Abstract
Nicotinamide phosphoribosyltransferase (NAMPT) is a pleiotropic protein implicated in the pathogenesis of acute respiratory distress syndrome, aging, cancer, coronary heart diseases, diabetes, nonalcoholic fatty liver disease, obesity, rheumatoid arthritis, and sepsis. However, the underlying molecular mechanisms of NAMPT in these physiological and pathological processes are not fully understood. Here, we provide experimental evidence that a Nampt gene homozygous knockout (Nampt−/−) resulted in lethality at an early stage of mouse embryonic development and death within 5–10 days in adult mice accompanied by a 25.24±2.22% body weight loss, after the tamoxifen induction of NamptF/F × Cre mice. These results substantiate that Nampt is an essential gene for life. In Nampt−/− mice versus Nampt+/+ mice, biochemical assays indicated that liver and intestinal tissue NAD levels were decreased significantly; histological examination showed that mouse intestinal villi were atrophic and disrupted, and visceral fat was depleted; mass spectrometry detected unusual higher serum polyunsaturated fatty acid containing triglycerides. RNA-seq analyses of both mouse and human pediatric liver transcriptomes have convergently revealed that NAMPT is involved in key basic cellular functions such as transcription, translation, cell signaling, and fundamental metabolism. Notably, the expression of all eight enzymes in the tricarboxylic acid cycle were decreased significantly in the Nampt−/− mice. These findings prompt us to posit that adult Nampt−/− mouse lethality is a result of a short supply of ATP from compromised intestinal absorption of nutrients from digested food, which leads to the exhaustion of body fat stores.
Nicotinamide phosphoribosyltransferase (NAMPT) was initially named pre-B-cell colony-enhancing factor (PBEF)1 as it synergized the pre-B-cell colony-formation activity of stem cell factor and interleukin 7. NAMPT catalyzes the condensation of nicotinamide with 5-phosphoribosyl-1-pyrophosphate to yield nicotinamide mononucleotide, an intermediate in the biosynthesis of NAD.2 In 2005, Fukuhara et al.3 reported Nampt as a Visfatin, which was enriched in visceral fat with insulin-mimetic function. The list of NAMPT's physiological roles is continuously growing. The dysregulation of NAMPT gene has been implicated in several human diseases.4 However, the underlying molecular mechanisms are not fully understood.
Loss-of-function experiments reveal the functions of a gene. The most dramatic phenotype is lethality, defining a gene as essential.5 Recently, three groups reported that there are about 2000 core genes essential for life in several types of human cells.6, 7, 8 They also found that the essentiality of some genes is context-dependent and affects viability in a cell-type-specific manner.9 It is more logical to establish any essential gene in an animal model, by exploring the terminal phenotypes and the various molecular mechanisms underlying lethality. The mouse has long been regarded as an ideal model system for this purpose. Conservation between humans and mice is underscored by strong one-to-one orthologous relationships between genes of the two species.10 We have previously reported that the Nampt gene DNA coding sequence between mouse and human is 96% identical.11 Hence, the homozygous Nampt gene knockout in mice would offer an attractive model to decipher fully the Nampt gene's pleiotropic roles, by probing molecular mechanisms underlying lethality in adult mice.
Here, we provide experimental evidence that the Nampt gene homozygous knockout (Nampt−/−) resulted in mouse embryonic lethality at an early stage. We also established Nampt gene conditional knockout mice and found that induced Nampt−/− adult mice died within 5–10 days, accompanied by a 25.24±2.22% body weight loss. These results substantiate that Nampt is an essential gene in vivo. RNA-seq analyses of both mouse and human pediatric liver transcriptomes revealed that Nampt is involved in key basic cellular functions such as transcription, translation, cell signaling, and fundamental metabolism. Particularly, expression of all eight enzymes in the tricarboxylic acid cycle(TAC) in Nampt−/− adult mice were decreased significantly, which suggests that depletion of cellular ATP energy sources may be the main culprit of adult Nampt−/− mouse death.
Results
Homozygous Nampt gene knockout mice are embryonically lethal
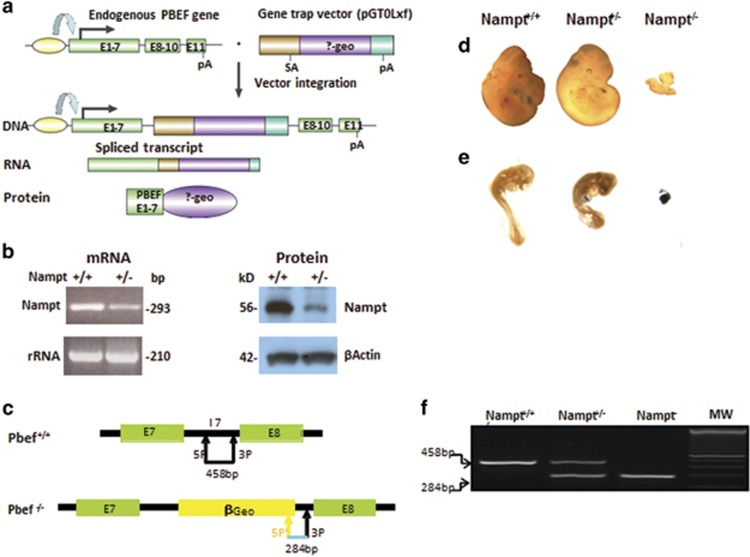
To study the biological functions of the Nampt gene, we established the Nampt gene-trapped mouse model (Figure 1a). To confirm the mutant ES cell line, RT-PCR results showed that the cell line only expresses about 50% Nampt mRNA of the Nampt+/+ wild-type control, and western blotting demonstrated that cellular Nampt protein in the Nampt+/− ES cell line is decreased significantly (Figure 1b). The validated ES cell lines were injected into blastocysts of pseudopregnant female C57BL/6N mice at the Johns Hopkins University transgenic mouse core facility to generate the Nampt+/− knockdown mouse model. To facilitate mouse genotyping needs, we sequenced the whole region of Nampt gene intron-7, including the inserted gene-trap cassette (Supplementary Figure 1), and designed three PCR primers (two forward and one reverse) to detect Nampt wild-type (Nampt+/+), Nampt heterozygous (Nampt+/−), and Nampt knockout (Nampt−/−) mice as indicated in Figure 1c. Representative genotyping images of Nampt+/+, Nampt+/−, and Nampt−/− mice are presented in Figure 1f. To generate Nampt−/− knockout mice, Nampt+/− male and female mice were bred. At embryonic day 11.5, Nampt+/+ and Nampt+/− embryos both have normal development with 40–45 pairs of somites, and normal embryo size. Nampt−/− embryos are much smaller in size, and display unclear abnormal embryo structure (Figure 1d). To establish the Nampt−/− embryonic lethality, we performed a β-gal staining of mouse embryos at 8.5 d.p.c. stage, as the inserted gene-trap vector β-Geo can express β-gal. A representative normal size Nampt+/− embryo has a diffused brown color staining as in the Nampt+/+ embryo, while an undeveloped Nampt−/− embryo shows strong dark blue staining (Figure 1e). These results corroborate that the Nampt−/− mice are lethal at an early embryonic stage.
Figure 1.
