Abstract
Dyslexia is a common specific learning disability with a substantive genetic component. Several candidate genes have been proposed to be implicated in dyslexia susceptibility, such as DYX1C1, ROBO1, KIAA0319, and DCDC2. Associations with variants in these genes have also been reported with a variety of psychometric measures tapping into the underlying processes that might be impaired in dyslexic people. In this study, we first conducted a literature review to select single nucleotide polymorphisms (SNPs) in dyslexia candidate genes that had been repeatedly implicated across studies. We then assessed the SNPs for association in the richly phenotyped longitudinal data set from the Dutch Dyslexia Program. We tested for association with several quantitative traits, including word and nonword reading fluency, rapid naming, phoneme deletion, and nonword repetition. In this, we took advantage of the longitudinal nature of the sample to examine if associations were stable across four educational time-points (from 7 to 12 years). Two SNPs in the KIAA0319 gene were nominally associated with rapid naming, and these associations were stable across different ages. Genetic association analysis with complex cognitive traits can be enriched through the use of longitudinal information on trait development.
Introduction
Reading ability is a complex behavioral trait. Several cognitive processes are involved in the acquisition of this skill.1 For example, successful reading of a novel word depends on phonological awareness, the ability to explicitly reflect on the internal sound structure of words, as well as phonological decoding, the ability to match phonetic units to their written equivalents (graphemes). The language in which reading is learned also plays an important role in the type of strategies learners use.1 In spite of the essential role reading plays in many human societies nowadays, about 5–7% of the population have trouble in acquiring reading skills and may be diagnosed with dyslexia.1
It is well known that a substantial amount of the variance in reading ability is explained by inherited factors: genetic variance explains about 20–80% of the total variation in reading skills.1 However, we still know little about the molecular underpinnings of this trait, since the genetic variants that have been identified so far only explain a tiny fraction of estimated heritability. Nevertheless, some dyslexia candidate loci have been identified through linkage and targeted association studies, leading to proposal of several potential susceptibility genes, including the axon guidance receptor ROBO1 in chromosome 3p12.3,2 DYX1C1 in chromosome 15q21.3,3 and KIAA0319 and DCDC2 in chromosome 6p22.3.4, 5
The candidate genes have been studied in relation to dyslexia affection status and/or other reading-related traits in multiple studies. Some associations have supporting evidence from independent samples, consistent with the hypothesis that they play a role in shaping the biology underlying the cognitive processes on which reading relies.
Despite this, the evidence supporting the relevance of specific genetic variants that have been proposed so far remains inconclusive: some studies have been unable to replicate previous findings; in other cases, associations showed an opposite direction of effect (ie, the risk allele of the original study was found to be protective in other studies). For example, the allele T of rs6935076, a SNP in the KIAA0319 gene, was originally reported to be associated with dyslexia affection status,6 and the same allele (T) was later associated with poorer performance on a language-standardized test and reading comprehension in a different sample.7 Nonetheless, multiple successive studies reported associations with the opposite direction of effect (ie, risk allele=C),8, 9, 10 and others did not find any association between this SNP and reading measures.11, 12
It is often argued that this lack of consistency could be at least partially explained by heterogeneity across studies,9, 11, 12 occurring at various levels: from study design (eg, sample size, language), to trait characterization (eg, ascertainment criteria, age at assesment), or the genetic background of the population being studied. It is also likely that some of the associations reflect false positive findings due to incomplete control of type-I error, a common challenge for complex trait genetics.
An additional source of heterogeneity might come from variation in the educational stage at which the diagnosis of dyslexia affection status or the quantitative trait measurement took place. It has been shown that there are changes in the relationship of reading-related traits (such as phonological awareness and rapid automized naming) with reading throughout development.13, 14 There is evidence from other fields of human genetics that an age-varying effect could be one issue underlying the non-replication of some association studies.15 Despite the use of normative scores to compare across grades and efforts to account for the effects of age on reading ability, it is possible that the variability of ages within and between data sets might have contributed to the inconsistency of results. Most studies reported so far on the genetics of reading ability have been cross-sectional, where association has been tested between the SNPs of interest and dyslexia status, or quantitative traits measured only at one educational time-point per subject. Apart from potentially providing higher statistical power than cross-sectional studies of equivalent sample sizes, longitudinal studies also allow evaluation of associations that change over time.15
Learning to read involves many cognitive processes, making it difficult to disentangle whether deficits that are used to characterize people with dyslexia (eg, lower phonological awareness) are the reason for the difficulty or the consequence of a reduced experience in reading.16 The direction of causality can be better studied by looking at longitudinal samples, because they enable comparison of developmental trajectories (even starting prior to reading instruction) between children that are eventually categorized as dyslexic, and those that are not.
In one of the few longitudinal studies of the genetics of reading skills, Zhang et al.17 tested association of three SNPs in DYX1C1 and orthographic skills in relation to children's development over time. They found that rs11629841 in DYX1C1 was associated with children's orthographic judgments at ages 7 and 8 years, but less-so at age 6 years.
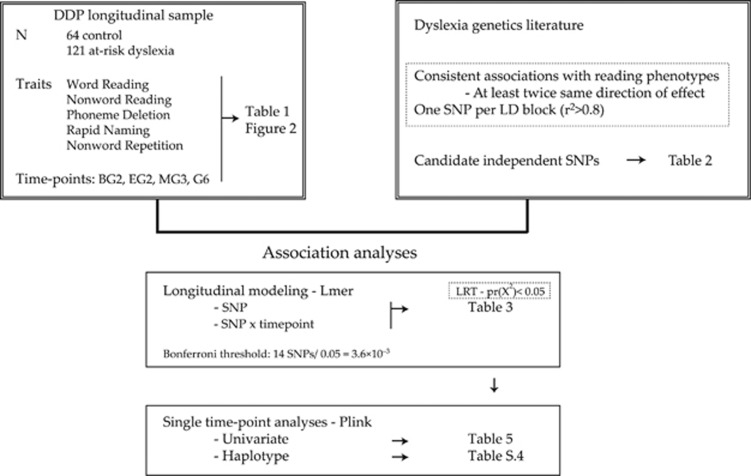
In the current study, we tested association of some of the most consistent dyslexia candidate genetic variants in an extensively characterized longitudinal sample that has not yet been studied for genetic effects (Figure 1). The Dutch Dyslexia Programme (DDP) cohort consists of children with and without familial risk for dyslexia that have been evaluated at multiple educational time-points, using psychometric measures related to the development of reading ability. In addition to studying genetic associations with measures of reading ability, the richness of this data set allowed us to look into specific endophenotypes linked to reading ability, such as speed of processing and phonological awareness.13, 18, 19 Importantly, some of these measures were taken across multiple educational time-points, allowing us to observe the trajectories of specific traits within this cohort. Two simultaneous questions could be asked about the association of a given genetic variant (SNP) and quantitative trait: (1) Does the SNP have an effect on the overall level of the trait? and (2) Does the effect of the SNP change over time?
