Abstract
Skin aging is a complicated physiological process and epigenetic feature, including microRNA-mediated regulation and DNA methylation, have been shown to contribute to this process. DNA methylation is regulated by DNA methyltransferase, of which DNA methyltransferase 1 (DNMT1) is the most abundantly known. But evidence supporting its role in skin aging remains scarce, and no report regards its specifical upstream-regulating molecules in the process of skin aging so far. Here, we found that DNMT1 expression was markedly higher in young human skin fibroblasts (HSFs) than that in passage-aged HSFs, and DNMT1 knockdown significantly induced the senescence phenotype in young HSFs. We predicted the upstream miRNAs which could regulate DNMT1 with miRNA databases and found miR-377 had high homology with a sequence in the 3′-UTR of human DNMT1 mRNA. We confirmed that miR-377 was a potential regulator of DNMT1 by luciferase reporter assays. miR-377 expression in passage-aged HSFs was markedly higher than that in the young HSFs. miR-377 overexpression promoted senescence in young HSFs, and inhibition of miR-377 reduced senescence in passage-aged HSFs. Moreover, these functions were mediated by targeting DNMT1. Microfluidic PCR and next-generation bisulfite sequencing of 24 senescent-associated genes' promoters revealed alterations of the promoter methylation levels of FoxD3, p53, and UTF1 in HSFs treated with miR-377 mimics or inhibitors. We also verified that the miR-377-mediated changes in p53 expression could be reversed by regulation of DNMT1 in HSFs. Similarly, there was a negative correlation between miR-377 and DNMT1 expression in young and photoaged HSFs, HSFs, or skin tissues from UV-unexposed areas of different aged donors. Our results highlight a novel role for miR-377-DNMT1-p53 axis in HSF senescence. These findings shed new light on the mechanisms of skin aging and identify future opportunities for its therapeutic prevention.
Aging is a complicated physiological process that occurs in all organisms. The aging process includes structure retrogression, dysfunction, adaptability, and reduced resistance.1 Skin aging is the most obvious sign of organism aging. In aging skin, physiological changes, including structural transformation, biochemical abnormalities, decreased repair capacity, and impaired permeability, appear gradually.1 Aged skin exhibits increased wrinkling, sagging, and decreased elasticity mainly because of impairment of connective tissue,2 an important component of the dermis. Fibroblasts are the main cells in the dermis and are primarily responsible for synthesizing collagen and extracellular matrix to maintain the structural integrity of the skin.3 During the process of skin aging, various biological changes occur in fibroblasts, including cell cycle abnormalities, DNA damage, apoptosis, and extracellular matrix synthesis and degradation, affecting the condition and function of the skin. Therefore, fibroblast senescence may play an important role in skin aging. The mechanisms through which senescence is induced in skin fibroblasts are complicated and include excessive oxidative stress,4 impaired mitochondria,5 deficient immune function,6 and alterations in genetic factors. Epigenetic alterations, for example, DNA methylation, histone modifications, and chromatin remodeling, are also thought to be critical in aging.7 However, the roles of epigenetic alterations in the senescence of skin fibroblasts are largely not known.
DNA methyltransferase 1 (DNMT1), which was first identified in 1988 by Bestor,8 primarily functions to maintain the methylated state after DNA replication and to ensure that this methylation state was passed on to the offspring cells. High-throughput analysis revealed the changes of global genome methylation levels and patterns during skin aging.9, 10, 11, 12 While roles and mechanisms of DNA methyltransferase in skin aging was rare, Qian H, et al. discovered that the expression of Dnmt3a, Dnmt3b, and Tet2 declined significantly in mouse skin during ageing.13 Notably, progressive alopecia appeared during aging of mice with epidermal loss of DNMT1, which was related to defeat in stem cell homeostasis maintaining.14 In addition, we found in our preliminary experiment that epidermis-specific DNMT1 knockdown in mice resulted in premature aging-like phenotypes, such as pachylosis, alopecia, and deep wrinkles (data not shown). So, we conferred DNMT1 might play a vital role in cellular senescence and skin aging. Nevertheless, its function in dermal fibroblast senescence remains unclear. Because of the important roles of DNMT1 in aging and other cellular processes, it will be important to elucidate the mechanisms that regulate the expression, stability, and activity of DNMT1, including transcriptional regulation, post-transcriptional auto-inhibitory controls, and post-translational modifications.15 The transcriptional promotion of DNMT1 gene expression is induced by signal transducer and activator of transcription 3 in malignant T cells,16 estrogen receptor α in human breast cancer MCF-7 cells,17 and Oct4 and Nanog in MSCs.18 HMG-box transcription factor 1 has been reported to be a transcriptional repressor of the DNMT1 gene in 2BS and WI-38 cells.19 Moreover, in zebrafish hepatocytes, overexpressing ubiquitin-like with PHD and ring finger domains 1 results in delocalization and destablization of DNMT1.20 Post-translational modifications, including acetylation, ubiquitination, phosphorylation, and methylation, regulate the stability of DNMT1 protein.21 Furthermore, various microRNAs (miRNAs),22 such as miR-29, miR-140, miR-148, miR-152, and miR-185, have been reported to target and modulate DNMT1 in various cancer cells. However, no studies have examined the roles of miRNAs in regulating DNMT1 in the context of cellular senescence or skin aging. In our preliminary work, we used computational miRNA target analysis from a miRNA database to identify the miRNA target sites of the 3′-untranslated region (UTR) of human DNMT1 mRNA and identified miR-377 as a DNMT1 regulator. miR-377 has been reported to enhance fibronectin protein production,23 regulate angiogenesis,24 suppress cell proliferation,25, 26 predict clinical outcomes in patients with gastric cancer, induce tumorigenesis,27 and promote oxidative stress.28 Owing to the pleiotropic functions and DNMT1 targeting potential of miR-377, miR-377 may regulate human skin fibroblast (HSF) senescence by targeting DNMT1.