Homozygous Nampt gene knockout mice are embryonically lethal. (a) The schematics of gene-trap strategy to create Nampt knockout mice. Gene-trap cassette were inserted in Nampt gene intron-7 and the transcription of the ‘trapped' Nampt gene results in a truncated mRNA that will translate into a Nampt exon 1–7 and β-geo fusion protein. (b) Phenotypic verification of Nampt gene-trapped embryonic stem (ES) cell line. Phenotypic verifications of an ES cell line (RRR084) with Nampt gene interruption (Nampt+/−) were carried out by RT-PCR for Nampt mRNA and western blotting for Nampt protein expression. Nampt+/+ represents wild-type ES cell line without a gene-trap vector insertion. (c) Genotyping strategy. Mouse yolk sac or tail DNA was used for genotyping by PCR. The locations of Nampt gene intron-7 and β-Geo specific primers used for genotyping and predicted size of the PCR products are shown. (d) Representative mouse embryos at 11.5 d.p.c. Timed pregnancies and embryo collection were carried out as described in the 'Materials and Methods' section. Mouse embryo images from representative Nampt+/+ , Nampt+/−, and Nampt−/− mice at 11.5 d.p.c. are displayed. (e) Representative mouse embryos at 8.5 d.p.c. Timed pregnancies and embryo collection were carried out as described in the 'Materials and Methods' section. Mouse embryo images from representative Nampt+/+, Nampt+/−, and Nampt−/− mice at 8.5 d.p.c. after β-Gal staining are displayed. (f) Representative genotyping images. Mouse genotyping was done using PCR amplification of genomic DNA isolated from mouse embryonic tissue or yolk sac with either primer pairs 5P and 3P or 5P' and 3P as depicted in c. The PCR products were subjected to 2% agarose electrophoresis and stained by ethidium bromide
Nampt is an essential gene for adult mouse survival
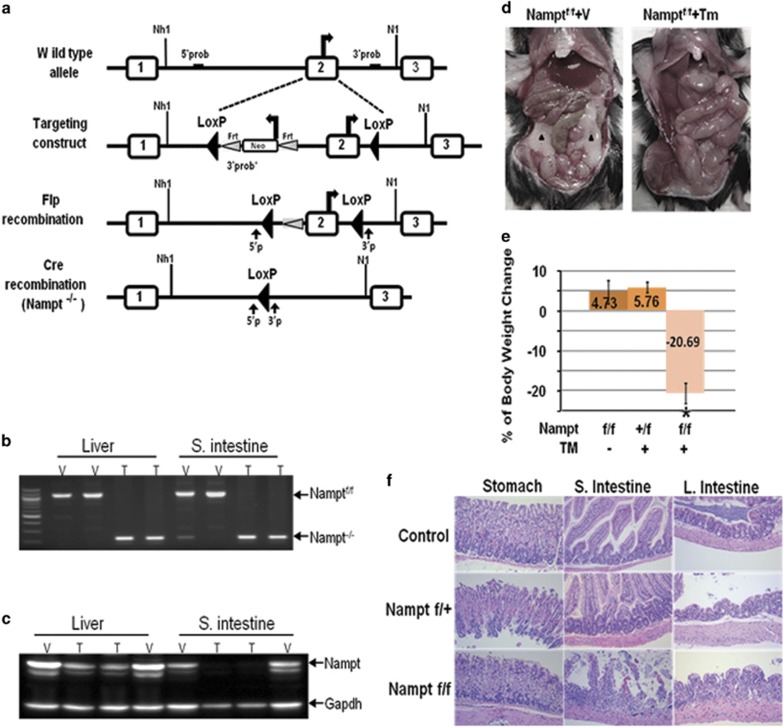
As Nampt−/− mice are embryonically lethal, we cannot use Nampt−/− mice for a complete ‘loss of function' study and hence we generated a Nampt conditional knockout mouse line (NampF/F). The Nampt gene spans over 36.9 kb and contains 11 exons and 10 introns, thus preventing the convenient floxing of its entire gene. We decided to introduce loxP sites into introns flanking exon 2. A Neo selection cassette flanked by FRT sites was inserted adjacent of the loxP site within intron-1 as well (Figure 2a). The Namptflox(Neo) targeting vector was introduced into mouse embryonic stem cells (mES), which were then injected into C57BL/6 host blastocysts to generate chimeric mice (Supplementary Figure 2). Namptflox(Neo)/+ heterozygous mice were crossed with the ACTB::FLPe deleter mouse line to remove the PGK-Neo cassette, which has been reported to interfere with gene expression (Figure 2a). Namptflox/+ mice were backcrossed with C57BL/6J wild-type mice for 10 generations before being considered as a pure C57BL/6 background, and subsequently intercrossed to obtain homozygous Namptflox/flox mice. All the genotypes were confirmed by PCR analysis (Figure 2b) and tissue expression levels of Nampt protein were verified by western blotting (Figure 2c). Namptflox/flox mice were born at a normal Mendelian ratio and were viable without displaying any obvious defect. To conditionally induce cre-loxP recombination, Namptflox/flox mice were mated to UbcCreERT2 mice. After 4-day intraperitoneal administration of tamoxifen (TMX), Namptflox/flox mice (n=10) displayed rapid weight loss. These mice became dehydrated, less active on day 3 after injection and reached >20% weight loss within 5–10 days before dying (Figure 2e). Dissection of dying Nampt−/− mice showed fat tissue depletion and intestinal abnormality in TMX-treated UbcCreERT2: Namptflox/flox mice (Figure 2d). The Nampt protein expression levels were dramatically reduced in TMX-treated UbcCreERT2: Namptflox/flox mice after day 5(Figure 2c). Histological examination revealed that Nampt depletion induced severe intestinal villus atrophy and the disruption of normal architecture (Figure 2f) compared with the wild-type control mice.
Figure 2.
