficiently achieved within 48 hours from ex vivo stimulation (not shown), supporting the notion that cells need to complete multiple cell divisions in order to establish the epigenetic changes underlying this phenotype (see Discussion).
Crebbp deficiency cooperates with BCL2 deregulation in lymphomagenesis in vivo
To investigate whether GC-specific haploinsufficiency of Crebbp promotes the development of lymphoma in vivo –either individually or in cooperation with other oncogenic lesions– we monitored the Cγ1-Cre and CD19-Cre cohorts for tumor incidence and survival.
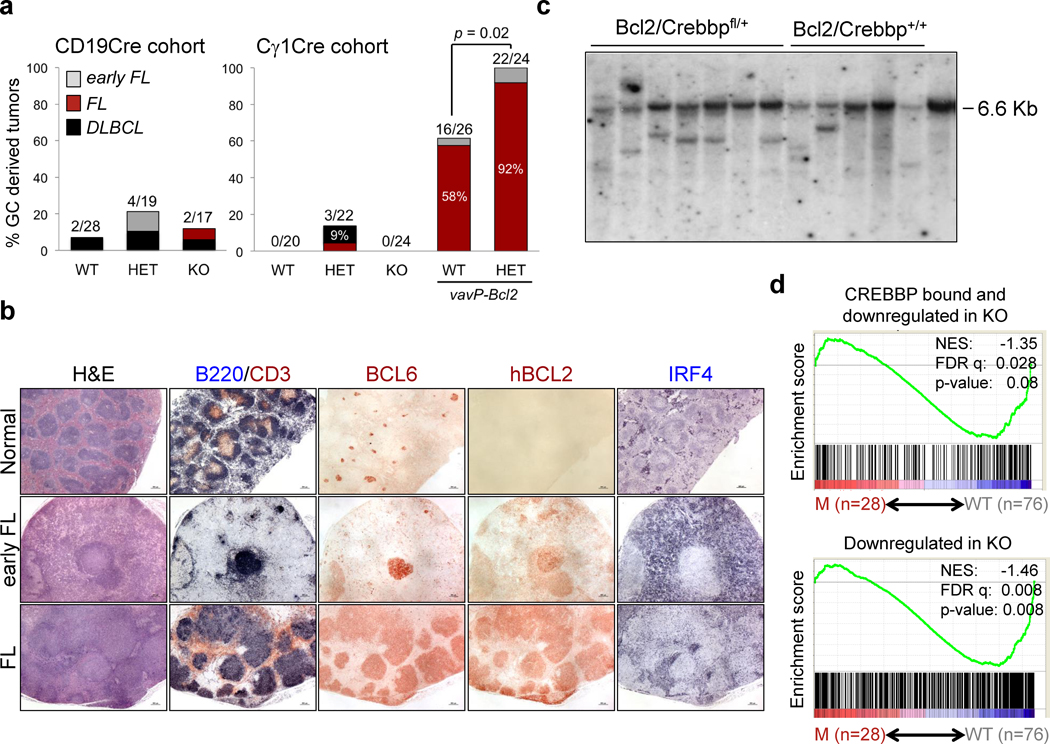
We did not observe significant differences in event free survival between Crebbpfl/fl and Crebbp+/+ Cγ1-Cre littermates over a follow up of 18 months and, while 3/22 (14%) heterozygous animals developed mature B cell lymphomas (one FL and two DLBCL), this difference did not reach statistical significance (Fig. 6A, right). Loss of Crebbp alone was not sufficient to drive oncogenic transformation even when engineered at earlier stages of B cell development, in the CD19-Cre mouse model (Fig. 6A, left), suggesting that additional oncogenic events are involved in the clonal expansion of cells carrying CREBBP/EP300 mutations.
Figure 6. Deletion of Crebbp in the GC cooperates with BCL2 deregulation to promote lymphomagenesis.
A, Incidence of B cell lymphoproliferative diseases in the two cohorts. The percentage of animals in each entity is given inside the bar, and the total number of animals diagnosed with GC-derived lymphomas, out of the total analyzed, is shown on top. p < 0.05, Fisher’s exact test, two tails. Only heterozygous null mice were included in the compound Crebbp floxed-BCL2 transgenic cohort because of the predominanty heterozygous pattern of mutations observed in human tumors (see Supplementary Fig. S6A–C). B, Histologic and immunohistochemical analysis of representative spleen sections from control mice and VavP-Bcl2/Crebbpfl/+/Cγ1-Cre mice (H&E staining; scale bar, 500µm). Double immunostaining for B220 and CD3 identifies the B and T cell zone. Bcl6 serves as a GC-specific marker. An anti-human BCL2 antibody that does not cross-react with the murine Bcl2 protein was used to specifically detect the expression of the VavP-Bcl2 transgene, and is thus not staining the tissue from the control Cγ1-Cre only animal. C, Southern blot analysis of EcoRI digested DNA from representative lymphomas, hybridized with a murine JH3-4 probe. Tail genomic DNA controls for the germline immunoglobulin heavy chain EcoRI fragment (6.5 kb). D, Enrichment plots of CREBBP core targets (top) or genes downregulated in Creebbp knock-out GC B cells (bottom) in the rank of genes differentially expressed between DLBCL biopsies carrying intact CREBBP/EP300 alleles (n=76; 30 GCB- and 46 ABC-type) and biopsies harboring CREBBP/EP300 genetic lesions (n=28; 13 GCB- and 15 ABC/unclassified).
