Abstract
Background
Epithelial to mesenchymal transition (EMT) contributes to metastases in various types of tumors, and is also the key step in the breast cancer metastatic cascade. In our previous study, a mouse model containing human-derived normal breast tissue was established and allowed EMT/MET process of human breast cancer cells to be mimicked in a humanized mammary microenvironment.
Material/Methods
Two-dimensional electrophoresis (2-DE) and mass spectrometry were used to detect different proteins between parental MDA-MB-231 and its variant sub-line obtained from tumors grown in transplanted normal human breast tissue (MDA-MB-231br). We knocked down the ezrin in 2 cell lines (MDA-MB-231 and SUM1315). The migration and invasion ability was assessed. EMT markers were examined by real-time reverse transcription PCR analysis and Western blot analysis. The relationship of ezrin with cortactin was tested by tissue microarray and co-immunoprecipitation.
Results
Proteomic analysis revealed 81 differentially expressed proteins between parental MDA-MB-231 and MDA-MB-231br. Among these proteins, the expression of ezrin and cortactin and the phosphorylation of ezrin were significantly correlated, accompanied with a group of classic EMT makers. Knockdown of ezrin reversed the expression of EMT markers and downregulated cortactin and EMT transcription factors. Ezrin silencing inhibited tumor cell migration and invasion. Breast cancer tissue microarray and immunohistochemistry showed a significant positive association between ezrin and cortactin.
Conclusions
These findings indicate that ezrin is correlated with cortactin in facilitating EMT in breast cancer. The interaction between ezrin and cortactin is a novel mechanism contributing to the EMT process in cancer metastases.
MeSH Keywords: Breast Neoplasms, Cortactin, Epithelial-Mesenchymal Transition, Neurofibromin 2
Background
According to the American Cancer Society, approximately 231 840 new cases of invasive breast cancer and 40 290 breast cancer deaths occurred among US women in 2015. Breast cancer is the second leading cause of cancer-related deaths in women [1]. The improvements in early diagnosis, surgical techniques, and adjuvant therapies have progressively decreased the mortality of breast cancer in the past 2 decades [2]. However, metastatic breast cancer is still incurable and fatal. Breast cancer metastasis is a multistep process associated with cellular motility, basement membrane degradation, epithelial to mesenchymal transition (EMT), and circulating and disseminating tumor cells [3,4]. EMT was first identified in embryonic development [5], and it participates in most cancer metastatic cascades. After EMT, tumor cells subsequently acquire invasiveness, penetrate to the basement membrane, gain access to the blood or lymphatic vascular systems, reach distant secondary sites, and eventually form macroscopic tumors [6–8]. Although transplantation models of breast cancer cells provide important mechanistic insights into the metastatic cascade, these models are limited by potential species incompatibilities in growth factor signaling pathways, which obscure the processes that are active in patients [9]. In terms of the primary lesion in transplantation models, the tumor cells are not in the same phase as in the EMT process. The transient nature of EMT adds to the complexity of the results and their interpretation. As one of the key components of tumor metastasis, EMT of tumor cells is intimately linked with the tumor microenvironment. Simulation of the special tumor microenvironment is therefore the central premise of EMT research.
A mouse model containing human-derived normal breast tissue was established in our previous study, which allowed human breast cancer cells to proliferate in the human mammary microenvironment and to finally metastasize to distant human tissues [10,11]. The initial microenvironment significantly influences the behavior of cancer cells. Homogeneous cells avoid interference by other cell types, and instead highlight changes in cells initiated by the microenvironment. MDA-MB-231br was obtained from the invasive front of a tumor in transplanted normal human breast tissue, and metastasized preferentially to human tissues when proliferating in the normal human mammary microenvironment [10]. Importantly, the invasive MDA-MB-231br cells participate in the EMT process. In a primary culture, MDA-MB-231br displayed EMT-like changes and was more invasive compared to parent MDA-MB-231 cells.
Ezrin, a member of the ezrin-radixin-moesin (ERM) family, acts as a crosslinker between membranes and the actin cytoskeleton. Ezrin also regulates a variety of cellular activities, such as actin cytoskeleton regulation, cellular morphology, cellular adherence, and modulation of intracellular signaling pathways [12–14]. It is expressed in most normal and cancer tissues, and is expressed at significantly higher levels in cancers of mesenchymal origin (sarcomas) [15,16]. The expression of cytoplasmic ezrin incrementally increases during the development of benign lesions into malignant breast tumors [17,18]. Although substantial evidence has linked ezrin to metastatic behavior in breast cancer, it is still unknown whether EMT is involved. Cortactin, a protein widespread in cellular locomotive organs (e.g., lamellipodia and invasive pseudopodia), regulates branched actin assembly, cell-cell adhesion, membrane trafficking, and ECM degradation [19]. It is able to promote tumor invasion and metastasis in several types of tumors [20–22]. However, details of ezrin-cortactin regulation, especially their potential function in EMT of breast cancer, have not yet been revealed.
Bioinformatic analysis suggests that ezrin and cortactin may be closely related to EMT. Both ezrin and cortactin are closely related to cell mobility, an important factor in the EMT process. Recent research has explored the regulatory effects of ezrin on EMT in cervical cancer and lung cancer [23,24]. The interaction between ezrin and cortactin might promote cancer cell metastasis [25]. The present work aimed to delineate the functional role and regulation of ezrin and cortactin during EMT of breast cancer. The interactive mechanisms between these 2 proteins were also investigated.
Material and Methods
Cell lines and culture
Human breast cancer cell lines (MDA-MB-231, MCF-7, and ZR-75-1) were purchased from ATCC (Manassas, VA, USA) and cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin, and 1% streptomycin. The cell line SUM1315 was kindly provided by Stephen Ethier (University of Michigan). The MDA-MB-231br cell line was established in a previous study [10]. All cells were incubated at 37°C in a humidified chamber supplemented with 5% CO2.
Two-dimensional electrophoresis (2-DE) and mass spectrometry
Two-dimensional electrophoresis analysis and protein identification by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF/TOF) mass spectrometry was performed as described previously [26]. Additional materials and methodologies are described in detail in the supporting materials and methods.
Plasmid construction and lentivirus packaging
Vectors for EZR (ezrin) knockdown (shEZR) or negative controls (NC) were cloned in PGLV3-h1-GFP-puro. Vectors for EZR overexpression or encoding a scrambled sequence (SCR) (GenePharma, Shanghai, China) were similarly cloned in pGLV5-h1-GFP-puro. All vectors were transfected into lentivirus packaging cells. Four shRNA plasmids were constructed against different EZR targets, including a negative control sequence.
sh1: 5′-GGAGTGAAATCAGGAACATCT-3′
sh2: 5′-GGACCCACAATGACATCATCC-3′
sh3: 5′-GCAACCATGAGTTGTATATGC-3′
sh4: 5′-GCAATGAGGAGAAGCGCATCA-3′
NC: 5′-TTCTCCGAACGTGTCACGT-3′
All plasmids were verified by sequencing (GenePharma, Shanghai, China). Retrovirus packaging and infection was conducted according to the manufacturer’s instructions. Stable pooled populations of breast cancer cells were selected in puromycin (3 μg/ml) containing medium for 2 weeks. For knockdown analysis, 2 constructs (sh1 and sh2) with ≥80% knockdown efficiency were used for further studies.
siRNA design and transfection
The siRNA (5′-GCUGAGGGAGAAUGUCUUUTT-3′) used for downregulation of cortactin as well as the negative control siRNA (5′-UUCUCCGAACGUGUCACGUdTdT-3′) were obtained from GenePharma, Shanghai, China. Transient transfection of the siRNA was performed using Lipofectamine 2000 according to the manufacturer’s instructions.
Western blot analysis
Cells were harvested using the total protein extraction kit (KeyGEN BioTECH, Nanjing, China). Samples were incubated for 0.5 h on ice with agitation and then centrifuged at 14 000×g for 15 min. Protein concentrations were determined using the bicinchoninic acid (BCA) protein assay kit (Pierce). Protein samples were subjected to electrophoresis on SDS-polyacrylamide gradient gels, transferred to a PVDF membrane, and blocked in 5% non-fat milk in TBST (phosphorylated proteins were blocked in 5% BSA in TBST) for 2 h at room temperature. Blots were incubated with primary antibodies to the following proteins: ezrin (Abcam), cortactin (Abcam), E-cadherin (CST), α-SMA (Abcam), Slug (CST), Snail (Abcam), Twist (Abcam), Twist2 (Abcam), phosphorylated ezrin at Y-567 (CST), and GAPDH (Beyotime). GAPDH was used on the same membrane as a loading control. The signal was detected after incubation with anti-rabbit or anti-mouse IgG secondary antibody (Bioworld) coupled to peroxidase, using ECL (Millipore). Protein expression levels were evaluated by densitometric analysis.
