Abstract
Fragile-X syndrome (FXS) is a frequent genetic form of intellectual disability (ID). The main recurrent mutagenic mechanism causing FXS is the expansion of a CGG repeat sequence in the 5′-UTR of the FMR1 gene, therefore, routinely tested in ID patients. We report here three FMR1 intragenic pathogenic variants not affecting this sequence, identified using high-throughput sequencing (HTS): a previously reported hemizygous deletion encompassing the last exon of FMR1, too small to be detected by array-CGH and inducing decreased expression of a truncated form of FMRP protein, in three brothers with ID (family 1) and two splice variants in boys with sporadic ID: a de novo variant c.990+1G>A (family 2) and a maternally inherited c.420-8A>G variant (family 3). After clinical reevaluation, the five patients presented features consistent with FXS (mean Hagerman's scores=15). We conducted a systematic review of all rare non-synonymous variants previously reported in FMR1 in ID patients and showed that six of them are convincing pathogenic variants. This study suggests that intragenic FMR1 variants, although much less frequent than CGG expansions, are a significant mutational mechanism leading to FXS and demonstrates the interest of HTS approaches to detect them in ID patients with a negative standard work-up.
Introduction
Fragile-X syndrome (FXS) is the most common known form of inherited intellectual disability (ID) and its neurodevelopmental phenotype often overlaps autism spectrum disorder (ASD).1, 2 FXS occurs in both genders with an estimated prevalence around 1/5000 in males and 1/8000 in females due to variable expressivity and incomplete penetrance. The high prevalence, the inconstant morphological phenotype especially in young children (long habitus, a long narrow face, prominent forehead, large prominent ears, hyperextensible finger joints, single palmar crease and macroorchidism at puberty) and the behavioral features, which are not always discriminative (hyperactivity and/or short attention span, tactile defensiveness, hand flapping and/or hand-biting, poor eye contact, perseverative speech),3 justify systematic FXS genetic testing in all patients with unexplained ID or autism spectrum disorder.4
FXS is caused by the absence of Fragile-X Mental Retardation Protein (FMRP), an intracellular RNA binding protein. This absence dysregulates dendritic signaling pathways and affects brain hippocampal synaptic plasticity5, 6 through incompletely understood mechanisms. The main recurrent mutagenic mechanism leading to the absence of FMRP is CGG-repeat expansion in the 5′-untranslated region (5′UTR) of FMR1,7 inducing abnormal methylation of this region followed by transcriptional silencing. This repeat expansion is believed to explain at least 99% of FXS cases.8 Around 1–2% of all children with developmental delay are found to have FXS caused by a CGG expansion.4 However, others disease-causing FMR1 variants have also been reported, including large gene deletions, 5′UTR or exon 1 deletions associated or not with a full FMR1 CGG expansion in the mother,9, 10, 11, 12, 13, 14, 15 as well as a very small number of single-nucleotide variants (SNVs) in the FMR1 coding regions. Direct sequencing of FMR1 was not frequently carried out, and only a few groups have screened for variants in FMR1 coding regions in clinically relevant cohorts presenting16, 17 or not18, 19, 20 alterations of FMRP at the protein level. Two other studies were conducted on unspecific ID patients.21, 22 This lack of FMR1 sequence screening has led to a marked deficit in the description of small exonic deletions or SNVs.
In this study, we report the clinical and molecular descriptions of three FMR1 variants identified by targeted high-throughput sequencing (HTS) of several hundred ID genes in 940 patients with unexplained ID and negative results following routine FXS screening: a maternally inherited deletion of the last exon of FMR1 previously reported in a family with presumed X-linked ID23 and two novel intronic substitutions, one de novo and one maternally inherited, affecting splicing in two sporadic male patients with ID. We also reviewed the other FMR1 variants described in the literature in patients with ID and concluded that six of them are convincingly pathogenic or likely pathogenic. Together with our results, there are now nine intragenic pathogenic variants in FMR1 outside the 5′UTR causing FXS.
Patients and methods
Patients
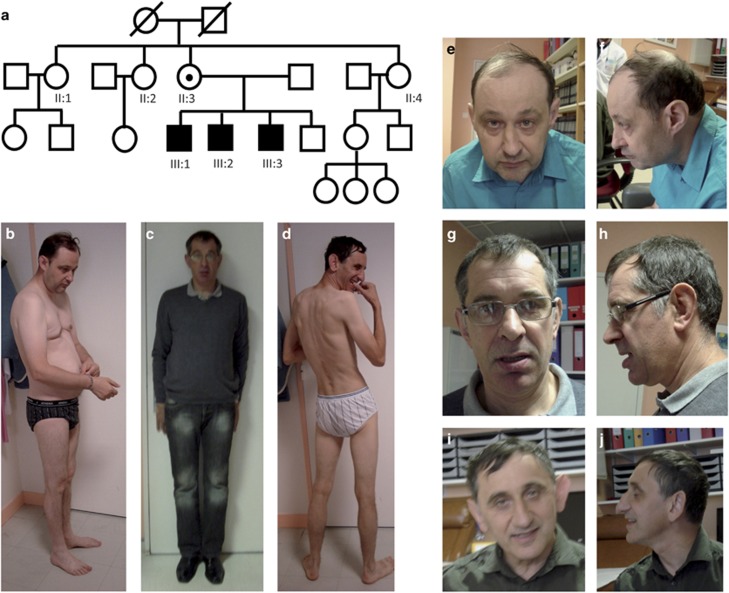
Patients III:1, III:2 and III:3 from pedigree 1 are three male siblings aged 50, 48 and 47 years. All display ID and were negative for routine FXS testing and array-CGH analysis, despite a suspected X-linked inheritance pattern (Figure 1a). They were part of the series of 100 patients with marfanoid habitus (MH) and an associated ID-phenotype published in 2013.24 Proband III:1 was included in a cohort of 106 ID patients who underwent targeted HTS of 217 ID genes and for whom we previously reported the sequencing results.23 Patients from pedigree 2 (Figure 3a) and from pedigree 3 (Figure 3b) presented undiagnosed ID and underwent routine FXS testing and array-CGH analysis. They were both included in second series of 834 additional ID patients for whom we sequenced coding regions of 275–451 ID genes. This study was approved by the local Ethics Committee of the Strasbourg University Hospital (Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale-CCPPRB). For all patients, written informed consent for genetic testing was obtained from their legal representative. For all the patients presented in this paper, families also gave consent for the publication of photographs.