Nampt is an essential gene for adult mouse survival. (a) Gene targeting strategy for production of mice for conditional inactivation of Nampt gene. Part of intron-1 of the Nampt gene was used as 5′ arm, part of intron-2 as 3′ arm, exon 2 floxed, Neo and TK used as positive and negative selection marker, respectively. In the process of homologous recombination, targeting construct replaced the exon 2 of Nampt gene. Neo was selectively removed by Flipase recombinase. Cre-Lox recombination leads to Nampt second exon excision. 5′P and 3′P represent mice genotyping PCR primers and the black arrows indicate the location of the primers. (b) Genotyping of Namptf/f and Nampt−/− mice. Representative images of genotyping of Namptf/f and Nampt−/− mice by PCR amplification of either liver or small intestine tissue DNAs are shown. T, tamoxifen; V, vehicle. (c) Nampt protein expression in Namptf/f and Nampt−/− mice. Western blotting detecting the Nampt protein expression in mouse liver and small intestine tissues are shown. Gapdh was included as a loading control. (d) Gross morphology of TMX-treated or control mouse abdominal visceral tissues. After TMX (0.04 mg/kg/day, ip) injection for 4 days, mice body weight was closely monitored. All the UbcCreERT2; Namptflox/flox mice treated with TMX will loss 20% the body weight within 5 days. Just before mouse morbid, mouse abdomen were dissected as shown. Black arrow head indicate the visceral adipose tissues in a vehicle control mouse. (e) Statistical analysis of mouse body weight changes. There are two groups of control mice, Namptf/f treated with vehicle and Nampt+/f treated with TMX, both are compared with Namptf/f mice treated with TMX; N=10 for each group. *P<0.01 Nampt−/−versus either Namptf/f or Nampt+/f group. (f) Histology of gastrointestinal track by H&E staining. Amplification, × 100; TMX, tamoxifen; Veh, vehicle
Abnormal serum lipid profile in Nampt−/− mice
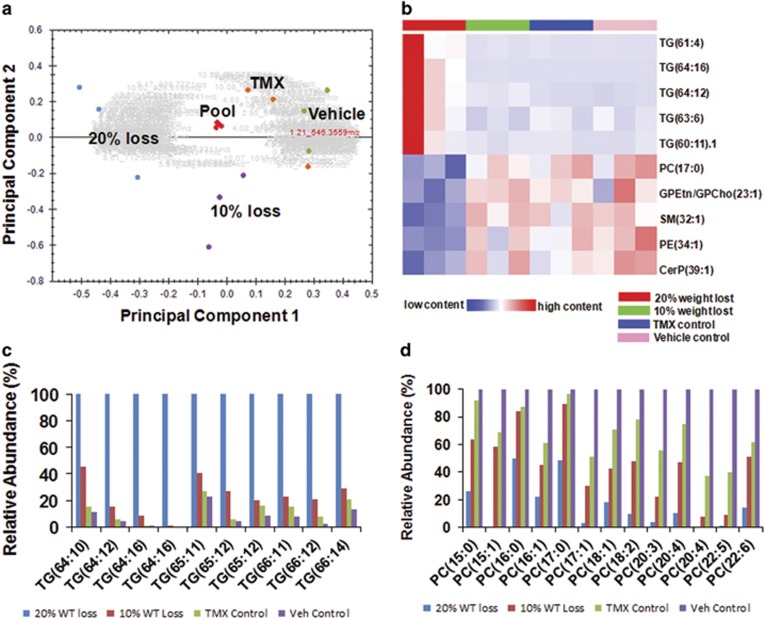
To characterize the rapid catabolism of visceral fat in Nampt−/− mice, we analyzed the serum lipid content in these mice (Supplementary Table 1) by UHPLC-qTOF MS method. Principal component analysis revealed that each experimental group clustered as a unique entity (Figure 3a). One hundred forty-one lipids were significantly and differentially expressed among the four experimental groups (Supplementary Table 1 and Supplementary Figure 3). A heatmap of the top 10 differentially presented lipids is displayed in Figure 3b. High molecular weight Triacylglyceride (TG) with long polyunsaturated carbon side chains (~4 or more double bonds) were more prevalent in the 10 and 20% weight loss group compared with the control groups (Figure 3c,Supplementary Table 1 and Supplementary Figure 3). Conversely, TGs with a lower number of side chain carbons and fewer double bonds were less abundant in the control mice than in the mice with weight loss, especially the 20% weight loss group. In contrast to TG levels, phosphatidylcholine lipid (PC) levels were less abundant in the Nampt knockout mice with 10 and 20% weight loss relative to the tamoxifen and vehicle control mice (Figure 3d, Supplementary Table 1 and Supplementary Figure 3).
Figure 3.
Serum lipid profiles in Nampt+/+ and Nampt−/− mice. (a) PCA analysis of mouse serum lipid profiles. PCA score plot is derived from MS spectrum of serum samples from vehicle control (vehicle •), tamoxifen control (TMX •), 10% weight loss (10% loss •), 20% weight loss (20% loss •), and pooled samples (Pool •). N=3 per group. (b) Heatmap of differentially expressed serum lipids in mice. Heatmap of the top five upregulated and the top five downregulated differentially expressed lipids between the Nampt knockdown (20% weight loss, 10% weight loss) and control (tamoxifen and vehicle) groups is presented. (c) Relative abundance of mouse serum triacylglycerides (TG). Relative abundance of TG with increased number side chain carbons (⩾64) and double bonds between the four groups is represented. (d) Relative abundance of mouse serum phosphatidylcholines (PC). Relative abundance of PC with 15 to 22 side chain carbons between the four groups is represented
Liver transcriptome profile in Nampt−/− mice
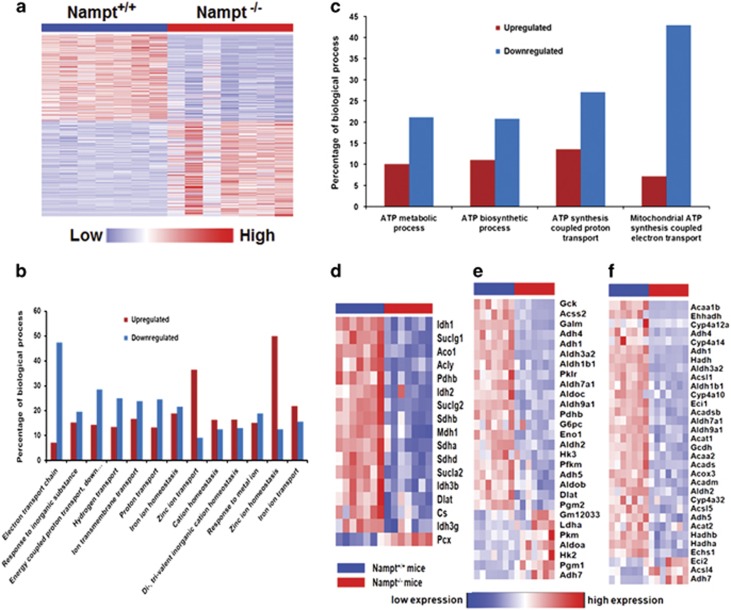
To comprehensively investigate the effect of the Nampt gene knockout on the mouse liver transcriptome profile, we used RNA-seq to determine and compare hepatic gene expression changes between Nampt−/− and wild-type Nampt+/+ mice. A total 4638 genes with significantly differential expression (P-value <0.05) in Nampt−/− mice were identified, of which 2434 genes were upregulated and 2204 genes were downregulated (Supplementary Table 2). A total 2443 genes (52.6%) had >2-fold change (Figure 4a). Most of the genes (97.5%) were protein-coding genes, 34 genes were lincRNAs, 19 were antisense RNAs, and 232 genes coded for transcription factors. GO biological process and KEGG pathway enrichment for the 4638 genes were calculated (Supplementary Table 3 and Supplementary Table 4). The results showed that 476 biological processes and 57 significant KEGG pathways were enriched (P-value <0.05). GO biological process showed that 1050 genes were involved in 106 metabolic processes and 272 genes were involved in 43 biosynthetic processes. In 91 metabolic processes and 40 biosynthetic processes, there were more downregulated genes than upregulated genes (Supplementary Table 5). The examined KEGG pathways include metabolism of xenobiotics by cytochrome P450, drug metabolism, fatty acid metabolism, glutathione metabolism, nitrogen metabolism, and the citrate cycle, glycolysis/gluconeogenesis, and nicotinate and nicotinamide metabolism (Supplementary Table 4), which are pertinent to liver and Nampt functions. These results suggest that most metabolic and biosynthetic processes have been slowed down in Nampt−/− mice.
Figure 4.
