Abstract
To identify known and novel CYP4V2 mutations in patients with Bietti crystalline cornea (BCD), expand the spectrum of CYP4V2 mutations, and characterize the population history of the c.802-8_810del17insGC mutation common in Asian populations, genomic DNA was isolated from peripheral blood samples from 58 unrelated patients with clinical diagnoses of BCD. Exons and flanking intronic regions of the CYP4V2 gene were dideoxy DNA sequenced. Nonpathogenic polymorphisms were excluded and known mutations were identified by sequencing 192 unaffected individuals from similar ethnic backgrounds and examination of online databases. The age of the c.802-8_810del17insGC mutation was estimated using three independent approaches. A total of 28 CYP4V2 mutations, 9 of which were novel, were detected in the 58 patients with BCD. These included 19 missense, 4 nonsense, 2 deletion, 2 splice site, and 1 insertion-deletion mutations. Two missense variants of uncertain significance were also detected. The age of the c.802-8_810del17insGC mutation was estimated to be 1040–8200 generations in the Chinese and 300–1100 generations in the Japanese populations. These results expand the mutation spectrum of CYP4V2, and provide insight into the origin of the c.802-8_810del17insGC mutation in the Chinese population and its transmission to the Japanese population.
Introduction
Bietti crystalline cornea-retinal dystrophy (BCD, MIM210370) is a progressive autosomal recessive retinal dystrophy first reported by Bietti in 1937. It is characterized by numerous small glittering yellow-white crystals at the posterior pole of the retina associated with progressive atrophy of the retinal pigment epithelium (RPE), pigment clumps, and choroidal sclerosis. Some patients have similar crystal deposits at the corneoscleral limbus and in circulating lymphocytes and skin fibroblasts.1 Most patients begin to show night blindness, decreased visual acuity, and paracentral scotomas between the second and fourth decades progressing to peripheral visual field loss and marked visual impairment.1 BCD is more common in East Asia, especially in China and Japan.2 Patients show abnormally high triglycerides and cholesterol storage in cultured cells from BCD patients, lack two fatty acid-binding protein activities,3 and have decreased metabolism of labeled fatty acid precursors into n-3 polyunsaturated fatty acids (n-3PUFA).4
Sequence variants that affect the function of CYP4V2 (MIM 608614), a member of the heme thiolate cytochrome P450 subfamily 4 (CYP4), predominantly active in fatty acid metabolism, cause BCD.5 The CYP4V2 gene spans 21 kb, comprising 11 exons encoding a 525 amino acid protein (Figure 1, top). Although CYP4V2 is expressed in almost all tissues, it is expressed at high levels in the retina and RPE, and at somewhat lower levels in the cornea tissues, which show the major clinical findings of BCD.5 CYP4V2 is a PUFA hydroxylase highly expressed P450 in the transformed human RPE cell line ARPE-19.6 Consistent with this, abnormalities in ω3-PUFAs and their metabolism have been demonstrated in patients with BCD.4, 7, 8 The retinal findings and systemic lipid abnormalities have been recapitulated in a knockout mouse model.9 Before this study, 73 mutations in CYP4V2 have been described in the literature and clinical databases. To expand the spectrum of these CYP4V2 mutations in patients with BCD, the CYP4V2 gene was sequenced in 58 patients diagnosed with BCD and their sequences were compared with those of 192 unrelated healthy controls as well as known mutations and sequence changes in online sequence variation databases, identifying 28 different mutations (9 novel) and 2 sequence variants of unknown significance, and delineating the origin of the common c.802-8_810del17insGC indel mutation.
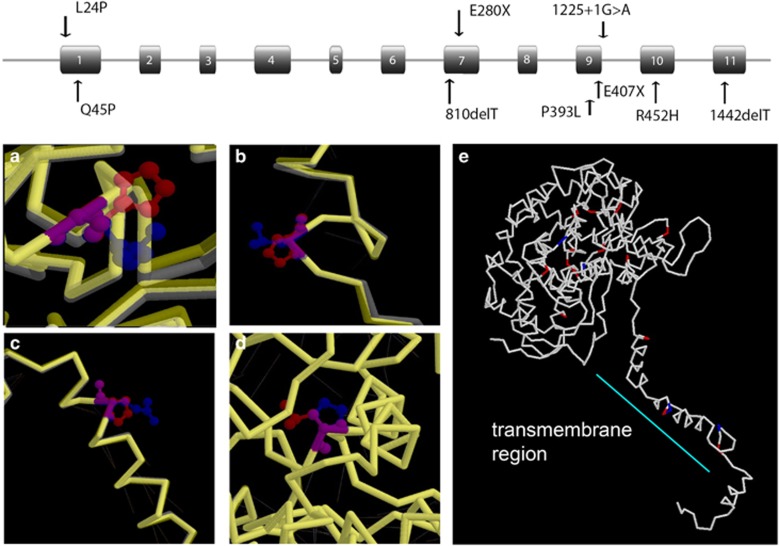
Figure 1.
Gene and mutation structural predictions of the effects of the four novel missense mutations. Top: gene structure and location of the 9 novel mutations in CYP4V2. Bottom: location of the four missense mutations shown in red: R452H, L24P, Q45P, and P393L. (a) Predicted effect of the R452H mutation. The wild-type R452 is shown in blue with the backbone shown in white, whereas the mutant H452 is shown in red with the backbone shown in yellow. There is both a local effect from the residue change and significant deformation of the protein backbone, which is near the active site. (b) Predicted effect of the L24P mutation. The wild-type L24 amino acid is shown in blue with the backbone shown in white, whereas the mutant P24 residue is shown in red with the backbone shown in yellow. There is minimal change in the backbone, and no hydrogen bonds are disrupted. (c) Predicted effect of the Q45P mutation. The wild-type Q45 is shown in blue with the backbone shown in white, and the mutant P45 is shown in red, with the backbone shown in yellow. Although the α-helix near the end of which the Q45 residue resides is only slightly distorted, two hydrogen bonds are disrupted and the helix is splayed slightly at the mutant residue. (d) Predicted effect of the P393L mutation. There is almost no distortion of the protein backbone in this region, and no hydrogen bonds are disrupted. (e) Summary of location of BCD missense mutations identified in CYP4V2. Novel mutations are indicated in blue, whereas previously identified mutations are shown in red. The mutations cluster in the transmembrane area and around the active site and its supporting α-helices. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Materials and methods
Patients
This study was approved by the CNS Institutional Review Board of the National Institutes of Health and consent obtained in accordance with the Declaration of Helsinki. Patients were diagnosed with BCD on the basis of clinical features,1 and were of European, Chinese, Palestinian Arabic, and Korean ancestry (Table 1). Control individuals were of matched ethnic backgrounds.