Figure 1.
Pedigree (a) and photographs of the first family of three affected brothers with ID (patient III:1 (b, e, f), patient III:2 (c, g, h), patient III:3 (d, i, j)).
Targeted high-throughput sequencing, bioinformatic pipeline and variant ranking
DNA was prepared as described previously.25 It was enriched in coding sequences of 275–451 genes known to cause ID, including FMR1, by target custom capture (SureSelect; Agilent, Santa Clara, CA, USA). Libraries were prepared separately for each patient. Parental DNA was pooled untagged by 14–16 (mothers and fathers separately) and each pool was treated as one sample for the library preparation. The enriched libraries were tagged and pooled by 32 in one lane of an Illumina sequencer (HiSeq2500) for a 100 bp paired-end run. Read mapping and variant calling were performed following standard procedures, and variants were annotated and filtered using VaRank via Alamut HT (Interactive Biosoftware, Rouen, France).23, 26 Candidate variants harboring a frequency compatible with the incidence of ID were selected.27 We retrieved candidate variants reported in EVS (http://evs.gs.washington.edu/EVS/) or ExAC (http://exac.broadinstitute.org/) databases in no more than one male for variants in X-linked genes (as we cannot exclude the possibility, although unlikely, that a particular carrier from the general population might have mild ID). Pools of untagged parental DNA (mothers and fathers separately) were analyzed to check for the presence of each candidate variation. Remaining variants, predicted as potentially pathogenic and matching the mode of inheritance associated with the affected gene, were further tested for validation. The potential effect of variants on splicing was analyzed using BDGP Splice Site Prediction by Neural Network (Nnsplice),28 MaxEntScan,29 Human Splicing Finder30 and GeneSplicer31 via Alamut HT. Putative heterozygous/homozygous/hemizygous copy number variants (CNVs) were highlighted using the previously described method based on a depth-of-coverage comparison between the index sample and eight other random samples from the same sequencing lane.25 For the X-chromosome, coverage was normalized according to the patient's gender. Confirmation of FMR1 SNVs by Sanger sequencing and analysis of the consequences of FMR1 variants at the RNA and protein levels are detailed in Supplementary Methods. Once confirmed, the three variants were submitted to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/): SCV000328672.1, SCV000328671.1 and SCV000328670.1.
Reverse clinical phenotyping
All patients were reevaluated using the Hagerman checklist.3 Patients from family 1 underwent clinical and behavioral evaluations after obtaining the molecular diagnosis in order to determine if they were a posteriori compatible with the FXS phenotype. The index patient III:1 had a standardized assessment by a multidisciplinary team, including a clinical geneticist, a psychiatrist and a neuropsychologist, in the presence of his referral caregiver, in order to provide a neurodevelopmental, psychocognitive, emotional and adaptive picture of his functioning. The clinical examination comprised a neurological evaluation, the examination of malformative or dysmorphic features (face, trunk, extremities and whole body; Figure 1). The interview concerned sleeping, feeding and digestive features. HistoFry data were collected using the Achenbach System of Empirically Based Assessment for Adult, ASEBA. The Wechsler Adult Intelligence Scale-Fourth Editionwas used. It included verbal and non-verbal tasks and four index scores: the Verbal Comprehension Index, the Perceptual Reasoning Index, the Working Memory Index and the Processing Speed Index. The Full Scale Intelligence Quotient was determined, as were specific index skills, significant gaps between subtests, and more generally strengths and weaknesses of the intellectual cognitive profile. Patient III:2 was also assessed for social cognitions using the Social Perception domain of the Neuropsychological Assessment-Second Edition, with an analysis of the recognition of facial emotions. Finally, the Vineland Adaptive Behavior Scales-Second Edition (Vineland-II, VABS) was used in a semistructured interview with the referral caregiver, to determine ages of development in each area.32 It assesses socialization (interpersonal relationships, games and leisure, coping), communication (receptive, expressive and written), autonomy in daily living activities (personal, domestic, community) and motor skills (gross, fine). Sibling patients III:1 and 3 were phenotyped in their specialized residential institution in the presence of the care and educational team by a psychiatrist and a clinical research assistant trained in administering the Vineland Adaptive Behavior Scales-Second Edition (Vineland-II, VABS).