Real-time reverse transcription PCR analysis
Total RNA was extracted using Trizol total RNA isolation reagent (TaKaRa), and cDNA was synthesized using PrimeScript RT Reagent (TaKaRa) according to the manufacturer’s instructions. Specific primers from Invitrogen (Shanghai, China) were used for transcript detection. All PCR reactions were performed with SYBR Green I (Roche) for detection. Real-time quantitative PCR was performed on StepOne Plus Real-Time PCR system (Applied Biosystems, USA). The following PCR primers were used:
ezrin forward, 5′-ACCAATCAATGTCCGATTACC-3′
ezrin reverse, 5′-GCCGATAGTCTTTACCACCTGA-3′
GAPDH forward, 5′-GCTGCGAAGTGGAAACCATC-3′
GAPDH reverse, 5′-CCTCCTTCTGCACACATTTGAA-3′
The average of 3 independent analyses for each gene and sample was calculated and normalized to the endogenous reference control gene GAPDH.
Matrigel invasion assay and migration assay
Matrigel was purchased from BD Biosciences and stored at −20°C. After thawing at 4°C overnight, the Matrigel was diluted in serum-free DMEM. For carrying out the invasion assay, 50 μl of the suspension was evenly inoculated onto the upper chamber of a Transwell membrane (8 μm pore size) and allowed to form a gel at 37°C. Cells (5×104) were overlaid with 200 μl of serum-free DMEM on Matrigel-coated Transwell membranes with 0.5 ml of complete medium in the lower chamber. After incubating for 48 h at 37°C in a humidified atmosphere of 5% CO2, the cells were fixed and stained with 0.1% crystal violet solution for 20 min, and the chamber was washed 3 times with phosphate-buffered saline (PBS). Non-invading cells on the top of the membrane were removed using cotton wool. Invading cells were counted under a microscope. In each Matrigel invasion experiment, 3 independent replicates were performed.
To carry out the migration assay, cells (3×104) were overlaid with 200 μl serum-free DMEM on Transwell membranes without Matrigel-coating, and incubated for 16 h. The remainder of this assay was performed as described in the invasion assay.
Growth curve by CCK8 assay
Cells (2×103) were grown in microtiter plates in a final volume of 100 μl of complete medium per well, at 37°C and 5% CO2. The growth curve was carried out over a period of 6 days. After the incubation period, 10 μl of the CCK8 (Dojindo) labeling reagent (0.5 mg/ml) was added to each well. The cells were subsequently analyzed by enzyme-labeled meter (Tecan) to measure their absorption at 450 nm. Each treatment was performed in triplicate.
Colony formation assay
Cells (5×102) were plated in a 6-well plate in complete medium. After incubation for 10–14 days, when the colonies were visible by eye, the culture was terminated by removing the medium and washing the cells twice with PBS. The colonies were fixed with 95% ethanol for 100 s, then dried and stained with 0.1% crystal violet solution for 10 min, and washed with PBS. Images were obtained and the number of colonies containing more than 50 cells was counted. Each treatment was performed in triplicate.
Tissue microarray (TMA) and immunohistochemistry (IHC)
TMAs were purchased from BioMax (USA). Sections were arranged in duplicate cores per case. TMAs were treated with xylene, then 100% ethanol, and then decreasing concentrations of ethanol. After antigen retrieval, they were blocked and stained with antibodies against ezrin or cortactin, followed by secondary antibody incubation and the standard avidin biotinylated peroxidase complex method. Hematoxylin was used for counterstaining and images were taken with an upright microscope Nikon system (Nikon, JAPAN).
A modified Histo-score was used to perform IHC scoring, which was assessed by both intensity of staining and percentage of positive area. The extent of the positive area was scored on a scale of 0–4 as follows: 0, 0%; 1, 1–10%; 2, 11–50%; 3, 51–80%; and 4, >80%. The final staining score from 0–12 for ezrin and cortactin was the product of the staining intensity and positive area scores. A score of 0–7 indicated low expression and between 8–12 indicated high expression in ezrin. In cortactin, scores of 0–3, 4–7, and 8–12 were considered indicative of low, moderate, and high expression, respectively.
Co-immunoprecipitation (Co-IP)
Extract from 1×107 cells was obtained. The protein was immunoprecipitated using anti-ezrin or rabbit IgG control according to the manufacturer’s instructions, and then subjected to immunoblotting.
Statistical analysis
Statistical significance between and within groups in the Transwell and proliferation assays was assessed by t test or one-way ANOVA, using SPSS 20.0. Statistical significance of differences between ezrin expression and other indexes in the TMA was determined by a non-parametric test, chi-square test, or Fisher exact test. For all tests, results at p values <0.05 were taken to represent a statistically significant difference.
Results
MDA-MB-231br has a strong EMT-like process, including invasion and migration
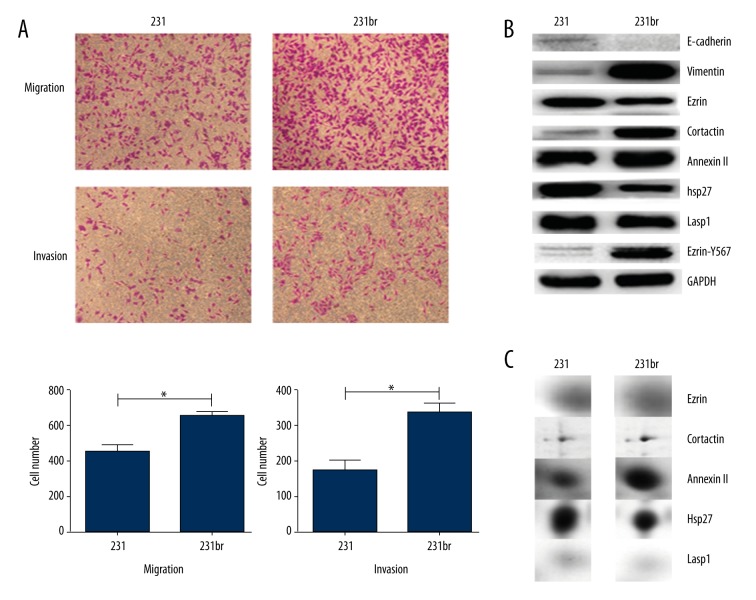
A Transwell assay was performed to measure the invasive and migratory ability of MDA-MB-231 and MDA-MB-231br cells. MDA-MB-231br cells had a significantly increased ability to migrate and invade compared to that of MDA-MB-231 cells (Figure 1A). Western blot analysis indicated that the level of E-cadherin expression was decreased in MDA-MB-231br compared to MDA-MB-231, while the expression pattern of vimentin was the inverse (Figure 1B).
Figure 1.
MDA-MB-231br cells are strongly capable of invasion and migration. (A) The novel human breast cancer cell line MDA-MB-231br isolated from the metastatic mouse model and MDA-MB-231 were stimulated to migrate and invade in Transwell chambers. Representative photographs (upper panel) and quantification (lower panel) are shown. Columns: average of 3 independent experiments, * p<0.05. Magnification ×100. (B) EMT markers, including E-cadherin, vimentin, ezrin, cortactin, annexin II, hsp27, lasp2, and ezrin-T567 were differently expressed in MDA-MB-231 and MDA-MB-231br cell lines, according to Western blotting analysis with GAPDH as the internal control. (C) Differentially expressed proteins were identified in MDA-MB-231 and MDA-MB-231br cell lines by two-dimensional electrophoresis (2-DE) and mass spectrometry.
Identification of differentially expressed proteins between MDA-MB-231 and MDA-MB-231br
Two-dimensional gel electrophoresis and mass spectrometry identified 81 differentially expressed proteins between MDA-MB-231 and MDA-MB-231br (Supplementary Figure 1, Supplementary Table 1). From these proteins, ezrin, cortactin, hsp27, lasp2, and annexin II were selected (Figure 1C) for validation by Western blot (Figure 1B). In MDA-MB-231, the level of ezrin expression decreased and cortactin expression increased compared to that in MDA-MB-231br (Figure 1B, 1C). However, the phosphorylation of ezrin at position Y-567 increased in MDA-MB-231br compared to that in MDA-MB-231 (Figure 1B).
