Abstract
Knowledge of variant pathogenicity is key to implementing genomic medicine. We describe variability between expert reviewers in assigning pathogenicity to sequence variants in LDLR, the causal gene in the majority of cases of familial hypercholesterolemia. LDLR was sequenced on the Illumina HiSeq platform (average read depth >200 × ) in 1013 Mayo Biobank participants recruited from 2012 to 2013. Variants with a minor allele frequency (MAF) <5% predicted to be functional or referenced in HGMD (Human Gene Mutation Database) or NCBI-ClinVar databases were reviewed. To assign pathogenicity, variant frequency in population data sets, computational predictions, reported observations and patient-level data including electronic health record-based post hoc phenotyping were leveraged. Of 178 LDLR variants passing quality control, 25 were selected for independent review using either an in-house protocol or a disease/gene-specific semi-quantitative framework based on the American College of Medical Genetics and Genomics-recommended lines of evidence. NCBI-ClinVar included interpretations for all queried variants with 74% (14/19) of variants with >1 submitter showing inconsistency in classification and 26% (5/19) appearing with conflicting clinical actionability. The discordance rate (one-step level of disagreement out of five classes in variant interpretation) between the reviewers was 40% (10/25). Two LDLR variants were independently deemed clinically actionable and returnable. Interpretation of LDLR variants was often discordant among ClinVar submitters and between expert reviewers. A quantitative approach based on strength of each predefined criterion in the context of specific genes and phenotypes may yield greater consistency between different reviewers.
Introduction
Determining disease-relevant DNA variants from genome sequencing is a significant challenge as false assignments of pathogenicity can have adverse consequences in clinical practice.1 Current mutation databases are not a reliable source for assigning variant pathogenicity and there is a need for accurate and up-to-date centralized repositories of potentially actionable variants based on rigorous evidence.2 The American College of Medical Genetics and Genomics (ACMG) recommends reporting actionable incidental findings from genome sequencing.3 It is therefore imperative to explore processes and criteria for assigning pathogenicity to specific variants and assess the frequency of such incidental findings in patients suspected to have disease as well as apparently healthy individuals from the general population.
Electronic health record (EHR)-based assessment of phenotypic correlates of variants in disease-related genes may facilitate interpretation of results of genome sequencing.4, 5 A common genetic disorder of major public health importance, familial hypercholesterolemia (FH) is associated with elevated low-density lipoprotein cholesterol (LDL-C) levels and if untreated, premature atherosclerotic cardiovascular disease (ASCVD) and significant decrease in life expectancy.6, 7 Pathogenic variants in one of three genes, that is, LDLR, APOB, PCSK9, leading to impaired LDL receptor function and elevated LDL-C levels, account for the majority of FH cases. The ACMG recommends returning incidental findings in these three genes implicated in FH.3, 8 The majority of FH patients with positive genetic testing results have rare pathogenic variants in LDLR9 which comprise 60% of the ~2000 LDLR genetic variants that have been submitted to the Human Gene Mutation Database (HGMD). Prior studies have highlighted inconsistency in variant classification between locus-specific databases as well as between interpreters.10, 11, 12, 13 In this report we describe discordance in reported classification among submitters to curated databases and among expert reviewers in assigning variant pathogenicity in individuals from the community who underwent sequencing of LDLR in a Clinical Laboratory Improvement Amendments-certified facility.
Materials and methods
Study participants
Participants for this investigation were sampled from the electronic MEdical Records and GEnomics Network (https://emerge.mc.vanderbilt.edu/) Pharmacogenomics (eMERGE-PGx) project in which a next-generation sequencing platform designed to assess sequence variation in 84 pharmacogenes was implemented in ~9000 patients likely to be prescribed drugs of interest in a 1–3 years' time frame across several clinical sites in US.14 The Mayo Clinic site's contribution included 1013 residents of Olmsted County who were participants in the Mayo biobank.15 The Right Drug, Right Dose, Right Time—Using Genomic Data to Individualized Treatment (RIGHT Protocol), investigated preemptive integration of actionable variants in these pharmacogenes in the EHR with linkage to clinical decision support. Baseline characteristics of study participants who were empaneled in the Mayo Clinic primary care practices (from 2012 to 2013), the sequencing methodology, and a prediction model to identify patients likely to be started on statin therapy within 3 years are described elsewhere.15 All subjects gave written informed consent, and the study protocol was approved by the Institutional Review Board at Mayo Clinic (Rochester, MN).