Thus, in this study, we examined whether miR-377 and DNMT1 were important molecules and miR-377 could directly target and inhibit DNMT1 during HSF senescence. We also explored the downstream effects of miR-377-dependent regulation of DNMT1, including p53 methylation and HSF senescence. Our data provided evidence for the role of the miR-377-DNMT1-p53 axis in HSF senescence.
Results
Expression and function of DNMT1 in HSF senescence
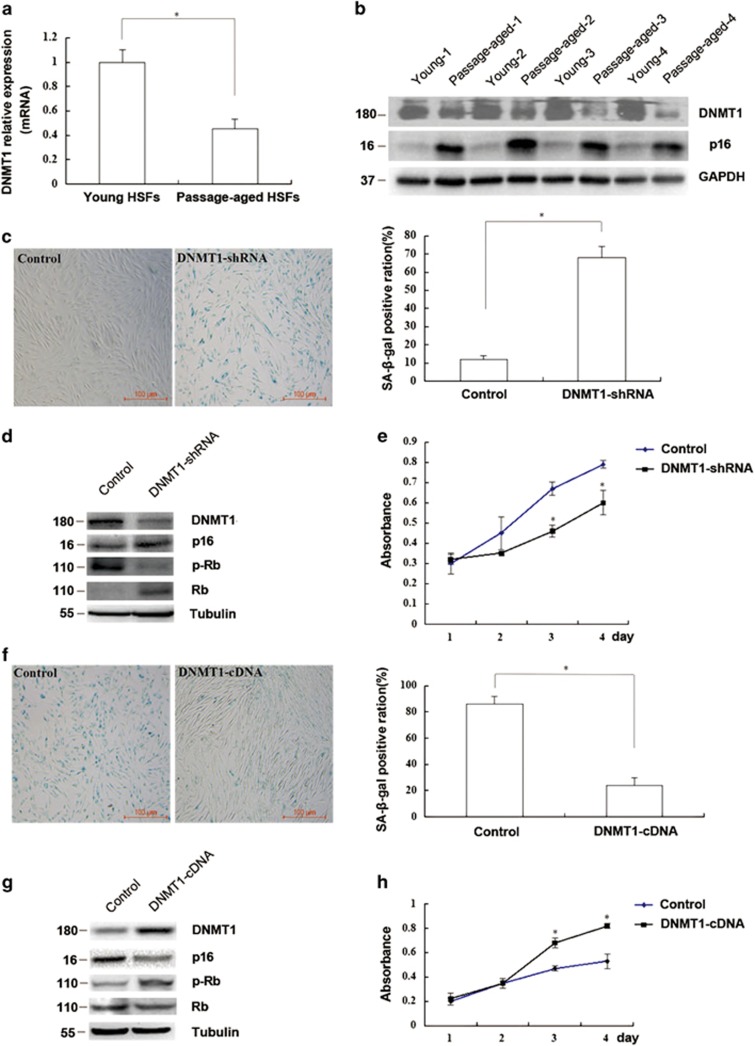
First, we analyzed the expression of DNMT1 in ten pairs of young (population doubling (PD) <10) and passage-aged (PD>50) HSFs by real time-quantitative PCR (RT-qPCR) and western blotting. DNMT1 expression in passage-aged HSFs (PD>50) with higher p16 expression was markedly lower than that in young HSFs (PD<10) at both the mRNA and protein levels (P<0.05; Figures 1a and b). In addition, DNMT1 knockdown increased the senescence-associated β-galactosidase (SA-β-gal) staining rate, p16 and Rb (Restinoblastoma tumor suppressor protein) expressions while reduced the phosphorylation level of Rb, and cell proliferation in young HSFs (PD<10) compared with that in the control group (P<0.05; Figures 1c–e). Besides, two DNMT1-shRNA (DNMT1-shRNA and DNMT-shRNA′) sequences were applied to eliminate the chance of off-target in gene silencing, while no alterations of DNMT(3a,3b) expression were detected in DNMT1 knockdown, which excluded the possibility that DNMT1 gene silencing may affect other DNMTs (Supplementary Figure S1). Inversely, upregulation of DNMT1 in passage-aged HSFs (PD>50) reduced the SA-β-gal staining rate, p16 and Rb expressions while increased the phosphorylation level of Rb and cell proliferation compared with that in the control group (P<0.05; Figures 1f–h). Taken together, these results suggested that DNMT1 may play an important role in the process of HSFs senescence.