Knockdown of ezrin changes cellular morphology, and membranous marker and EMT indicator expression
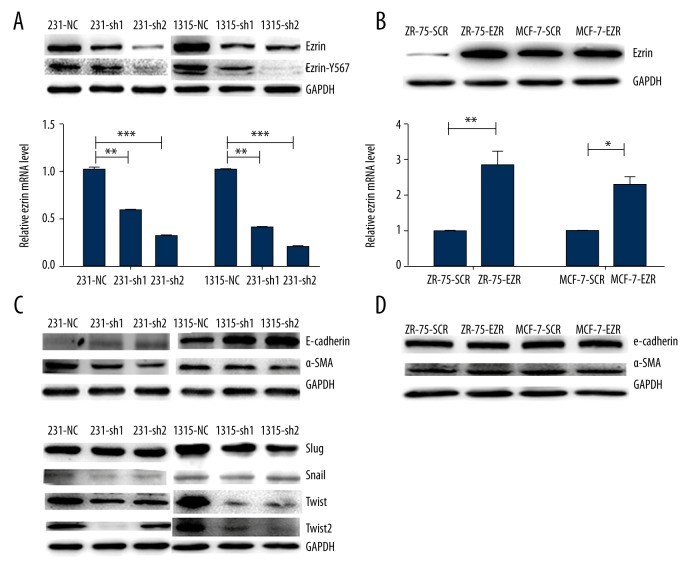
MDA-MB-231 and SUM1315 breast cancer cells, which have high invasive potential and highly-expressed ezrin, were used for the next study. Knockdown efficiency of ezrin was confirmed by real-time PCR and immunoblotting (Figure 2A). The phosphorylation of ezrin at position Y-567 was also decreased in ezrin knockdown cells (Figure 2A). Alternatively, MCF-7 and ZR-75-1 breast cancer cells, which rarely metastasize [27], were transfected with an ezrin overexpression vector. In the cells overexpressing ezrin, marked upregulation of ezrin was confirmed by real-time PCR and immunoblotting (Figure 2B).
Figure 2.
Downregulation of ezrin reverses EMT in MDA-MB-231 and SUM1315 cells. (A) The expression levels of ezrin were verified in MDA-MB-231 and SUM1315 after transfection of shRNA against ezrin or upon ezrin overexpression, by Western blot and qRT-PCR, respectively. ** p<0.01, *** p<0.001. The phosphorylation of ezrin at position Y-567 was decreased with decreased ezrin expression. (B) The expression of ezrin was verified in MCF-7 and ZR-75-1 cells after ezrin overexpression, by Western blots and qRT-PCR, respectively. ** p<0.01, * p<0.05. (C) Western blot analysis revealed that knockdown of ezrin resulted in increased expression of an epithelial marker (E-cadherin) and decreased expression of a mesenchymal marker (α-SMA) in MDA-MB-231 and SUM1315 cells. Snail, Twist, and Twist2, which are key transcription factors that promote EMT, were significantly decreased in ezrin-downregulated MDA-MB-231 and SUM1315 cells. (D) The overexpression of ezrin did not alter the expression of E-cadherin and α-SMA in MCF-7 and ZR-75-1 cells.
The expression levels of an epithelial marker (E-cadherin) and a mesenchymal marker (α-SMA) were examined by immunoblotting. The expression level of E-cadherin was significantly increased, whereas that of α-SMA was decreased in ezrin-knockdown cells compared to that in control cells (Figure 2C). In MCF-7 and ZR-75-1 cell lines, overexpressing ezrin had no effect on the expression of E-cadherin and α-SMA (Figure 2D). The expression levels of Slug, Snail, Twist, and Twist2, which are key transcription factors that promote EMT, were also determined by immunoblotting. Significantly reduced expression of Snail, Twist, and Twist2 was observed in ezrin-knockdown cells compared with that in the control cells (Figure 2C).
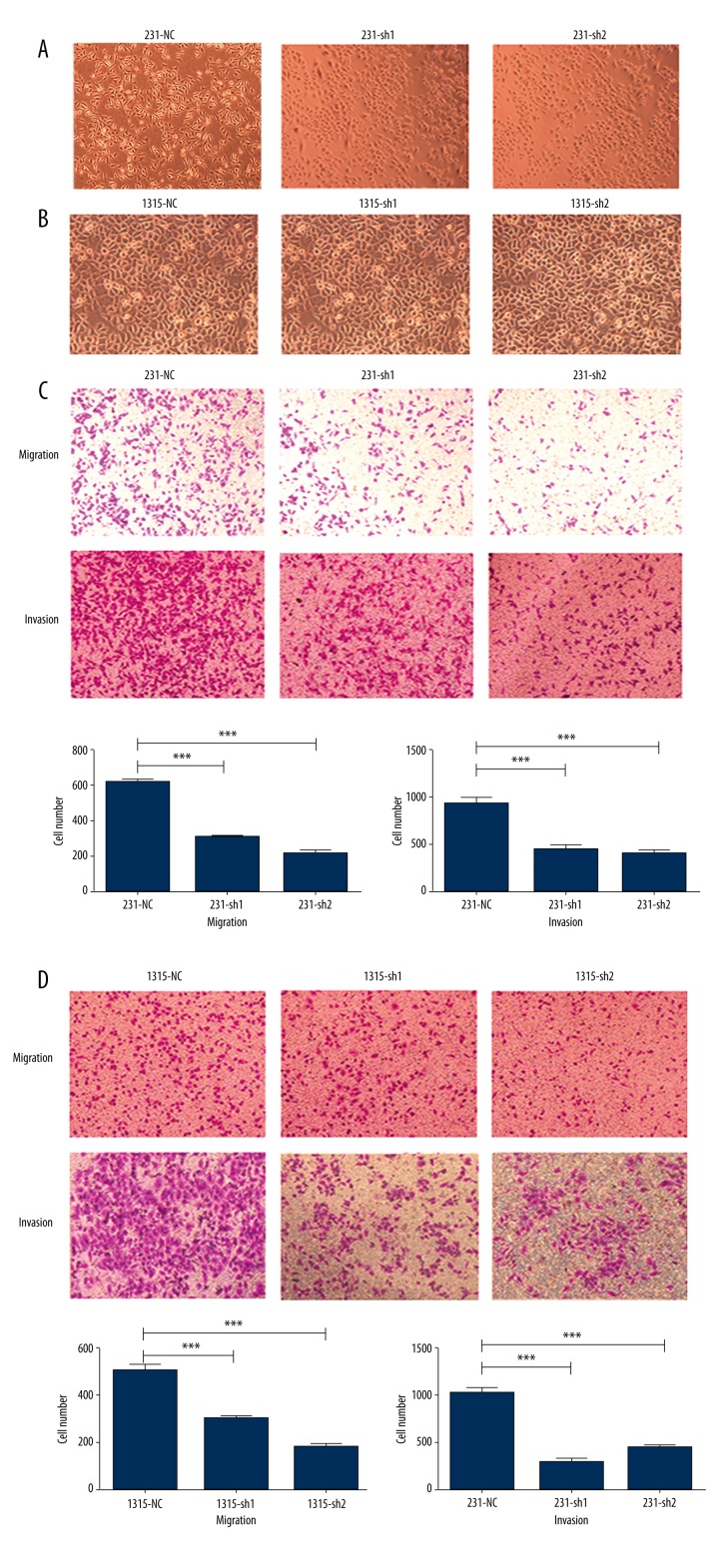
Ezrin knockdown reduced the migration and invasion of breast cancer cells
Ezrin knockdown in MDA-MB-231 cells changed the cell morphology remarkably (Figure 3A). MDA-MB-231 cells are widely accepted as having a mesenchymal phenotype, while ezrin-knockdown cells presented an epithelial phenotype (Figure 3A). However, the shape of SUM1315 after ezrin knockdown did not visibly change (Figure 3B). Increased motility and invasion of breast cancer cells are key steps in the EMT process. Ezrin knockdown was associated with significantly reduced migratory and invasive ability in MDA-MB-231 and SUM1315 cells (Figure 3C, 3D).
Figure 3.
Knockdown of ezrin changes the phenotype and significantly decreases the migratory and invasive potential of breast cancer cells. (A) The phenotype of MDA-MB-231 cells after ezrin knockdown changed from spindle to oval. Magnification ×100. (B) The shape of SUM1315 after ezrin knockdown had no visible change. Magnification ×100. (C, D) MDA-MB-231 and SUM1315 cells transfected with shRNA against ezrin were subsequently subjected to Transwell migration assay and Matrigel invasion assay. Representative photographs (upper panel) and quantification (lower panel) are shown. Columns: average of 3 independent experiments, * p<0.05, ** p<0.01. Magnification ×100.