LDLR sequencing
LDLR was sequenced using the PGRN-Seq16 capture reagent coupled with a high-throughput next-generation sequencing (NGS) platform in the Mayo Clinical Genome Sequencing Laboratory which is Clinical Laboratory Improvement Amendments-certified and College of American Pathologists accredited. The complete coding region plus 2 kb upstream and 1 kb downstream was sequenced on the Illumina HiSeq 2500 (Illumina, San Diego, CA, USA). A number of filters were implemented to remove false-positive calls, and data quality and error rates were carefully evaluated. All samples had at least 30 × coverage in all exons with nearly 25% of samples having 600 × coverage in the exons. The per-exon average coverage for LDLR varied from 251 to 862 reads. Following standard bioinformatics processing and variant calling, we employed SnpEff17 to annotate and predict the effects of genetic variations. Our choice of SnpEff was determined based on its ability to accept Variant Call Format files as input, continually updated transcript databases and support for the Genome Analysis Toolkit.18 Cohort description, phenotyping and associated quality control analysis workflow are available in the online Supplementary Materials and Methods.
Variant selection
Variant selection was completed using bioinformatics prediction tools, DNA variation databases and expert review. A tiered strategy for selecting variants for further expert review was employed. Variants with a minor allele frequency (MAF) <5% were selected for further analysis if predicted to be high/moderate/low impact based on SnpEff or reported in the HGMD Professional 2015.4 or NCBI-ClinVar in association with FH phenotype. Sequence data and read-alignments for selected variants were reviewed by a bioinformatics specialist. Further manual review of filtered variants was performed by two independent laboratory specialists involved in interpretation of genetic testing results for FH.
Variant annotation
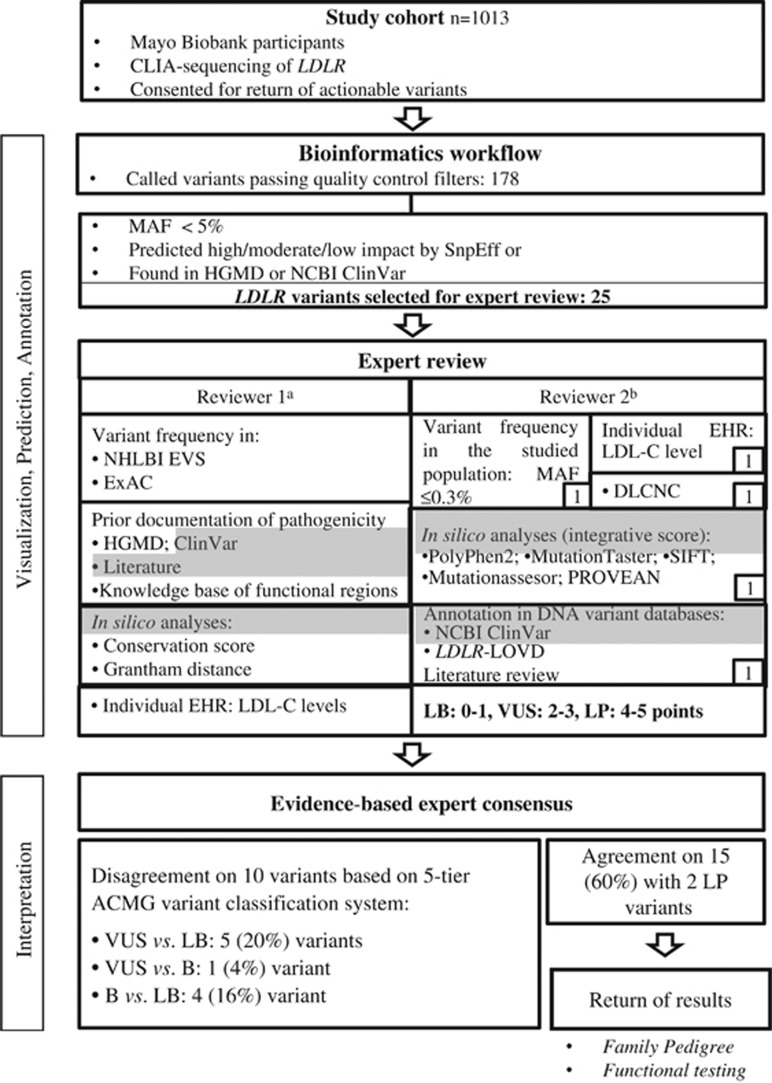
An average of 22.7 variants were called in LDLR with a variant density of 4.3 variants/kb (5.3 kb of sequence, Supplementary Figure 1). An outline of the workflow for LDLR variant annotation is depicted in Figure 1. One reviewer used a conventional assessment protocol and classification system for variant designation adopted by Mayo Clinic's Department of Laboratory Medicine and Pathology while the second reviewer annotated variants using scoring criteria tailored based on the lines of evidence recommended by the ACMG guidelines.