Figure 1.
The expression and role of DNMT1 in HSFs senescence. (a) DNMT1 mRNA in passage-aged (PD>50) and young (PD<10) HSFs was detected by RT-qPCR (Data represented as the mean±S.E.M. n=10, *P<0.05). Representative data was shown. (b) DNMT1 and p16 protein levels in passage-aged (PD>50) and young (PD<10) HSFs were detected by western blot (n=10, *P<0.05). Representative data was shown. (c) SA-β-gal-positive cells in young HSFs (PD<10) transfected with control shRNA or DNMT1-shRNA were detected by using kit (left). The SA-β-gal-positive ratio was shown (right; Data represented as the mean±S.E.M. n=3, *P<0.05). (d) DNMT1, p16, and Rb protein expressions and phosphorylation of Rb in young HSFs (PD<10) transfected with control shRNA or DNMT1-shRNA were detected by western blot (*P<0.05). Representative data was shown. (e) Absorbance at 490 nm in young HSFs (PD<10) transfected with control shRNA or DNMT1-shRNA was detected by MTS assays (Data represented as the mean±S.E.M. n=3 at each time point, *P<0.05). (f) SA-β-gal-positive cells in the passage-aged HSFs (PD>50) transfected with control cDNA or DNMT1 cDNA were detected by using kit (left). The SA-β-gal-positive ratio was shown (right; data represented as the mean±S.E.M. n=3, *P<0.05). (g) DNMT1, p16, and Rb protein expressions and phosphorylation of Rb were detected by western blot in the passage-aged HSFs (PD>50) after being transfected with control cDNA or DNMT1 cDNA (*P<0.05). Representative data was shown. (h) Absorbance at 490 nm was detected in the passage-aged HSFs (PD>50) after being transfected with control cDNA or DNMT1 cDNA by MTS assays. (Data represented as the mean±S.E.M. n=3 at each time point, *P<0.05)
miR-377 regulated DNMT1 expression by directly targeting DNMT1 in HSFs
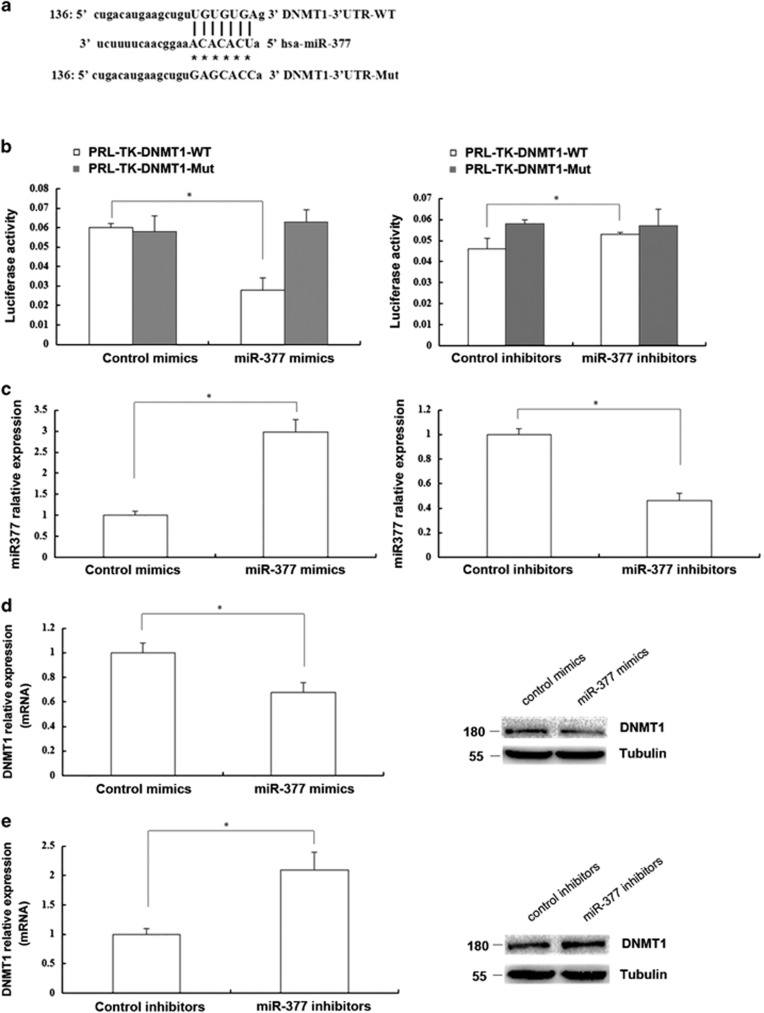
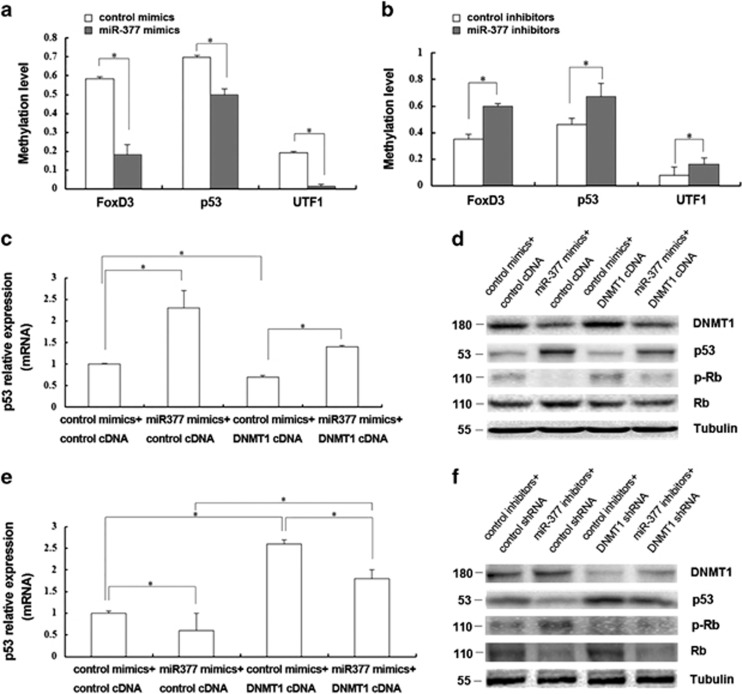
Computational miRNA target analysis from a miRNA database demonstrated that miR-377 had high homology with a sequence in the 3′-UTR of human DNMT1 mRNA (Figure 2a). To confirm whether miR-377 directly target DNMT1, we constructed a wild-type (WT) DNMT1 3′-UTR luciferase reporter vector and a miR-377 homologous sequence mutant DNMT1 3′-UTR luciferase reporter