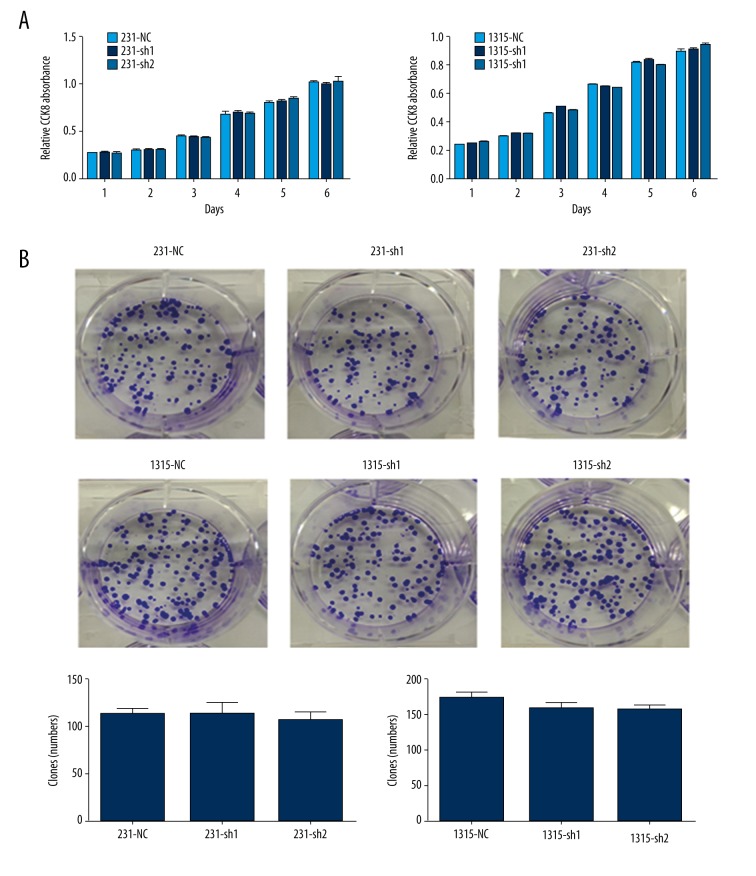
In general, cells with high invasive ability had enhanced ability for proliferation. However, the CCK8 assay and colony formation assay suggested that decreased expression of ezrin in breast cancer cells had no effect on cell proliferation (Figure 4A, 4B).
Figure 4.
Effect of ezrin on proliferation and growth of breast cancer cell lines. (A) The growth of MDA-MB-231 and SUM1315 cells cultured for 6 days was measured by use of a cell counting kit (CCK-8) assay. There was no obvious change between MDA-MB-231 or SUM1315 and the corresponding control cells with regards to the rate of proliferation. (B) The colony formation assay was also performed to measure the growth of cells. There was also no obvious change between MDA-MB-231 or SUM1315 cells compared to the rate of proliferation of their respective control cells.
Ezrin is positively correlated with cortactin in breast cancer
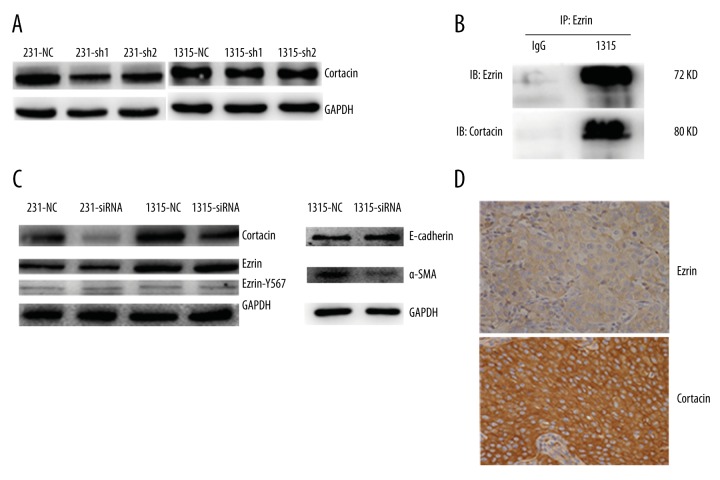
The expression of cortactin decreased with the reduction of ezrin expression (Figure 5A). Co-IP analysis suggested a connection between the expression of ezrin and cortactin in SUM1315 cells (Figure 5B), but not in MDA-MB-231 cells (data not shown). The expression of cortactin was knocked down and confirmed by immunoblotting (Figure 5C). In cortactin-knockdown cells, the expression of ezrin and p-ezrin did not change obviously. In cortactin-knockdown cells, the level of E-cadherin expression was significantly increased, whereas that of α-SMA was decreased (Figure 5C).
Figure 5.
Ezrin and cortactin express a similar pattern in breast cancers. (A) In MDA-MB-231 or SUM1315 cells, the reduction in ezrin expression occurred alongside the reduction in cortactin. (B) Co-IP analysis suggested a connection between ezrin and cortactin in SUM-1315 cells. (C) The expression and phosphorylation of ezrin did not change in MDA-MB-231 or SUM1315 cells transfected with siRNA against cortactin. Knockdown of cortactin resulted in increased expression of E-cadherin and decreased expression α-SMA in SUM1315 cells. (D) Immunohistochemistry revealed significant correlations between ezrin expression and cortactin expression. Magnification ×400.
In clinical breast cancer tissues, a significant positive association was observed between ezrin and cortactin expression (Table 1, Figure 5D).
Table 1.
The association of ezrin with cortactin expression and clinicopathologic features of breast cancer.
| Clinicopathologic parameters | Number of case | Ezrin expression | P-value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | 0.992 | |||
| ≤55 | 55 | 38 | 17 | |
| >55 | 13 | 9 | 4 | |
| Grade | 0.325 | |||
| I | 3 | 1 | 2 | |
| II | 37 | 20 | 17 | |
| III | 9 | 6 | 3 | |
| Unclear | 19 | 13 | 6 | |
| TNM stage | 0.356 | |||
| I+II | 52 | 29 | 23 | |
| III+IV | 16 | 11 | 5 | |
| Cortactin | 0.001 | |||
| Low | 4 | 4 | 0 | |
| Middle | 11 | 11 | 0 | |
| High | 53 | 25 | 28 | |
TNM classification according to the International Union Against Cancer criteria.
Discussion
In this study, evidence for MDA-MB-231br undergoing an EMT-like process included increased spindle morphology, stronger invasive ability, and decreased expression of E-cadherin. Eighty-one differentially expressed proteins were identified following proteomic analysis comparing MDA-MB-231br and MDA-MB-231 cells. Along with protein-level changes in expression of a group of classic EMT markers, ezrin and cortactin were significantly regulated. The changes observed in these cells may be attributed to a response to an EMT-inducing microenvironmental signal. We hypothesized that these proteins included novel protein signatures of EMT, and demonstrated the function of ezrin and cortactin in breast cancer. In MDA-MB-231br cells with an EMT-like phenotype, the expression of ezrin decreased, whereas the phosphorylation of ezrin at position Y-567 changed in the opposite way. Ezrin and phosphorylated ezrin were decreased synchronously, and cell morphology varied from spindle to oval. Reduced ezrin expression reversed the EMT process through changes in the expression of EMT markers and downregulation of transcription factors. Conversely, overexpression of ezrin had no obvious effect on the EMT process. This implies that ezrin-knockdown could reverse the EMT cascade, which might affect its phosphorylation state. The phosphorylation state of ezrin might be more important than its expression level. The role of ezrin expression in metastasis has been demonstrated in animal models, while no change has been observed in primary tumor growth rate [17]. The expression level of ezrin is significantly higher in lymphatic metastases than in primary breast cancer tissues [28]. Cytoplasmic ezrin is regarded as a single characteristic marker in breast cancer [29]. The phosphorylation of ezrin at position Y-567 disrupts its amino- and carboxyl-terminal associations, and thus plays a key role in modulating the conformation and function of ERM proteins [30] It is reported that the phosphorylation of ezrin regulates invasion and metastasis of breast cancer cells [31].
In the present study, increased expression of cortactin was observed in MDA-MB-231br cells with increased expression of phosphorylated ezrin. Conversely, decreased phosphorylation of ezrin reduced the expression of cortactin. Cortactin knockdown had no effect on the expression of ezrin and its phosphorylation. Ezrin also acts as a membrane-cytoskeleton crosslinker, which can affect structural modifications under the control of several upstream or downstream regulators. Paladin and S100P are known to form large protein complexes with ezrin [32,33], and cortactin is thought to cooperate with ezrin in tumor progression [25]. Cortactin, which is localized to cortical actin structures, not only plays a crucial role as a regulator of actin cytoskeletal dynamics, but also acts as a key player in aggressive cancers [31].