Literature and database review of FMR1 variations
We compiled the rare non-synonymous variations previously identified in the different studies, which screened for FMR1 coding sequences in patients with ID over the last 20 years. These studies can be classified into three groups. The first group concerned studies conducted on a cohort of ID patients with a phenotype evocative of FXS but negative after CGG expansion testing.16, 17, 18, 19, 20 FMR1 coding regions were screened for single-nucleotide SNV (using heteroduplex analysis, single-strand conformation analysis, genomic or cDNA Sanger sequencing) either directly or only in patients in whom the FMRP protein was found to be absent (by western blot). The second category of studies analyzed FMR1 coding sequences in male ID patients, with no specific clinical orientation, using high-resolution melting, Sanger or HTS).21, 22, 23 The third one included large-scale studies in which FMR1 was sequenced together with other genes in unspecific ID (large panels, X-exome, exome, genome).33, 34, 35, 36, 37, 38, 39, 40, 41 For each non-synonymous variant, we compiled genetic and functional evidence of its pathogenicity and its frequency in general population database such as Exome Variant Server. The clinical neurodevelopmental and morphological data available for patients were also compiled and classified as highly, moderately, poorly or not evocative of FXS. HTS studies were also considered: 1228 ID individuals from large ID panels including FMR1,39, 40, 41 627 patients who underwent X-exome sequencing 35, 36, 38 and 1325 patients analyzed by WES.33, 34, 37, 42
Results
FMR1 exon 17 deletion previously identified in three brothers with ID leads to a truncated FMRP protein (family 1)
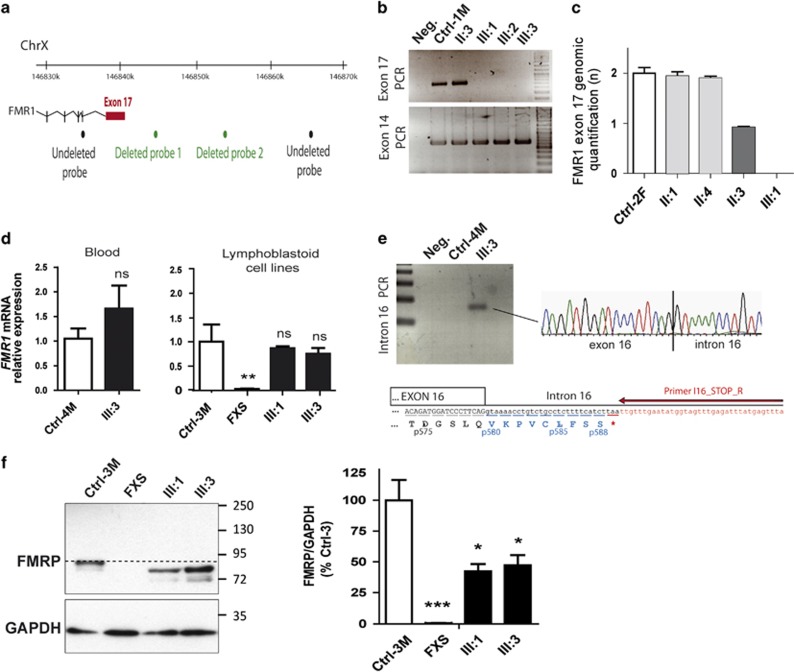
In family 1 (Figure 1a), deletion of the last exon of FMR1 (exon 17) was previously detected by targeted HTS of 217 genes performed on patient III:1.23 No reads were obtained for this exon in the patient while a high depth of coverage was observed for the other samples of the same sequencing batch (depth of coverage of 210±92 reads for males; Supplementary Table S1). An array-CGH previously performed on the patient was reanalyzed a posteriori and confirmed this result, revealing two consecutive probes (below the detection threshold of three) deleted in this region (Figure 2a). The deletion of exon 17 was confirmed by PCR using primers located at either side of the exon revealing no amplification in the three affected brothers (III:1, III:2 and III:3; Figure 2b). Genomic qPCR analysis revealed that this deletion was present at the heterozygous state in their unaffected mother and was not found in their two maternal aunts, leading to a reassuring genetic counseling for the other family members (Figure 2c). RT-qPCR showed that FMR1 RNA was expressed at a normal level in blood extract and lymphoblastoid cell lines from patient III:3 (Figure 2d), suggesting it escapes nonsense-mediated mRNA decay. We investigated the consequences of the loss of exon 17 and its acceptor splice site on FMR1 mRNA. We observed a retention of the beginning of intron 16 in FMR1 mRNA from patient III:3 while this was not observed in mRNA from an unrelated individual 4 (Figure 2e). This insertion of intronic sequence, r.1737_1738ins1737+1_?, leads to a frameshift resulting in a premature stop codon at position 589 instead of 633: p.Ile580fs*9. Western blot analysis confirmed the existence of a truncated form of FMRP protein in patient's cells with an ~10% reduction in size (~75 vs ~85 kDa) consistent with the loss of 44 amino acids (Figure 2f). The expression of this truncated form in patients was lower that the expression of normal FMRP protein observed in controls (~50% lower levels of truncated FMRP in lymphoblastoid cells from patients III:1 and III:3), suggesting reduced stability of the truncated protein.
Figure 2.
Identification of a deletion of exon 17 of FMR1 leading to a truncated FMRP protein in the three brothers (first family). (a) Location of the deleted array-CGH probes. (b) PCR amplification of FMR1 exon 14 and exon 17 (Ctrl-1M, unrelated male individual 1; Neg, experiment negative control). (c) Relative quantification of genomic DNA by qPCR amplification of exon 17 (Ctrl-2F, unrelated female individual 2). (d) Relative quantification of FMR1 mRNA by qPCR analysis on blood cDNA and on mRNA extracted from lymphoblastoid cell lines (n=6) (Ctrl-3M and 4M, unrelated males individuals 3 and 4). (e) PCR amplification of blood cDNA using a reverse primer located at the beginning of intron 16. (f) Western blot analysis of FMRP protein expression on lymphoblastoid cell lines from FXS, patients III:1 and III:3 and unrelated male control individual 3. Normal size of FMRP protein is indicated by the dotted line. Semiquantification of FMRP protein level (normalized to GAPDH) shows a reduction in FMRP expression in patients' cell lines compared with controls (n=5; Student's t-test: ns, nonsignificant; *P<0.05; **P<0.01; ***P<0.001).
The three affected brothers of this family experienced a diagnostic odyssey lasting many years. Patient III:1, aged 50, is a protected adult under tutorship, living in a specialized institution with patient III:3. He is the most severely affected of the siblings. He has a narrow, elongated face with a prominent forehead (Figure 1), macroorchidia and mild gynecomastia. When examined, he was non-verbal pronouncing only syllables and a few simple words. He suffered from currently controlled epilepsy. He presents behavioral problems (long-standing food selectivity, opposition attitudes, one episode of aggressiveness towards others and himself). Patient III:2, aged 48, presents moderate ID and is a protected adult under tutorship, living and working in a specialized institute. He has a long, thin habitus in spite of standard measurements, and an elongated face with thick lower lip (Figure 1). He was described as fearful, avoided eye contact and had an anxious bearing. He did not express himself spontaneously and answered questions briefly in a few words. He had developed coping skills for daily life situations. Patient III:3, aged 47, was a protected adult under tutorship. He had a long, thin habitus in spite of standard measurements, a narrow, elongated face and prominent ears and forehead. He had macroorchidia. He was non-verbal. He did not present any aggressiveness towards others or self-injurious behavior. He developed social relationships despite some non-pervasive rituals and stereotypies. Complete neuropsychological evaluations were performed to assess the severity of impairments and their impact on intellectual functioning and adaptative behavior (Vineland, Wechsler Adult Intelligence Scale-Fourth Edition, Neuropsychological Assessment-Second Edition, Supplementary Information and Table 1). Hagerman scores for the three brothers were calculated. Two of the three brothers had a score above 16 (Table 2).