Figure 1.
Outline of the workflow and data analyses for LDLR variant interpretation. B, benign; CLIA, Clinical Laboratory Improvement Amendments; DLCNC, Dutch Lipid Clinic Network criteria; LB, likely benign; LP, likely pathogenic; P, pathogenic; VUS, variant of uncertain significance. aReviewer# 1, a laboratory medicine specialist–clinical molecular geneticist with expertise in cardiovascular genetics, including FH, assisted by a genetic counselor, used a conventional assessment protocol and classification system for variant designation adopted by Mayo Clinic's Department of Laboratory Medicine and Pathology. bReviewer #2, a cardiologist with expertise in heritable lipid disorders, annotated variants using scoring criteria for incidental findings in LDLR based on the ACMG guidelines. Criteria highlighted in gray overlap between the two protocols/algorithms.
Variant pathogenicity assignments have been submitted to the NCBI-ClinVar under the ‘Cardiovascular Biomarker Research Laboratory, Mayo Clinic, Organization ID 505718, under accession numbers SCV000266306.1, SCV000266307.1, SCV000266308.1, SCV000266309.1, SCV000266310.1, SCV000266311.1, SCV000266312.1, SCV000266313.1, SCV000266314.1, SCV000266315.1, SCV000266316.1, SCV000266317.1, SCV000266318.1, SCV000266319.1, SCV000266320.1, SCV000266321.1, SCV000266322.1, SCV000266323.1, SCV000323101, SCV000323102, SCV000323103, SCV000323104, SCV000323105, SCV000323106 and SCV000323107 (http://www.ncbi.nlm.nih.gov/clinvar/submitters/505718/).
Results
Study participants
The median age of the study cohort (n=1013) was 56 years, 47% were men and 86% were white. Twenty-five putatively functional pathogenic variants in LDLR were identified in 124 individuals (12%). The mean age of this sample was 57.9±5.5 years, 53% were women, and mean LDL-C level was 3.5±0.8 mmol/l, 98% white. Of these 124 participants, five (4%) patient had severe hypercholesterolemia (LDL-C ≥4.9 mmol/l), 27 (22%) had high LDL-C levels (4.1–4.9 mmol/l), and 28 (23%) had a borderline high levels (3.4–4.1 mmol/l). The LDL-C levels were extracted from structured laboratory databases. Dutch Lipid Clinic Network (DLCN) criteria19 were used to identify patients likely to have an FH-causing genetic variant. Supplementary Table 1 details clinical and sequencing characteristics pertinent to the 25 LDLR variants. LDL-C levels were ascertained when participants were not on lipid-lowering therapy.
Each participant filled a questionnaire designed by Mayo Biobank investigators to ascertain risk factor profile and family and personal medical history. An EHR review provided comprehensive information on individual's personal or family history of ASCVD (Supplementary Table 1). We reviewed ‘family history' and ‘patient provided information' sections of the EHR and noted a positive family history of early-onset ASCVD in 22 cases (18% Supplementary Material). A clinical diagnosis of possible FH (DLCN score of 3–5 points) was established in 5 subjects, carriers of c.1432G>A (p.(Gly478Arg)) and c. c.1171G>A (p.(Ala391Thr)). On follow-up within the EHR surveillance timeline, 21 individuals were started on a statin therapy and in 2 individuals ezetimibe or niacin were initiated. We observed a lower than predicted reduction in the LDL-C levels in 4 out of 12 treated carriers of c.1171G>A (p.(Ala391Thr)) and in a carrier of two variants c.58G>A (p.(Gly20Arg)) and c.2177C>T (p.(Thr726Ile)). Expected lipid-lowering effect of medications based on the type and dosage has been reviewed elsewhere20 and is summarized in Supplementary Table 1.
Variant annotation
The algorithms integrated sequencing alignment and variant calling by the bioinformatics specialist, variant database review and expert review using two different protocols.
Bioinformatic analyses
A customized NGS variant analysis pipeline was used to assess putative functional effects of each LDLR variant. Supplementary Figure 2 describes the distribution of MAFs of the 25 LDLR variants among 124 individuals. Seventeen variants were each present in a single patient. There were 15 (60%) missense, 5 (20%) synonymous, 3 (12%) splice site and 2 intronic variants.