Liver transcription profiles in Nampt−/− mice. (a) Heatmap of 4638 significantly differentially expressed liver genes in Nampt−/− mice versus Nampt+/+ mice. Each column represents a mouse sample. Each row represents a gene. Red color indicates an increased expression. Blue color indicates a decreased expression. N=7 for each group. (b) Expression profile of significantly differentially expressed liver genes in 13-ion-associated transport biological processes of Nampt−/− mice versus Nampt+/+ mice. Percentage value is derived from the number of significantly differentially expressed expression genes in each process divided by total genes involved in 13-ion-associated transport processes. (c) Expression profile of significantly differentially expressed liver genes in four ATP-associated metabolic processes of Nampt−/− mice versus Nampt+/+ mice. Percentage value is derived from the number of significantly differentially expressed expression genes in each process divided by total genes involved in four ATP-associated metabolic processes. (d) Heatmap for significantly differentially expressed genes in citrate cycle in Nampt−/− mice versus Nampt+/+ mice. (e) Heatmap for significantly differentially expressed genes in glycolysis and gluconeogenesis in Nampt−/− mice versus Nampt+/+ mice. (f) Heatmap for significantly differentially expressed genes in fatty acid metabolism in Nampt−/− mice versus Nampt+/+ mice. In all heat maps, red color indicates high expression, blue color indicates low expression. N=7 in each group. The expression of all genes in (d, e and f) were significantly changed between Nampt−/− and Nampt+/+ (P-value <0.05)
As adult Nampt−/− mice showed the similar intestinal villus atrophy and damage (Figure 2f) as adult replication protein A subunit3 (Rpa3, another proven essential gene) knockout mice, we compared expression levels of the liver Rpa3 gene and three other genes(Trp53, Atr and Mapk7), whose conditional homozygous knockouts caused the death,5, 12, 13 between Nampt−/− versus Nampt+/+ mice, to determine whether the Nampt−/− caused lethality is due to downregulation of these genes. Only the Rpa3 gene expression was significantly downregulated in Nampt−/− mice versus Nampt+/+ mice (2.40±0.75 versus 5.19±0.87, Supplementary Figure 4).
As intestinal swelling (Figure 2d), villus atrophy, and damage (Figure 2f) occurred in Nampt−/− mice, we surveyed significantly differentially expressed genes in 13-ion-associated transport processes of Nampt−/− mouse liver tissues, which may provide information about the gastrointestinal absorption or transport of ions and nutrients from the food supply. As presented in Figure 4b, 8 out of 13-ion-associated transport processes were downregulated. Furthermore, 47.3% of genes in the whole electron transport chain (53 genes in total) were downregulated, indicating that the normal function of the electron transport chain has been compromised (Supplementary Table 6). In mitochondrial ATP biosynthesis coupled electron transport, more genes were inhibited. The same trend was true of three other ATP involved processes (Figure 4c). Of note is that the expression of 16 enzymes in TCA were downregulated, except Pcx, which catalyzes the carboxylation of pyruvate to oxaloacetate involved in gluconeogenesis (Figure 4d).
We further focused on the analysis of those genes involved in carbohydrate and lipid metabolism (Supplementary Table 7). Most genes in glycolysis were downregulated, but most genes in gluconeogenesis were upregulated (Figure 4e). Expressions of 29 of 32 enzymes involved in fatty acid metabolism were downregulated (Figure 4f). These findings suggest that the Nampt gene knockout may result in a decrease of glucose and fatty acid metabolism and an augmentation of glucose synthesis by gluconeogenesis in mouse livers. We also noticed that the expression of the lipase-encoding genes (Lipc, Lipg), 2, 4-dienoyl CoA reductase genes (Decr1, Decr2), and the enoyl-CoA isomerase (Eci1) were all downregulated in Nampt−/− mice compared with the Nampt+/+ control mice. This may explain the increased accumulation of serum high polyunsaturated fatty acid containing TGs in Nampt−/− mice compared with the Nampt+/+ control mice (Figure 3c). To investigate whether the reduced circulating phosphatidylcholine (PC) levels in Nampt−/− mice (Figure 3d) might be explained by changes in transport, we examined the expression profiles for genes encoding PC transporters in our RNA-seq data (Supplementary Table 8). The expressions of several PC transporters, including Abca3, Abcb4, Pcyt1a, and Stard7, were decreased significantly in Nampt−/− mice compared with the Nampt+/+ mice.
Effect of Nampt gene knockout on NAD metabolism and NAD-involved gene expression
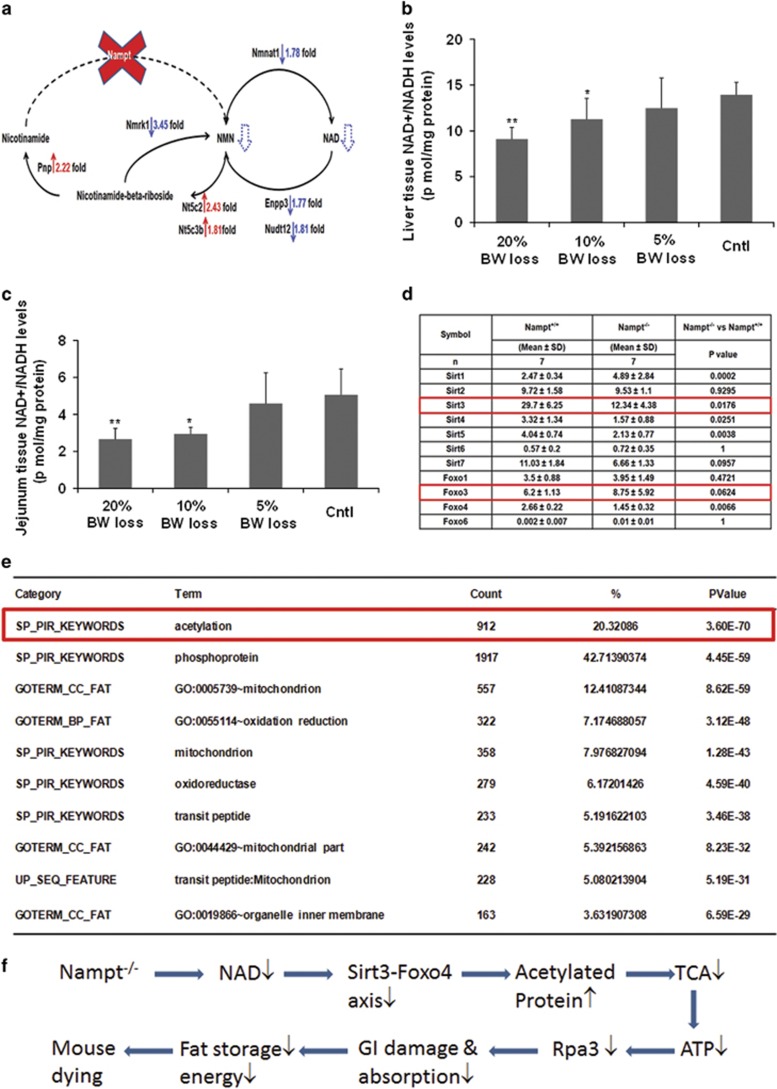
Nampt is a key enzyme in the cellular NAD synthesis of a mammalian salvage pathway from nicotinamide. However, the global effect and full repertoire of gene pools affected by the Nampt gene knockout are not fully characterized. The adult Nampt−/− mice have afforded us the opportunity to realize this goal. Figure 5a displays that in Nampt−/− mice, expression of nicotinamide nucleotide adenylyltransferase 1(Nmnat1), which catalyzes the formation of NAD+ from nicotinamide mononucleotide (NMN), was decreased significantly. Ectonucleotide pyrophosphatase/phosphodiesterase 3(Enpp3), which catalyzes the hydrolysis of pyrophosphate bonds in NAD+ to regenerate NMN, Nudix (nucleoside diphosphate linked moiety X)-type motif 12 (Nudt12), which cleaves NAD+ to AMP and NMN, and Nicotinamide riboside kinase 1 (Nmrk1), which catalyzes the nicotinamide-β-riboside to form NMN were all downregulated. Expression of three enzyme genes, Nt5c2: 5′-nucleotidase, cytosolic II (Nt5c2), 5′-nucleotidase, cytosolic IIIB (Nt5c3b), and Purine-nucleoside phosphorylase (Pnp), were all upregulated. Activation of these genes contributes to the generation of nicotinamide from NMN. All these interactions led to the decreased synthesis of NMN and hence NAD. Indeed, we found that NAD levels in liver tissue and jejunum tissue were decreased significantly in Nampt−/− mice versus Nampt+/+ mice and paralleled to their body weight loss. In liver tissues, NAD levels are significantly lower in 20 and 15% body weight loss groups than the control group (9.10±1.27 and 11.27± 2.27 versus 13.94±1.35 pmole/mg tissue protein; Figure 5b). In jejunum tissue, NAD levels are significantly lower in 20 and 15% body weight loss groups than the control group (2.66±0.59 and 2.93±0.38 versus 5.06±1.40 pmole/mg tissue protein). In both tissues, NAD levels are decreased in 5% body weight loss group too.