Table 1. Results of Vineland scales in the four domains, communication, autonomy, socialization and motricity, in the three siblings of family 1.
| III:1 | III:2 | III:3 | |
|---|---|---|---|
| Communication | 1 year 6 months | 3 years 4 months | 1 year 6 months |
| Receptive | 1 year 4 months | 3 years 11 months | 1 year 6 months |
| Expressive | 1 year 8 months | 3 years 3 months | 1 year 7 months |
| Written | 1 year 6 months | <3 years | 1 year 6 months |
| Autonomy | 3 years 7 months | 6 years 2 months | 4 years 6 months |
| Personal | 3 years 7 months | 7 years 8 months | 4 years 6 months |
| Familial | 4 years 1 month | 6 years 5 months | 4 years 9 months |
| Social | 3 years 1 month | 5 years 6 months | 4 years 1 month |
| Socialization | 1 year 6 months | 3 years 1 month | 3 years 3 months |
| Interpersonal relationships | 1 year 11 months | 2 years 4 months | 2 years 11 months |
| Games and leisure | <1 year | 1 year 5 months | 8 years |
| Adaptation | 4 years 9 months | 1 year 4 months | 7 years 5 months |
| Motricity | 2 years | 5 years 3 months | 2 years 7 months |
| Global | 1 year 10 months | >5 years 11 months | 2 years 2 months |
| Fine | 1 year 4 months | 5 years 2 months | 3 years |
Table 2. Hagerman scores for the five patients (according to Hagerman et al., 1991).
| Patient III:1 Family 1 | Patient III:2 Family 1 | Patient III:3 Family 1 | Proband Family 2 | Proband Family 3 | |
|---|---|---|---|---|---|
| Intellectual deficiency | 2 | 2 | 2 | 2 | 2 |
| Hyperactivity | 1 | 1 | 2 | 0 | 0 |
| Short attention span | 2 | 2 | 2 | 0 | 2 |
| Tactilely defensive | 2 | 1 | 1 | 2 | 1 |
| Hand flapping | 0 | 0 | 0 | 2 | 2 |
| Hand biting | 0 | 0 | 0 | 2 | 2 |
| Poor eye contact | 2 | 2 | 2 | 2 | 2 |
| Perseverative speech | 2 | 1 | 2 | 0 | 2 |
| Hyperextensible MP joints | 1 | 1 | 1 | 2 | 1 |
| Large or prominent ears | 1 | 1 | 2 | 1 | 0 |
| Large testicles | 2 | 1 | 2 | 0 | 0 |
| Simian crease or sydney line | 0 | 0 | 0 | 2 | 0 |
| Family history of ID | 2 | 2 | 2 | 0 | 0 |
| Total score | 17 | 14 | 18 | 15 | 14 |
0, not present; 1, borderline or present in the past; 2, definitely present.
Identification of two novel intronic variants affecting FMR1 splicing (families 2 and 3)
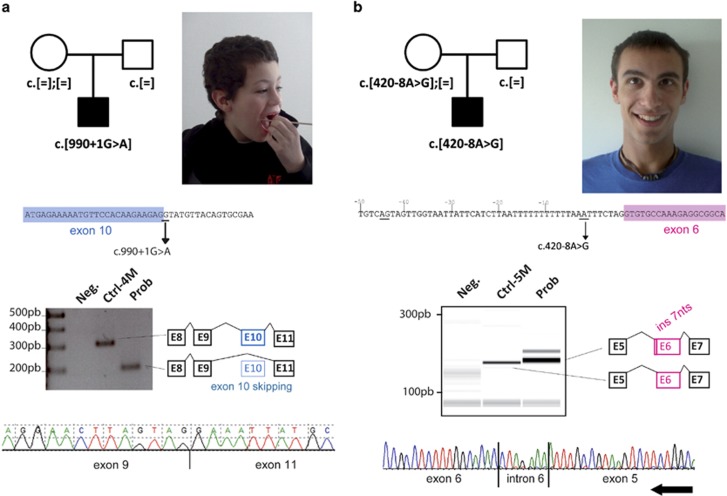
In the second family (Figure 3a), targeted HTS of 451 ID genes revealed a substitution c.990+1G>A, predicting to disrupt the splice donor site of exon 10, in one boy presenting sporadic ID. This substitution was not present in maternal blood DNA suggesting a de novo inheritance. Analysis of RNA from the patient's blood demonstrated that this variant led to an aberrant splicing event, skipping exon 10 entirely. The loss of this 110pb exon (r.881_990del) leads to a frameshift after amino acid 294 and to the appearance of a premature stop codon: p.Lys295Asnfs*11. The resulting truncated protein lacks essential domains such as the nuclear export signal and the RGG domain responsible for the interaction of FMRP with target mRNAs. The proband, aged 10 years, was the first child of healthy parents, with no other case of ID in the family. A younger brother had normal development. The pregnancy and delivery were uneventful. His birth length (50 cm), weight (3350 g) and head circumference (34.5 cm) were normal. His motor skills were delayed as he could not walk before the age of 24 months. At age 3 years, he had no language and normal hearing. At that time, he began to show features of repetitive behavior as well as hand flapping, and avoided physical contact. Physical examination revealed hypotonia with valgus feet and knees. He had a high forehead, a coarse face, a wide mouth with an everted lower lip. His ears were of normal size, but abnormally shaped (Figure 3a). He had clinodactyly of the fifth fingers, square feet with short toes and a shawl scrotum. At age 10 years, his height was at +1 SD, weight at −0.5 SD, and head circumference at the mean. His behavior was marked by autistic traits, making physical examination difficult. He did not have any neuropsychological evaluation. He needed to be educated in a specialized school. His shawl-shaped scrotum could have suggested an Aarskog syndrome, but the lack of other major signs as facial dysmorphism and growth retardation excluded this diagnosis. Metabolic analyses, electroencephalogram and cerebral magnetic resonance imaging were normal, as well as CGG-repeat test of the FMR1 gene, DMPK (type 1 dystrophic myotonia) and ARX genes, and array-CGH. No pathogenic variant was found in FGD1. His Hagerman score was calculated as high (15/26) considering his young age (no testicular phenotype observable) and the absence of familial history (consistent with de novo status of the variant), but he presented no attention deficit disorder (Table 2).