Variant database review
We further reviewed the classification of these variants in three databases, that is, NCBI-ClinVar, LDLR Leiden Open Variation Database (LOVD) and HGMD. As of August 2016 all 25 variants were present in ClinVar. In ClinVar, the review status for these variants ranged from (i) one submitter providing an interpretation with assertion criteria or multiple submitters providing conflicting interpretations to (ii) two or more submitters providing assertion criteria with the same interpretation. In 76% (19/25) of variants with more than one submitter, 74% (14/19) of interpretations differed across a three-tier classification system spanning from pathogenic/likely pathogenic to uncertain significance to likely benign/benign. Five variants (5/19, 26%) had significantly discordant labels (likely benign/benign versus likely pathogenic/pathogenic). We found that in the HGMD variants were classified as disease-causing more frequently than in ClinVar and literature reports not cited in the context of ClinVar (P<0.01; Supplementary Table 1; ‘Variant Categorization').
Expert review
There were 50 unique interpretations based on two different algorithms (Table 1). Supplementary Table 1 depicts results of comprehensive computational analyses of variant effect prediction, database and literature-derived annotation as well as clinical characteristics from the EHR. Reviewers #1 and #2 classified 25 variants according to prespecified procedures (Figure 1). Categorization of LDLR variants by reviewer #1 was based on variant frequency in NHLBI-exome variant server and Exome Aggregation Consortium, literature and HGMD reports, ClinVar data entries, in-silico prediction tools, including amino acid conservation and Grantham distance, and knowledge of LDLR functional regions. Classification of sequence variants was initially blinded to patient phenotypic data such as lipid levels and family or personal history of ASCVD. Post hoc phenotyping was performed only for three variants by requesting LDL-C levels.
Table 1. Comparison of assignment of variant pathogenicity among submitters to DNA variation databases and among expert reviewers.
| Sequencing data | Clinical characteristics | Variant databases and Literature | Expert review | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # | rsID | cDNA position | Amino acid change | GVS function | DLCNC | Max LDL-Ca | MAF, % | In-silico analyses | ClinVar | Reports | #1 | #2 |
| 1 | rs758194385 | c.1691A>G | p.(Asn564Ser) | Missense | 2 | 4.7 | 0.05 | LP | LP [1] | P | LP | LP |
| 2 | rs144614838 | c.1432G>A | p.(Gly478Arg) | Missense | 3 | 6.4 | 0.05 | LP | P; LP; VUS [1] | VUS | LP | LP |
| 3 | rs137853962 | c.1875C>T | p.(Asn625=) | Synonymous | 0 | 3.6 | 0.05 | LB | LB [1] | LB | LB | LB |
| 4 | rs137853960 | c.148G>T | p.(Ala50Ser) | Missense | 0 | 3.1 | 0.05 | LB | B/VUS [1] | VUS | LB | LB |
| 5 | rs148698650 | c.829G>A | p.(Glu277Lys) | Missense | 1 | 4.0 | 0.05 | VUS | B/LB [2] | B | LB | LB |
| 6 | rs368562025 | c.1238C>T | p.(Thr413Met) | Missense | 1 | 4.0 | 0.05 | LP | P, LP, VUS [1] | VUS | VUS | VUS |
| 7 | rs150673992 | c.757C>T | p.(Arg253Trp) | Missense | 1 | 2.6 | 0.05 | LP | LP, VUS, LB [1] | LB/B | VUS | VUS |
| 8 | rs200142970 | c.2252G>A | p.(Arg751Gln) | Missense | 1 | 4.8 | 0.05 | LB | LB, VUS [1] | VUS | VUS | VUS |
| 9 | rs148181903 | c.1837G>A | p.(Val613Ile) | Missense | 0 | 3.4 | 0.05 | LB | LB, VUS [1] | VUS | LB | LB |
| 10 | rs139089530 | c.508G>A | p.(Asp170Asn) | Missense | 2 | 4.6 | 0.05 | LP | VUS [1] | — | VUS | VUS |
| 11 | rs143992984 | c.806G>A | p.(Gly269Asp) | Missense | 1 | 3.9 | 0.05 | LP | LB, VUS [1] | VUS/B | LB | LB |
| 12 | rs146354103 | c.507C>T | p.(Asn169Asn) | Synonymous | 1 | 4.8 | 0.05 | LB | B/LB [2] | — | VUS | VUS |
| 13 | rs143872778 | c.1836C>T | p.(Ala612=) | Synonymous | 1 | 4.3 | 0.05 | LB | LB [1] | — | VUS | VUS |
| 14 | rs116405216 | c.941-4G>A | — | Splice region | 0 | 3.5 | 0.01 | LP | LB [2] | P | VUS | VUS |