Figure 5.
NAD metabolism and NAD-involved gene expression. (a) NAD synthesis from Nampt catalyzed salvage pathway and its metabolism in Nampt−/− mice. Solid red arrows indicate gene upregulation while blue arrows indicate gene downregulation. Blue dotted arrows indicate reduced metabolite production. (b) Mouse liver NAD levels. Mouse liver tissues (30 mg) were homogenized and the supernatants were taken for NAD+/NADH assays as described in the 'Materials and Methods' section using a fluorimetric NAD+/NADH assay kit; 20, 10, and 5% BW loss groups versus tamoxifen-treated control (cntl) group, respectively (Bar=mean±S.D., each group n=5; *P<0.05, **P<0.01). BW, body weight. (c) Mouse jejunum tissue NAD levels. Mouse jejunum tissues (30 mg) were homogenized and the supernatants were taken for NAD+/NADH assays as described in the 'Materials and Methods' section using a fluorimetric NAD+/NADH assay kit; 20, 10, and 5% BW loss groups versus Tamoxifen-treated control (cntl) group, respectively (Bar=mean±S.D., each group n=5; *P<0.05, **P<0.01). BW, body weight. (d) Liver Sirt and Foxo gene expression in both Nampt−/− versus Nampt+/+ mice. RNA-seq was carried out as described in the 'Materials and Methods' section. Gene expression levels are expressed as mean±S.D. of FPKM in each group. (e) Top 10 function enrichment for 4638 significantly changed genes in Nampt−/− versus Nampt+/+ mice. (f) Schema of possible dying causes in Nampt−/− mice. The explanation is provided in the 'Discussion' section
NAD is a critical cofactor for sirtuin protein deacetylase. We examined the Nampt gene knockout on the expression of seven sirtuin genes. RNA-seq revealed that expression of Sirt3, 4, and 5 in mitochondria were decreased significantly while expression of Sirt1 in nuclei was increased significantly in Nampt−/− mice versus Nampt+/+ mice (Figure 5d). Sirtuin protein controls mitochondrial function through the deacetylation of targets that include the FOXO protein family. We then examined the effect of sirtuin downregulation on the expression status of the liver-expressed FOXO protein family. Liver Foxo4 gene expression was inhibited significantly in the Nampt−/− mice (Figure 5d). Sirtuin–Foxo signaling may control the expression of a number of genes. Figure 5e shows that acetylated proteins are one of the two top affected protein families, accounting for 20% of 4638 significantly differently expressed genes in Nampt−/− versus Nampt+/+ mice.
Co-expressed genes of human NAMPT gene in human prenatal and pediatric livers
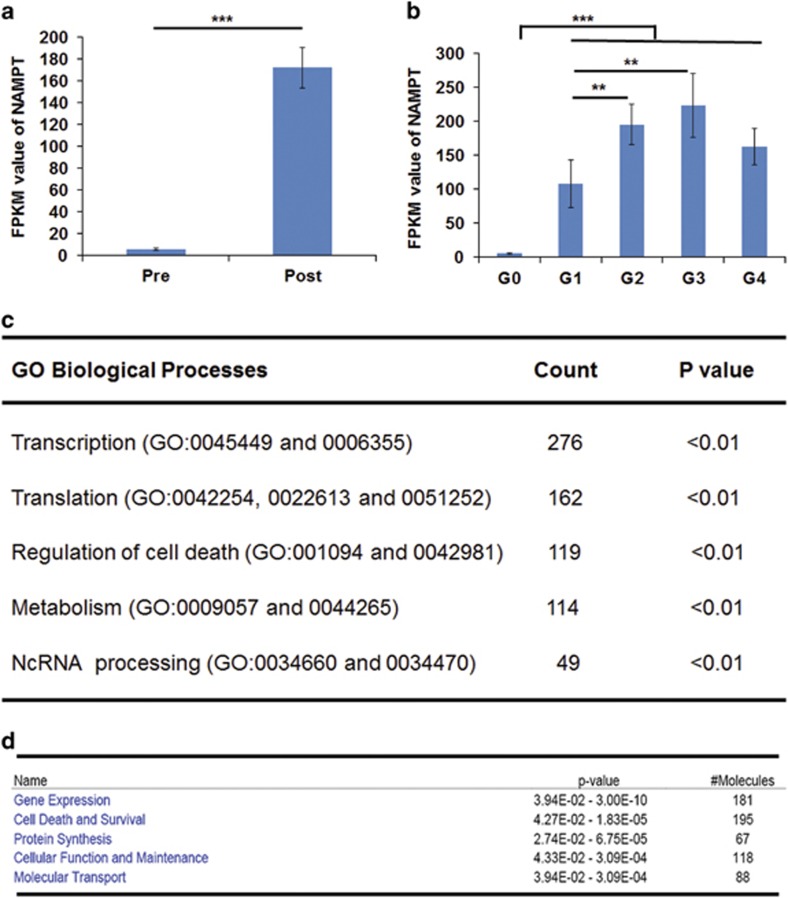
To relate what we learned about potential functions of the Nampt gene in mouse liver to NAMPT's role in human liver, we examined the expression patterns of the NAMPT gene and its co-expressed genes in human prenatal and pediatric liver from an existing RNA-seq data set. The goal of this ‘guilt-by-association' coexpression relationship analysis was to provide insights into potential functional roles of NAMPT and associated genes. After birth, expression of the NAMPT gene was upregulated significantly (171.98±18.47 FPKM postnatally, n=52 versus 5.69±1.34 FPKM prenatally, n=10, P<0.01; Figure 6a). Expression of NAMPT genes positively correlated with the children's development and growth until puberty (Figure 6b). A total 1064 genes displayed the same expression pattern as the NAMPT gene (Supplementary Table 9). GO biological process enrichment of these co-expressed genes found that they are involved in several important processes: transcription, translation, regulation of cell death, metabolism, and noncoding RNA processing (Figure 6c). The top five involved molecular and cellular functions of these co-expressed genes include gene expression, cell death and survival, protein synthesis, cellular functions and maintenance, and molecular transport (Figure 6d). These results suggest that NAMPT may have similar roles in these processes and functional categories.
Figure 6.