vector. Expression of miR-377 mimics decreased the relative luciferase activity of the wild-type reporter (P<0.05) but had no effect on the mutant reporter compared with the control in HEK293T cells (P>0.05). Moreover, expression of miR-377 inhibitors increased the relative luciferase activity of the wild-type reporter (P<0.05) but did not affect that of the mutant reporter compared with the control in HEK293T cells (P>0.05; Figure 2b).
Figure 2.
miR-377 could regulate DNMT1 expression by directly targeting DNMT1 in HSFs. (a) Though bioinformatics prediction, the sequence of the miR-377 binding site in the 3′-UTR of DNMT1 was shown at the upper site. Mutated residues were shown at the lower site. (b) Luciferase activity change of the wild-type 3′-UTR reporters and the mutant 3′-UTR reporters in 293T cells treated with control mimics or miR-377 mimics (left) and 293T cells treated with control inhibitors or miR-377 inhibitors (right) was shown, respectively (Data represented as the mean±S.E.M. n=3, *P<0.05, respectively). (c) miR-377 level in young HSFs (PD<10) treated with control mimics or miR-377 mimics (left) and in passage-aged HSFs (PD>50) treated with control inhibitors or miR-377 inhibitors (right) was respectively detected by RT-qPCR (Data represented as the mean±S.E.M. n=3, *P<0.05, respectively). (d) DNMT1 mRNA and protein expression in the young HSFs (PD<10) treated with control mimics or miR-377 mimics was detected by RT-qPCR and western blot, respectively (Data represent the mean±S.E.M. n=3, *P<0.05). (e) DNMT1 mRNA and protein expression in the passage-aged HSFs (PD>50) treated with control inhibitors or miR-377 inhibitors was detected by RT-qPCR and western blot, respectively (Data represent the mean±S.E.M. n=3, *P<0.05)
Next, we examined the effects of miR-377 on the expression of DNMT1 in HSFs. We treated HSFs with miR-377 mimics or inhibitors and measured the DNMT1 expression (Figure 2c). DNMT1 mRNA and protein expression levels were significantly decreased by the miR-377 mimics (P<0.05; Figure 2d) and elevated by the miR-377 inhibitors (P<0.05; Figure 2e). Collectively, our findings indicated that miR-377 suppressed DNMT1 expression by directly targeting DNMT1 in HSFs.