| 15 | rs138315511 | c.1085A>C1 | p.(Asp362Ala) | Missense | 1 | 3.8 | 0.10 | LP | LP, LB [1] | P, LB | VUS | VUS |
| 16 | rs147509697 | c.58G>A | p.(Gly20Arg) | Missense | 0 | 3.8 | 0.10 | LB | LB, VUS [1] | P, VUS, B | LB | LB |
| 17 | rs13306498 | c.1194C>T | p.(Ile398=) | Synonymous | 2 | 2.5 | 0.15 | LB | B [1] | B | LB | LB |
| 18 | rs137853963 | c.2231G>A | p.(Arg744Gln) | Missense | 2 | 3.9 | 0.10 | LB | LB, LP [1] | LP, B | LB | LB |
| 19 | rs11669576 | c.1171G>A | p.(Ala391Thr) | Missense | 4 | 3.3 | 3.95 | LB | B [1] | — | LB | LB |
| 20 | rs12710260 | c.1060+10G>C | — | Intron | 0 | 3.0 | 0.05 | LB | B [2] | — | LB | LB |
| 21 | rs17248882 | c.1706-10G>A | — | Intron | 2 | 4.2 | 0.01 | LB | B, LB, VUS [1] | — | VUS | VUS |
| 22 | rs45508991 | c.2177C>T | p.(Thr726Ile) | Missense | 2 | 3.8 | 0.94 | LP | B/LB/VUS [1] | B | LB | LB |
| 23 | rs5926 | c.1959T>C | p.(Val653Val) | Synonymous | 2 | 3.1 | 0.30 | LB | B [2] | B | LB | LB |
| 24 | rs72658861 | c.1061-8T>C | — | Splice region | 0 | 3.2 | 0.05 | LB | B/LB/VUS [1] | B | LB | LB |
| 25 | rs72658867 | c.2140+5G>A | — | Splice region | 0 | 3.0 | 0.05 | VUS | LB/VUS [1] | B | LB | LB |
Abbreviations: B, benign; DLCNC, Dutch Lipid Clinic Network Criteria 19,26 LDL-C, low-density lipoprotein cholesterol; LB, likely benign; LP, likely pathogenic; MAF, minor allele frequency; P, pathogenic; VUS, variant of uncertain significance.
LDL-C levels are given in mmol/L.
The NCBI-ClinVar database's levels of evidence: 0 – no assert criteria or clinical significance not provided; 1 – single source with criteria provided; 2 – multi-source consistency; 3 – expert panel; 4 – based on practice guidelines of ACMG, CPIC. Reference sequence: NM_000527.4, NP_000518.1, NG_009060.1, LRG_274.
Highlighted in bold are LDLR variants identified as likely pathogenic and reportable based on final expert review and consensus.
Five variants (c.1691A>G, c.941-4G>A4, c.1085A>C2, c.58G>A and c.1194C>T) were reported at least once in the literature either in the disease setting, as part of FH case descriptions or FH registry analysis. Supplementary Table 3 summarizes the analysis of the original contributions categorizing variants reported herein, including incidental findings in whole-exome sequencing of ACMG genes.
Reviewer #2 annotated variants using a framework based on: (i) review of variant-level data, such as variant frequency, variant repositories and in-silico pathogenicity scores; (ii) review of primary literature (Supplementary Table 2) for the reported variants in the context of FH; dbSNP and dbVar were queried for LDLR variants and PubMed and Google Scholar searched using the following search terms: rsID and cDNA position for each variant, FH, secondary, and incidental findings; (iii) extensive EHR review including assessment of demographic data, LDL-C level and ascertaining DLCN criteria for FH comprising lipid levels, presence of personal or family history of premature ASCVD and hypercholesterolemia, arcus cornealis and xanthomas. Structured EHR data were mined for the highest untreated LDL-C levels. Family history was defined as occurrence of ASCVD before age 55 in men and 65 years in women. When taking into account FH-specific clinical factors in individuals that shared a variant we considered the highest DLNC score and the median of LDL-C levels. DNA variation databases included NCBI-ClinVar and LDLR-LOVD. For each variant the likelihood of altered LDL receptor activity was determined by an integrative score unifying different annotations from Polymorphism Phenotyping v2, SIFT, MutationTaster, Mutationassesor, Protein Variation Effect Analyzer. Supplementary Table 3 provides an outline for variant categorization and references the utilized tools. A total score was computed for each selected variant (Supplementary Table 1, ‘Variant Categorization'). Variants scored 4–5 were defined as likely pathogenic, 2–3 as variants of uncertain significance and 0–1 as likely benign.