Figure 1.
DDP longitudinal association study design. BG2, Beginning Grade 2; EG2, End Grade 2; MG3, Middle Grade 3; G6, Grade 6.
Materials and Methods
Data set
The DDP data set comprises children from families that were identified along two sets of diagnostic criteria. Some of the children were recruited based on family risk for dyslexia, that is, the child had at least one parent and another first-degree relative with self-reported dyslexia, which was confirmed by tests measuring word and pseudoword reading fluency, as described by Koster et al.20 (Nrisk=121). The remainder comprised control children without any family history of reading disability, according to the same criteria (Ncontrol=64). All children had been followed from 0 to 12 years of age within the DDP longitudinal study. Participants had Dutch as their first language, but information on ethnicity was not collected.
The present study focused on a number of reading- and language-related traits that had been measured at several educational time-points over 4 years (Table 1). This study included 185 children with both behavioral measurements and available DNA (collected through Oragene saliva kits (DNA-Genotek, Ottawa, ON, Canada)), from 180 unrelated families. Therefore, most children were unrelated, but there were three sibships of two children, and one sibship of three. Some of the children (52 at risk, 14 controls) fulfilled the dyslexia definition at time-point MG3 (mean age=8.93 years) according to the DDP criteria (ie, performing below the 10th percentile for that age on the word or nonword reading tests, or below the 25th percentile on both tests).
Table 1. Summary of the sample characteristics and longitudinal phenotypic measures available at different educational time-points.
| Educational time points (N) | BG2 (169) | EG2 (170) | MG3 (170) | G6 (116) | |
| Average age (SD) | 7.47 (0.41) | 8.14 (0.37) | 8.93 (0.36) | 12.13 (0.40) | |
| Males : Females | 102:67 | 102:68 | 103:67 | 67:49 | |
| Parental education (Na) | 3.64 (130) | 3.66 (132) | 3.66 (132) | 3.63 (91) | |
| Trait | Description | Tests | |||
| WRF | Word reading fluency | DMT | DMT | EMT | EMT |
| NWRF | Nonword reading fluency | – | Klepel | Klepel | Klepel |
| RAN | Rapid naming | RANdig | RANdig | RANdig | RANdig |
| PD | Phoneme deletion | PD1 | PD1,PD2 | PD1,PD2 | PDAKT |
| NWR | Nonword repetition | – | – | NWR | – |
Abbreviations: BG2, beginning grade 2; EG2, end grade 2; MG3, middle grade 3; G6, grade 6; Na, sample size for analysis regarding mean parental education (scale of 1–5, where 1=primary education, 5=university degree).
The acronym and description for each trait of interest is indicated, together with the name of the test that has been used to measure it in each time-point. DMT and EMT: word reading fluency tests; Klepel: nonword reading fluency test; RANdig: rapid naming of digits test; PD1, PD2, and PDAKT: phoneme deletion tests. ‘–' indicates an absence of measurement at that time-point.
The genotypic and phenotypic data have been deposited at The Language Archive (TLA, https://corpus1.mpi.nl/ds/asv/?6), under node ID: MPI2269116# (https://hdl.handle.net/1839/1C0FA0F0-1848-4890-A543-FF0329E531BE@view).
Phenotypes
A subset of measures available from the DDP was selected for testing in relation to dyslexia candidate gene variants (Table 1): word reading fluency (WRF), nonword reading fluency (NWRF), rapid naming (RAN), phoneme deletion (PD), and nonword repetition (NWR). Test reliabilities ranged from 0.73 to 0.97.21 Details of all traits measured in the DDP sample can be found in van Bergen et al.21 Datapoints were excluded as outliers, if they deviated more than 3 SDs from the relevant trait mean within the educational time-point.
Word reading fluency
WRF was measured using standard Dutch reading tests that consist of reading aloud from a list as many words as possible within one minute. Two different tests were administered depending on the grade (see Table 1): the ‘Drie Minuten Test' (DMT: three minute test: three lists, one minute each22) and the ‘Een Minuut Test' (EMT: one minute test,23). In both cases, the number of correctly read items per minute was taken as the outcome. These tests assess reading accuracy as well as fluency.
Nonword reading fluency
NWRF was measured using the ‘Klepel' nonword reading test.24 Similarly to the word reading tests, a list of 50 nonwords must be read within a time limit, in this case two minutes. The outcome measure is the number of items read correctly.
Rapid naming
Serial rapid automatized naming (RANdig)25 measures the speed of naming over-learned information. Children were asked to name five different digits, each occurring 10 times, as quickly as possible. The outcome measure is expressed as the number of digits named per second (ie, 50 item/time in seconds).
Phoneme deletion
Phonological awareness was measured using a PD task, in which a phoneme (always a consonant) had to be deleted from a nonword, resulting in another nonword.13 There was no time limit for completing this task. The task was divided in two parts (PD1 and PD2), which differed in the type of tested nonwords. In PD1, nine monosyllabic and nine disyllabic nonwords were included. In PD2, the items were nine disyllabic nonwords, in which the phoneme to be deleted occurred twice. The outcome measure for each part was the proportion of correct items. We then calculated at each educational time-point a composite score, the proportion of correct items for all available parts (PDtot).
A different PD task was used in grade 6: PDAKT (Amsterdam Phoneme Deletion Test),26 which consisted of 12 items. The outcome measure was the proportion of correct items.
Nonword repetition
NWR consisted of a test in which children had to repeat a list of 27 nonwords that were presented to them auditorily. There was no time limit for completing this task. The outcome measure was the number of correctly repeated items.