Our results elucidate the interaction between ezrin and cortactin. The association between ezrin and cortactin was explored in SUM1315 cells. There was no observed co-precipitation between ezrin and cortactin in the MDA-MB-231 cell line, which might be because ezrin expression was not high enough to show this change in these cells. The positive correlation between these 2 proteins was demonstrated in the breast cancer tissue microarray.
Conclusions
In summary, ezrin, based mainly on its phosphorylation state, appears to interact with cortactin to promote the EMT process in breast cancer. Ezrin and cortactin may therefore be potential therapeutic targets and prognostic indicators of breast cancer.
Supplementary Files
Two-dimensional gel electrophoresis(2-DE) analysis of proteins extracted from different cell lines species: (A) MDA-MB-231; (B) MDA-MB-231br.
Supplementary Table 1.
Identification of up- and down-regulated proteins in MDA-MB-231br cells compared to MDA-MB-231 cells.
| No. | Accession No | AC | Theor. mol | Measured | Sequence | Peptides a | Fold chang | Student T |
|---|---|---|---|---|---|---|---|---|
| 1 | gi|7706425 | LSM8 | 10.4/4.34 | 11.5/4.11 | 0.82 | 4 | 0 | 6.9501 |
| 2 | gi|38511503 | LOC400581 | 13.2/7.88 | 14.4/7.73 | 0.57 | 11 | 0 | 3.32884 |
| 3 | gi|5453740 | MRCL3 | 19.8/4.67 | 21.0/4.46 | 0.63 | 12 | 0 | 22.6371 |
| 4 | gi|11957604 | GLO1 | 21.0/5.12 | 23.1/5.38 | 0.29 | 19 | 0.1458951 | 5.8117 |
| 5 | gi|11958606 | PRDX6 | 22.8/5.98 | 20.6/5.65 | 0.56 | 14 | 0.3751647 | 3.13876 |
| 6 | gi|4504517 | HSPB1 | 22.8/5.98 | 21.5/6.26 | 0.49 | 11 | 1.7085594 | 6.85507 |
| 7 | gi|4506195 | PSMB2 | 23.0/6.51 | 17.7/5.28 | 0.37 | 11 | 0.5424243 | 3.47661 |
| 8 | gi|11962339 | RANBP1 | 23.5/5.19 | 26.9/4.31 | 0.42 | 16 | 0.5717237 | 5.58285 |
| 9 | gi|31645 | GAPDH | 31.7/7.15 | 35.9/4.79 | 0.57 | 27 | 2.3023371 | 3.26693 |
| 10 | gi|914957 | GRB2 | 25.3/5.89 | 28.4/5.48 | 0.51 | 13 | ND | 5.22913 |
| 11 | gi|20149675 | TUBB | 26.8/5.15 | 31.2/4.82 | 0.46 | 12 | 3.1617546 | 9.1124 |
| 12 | gi|16357477 | CDC34 | 26.9/4.41 | 28.0/4.2 | 0.32 | 9 | 0.3679367 | 3.0581 |
| 13 | gi|44979607 | IMPDH2 | 27.3/5.84 | 29.4/6.1 | 0.6 | 9 | 0.4022986 | 5.13788 |
| 14 | gi|48145973 | NDUFV2 | 27.7/8.22 | 26.3/8.67 | 0.33 | 8 | 0.3474179 | 2.92579 |
| 15 | gi|19298714 | PGLS | 27.8/5.7 | 25.6/5.37 | 0.37 | 9 | 0.4460076 | 3.16474 |
| 16 | gi|4758484 | GSTO1 | 27.8/6.23 | 26.5/6.51 | 0.48 | 15 | 0.2449022 | 4.34464 |
| 17 | gi|19378506 | PSPHL;ACTG1 | 28.5/5.2 | 23.2/3.97 | 0.45 | 12 | 0.4615403 | 3.08667 |
| 18 | gi|17402904 | EXOSC6 | 28.5/6.06 | 31.9/5.18 | 0.3 | 12 | 0.2836561 | 4.7922 |
| 19 | gi|62897753 | hCG_2015269 | 29.0/6.67 | 26.8/6.34 | 0.79 | 19 | 0.3179457 | 3.64943 |
| 20 | gi|19298714 | ERP29 | 29.0/6.77 | 27.7/7.05 | 0.39 | 10 | 0.4765053 | 3.36723 |
| 21 | gi|4826659 | CAPZB | 29.6/6.45 | 24.3/5.22 | 0.64 | 26 | 0.4184514 | 2.90687 |
| 22 | gi|19438056 | LASP1 | 30.1/6.61 | 33.5/5.73 | 0.58 | 8 | 0.2488259 | 5.5738 |
| 23 | gi|22652991 | TPI1 | 31.1/5.65 | 35.3/3.29 | 0.56 | 14 | 0.4883441 | 7.47102 |
| 24 | gi|21361454 | PYCR2 | 34.4/8.48 | 37.5/8.07 | 0.54 | 24 | 0.3732397 | 5.90677 |
| 25 | gi|18906542 | HNRPC | 32.4/4.94 | 36.8/4.61 | 0.53 | 20 | 0.5408701 | 6.3532 |
| 26 | gi|12803709 | CK14 | 32.4/4.97 | 33.5/4.76 | 0.33 | 17 | 0.4598442 | 3.63144 |
| 27 | gi|11056044 | PPA1 | 33.1/5.54 | 35.3/5.8 | 0.5 | 17 | 0.4610924 | 2.78434 |
| 28 | gi|5174447 | TST | 33.6/6.77 | 32.3/7.22 | 0.37 | 14 | 0.4618559 | 2.78215 |
| 29 | gi|10437514 | SUHW4 | 35.1/8.68 | 32.9/8.35 | 0.42 | 14 | ND | 11.2023 |
| 30 | gi|11962858 | DNAH3 | 473.8/6.04 | 472.5/6. | 0.27 | 14 | 0.3843506 | 2.9416 |
| 31 | gi|4826643 | ANXA3 | 36.5/5.63 | 31.2/4.4 | 0.56 | 23 | 0.5335611 | 5.71278 |
| 32 | gi|19438981 | CNN3 | 36.6/5.69 | 35.2/6.14 | 0.49 | 17 | 3.3243097 | 7.20813 |
| 33 | gi|2119276 | TUBB | 48.1/4.7 | 45.9/4.37 | 0.25 | 14 | 1.8615634 | 3.20717 |
| 34 | gi|73976124 | HNRPA2B1 | 37.5/8.97 | 36.1/9.25 | 0.46 | 12 | 2.5249017 | 6.79964 |
| 35 | gi|17402893 | PSAT1 | 40.8/7.56 | 35.5/6.33 | 0.55 | 26 | 1.6654277 | 3.88025 |
| 36 | gi|19437787 | ARFIP2 | 37.9/5.72 | 41.4/4.84 | 0.38 | 12 | 1.7420536 | 2.80585 |
| 37 | gi|22104024 | PPP1CB | 38.0/5.84 | 42.2/3.48 | 0.22 | 8 | 0.3430111 | 4.85873 |
| 38 | gi|11960877 | PPP1CB | 38.0/5.84 | 41.1/5.43 | 0.4 | 23 | 0.4678427 | 3.05192 |
| 39 | gi|6912280 | AHSA1 | 38.4/5.41 | 33.1/4.18 | 0.19 | 8 | 4.3644831 | 5.00035 |
| 40 | gi|7305503 | STOML2 | 38.6/6.88 | 42.1/6 | 0.52 | 18 | 0.2565899 | 4.2268 |
| 41 | gi|11959799 | ANXA2 | 38.8/7.57 | 36.6/7.24 | 0.6 | 20 | 2.2824303 | 4.00567 |
| 42 | gi|4506127 | PRPS1 | 35.3/6.51 | 34.0/6.79 | 0.47 | 21 | 1.9003629 | 5.43115 |
| 43 | gi|93277122 | BUB3 | 32.1/8.4 | 26.8/7.17 | 0.43 | 15 | 3.4236124 | 10.8789 |
| 44 | gi|4503895 | GALK1 | 42.7/6.04 | 46.9/3.68 | 0.35 | 17 | 6.5938433 | 3.2962 |
| 45 | gi|11960222 | CKB | 42.9/5.34 | 46.0/4.93 | 0.45 | 15 | 2.6276618 | 3.86433 |
| 46 | gi|15825518 | SERPINB7 | 43.2/6.34 | 47.6/6.01 | 0.34 | 12 | 0.2890603 | 11.6406 |
| 47 | gi|19438418 | HSPA8 | 71.1/5.37 | 72.2/5.16 | 0.24 | 14 | 1.8781699 | 3.44432 |
| 48 | gi|5174529 | MAT2A | 44.0/6.02 | 46.1/6.28 | 0.32 | 14 | 0.3873342 | 3.38533 |
| 49 | gi|19378506 | ACTB | 45.5/5.82 | 44.1/6.27 | 0.27 | 7 | 2.5188648 | 4.86371 |
| 50 | gi|5729991 | PSMC4 | 47.5/5.09 | 45.2/4.76 | 0.45 | 19 | 2.9751505 | 7.05399 |
| 51 | gi|20328236 | ENO1 | 47.5/7.01 | 50.9/6.13 | 0.33 | 14 | 0.4514455 | 5.84011 |