Among variants classified discordantly, use of an LDLR-tailored framework based on the ACMG/Association for Molecular Pathology (AMP) guidelines more frequently tended to categorize variants as uncertain significance (10 versus 7). Intra-laboratory comparison of the in-house laboratory process and the proposed framework for the 25 paired variant assessments showed a disagreement rate of 40% (10/25) in variant classification when reviewers used a five-level classification (Supplementary Figure 3A). Using a three-tier classification system (pathogenic/likely pathogenic versus uncertain significance versus likely benign/benign) 24% (6/25) of variants were assessed discordantly by the two reviewers (Supplementary Figure 3B). We found 20% (5/25) variants to be reported in the literature at least once either in the disease setting, as part of FH case descriptions or FH registry analysis. A final discussion of which of the variants to return included review of the aforementioned information leading to a unanimous consensus that two variants (c.1691A>G, c.1432G>A) should be returned. There was a significant difference in the LDL-C levels between individuals with variants deemed to be actionable (n=2, 1 European American and 1 Asian Chinese, median LDL-C, 5.4 mmol/l) compared to the rest of the samples (n=122, 97% European Americans and 3% African Americans, median LDL-C, 3.5 mmol/l; Wilcoxon rank-sum test, P=0.045).
Discussion
We found 25 putatively disruptive low-frequency variants in the LDLR sequences of 1013 Biobank participants. Assignment of LDLR variant pathogenicity was often discordant between ClinVar submitters and two expert reviewers, highlighting the challenge of determining which variants should be returned to patients. Based on independent review, two LDLR variants were deemed clinically actionable and returnable. Our report provides a conceptual framework for a gene/disease-tailored approach to variant classification. We estimated the burden of actionable incidental findings after Clinical Laboratory Improvement Amendments sequencing of LDLR in a community based cohort.
A number of groups, including the ACMG and Human Variome Project, have developed standards for curation and interpretation of genetic variants identified by genome sequencing. Although LDLR variant databases are close to meeting ACMG-recommended levels of curation,21 a third of the variants have discordant interpretation at the level of clinical actionability, supporting data from a recent report of inconsistency in variant classification between genetics laboratories.13 A recent review of HGMD and ClinVar databases revealed that almost half of the variants in the genes listed by the ACMG are left unclassified.12 Although we integrated available pathogenicity assignments into the variant annotation framework, discrepancies between databases and lack of evidence required by ACMG guidelines as well as a paucity of published data on interpretation of incidental findings in LDLR hampered the process of assigning pathogenicity. Together with the reported discordance across laboratories in categorizing Mendelian-disease variants these findings reinforce a need for a consistent and transparent approach to variant classification.22
The two independent reviewers disagreed on almost half of the variants with inter-rater reliability of 60% within the ACMG/AMP-recommended classification system but were fully concordant in reporting actionable variants. It is recommended that various lines of evidence should be combined when assessing the pathogenicity of genetic variants. It is worth noting that pathogenicity for the vast majority of genetic variants has yet to be validated and therefore remains unknown. Interestingly, the experiment integrating the ACMG/AMP system23 in classifying 347 variants sequenced in genes associated with Mendelian disorders did not increase the agreement rate among nine independent laboratories when compared to internal protocols.13 In our study we observed that the variability in weighting the impact of available components for variant classification was the main cause of discordance between the two reviewers. In this cohort of individuals without clinical indications for genetic testing, absence of a ‘gold standard' for likelihood of pathogenicity, such as quantification of LDL receptor activity (available only for seven variants in our testing set), the proposed framework recognized three levels of certainty of pathogenicity ranging from likely pathogenic to likely benign. This introduced another reason for disagreement involving four variants among the reviewers. These findings suggest that a standardized approach built on strength of each predefined criterion in the context of specific genes and syndromes might have yielded greater consistency in variant classification.
Interpretation of genetic sequencing results is a rapidly growing need; by the time of completion of the PGx sequencing project, classification of 33% of LDLR variants was not available in the databases but within a year annotations for the manually ascertained variants in LDLR were available for review in databases (details presented in Supplementary Material Online). We approached the pitfalls stemming from inconsistency in the reported variant classification by assessing each publication's assertion criteria. We assigned greater weight to variants with functional validation, followed by variants with interpretations based on more than one line of evidence and lastly variants without assertion criteria in ClinVar.
We identified a strong agreement in assigning clinical actionability between two independent reviewers (Cohen's Kappa of 0.1) when compared with the reported variant categorization. Following a discussion of discordant assignments a consensus was achieved between the reviewers and 1.1% (2/178) variants were considered reportable and actionable. This finding is consistent with prior reports demonstrating frequency of 1.4% for actionable incidental findings from disease-related genes,24, 25 varying from 0.912 to 5%10 for the 56 ACMG-approved gene set.