To test this hypothesis, Crebbpfl/fl Cγ1-Cre mice were crossed with VavP-Bcl2 transgenic mice (37), which carry a deregulated BCL2 allele mimicking the translocation that frequently co-occurs with CREBBP mutations in human FL and DLBCL (∼50% of mutated cases; Supplementary Fig. S6A–C). The compound animals were undistinguishable from VavP-Bcl2 littermates in terms of overall survival (Supplementary Fig. S7A,B), likely due to the glomerulonephritis and autoimmune diseases developing in this strain at nearly 100% penetrance, as reported (37). However, when combined with BCL2 deregulation, Crebbp haploinsufficiency led to a significant increase in the incidence of B cell malignancies recapitulating molecular and phenotypic features of the human FL, including their derivation from GC B cells, as shown by the expression of the GC-specific marker BCL6 in the absence of post-GC markers (IRF4 and CD138) (Fig. 6B), and the evidence of clonally rearranged immunoglobulin genes harboring somatic mutations (Fig. 6C and Supplementary Table S7). Overall, 92% (22/24) Crebbpfl/+ VavP-Bcl2 compound animals presented with “in situ” or overt FL in the spleen, lymph nodes, and frequently in the Peyer’s Patches, as opposed to 16/26 (61.5%) VavP-Bcl2 mice. Also of interest was the observation of rare B220−PAX5−BCL6− lymphoproliferations showing plasmacytic morphology and IRF4 positivity (Supplementary Fig. S7C) in the VavP-Bcl2 mice (n=3/26), but not in their compound Crebbp fl/+ littermates, lending further support to the hypothesis that reduced dosage of Crebbp disturbs terminal differentiation.
Interestingly, GSEA of human DLBCLs using the Crebbp signature identified by our integrated approach showed a significant under-representation of these genes in tumors carrying CREBBP or EP300 genetic lesions, compared to CREBBP-wild type biopsies, independent of molecular subtype (Fig. 6D), supporting the relevance of our model to the human disease and confirming a role for perturbations of this program in DLBCL pathogenesis.
Collectively, these data document a role for Crebbp as a bona fide haploinsufficient tumor suppressor gene in GC B cells, whose reduced levels cooperates with Bcl2 deregulation to facilitate the development of B cell NHL.
DISCUSSION
CREBBP is one of the most commonly mutated genes in GC-derived B-NHL (3–9,11), but the specific role of this acetyltransferase in the GC B cell compartment and the mechanism by which its inactivation contributes to lymphomagenesis remain unknown. The results reported here establish CREBBP as a major regulator of GC enhancer networks, identify the transcriptional program that is sensitive to its dose reduction in this compartment, and demonstrate that CREBBP is a haploinsufficient tumor suppressor of GC-derived lymphomagenesis. These findings have implications for normal GC B cell physiology, for the mechanisms leading to FL and DLBCL, and for the therapeutic targeting of these cancers.
Our results show that CREBBP binds extensively to the genome of GC B cells (∼16,000 peaks), and that most of these regions exhibit features of transcriptionally active (or poised) enhancers (38), including virtually all predicted GC-specific super-enhancers. These clusters of enhancers are known to provide cell-specific sites of open chromatin that can be accessed by signal-dependent transcription factors, and are thus primarily responsible for cell-type-specific transcriptional responses (25,39). Thus, CREBBP may act as a major regulator of enhancer networks in the specialized microenvironment of the GC, analogous to what has been established for the CREBBP/EP300 complex in embryonic stem cells (25).
In apparent contrast with this genome-wide distribution, deletion of Crebbp had modest consequences on global gene expression, both in terms of number of differentially expressed genes and of magnitude of the observed transcriptional changes, suggesting that most genes can tolerate reduced availability of this protein, at least in “non-stress” conditions. This finding may reflect in part the compensatory role of the CREBBP paralogue EP300. Indeed, while the single GC-specific Ep300 knock-out model shows an analogous modest phenotype, Crebbp/Ep300 double knock-out animals are completely unable to form GCs upon antigenic stimulation (data not shown). Nonetheless, we were able to identify a restricted set of genes that were sensitive to Crebbp depletion in vivo, and this network was preferentially enriched in signal transduction programs that are normally suppressed by BCL6 in the GC DZ but become engaged in the LZ of the GC (e.g. BCR, NF-κB) (27,32), supporting a role for CREBBP dosage in cell fate specification and terminal B cell differentiation in this compartment.
In this context, the almost complete overlap between the genome-wide mapping of BCL6 (a transcriptional repressor) and CREBBP (a positive regulator of transcription) suggests that a major role of CREBBP in the GC is to counterbalance BCL6 suppressive effects by directly binding to its targets and licensing their transcription. This model is further reinforced by the known ability of CREBBP to regulate BCL6 activity through acetylation-mediated inactivation of its protein (6,13). Together, these two mechanisms may ensure a continued modulation on BCL6 activity, allowing signal dependent rapid re-induction of BCL6 target genes during GC exit. Nonetheless, the fact that multiple proteins have been identified as substrates of the CREBBP enzymatic activity (e.g. TP53), and the observation of reduced H3K27 and H3K18 acetylation levels in CREBBP deficient B cells (6) and Supplementary Fig. S7D,E) are consistent with the involvement of both BCL6-dependent and BCL6-independent effects in the tumor suppressor activity of CREBBP.