Genetic variants
Fourteen candidate SNPs were tested for association in the DDP longitudinal sample. The choice of SNPs was based on a literature review at the time of designing the study (see Table 2).4, 5, 6, 7, 8, 9, 10, 11, 12, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40 We first identified 18 SNPs that had been associated in at least two separate studies with reading-related traits (ie, with at least one of the following in a given study: dyslexia affection status, reading fluency, NWR, orthographic choice, spelling ability, phonological awareness, or discriminant score), and in a consistent manner, that is, with the same directions of allelic effect across studies. Since the data sets used in previous studies were assessed using several different measures, we considered a wide range of ‘reading-related' phenotypes to identify our candidate SNPs. However, some traits (ie, orthographic choice, spelling) were not available in our data set, and therefore we did not perform association testing with these measures, but rather with the reading-related measures that were available in the DDP data set.
Table 2. SNPs analyzed in the current study.
| SNP | Position (hg19) | Gene | Linkage region | Identified by (refs) | Risk allele | Alleles | MAF | Associated phenotype a | Consistent associations (refs) | Inconsistent association (refs) | Lack of associations (refs) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs793862 | chr6:g.24207200A>G | DCDC2 | DYX2 | 11 | A | G/A | 0.41 | Dyslexia status | 27,28,10 | 9,29 | |
| rs807701 | chr6:g.24273791G>A | DCDC2 | DYX2 | 11 | G | A/G | 0.42 | Dyslexia status | 28,10,9 | 29 | |
| rs807724 | chr6:g.24278869C>T | DCDC2 | DYX2 | 30 | C | T/C | 0.24 | Discriminant score | 28,10,9 | 29 | 12 |
| rs4504469 | chr6:g.24588884C>T | KIAA0319 | DYX2 | 4 | C | C/T | 0.23 | Single word reading | 5,6,31,7,9 | 32 | 11,33 |
| rs761100 | chr6:g.24632642A>C | KIAA0319 | DYX2 | 6 | C | C/A | 0.32 | Single word reading | 7 | 9 | |
| rs6935076 | chr6:g.24644322C>T | KIAA0319 | DYX2 | 5 | T | C/T | 0.26 | Dyslexia status | 7,34 | 8,9,10 | 11,12 |
| rs2038137 | chr6:g.24645943T>G | KIAA0319 | DYX2 | 4 | G | G/T | 0.25 | Single word reading | 5,6,31 | ||
| rs9467247 | chr6:g.24647219C>A | KIAA0319 | DYX2 | 4 | A | C/A | 0.33 | Single word reading | 35 | ||
| rs2143340 | chr6:g.24659071A>G | TTRAP | DYX2 | 4 | G | A/G | 0.15 | Single word reading | 31,9,10 | 33,29 | |
| rs759178 | chr7:g.147575112T>G | CNTNAP2 | 36 | G | G/T | 0.4 | Nonword repetition | 37 | |||
| rs17236239 | chr7:g.147582305A>G | CNTNAP2 | 36 | G | A/G | 0.24 | Nonword repetition | 37 | |||
| rs2710117 | chr7:g.147601772T>A | CNTNAP2 | 36 | A | A/T | 0.37 | Nonword repetition | 37 | |||
| rs3743204 | chr15:g.55790310G>T | DYX1C1 | DYX1 | 38 | T | G/T | 0.33 | Short-term memory | 39 | 34 | |
| rs16955705 | chr16:g.81673350A>C | CMIP | SLI1 | 40 | A | A/C | 0.36 | Nonword repetition | 10,40 |
Position (hg19): genomic coordinate in hg19, with the reference>variant alleles. Risk: allele associated with lower scores/affection status in the first study that reported the association. Alleles: major/minor. Consistent associations: significant association of the marker SNP with a reading-related trait with the same direction of effect. Inconsistent associations: significant association of the SNP marker with a reading-related trait with the opposite direction of effect. Lack of associations: tested the association of the SNP with a reading-related trait but did not replicate the association.
Strongest signal (if several) on the first study reporting the association of the SNP with reading-related abilities.
We pruned the SNP list to reduce redundancy, based on linkage disequilibrium (LD), by selecting only one SNP per LD block (EQ r2>0.8) using SNAP (CEU population).41 As a result, four SNPs were excluded (rs2179515, rs2235676, rs3212236, and rs9461045). The list of the 14 selected SNPs after pruning is summarized in Table 2.
The DDP children and their parents (N=555) were directly genotyped in-house using KASP assays (LGC Ltd., Teddington, UK). We excluded nine individuals with more than 3/14 missing genotypes (ie, a missing genotype rate exceeding 20%) from analyses. Mendelian inconsistencies were flagged using PLINK v1.07(ref. 42) and, as a result, the genotypes for one SNP were excluded in one family. The total genotyping rate in the remaining individuals was 98.8%, with a missing rate <5% for all the SNPs. All SNPs were in Hardy–Weinberg equilibrium in the unrelated parents (P>0.05). Subsequent analysis was carried out using only child trait measurements and genotypes.
Statistical analyses
Phenotypic correlations
Pearson's correlations between traits, within educational time-points, were computed using R statistical software.43 We also computed correlations for each trait across educational time-points (Supplementary Figure S1).
Longitudinal modeling of SNP effects
To examine the longitudinal dimension of the genetic effects, a linear mixed model was fitted to each trait in R using the ‘lme4' package.44 First, we fitted a null model for each standardized trait (using Blom's transformation), which consisted of a fixed-effect part and a random-effect part. All models contained the same fixed-effect terms: age, educational time-point, sex, cohort (ie, recruitment site), and group (risk and non-risk; Supplementary Table S1). They also all contained a random effect for family intercept to account for the relatedness of some of the samples. The other random effects varied per trait (see Supplementary Table S2), since the models were fitted depending on the number of repeated observations per subject that were available (ie, same trait across time-points as indicated in Table 1).
For each of Klepel, RANdig, and PDtot, three or more educational time-points were available. Thus, we included a random effect for subject intercept and a slope for age per subject, to allow children to differ in their rates of development. For each of DMT and EMT (reading fluency measures), only two educational time-points were available. Hence, it was not possible to include a random effect for slope, and we only included a random intercept per subject. Both WRF measurements (DMT and EMT) were analyzed in separate models. When a trait had only been measured at one educational time-point (ie, NWR and PDAKT), the time-point term was dropped, and the random effect part only contained the intercept for the family.
For a subset of the data set (n=132) that had information on parental education (a five-point scale ranging from ‘1' for primary school only, to ‘5' for a university degree, Table 1), we evaluated whether this factor was a significant predictor of the traits after having accounted for the other covariates specified above. Since it was not (P(χ2) >0.2 for all traits), and because including this covariate would reduce our sample size, the rest of the analyses were performed on the whole data set without accounting for parental education. Data on socioeconomic status were not collected in the DDP data set.