Table 1. Mutation data for BCD patients.
| Ptnt | Allele 1 | Allele 2 | Refs | Pop | PP2 | P | Sift | Condel | |
|---|---|---|---|---|---|---|---|---|---|
| A. Patients with novel mutations in CYP4V2 | |||||||||
| 1 | DDL934 | c.71T>C, p.(Leu24Pro)a | Homozygous | U | B | Del | Dam | N | |
| 2 | 6295 | c.134A>C, p.(Gln45Pro)a | Homozygous | E | B | N | Dam | N | |
| 3 | 6296 | c.134A>C, p.(Gln45Pro)a | Homozygous | E | B | N | Dam | N | |
| 4 | W155 | c.810delT, p.(Glu271Asnfs*6)a | c.1091-2A>G, p.(Gly364_Val408del)b,c,d | 5 | C | ||||
| 5 | W172 | c.810delT, p.(Glu271Asnfs*6)a | c.1091-2A>G, p.(Gly364_Val408del)b,c,d | 5 | C | ||||
| 6 | W76 | c.810delT, p.(Glu271Asnfs*6)a | c.694C>T, p.(Arg232Ter)c,d | 37 | C | ||||
| 7 | W69 | c.810delT, p.(Glu271Asnfs*6)a | c.958C>T, p.(Arg320Ter)c,d | 30 | C | ||||
| 8 | 13805 | c.928G>T, p.(Glu310Ter)a,c | c.130T>A, p.(Trp44Arg)d | 5 | E | ||||
| 9 | 10938 | c.1178C>T, p.(Pro393Leu)a | Homozygous | U | PD | Del | Dam | Del | |
| 10 | DDL686 | c.1219G>T, p.(Glu407Ter)a,c | Homozygous | U | |||||
| 11 | 11240 | c.1355G>A, p.(Arg452His)a | c.802-8_810del17insGC, p.(Val268_Glu329del)d | 5 | K | Dam | Del | Dam | Del |
| 12 | 6298 | c. 1225+1G>A, p.(Gly364_V408del)a,b,c | Homozygous | A | |||||
| 13 | W180 | c.1442delT, p.(Ser482Argfs*4)a | c.802-8_810del17insGC, p.(Val268_Glu329del)d | 5 | C | ||||
| B. Patients with previously identified mutations in CYP4V2 | |||||||||
| 14 | 14335 | c.64C>G, p.(Leu22Val)e | Homozygous | 24 | E | ||||
| 15 | 13804 | c.64C>G, p.(Leu22Val)e | Homozygous | 19 | E | ||||
| 16 | 14331 | c.64C>G, p.(Leu22Val)e | Homozygous | 19 | E | ||||
| c.775A>C,p(Lys259Gln)e | Homozygous | 25 | |||||||
| 17 | DDL1056 | c.197T>G, p.(Met66Arg) | Homozygous | 38 | U | ||||
| 18 | 10939 | c.283G>A, p.(Gly95Arg) | c.1393A>G, p.(Arg465Gly) | 24, 37 | U | ||||
| 19 | 8229 | c.332T>C, p.(Ile111Thr) | Homozygous | 5 | E | ||||
| 20 | 8230 | c.332T>C, p.(Ile111Thr) | Homozygous | 5 | E | ||||
| 21 | 8231 | c.332T>C, p.(Ile111Thr) | Homozygous | 5 | E | ||||
| 22 | 11208 | c.332T>C, p.(Ile111Thr) | Homozygous | 5 | E | ||||
| 23 | 11209 | c.332T>C, p.(Ile111Thr) | Homozygous | 5 | E | ||||
| 24 | 11210 | c.332T>C, p.(Ile111Thr) | Homozygous | 5 | E | ||||
| 25 | 13806 | c.332T>C, p.(Ile111Thr) | Homozygous | 5 | E | ||||
| 26 | DDL323 | c.677T>A, p.(Met226Lys) | Homozygous | 38 | E | ||||
| 27 | W50-1 | c.694C>T, p.(Arg232Ter)c | c.1027T>G, p.(Tyr343Asp) | 37, 39 | C | ||||
| 28 | 6139 | c.694C>T, p.(Arg232Ter)c | c.1199G>A, p.(Arg400His) | 24, 37 | E | ||||
| 29 | 14332 | c.775A>C,p(Lys259Gln)e | Heterozygote | 25 | E | ||||
| 30 | 14333 | c.775A>C,p(Lys259Gln)e | Heterozygote | 25 | E | ||||
| 31 | 14334 | c.775A>C,p(Lys259Gln)e | Heterozygote | 25 | E | ||||
| 32 | 6090 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | Homozygous | 5 | K | ||||
| 33 | 13151 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | Homozygous | 5 | C | ||||
| 34 | DDL708 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | Homozygous | 5 | C | ||||
| 35 | DDL1080 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | Homozygous | 5 | C | ||||
| 36 | W145-1 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | Homozygous | 5 | C | ||||
| 37 | 11196 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | c.992A>C, p.(His331Pro) | 5 | K | ||||
| 38 | 11242 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | c.992A>C, p.(His331Pro) | 5 | K | ||||
| 39 | DDL591 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | c.1091-2A>G, p.(Gly364_Val408del)b,c | 5 | U | ||||
| 40 | DDL593 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | c.992A>C, p.(His331Pro) | 5 | C | ||||
| 41 | DDL933 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | c.992A>C, p.(His331Pro) | 5 | C | ||||
| 42 | F49-1 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | c.1091-2A>G, p.(Gly364_Val408del)b c | 5 | C | ||||
| 43 | W150-1 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | c.992A>C, p.(His331Pro) | 5 | C | ||||
| 44 | W46-1 | c.802-8_810del17insGC, p.(Val268_Glu329del)f | c.1199G>A, p.(Arg400His) | 5, 24 | C | ||||
| 45 | W20-1 | c.992A>C, p.(His331Pro) | Homozygous | 5 | C | ||||
| 46 | DDL495 | c.992A>C, p.(His331Pro) | Homozygous | 5 | U | ||||
| 47 | DDL940 | c.992A>C, p.(His331Pro) | c.1187C>T, p.(Pro396Leu) | 5, 28 | U | ||||
| 48 | W27-1 | c.992A>C, p.(His331Pro) | c.1091-2A>G, p.(Gly364_Val408del)b,c | 5 | C | ||||
| 49 | W9-1 | c.992A>C, p.(His331Pro) | c.1091-2A>G, p.(Gly364_Val408del)b,c | 5 | C | ||||
| 50 | W15-1 | c.1091-2A>G, p.(Gly364_Val408del)b,c | Homozygous | 5 | C | ||||
| 51 | DDL1076 | c.1168C>T, p.(Arg390Cys) | Homozygous | 33 | U | ||||
| 52 | DDL942 | c.1169G>A, p.(Arg390His) | Homozygous | 30 | U | ||||
| 53 | 13973 | c.1198C>T, p.(Arg400Cys) | Homozygous | 28 | E | ||||
| 54 | 6272 | c.1328G>A, p.(Arg443Gln) | Homozygous | 37 | E | ||||
| 55 | 6271 | c.1328G>A, p.(Arg443Gln) | Homozygous | 37 | E | ||||
| 56 | 11203 | c.1328G>A, p.(Arg443Gln) | c.1372G>A, p.(Val458Met) | 37, 40 | E | ||||
| 57 | 11204 | c.1328G>A, p.(Arg443Gln) | c.1372G>A, p.(Val458Met) | 37, 40 | E | ||||
| 58 | 6829 | c.1393A>G, p.(Arg465Gly) | Homozygous | 37 | E | ||||
Abbreviations: A, Arabic; C, Chinese; E, European; K, Korean; P, Provean; PP2, PolyPhen-2; U, unknown.
Novel mutation.
Skips exon 9 (predicted).
Subject to nonsense-mediated decay.
Previously reported.
Probably not pathogenic.
Skips exon 7 (verified experimentally).
DNA amplification and mutation detection
Genomic DNA was extracted directly from blood or transformed lymphoblastoid cell lines by standard phenol–chloroform protocols.10 PCR amplification of CYP4V2 exons 1–11 including intron–exon boundaries and 50 bp of flanking sequence was performed as reported previously.5 PCR products were purified using Agencourt CleanSEQ (Beckman Coulter, Biomek NX, Brea, CA, USA). Sequencing was performed on an ABI PRISM 3130 Automated sequencer (Applied Biosystems, Foster City, CA, USA) and analyzed using Mutation Surveyor v3.30 (Soft Genetics, State College, PA, USA) or Lasergene 8.0, (DNASTAR, Madison, WI, USA). Control DNA samples from 192 unrelated individuals of Chinese, Japanese, or European ethnic origin were also analyzed and the 1000 Genomes Database (http://www.1000genomes.org/), NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/), NHLBI Exome Variant Server (http://evs.gs.washington.edu/EVS/), Biobase (https://portal.biobase-international.com) and ExAC Database (http://exac.broadinstitute.org/) were also searched for all novel sequence variations. Effects of potential splice mutations were modeled using online resources including neural networking through The Berkeley Drosophila Genome Project (http://www.fruitfly.org/seq_tools/splice.html) and the Center for Biological Sequence Analysis (http://www.cbs.dtu.dk/services/NetGene2/), consensus values at the Inserm Human Splicing Finder (http://umd.be/HSF/), the Alternative Splice Site Predictor (ASSP, http://wangcomputing.com/assp/index.html), and MaxEntScan (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html). Mutations are described with reference to the CYP4V2 mRNA sequence NM_207352.3 and gene sequence NG_007965.1. Chromosomal positions are based on hg19/GRCh37. Variants were submitted to the LOVD (http://databases.lovd.nl/shared/genes/CYP4V2) with screening (submission) numbers 0000081248- 0000081331.