Co-expressed genes and pathway analysis of human Nampt gene in human pediatric liver samples (a) Nampt expression levels in prenatal and postnatal pediatric liver samples. RNA-seq analysis of these samples were carried out as described in the 'Materials and Methods'. Nampt expression levels are presented as FPKM. They were derived from 10 prenatal samples and 52 postnatal samples with 0–17 ages. (b) Nampt expression levels in prenatal (G0) and four different groups (G1–4) of postnatal pediatric liver samples. Nampt was expressed with 5.69±1.34 FPKM in G0 group. Nampt expression in G1 to G4 were 108.26±35.08 (G1, 0–1 year old, n=14), 195.06±29.80 (G2, 1–5 years old, n=14), 223.11±46.85 (G3, 6–11 years old, n=13), and 163.25±26.65 (G4, 12–17 years old, n=11). **P<0.01, ***P<0.001. (c) Top five GO biological process enrichment of co-expressed genes of human Nampt gene in human pediatric liver samples. (d) Top five molecular and cellular functions of co-expressed genes of human Nampt gene in human pediatric liver samples
Discussion
Our study represents the first scientific report, with concrete experimental evidence in a C57BL/6J mouse model, that Nampt is an essential gene in vivo. This claim is supported by the findings that the Nampt gene homozygous knockout (Nampt−/−) mice were embryonically lethal and Nampt−/− adult mice died within 5–10 days with 25.24±2.22% body weight loss. Although several groups – including ours – previously noted that Nampt gene homozygous knockout mice were embryonic lethal, none of them reported physical evidence.3, 14, 15, 16, 17 Here, we have presented the morphological and genotyping evidence of mouse embryos at both 8.5 and 11.5 d.p.c. (Figures 1d and e), clearly demonstrating that Nampt−/− mouse embryos had significantly smaller sizes and abnormal gross structures, and died before organogenesis. This ‘gold standard' of evidence authenticates that the Nampt gene is an essential gene in living organisms.
In adult Nampt−/− mice, we found seven phenotypes: death at 5–10 days after the induction >20% body weight loss (Figure 2); decreased cell NAD levels of both liver and intestinal tissues (Figures 5b and c); intestinal swelling (Figure 2d); atrophic intestinal villi (Figure 2f); visceral fat depletion (Figure 2d); higher serum TG and lower serum PC levels (Figure 3). To unravel the molecular underpinnings of these processes, we utilized RNA-seq18 to profile mouse liver transcriptomes to gain transcriptional insights into the essential role of Nampt in living organisms. Several important themes have emerged. First, 4638 differentially expressed genes in Nampt−/− mouse livers (Figure 4a and Supplementary Table 2) affect 476 biological processes and 57 significant KEGG pathways, corroborating Nampt's pleiotropic functions. Second, our study found that liver tissue and jejunum tissue NAD levels were decreased significantly in Nampt−/− mice versus Nampt+/+ mice (Figures 5b and c), confirming Nampt's key role in cell NAD synthesis.2, 16, 19 RNA-seq revealed that in Nampt−/− mice, key enzymes in two other salvage pathways and the de novo synthesis pathway of NAD synthesis were significantly decreased. It would be of interest to determine whether NAMPT directly or indirectly inhibits expression of these genes. Third, RNA-seq analysis of mouse liver transcriptomes in Nampt−/− mice versus Nampt+/+ mice have provided a number of novel insights into the essential role of the Nampt gene. In Nampt−/− mice, genes encoding acetylated proteins and genes in gluconeogenesis were upregulated while genes in glycolysis, lipase and tricarboxylic acid cycle were downregulated (Figure 4d). TCA shutdown would decrease ATP synthesis. Without enough ATP, those ATP-dependent transporters – such as those in intestine involved in nutrient absorption or transport – would cease to function. RNA-seq analysis indicated that expression of most ion transporter genes were inhibited significantly in Nampt−/− mice (Figure 4b). In addition, ATP is needed to maintain normal tissue structural integrity. In Nampt−/− mice, the short supply of ATP may be responsible for disrupted intestinal architecture and villus atrophy (Figure 2f). This may also be due to the high rate of cell turnover in this tissue, which makes it more susceptible to energy deficiency. This could explain why, in clinical trials of cancer treatment, oral administration of FK866 or CHS828,20, 21 which inhibits NAMPT enzymatic activity, caused unwanted side effects dominated by gastrointestinal symptoms. It suggests that alternative routes of drug administration may avoid or minimize gastrointestinal side effects, and achieve better therapeutic efficacy. Of interest is that we found that Rpa3 gene expression was decreased by more than 50% in Nampt−/− mice (Supplementary Figure 4). The phenotype of disrupted intestinal architecture and villus atrophy in Nampt−/− mice looks similar to those in Rpa3−/− mice.5 It remains to be elucidated whether this effect is due to the direct regulation of the Nampt gene knockout on Rpa3 gene expression.
NAD is a critical cofactor for sirtuin protein deacetylase. In Nampt−/− mice, expression of mitochondrial Sirt3, Sirt4, and Sirt 5, especially Sirt3, were all diminished significantly while nuclear Sirt1 was augmented (Figure 5f), suggesting that Sirt3, instead of Sirt1, may have a key role in life among seven sirtuin proteins. This is in line with previous evidence by Yang et al.22 Sirtuins can regulate transcription by deacetylating transcription factors such as FOXO family. Among several common FOXO genes, expression of FOXO4 was inhibited significantly. The dysregulated Sirt3–Foxo4 axis may set up a chain of reaction to engender the dysregulation of a number of genes in Nampt−/− mice, becoming a death axis in Nampt−/− mice. Based on the transcriptional insights, we sum up a tentative schema in Figure 5f to explain Nampt−/−-induced death process. Briefly, the homozygous Nampt gene knockout causes a decrease in cell NAD synthesis, which leads to the dysregulation of NAD-dependent genes such as those encoding acetylated proteins including Sirt3–Foxo4 axis and especially the suppression of eight enzymes in TCA cycle. The resultant decreased ATP output may adversely affect other genes including Rpa3, ion and nutrient transporters, as well as disrupt normal intestinal tissue architecture, which hinders normal nutrient absorption and transport into the visceral organs and forces the mobilization and exhaustion of fat storage and all energy sources, culminating in mouse death.
It should be pointed out that Nampt's functional roles may not solely depend on its enzymatic activity on NAD synthesis.23, 24, 25 In Nampt−/− mice, liver and intestinal NAD levels were only 20–30% lower than those in surviving adult Nampt+/+ mice. Thus, it is warranted to explore whether the disruption of non-enzymatic functions of Nampt also has a critical role in the dying phenotype of adult Nampt−/− mice.
Although most data in this study were derived from mice, transcriptional insights from the mouse liver transcriptome are largely confirmed by data derived from co-expressed genes of the NAMPT gene in human prenatal and pediatric liver samples (Figures 6a–d and Supplementary Table 9). These results strongly indicate that our findings are applicable to human studies. NAMPT has been implicated in the pathogenesis or therapeutic targets of a number of human diseases or conditions,4 such as acute respiratory distress syndrome,26 aging,27 cancer,28 coronary artery disease,29 diabetes, nonalcoholic fatty liver disease and obesity,30 rheumatoid arthritis, and sepsis.31 Recently, a small chemical molecule, P7C3 and its derivatives, has been found to exert neuroprotective activity in animal models of Parkinson's disease, amyotrophic lateral sclerosis, and concussive injury.32 This compound was identified as an allosteric activator of NAMPT.33 Our study contributes global metabolic and transcriptional insights into Nampt's essential roles and also provides potential leads to Nampt's pleiotropic physiological functions and pathological roles applicable to disease in humans.