The effect of a SNP on the overall level of a trait was then assessed by comparing the null model with a full model, in which SNP allele dosage was included as a fixed effect (ie, additive model). There was no background information that would support modeling dominant or recessive effects for these SNPs, and multiple testing would have been increased by investigating this. Clustering rarer homozygotes together with heterozygotes assumes a particular direction of recessive/dominance relationship, which can fit the data in some instances, but in other instances will be the opposite of any true dominance/recessivity. The effect of a SNP on the trajectory was assessed by comparing the model including the SNP with a model that included the SNP and SNP × time-point interaction terms. A likelihood ratio test (LRT) between the nested models (see Equations in Supplementary B), was used to assess the significance of the term of interest (ie, ‘SNP' or ‘time-point × SNP'). The significances of the estimates were calculated using Satterthwaite approximations to determine denominator degrees of freedom, in the package ‘lmerTest'.45
For the 14 SNPs tested, the application of Bonferroni correction would set a conservative threshold for significance at P=3.6 × 10−3 (conservative because of the partial dependence of some of the SNPs due to LD). Since the five traits were not independent of each other (due to substantial correlations between traits, see Figure 2), we did not consider a further correction of P-values for multiple testing across the five traits.
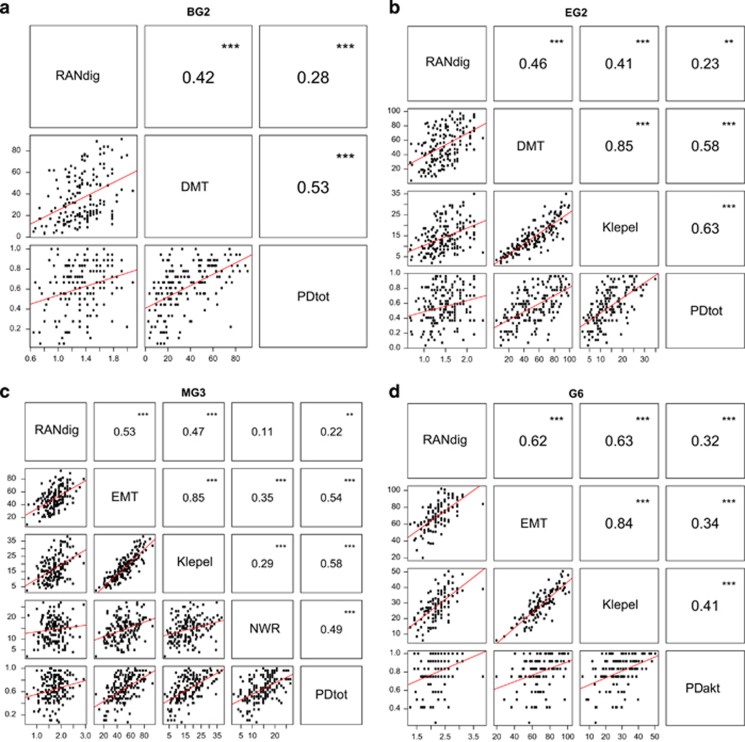
Figure 2.
Correlation panel of phenotypes of interest per educational time-point. The lower panel contains the scatter plot of the raw data for each pair of phenotypes, with a linear regression line. The values in the upper panel correspond to the Pearson's correlation coefficient, and the significance of the correlation. (a) BG2: Beginning Grade 2; (b) EG2: End Grade 2; (c) MG3: Middle Grade 3; (d) G6: Grade 6. DMT and EMT: WRF tests; Klepel: NWRF test; PD1, PD2, and PDAKT: PD tests; NWR: nonword repetition test; RANdig: rapid naming test.
Single time-point analyses
SNPs that showed significant association in the longitudinal analysis (for SNP or time-point × SNP) were further explored by testing additive linear association at each separate educational time-point using PLINK v1.07(ref. 42) (–qfam-total and permutations to correct for the sibship structure of a small minority of families). For these analyses, we first adjusted the traits for covariate effects with a predictive linear model (separately for each educational time-point). We considered age (centered by substracting the mean age) as a variable, and sex, cohort (ie, recruitment site), and group (risk and non-risk) as factors for each trait at each time-point (see Supplementary Table S1). Although not all covariates were significant predictors of all traits, we kept them in order to be consistent in the way we analyzed the different traits. Blom's transformation was used to rank normalize residuals and attain normality within each time-point.
To assess whether trait-associations of several neighbouring SNPs were independent, we performed conditional association analysis using the condition option in PLINK v1.07.
We also evaluated haplotypes for two SNPs in KIAA0319 in relation to RAN using PLINK v1.07 (–hap-assoc).
To investigate population stratification as a possible confounding factor, tests were performed assessing the equivalence of the ‘within-family' and ‘between-family' mean allelic effects using the –ap model in QTDT (Linkage Disequilibrium Analyses for Quantitative and Discrete Traits) 2.6.1(ref. 46) for all SNP-trait combinations (single time-point) that were tested with univariate analysis.
Results
The five traits were substantially inter-correlated within each educational time-point (Figure 2). Overall, the two reading fluency measures (word and nonword reading) were most highly correlated with each other (r=0.85), whereas the correlations of the other phenotypes were more moderate (r=0.11–0.62). The lowest correlations were seen between RAN and PD (PDtot and PDAKT, r=0.22–0.32), and between RAN and NWR (r=0.11). The correlation structure was largely stable across time-points, although there was some variation, for example the correlation between reading fluency and RAN increased over the educational stages.
The longitudinal assessments of the SNP effects and time-point × SNP interactions are summarized in Table 3, showing associations for which Pr(χ2)<0.05 in likelihood ratio tests (see Supplementary Table S3 for full LRT tables and Supplementary Tables S6–S16 for LME model estimates). For the SNPs that showed significant effects (either main SNP effect or time-point × SNP interaction effect), follow-up univariate association analyses per time-point are shown in Table 4. Five out of the 14 SNPs tested showed evidence of association with RAN (rs761100 and rs2038137), WRF (rs6935076), or NWR (rs17236239). A comprehensive analysis of the results for association signals is given below.