Figure 3.
Identification of two novel FMR1 splice variants in two boys with sporadic ID (second and third families). (a) Pedigree and photograph of the patient from the second family carrying a de novo c.990+1G>A variant. This variant affects the donor splice site of exon 10 and leads to the skipping of the entire exon in FMR1 mRNA. (b) Pedigree and photograph of the patient from the third family carrying a c.420-8A>G variant. This variant alters the splice acceptor site of exon 6 and leads to the use of alternative intronic cryptic sites.
In the third family, targeted HTS of 451 ID genes revealed an intronic substitution c.420-8A>G (Figure 3b). This substitution was present in maternal blood. This variation was predicted to decrease the use of the normal splice acceptor site according to three prediction programs (score decrease for SpliceSite Finder: −100%, Genesplicer: −100% and MaxEnt: −52%, no change for Nnsplice and Human Splicing Finder). It also created a cryptic intronic acceptor site with high prediction scores of being used (SpliceSite Finder: 83.8 (0–100), MaxEnt: 8.1 (0/16), Nnsplice: 1 (0–1), Genesplicer: 5 (0–15) and Human Splicing Finder: 84 (0–100)). Analysis of mRNA in the patient's blood demonstrated that this variant led to at least one aberrant splicing event (Figure 3c) with the use of this cryptic intronic acceptor site, leading to the retention of seven nucleotides of intron 5 (r.419_420ins420-7_420-1). This leads to a frameshift after amino acid 140 with the appearance of a premature stop codon three amino acids downstream, p.Met140Ilefs*3. The resulting truncated protein lacks essential domains such as the nuclear export signal and the RGG domain responsible for the interaction of FMRP with target mRNA. This proband, aged 20 years, was the second child of unrelated healthy parents. The older sister does not present any cognitive or health problems. The pregnancy was characterized by maternal depression without exposure to teratogens; the delivery was uneventful. His birth measurements and APGAR scores were normal (length: 52 cm, weight: 3310 g, head circumference: 33.5 cm, APGAR 10/10). He presented a strabismus that was corrected by surgery at the age of 7 years. He had axial hypotonia in his first year and walked at the age of 23 months. Language delay was noticed without hearing impairment. He presented stereotypies and features of repetitive behavior such as rocking and hand flapping, and a high level of change-related anxiety. He went to a specialized school. At age 20.5 years, he had a normal height, weight and head circumference (176.6 cm, 59.2 kg and 57.5 cm). A long face and mild clinodactyly of the third and fifth fingers were noted. Karyotype, array-CGH and the FMR1 CGG expansion test were negative. Standard metabolic tests did not reveal any anomalies and investigations for ARX recurrent 24 bp duplication or 22q11 microdeletion were returned negative. No X-inactivation bias was identified in the patient's mother. Brain magnetic resonance imaging, performed at 21 months, did not reveal any significant abnormalities: a mildly short splenium was noted. His Hagerman score was 14/26 (Table 2).
Other FMR1 point variations reported in individuals with ID and in the general population
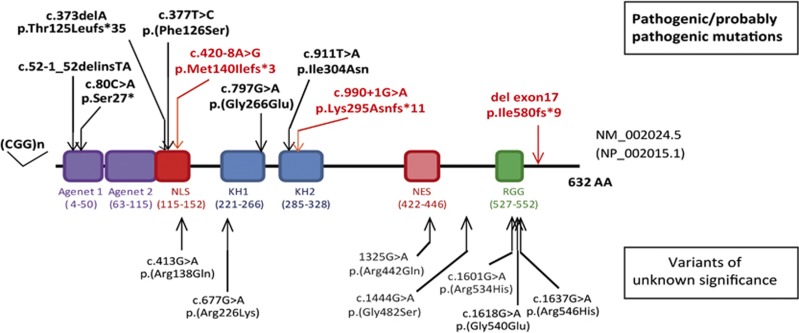
Whereas it is clear that CGG-repeat expansion in the FMR1 promoter is by far the most frequent cause of FXS, other types of variations, particularly FMR1 deletions, have also been reported in FXS.9 Deletions of the entire gene or of its first exon,10, 11, 12 associated or not with full FMR1 CGG expansion in the mother,13, 14, 15 have been found by conventional CGG expansion testing or by microarray analysis. Since 1992, screening for variations in the FMR1 coding sequence, performed in Fragile-X-evocative or nonspecific ID patients without expanded CGG-repeats and subsequent methylation,16, 17, 18, 19, 20, 22, 23 has led to the identification of several rare intragenic non-synonymous variations within the FMR1 locus (Table 3 and Supplementary Table S2). We have also compiled the rare non-synonymous variants identified in FMR1 during large-scale studies (large panel, X-exome or whole-exome sequencing) performed on ID patients by other teams: only missense variants but no truncating variants were identified in the 3180 individuals tested.33, 34, 35, 36, 37, 38, 39, 40, 41, 42 We reanalyzed all these variants in FMR1 in the light of new public sequencing data (Exome Variant Server and the ExAC project, supposed to be free or at least not enriched in severe developmental disorders), prediction tools and functional evidence. Altogether, six previously reported variants could also be reclassified as disease-causing (pathogenic-class 5 or likely pathogenic-class 4 variants, summarized in Table 3 and in Figure 4). Half of them are truncating changes (p.Thr125Leufs*35, c.52-1_52delinsTA and p.Ser27*) and the other half are missense changes (p.(Phe126Ser), p.(Gly266Glu) and p.Ile304Asn) not reported in EVS nor ExAC, occurring de novo and/or and having a functional impact in vitro (neurons) or in vivo (knock-in mice).16, 17, 18, 19 For the other missense variants reported, some are predicted to be damaging (SIFT, Polyphen2), but neither the amino acid changes by themselves nor the phenotypes of patients make it possible to draw any clear conclusions about their pathogenicity and we thus considered them ‘variants of uncertain significance' (VUS).11, 20, 21, 22 Some missense variants were even classified as ‘likely benign'. One of them, p.Asp438Asn, located in the nuclear localization signal of FMR1, was identified by targeted HTS of 217 ID genes in two brothers with no morphological symptoms of FXS but with FXS neurodevelopmental phenotype: further familial segregation analysis showed that one unaffected half-brother also carries this variation too thus suggesting that it is a benign variant.23 Besides these non-synonymous variations, an intronic variant, c.990+14C>T, first described as altering the splicing of exon 10 was reported 20 years ago in three unrelated ID males,20 but this result was controversial since this variation was also identified in healthy control individuals.43 Data from ExAC confirmed the polymorphic status of this variation, present at the hemizygous state in more than 4000 males of this general population.