Molecular modeling
Modeling of the novel CYP4V2 mutant proteins was performed by homology modeling based on predicted coordinates for the wild-type CYP4V2 protein5 using the SWISSMODEL Workspace web-based environment with default values,11, 12, 13 and visualized using RASMOL.14
Haplotype analysis and age estimation
Delineation of SNP haplotypes in individuals carrying the c.802-8_810del17insGC mutation was done by examination as most individuals were homozygous. The haplotype was extended until half of the individuals in the Chinese or Japanese populations diverged. Haplotypes were grouped by country and were sorted into those requiring one or two recombination events in their descent from the ancestral haplotype by examination to minimize the number of recombination events required.
The mutation age was estimated as described in Equation (1) of Risch et al,15, 16
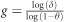 , where δ is the linkage disequilibrium constant, δ=(PD−PN)/(1−PN), with PD being the frequency of the allele on chromosomes of mutation carriers and PN being the frequency of the allele in the control population. Genotypes of unaffected individuals were taken from the 1000 Genomes database (103 Han Chinese in Beijing: CHB and 105 Southern Han Chinese: CHS for the Chinese population; 104 Japanese in Tokyo: JPT for the Japanese population). Haplotypes were estimated using the CHM method as implemented in the Golden Helix SVS (Golden Helix, Bozeman, MT, USA). The map distances were inferred on the basis of the physical distances as given in GRCh37/hg19 from the UCSC Genome Browser assuming 1 Mb=1 cM. Alleles at SNPs showing no recombination in cases also were collapsed into a single haplotype and analyzed as a single marker in the two marker approach described in Equation (2) of Risch et al. In addition, marker specific values of g were estimated using the method described in Equation 1 of Goldstein et al.17
K=cR+μM+(1-c-μ)I, where c and μ are the recombination and mutation rates, respectively, R is a 2 × 2 matrix with R11=R12=a, and R21=R22=1−a, where a is the frequency of the ancestral allele in the control population, M is a 2 × 2 matrix with M11=0, M12=1/3, M21=1, and M22=2/3, to account for the frequency with which a mutation might remove an ancestral allele (all cases) or move to an ancestral allele (1 of 3 possible bases), and I is the 2 × 2 identity matrix. The original frequency vector is (1, 0) as it occurs on the founder haplotype, and this association is reduced by multiplying by K at each generation (g) until the current frequency of the ancestral allele is reached. Finally, the marginal posterior probability distribution of the age18 of the c.802-8_810del17insGC was estimated using the DMLE+2.2 software developed by Reeve and Rannala.19 This program estimates the age in generations by comparing the observed haplotypes in chromosomes from affected and unaffected sample sets considering the map distances, the population growth rate (genr), and the proportion of the mutation bearing chromosomes sampled, but has a strong dependence on assumptions regarding the population history of China and Japan. Somewhat arbitrarily, population growth was modeled since 2300 BC, the earliest time at which reliable figures were available for both populations, although not as old as estimates of the mutation age, assuming 25 year generations.
, where δ is the linkage disequilibrium constant, δ=(PD−PN)/(1−PN), with PD being the frequency of the allele on chromosomes of mutation carriers and PN being the frequency of the allele in the control population. Genotypes of unaffected individuals were taken from the 1000 Genomes database (103 Han Chinese in Beijing: CHB and 105 Southern Han Chinese: CHS for the Chinese population; 104 Japanese in Tokyo: JPT for the Japanese population). Haplotypes were estimated using the CHM method as implemented in the Golden Helix SVS (Golden Helix, Bozeman, MT, USA). The map distances were inferred on the basis of the physical distances as given in GRCh37/hg19 from the UCSC Genome Browser assuming 1 Mb=1 cM. Alleles at SNPs showing no recombination in cases also were collapsed into a single haplotype and analyzed as a single marker in the two marker approach described in Equation (2) of Risch et al. In addition, marker specific values of g were estimated using the method described in Equation 1 of Goldstein et al.17
K=cR+μM+(1-c-μ)I, where c and μ are the recombination and mutation rates, respectively, R is a 2 × 2 matrix with R11=R12=a, and R21=R22=1−a, where a is the frequency of the ancestral allele in the control population, M is a 2 × 2 matrix with M11=0, M12=1/3, M21=1, and M22=2/3, to account for the frequency with which a mutation might remove an ancestral allele (all cases) or move to an ancestral allele (1 of 3 possible bases), and I is the 2 × 2 identity matrix. The original frequency vector is (1, 0) as it occurs on the founder haplotype, and this association is reduced by multiplying by K at each generation (g) until the current frequency of the ancestral allele is reached. Finally, the marginal posterior probability distribution of the age18 of the c.802-8_810del17insGC was estimated using the DMLE+2.2 software developed by Reeve and Rannala.19 This program estimates the age in generations by comparing the observed haplotypes in chromosomes from affected and unaffected sample sets considering the map distances, the population growth rate (genr), and the proportion of the mutation bearing chromosomes sampled, but has a strong dependence on assumptions regarding the population history of China and Japan. Somewhat arbitrarily, population growth was modeled since 2300 BC, the earliest time at which reliable figures were available for both populations, although not as old as estimates of the mutation age, assuming 25 year generations.
Results
Mutation screening of the CYP4V2 gene
Mutation screening of CYP4V2 was performed in 58 patients with BCD. A total of 28 mutations and 2 variants of uncertain significance were identified (Table 1 and Figure 1). Of the nine novel mutations, five were in homozygous and 4 in compound heterozygous cases (Table 1A). Patients 6295 and 6296 were twins; the other 10 cases were sporadic affected individuals. Clinical details of cases with novel mutations are given in Supplementary Table S1. Structural predictions of the effects of the four novel missense mutations were analyzed by homology based on coordinates for the wild-type CYP4V2 protein (Figure 1, bottom).
c.71T>C, p.(Leu24Pro) produces minimal change in the backbone of the protein, and no hydrogen bonds are disrupted. However, the potential of this variant to affect CYP4V2 function and its high conservation across species possibly relates to its position in the transmembrane region, which would be highly sensitive to amino acid changes, and especially a change to a proline, which would disrupt the α-helical structure of this region (Figures 1b and e). Leu24 is conserved among seven of the aligned species, although absent from the horse and zebrafish (Table 2), and p.(Leu24Pro) has blosum 62 and Grantham scores of −3 and 98, respectively. 20, 21
c.134A>C, p.(Gln45Pro) Gln45 is not completely conserved in all nine species (Table 2), although the substitutions are conservative in all species except the chicken. It has a blosum 62 and Grantham scores of −1 and 76, respectively. As with the c.71T>C, p.(L24P) mutation, the disagreement in the predictive programs and high conservation of this residue might relate to its position in the transmembrane segment, where it distorts the α-helix, slightly disrupting two hydrogen bonds (Figures 1c and h).
c.810delT, p.(Glu271Argfs*34) results in a premature stop in an internal exon, so that the mRNA is predicted to undergo nonsense-mediated decay (NMD).
c.838G>T (GAA>TAA), p.(Glu280*) also should result in NMD.