Materials and Methods
Generation of Nampt+/− mice and their timed pregnancy
A mouse (129/Ola) embryonic stem (ES) cell line (RRR084) harboring characterized insertional mutation in the intron-7 of mouse Nampt gene via gene-trap vector inactivation was obtained from BayGenomics (http://baygenomics.ucsf.edu), a consortium of research groups in the San Francisco Bay Area funded as part of the NHLBI's Programs for Genomic Applications.34 The verified RRR084 clone was injected into C57BL/6N (NCI) blastocysts to generate chimeras in the Johns Hopkins University School of Medicine (Baltimore, MD, USA) transgenic facility. The generated chimeric Nampt+/− mice were genotyped by PCR of their tail DNAs. The chimeric Nampt+/− mice were backcrossed with C57BL/6J for 10 generations before their usage. All mice were fed an AIN-76 A diet and water ad libitum. They were housed under controlled conditions (25±2 °C; 12 h light–dark periods). All mouse experiments were conducted in accordance with the NIH guidelines and were approved by the University of Missouri Kansas City Animal Care and Use Committee.
The early stages of Nampt gene mutant mouse embryos were obtained by timed pregnancy after breeding between Nampt+/− male and Nampt+/− female mice. Visual observation of a vaginal plug in female Nampt+/− mice after the mating was counted 0.5 days post coitum (d.p.c.). Mouse embryos at 8.5 or 11.5 d.p.c. were harvested and fixed in 4% paraformaldehyde for 24 h. The genotyping of mouse embryos was done by PCR amplification of yolk sac DNA. Mouse embryos at 8.5 d.p.c. were also subjected to β-Gal staining.
Generation of Nampt gene exon 2 floxed (NamptF/F) and Nampt−/− mice
A Nampt gene target vector prepared in our lab, in which the exon 2 of Nampt gene was floxed, was electroporated into a B6-White Murine ES Cell Line (cat. no. SCR011, EMD Millipore, Billerica, MA, USA) and injected into C57BL/6J mouse blastocysts in The Transgenic Animal Core, University of Missouri, Columbia, USA. The generated chimeric NamptF/F mice were verified by PCR analyses of their tail DNAs.
Adult Nampt−/− mice were generated by mating NamptF/F mice with B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J(Stock No:004682, Jax Lab, Bar Harbor, ME, USA) followed by administering tamoxifen induction.
Histological examination of mouse intestinal tissues
Formalin-fixed mouse intestinal tissue samples were embedded in paraffin, and 5-micron-thick sections were cut. Replicate sections were stained with hematoxylin and eosin (H & E) according to standard methods.
Mass Spec analysis of mouse serum lipid levels
The serum samples were collected and stored at −80 °C until analyzed. The samples were extracted and subjected to the ultra-high performance liquid chromatography – high resolution mass spectrometry UHPLC-HRMS procedure as outlined by Oliver Fiehn's group35 and (http://www.metabolomicsworkbench.org/protocols/getprotocol.php?file_id=163). The analysis was conducted on a Waters Acquity UPLC coupled to a Waters Xevo G2-qTOF (quadrupole time-of-flight) mass spectrometer. The samples were analyzed in positive electrospray mode, and the data were acquired using the MSe acquisition format with a scan rate of 2 Hz. High-energy MSe scans were ramped from 15 eV to 30 eV. A leucine enkaphlin (2 μg/ml) solution was used as lockspray solution. Data processing, including chromatographic alignment and peak picking occurred in Progenesis QI 2.0. Potential markers were identified by matching the molecular ion (5 p.p.m. error) and fragment ions (50 p.p.m. error) to the lipidblast database using the database search feature within progenesis QI. Further processing occurred by exporting the data to EZinfo 3.0.
Mouse liver RNA isolation
Total mouse liver RNA was isolated from either the wild-type mice or Nampt−/− mice using the MirVana kit (cat. no. AM1560, ThermoFisher Scientific, Grand Island, NY, USA) according to the manufacturer's instructions.
RNA sequencing of mouse liver transcriptomes
RNA-seq of mouse liver RNAs was carried out in the Core of Genetic Research, Children's Mercy, Kansas City, KS, USA, according to the previously published protocol36 except that sequencing library from 1 μg total mouse liver RNA of each sample was prepared using the Illumina TruSeq Stranded Total RNA Sample Prep Kit (cat. No. RS-122-2201) and 2 × 101 paired-end sequencing of high output run mode was performed using the HiSeq 1500 instrument. Each sample was sequenced to an average depth of ~63 million reads with >80% of bases above Q30. The resulting base calling (.bcl) files were converted to FASTQ files using Illumina's bcl2fastq v2.17.1.14 software. Mapping of RNA-seq reads and transcript assembly and abundance estimation were conducted using Tuxedo Suite pipeline (TopHat v1.3.0/Bowtie v0.12.7/Cufflinks v1.0.3) and reported in Fragments Per Kilobase of exon per Million fragments mapped (FPKM).
Functional analysis of differentially expressed gene lists using Ingenuity Pathway Analysis
The pathway analysis of differentially expressed gene lists in Nampt−/− mice were analyzed using DAVID (https://david.ncifcrf.gov/)37 and Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Inc., Redwood City, CA, USA) as we described before.38, 39 Briefly, genes with a significant expression change (P<0.05) in mouse liver expression in Nampt−/− mice versus wild-type mice were loaded into IPA and they were associated with various pathways in Ingenuity's Knowledge Base for the analysis. The significance of the association between the data set and any pathway was measured in two ways: (i) the ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the pathway, and (ii) Fisher's exact test was used to calculate a P-value determining the probability that the association between the genes in the data set and the pathway in IPA is explained by chance alone. Transcription factor prediction database (DBD)40 and Database of Essential Genes (DEG)41 were used to annotate mouse transcript factors and essential genes for differentially expressed gene list.
Human pediatric liver samples and RNA-seq analysis
Forty-eight, anonymized pediatric liver samples, ages 0 days to 17 years, were received from the following sources: the NICHD Brain and Tissue Bank at the University of Maryland (funded by NIH contract HHSN275200900011C, Ref. No. #N01-HD-9-0011); the Liver Tissue Cell Distribution System (funded by NIH Contract #N01-DK-7-0004/HHSN267200700004C); and six samples were received from XenoTech, LLC (Lenexa, KS, USA), to whom we are grateful for their generous gift of tissue samples. Thirty-four prenatal human liver samples (57–196 days, post-conception) were received from an additional NICHD-funded tissue retrieval center, the Central Laboratory for Human Embryology (University of Washington, Seattle, WA, USA). All tissues were flash frozen and maintained at −80 °C before isolation of DNA and RNA. All the samples were from anonymous, deceased donors, and the use of these tissues was declared as nonhuman subjects research by the University of Missouri-Kansas City Pediatric Health Sciences Review Board.
RNA-seq analyses of human pediatric liver samples were carried out as described above for mouse liver transcriptome analyses.
Measurement of mouse liver and jejunum NAD+/NADH levels
Mouse liver and jejunum tissues (10–30 mg) were homogenized and the supernatants were applied for NAD+/NADH assays using a Fluorimetric NAD+/NADH assay kit (Cat. No. 15263; AAT Bioquest. Inc. Sunnyvale, CA, USA).