Table 3. Nominally significant associations for the SNP-fixed effect terms and time-point*SNP interaction terms from the linear mixed models.
| Time | points | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Test | Term | BG2 | EG2 | MG3 | G6 | Nind | Nfam | Alleles | Risk | Estimate | SE | df | T | Pr(>T) |
| Maj/Min | |||||||||||||||
| NWR | NWR | rs17236239 | x | 159 | 155 | A/G | A | 0.294 | 0.118 | 151.36 | 2.491 | 0.014 | |||
| WRF | EMT | rs6935076 | x | x | 161 | 154 | C/T | C | 0.217 | 0.100 | 150.54 | 2.177 | 0.031 | ||
| RAN | RANdig | rs2038137 | x | x | x | x | 167 | 163 | G/T | T | −0.187 | 0.074 | 163.67 | −2.526 | 0.012 |
| RAN | RANdig | rs761100 | x | x | x | x | 165 | 161 | C/A | A | −0.187 | 0.072 | 162.01 | −2.610 | 0.010 |
| NWRF | Klepel | x | x | x | 164 | 160 | C/A | ||||||||
| rs759178 | 0.054 | 0.084 | 174.20 | 0.601 | 0.548 | ||||||||||
| MG3:rs759178 | −0.083 | 0.037 | 180.30 | −2.231 | 0.027 | ||||||||||
| G6:rs759178 | 0.024 | 0.072 | 129.50 | 0.325 | 0.745 |
Abbreviations: NWRF, nonword reading fluency; WRF, word reading fluency; RAN, rapid naming; NWR, nonword repetition.
Test: dependent variable in the model. Risk: allele associated with lower scores. The estimates for the SNP are for the centered-dependent variables specified in the model. For the time-point*SNP interaction term, estimates for the centered-dependent variables specified on the model are given for the marker per each level of the time-point (except for the baseline, ie, EG2). T-values for the coefficient estimates and associated P-values are shown. The degrees of freedom (df) are estimated with Satterthwaite's approximation.
Table 4. Plink univariate association results per educational time-point (BG2, EG2, MG3, G6).
| BG2 | EG2 | MG3 | G6 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Test | SNP | N | β | EMP1 | N | β | EMP1 | N | β | EMP1 | N | β | EMP1 |
| NWRF | Klepel | rs759178 | 160 | −0.162 | 0.167 | 160 | 0.011 | 0.929 | 110 | −0.076 | 0.584 | |||
| WRF | DMT | rs6935076 | 156 | 0.243 | 0.058 | 157 | 0.243 | 0.042 | ||||||
| EMT | rs6935076 | 157 | 0.245 | 0.047 | 110 | 0.280 | 0.054 | |||||||
| RAN | RANdig | rs761100 | 158 | −0.339 | 0.001 | 161 | −0.294 | 0.009 | 162 | −0.286 | 0.009 | 111 | −0.256 | 0.062 |
| RANdig | rs2038137 | 160 | −0.302 | 0.006 | 163 | −0.290 | 0.014 | 164 | −0.307 | 0.005 | 112 | −0.248 | 0.063 | |
| NWR | NWR | rs17236239 | 159 | 0.283 | 0.025 |
Abbreviations: N: number of individuals in the analysis. β: regression coefficient for the major allele; EMP1, empirical P-values (10 000 permutations); BG2, beginning grade 2; EG2, end grade 2; MG3, middle grade 3; G6, grade 6; NWRF, nonword reading fluency; WRF, word reading fluency, RANdig, rapid naming; NWR, nonword repetition.
The tests for population stratification did not show significant differences of the between-family and within-family components of association for most of these SNPs (rs759178, rs761100, rs2038137, and rs17236239: P>0.05), suggesting that population stratification was unlikely to be a substantial confounding factor in the analysis for these SNPs. However, these tests yielded nominally significant stratification effects for rs6935076 and WRF tests at two time-points (DMT: pBG2=0.022; EMT: pG6=0.029). These specific SNP-trait combinations had not shown significant associations in these two time-points (DMT: pBG2=0.058; EMT: pG6=0.054) suggesting that if stratification played any role in our analyses, it was to mask possible effects rather than create spurious signals.
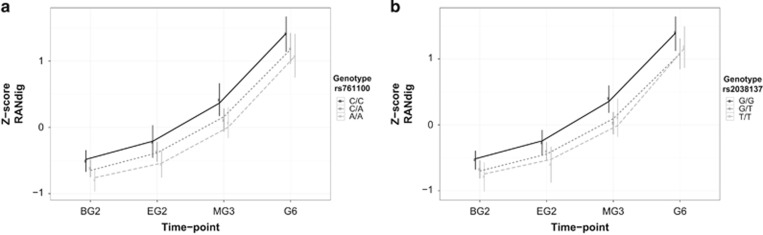
RAN of digits was nominally associated with two neighbouring SNPs in KIAA0319 (Table 3: rs761100 χ2(1)=6.927, P=0.009; rs2038137 χ2(1) =6.496, P=0.011), the minor alleles being associated with slower naming. These results were independent of educational time-point, since the interactions between the SNPs and time-point were not significant. The single time-point analysis reflected the same signal, showing significant associations of these two SNPs at multiple educational time-points, the minor alleles consistently yielding lower scores (Table 4, Figure 3); the most significant association was at beginning of grade 2 for rs761100 (pEG2=0.001) and at the middle of grade 3 for rs2038137 (pEG2=0.005).
Figure 3.
Z-standardized performance on RAN (RANdig) per educational time-point and genotypic groups of (a) rs761100 and (b) rs2038137. The lines represent the fitted residuals to the mixed linear model including the allelic state of the SNP as a variable, and the points and error bars are the mean and SD of the mean per time-point.
Rs761100 and rs2038137 are located 13 kb apart in the 5′ untranslated region (UTR) of the KIAA0319 gene, and they are in LD with each other (r2=0.735, for the 1000G CEU population).41 When both SNPs were modeled together as fixed effects for association with RAN, the second SNP was not a significant predictor. Similarly, the association was no longer significant, when conditioning the test of one of the SNPs on the other (Supplementary Table S5). Haplotype analyses per educational time-point indicated that the two minor alleles rs761100-A and rs2038137-T form the risk haplotype (P≤0.01 for all time-points except G6, Supplementary Table S4).