Differences in methodologies, subjectivity in processing and weighting the evidence for variant classification as well as discrepancies among bioinformatics prediction tools and DNA variation databases reinforce the need for a phenotype-centered system for variant classification followed by systematic aggregation of evidence-based set of rules to define whether variants are returnable/actionable. Despite differences in approaches used for assignment of variant pathogenicity we found reasonable concordance in interpretation between two independent reviewers with distinct backgrounds. When an ACMG-recommended five-tier classification was applied a greater inter-reviewer variability was observed, but even this discrepancy was less than a 53% disagreement rate described recently for a 153 variant set.11
We employed a disease-specific criterion of a MAF ≤0.3% to screen for likely disease associated variants in LDLR based on the reported prevalence of FH in the general population excluding founder populations.6 Frequency of likely pathogenic variants identified in our study (1:507; 0.20%) matches the common estimations of FH prevalence in the general population (1:500).26 LDLR sequence lends itself to evaluation of approaches for annotation of incidental findings given the objective phenotypic readout of LDL-C levels. Over 1200 variants in LDLR are responsible for ~90% of FH cases with an identifiable genetic etiology. However a few caveats need to be considered: (i) in nearly 25% of patients with clinical diagnosis of FH a specific pathogenic variant in any of three FH-causing genes is not detected,9 and (ii) certain individuals with a causal variant may not present with the phenotype (reduced penetrance), highlighting challenges associated with counseling and communicating results to patients. An overlap in the distributions of untreated LDL-C levels in mutation-positive and mutation-negative relatives of heterozygote FH patients implies that diagnostic criteria that rely solely on LDL-C levels may result in misdiagnosis.27 This observation underscores the utility of DNA-based tests for efficient cascade testing and the need for standardized variant annotation protocols across laboratories. To date, the Exome Aggregation Consortium resource provides the largest database for the estimation of allele frequency for variations in exomes and was our primary source for the MAF-estimates. A paucity of diverse control data may lead to overcalling variant pathogenicity,2, 28 and it is therefore imperative to further expand the catalog of human genetic diversity currently being generated as part of the Exome Aggregation Consortium29 to aid automated gene/disease-specific filtering of plausibly pathogenic variants.
Several limitations of our study need to be mentioned. Testing of family members, knowledge of additional genetic and environmental factors and assays for LDL receptor functional activity will facilitate assigning pathogenicity to LDLR variants but were outside the scope of our study. Expanding the sample size and the number of interpreters would have allowed a more robust evaluation of variability in assigning pathogenicity. In order to identify those individuals with other than non-LDLR single-gene effects affecting lipid metabolism, a more comprehensive genetic assessment for APOB and PCSK9 variants would be needed. Integration of a Bayesian quantitative genomics approach for weighing levels of evidence and variant categorization may unify the assigned weights and assist in automated classification of incidental findings.
Conclusion
We report the extent of discordant interpretation of variant pathogenicity in LDLR among submitters to public databases and expert reviewers from a single center. Our report highlights challenges in identifying potentially actionable genetic variants from sequencing of disease-related genes and the need for a repository of annotated variants to facilitate implementation of precision medicine.
Acknowledgments
We thank the RIGHT Protocol and Mayo Clinic Biobank study investigators, field staff, and study participants. We thank Luanne F Wussow for assistance in preparation of the manuscript. We appreciate thoughtful comments on this contribution and valuable discussions with Xiao Fan PhD (Cardiovascular Biomarkers Research Laboratory, Mayo Clinic, Rochester, MN, USA). Dr Safarova is supported by AHA Postdoctoral Fellowship Award 16POST27280004. This study was funded as part of the National Human Genome Research Institute's electronic Medical Records and Genomics Network grants to Mayo Clinic (HG04599 and HG006379), R01 GM28157, U01 HG005137, R01 CA138461, R01 AG034676 (The Rochester Epidemiology Project), and the Mayo Clinic Center for Individualized Medicine. The sequencing platform was developed by the next-generation sequencing centers of the Pharmacogenomics Research Network supported by NIH grants U19GM61388, U19HLO69757 and U01GMO97119. The National Human Genome Research Institute and American Heart Association had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review or approval of the manuscript, including the decision to submit the manuscript for publication. Discussion of this paper by Drs Kullo and Safarova is available in the Supplementary Video.