As previously observed with KMT2D, a methyltransferase whose activity is also genetically reduced in FL and DLBCL (4,6,40), deletion of Crebbp early during B cell development led to a more pronounced phenotype compared to that of mice where deletion occurs upon initiation of the GC. This may in part be explained by the fact that multiple rounds of cell division are thought to be necessary to implement the substitution of modified histones with non-modified histones upon loss of CREBBP, or that the cumulative effect of small changes in gene expression may be amplified by additional time and cell divisions, and is consistent with a model whereby Crebbp loss occurs early in the tumor ancestor clone.
Indeed, while individual loss of Crebbp upon initiation of the GC reaction or in pro B cells was insufficient to induce the development of lymphoma, its heterozygous deletion accelerated tumor formation when combined with BCL2 deregulation, establishing this acetyltransferase as a bona fide haploinsufficient tumor suppressor gene in FL and DLBCL. These findings are again analogous to what we previously observed with KMT2D (40) and, together with the observation that both KMT2D and CREBBP inactivating mutations represent early events in lymphomagenesis, suggest that CREBBP acts as a non-conventional tumor suppressor, the loss of which contributes to oncogenic transformation by establishing a permissive epigenetic environment for the accumulation of additional oncogenic perturbations.
In the context of the GC, our in vitro data suggest that one mechanism by which dose reduction of Crebbp may contribute to lymphomagenesis is by favoring proliferation at the expense of differentiation. These alterations are reflected by the in vivo phenotype of CREBBP–depleted GCs, which show an expansion of the DZ (i.e. the site of intense proliferation), at the relative expense of the LZ, where cells are induced to differentiate. Additionally, CREBBP inactivation may facilitate malignant transformation by weakening the transcriptional responses of several tumor suppressors associated with FL/DLBCL pathogenesis, including TNFAIP3, GNA13, and S1PR2, while perturbing antigen presentation processes that are mediated by MHC class II molecules (Supplementary Fig. S1).
The reason why FL and DLBCL almost invariably retain one normal CREBBP allele is unclear, but may be related to the need for a minimum dose of CREBBP/EP300 activity in the cell. This notion is supported by the analogous monoallelic distribution of EP300 somatic mutations (found in a smaller subset of DLBCL and FL)(6) and germline alterations of CREBBP/EP300 in patients with Rubinstein-Taybi syndrome (10), as well as by their mutually exclusive pattern in these diseases. Interestingly, monoallelic CREBBP/EP300 alterations are often concurrent with inactivating mutations of the methyltransferase KMT2D (∼60% of FL and 40% of mutated DLBCL), and these two histone modifiers are often found at the same genomic locations in GC B cells (not shown), suggesting a crosstalk between their enzymatic activity, as reported for EP300 and the SET1 methyltransferase (41).
Finally, the identification of the transcriptional program perturbed by reduced dosage of CREBBP may provide a framework for the development of combinatorial therapies aimed at re-establishing physiologic acetylation levels (e.g. by using deacetylase inhibitors, or compounds that can stimulate histone acetylation) while modulating signaling pathways (e.g. BCR, NF-κB) that are concurrently dysregulated in these tumors. Moreover, the EP300-addiction presumably created by CREBBP loss in tumor cells could be exploited for therapeutic targeting. In this context, the animal model described here represents a useful tool for the pre-clinical testing of novel therapeutic approaches.
METHODS
Isolation of human centroblasts
Tonsils were obtained as residual material from routine tonsillectomies at the Children’s Hospital of Columbia-Presbyterian Medical Center, in compliance with Regulatory Guideline 45 CFR 46.101 (b)(4) for Exempt Human Research Subjects of the US Department of Health and Human Services and according to protocols approved by the Institutional Ethics Committee. Human GC centroblasts, naïve B cells and memory B cells were isolated from reactive tonsils as described (42)
Chromatin Immunoprecipitation and sequencing (ChIP)
Purified GC B cells were cross-linked with 1% formaldehyde for 10 min at RT, quenched by the addition of glycine to a final concentration of 0.125 M, and frozen. To minimize potential biases between individual donors, ChIP was performed on two independent biological replicates each consisting of 3–5 GC B cell isolations. Cell lysis and nuclei isolation was performed using the TruChIP Chromatin Shearing Kit High Cell SDS (Covaris). Nuclei were sonicated using the S220 Ultrasonicator (Covaris) in order to obtain chromatin fragments of 200–500bp. Sheard chromatin was incubated overnight with 4µg of anti-CREBBP (A-22 rabbit polyclonal, Santa Cruz Biotechnology), anti-H3K27Ac (Active Motif), anti-H3K4me3 (Abcam), or 2µg of anti-H3K4me1 (Abcam). Protein A magnetic beads were then added for 4hr incubation followed by sequential washes at increasing stringency and reverse cross-linking. Upon RNAse and proteinase K treatment, ChIP DNA was purified using the MiniElute Reaction Clean Up Kit (Qiagen) and quantified by Quant-iT PicoGreen dsDNA Reagent (Life Technologies). The specificity of the antibodies against H3K4 methylation and H3K27 acetylation has been previously documented (40).
ChIP-seq library preparation and Illumina sequencing
ChIP-seq libraries were constructed starting from 4ng of ChIP or Input DNA as reported in (40). Libraries were quantified using the KAPA SYBR FAST Universal qPCR Kit (KAPA Biosystems), normalized to 10nM, pooled and sequenced in an Illumina HiSeq 2000 instrument as single-end 100 bp reads, obtaining on average 25×106 reads/sample.