Table 3. Review of the intragenic FMR1 variants previously identified in ID patients.
| Variation | Exon affected | Reference | Pathogenicity | Inheritance | FXS phenotype |
|---|---|---|---|---|---|
| c.52-1_52delinsTA; p.? | Exon 2 | 17 | Pathogenic | Maternally inherited | +++ |
| c.80C>A; p.Ser27* | Exon 2 | 16 | Pathogenic | Maternally inherited | ++/+++ |
| c.373delA; p.Thr125Leufs*35 | Exon 5 | 17 | Pathogenic | De novo | ++ |
| c.420-8A>G; p.Met140Ilefs*3 | Exon 6 | This report | Pathogenic | Maternally inherited | ++ |
| c.990+1G>A; p.Lys295Asnfs*11 | Exon 10 | This report | Pathogenic | De novo | ++ |
| g.(?_ 147030202)_(147046357 _?) del; p.Ile580fs*9 | Exon 17 | 23This report | Pathogenic | Maternally inherited | +++ |
| c.377T>C; p.(Phe126Ser) | Exon 5 | 42 | Likely pathogenic | De novo | NA |
| c.797G>A; p.(Gly266Glu) | Exon 8 | 19 | Likely pathogenic | NA | +++ |
| c.911T>A; p.Ile304Asn | Exon 10 | 18 | Likely pathogenic | De novo | ++ |
| c.413G>A; p.(Arg138Gln) | Exon 5 | 21 | Unknown significance | NA | − |
| c.677G>A; p.(Arg226Lys) | Exon 8 | 41 | Unknown significance | NA | NA |
| c.1325G>A; p.(Arg442Gln) | Exon 14 | 42 | Unknown significance | Maternally inherited | NA |
| c.1444G>A; p.(Gly482Ser) | Exon 14 | 22 | Unknown significance | NA | NA |
| c.1601G>A; p.(Arg534His) (2x) | Exon 15 | 22 | Unknown significance | Maternally inherited | NA |
| Maternally inherited | + | ||||
| c.1618G>A; p.(Gly540Glu) | Exon 15 | 35 | Unknown significance | NA | NA |
| c.1637G>A; p.(Arg546His) | Exon 15 | 20 | Unknown significance | NA | NA |
Abbreviations: ASD, autism spectrum disorder; CNV, Copy Number variant; FMRP, Fragile-X mental retardation protein; FSIQ, Full Scale Intelligence Quotient; FXS, Fragile-X syndrome; HTS, high-throughput sequencing; ID, intellectual disability; NA, clinical data not available. PRI, Perceptual Reasoning Index; PSI, Processing Speed Index; VCI, Verbal Comprehension Index; WMI, Working Memory Index. Phenotype highly (+++), moderately (+), poorly (+) or not (−) evocative of an FXS. Nomenclatures c are indicated in NM_002024.5 isoform; g, references to Hg19 human genome version.
Figure 4.
Representation of the FMRP protein with the different FMR1 pathogenic variants and variants of unknown significance identified in individuals with ID. KH, K homology domain; NES, nuclear export signal; NLS, nuclear localization signal; RGG, glycine–arginine rich domain.
In this ExAC general population, three variations predicted to affect splice sites are reported at the hemizygous state: a recurrent disruption of the exon 12 splice acceptor site present in 44 hemizygous males, and two unique substitutions of either the exon 12 splice donor site or the exon 16 splice acceptor site. The skipping of these exons is thought to lead to in-frame deletions in FMRP protein sequence (21 amino acids lost for exon 12 and 8 amino acids for exon 16). Moreover, exon 12 is found alternatively spliced and included in only some of the different isoforms. This suggests that exon 12 and its encoded amino acids (a.a. 376-396 in NM_002024.5) might be dispensable. A total of 29 missense variants affecting the longest isoform NM_002024.5 of FMR1 predicted to have benign or damaging effects are also reported at the hemizygous state in 236 males of the ExAC population (Supplementary Table S3). Therefore, data from this general population underline that one should interpret non-synonymous variations identified in the FMR1 gene with caution, even if they affect conserved amino acids and are predicted as damaging, and especially if patients are not clinically evocative of FXS.
Discussion
We report here the cases of three brothers with ID of presumed X-linked inheritance and a phenotype compatible with FXS and two sporadic male patients from other families, all negative for routine FXS testing. In the first family, deletion of the last exon of FMR1 (exon 17) detected by targeted HTS of 217 genes performed on index patient23 was confirmed in the two affected brothers by PCR. This deletion led to a decreased expression level of a truncated form of FMRP. These are the first cases of a single exon deletion not comprising the 5′UTR or exon 1 of the FMR1 gene, and therefore the first deletion to escape both expansion testing and CNV search by array-CGH. This illustrates the power of HTS in identifying small CNVs. In the second and third families, variants that altered splicing and led to frameshifts and early truncated proteins were identified.
FXS is almost exclusively diagnosed by molecular testing for CGG repeat expansion. Indeed, as FXS is not always clinically obvious, especially for prepubertal children, the CGG-repeat test, which is cheap and fast, is the standard of care for any child not meeting developmental milestones and is one of the most frequently ordered genetic tests. Among all children with developmental delay, the actual diagnostic yield of FXS testing is only 1–2%.4 CGG-repeat expansion in the FMR1 promoter is the most frequent cause of FXS, but deletions of the entire FMR1 gene10, 11, 12 or of the 5′UTR have also been reported in FXS.9 Rare intragenic non-synonymous variations have also been identified within the FMR1 locus16, 17, 18, 19, 20, 22, 23, 35, 41, 42 but after reanalysis in the light of new public sequencing data, prediction tools and functional evidence, we showed that only six could be reclassified as convincing.