miR-377 mediated senescence in HSFs
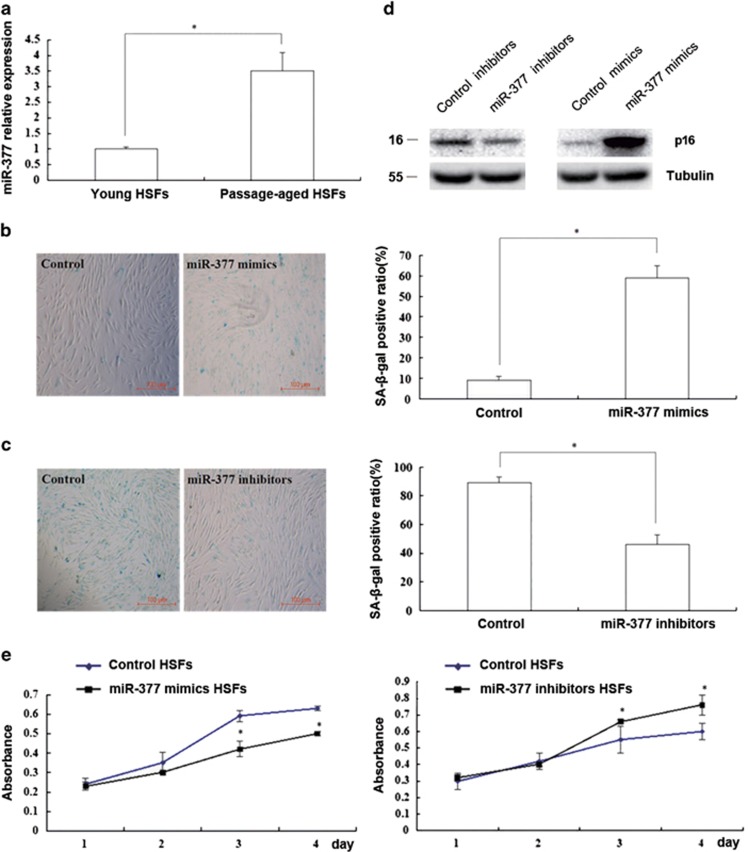
To investigate the role of miR-377 in HSF senescence, we analyzed the expression of miR-377 in HSFs. We found that miR-377 expression was significantly higher in passage-aged HSFs (PD>50) than in the young HSFs (PD<10; P<0.05; Figure 3a). Moreover, overexpression of miR-377 increased the SA-β-gal-positive ratio and p16 expression while decreased HSF proliferation in young HSFs (PD<10; P<0.05; Figures 3b, d and e). In contrast, inhibition of miR-377 reduced the SA-β-gal-positive ratio and p16 expression while increased cell proliferation in passage-aged HSFs (PD>50; P<0.05; Figures 3c–e). Taken together, our data indicated that miR-377 played an important role in HSF senescence.
Figure 3.
miR-377 mediated senescence in HSFs. (a) miR-377 level in the young (PD<10) and passage-aged (PD>50) HSFs was detected by RT-qPCR (Data represented as the mean±S.E.M. n=10, *P<0.05). (b) SA-β-gal-positive cells in young HSFs (PD<10) treated with control mimics or miR-377 mimics were detected by using kit (left). The SA-β-gal-positive ratio was shown (right; data represented as the mean±S.E.M. n=3, *P<0.05). (c) SA-β-gal-positive cells in passage-aged HSFs (PD>50) treated with control inhibitors or miR-377 inhibitors were detected by using kit (left). The SA-β-gal-positive ratio was shown (right; data represented as the mean±S.E.M. n=3, *P<0.05). (d) p16 protein expression was respectively detected by western blot in passage-aged HSFs (PD>50) treated with control inhibitors or miR-377 inhibitors (left) and in young HSFs treated with control mimics or miR-377 mimics (right; n=3, *P<0.05). Representative data was shown. (e) Absorbance at 490 nm was respectively detected by MTS assays in young HSFs (PD<10) treated with control mimics or miR-377 mimics (left) and in passage-aged HSFs treated with control inhibitors or miR-377 inhibitors (right; data represented as the mean±S.E.M. n=3 at each time point, *P<0.05)
miR-377 promoted senescence by suppressing DNMT1 expression in HSFs
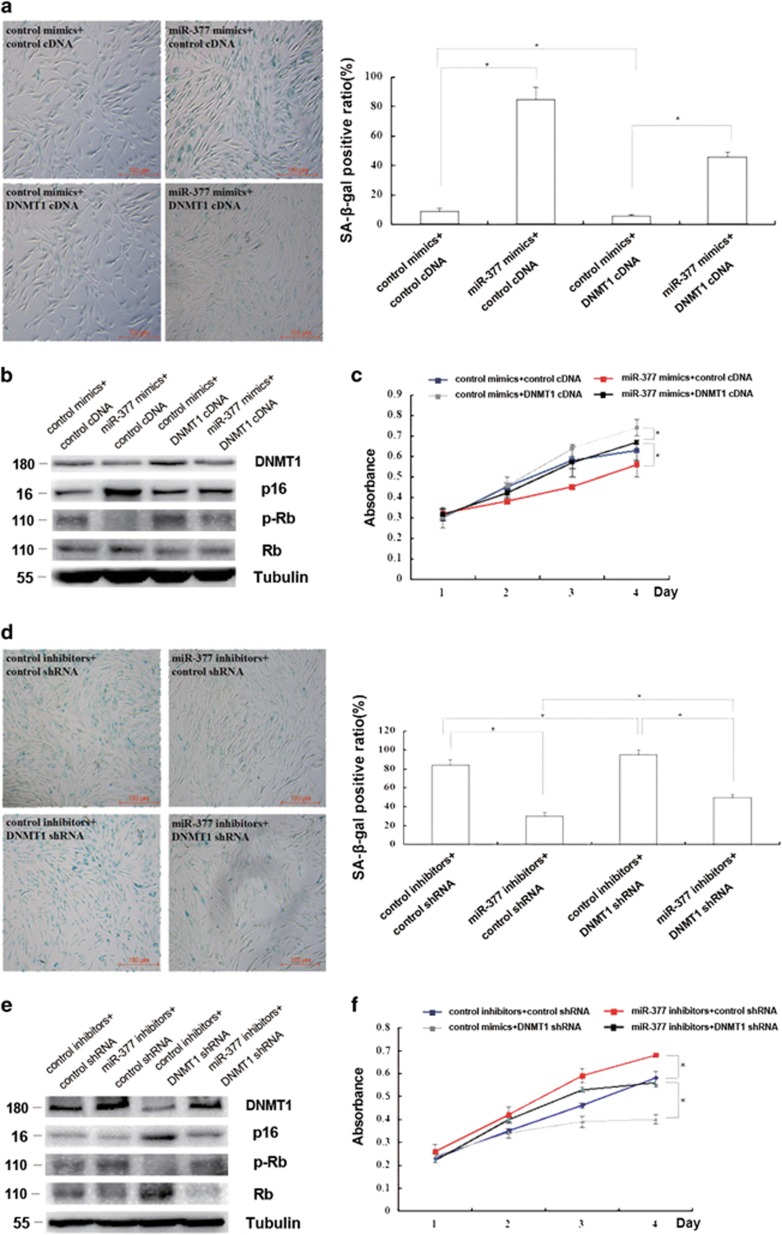
To examine how miR-377 exerts its functions, we altered DNMT1 expression in HSFs with miR-377 overexpressed or knocked-down. Enhancement of DNMT1 expression partly blocked the increased SA-β-gal activity, p16 and Rb levels together with the decreased phosphorylation of Rb and the proliferation in young HSFs (PD<10) overexpressing miR-377 (P<0.05; Figures 4a–c). Conversely, suppression of DNMT1 expression partly restored the decreased SA-β-gal activity, p16 and Rb levels together with the increased phosphorylation of Rb and the proliferation in passage-aged HSFs (PD>50) in the presence of miR-377 inhibitors (P<0.05; Figures 4d–f). Collectively, these findings indicated that miR-377 regulated HSF senescence, potentially through modulation of DNMT1 expression.
Figure 4.