Web resources
NCBI-ClinVar database, http://www.ncbi.nlm.nih.gov/clinvar first assessed July 2015, last assessed August 2016, LDLR Leiden Open Variation, LOVD; versions 1.1.0 build 12, 2.0 build 36, and 3.0 build 13; http://www.ucl.ac.uk/ldlr/LOVDv.1.1.0/index.php?select_db=LDLR, http://www.ucl.ac.uk/ldlr, https://grenada.lumc.nl/LOVD2/UCL-Heart/home.php?select_db=LDLR, http://databases.lovd.nl/whole_genome/genes/LDLR assessed December 2015, NHLBI-Exome Variant Server, EVS; http://evs.gs.washington.edu/EVS/ assessed December 2015.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
A supplementary video accompanies this article on European Journal of Human Genetics website
The authors declare no conflict of interest.
Supplementary Material
References
- Fabsitz RR, McGuire A, Sharp RR et al: Ethical and practical guidelines for reporting genetic research results to study participants: updated guidelines from a National Heart, Lung, and Blood Institute working group. Circ Cardiovasc Genet 2010; 3: 574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP et al: Guidelines for investigating causality of sequence variants in human disease. Nature 2014; 508: 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW et al: ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013; 15: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullo IJ, Haddad R, Prows CA et al: Return of results in the genomic medicine projects of the eMERGE network. Front Genet 2014; 5: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driest SL, Wells QS, Stallings S et al: Association of arrhythmia-related genetic variants with phenotypes documented in electronic medical records. JAMA 2016; 315: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safarova MS, Kullo IJ: My Approach to the patient with familial hypercholesterolemia. Mayo Clin Proc 2016; 91: 770–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjouke B, Kusters DM, Kindt I et al: Homozygous autosomal dominant hypercholesterolaemia in the Netherlands: prevalence, genotype-phenotype relationship, and clinical outcome. Eur Heart J 2015; 36: 560–565. [DOI] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB et al: ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med 2008; 10: 294–300. [DOI] [PubMed] [Google Scholar]
- Futema M, Plagnol V, Li K et al: Whole exome sequencing of familial hypercholesterolaemia patients negative for LDLR/APOB/PCSK9 mutations. J Med Genet 2014; 51: 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence L, Sincan M, Markello T et al: The implications of familial incidental findings from exome sequencing: the NIH Undiagnosed Diseases Program experience. Genet Med 2014; 16: 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola LM, Dorschner MO, Robertson PD et al: Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res 2015; 25: 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens J, Ling H, Hetrick K et al: Assessment of incidental findings in 232 whole-exome sequences from the Baylor-Hopkins Center for Mendelian Genomics. Genet Med 2015; 17: 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola LM, Jarvik GP, Leo MC et al: Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet 2016; 98: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen-Torvik LJ, Stallings SC, Gordon AS et al: Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther 2014; 96: 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinski SJ, Olson JE, Pathak J et al: Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc 2014; 89: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AS, Fulton RS, Qin X, Mardis ER, Nickerson DA, Scherer S: PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet Genomics 2016, e-pub ahead of print 5 January 2016 doi:10.1097/FPC.0000000000000202. [DOI] [PMC free article] [PubMed]
- Cingolani P, Platts A, Wang le L et al: A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012; 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health OrganizationFamilial hypercholesterolemia: report of a second WHO Consultation. World Health Organization: Geneva, Switzerland, 1999 WHO publication No. WHO/HGN/FH/CONS/99.2.
- Haralambos K, Whatley SD, Edwards R et al: Clinical experience of scoring criteria for Familial Hypercholesterolaemia (FH) genetic testing in Wales. Atherosclerosis 2015; 240: 190–196. [DOI] [PubMed] [Google Scholar]
- Rehm HL, Bale SJ, Bayrak-Toydemir P et al: ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013; 15: 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JE, Irving SA, Biesecker LG et al: A standardized, evidence-based protocol to assess clinical actionability of genetic disorders associated with genomic variation. Genet Med 2016; 18: 1258–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S et al: Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner MO, Amendola LM, Turner EH et al: Actionable, pathogenic incidental findings in 1000 participants' exomes. Am J Hum Genet 2013; 93: 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Rubinstein WS, Facio FM et al: Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet 2012; 91: 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins PN, Toth PP, Ballantyne CM, Rader DJ: Familial hypercholesterolemias: prevalence, genetics, diagnosis and screening recommendations from the National Lipid Association Expert Panel on Familial Hypercholesterolemia. J Clin Lipidol 2011; 5: S9–17. [DOI] [PubMed] [Google Scholar]
- Gidding SS, Ann Champagne M, de Ferranti SD et al: The agenda for familial hypercholesterolemia: a scientific statement from the American Heart Association. Circulation 2015; 132: 2167–2192. [DOI] [PubMed] [Google Scholar]
- Manrai AK, Funke BH, Rehm HL et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med 2016; 375: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV et al: Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.