Rs6935076, another SNP in KIAA0319, was associated with WRF (Table 3: DMT: χ2(1)=3.568, P=0.059; EMT: χ2(1)=4.861, P=0.027). This association was confirmed in two of the educational stages by the single time-point analysis (Table 4: DMT: pEG2=0.042; EMT: pMG3=0.047). Although this SNP is located between rs761100 and rs2038137, the two SNPs that were associated with RAN, it is in low LD with these (r2= 0.21–0.28) and did not itself show any association with RAN (RANdig: χ2(1)=1.042, P=0.307).
The longitudinal analysis showed an educational time-point-dependent association between the CNTNAP2 variant rs759178 and NWRF (χ2(2)=7.131, P=0.028; Table 3, Supplementary Figure S2). Of note, there were no significant differences in nonword reading scores between the genotypes at the individual time-points themselves (Table 4).
We found that rs17236239, a second SNP in CNTNAP2, was associated with NWR, both via linear regression in R (Table 3, χ2(1)=6.380, P=0.012) and PLINK (Table 4, P=0.025).
We did not find significant associations between PD measures (PDtot and PDAKT) and any of the SNPs tested.
Discussion
In this study, we performed candidate gene association analyses in a Dutch sample with longitudinal measures for several reading-related measures. This data set consisted of a richly phenotyped sample, in which genetic associations could be detected via intermediate measures related to the cognitive processes involved in reading (such as PD and RAN) collected at multiple educational time-points. Based on a literature search, we selected and genotyped 14 SNPs that had been associated with dyslexia and/or relevant quantitative traits, consistently in at least two separate studies, and found in the prominent candidate genes DYX1C1, DCDC2, KIAA0319, CMIP, and CNTNAP2. We modeled the data longitudinally to assess overall and educational time-dependent effects of these SNPs on WRF and NWRF, NWR, RAN, and PD. A number of nominally significant associations were observed, and detailed single time-point analyses of these associations confirmed that most were consistent across educational time-points. Below, we discuss the results across the different analyses, considering them in relation to the pool of existing data currently available in the field of dyslexia genetics.
The most significant association that we found in the DDP sample was between RAN and two SNPs, rs761100 and rs2038137, in the 5′UTR of the KIAA0319 gene. The ability to rapidly name a limited set of well-known items is considered a measure of processing speed; it tackles the timing mechanisms necessary for the automaticity required in advanced stages of reading development1, 18 and is one of the strongest predictors of reading ability in pre-literate children.18 Moreover, superior parental RAN proficiency has been shown to be a protective factor for children at familial risk for dyslexia in the DDP sample,18, 21 suggesting that it is an important intergenerational precursor of reading.47 However, the effect of these SNPs on WRF in our samples is at best only marginally significant (DMT, rs761100, χ2(1)=2.697, P=0.1). This could reflect heterogeneity in reading strategies, since the overall variance in reading is explained by other factors beyond RAN, and those other factors might not be affected by these SNPs. On the other hand, we also found that rs6935076, another SNP in the same region of KIAA0319, was associated with WRF in the DDP sample, although to a lesser extent and not at all educational time-points, but was not associated with RAN.
RAN has not often been investigated in previous genetic studies of reading ability; it has been included in only three linkage studies48, 49, 50 and a small number of recent association studies.51 One of these studies found evidence of linkage for the composite score of RAN on 6p21.48 close to the DYX2 locus spanning KIAA0319. The linkage was not found in two other studies that also included several RAN measures.49, 50 The lack of consistency across studies is a long-standing problem for the field, in part due to the heterogeneity across studies at various levels, such as study design, sample size, ascertainment scheme, and population.52
A recent study that tested for association between RAN of digits and dyslexia candidate SNPs in a Chinese population, found that this trait was nominally associated with several SNPs in KIAA0319,51 including rs2038137 (P=0.025), one of the associated SNPs in the present study. This same SNP was also associated with scores on Chinese dictation and phonological awareness in the Chinese sample. However, the direction of effect of this SNP in the present study was not congruent with previous reports. We found the minor allele T to be associated with lower scores, but it was originally reported that the major allele G was associated with reduced performance on word reading, orthographic choice, and spelling,4 and this association has been observed in additional studies with this same direction of effect (ie, risk allele=G).5, 6
The other SNP that we found to be associated with RAN was rs761100, also in KIAA0319. This SNP was first found associated with several quantitative traits including reading and spelling, and the risk allele was reported to be the major allele C,10 opposite to our direction of effect (risk allele=minor T). However, another study found that the minor allele was associated with reduced expressive language in a sample of children with specific language impairment.9 This SNP was also included in a recent cross-linguistic meta-analysis across several European samples, and its minor allele T was nominally associated with lower spelling scores in the meta-analysis, although not in any separate subsamples.53 This SNP was not included in the Chinese study that investigated RAN.51
We observed association between NWR and rs17236239, a CNTNAP2 SNP that was selected based on its previous association with this trait. However, the DDP sample showed the opposite direction of effect to that previously reported.36, 37
We did not find association between any of the SNPs and PD, which is a measure of phonological awareness that has been repeatedly associated with candidate SNPs in prior literature.4, 51
Another question that we asked concerned the stability of associations across different educational time-points. Overall, most associations that we found were time-independent. When looking at the single time-point analysis, we did observe that some of the association signals differed at distinct educational stages, mainly becoming less significant at the latest time-point (G6, mean age=12.1 years). This drop in significance may relate to the drop of sample size in the latest stages of the project, rather than indicating a decrease of the genetic effect on these traits as the age increases. Moreover, our time-span ranged only from age 7 to 12 years, and for some traits of interest the measurement instrument was not constant across educational time-points (eg, reading fluency with DMT and EMT), which broke the longitudinal analysis into two steps. These factors, together with our moderate sample size, might have reduced our chances of detecting genetic effects that are sensitive to reading experience. Nevertheless, we did observe one suggestive finding in our longitudinal analysis: an interaction of rs759178, NWRF, and educational time-point. This interaction involved a smaller difference across genotypes in the middle time-point (MG3, mean age=8.9 years) compared with the other earlier and later ages (Supplementary Figure S2). This result is difficult to interpret biologically, and a wider time-span might be required to understand the effect that rs759178 has on the trajectory of NWRF development. Alternatively, this result may be a false positive association. Nevertheless, it also illustrates how cross-sectional studies could miss associations that are present only at certain educational stages. Longitudinal analysis of genetic effects in reading ability and related quantitative traits is a potentially powerful method that has been underexploited so far, and should be considered whenever this type of data is available, as in the DDP cohort.