Although c.1178C>T, p.(Pro393Leu) produced little distortion of the protein backbone in this region and disrupted no hydrogen bonds (Figure 1d), it predicted to be damaging because of its position near the porphyrin ring and heme group of the active site of the protein. Pro393 is highly conserved and has blosum 62 and Grantham scores of −3 and 98, respectively.
c.1219G>T, p.(Glu407*) should result in NMD.
c.1225+1 G>A, p.(Gly364_V408del) is predicted to skip exon 9.
c.1355G>A, p.(Arg452His) is predicted to interfere with coordination of the heme ring and binding of the fatty acid substrate (Figures 1a and e). Arg452 is conserved in all species examined from human to zebrafish (Table 2).
c.1441delT, p.(Ser481Argfs*4) would not result in nonsense-mediated decay, but would substitute 4 random amino acids for the last 44, destabilizing the active site.
Table 2. Conservation across species of amino acids in missense mutations.
| Human | A38 | S39 | Y40 | A41 | R42 | K43 | W44 | Q45 | Q46 | M47 | R48 | P49 | I50 | P51 | T52 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP4V2 c.134A>C, p.Q45P | |||||||||||||||
| Chimp | A | T | Y | A | R | K | W | Q | Q | Q | R | P | I | P | T |
| Mouse | V | S | Y | A | R | K | W | Q | Q | Q | R | S | I | P | S |
| Rabbit | A | S | Y | A | R | K | W | Q | Q | Q | R | P | I | P | T |
| Cow | A | S | Y | A | R | K | W | R | Q | R | R | P | V | P | T |
| Horse | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Dog | A | S | Y | A | Q | K | C | H | Q | H | R | A | F | P | S |
| Chicken | L | N | Y | W | W | W | W | W | V | W | K | P | I | P | G |
| Zebrafish | L | S | Y | M | M | W | W | K | E | K | K | P | I | P | G |
| CYP4V2 c.1178C>T, p.P393L | |||||||||||||||
| Human | K386 | E387 | T388 | L389 | R390 | L391 | F392 | P393 | S394 | V395 | P396 | L397 | F398 | A399 | R400 |
| Chimp | K | E | T | L | R | L | F | P | S | V | P | L | F | A | R |
| Mouse | K | E | T | L | R | V | F | P | S | V | P | L | F | A | R |
| Rabbit | K | E | T | L | R | L | F | P | S | V | P | L | F | A | R |
| Cow | K | E | S | L | R | L | F | P | S | V | P | F | F | A | R |
| Horse | K | E | T | L | R | L | F | P | S | V | P | L | F | A | R |
| Dog | K | E | S | L | R | L | F | P | S | V | P | L | F | A | R |
| Chicken | K | E | A | L | R | L | F | P | S | V | P | M | F | A | R |
| Zebrafish | K | E | S | L | R | L | F | P | S | V | P | F | F | A | R |
| CYP4V2 c.1355G>A, p.R452H | |||||||||||||||
| Human | F445 | P446 | E447 | N448 | A449 | Q450 | G451 | R452 | H453 | P454 | Y455 | A456 | Y457 | V458 | P459 |
| Chimp | F | P | K | N | A | Q | G | R | H | P | Y | A | Y | V | P |
| Mouse | F | P | E | N | S | Q | G | R | H | P | Y | A | Y | V | P |
| Rabbit | F | P | E | N | A | Q | G | R | H | P | Y | A | Y | V | P |
| Cow | F | P | E | N | L | K | G | R | H | T | Y | A | Y | V | P |
| Horse | F | P | E | N | S | Q | G | R | H | P | Y | A | Y | V | P |
| Dog | F | P | E | N | L | Q | G | R | H | P | Y | A | Y | I | P |
| Chicken | F | P | E | N | S | K | G | R | H | P | Y | A | Y | V | P |
| Zebrafish | L | P | E | N | C | V | G | R | H | P | Y | A | Y | I | P |
| CYP4V2 c.71T>C, p.L24P | |||||||||||||||
| Human | G17 | A18 | A19 | S20 | A21 | L22 | S23 | L24 | A25 | G26 | A27 | S28 | L29 | V30 | L31 |
| Chimp | G | A | A | S | A | V | S | L | A | G | A | S | L | V | L |
| Mouse | G | A | A | S | A | V | S | L | A | G | A | T | I | L | I |
| Rabbit | G | A | A | S | A | V | S | L | A | G | A | S | L | F | L |
| Cow | S | G | L | C | A | V | S | L | A | G | A | T | L | T | L |
| Horse | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Dog | G | A | L | G | F | P | S | L | G | G | P | P | L | V | V |
| Chicken | V | A | G | A | I | T | S | L | A | A | D | G | V | T | V |
| Zebrafish | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
Alignment of novel mutations (bold) identified in the CYP4V2 gene in nine species ranging from human to zebrafish.
The remaining 28 mutations, including 19 (68%) missense, 4 (14%) nonsense, 2 (7%) splice, 2 (7%) deletions, and a single indel (4%), have been reported previously (Table 1B). Although the nonsense, splice, deletion, and indel mutations would be predicted to have a severe effect wherever they occurred, the missense mutations might be expected to identify parts of the protein structure particularly susceptible to perturbation. As can be seen in Figure 1e, the missense mutations taken as a whole cluster in two regions: the transmembrane region, without which the CYP4V2 protein would not be expected to insert into the membrane, and the area around the porphyrin ring, including residues that might influence coordination of the heme group and those in the α-helices that support and position them.
Ethnic origins of CYP4V2 mutations in BCD patients
Although the number of mutations identified in this study and Li et al,5 40, is relatively small, some patterns do emerge when the ethnicity of the patients displaying those mutations is examined, especially the more common mutations. Of the 99 combined unrelated patients and families examined, the ethnicity of 13 of them was unknown, leaving 86: 31 of European origin, 38 of Chinese origin, 1 of Arabic origin, 8 of Korean origin, and 8 of Japanese origin (Table 3). Although there is some overlap, specific mutations are generally restricted to Asian (Chinese, Korean, and Japanese) or European populations.
Table 3. Summary of mutations and their ethnicity.