Altogether, in the cohort of 940 French individuals with nonspecific ID and a negative FMR1 CGG expansion test, three intragenic pathogenic FMR1 variants were identified, thus accounting for a non-negligible proportion of ID cases (0.3%). However, if we consider the other HTS studies performed on nonspecific ID patients, the mutation yield is lower: only one convincing disease-causing missense variant p.(Phe126Ser) in 3180 individuals with ID (plus four variants of unknown significance). Therefore, except for recurrent CGG expansion, the FMR1 gene is infrequently mutated in ID as compared with other genes of the same or a smaller size, such as RPS6KA3 or MECP2.44
The six patients reported in the literature and carrying convincing pathogenic variants in FMR1 mainly come from series of patients with clinical features evocative of FXS. They all present a long face and large ears, and except for the prepubertal patient, macroorchidism. We describe here five other patients with unexplained ID, who were negative for Fragile-X testing of the CGG expansion, who carry FMR1 changes. Fine reverse phenotyping of the three adult siblings of the first family carrying the exon 17 deletion revealed the dysmorphism and behavioral phenotype of the FXS spectrum. They present various dysmorphic features that have been reported in FXS, although more typical in patient III:2 than in his siblings, and two of them had macroorchidism. Although nonspecific, their cognitive evaluations were also compatible with FXS. Two of the brothers obtained high scores for the FXS evaluation established by Hagerman et al.3 (17/26 and 18/26 for individual III:1 and III:3, respectively), corresponding to what is observed in patients carrying the CGG-repeat expansion (mean score of 17.3). The third brother had a lower score (14/26), as his dysmorphic features were less pronounced, but his score was still above the mean score obtained for non-FXS ID patients.3 Reverse phenotyping was more difficult in the young boy from the second family and the young adult of the third family, since the dysmorphic features and macroorchidism tend to appear or increase with age, but they had some neuropsychological features, especially autistic traits, consistent with FXS. In contrast, they both presented without hyperactivity and without macroorchidism and had moderately high Hagerman scores (15/26 and 14/26 respectively). It was particularly difficult to suspect FXS in the patient of the second family and, due to the de novo occurrence of this variant, there was no history of ID in his family. Together, considering the five patients described here and the six from the literature carrying pathogenic variants in FMR1, we found no obvious clinical differences between patients with intragenic changes compared and those with classic CGG expansions. The two boys of the second and third families present a milder phenotype than do the three affected brothers of the first family, even though they carry variants leading to more prematurely truncated FMRP protein, lacking several essential functional domains. This study confirmed that other types of FMR1 changes, besides classic CGG expansions, which could not be diagnosed through standard tests, may cause an FXS phenotype.
In conclusion, this report enlarges the molecular description of FXS, with the description of five individuals carrying intragenic pathogenic variants of FMR1. In total, nine pathogenic non-synonymous changes (loss of function and missense variants) are now described in FMR1 in 11 individuals with a compatible FXS phenotype. We demonstrate the interest of HTS in identifying FMR1 intragenic deletions and SNVs in ID patients with a negative standard work-up. The increasing use of HTS in CGG expansion negative patients with ID will lead to the identification of all types of variants (SNVs and CNVs) affecting FMR1 coding sequences and determine the true frequency of FXS in ID. However, classical CGG expansion will still need to be tested separately, as current HTS methods fail to detect them. Making a diagnosis of FXS is of particular importance at two levels. First, genetic counseling for patients' families is essential due to the high risk of recurrence of this dominant X-linked form of ID. Second, numerous targeted therapeutics for FXS are being developed,45, 46 and some FXS patients are already benefiting from some of these treatments.47, 48
Acknowledgments
We thank the families for their participation to the study. We also thank the French Ministry of Health (PHRC National 2008) as well as the Regional Council of Burgundy, the Fondation Jerome Lejeune, the Agence de la Biomédecine, CREGEMES, the Agence Nationale de la Recherche for their financial support. IGBMC is supported by the ANR-10-IDEX-0002-02. Angelique Quartier was supported by MENESR et Fondation Recherche Médicale. We thank Ricardos Tabet and people from the IGBMC sequencing platform for their technical and bioinformatic support. We also thank the Centre National de Génotypage (Jean-François Deleuze, Robert Olaso, Anne Boland and the technicians and bioinformaticians) for their participation in library preparation and DNA sequencing. We are very grateful to Philip Bastable for language editing.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Boyle L, Kaufmann WE: The behavioral phenotype of FMR1 mutations. Am J Med Genet C Semin Med Genet 2010; 154C: 469–476. [DOI] [PubMed] [Google Scholar]
- Tranfaglia MR: The psychiatric presentation of fragile x: evolution of the diagnosis and treatment of the psychiatric comorbidities of fragile X syndrome. Dev Neurosci 2011; 33: 337–348. [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Amiri K, Cronister A: Fragile X checklist. Am J Med Genet 1991; 38: 283–287. [DOI] [PubMed] [Google Scholar]
- Rauch A, Hoyer J, Guth S et al: Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A 2006; 140: 2063–2074. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST: Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron 2008; 60: 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ et al: Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci USA 1997; 94: 5401–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberle I, Vincent A, Abbadi N et al: New polymorphism and a new chromosome breakpoint establish the physical and genetic mapping of DXS369 in the DXS98-FRAXA interval. Am J Med Genet 1991; 38: 336–342. [DOI] [PubMed] [Google Scholar]
- Jacquemont S, Birnbaum S, Redler S, Steinbach P, Biancalana V: Clinical utility gene card for: fragile X mental retardation syndrome, fragile X-associated tremor/ataxia syndrome and fragile X-associated primary ovarian insufficiency. Eur J Hum Genet 2011; 19: e-pub ahead of print 4 May 2011 doi:10.1038/ejhg.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RD: Mutation spectra in fragile X syndrome induced by deletions of CGG*CCG repeats. J Biol Chem 2009; 284: 7407–7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Ikeda M, Budimirovic DB, Hjelm LN, Kaufmann WE, Warren ST: Mosaic FMR1 deletion causes fragile X syndrome and can lead to molecular misdiagnosis: a case report and review of the literature. Am J Med Genet A 2008; 146A: 1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Huang W, Xia Q, Du Q, Wu L, Duan R: Mutational analyses of the FMR1 gene in Chinese pediatric population of fragile x suspects: low tolerance for point mutation. J Child Neurol 2015; 30: 803–806. [DOI] [PubMed] [Google Scholar]