| 52 | gi|20328236 | ENO1 | 47.5/7.01 | 51.7/4.65 | 0.38 | 14 | 2.1757631 | 2.78494 |
| 53 | gi|28872725 | PSMD11 | 47.8/6.08 | 50.9/5.67 | 0.25 | 12 | 2.2969517 | 4.67432 |
| 54 | gi|35071 | MTHFD2 | 38.0/8.86 | 42.4/8.53 | 0.02 | 11 | 3.0997653 | 3.33509 |
| 55 | gi|21623852 | PSMC3 | 49.5/5.13 | 48.1/5.58 | 0.22 | 19 | 3.0467243 | 3.32863 |
| 56 | gi|21623852 | PSMC3 | 49.5/5.13 | 47.3/4.8 | 0.47 | 24 | 1.9425454 | 4.2506 |
| 57 | gi|20328236 | ENO1 | 47.5/7.01 | 46.2/7.29 | 0.63 | 29 | 1.5667183 | 4.24397 |
| 58 | gi|16507237 | VIM | 49.7/5.19 | 44.4/3.96 | 0.4 | 20 | 2.3352391 | 3.4501 |
| 59 | gi|14043072 | HNRPA2B1 | 37.5/8.97 | 40.9/8.09 | 0.5 | 25 | 2.118541 | 2.97696 |
| 60 | gi|11960877 | TUBB2A | 50.3/4.78 | 54.5/2.42 | 0.42 | 16 | 2.5802857 | 3.58321 |
| 61 | gi|28948869 | VIL2 | 69.5/5.94 | 72.6/5.53 | 0.39 | 18 | 2.6162373 | 3.95062 |
| 62 | gi|11960683 | GDI2 | 51.1/6.11 | 55.5/5.78 | 0.38 | 16 | 4.2414899 | 8.61448 |
| 63 | gi|38569475 | MTX1 | 51.7/9.8 | 46.4/8.57 | 0.36 | 14 | ND | 10.9218 |
| 64 | gi|449441 | UGP2 | 55.8/7.69 | 59.2/6.81 | 0.39 | 24 | 3.9755451 | 4.09809 |
| 65 | gi|48255968 | UGP2 | 55.8/7.69 | 60.0/5.33 | 0.52 | 27 | 2.5612084 | 6.32672 |
| 66 | gi|66933016 | IMPDH2 | 56.2/6.44 | 59.4/6.03 | 0.42 | 20 | 0.4828852 | 10.5545 |
| 67 | gi|11961763 | CCT2 | 57.8/6.01 | 62.2/5.68 | 0.39 | 15 | 4.6695429 | 4.39079 |
| 68 | gi|4557976 | ALDOA | 39.9/8.3 | 41.0/8.09 | 0.47 | 20 | 2.0385729 | 3.2317 |
| 69 | gi|22104614 | CCT5 | 60.1/5.45 | 62.2/5.71 | 0.53 | 26 | 2.3538723 | 4.52031 |
| 70 | gi|20151189 | GLUD1 | 61.7/7.66 | 60.3/8.11 | 0.33 | 13 | 0.3442582 | 9.36776 |
| 71 | gi|19437434 | HSPA5 | 72.5/5.07 | 70.3/4.74 | 0.71 | 24 | 5.0750799 | 3.96361 |
| 72 | gi|11463825 | STIP1 | 63.2/6.4 | 66.7/5.52 | 0.51 | 37 | 3.6341375 | 2.79519 |
| 73 | gi|16915936 | MGAT4B | 65.1/8.38 | 69.3/6.02 | 0.29 | 10 | 2.8100682 | 3.95651 |
| 74 | gi|9910460 | ZNF714 | 69.3/9.16 | 72.4/8.75 | 0.3 | 9 | 2.8886894 | 3.2727 |
| 75 | gi|182087 | CTTN | 61.9/5.28 | 66.3/4.95 | 0.32 | 21 | 7.0312418 | 6.24639 |
| 76 | gi|13676857 | HSPA2 | 70.3/5.56 | 68.9/6.01 | 0.27 | 15 | 2.6325321 | 3.0962 |
| 77 | gi|13676857 | HSPA2 | 70.3/5.56 | 68.1/5.23 | 0.44 | 29 | ND | 37.1179 |
| 78 | gi|1017722 | ZNF85 | 70.9/9.48 | 69.6/9.76 | 0.26 | 14 | 3.6214245 | 3.22252 |
| 79 | gi|57014047 | LMNA | 70.9/8.55 | 65.6/7.32 | 0.5 | 31 | 5.398146 | 3.33403 |
| 80 | gi|29645294 | SNIP | 112.7/9.32 | 116.1/8. | 0.24 | 16 | 2.8558356 | 3.07003 |
| 81 | gi|542991 | ROR2 | 80.8/7.3 | 85.1/4.94 | 0.51 | 12 | ND | 11.6562 |
VIL2 – Chain A, structure of the active ferm domain of ezrin; CTTN – cortactin.
Footnotes
Conflict of Interest
The authors declare that they have no competing interests.
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Source of support: This study was funded by the Natural Science Foundation of China (81202077, 81572607, 81172502, 81272916, 81572595, 81472703) “Qinglan” and “Six talent peaks” projects to Tiansong Xia, a project Funded by the Priority Academic Program Development of Jiangsu higher Education Institutions (PAPD)
References
- 1.DeSantis CE, Fedewa SA, Goding Sauer A, et al. Breast cancer statistics, 2015. Convergence of incidence rates between black and white women. Cacer J Clin. 2016;66:31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. Cacer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 3.Geng SQ, Alexandrou AT, Li JJ. Breast cancer stem cells: Multiple capacities in tumor metastasis. Cancer Lett. 2014;349:1–7. doi: 10.1016/j.canlet.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen DX, Bos PD, Massague J. Metastasis: From dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–84. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 5.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154:13. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–72. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 8.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science (New York, NY) 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 9.Bill R, Christofori G. The relevance of EMT in breast cancer metastasis: Correlation or causality? FEBS Lett. 2015;589:1577–87. doi: 10.1016/j.febslet.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Xia TS, Liu XA, et al. A novel orthotopic and metastatic mouse model of breast cancer in human mammary microenvironment. Breast Cancer Res Treat. 2010;120:337–44. doi: 10.1007/s10549-009-0391-4. [DOI] [PubMed] [Google Scholar]
- 11.Xia TS, Wang J, Yin H, et al. Human tissue-specific microenvironment: An essential requirement for mouse models of breast cancer. Oncol Rep. 2010;24(1):203–11. doi: 10.3892/or_00000847. [DOI] [PubMed] [Google Scholar]
- 12.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: Integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–99. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 13.Hughes SC, Fehon RG. Understanding ERM proteins – the awesome power of genetics finally brought to bear. Curr Opin Cell Biol. 2007;19:51–56. doi: 10.1016/j.ceb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Clucas J, Valderrama F. ERM proteins in cancer progression. J Cell Sci. 2014;127:267–75. doi: 10.1242/jcs.133108. [DOI] [PubMed] [Google Scholar]
- 15.Bruce B, Khanna G, Ren L, et al. Expression of the cytoskeleton linker protein ezrin in human cancers. Clin Exp Metastasis. 2007;24:69–78. doi: 10.1007/s10585-006-9050-x. [DOI] [PubMed] [Google Scholar]