| Mutation | AA change | European | Chinese | Arabic | Korean | Japanese | Unknown |
|---|---|---|---|---|---|---|---|
| c.64C>G | p.(Leu22Val) | 3 | 0 | 0 | 0 | 0 | 0 |
| c.71T>C | p.(Leu24Pro) | 0 | 0 | 0 | 0 | 0 | 1 |
| c.130T>A* | p.(Trp44Arg) | 1 | 0 | 0 | 0 | 0 | 0 |
| c.134A>C | p.(Gln45Pro) | 1 | 0 | 0 | 0 | 0 | 0 |
| c.181G>A* | p.(Gly61Ser) | 0 | 1 | 0 | 0 | 0 | 0 |
| c.197T>G | p.(Met66Arg) | 0 | 0 | 0 | 0 | 0 | 1 |
| c.237G>A* | p.(Glu79Asp) | 0 | 1 | 0 | 0 | 0 | 0 |
| c.283G>A | p.(Gly95Arg) | 1 | 0 | 0 | 0 | 0 | 0 |
| c.327+1G>A* | p.(Glu72Glyfs*5) | 0 | 0 | 0 | 0 | 1 | 0 |
| c.328-1G>A* | p.G(lu109Tyrfs*30) | 1 | 0 | 0 | 0 | 0 | 0 |
| c.332T>C | p.(Ile111Thr) | 8 | 0 | 0 | 0 | 0 | 0 |
| c.367A>G* | p.(Met123Val) | 3 | 1 | 0 | 0 | 0 | 0 |
| c.400G>T* | p.(Gly134*) | 1 | 0 | 0 | 0 | 0 | 0 |
| c.677T>A | p.(Met226Lys) | 0 | 0 | 0 | 0 | 0 | 1 |
| c.694C>T | p.(Arg232*) | 1 | 2 | 0 | 0 | 0 | 0 |
| c.802-8_810del17insGC | p.(Val268_Glu329del) | 0 | 10 | 0 | 4 | 7 | 1 |
| c.810delT | p.(Glu271Asnfs*6) | 0 | 4 | 0 | 0 | 0 | 0 |
| c.838G>T | p.(Glu280*) | 1 | 0 | 0 | 0 | 0 | 0 |
| c.958C>T | p.(Arg320*) | 0 | 1 | 0 | 0 | 0 | 0 |
| c.992A>C* | p.(His331Pro) | 0 | 6 | 0 | 2 | 0 | 2 |
| c.1021T>C* | p.(Ser341Pro) | 1 | 0 | 0 | 0 | 0 | 0 |
| c.1027T>G | p.(Tyr343Asp) | 0 | 1 | 0 | 0 | 0 | 0 |
| c.1091-2A>G* | p.(Gly364_Val408del) | 0 | 9 | 0 | 0 | 0 | 1 |
| c.1168C>T | p.(Arg390Cys) | 0 | 0 | 0 | 0 | 0 | 1 |
| c.1169G>A | p.(Arg390His) | 0 | 0 | 0 | 0 | 0 | 1 |
| c.1178C>T | p.(Pro393Leu) | 0 | 0 | 0 | 0 | 0 | 1 |
| c.1187C>T | p.(Pro396Leu) | 0 | 0 | 0 | 0 | 0 | 1 |
| c.1198C>T | p.(Arg400Cys) | 1 | 0 | 0 | 0 | 0 | 0 |
| c.1199G>A | p.(Arg400His) | 1 | 1 | 0 | 0 | 0 | 0 |
| c.1219G>T | p.(Glu407*) | 0 | 0 | 0 | 0 | 0 | 1 |
| c.1225+1G>A | p.(Gly364_Val408del) | 0 | 0 | 1 | 0 | 0 | 0 |
| c.1328G>A | p.(Arg443Gln) | 4 | 0 | 0 | 1 | 0 | 0 |
| c.1355G>A | p.(Arg452His) | 0 | 0 | 0 | 1 | 0 | 0 |
| c.1372G>A | p.(Val458Met) | 2 | 0 | 0 | 0 | 0 | 0 |
| c.1393A>G | p.(Arg465Gly) | 0 | 0 | 0 | 0 | 0 | 1 |
| c.1442delT | p.(Ser482Argfs*4) | 0 | 1 | 0 | 0 | 0 | 0 |
| c.1573G>A* | p.(Arg508His) | 1 | 0 | 0 | 0 | 0 | 0 |
Of these, 22 patients showed the c.802-8_810del17insGC indel mutation distributed among the Chinese, Korean, and Japanese patients. This mutation was not seen in European or Arabic populations. The next two most common mutations (seen in 10 patients each) were the c.992A>C mutation, seen in Chinese and Korean populations, and the c.1091-2A>G mutation, restricted to Chinese patients. In contrast, the c.332T>C mutation, seen in 8 patients and the c.64C>G mutation were restricted to 3 Europeans. The c.367A>G, c.694C>T, c.1199G>A, and c.1328G>A mutations were seen in both European and Chinese populations, whereas the remaining mutations, although reported previously, occurred only a single time in this study.
Haplotype analysis and history of the c.802-8_810del17insGC indel mutation
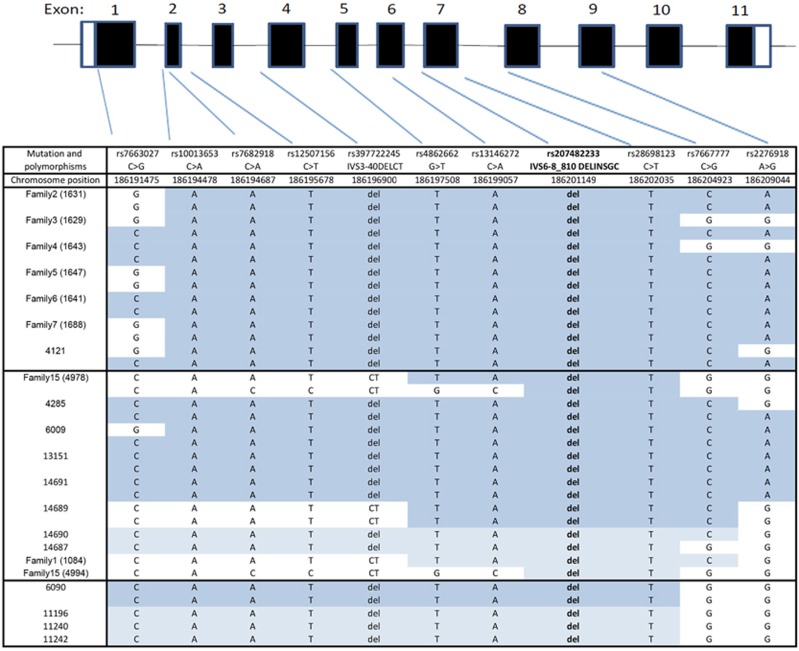
Intragenic SNP haplotypes from the Chinese and Japanese BCD patients harboring the c.802-8_810del17insGC indel mutation included in this study were aligned along with those from Li et al,5 Lin et al,22 and Lee et al23 as shown in Figure 2. The CAAT(delCT)TA(indel)TCA haplotype shaded in dark blue in homozygotes was the most common in Chinese, Japanese, and Koreans, and was assumed to be the ancestral haplotype for all three population groups.22 Individual alleles of the SNPs composing the founder haplotype were highly conserved among patients: rs7663027 (74% C), rs10013653 (100% A), rs7682918 (94% A), rs12507156 (94%T), rs397722245 (83% deletion), rs4862662 (94% T), rs13146272 (94% A), rs207482233 (100% del—the c.802-8_810del17insGC indel), rs28698123 (100%T), rs7667777 (68% C), and rs2276918 (51% A), although some lie beyond divergent alleles closer to the mutation and thus are identical by state rather than descent. The entire founder haplotype was found in 0 and 30% of Chinese controls and patients and 0 and 21% of Japanese controls and patients, respectively, for a P=0.0039 and 0.00676 in Chinese and Japanese populations. In fact, none of the partial founder haplotypes shown in Figure 3 were estimated to be present in the Japanese population, whereas only the partial founder haplotype CAAT(CT)TATCG, was estimated to occur in 4% of the Chinese control population. Because of the availability of pedigree information and the assumptions made in Haplotyping the families, the EM-estimated frequencies of the founder haplotype differ from those seen in our study (44 and 29% in Chinese and Japanese affected, respectively).
Figure 2.
Schematic of the CYP4V2 gene showing the position of SNPs and the risk haplotypes. The alleles in various families are shown below the schematic. Families and individuals are grouped by population with Japanese in the top panel, Chinese in the middle panel, and Koreans in the bottom panel. The risk haplotype identified unambiguously in individuals homozygous for the c.802-8_810del17insGC indel mutation is shaded dark blue. The risk haplotype predicted in individuals heterozygous for the c.802-8_810del17insGC indel mutation is shaded light blue. Alleles showing or beyond recombination events and not included in the risk haplotype are not shaded. A full color version of this figure is available at the European Journal of Human Genetics journal online.