ChIP-seq analysis
Sequencing data were acquired through the default Illumina pipeline using Casava V1.8. Reads were aligned to the human genome (UCSC hg19) using the Bowtie2 aligner v2.1.0 (43), allowing up to two mismatches to cope with human variation. Duplicate reads (i.e., reads of identical length mapping to exactly the same genomic locations) were removed with SAM tools v0.1.19 using the rmdup option (44). Reads were normalized to total reads aligned (counts per million). Peak detection was done using ChIPseeqer v2.0 (45) enforcing a minimum fold change of 2 between ChIP and input reads, a minimum peak width of 100 bp, and a minimum distance of 100 bp between peaks. The threshold for statistical significance of peaks was set at 10−12 for CREBBP and BCL6, and 10−15 for H3K4me1, H3K4me3 and H3K27Ac. The significance of the overlap between CREBBP occupied regions and H3K27 acetylated regions was assessed by binomial test considering the total size (in bp) of the significant peaks identified in the two ChIP-seq experiments (e.g., CREBBP vs H3K27Ac) and the size of the overlapping region between the two experiments, relative to total genome size of the UCSC hg19 assembly. The BCL6 ChIP-seq analysis is described in (46). The ChIP-Seq data has been deposited in the GEO database under accession number GSE89688.
Definition of active promoters and enhancers
Significant CREBBP bound regions (peaks) were classified as active promoters if located within −2/+1 kb from the transcription start site (TSS) of an annotated gene and enriched in H3K4me3, but not H3K27me3. Intergenic and intragenic (exonic or intronic) regions were considered as active enhancers if located distal to TSS sites and enriched in H3K4me1 and H3K27Ac, but not in H3K4me3 (38). For the identification of super-enhancers in GC B cells, we applied the ROSE algorithm (25) to our H3K27Ac ChIP-Seq dataset. Occupancy of CREBBP at super-enhancers was then determined based on the overlap between CREBBP peaks and genomic regions identified by ROSE.
Assignment of active enhancers to genes
CREBBP bound enhancers, identified as described above, were assigned to the nearest expressed and transcriptionally active gene (distance from enhancer center to TSS) as the most likely candidate target gene (40).
Mice
The conditional Crebbp knock-out mouse model (mixed C57BL/6 × Sv129 background) has been reported (28). Deletion of Crebbp was directed to GC B cells or precursor B cells by breeding Crebbpfl/+ mice with the Cγ1-Cre (29) or CD19-Cre (35) deletor strain (both in pure C57BL/6 background), followed by offspring intercrossing. Compound mice carrying the conditional Crebbp × Cγ1-Cre allele and the VavP-Bcl2 transgene were obtained by breeding with VavP-Bcl2 mice (C57BL/6 background)(37). A sample size of ≥23 animals/genotype was calculated to ensure over 80% power to detect an increase in tumor formation in the Crebbp compound lines at a p<0.05 significance. No randomization was used. All animal work was performed under National Cancer Institute and Columbia University Institutional Animal Care and Use Committee-approved protocols. Genotyping was performed by PCR analysis and the protocol is available upon request. Animals were monitored for tumor incidence and survival twice/week over a period of 18 months, and sacrificed for analysis when visibly ill or when they reached end point. Kaplan-Meier event-free survival curves were generated using the GraphPad Prism 5 software (GraphPad Sofware), and statistical significance was calculated using the log-rank (Mantel-Cox) test. The Fisher’s exact test (two-tailed) was used to assess whether differences in the incidence of lymphoid malignancies between Crebbp deficient mice and control wild-type littermates were significant.
Mouse Immunizations
For the analysis of T cell dependent responses, age-matched 10–16-week-old mice were immunized by intraperitoneal injection of 500 million sheep red blood cells (SRBC) (Cocalico Biologicals) in PBS and analyzed after 10 days (12 days for the BrdU in vivo labeling experiment). Alternatively, mice were immunized by 0.5mg/ml 4-Hydroxy-3-nitrophenylacetyl hapten conjugated to Keyhole Limpet Hemocyanin (NP-KLH) (Biosearch Technologies) along with Complete Freund’s Adjuvant (Sigma-Aldrich), and analyzed after 12 and 28 days. To achieve a higher yield of GC B cells for gene expression profile analysis, mice were immunized with two sequential SRBC injections (day 0, 1×108; day 5, 1×109) and sacrificed at day 12.
Flow cytometric analysis
Multi-color flow cytometric analysis of the B and T cell lymphoid compartment was performed at 3 and 6 months of age as previously reported (40), using 3–4 mice/genotype/experiment. The B cell compartment was also examined in all animals from the tumor-watch cohort, when sacrificed because visibly ill or at end of the study. Briefly, single cell suspensions prepared from various lymphoid organs and from the peritoneal cavity were stained using different combinations of the following fluorescent-labeled antibodies: anti-B220 (RA3-6B2), -IgM, -IgD, -IgK , -CD21, -CD23, -IgG1, -CD138, -CD95, -Mouse Early B Lineage, -CD5, -Mac1, -CD4, -CD8, -CD43, -BCL6 (all from BD Biosciences); anti-CXCR4 and anti-CD86 (eBioscience); and fluorescein labeled Peanut Agglutinin (PNA) (Vector). The complete list of antibodies used for FACS analysis can be found in Supplementary Table S8. Samples were acquired on a FACSCanto™ II or on a FACSCalibur™ (BD Biosciences), and analyzed using the FlowJo software (Tree Star).