- 16.Chand K, Wan XL, Seuli B, et al. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10:5. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 17.Elliott BE, Meens JA, SenGupta SK, et al. The membrane cytoskeletal crosslinker ezrin is required for metastasis of breast carcinoma cells. Breast Cancer Res. 2005;7(3):R365–73. doi: 10.1186/bcr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gschwantler-Kaulich D, Natter C, Steurer S, et al. Increase in ezrin expression from benign to malignant breast tumours. Cell Oncol (Dordr) 2013;36(6):485–91. doi: 10.1007/s13402-013-0153-5. [DOI] [PubMed] [Google Scholar]
- 19.Sung BH, Zhu X, Kaverina I, Weaver AM. Cortactin controls cell motility and lamellipodial dynamics by regulating ECM secretion. Cell Oncol (Dordr) 2013;36(6):485–91. doi: 10.1016/j.cub.2011.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver AM. Cortactin and tumor invasiveness. Cancer Lett. 2008;265:157–66. doi: 10.1016/j.canlet.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li A, Zhang L, Zhang X, et al. Expression and clinical significance of cortactin protein in ovarian neoplasms. Clin Transl Oncol. 2016;18:220–27. doi: 10.1007/s12094-015-1360-5. [DOI] [PubMed] [Google Scholar]
- 22.Ni QF, Yu JW, Qian F, et al. Cortactin promotes colon cancer progression by regulating ERK pathway. Int J Oncol. 2015;47:1034–42. doi: 10.3892/ijo.2015.3072. [DOI] [PubMed] [Google Scholar]
- 23.Kong J, Di C, Piao J, et al. Ezrin contributes to cervical cancer progression through induction of epithelial-mesenchymal transition. Oncotarget. 2016;7(15):19631–42. doi: 10.18632/oncotarget.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen MJ, Gao XJ, Xu LN, et al. Ezrin is required for epithelial-mesenchymal transition induced by TGF-beta1 in A549 cells. Int J Oncol. 2014;45:1515–22. doi: 10.3892/ijo.2014.2554. [DOI] [PubMed] [Google Scholar]
- 25.Kocher HM, Sandle J, Mirza TA, et al. Ezrin interacts with cortactin to form podosomal rosettes in pancreatic cancer cells. Gut. 2009;58:271–84. doi: 10.1136/gut.2008.159871. [DOI] [PubMed] [Google Scholar]
- 26.Zhu YF, Cui YG, Guo XJ, et al. Proteomic analysis of effect of hyperthermia on spermatogenesis in adult male mice. J Proteome Res. 2006;5:2217–25. doi: 10.1021/pr0600733. [DOI] [PubMed] [Google Scholar]
- 27.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Wu M, Wang H, et al. Ezrin silencing by small hairpin RNA reverses metastatic behaviors of human breast cancer cells. Cancer Lett. 2008;261:55–63. doi: 10.1016/j.canlet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 29.Arslan AA, Silvera D, Arju R, et al. Atypical ezrin localization as a marker of locally advanced breast cancer. Breast Cancer Res Treat. 2012;134:981–88. doi: 10.1007/s10549-012-2017-5. [DOI] [PubMed] [Google Scholar]
- 30.Mangeat P, Roy C, Martin M. ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol. 1999;9:187–92. doi: 10.1016/s0962-8924(99)01544-5. [DOI] [PubMed] [Google Scholar]
- 31.MacGrath SM, Koleske AJ. Cortactin in cell migration and cancer at a glance. J Cell Sci. 2012;125(Pt 7):1621–26. doi: 10.1242/jcs.093781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Missiaglia E, Blaveri E, Terris B, et al. Analysis of gene expression in cancer cell lines identifies candidate markers for pancreatic tumorigenesis and metastasis. Int J Cancer. 2004;112:100–12. doi: 10.1002/ijc.20376. [DOI] [PubMed] [Google Scholar]
- 33.Kay LP, Ru C, Mary PB, et al. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PloS One. 2006;3:13. doi: 10.1371/journal.pmed.0030516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Two-dimensional gel electrophoresis(2-DE) analysis of proteins extracted from different cell lines species: (A) MDA-MB-231; (B) MDA-MB-231br.
Supplementary Table 1.
Identification of up- and down-regulated proteins in MDA-MB-231br cells compared to MDA-MB-231 cells.
| No. | Accession No | AC | Theor. mol | Measured | Sequence | Peptides a | Fold chang | Student T |
|---|---|---|---|---|---|---|---|---|
| 1 | gi|7706425 | LSM8 | 10.4/4.34 | 11.5/4.11 | 0.82 | 4 | 0 | 6.9501 |
| 2 | gi|38511503 | LOC400581 | 13.2/7.88 | 14.4/7.73 | 0.57 | 11 | 0 | 3.32884 |
| 3 | gi|5453740 | MRCL3 | 19.8/4.67 | 21.0/4.46 | 0.63 | 12 | 0 | 22.6371 |
| 4 | gi|11957604 | GLO1 | 21.0/5.12 | 23.1/5.38 | 0.29 | 19 | 0.1458951 | 5.8117 |
| 5 | gi|11958606 | PRDX6 | 22.8/5.98 | 20.6/5.65 | 0.56 | 14 | 0.3751647 | 3.13876 |
| 6 | gi|4504517 | HSPB1 | 22.8/5.98 | 21.5/6.26 | 0.49 | 11 | 1.7085594 | 6.85507 |
| 7 | gi|4506195 | PSMB2 | 23.0/6.51 | 17.7/5.28 | 0.37 | 11 | 0.5424243 | 3.47661 |
| 8 | gi|11962339 | RANBP1 | 23.5/5.19 | 26.9/4.31 | 0.42 | 16 | 0.5717237 | 5.58285 |
| 9 | gi|31645 | GAPDH | 31.7/7.15 | 35.9/4.79 | 0.57 | 27 | 2.3023371 | 3.26693 |
| 10 | gi|914957 | GRB2 | 25.3/5.89 | 28.4/5.48 | 0.51 | 13 | ND | 5.22913 |
| 11 | gi|20149675 | TUBB | 26.8/5.15 | 31.2/4.82 | 0.46 | 12 | 3.1617546 | 9.1124 |
| 12 | gi|16357477 | CDC34 | 26.9/4.41 | 28.0/4.2 | 0.32 | 9 | 0.3679367 | 3.0581 |
| 13 | gi|44979607 | IMPDH2 | 27.3/5.84 | 29.4/6.1 | 0.6 | 9 | 0.4022986 | 5.13788 |
| 14 | gi|48145973 | NDUFV2 | 27.7/8.22 | 26.3/8.67 | 0.33 | 8 | 0.3474179 | 2.92579 |
| 15 | gi|19298714 | PGLS | 27.8/5.7 | 25.6/5.37 | 0.37 | 9 | 0.4460076 | 3.16474 |
| 16 | gi|4758484 | GSTO1 | 27.8/6.23 | 26.5/6.51 | 0.48 | 15 | 0.2449022 | 4.34464 |
| 17 | gi|19378506 | PSPHL;ACTG1 | 28.5/5.2 | 23.2/3.97 | 0.45 | 12 | 0.4615403 | 3.08667 |
| 18 | gi|17402904 | EXOSC6 | 28.5/6.06 | 31.9/5.18 | 0.3 | 12 | 0.2836561 | 4.7922 |