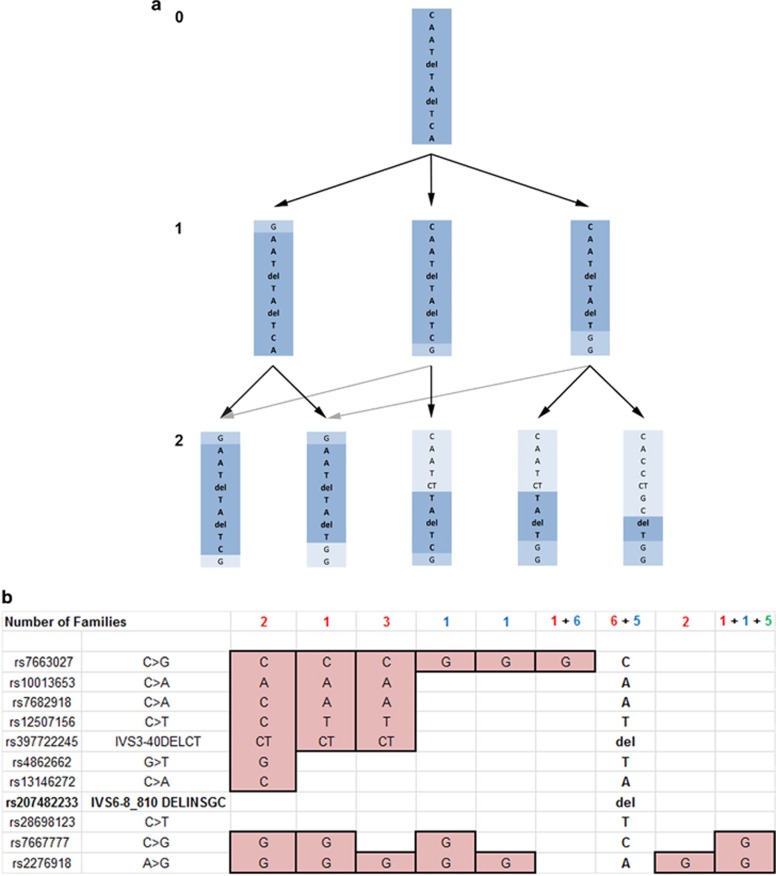
Figure 3.
Evolution and population occurrence of risk haplotypes for the c.802-8_810del17insGC indel mutation. (a) Identified risk haplotypes are shown ordered by the number of recombination events required to generate them from the founder risk haplotype: 0 (top), 1 (middle) and 2 (bottom). The founder risk haplotype is shaded in dark blue, probable initial recombination events are shaded in medium blue, and probable second recombinant events are shown in light blue. Black arrows show probable paths for the recombination events, and gray arrows show alternative orders of recombination events. (b) Identified risk haplotypes ordered by distance of recombination events from the c.802-8_810del17insGC indel mutation. Alleles beyond recombination events are shaded pink. The numbers of individuals or families with each haplotype are shown above the haplotype with the number of Chinese individuals shown in red, Japanese individuals shown in blue, and Korean individuals shown in green. A full color version of this figure is available at the European Journal of Human Genetics journal online.
In Figure 3a the BCD-associated haplotypes from all three populations are ordered into three levels based on the number of recombination events required to derive each haplotype from the founder, which is shown at the top (level 0). A single recombination event can generate the three haplotypes shown in level 1 (one above and two different events below the mutation). From these three haplotypes the remainder, seen on level 2, can be generated by a second recombination on the opposite side of the mutation from the first. An alternative, although more complex, pathway to the two haplotypes on the left side of level 2 from the two haplotypes on the right of level 1 is shown with gray arrows. Thus, all haplotypes bearing the c.802-8_810del17insGC mutation can be generated from the founder haplotype by a maximum of two recombination events, although additional recombination events occurring beyond those shown in Figure 3a would not be identifiable from this analysis, and mutation of a SNP, while infrequent, could not be differentiated from a recombination event.
In Figure 3b the haplotypes are arranged in order of their divergence from the founder haplotype with the number of individuals (or families) and the population to which they belong shown above each haplotype. The founder haplotype is the most common in the Chinese population, whereas the founder haplotype and a second haplotype with a single C>G change are both common in the Japanese population. Finally, a haplotype differing from the founder by having G alleles for the final two SNPs, rs7667777 and rs2276918, accounts for all c.802-8_810del17insGC mutations seen in the Korean population. This haplotype is also seen in a single family in both the Japanese and Chinese populations. The preservation of the founder haplotype in all three populations strongly suggests a single common origin for the c.802-8_810del17insGC mutation, as has been suggested by Lin et al.22 In addition, the greater diversity (12 obligate recombination events in the Chinese vs 6 in the Japanese haplotypes) and occurrence of recombination events closer to the mutation itself suggest that the mutation might have existed in the Chinese population longer than it has in the Japanese population. Although the available sample set does not include enough individuals of Korean origin to provide strong support, the existence of a single haplotype would suggest that the c.802-8_810del17insGC mutation might have been introduced into this population relatively recently. As this haplotype is present in both Chinese and Japanese populations, it is not possible to determine from which it originates.
To examine the origin of the c.802-8_810del17insGC mutation more closely, the mutation age was estimated in the Chinese and Japanese populations using the approach described by Risch et al,15, 16 and Goldstein et al,17 as well as Bayesian disequilibrium estimates of the marginal posterior density as instituted in the DMLE+ program.19 A summary of these results with the age given in generations is shown in Table 4. Using the method described by Risch et al for single markers, estimates ranged from 1150 to 7044 generations, whereas the multiple marker approach using markers combined into haplotypes gave estimates of 3400 and 4500 generations with left and right flanking markers, respectively. Using the method described by Goldstein et al, a similar range was obtained for the single markers, 1841–8200, and 1900 for the rs10013653-rs12507156-rs12507156-rs397722245 haplotype. With both methods, the estimates obtained with rs7663027 is significantly higher than that obtained with the other markers. One possible explanation for this is that the founder allele frequency at this marker, 0.5625, is beginning to approach the allele frequency in the control population, 0.475, suggesting that the distance between this marker and the mutation might be on the upper limits for usefulness in this estimation. Finally, the estimate using DMLE2 assuming population growth rates from 2300 BC, the earliest time reliable estimates are available for this population, is 1040 with 95% confidence limits of 931–1237.
Table 4. Mutation age estimates of the c.802-510 8_810del17insGC indel mutation in the Chinese and Japanese populations.
| Method | rs7663027 | rs10013653 | rs7682918 | rs12507156 | rs397722245 | rs4862662 | rs13146272 | * | rs28698123 | rs7667777 | rs2276918 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C>G | C>A | C>A | C>T | 605-40DELCT | G>T | C>A | C>T | C>G | A>G | ||
| Chinese | |||||||||||
| Risch et al15 | |||||||||||
| Single marker | 7044 | 2605 | 3306 | 4986 | 1510 | 1150 | 2278 | – | 2517 | 5208 | |
| Multiple marker haplotypea,b | 3400 | 4500 | |||||||||
| Goldstein et al17 | 8200 | 2245 | 2277 | 2250 | 1841 | 3985 | |||||
| Haplotype | 1900 | ||||||||||
| DMLE2: 2300 BC | 1040 (95% CI: 931–1237) | ||||||||||
| Japanesea | |||||||||||
| Risch et al | |||||||||||
| Single marker | – | – | – | – | – | – | – | – | 933 | 708 | |
| Multiple marker haplotypeb | 1100 | ||||||||||
| Goldstein et al | – | – | – | – | – | – | – | – | 933 | 300 | |
| DMLE2:2300 BC | 624 (95% CI: 538–738) | ||||||||||
*rs207482233 (c.802-8_10 DELINSGC).
Left haplotype uses CHM estimates of markers rs10013653-rs13146272 and markers 424862662-rs13146272 for multiple marker calculation.
Right haplotype uses CHM estimates of rs28698123-rs2276918 and rs7667777-rs2276918 for multiple marker calculation.
When the age of the mutation in the Japanese population was estimated, values of 708 and 933 generations were obtained with rs7667777 and rs2276918, respectively. A multi-marker analysis of the rs28698123-rs7667777 and rs28698123-rs7667777-rs2276918 haplotypes yielded an estimate of 1100 generations, and the method described by Goldstein et al provided estimates of 933 and 300 generations for rs7667777 and rs2276918, respectively. The estimate from DMLE2 using population growth from 2300 BC was 624 generations with 95% confidence intervals of 538–738 generations. In the Japanese population, estimates could not be made with many markers because there was no recombination within the haplotype or, for rs7663027 the founder allele frequency was actually below that in the general population. The population age in the Korean population could not be calculated because of the small number of risk haplotypes available and the absence of available control haplotype data.