Another strength of the DDP cohort is the richness of the assessment, involving several quantitative traits. Even when the effects that we observed were stable in time, the availability of multiple reading- and language-related traits permitted a detailed understanding of the type of process that the genetic variation could be affecting.
The literature on candidate genetic variants for reading is difficult to interpret, as reflected by the summary in Table 2. Recent efforts have tried to integrate evidence across studies, to get insights into the relevance of these candidate SNPs for dyslexia. For example, the NeuroDys consortium meta-analyzed association results for 19 SNPs (including eight that we analyzed in the present study) across several European samples, but did not find any significant association after correcting for multiple comparisons.53 Such efforts have been highly constrained by heterogeneity and limited availability of any given trait measurement across studies. One source of study heterogeneity is the orthography of the language (eg, more transparent orthography in Dutch versus a more complex orthography in English). It is thought that the relationship between reading-related cognitive abilities and reading skills varies depending on the orthographic system. For example, it has been proposed that PD and RAN digits have a stronger impact for predicting developmental dyslexia in more complex orthographies.19 Thus, it might be important to reconsider the available data on the genetic studies of reading, taking into account factors such as orthographic complexity.
The main limitation of the present study is its moderate sample, which is not well powered to detect small effect sizes. However, the DDP data set consists of a very well-characterized sample at the phenotypic level, and we have evaluated some of the most intensively studied candidate SNPs for dyslexia in a longitudinal data set for the first time, while the previously reported effect sizes for many of these SNPs were large enough to be detected in comparatively sized data sets.
Future genetic studies of reading-related traits will probably depend on increasing power by meta-analyzing many of the available samples, an approach that has proven successful for other complex traits. The present longitudinal study reminds us that there are also non-genetic dimensions that should be accounted for, including the educational time-point.
Acknowledgments
The genetic studies of the DDP were funded by grant 200-62-305 from the Netherlands Organization for Scientific Research (NWO) as part of the research programme ‘The genetic dissection of developmental dyslexia'. The longitudinal study of the DDP was funded by grants 200-62-302/303/304 from The Netherlands Organization for Scientific Research (NWO) under the title ‘Early Precursors of Familial Dyslexia: A Prospective Longitudinal Study' as part of the research programme ‘Identifying the core features of developmental dyslexia'. AC-C, CF, and SEF are supported by the Max Planck Society (Germany). BF is supported by a Vici grant from the Netherlands Organization for Scientific Research (NWO; grant 016-130-669). We thank Britt Hakvoort and Ellie van Setten for testing the participants. Many thanks to all of the participants in the study.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Barbara Franke received an educational speaking fee from Merz.
Supplementary Material
References
- Pennington BF: From single to multiple deficit models of developmental disorders. Cognition 2006; 101: 385–413. [DOI] [PubMed] [Google Scholar]
- Hannula-Jouppi K, Kaminen-Ahola N, Taipale M et al: The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet 2005; 1: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nopola-Hemmi J, Taipale M, Haltia T, Lehesjoki AE, Voutilainen A, Kere J: Two translocations of chromosome 15q associated with dyslexia. J Med Genet 2000; 37: 771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francks C, Paracchini S, Smith SD et al: A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am J Hum Genet 2004; 75: 1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope N, Harold D, Hill G et al: Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet 2005; 76: 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Paracchini S, Scerri T et al: Further evidence that the KIAA0319 gene confers susceptibility to developmental dyslexia. Mol Psychiatry 2006; 11: 1085–1091. [DOI] [PubMed] [Google Scholar]
- Rice ML, Smith SD, Gayan J: Convergent genetic linkage and associations to language, speech and reading measures in families of probands with specific language impairment. J Neurodev Disord 2009; 1: 264–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto JM, Livne-Bar I, Huang K et al: Association of reading disabilities with regions marked by acetylated H3 histones in KIAA0319. Am J Med Genet B 2010; 153B: 447–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury DF, Paracchini S, Scerri TS et al: Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. Behav Genet 2011, Jan 41: 90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scerri TS, Morris AP, Buckingham LL et al: DCDC2, KIAA0319 and CMIP are associated with reading-related traits. Biol Psychiatry 2011; 70: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Anthoni H, Dahdouh F et al: Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am J Hum Genet 2006; 78: 52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkanac Z, Chapman NH, Matsushita MM et al: Evaluation of candidate genes for DYX1 and DYX2 in families with dyslexia. Am J Med Genet B 2007; 144B: 556–560. [DOI] [PubMed] [Google Scholar]
- de Jong P, van der Leij A: Developmental changes in the manifestation of a phonological deficit in dyslexic children learning to read a regular orthography. J Educ Psychol 2003; 95: 22–40. [Google Scholar]
- Wagner RK, Torgesen JK, Rashotte CA et al: Changing relations between phonological processing abilities and word-level reading as children develop from beginning to skilled readers: a 5-year longitudinal study. Dev Psychol 1997; 33: 468–479. [DOI] [PubMed] [Google Scholar]
- Lasky-Su J, Lyon HN, Emilsson V et al: On the replication of genetic associations: timing can be everything!. Am J Hum Genet 2008; 82: 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami U: Sensory theories of developmental dyslexia: three challenges for research. Nat Rev Neurosci 2015; 16: 43–54. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li J, Tardif T et al: Association of the DYX1C1 dyslexia susceptibility gene with orthography in the Chinese population. PLoS ONE 2012; 7: e42969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Leij A, van Bergen E, van Zuijen T: de Jong P, Maurits N, Maassen B: Precursors of developmental dyslexia: an overview of the longitudinal Dutch Dyslexia Programme study. Dyslexia 2013; 19: 191–213. [DOI] [PubMed] [Google Scholar]