Enzyme-linked immunosorbent assay (ELISA)
Total IgM, IgG1, IgG2b, IgG3 and IgA serum titers were measured by ELISA as previously described (47), using the following antibodies: anti–mouse Ig H+L (1010-01, Southern Biotech), anti–mouse IgM–AP (1020-04, Southern Biotech), anti–mouse IgG1–AP (1070-04, Southern Biotech) anti-mouse IgG2b-AP (1090-04, Southern Biotech), anti-mouse IgG3-AP (1100-04, Southern Biotech) and anti-mouse IgA-AP (1040-04, Southern Biotech). To quantify NP-specific antibodies, serum was collected at day 12 after a single intraperitoneal injection of NP-KLH (100 µg) in Complete Freund’s Adjuvant (Sigma). Analysis in Figure S5D was performed on 5 Crebbp+/+, 6 Crebbpfl/+ and 6 Crebbpfl/fl CD19 Cre mice; for IgG3 and IgA, one Crebbpfl/fl animal was censored from the analysis due to insufficient material. NP-specific high-affinity antibodies were captured on plates coated with 2.5 µg/ml NP25-BSA (SouthernBiotech), and bound antibodies were detected with alkaline phosphatase–conjugated IgM-, IgG1-, IgG2b,IgG3 or IgA specific antibodies (Southern Biotech). To quantify the total antibody serum levels in unimmunized mice, serum was collected at d0 from Crebbpfl/fl, Crebbpfl/+ and Crebbp+/+ CD19Cre mice (n=3 per genotype). Pan-mouse immunoglobulins were captured on plates coated with 2.5 µg/ml of goat anti-mouse Ig (H+L) (Southern Biotech). Each assay was performed in duplicates using 12 (NP-KLH - immunized mice) or 7 (unimmunized mice) serial dilutions.
Ex vivo stimulation of splenic lymphocytes with anti-CD40+IL4 or LPS+IL4
Splenic murine B-cells were isolated from 10–16 weeks old mice using the mouse B-cell isolation kit (Miltenyi Biotec) according to the manufacturer’s instructions, plated at 2 × 106 cells/ml and cultured for 4 days in RPMI 1640 medium supplemented with 15% FBS, 55 µM β-mercaptoethanol, 50ng/ml recombinant mouse IL4 (R&D systems) and either 1µg/ml Hamster monoclonal anti-CD40 (clone HM40-3, BD Pharmingen) or 5µg/ml LPS (L263, Sigma). Cells were harvested at day 2, 3 and 4 and processed for flow cytometric analysis and RNA or protein isolation.
Cell viability and proliferation assays
The number of viable cells in the ex vivo cultures was determined by measuring the levels of ATP, which signals the presence of metabolically active cells, using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega), according to the manufacturer’s instructions. Cell proliferation was analyzed with the CellTrace™ Violet Cell Proliferation Kit (Life Technologies), which monitors distinct generations of proliferating cells by a fluorescent dye dilution. Data were acquired at day 2–4 on a FACSCanto™ II (BD Biosciences) flow cytometer with 405 nm excitation and an emission filter in the 450nm range. All experiments were performed at least twice in triplicate, and the genetic background of the animals was verified by amplification and sequencing of a fragment in the IGH locus, which encompasses a known SNP discriminating between the 129Sv and the C57BL6 strain. Analysis of BrdU incorporation was performed using the APC BrdU Flow Kit (BD Pharmingen), according to the manufacturer’s protocol.
Gene expression profile analysis of mouse GC B cells
GC B-cells (B220+CD95+PNAhi) were sorted from Crebbpfl/fl, Crebbpfl/+ and Crebbp+/+ Cγ1-Cre splenocytes (n=4–5 mice/genotype) in a BD FACSAria III cell sorter (BD Biosciences). Total RNA was extracted using the NucleoSpin RNA II kit according to the manufacturer’s instructions (MACHEREY-NAGEL), and samples with RNA integrity numbers (RIN) >9, as assessed on a BioAnalyzer 2100 (Agilent), were processed for cDNA synthesis. From each sample, twenty nanograms were reverse transcribed and amplified using the Ovation RNA Amplification System (NuGEN), followed by labeling with the Encore Biotin Module (NuGEN). Labeled samples were hybridized to Affymetrix MG 430 2.0 arrays according to the manufacturer’s protocol. Raw expression values were normalized using the Robust Multiarray Averaging (RMA) algorithm in GenePattern (48), and multiple probes corresponding to the same gene were collapsed to a single probe based on maximum t-statistic. Differentially expressed genes were determined by the Student’s t-test using a P<0.05, absolute fold change ≥ 1.2, and FDR <0.5 after Benjamini-Hochberg correction. This lenient cutoff was allowed because the transcriptional changes imposed by CREBBP loss are expected to be modest, as reported in a number of studies performed in other cells/tissues, and intersection of the gene expression data with the list of genes bound by CREBBP provided further filtering to the analysis. The GEO accession number for these data is GSE88799. For visualization of gene-expression intensity, all gene expression data were normalized by gene and row (z-score transformation) with the GenePattern tool.