miR-377 promoted senescence by suppressing DNMT1 expression in HSFs. (a) Cellular senescence was detected by evaluating SA-β-gal-positive cells in young HSFs (PD<10) treated with miR-377 mimics together with control cDNA or DNMT1 cDNA as indicated (left). The positive cell quantification was shown (right; data represent the mean±S.E.M. n=3, *P<0.05). (b) DNMT1, p16, and Rb expressions and phosphorylation level of Rb were detected by western blot in young HSFs (PD<10) treated with miR-377 mimics together with control cDNA or DNMT1 cDNA as indicated (*P<0.05). Representative data was shown. (c) Absorbance at 490 nm was detected by MTS assays in young HSFs (PD<10) treated with miR-377 mimics together with control cDNA or DNMT1 cDNA as indicated (data represented as the mean±S.E.M. n=3 at each time point, *P<0.05). (d) Cellular senescence was detected by evaluating SA-β-gal-positive cells in passage-aged HSFs (PD>50) treated with miR-377 inhibitors together with control shRNA or DNMT1 shRNA as indicated (left). The SA-β-gal-positive ratio was shown (right; data represented as the mean±S.E.M. n=3, *P<0.05). (e) DNMT1, p16, and Rb expressions and phosphorylation level of Rb were detected by western blot in passage-aged HSFs (PD>50) treated with miR-377 inhibitors together with control cDNA or DNMT1 cDNA as indicated (*P<0.05). Representative data was shown. (f) Absorbance at 490 nm was detected by MTS assays in passage-aged HSFs (PD>50) treated with miR-377 inhibitors together with control cDNA or DNMT1 cDNA as indicated (data represented as the mean±S.E.M. n=3 at each time point, *P<0.05)
Role of miR-377 in modulating promoter methylation levels of senescent-associated genes in HSFs
DNMT1 functions by methylating DNA, thereby regulating gene expression and other processes. To determine whether miR-377-mediated changes in DNMT1 expression influence the methylation of the promoters of senescence-associated genes, we treated HSFs with the miR-377 mimics or inhibitors and analyzed promoter methylation after 48 h. Promoter methylation levels of 24 senescent-associated genes (Supplementary Table 1) were analyzed through microfluidic PCR and next-generation bisulfite sequencing. From these analyses, the promoters of three genes were found to be significantly methylated (Figures 5a and b). Of these three genes, p53 was further confirmed to be regulated by miR-377 using RT-qPCR and western blotting. Increased p53 mRNA, p53 and Rb expressions together with diminished phosphorylation of Rb were induced by the miR-377 mimics and reverse changes were induced by the miR-377 inhibitors compared with that of the respective controls (P<0.05; Figures 5c–f). Furthermore, modulation of DNMT1 expression could partly reverse the changes in protein levels of p53, Rb and phosphorylation of Rb induced by miR-377 (Figures 5c–f). These data suggested that miR-377 could regulate the methylation of the p53 promoter and thus influence HSF senescence through modulation of DNMT1 expression.