- Landerl K, Ramus F, Moll K et al: Predictors of developmental dyslexia in European orthographies with varying complexity. J Child Psychol Psychiatry 2013; 54: 686–694. [DOI] [PubMed] [Google Scholar]
- Koster C, Been PH, Krikhaar EM, Zwarts F, Diepstra HD, Van Leeuwen TH: Differences at 17 months: productive language patterns in infants at familial risk for dyslexia and typically developing infants. J Speech Lang Hear Res 2005; 48: 426–438. [DOI] [PubMed] [Google Scholar]
- van Bergen E, de Jong PF, Plakas A, Maassen B, van der Leij A: Child and parental literacy levels within families with a history of dyslexia. J Child Psychol Psychiatry 2012; 53: 28–36. [DOI] [PubMed] [Google Scholar]
- Verhoeven L: Drie-Minuten-Toets (DMT). Arnhem, The Netherlands: CITO, 1995. [Google Scholar]
- Brus BT, Voeten MJM: Een-Minuut-Test [One-Minute-Test]. Swets & Zeitlinger: Lisse, The Netherlands, 1972. [Google Scholar]
- van den Bos KP, Lutje Spelberg HC, Scheepstra AJM, de Vries JR: De klepel: Een test voor de leesvaardigheid van pseudowoorden [The Klepel: A Test for the Reading Skills of Pseudowords]. Swets & Zeitlinger: Lisse, The Netherlands, 1994. [Google Scholar]
- van den Bos KP: Serieel benoemen en woorden lezen [Serial naming and Word Reading]. Rijksunversiteit Groningen: Groningen, The Netherlands, 2003. [Google Scholar]
- van Bergen E, Bishop D, van Zuijen T: de Jong PF: How does parental reading influence children's reading? A study of cognitive mediation. Sci Stud Read 2015; 19: 325–339. [Google Scholar]
- Deffenbacher KE, Kenyon JB, Hoover DM et al: Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: linkage and association analyses. Hum Genet 2004; 115: 128–138. [DOI] [PubMed] [Google Scholar]
- Wilcke A, Weissfuss J: Kirsten H, Wolfram G, Boltze J, Ahnert P: The role of gene DCDC2 in German dyslexics. Ann Dyslexia 2009; 59: 1–11. [DOI] [PubMed] [Google Scholar]
- Cope N, Eicher JD, Meng H et al: Variants in the DYX2 locus are associated with altered brain activation in reading-related brain regions in subjects with reading disability. Neuroimage 2012; 63: 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Smith SD, Hager K et al: DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci USA 2005; 102: 17053–17058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paracchini S, Steer CD, Buckingham LL et al: Association of the KIAA0319 dyslexia susceptibility gene with reading skills in the general population. Am J Psychiatry 2008; 165: 1576–1584. [DOI] [PubMed] [Google Scholar]
- Venkatesh SK, Siddaiah A, Padakannaya P, Ramachandra NB: Analysis of genetic variants of dyslexia candidate genes KIAA0319 and DCDC2 in Indian population. J Hum Genet 2013; 58: 531–538. [DOI] [PubMed] [Google Scholar]
- Luciano M, Lind PA, Duffy DL et al: A haplotype spanning KIAA0319 and TTRAP is associated with normal variation in reading and spelling ability. Biol Psychiatry 2007; 62: 811–817. [DOI] [PubMed] [Google Scholar]
- Darki F, Peyrard-Janvid M, Matsson H, Kere J, Klingberg T: Three dyslexia susceptibility genes, DYX1C1, DCDC2, and KIAA0319, affect temporo-parietal white matter structure. Biol Psychiatry 2012; 72: 671–676. [DOI] [PubMed] [Google Scholar]
- Dennis MY, Paracchini S, Scerri TS et al: A common variant associated with dyslexia reduces expression of the KIAA0319 gene. PLoS Genet 2009; 5: e1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Newbury DF, Abrahams BS et al: A functional genetic link between distinct developmental language disorders. N Engl J Med 2008; 359: 2337–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse AJ, Bishop DV, Ang QW, Pennell CE, Fisher SE: CNTNAP2 variants affect early language development in the general population. Genes Brain Behav 2011; 10: 451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahdouh F, Anthoni H, Tapia-Paez I et al: Further evidence for DYX1C1 as a susceptibility factor for dyslexia. Psychiatr Genet 2009; 19: 59–63. [DOI] [PubMed] [Google Scholar]
- Bates TC, Lind PA, Luciano M, Montgomery GW, Martin NG, Wright MJ: Dyslexia and DYX1C1: deficits in reading and spelling associated with a missense mutation. Mol Psychiatry 2010; 15: 1190–1196. [DOI] [PubMed] [Google Scholar]
- Newbury DF, Winchester L: Addis L et al: CMIP and ATP2C2 modulate phonological short-term memory in language impairment. Am J Hum Genet 2009; 85: 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ: de Bakker PI: SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008; 24: 2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team A Language and Environment for Statistical Computing. Vienna, Austria, 2014, http://www.R-project.org.
- Bates D, Maechler M, Bolker B, Walker S: lme4: Linear Mixed-Effects Models Using Eigen and S4. R package version 1: 1–82015, http://CRAN.R-project.org/package=lme4.
- Kuznetsova A, Bruun Brockhoff P, Haubo Bojesen Christensen R: lmerTest: Tests in Linear Mixed Effects Modelsc. R package version 2: 0–252015, http://CRAN.R-project.org/package=lmerTest. [Google Scholar]
- Abecasis GR, Cardon LR, Cookson WO: A general test of association for quantitative traits in nuclear families. Am J Hum Genet 2000; 66: 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bergen E, van der Leij A: de Jong PF: The intergenerational multiple deficit model and the case of dyslexia. Front Hum Neurosci 2014; 8: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig IR, Schumacher J, Hoffmann P et al: Mapping for dyslexia and related cognitive trait loci provides strong evidence for further risk genes on chromosome 6p21. Am J Med Genet B 2011; 156B: 36–43. [DOI] [PubMed] [Google Scholar]
- Rubenstein KB, Raskind WH, Berninger VW, Matsushita MM, Wijsman EM: Genome scan for cognitive trait loci of dyslexia: rapid naming and rapid switching of letters, numbers, and colors. Am J Med Genet B 2014; 165B: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CG, Franke B, Hol FA et al: Confirmation of dyslexia susceptibility loci on chromosomes 1p and 2p, but not 6p in a Dutch sib-pair collection. Am J Med Genet B 2008; 147: 294–300. [DOI] [PubMed] [Google Scholar]
- Lim CK, Wong AM, Ho CS, Waye MM: A common haplotype of KIAA0319 contributes to the phonological awareness skill in Chinese children. Behav Brain Funct 2014; 10: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, DeFries JC: Developmental dyslexia: genetic dissection of a complex cognitive trait. Nat Rev Neurosci 2002; 3: 767–780. [DOI] [PubMed] [Google Scholar]
- Becker J, Czamara D, Scerri TS et al: Genetic analysis of dyslexia candidate genes in the European cross-linguistic NeuroDys cohort. Eur J Hum Genet 2014; 22: 675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.