Gene Set Enrichment Analysis (GSEA)
Gene expression profile data from Crebbpfl/fl, Crebbpfl/+ and Crebbp+/+ Cγ1Cre GC B cells were analyzed for enrichment in pre-defined sets of genes with GSEA-2.0 (49), using 1000 gene set permutations and the Canonical Pathway (CP) Molecular Signature Database gene sets collection, or in the Lymphoid Signature Database (30). Multiple probes corresponding to the same gene were collapsed to a single probe based on maximum t-statistic. Enrichments were considered significant if FDR≤0.15 after correction for multiple hypothesis. GSEA of the CREBBP core-targets in human and mouse DZ vs LZ GC B cells was performed using a list of 243 genes that were co-bound by CREBBP and H3K27Ac in human tonsils, conserved in the mouse and down-regulated in Crebbpfl/fl vs Crebbp+/+ Cγ1Cre cells. This list is reported in Supplementary Table S3, and the gene expression data of DZ and LZ GC B cells have been previously published (33). To interrogate the enrichment of LZ-upregulated genes in the rank of genes differentially expressed between Crebbpfl/fl and Crebbp+/+ Cγ1Cre GC B cell, we used the list of genes differentially expressed in DZ vs LZ subpopulations, identified as described in (33), while enrichment in FOXO1 bound genes was performed using separately the list of the top 300 genes bound by FOXO1 in human GC B cells as well as the top 300 promoter-bound genes and the integrated list of genes bound by FOXO1 and down-regulated in FOXO1-deficient GC B cells (46).
DAVID and Computation of Overlaps
To determine whether genes bound by CREBBP at active or poised promoters/enhancers and down-regulated in Crebbp-deficient GC B cells were enriched in annotated functional categories, we overlapped the list of down-regulated genes obtained by supervised analysis of mouse GC B cells (p<0.05 and fold change≥1.2) with genes identified by the integrated CREBBP/H3K27Ac ChIP-Seq analysis of human GC B cells. This unique list of overlapping genes was then analyzed in the GSEA software (49) and the DAVID software (50). Genes were interrogated for enrichment in all Canonical Pathways and Gene Ontology annotation for biological processes, and clusters with a significant p-value (<0.05 after Benjamini-Hochberg correction) were retained. Pathway analysis using the DAVID software was also performed with the list of genes co-bound by CREBBP and BCL6.
Protein extraction
Whole cell extracts were obtained from purified mouse B cells using NP-40 lysis buffer according to a previously described protocol (13). For the analysis of histone modifications, the chromatin pellet was subsequently resuspended in RIPA-coIP buffer (Tris-HCl pH 8.0 50mM, NaCl 200mM, Glycerol 5%, MgCl2 2.5mM, sodium deoxycholate 0.05%, SDS 0.05%) and mobilized by sonication in a Bioruptor Standard sonication device (Diagenode). For the analysis of histone modifications from mouse frozen tumor biopsies, the chromatin pellet was resuspended in 0.2 N HCl and incubated overnight at 4°C. Extracts were cleared by centrifugation at 12,000 r.p.m. for 10 min.
Immunoblot analysis
Protein extracts were resolved on NuPAGE Tris-acetate 3–8% gels (for CREBBP and EP300) or Tris-glycine 4–20% gels (for histone H3) (Life Technologies) and transferred to nitrocellulose membranes (GE Healthcare) according to the manufacturer’s instructions. Antibodies used were: rabbit polyclonal anti-CREBBP (A22, Santa Cruz Biotechnology), rabbit polyclonal anti-EP300 (N15, Santa Cruz Biotechnology), rabbit polyclonal anti-BCL6 (Cell Signaling, #4242), mouse monoclonal anti-β-Actin (A5441, Sigma-Aldrich mouse monoclonal anti-α-tubulin (clone DM1A, Sigma-Aldrich), rabbit polyclonal anti-H3K18Ac (Abcam, cat#ab1191), anti-H3K27Ac (Abcam, cat#ab4729), and rabbit monoclonal anti-Histone H3 (clone D1H2, Cell Signaling Technology). Loading of chromatin extracts (typically 0.5–3 µg) was adjusted to ensure comparable amounts of total histone H3. Quantitation of signal intensity was obtained in the ImageJ software by subtracting the background signal measured above each band from the signal measured in each band; areas of the same size (set on the image of the wild-type protein) were used for all measurements. Values are expressed as fold differences relative to the wild-type protein sample, set at 1, after normalization for the loading control.
Histological and immunohistochemical analysis of mouse tissues
Histological analysis of mouse organs was performed on 4µm-thick FFPE tissue sections, stained with Hematoxylin & Eosin (Thermo Scientific) following standard procedures. The following primary antibodies were used for immunohistochemical analysis: anti-Bcl6 (1:300) (N3, rabbit polyclonal, Santa Cruz Biotechnology), anti-PAX5 (1:400) (rabbit polyclonal, Neomarker), anti-CD3 (1:800) (rabbit monoclonal, clone SP7, Neomarker), anti-B220 (1:400) (rat monoclonal, clone RA3-6B2, BD Biosciences), anti-IRF4 (1:200) (M-17, goat polyclonal, Santa Cruz Biotechnology), anti-BCL2 human (1:100) (rabbit polyclonal, Santa Cruz Biotechnology), and anti-CD138 (1:200) (rat monoclonal, clone 281-2, BD Biosciences).
Immunofluorescence analysis
Double-immunofluorescence analysis of CREBBP and PNA was performed on formalin-fixed paraffin-embedded (FFPE) material from mouse spleens, using a combination of two anti-CREBBP antibody (each at 1:400 dilution)(A22 and C20, Santa Cruz Biotechnologies) and biotinylated PNA (1:300 dilution)(Vector Laboratories, cat#B-1075). A serial section was stained with anti-EP300 (1:200 dilution)(N15, Santa Cruz Biotechnologies). Detection of CREBBP and EP300 was obtained using the EnVision System–HRP-Rabbit antibody (Dako) followed by Tyramide Signal Amplification system (PerkinElmer); NeutrAvidin®, FITC conjugate (cat#A2662, Invitrogen) was used at 1:300 dilution to detect PNA.