- Vengoechea J, Parikh AS, Zhang S, Tassone F: De novo microduplication of the FMR1 gene in a patient with developmental delay, epilepsy and hyperactivity. Eur J Hum Genet 2012; 20: 1197–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond LS, Macias MM, Tarleton JC, Shashidhar Pai G: Fragile X syndrome and deletions in FMR1: new case and review of the literature. Am J Med Genet 1997; 72: 430–434. [PubMed] [Google Scholar]
- Luo S, Huang W, Xia Q et al: Cryptic FMR1 mosaic deletion in a phenotypically normal mother of a boy with fragile X syndrome: case report. BMC Med Genet 2014; 15: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveiros MT, Santos MD, Dos Santos JM et al: Screening for fragile X syndrome in males from specialized institutions in the northeast region of Brazil. Genet Mol Res 2015; 14: 6897–6905. [DOI] [PubMed] [Google Scholar]
- Gronskov K, Brondum-Nielsen K, Dedic A, Hjalgrim H: A nonsense mutation in FMR1 causing fragile X syndrome. Eur J Hum Genet 2011; 19: 489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugenbeel KA, Peier AM, Carson NL, Chudley AE, Nelson DL: Intragenic loss of function mutations demonstrate the primary role of FMR1 in fragile X syndrome. Nat Genet 1995; 10: 483–485. [DOI] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E et al: A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet 1993; 3: 31–35. [DOI] [PubMed] [Google Scholar]
- Myrick LK, Nakamoto-Kinoshita M, Lindor NM, Kirmani S, Cheng X, Warren ST: Fragile X syndrome due to a missense mutation. Eur J Hum Genet 2014; 22: 1185–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Lin ML, Lin SJ, Li YC, Li SY: Novel point mutation within intron 10 of FMR-1 gene causing fragile X syndrome. Hum Mutat 1997; 10: 393–399. [DOI] [PubMed] [Google Scholar]
- Collins SC, Bray SM, Suhl JA et al: Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am J Med Genet A 2010; 152A: 2512–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handt M, Epplen A, Hoffjan S, Mese K, Epplen JT, Dekomien G: Point mutation frequency in the FMR1 gene as revealed by fragile X syndrome screening. Mol Cell Probes 2014; 28: 279–283. [DOI] [PubMed] [Google Scholar]
- Redin C, Gerard B, Lauer J et al: Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J Med Genet 2014; 51: 724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callier P, Aral B, Hanna N et al: Systematic molecular and cytogenetic screening of 100 patients with marfanoid syndromes and intellectual disability. Clin Genet 2013; 84: 507–521. [DOI] [PubMed] [Google Scholar]
- Redin C, Le Gras S, Mhamdi O et al: Targeted high-throughput sequencing for diagnosis of genetically heterogeneous diseases: efficient mutation detection in Bardet-Biedl and Alstrom syndromes. J Med Genet 2012; 49: 502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy V, Pizot C, Redin C et al: VaRank: a simple and powerful tool for ranking genetic variants. PeerJ 2015; 3: e796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton A, Redin C, Mandel JL: XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am J Hum Genet 2013; 93: 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D: Improved splice site detection in Genie. J Comput Biol 1997; 4: 311–323. [DOI] [PubMed] [Google Scholar]
- Eng L, Coutinho G, Nahas S et al: Nonclassical splicing mutations in the coding and noncoding regions of the ATM Gene: maximum entropy estimates of splice junction strengths. Hum Mutat 2004; 23: 67–76. [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C: Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009; 37: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Lin X, Salzberg SL: GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res 2001; 29: 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow SS, Cicchetti DV: The behavior inventory for rating development (BIRD): assessments of reliability and factorial validity. Appl Res Ment Retard 1984; 5: 219–231. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BW et al: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Srour M, Capo-Chichi JM et al: De novo mutations in moderate or severe intellectual disability. PLoS Genet 2014; 10: e1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Haas SA, Chelly J et al: X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry 2016; 21: 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips AK, Siren A, Avela K et al: X-exome sequencing in Finnish families with intellectual disability – four novel mutations and two novel syndromic phenotypes. Orphanet J Rare Dis 2014; 9: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E et al: Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012; 380: 1674–1682. [DOI] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E et al: A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet 2009; 41: 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F, Caro-Llopis A, Rosello M et al: High diagnostic yield of syndromic intellectual disability by targeted next-generation sequencing. J Med Genet, e-pub ahead of print 12 September 2016 doi:10.1136/jmedgenet-2016-103964. [DOI] [PubMed]
- Tzschach A, Grasshoff U, Beck-Woedl S et al: Next-generation sequencing in X-linked intellectual disability. Eur J Hum Genet 2015; 23: 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozeva D, Carss K, Spasic-Boskovic O et al: Targeted next-generation sequencing analysis of 1,000 individuals with intellectual disability. Hum Mutat 2015; 36: 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD et al: Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 2015; 385: 1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JB, Gurling HM: Point mutation in intron 10 of FMR1 is unlikely to be a cause of fragile X syndrome. Hum Mutat 1998; 12: 431–432. [DOI] [PubMed] [Google Scholar]
- Chelly J, Mandel JL: Monogenic causes of X-linked mental retardation. Nat Rev Genet 2001; 2: 669–680. [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Des-Portes V, Gasparini F, Jacquemont S, Gomez-Mancilla B: Translating molecular advances in fragile X syndrome into therapy: a review. J Clin Psychiatry 2014; 75: e294–e307. [DOI] [PubMed] [Google Scholar]
- Jacquemont S, Berry-Kravis E, Hagerman R et al: The challenges of clinical trials in fragile X syndrome. Psychopharmacology (Berl) 2014; 231: 1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiss Hess L, Fitzpatrick SE, Nguyen DV et al: A randomized, double-blind, placebo-controlled trial of low-dose sertraline in young children with fragile X syndrome. J Dev Behav Pediatr 2016; 37: 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh MJ, Nguyen DV, Mu Y et al: A randomized double-blind, placebo-controlled trial of minocycline in children and adolescents with fragile x syndrome. J Dev Behav Pediatr 2013; 34: 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.