Figure 5.
Role of miR-377 in modulating promoter methylation levels of senescent-associated genes in HSFs. (a) The promoter methylation levels of FoxD3, p53 and UTF1 were analyzed in young HSFs (PD<10) transfected with control mimics or miR-377 mimics through microfluidic PCR and next-generation bisulfite sequencing (data represented as the mean±S.E.M. *P<0.05). (b) The promoter methylation levels of FoxD3, p53 and UTF1 were analyzed in passage-aged HSFs (PD>50) transfected with control inhibitors or miR-377 inhibitors through microfluidic PCR and next-generation bisulfite sequencing (data represented as the mean±S.E.M. *P<0.05). (c) p53 mRNA was detected by RT-qPCR in young HSFs (PD<10) treated with miR-377 mimics together with control cDNA or DNMT1 cDNA as indicated (data represented as the mean±S.E.M. n=3, *P<0.05). (d) DNMT1, p53, and Rb expressions and phosphorylation of Rb were detected by western blot in young HSFs (PD<10) treated with miR-377 mimics together with control cDNA or DNMT1 cDNA as indicated (*P<0.05). Representative data was shown. (e) p53 mRNA was detected by RT-qPCR in passage-aged HSFs (PD>50) treated with miR-377 inhibitors together with control cDNA or DNMT1 cDNA as indicated (data represented as the mean±S.E.M. n=3, *P<0.05). (f) DNMT1, p53, and Rb expressions and phosphorylation of Rb were detected by western blot in passage-aged HSFs (PD>50) treated with miR-377 inhibitors together with control shRNA or DNMT1 shRNA as indicated (*P<0.05). Representative data was shown
miR-377 and DNMT1 expression in intrinsic aging of skin in vivo and in photoaged HSFs
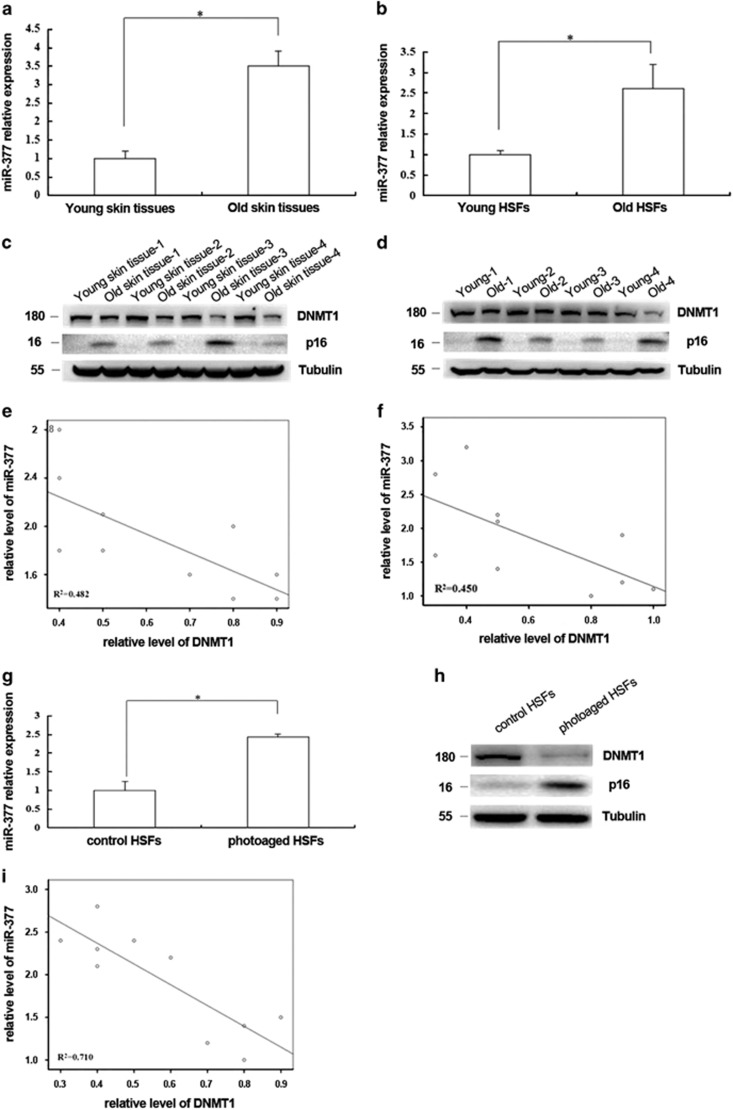
To evaluate the potential roles of miR-377 and DNMT1 in the intrinsic aging of skin, we examined miR-377 and DNMT1 expression levels in primary cultured HSFs or skin tissues from ultraviolet (UV)-unexposed areas of differently aged donors (<10 years old and >65 years old) by RT-qPCR and western blotting. The results showed that HSFs and skin tissues from old donors with higher level of p16 expression had significantly higher levels of miR-377 and lower levels of DNMT1 than those from young donors with lower p16 expression (Figures 6a–d). Negative correlations were identified between miR-377 and DNMT1 expression in HSFs or skin tissues collected from UV-unexposed areas of differently aged individuals (Figures 6e and f; R2=0.482, R2=0.45, respectively; P<0.05). In addition, miR-377 was also increased and DNMT1 was decreased in UVA-induced senescent HSFs (P<0.05; Figures 6g and h) and a similar negative correlation was found between miR-377 and DNMT1 expression in UVA-induced senescent HSFs (Figure 6i, R2=0.710, P<0.05).