Diagnosis of murine lymphoid tumors
Lymphoproliferative diseases developing in the VavP-Bcl2/Crebbp/Cγ1Cre cohort were diagnosed based on morphology and phenotype according to the following criteria, in analogy to the classification of human lymphoma: i) early stages of FL (FL in situ), defined by the presence of oversized, often coalescent follicles with partial or absent mantle zone and loss of confinement in the context of a yet preserved tissue architecture; ii) overt FL of various grades, characterized by the effacement of the nodal and/or splenic architecture by a proliferation of follicle center B cells with a follicular growth pattern occupying the medullary and/or paracortical areas; iii) DLBCL, defined by the effacement of the lymphoid organ architecture due to the expansion of large cells, with occasional infiltration beyond the capsule into surrounding soft tissues. Lymphoid tumors with plasmacytoid features were also occasionally observed in the VavP-Bcl2 mice; these lymphomas did not express B220 and PAX5 as well as BCL6, but stained positive for IRF4 and showed clonally rearranged Ig genes, indicating their B lymphoid origin. The genotype of the animal was not disclosed to the pathologist (S.H.). Immunohistochemical analysis of B220/CD3, BCL6, PAX5, IRF4, BCL2, CD138 and Ki67 was performed as described in the previous section.
Southern blot analysis of mouse tumor samples
To assess the presence of clonal immunoglobulin gene rearrangements in lymphoid tissues presenting morphologic and histological evidence of lymphoma, high molecular weight genomic DNA was extracted from frozen tissues by phenol-chloroform extraction and digested with EcoRI overnight at 37°C (4µg). The digestion reaction was resolved on 0.8% agarose gel followed by transfer to nitrocellulose membrane according to standard procedures. Hybridization was performed at 37°C overnight using a JH4 specific probe, as reported (40).
Mutation analysis of rearranged immunoglobulin genes
The rearranged immunoglobulin VH sequences were amplified from mouse tumor DNAs by PCR as described (40). The Sµ region, a target of AID-mediated activity in cells undergoing class switch recombination, was also analyzed by PCR amplification and direct sequencing, using primers SµA (ATTCCACACAAAGACTCTGGACC) and SµD (CAGTCCAGTGTAGGCAGTAGA), and the following PCR conditions: 94°C for 30 sec, 58°C for 30 sec and 72°C for 1min, for 30 cycles.
RNA extraction, cDNA synthesis and quantitative real-time PCR
Total RNA was extracted from primary mouse lymphocytes by TRIzol (Life Technologies) and treated with DNAse prior to cDNA synthesis, which was performed using the SuperScript® First-Strand Synthesis System (Life Technologies), according to the manufacturer’s instructions. The ABsolute QPCR SYBR green mix (Thermo Scientific) was then utilized to amplify specific cDNA fragments in the 7300 Real Time PCR system (Applied Biosystems), with the following oligonucleotides: mGapdh_qPCR_F1 (5’-CATGGCCTTCCGTGTTCCTA-3’); mGapdh_qPCR_R1 (5’-GCGGCACGTCAGATCCA-3’); mPrdm1_qPCR_F2 (5’-GGCATTCTTGGGAACTGTGT-3’); mPrdm1_qPCR_R2 (5’-ACCAAGGAACCTGCTTTTCA-3’). Data were analyzed by the change-in-threshold (2−ΔΔCT) method, using Gapdh as “housekeeping” reference gene. Results are represented as fold changes relative to the expression levels of the wild type animals, arbitrarily set as 1.
CREBBP genetic data
The CREBBP mutation pattern in DLBCL, tFL and FL was assessed by integrating data generated in our previous studies (6–8) with published information from exome sequencing and/or transcriptome sequencing studies (3–5,11), resulting in a total of 243 DLBCL primary cases, 31 DLBCL cell lines, and 71 FL. Co-occurrence of CREBBP mutations/deletions and BCL2 translocations was assessed using only samples for which both data were available. Gene expression profile data were available for 104 DLBCL primary cases and were used for the GSEA analysis shown in Fig. 6D.
Statistical analyses
P values were calculated with the Student’s t-test or the one-way ANOVA in the Graphpad Prism software, unless described otherwise. The significance of the overlap between genes down-regulated in Crebbp-deficient GC B cells (n=681) and genes bound by CREBBP, decorated by H3K27Ac and represented in the mouse MG430.2 expression array (n=243) was calculated by the hypergeometric distribution considering that the total number of unique genes represented in the MG430_2 array and mapped to human genes is 16,567.
Data Deposition
The gene expression profile data of mouse GC B cells and the ChIP-seq data of human GC B cells have been deposited in the GEO database under accession no. GSE67388, GSE67494, GSE88799 and GSE89688.
Supplementary Material
Statement of Significance.
Loss-of-function mutations of CREBBP are common and early lesions in FL and DLBCL, suggesting a prominent role in lymphoma initiation. Our studies identify the cellular program by which reduced CREBBP dosage facilitates malignant transformation, and have direct implications for targeted lymphoma therapy based on available drugs affecting CREBBP-mediated chromatin acetylation.