Discussion
In the current study, we report the results of analysis of 58 unrelated patients with BCD, identifying 9 novel mutations (Table 1), a significant addition to the 82 mutations currently listed in HGMD. One of the limitations of this study is the paucity of precise clinical data sufficient to completely characterize some of the patients. However, the available data suggest that all the patients have typical BCD, which is perhaps related to their being referred specifically for sequencing of the CYP4V2 gene. Similarly, from the clinical data available it was not possible to correlate any specific genotype with a phenotypic characteristic or severity. This is consistent with previous studies showing a high degree of clinical variability within and between families, without an obvious correlation to a specific mutation, and also is not surprising, as each of the identified mutations is predicted to result in complete absence of function in CYP4V2.
Overall, 19 missense (68%), 1 indel (4%), 4 nonsense (14%), 2 deletions (7%), and 2 splice site mutations (7%) were identified in this study (28 mutations in all). These results are very similar to mutation frequencies tabulated in HGMD (missense and nonsense, 68% indels, 1% deletions, 12% insertions 3% gross deletions 2% and splice site mutations, 14%). The CYP4V2 protein structure, as estimated by homology modeling, is predicted to have a transmembrane segment at the amino terminal end, followed by a link to the globular domain, consisting of 18 α-helices connected by β-sheet and random coil structures.5 Although the splice, frameshift, and nonsense mutations are predicted to cause gross structural deformities of the protein fold, the missense mutations are seen to cluster in two areas. One is the transmembrane segment, which should interfere with insertion of the enzyme into the membrane. The other is the active site, in which mutations would disrupt coordination of the porphyrin ring required for enzymatic activity or an α-helix supporting this ring.
The c.1355G>A (p. Arg452His) mutation is listed as novel, because although it is present in heterozygous form in the ExAC database (mostly in Europeans), it has not previously been associated with BCD or described in any BCD patients. The previously described missense variants c.64C>G, p.(L22V), and c.775A>C, p(Lys259Gln) were both found in both homozygous and heterozygous individuals (Table 1B). They are predicted to be tolerated by SIFT and benign by PolyPhen2. They also have frequencies of 27–52% and 9–25%, respectively, in various populations on the 1000 Genomes database, so that they seem unlikely to affect CYP4V2 function. This has been noted in previous publications although they do turn up frequently in BCD cases, and the c.775A>C, p(Lys259Gln) variant has been associated with deep vein thrombosis.22, 24, 25
Estimating the likelihood of novel missense variants to affect function is certainly less certain than null alleles, especially where there is disagreement between the predictive programs. In part, this disagreement results from the different predictive algorithms used by each program. Polyphen-2 uses specific sequence (eg, active site or known motif), phylogenetic (species alignment with position-specific independent counts or PSIC), and structural information to estimate the effect on the protein structure and function. PROVEAN predictions are based on conservation of closely related sequences, whereas Condel combines the results of SIFT, PolyPhen2, MutationAssessor and FatHMM to predict the effects of an amino acid change. Thus, each of these programs uses a slightly different combination of factors normalized to different databases, with varying degrees of sensitivity and specificity, so that some inconsistencies are to be expected. However, their predictive accuracy certainly increases when all agree. Although the identified changes seem likely to be causative, it is possible that mutations in regulatory regions outside the coding sequence cause the disease. We have tried to minimize this problem by sequencing the core promoter region (~100 bp) of CYP4V2 in all patients with novel missense mutations, and no changes were identified (data not shown). Although it is theoretically possible that mutations in a different gene can cause BCD in these cases, no other gene or linkage region has been suggested in multiple previous studies, many of which include linkage data, and two mutations have been found in ~92% of BCD patients.5, 22, 23, 24, 26, 27, 28, 29, 30, 31, 32, 33, 34
The c.802-8_810del17insGC change, the most common mutation of CYP4V2 in East Asian BCD patients, was found in 15 cases of 58 BCD patients in this study, all of East Asian origin. Overall, we have found the c.802-8_810del17insGC change in 14 individuals of Chinese, 1 of Japanese and 4 of Korean origin,5 consistent with estimates from the 1000 Genomes database, in which the allele frequency of this mutation is 0.01 in Japan and 0.005 in China, but only 0.002 in Europe and was not seen in African, South Asian, or American populations. In this regard, BCD overall has been reported to be more common in East Asia populations with an estimated gene frequency of 0.005 in China.35
Estimation of the population age, origin, and population history of found the c.802-8_810del17insGC mutation were based on our results and those of previous studies in which the risk haplotype was determined in Chinese and Japanese Bietti cases.5, 22, 23 The age of the mutation was estimated using three approaches, an analytical approach,15 an iterative approach,17 and a posterior Bayesian distribution approach as implemented in the program DMLE+2.2.36 On the basis of their analysis of the intragenic SNP haplotypes, Lin et al previously suggested that the c.802-8_810del17insGC had a common origin in the Japanese and Chinese populations. Our analysis is completely consistent with that suggestion, especially as the ancestral risk haplotype was not identified in either Chinese or Japanese control individuals, suggesting that it is quite rare in both populations. Although the age estimates vary widely, two findings are apparent: (1) the mutation is extremely old, occurring millennia ago, and (2) the temporal precedence of the c.802-8_810del17insGC mutation in the Chinese population further suggests that the mutation originated in ancient China and after some time, perhaps tens of thousands of years, was introduced into the Japanese population where it eventually attained the current allele frequency. It is difficult to know whether the Chinese or Japanese population served as the source of this mutation in the Korean population, as there have been interchanges with both in recent and ancient history. The identical haplotype found in Korea is found in a single instance in both the Chinese and Japanese patients, and we have only a limited number of Korean risk and no control haplotypes on which to base an age estimate.
However, these results must be interpreted with some caution, as they are sensitive to the parameters used in the estimation. Although physical distance between the SNPs is accurately known, the associated recombination frequency was estimated from an average value for the genome. This probably would have minimal effects on the relative age of the mutation in the Japanese and Chinese populations, as the same values were used for both estimates. Similarly, negative selection would probably not be a major problem in this recessive disease, especially as most BCD patients maintain functional vision through the fourth or fifth decade, well beyond reproductive age.23, 26 Estimates of haplotype and genotype frequencies of the risk population should also be robust, and the risk haplotypes include all those studied and published so far. The results are also consistent with the greater diversity of the risk haplotypes and presence of recombination events closer to the mutation in the Chinese population.
In conclusion, we report 28 CYP4V2 mutations, 9 of which were novel. These expand the spectrum of CYP4V22 mutations associated with BCD, assisting with molecular diagnosis and providing additional insight into the structural importance of the variant amino acids. Although there is some overlap, most mutations are confined either to the European or Asian population groups. In addition, these data suggest a single origin of the common c.802-8_810del17insGC mutation in the Chinese population, followed by spread to Japan and then Korea.
Acknowledgments
We would like to thank the families and patients who participated in this work. In addition, we are grateful to NEI project EYE000272 for support. MBG has received support from the Department of Ophthalmology at UCLA, the Harold and Pauline Price Foundation, and from Research to Prevent Blindness, New York, NY. ZBJ has received support from the National Natural Science Foundation of China (81522014, 81371059). AI is supported by the Research to Prevent Blindness, New York, NY (Physician Scientist Award to AI and unrestricted grant to the UTHSC Department of Ophthalmology).