Statistical analyses
Statistical analyses were performed using the Sigma Stat (ver 13.0, Systat Software, Inc., San Jose, CA, USA). Results are expressed as means±S.D. of more than three samples for each group from at least two independent experiments. Two group comparisons were done by unpaired t-test. Three or more group comparisons were carried out using ANOVA followed by a Holm–Sidak test, P<0.05 was considered statistically significant.
Acknowledgments
We thank Marianne N Nsumu and Suman Chaudhary for excellent technical assistance. Our research has been in part supported by National Institutes of Health Grant HL 080042 (SQY), the start-up fund and William R Brown/Missouri Endowment of The Children's Mercy Hospitals and University of Missouri at Kansas City (SQY).
Author contributions
LQZ, SQY, D-YL, and JSL conceived the project. LQZ, DPH, MX, DNG, LG, CB, and SQY analyzed the data and wrote the manuscript. LQZ, DPH, LVH, PH, XJ, and RG performed the experiments. GW, LL, and MW had inputs on the experimental designs and critically read the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by Shui Qing Ye
The authors declare no conflict of interest.
Supplementary Material
References
- Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol 1994; 14: 1431–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PR, Shea RJ, Mulks MH. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J Bacteriol 2001; 183: 1168–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara A, Matsuda M, Nishizawa M, Segawa K, Tanaka M, Kishimoto K et al. Visfatin: a protein secreted by visceral fat that mimics the effects of insulin. Science 2005; 307: 426–430. [DOI] [PubMed] [Google Scholar]
- Zhang LQ, Heruth DP, Ye SQ. Nicotinamide phosphoribosyltransferase in human diseases. J Bioanal Biomed 2011; 3: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K, Mazurek A, Premsrirut PK, Zuber J, Dow LE, Simon J et al. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proc Natl Acad Sci USA 2011; 108: 7113–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J et al. Gene essentiality and synthetic lethality in haploid human cells. Science 2015; 350: 1092–1096. [DOI] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ et al. Identification and characterization of essential genes in the human genome. Science 2015; 350: 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 2015; 163: 1515–1526. [DOI] [PubMed] [Google Scholar]
- Boone C, Andrews BJ. HUMAN GENOME. The indispensable genome. Science 2015; 350: 1028–1029. [DOI] [PubMed] [Google Scholar]
- Wallace HA, Marques-Kranc F, Richardson M, Luna-Crespo F, Sharpe JA, Hughes J et al. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell 2007; 128: 197–209. [DOI] [PubMed] [Google Scholar]
- McGlothlin JR, Gao L, Lavoie T, Simon BA, Easley RB, Ma SF et al. Molecular cloning and characterization of canine pre-B-cell colony-enhancing factor. Biochem Genet 2005; 43: 127–141. [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Schoppy DW, Asare A, Clark CE, Vonderheide RH, Brown EJ. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat Genet 2009; 41: 1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Kim SW, Imanaka-Yoshida K, Yoshida T, Abel ED, Eliceiri B et al. Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J Clin Invest 2004; 113: 1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Huang Y, Moreno-Vinasco L, Sammani S, Moitra J, Barnard JW et al. Essential role of pre-B-cell colony enhancing factor in ventilator-induced lung injury. Am J Respir Crit Care Med 2008; 178: 605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo JR, Korner A, Mills KF, Satoh A, Wang T, Garten A et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab 2007; 6: 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O et al. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol 2002; 32: 3225–3234. [DOI] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Blake JA, Kadin JA, Richardson JE. Mouse genome database 2016. Nucleic Acids Res 2015; 4: D840–D847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem 2004; 279: 50754–50763. [DOI] [PubMed] [Google Scholar]
- von Heideman A, Berglund A, Larsson R, Nygren P. Safety and efficacy of NAD depleting cancer drugs: results of a phase I clinical trial of CHS 828 and overview of published data. Cancer Chemother Pharmacol 2010; 65: 1165–1172. [DOI] [PubMed] [Google Scholar]
- Holen K, Saltz LB, Hollywood E, Burk K, Hanauske AR. The pharmacokinetics, toxicities, and biologic effects of FK866, a nicotinamide adenine dinucleotide biosynthesis inhibitor. Invest New Drugs 2008; 26: 45–51. [DOI] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 2007; 130: 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Y, Dorweiler B, Cui D, Wang T, Woo CW et al. Extracellular Nampt promotes macrophage survival via a nonenzymatic interleukin-6/STAT3 signaling mechanism. J Biol Chem 2008; 283: 34833–34843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Li H, Cepeda J, Xia Y, Kempf JA, Ye H et al. Regulation of inflammatory cytokine expression in pulmonary epithelial cells by pre-B-cell colony-enhancing factor via a nonenzymatic and AP-1-dependent mechanism. J Biol Chem 2009; 284: 27344–27351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malam Z, Parodo J, Waheed F, Szaszi K, Kapus A, Marshall JC. Pre-B cell colony-enhancing factor (PBEF/Nampt/visfatin) primes neutrophils for augmented respiratory burst activity through partial assembly of the NADPH oxidase. J Immunol 2011; 186: 6474–6484. [DOI] [PubMed] [Google Scholar]
- Ye SQ, Simon BA, Maloney JP, Zambelli-Weiner A, Gao L, Grant A et al. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med 2005; 171: 361–370. [DOI] [PubMed] [Google Scholar]
- Stein LR, Imai S. Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. EMBO J 2014; 33: 1321–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res 2003; 63: 7436–7442. [PubMed] [Google Scholar]
- Hsu CP, Oka S, Shao D, Hariharan N, Sadoshima J. Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ Res 2009; 105: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten A, Schuster S, Penke M, Gorski T, de Giorgis T, Kiess W. Physiological and pathophysiological roles of NAMPT and NAD metabolism. Nat Rev Endocrinol 2015; 11: 535–546. [DOI] [PubMed] [Google Scholar]
- Jia SH, Li Y, Parodo J, Kapus A, Fan L, Rotstein OD et al. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest 2004; 113: 1318–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper AA, Xie S, Capota E, Estill SJ, Zhong J, Long JM et al. Discovery of a proneurogenic, neuroprotective chemical. Cell 2010; 142: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Han T, Nijhawan D, Theodoropoulos P, Naidoo J, Yadavalli S et al. P7C3 neuroprotective chemicals function by activating the rate-limiting enzyme in NAD salvage. Cell 2014; 158: 1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryke D, Kawamoto M, Huang CC, Johns SJ, King LA, Harper CA et al. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res 2003; 31: 278–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 2008; 49: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheranova D, Gibson M, Chaudhary S, Zhang LQ, Heruth DP, Grigoryev DN et al. RNA-seq analysis of transcriptomes in thrombin-treated and control human pulmonary microvascular endothelial cells. J Vis Exp 2013; pii: 4393. [DOI] [PMC free article] [PubMed]
- Huang, da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Zhang LQ, Cheranova D, Gibson M, Ding S, Heruth DP, Fang D et al. RNA-seq reveals novel transcriptome of genes and their isoforms in human pulmonary microvascular endothelial cells treated with thrombin. PLoS ONE 2012; 7: e31229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heruth DP, Gibson M, Grigoryev DN, Zhang LQ, Ye SQ. RNA-seq analysis of synovial fibroblasts brings new insights into rheumatoid arthritis. Cell Biosci 2012; 2: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummerfeld SK, Teichmann SA. DBD: a transcription factor prediction database. Nucleic Acids Res 2006; 34: D74–D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Lin Y, Gao F, Zhang CT, Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res 2014; 42: D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.