Acknowledgments
We would like to thank U. Klein for discussions and critical reading of the manuscript, C. Scuoppo and N. De Silva for suggestions, W. Zheng for help with the initial analysis of CD19-Cre mice, H. Tang for assistance with the mouse analysis; the Flow Cytometry Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University for assistance with cell sorting procedures; and Roxanne Ko at the JP Sulzberger Genome Center of Columbia University for help with high throughput sequencing. We also thank S. Cory (Walter and Eliza Hall Institute of Medical Research, Parkville, Australia) for the VavP-BCL2 mice, and K. Rajewsky (Max Delbruck Center for Molecular Medicine, Berlin, Germany) for the CD19-Cre and Cγ1-Cre mice.
Financial support: LP is supported by National Institutes of Health (NIH) Grants RO1-CA172492 and R21-CA192854, and a Columbia University Interprogrammatic Pilot Project award. RDF is supported by NIH grants R01-CA164152, R35-CA210105, and P50-CA192937. JZ was a Fellow of the Lymphoma Research Foundation.
Abbreviations list
- DLBCL
Diffuse large B-cell lymphoma
- FL
follicular lymphoma
- GC
germinal center
- BCR
B cell receptor
- SRBC
sheep red blood cells
- DZ
dark zone
- LZ
light zone
Footnotes
No potential conflicts of interests were disclosed by the authors
References
- 1.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nature reviews Cancer. 2010;10(7):457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan MA, Shilatifard A. Chromatin signatures of cancer. Genes Dev. 2015;29(3):238–249. doi: 10.1101/gad.255182.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46(2):176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nature genetics. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6(1):130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roelfsema JH, Peters DJ. Rubinstein-Taybi syndrome: clinical and molecular overview. Expert Rev Mol Med. 2007;9(23):1–16. doi: 10.1017/S1462399407000415. [DOI] [PubMed] [Google Scholar]
- 11.Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A. 2015;112(10):E1116–E1125. doi: 10.1073/pnas.1501199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5(1):9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32(4):606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 14.Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89(7):1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 15.Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387(6635):819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 16.Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387(6635):823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 17.Kalkhoven E. CBP and p300: HATs for different occasions. Biochemical pharmacology. 2004;68(6):1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 18.Kasper LH, Boussouar F, Ney PA, Jackson CW, Rehg J, van Deursen JM, et al. A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature. 2002;419(6908):738–743. doi: 10.1038/nature01062. [DOI] [PubMed] [Google Scholar]
- 19.Fukuyama T, Kasper LH, Boussouar F, Jeevan T, van Deursen J, Brindle PK. Histone acetyltransferase CBP is vital to demarcate conventional and innate CD8+ T-cell development. Mol Cell Biol. 2009;29(14):3894–3904. doi: 10.1128/MCB.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kung AL, Rebel VI, Bronson RT, Ch’ng LE, Sieff CA, Livingston DM, et al. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14(3):272–277. [PMC free article] [PubMed] [Google Scholar]
- 21.Xu W, Fukuyama T, Ney PA, Wang D, Rehg J, Boyd K, et al. Global transcriptional coactivators CREB-binding protein and p300 are highly essential collectively but not individually in peripheral B cells. Blood. 2006;107(11):4407–4416. doi: 10.1182/blood-2005-08-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmqvist PH, Mannervik M. Genomic occupancy of the transcriptional co-activators p300 and CBP. Transcription. 2013;4(1):18–23. doi: 10.4161/trns.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basso K, Dalla-Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- 27.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nature reviews Immunology. 2015;15(3):137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang-Decker N, Tong C, Boussouar F, Baker DJ, Xu W, Leontovich AA, et al. Loss of CBP causes T cell lymphomagenesis in synergy with p27Kip1 insufficiency. Cancer Cell. 2004;5(2):177–189. doi: 10.1016/s1535-6108(04)00022-4. [DOI] [PubMed] [Google Scholar]
- 29.Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Hao Z, et al. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci U S A. 2006;103(19):7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaffer AL, Wright G, Yang L, Powell J, Ngo V, Lamy L, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 31.http://software.broadinstitute.org/gsea/msigdb
- 32.Victora GD, Nussenzweig MC. Germinal centers. Annual review of immunology. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 33.Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, Nussenzweig MC. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012;120(11):2240–2248. doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nature reviews Genetics. 2010;11(4):285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25(6):1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasbold J, Corcoran LM, Tarlinton DM, Tangye SG, Hodgkin PD. Evidence from the generation of immunoglobulin G-secreting cells that stochastic mechanisms regulate lymphocyte differentiation. Nat Immunol. 2004;5(1):55–63. doi: 10.1038/ni1016. [DOI] [PubMed] [Google Scholar]
- 37.Egle A, Harris AW, Bath ML, O’Reilly L, Cory S. VavP-Bcl2 transgenic mice develop follicular lymphoma preceded by germinal center hyperplasia. Blood. 2004;103(6):2276–2283. doi: 10.1182/blood-2003-07-2469. [DOI] [PubMed] [Google Scholar]
- 38.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nature reviews Molecular cell biology. 2015;16(3):144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21(10):1190–1198. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Z, Chen WY, Shimada M, Nguyen UT, Kim J, Sun XJ, et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013;154(2):297–310. doi: 10.1016/j.cell.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, et al. Transcriptional analysis of the B cell germinal center reaction. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(5):2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC Bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dominguez-Sola D, Kung J, Holmes AB, Wells VA, Mo T, Basso K, et al. The FOXO1 Transcription Factor Instructs the Germinal Center Dark Zone Program. Immunity. 2015;43(6):1064–1074. doi: 10.1016/j.immuni.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 47.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7(7):773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 48.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 49.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23(23):3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 50.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.