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Kaiser-Kupfer MI, Chan CC, Markello TC et al: Clinical biochemical and pathologic correlations in Bietti's crystalline dystrophy. Am J Ophthalmol 1994; 118: 569–582. [DOI] [PubMed] [Google Scholar]
- Hu DN: Prevalence and mode of inheritance of major genetic eye diseases in China. J Med Genet 1987; 24: 584–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Jiao X, Hejtmancik JF, Kaiser-Kupfer M, Chader GJ: Identification, isolation, and characterization of a 32-kDa fatty acid-binding protein missing from lymphocytes in humans with Bietti crystalline dystrophy (BCD). Mol Genet Metab 1998; 65: 143–154. [DOI] [PubMed] [Google Scholar]
- Lee J, Jiao X, Hejtmancik JF et al: The metabolism of fatty acids in human Bietti crystalline dystrophy. Invest Ophthalmol Vis Sci 2001; 42: 1707–1714. [PubMed] [Google Scholar]
- Li A, Jiao X, Munier FL et al: Bietti crystalline corneoretinal dystrophy is caused by mutations in the novel gene CYP4V2. Am J Hum Genet 2004; 74: 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Kelly EJ, Rettie AE: Expression and characterization of CYP4V2 as a fatty acid {omega}-hydroxylase. Drug Metab Dispos 2009; 37: 2119–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Kelly EJ, Wiek C, Hanenberg H, Rettie AE: CYP4V2 in Bietti's crystalline dystrophy: ocular localization, metabolism of omega-3-polyunsaturated fatty acids, and functional deficit of the p.H331P variant. Mol Pharmacol 2012; 82: 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Chu KO, Chan KP et al: Alterations in serum fatty acid concentrations and desaturase activities in Bietti crystalline dystrophy unaffected by CYP4V2 genotypes. Invest Ophthalmol Vis Sci 2010; 51: 1092–1097. [DOI] [PubMed] [Google Scholar]
- Lockhart CM, Nakano M, Rettie AE, Kelly EJ: Generation and characterization of a murine model of Bietti crystalline dystrophy. Invest Ophthalmol Vis Sci 2014; 55: 5572–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJH, Holcomb JD, Daiger SP et al: Exclusion of Usher syndrome gene from much of chromosome 4. Cytogenet Cell Genet 1989; 50: 102–106. [DOI] [PubMed] [Google Scholar]
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T: Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 2009; 4: 1–13. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T: The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 2006; 22: 195–201. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A et al: SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 2014; 42: W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle RA, Milner-White EJ: RASMOL: biomolecular graphics for all. Trends Biochem Sci 1995; 20: 374. [DOI] [PubMed] [Google Scholar]
- Risch N, de Leon D, Ozelius L et al: Genetic analysis of idiopathic torsion dystonia in Ashkenazi Jews and their recent descent from a small founder population. Nat Genet 1995; 9: 152–159. [DOI] [PubMed] [Google Scholar]
- Colombo R: Age and origin of the PRNP E200K mutation causing familial Creutzfeldt–Jacob disease in Libyan Jews. Am J Hum Genet 2000; 67: 528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Reich DE, Bradman N, Usher S, Seligsohn U, Peretz H: Age estimates of two common mutations causing factor XI deficiency: recent genetic drift is not necessary for elevated disease incidence among Ashkenazi Jews. Am J Hum Genet 1999; 64: 1071–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Rannala B: Estimating allele age. Annu Rev Genomics Hum Genet 2000; 1: 225–249. [DOI] [PubMed] [Google Scholar]
- Reeve JP, Rannala B: DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics 2002; 18: 894–895. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG: Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 1992; 89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R: Amino acid difference formula to help explain protein evolution. Science 1974; 185: 862–864. [DOI] [PubMed] [Google Scholar]
- Lin J, Nishiguchi KM, Nakamura M, Dryja TP, Berson EL, Miyake Y: Recessive mutations in the CYP4V2 gene in East Asian and Middle Eastern patients with Bietti crystalline corneoretinal dystrophy. J Med Genet 2005; 42: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Koh AH, Aung T et al: Characterization of Bietti crystalline dystrophy patients with CYP4V2 mutations. Invest Ophthalmol Vis Sci 2005; 46: 3812–3816. [DOI] [PubMed] [Google Scholar]
- Shan M, Dong B, Zhao X et al: Novel mutations in the CYP4V2 gene associated with Bietti crystalline corneoretinal dystrophy. Mol Vis 2005; 11: 738–743. [PubMed] [Google Scholar]
- Bezemer ID, Bare LA, Doggen CJ et al: Gene variants associated with deep vein thrombosis. JAMA 2008; 299: 1306–1314. [DOI] [PubMed] [Google Scholar]
- Meng XH, Guo H, Xu HW et al: Identification of novel CYP4V2 gene mutations in 92 Chinese families with Bietti's crystalline corneoretinal dystrophy. Mol Vis 2014; 20: 1806–1814. [PMC free article] [PubMed] [Google Scholar]
- Yin H, Jin C, Fang X et al: Molecular analysis and phenotypic study in 14 Chinese families with Bietti crystalline dystrophy. PLoS One 2014; 9: e94960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai TY, Ng TK, Tam PO et al: Genotype phenotype analysis of Bietti's crystalline dystrophy in patients with CYP4V2 mutations. Invest Ophthalmol Vis Sci 2007; 48: 5212–5220. [DOI] [PubMed] [Google Scholar]
- Tian R, Wang SR, Wang J, Chen YX: Novel CYP4V2 mutations associated with Bietti crystalline corneoretinal dystrophy in Chinese patients. Int J Ophthalmol 2015; 8: 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Mai G, Li S, Guo X, Zhang Q: Identification of CYP4V2 mutation in 21 families and overview of mutation spectrum in Bietti crystalline corneoretinal dystrophy. Biochem Biophys Res Commun 2011; 409: 181–186. [DOI] [PubMed] [Google Scholar]
- Hayakawa M, Fujiki K, Kanai A et al: Multicenter genetic study of retinitis pigmentosa in Japan: I. Genetic heterogeneity in typical retinitis pigmentosa. Jpn J Ophthalmol 1997; 41: 1–6. [DOI] [PubMed] [Google Scholar]
- Wada Y, Itabashi T, Sato H, Kawamura M, Tada A, Tamai M: Screening for mutations in CYP4V2 gene in Japanese patients with Bietti's crystalline corneoretinal dystrophy. Am J Ophthalmol 2005; 139: 894–899. [DOI] [PubMed] [Google Scholar]
- Yokoi Y, Sato K, Aoyagi H, Takahashi Y, Yamagami M, Nakazawa M: A novel compound heterozygous mutation in the CYP4V2 gene in a Japanese patient with Bietti's crystalline corneoretinal dystrophy. Case Rep Ophthalmol 2011; 2: 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin ZB, Ito S, Saito Y, Inoue Y, Yanagi Y, Nao-i N: Clinical and molecular findings in three Japanese patients with crystalline retinopathy. Jpn J Ophthalmol 2006; 50: 426–431. [DOI] [PubMed] [Google Scholar]
- Hu DN: Ophthalmic genetics in China. Ophthalmic Paediatr Genet 1983; 2: 39–45. [Google Scholar]
- Rannala B, Reeve JP: High-resolution multipoint linkage-disequilibrium mapping in the context of a human genome sequence. Am J Hum Genet 2001; 69: 159–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S, Testa F, Li A et al: Clinical and genetic features in Italian Bietti crystalline dystrophy patients. Br J Ophthalmol 2013; 97: 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S, Liew G, Mackay DS et al: Detailed phenotypic and genotypic characterization of bietti crystalline dystrophy. Ophthalmology 2014; 121: 1174–1184. [DOI] [PubMed] [Google Scholar]
- Wang F, Wang H, Tuan HF et al: Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet 2014; 133: 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad NM, Waked N, Bejjani R et al: Clinical and molecular findings in three Lebanese families with Bietti crystalline dystrophy: report on a novel mutation. Mol Vis 2012; 18: 1182–1188. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.