| 19 | gi|62897753 | hCG_2015269 | 29.0/6.67 | 26.8/6.34 | 0.79 | 19 | 0.3179457 | 3.64943 |
| 20 | gi|19298714 | ERP29 | 29.0/6.77 | 27.7/7.05 | 0.39 | 10 | 0.4765053 | 3.36723 |
| 21 | gi|4826659 | CAPZB | 29.6/6.45 | 24.3/5.22 | 0.64 | 26 | 0.4184514 | 2.90687 |
| 22 | gi|19438056 | LASP1 | 30.1/6.61 | 33.5/5.73 | 0.58 | 8 | 0.2488259 | 5.5738 |
| 23 | gi|22652991 | TPI1 | 31.1/5.65 | 35.3/3.29 | 0.56 | 14 | 0.4883441 | 7.47102 |
| 24 | gi|21361454 | PYCR2 | 34.4/8.48 | 37.5/8.07 | 0.54 | 24 | 0.3732397 | 5.90677 |
| 25 | gi|18906542 | HNRPC | 32.4/4.94 | 36.8/4.61 | 0.53 | 20 | 0.5408701 | 6.3532 |
| 26 | gi|12803709 | CK14 | 32.4/4.97 | 33.5/4.76 | 0.33 | 17 | 0.4598442 | 3.63144 |
| 27 | gi|11056044 | PPA1 | 33.1/5.54 | 35.3/5.8 | 0.5 | 17 | 0.4610924 | 2.78434 |
| 28 | gi|5174447 | TST | 33.6/6.77 | 32.3/7.22 | 0.37 | 14 | 0.4618559 | 2.78215 |
| 29 | gi|10437514 | SUHW4 | 35.1/8.68 | 32.9/8.35 | 0.42 | 14 | ND | 11.2023 |
| 30 | gi|11962858 | DNAH3 | 473.8/6.04 | 472.5/6. | 0.27 | 14 | 0.3843506 | 2.9416 |
| 31 | gi|4826643 | ANXA3 | 36.5/5.63 | 31.2/4.4 | 0.56 | 23 | 0.5335611 | 5.71278 |
| 32 | gi|19438981 | CNN3 | 36.6/5.69 | 35.2/6.14 | 0.49 | 17 | 3.3243097 | 7.20813 |
| 33 | gi|2119276 | TUBB | 48.1/4.7 | 45.9/4.37 | 0.25 | 14 | 1.8615634 | 3.20717 |
| 34 | gi|73976124 | HNRPA2B1 | 37.5/8.97 | 36.1/9.25 | 0.46 | 12 | 2.5249017 | 6.79964 |
| 35 | gi|17402893 | PSAT1 | 40.8/7.56 | 35.5/6.33 | 0.55 | 26 | 1.6654277 | 3.88025 |
| 36 | gi|19437787 | ARFIP2 | 37.9/5.72 | 41.4/4.84 | 0.38 | 12 | 1.7420536 | 2.80585 |
| 37 | gi|22104024 | PPP1CB | 38.0/5.84 | 42.2/3.48 | 0.22 | 8 | 0.3430111 | 4.85873 |
| 38 | gi|11960877 | PPP1CB | 38.0/5.84 | 41.1/5.43 | 0.4 | 23 | 0.4678427 | 3.05192 |
| 39 | gi|6912280 | AHSA1 | 38.4/5.41 | 33.1/4.18 | 0.19 | 8 | 4.3644831 | 5.00035 |
| 40 | gi|7305503 | STOML2 | 38.6/6.88 | 42.1/6 | 0.52 | 18 | 0.2565899 | 4.2268 |
| 41 | gi|11959799 | ANXA2 | 38.8/7.57 | 36.6/7.24 | 0.6 | 20 | 2.2824303 | 4.00567 |
| 42 | gi|4506127 | PRPS1 | 35.3/6.51 | 34.0/6.79 | 0.47 | 21 | 1.9003629 | 5.43115 |
| 43 | gi|93277122 | BUB3 | 32.1/8.4 | 26.8/7.17 | 0.43 | 15 | 3.4236124 | 10.8789 |
| 44 | gi|4503895 | GALK1 | 42.7/6.04 | 46.9/3.68 | 0.35 | 17 | 6.5938433 | 3.2962 |
| 45 | gi|11960222 | CKB | 42.9/5.34 | 46.0/4.93 | 0.45 | 15 | 2.6276618 | 3.86433 |
| 46 | gi|15825518 | SERPINB7 | 43.2/6.34 | 47.6/6.01 | 0.34 | 12 | 0.2890603 | 11.6406 |
| 47 | gi|19438418 | HSPA8 | 71.1/5.37 | 72.2/5.16 | 0.24 | 14 | 1.8781699 | 3.44432 |
| 48 | gi|5174529 | MAT2A | 44.0/6.02 | 46.1/6.28 | 0.32 | 14 | 0.3873342 | 3.38533 |
| 49 | gi|19378506 | ACTB | 45.5/5.82 | 44.1/6.27 | 0.27 | 7 | 2.5188648 | 4.86371 |
| 50 | gi|5729991 | PSMC4 | 47.5/5.09 | 45.2/4.76 | 0.45 | 19 | 2.9751505 | 7.05399 |
| 51 | gi|20328236 | ENO1 | 47.5/7.01 | 50.9/6.13 | 0.33 | 14 | 0.4514455 | 5.84011 |
| 52 | gi|20328236 | ENO1 | 47.5/7.01 | 51.7/4.65 | 0.38 | 14 | 2.1757631 | 2.78494 |
| 53 | gi|28872725 | PSMD11 | 47.8/6.08 | 50.9/5.67 | 0.25 | 12 | 2.2969517 | 4.67432 |
| 54 | gi|35071 | MTHFD2 | 38.0/8.86 | 42.4/8.53 | 0.02 | 11 | 3.0997653 | 3.33509 |
| 55 | gi|21623852 | PSMC3 | 49.5/5.13 | 48.1/5.58 | 0.22 | 19 | 3.0467243 | 3.32863 |
| 56 | gi|21623852 | PSMC3 | 49.5/5.13 | 47.3/4.8 | 0.47 | 24 | 1.9425454 | 4.2506 |
| 57 | gi|20328236 | ENO1 | 47.5/7.01 | 46.2/7.29 | 0.63 | 29 | 1.5667183 | 4.24397 |
| 58 | gi|16507237 | VIM | 49.7/5.19 | 44.4/3.96 | 0.4 | 20 | 2.3352391 | 3.4501 |
| 59 | gi|14043072 | HNRPA2B1 | 37.5/8.97 | 40.9/8.09 | 0.5 | 25 | 2.118541 | 2.97696 |
| 60 | gi|11960877 | TUBB2A | 50.3/4.78 | 54.5/2.42 | 0.42 | 16 | 2.5802857 | 3.58321 |
| 61 | gi|28948869 | VIL2 | 69.5/5.94 | 72.6/5.53 | 0.39 | 18 | 2.6162373 | 3.95062 |
| 62 | gi|11960683 | GDI2 | 51.1/6.11 | 55.5/5.78 | 0.38 | 16 | 4.2414899 | 8.61448 |
| 63 | gi|38569475 | MTX1 | 51.7/9.8 | 46.4/8.57 | 0.36 | 14 | ND | 10.9218 |
| 64 | gi|449441 | UGP2 | 55.8/7.69 | 59.2/6.81 | 0.39 | 24 | 3.9755451 | 4.09809 |
| 65 | gi|48255968 | UGP2 | 55.8/7.69 | 60.0/5.33 | 0.52 | 27 | 2.5612084 | 6.32672 |
| 66 | gi|66933016 | IMPDH2 | 56.2/6.44 | 59.4/6.03 | 0.42 | 20 | 0.4828852 | 10.5545 |
| 67 | gi|11961763 | CCT2 | 57.8/6.01 | 62.2/5.68 | 0.39 | 15 | 4.6695429 | 4.39079 |
| 68 | gi|4557976 | ALDOA | 39.9/8.3 | 41.0/8.09 | 0.47 | 20 | 2.0385729 | 3.2317 |
| 69 | gi|22104614 | CCT5 | 60.1/5.45 | 62.2/5.71 | 0.53 | 26 | 2.3538723 | 4.52031 |
| 70 | gi|20151189 | GLUD1 | 61.7/7.66 | 60.3/8.11 | 0.33 | 13 | 0.3442582 | 9.36776 |
| 71 | gi|19437434 | HSPA5 | 72.5/5.07 | 70.3/4.74 | 0.71 | 24 | 5.0750799 | 3.96361 |
| 72 | gi|11463825 | STIP1 | 63.2/6.4 | 66.7/5.52 | 0.51 | 37 | 3.6341375 | 2.79519 |
| 73 | gi|16915936 | MGAT4B | 65.1/8.38 | 69.3/6.02 | 0.29 | 10 | 2.8100682 | 3.95651 |
| 74 | gi|9910460 | ZNF714 | 69.3/9.16 | 72.4/8.75 | 0.3 | 9 | 2.8886894 | 3.2727 |
| 75 | gi|182087 | CTTN | 61.9/5.28 | 66.3/4.95 | 0.32 | 21 | 7.0312418 | 6.24639 |
| 76 | gi|13676857 | HSPA2 | 70.3/5.56 | 68.9/6.01 | 0.27 | 15 | 2.6325321 | 3.0962 |
| 77 | gi|13676857 | HSPA2 | 70.3/5.56 | 68.1/5.23 | 0.44 | 29 | ND | 37.1179 |
| 78 | gi|1017722 | ZNF85 | 70.9/9.48 | 69.6/9.76 | 0.26 | 14 | 3.6214245 | 3.22252 |
| 79 | gi|57014047 | LMNA | 70.9/8.55 | 65.6/7.32 | 0.5 | 31 | 5.398146 | 3.33403 |
| 80 | gi|29645294 | SNIP | 112.7/9.32 | 116.1/8. | 0.24 | 16 | 2.8558356 | 3.07003 |
| 81 | gi|542991 | ROR2 | 80.8/7.3 | 85.1/4.94 | 0.51 | 12 | ND | 11.6562 |
VIL2 – Chain A, structure of the active ferm domain of ezrin; CTTN – cortactin.