Figure 6.
miR-377 and DNMT1 expression in vivo and photoaged HSFs. (a) miR-377 level was detected by RT-qPCR in the young and the old skin tissues. (Data represented as the mean±S.E.M. n=10, *P<0.05). (b) miR-377 level was detected by RT-qPCR in HSFs from the young and the old skin tissues (Data represented as the mean±S.E.M. n=10, *P<0.05). (c) DNMT1 and p16 expressions were detected by western blot in the young and the old skin tissues (n=10, *P<0.05). Representative data was shown. (d) DNMT1 and p16 expressions were detected by western blot in HSFs from the young and the old skin tissues (n=10, *P<0.05). Representative data was shown. (e and f) The correlations between miR-377 and DNMT1 levels in different couples of young and old skin tissues and in HSFs from young and old skin tissues were shown respectively (n=10, R2=0.482,0.450, respectively, *P<0.05). (g) miR-377 level was detected in control and photoaged HSFs by RT-qPCR (Data represented as the mean±S.E.M. n=3, *P<0.05). (h) DNMT1 and p16 expression levels were detected by western blot in control and photoaged HSFs (*P<0.05). Representative data was shown. (i) The correlation between miR-377 and DNMT1 levels was analyzed in several couples of UVA-untreated and UVA-treated HSFs (n=10, R2=0.471, *P<0.05)
Discussion
In this study, we examined the roles of miR-377 and DNMT1 in senescence in HSFs. Our results showed that miR-377 regulated the methylation of p53 through direct targeting DNMT1, thereby mediating HSF senescence. These findings may have implications in the therapeutic prevention of skin aging.
Skin aging is a complicated physiological process. Older monozygotic twins have been shown to exhibit marked differences in skin aging phenotypes, despite sharing a common genotype; these differences may be explained by distinct profiles of DNA methylation in the twins.29 DNA methylation is regulated by DNMTs, of which DNMT1 is the most abundant. Knockdown of DNMT1 by siRNA has been shown to alter the methylation of various CpG islands and induce senescence in human umbilical cord blood-derived stem cells.30 Moreover, mutations in DNMT1 can cause both central and peripheral neurodegeneration through aberrant methylation.31 Studies on Dnmt1+/− mice have shown that changes in DNA methylation may contribute to some forms of aging-related amyloidosis.32 In our present study, we found, for the first time, that DNMT1 expression decreased with age in passage-aged HSFs and that downregulation of DNMT1 aggravated the senescent phenotype in young HSFs. Our data were consistent with previous studies, which indicated that DNMT1 is involved in the process of cellular senescence in the skin.
Many studies have suggested that epigenetic modifications are deeply involved in the modulation of DNMT1, including miRNA regulation.33, 34 No reports have described the targeting of DNMT1 by miRNAs in the context of aging. Bioinformatics analysis of the DNMT1 promoter showed that miR-377, with 7 bases predicted complementarily pairing with DNMT1 3′UTR, which has been shown to function as a tumor suppressor in various types of cancer cells, including clear cell renal cell carcinoma,35 malignant melanoma,36 hepatocellular carcinoma,25 osteosarcoma,26 glioblastoma,37 and prostate cancer cells,38 may regulate DNMT1. Furthermore, miR-377 is involved in angiogenesis24 through targeting of vascular endothelial growth factor. Interestingly, miR-377 can promote oxidative stress by suppressing superoxide dismutase (SOD) 1 and SOD2, increasing p38 mitogen-activated protein kinase (MAPK) phosphorylation and thioredoxin-interacting protein expression, and activating the NLR family, pyrin domain-containing 3 (NLRP3) inflammasome pathway,28 indicating that miR-377 can influence important aging-related factors. In our study, we found that miR-377 expression was significantly increased in passage-aged HSFs and that inhibition of miR-377 could reverse the passage-induced senescence of HSFs; this effect could be partly rescued by knockdown of DNMT1. We confirmed, for the first time, that miR-377 acted as a senescence inducer by regulating DNMT1 in HSFs. Moreover, negative correlations were observed between miR-377 and DNMT1 expression in passage-aged HSFs, photoaged HSFs, and intrinsic aged HSFs and skin tissues in our present study, supporting that miR-377 and DNMT1 played important roles in various types of cellular senescence and aging processes in the skin.
DNA methylation by DNMT1 is an important epigenetic mechanism that functions to maintain correct gene expression and stability. Dysregulation of DNMT1 is involved in a variety of diseases.39 Previous studies have shown that DNMT1 can regulate the methylation level of the PTEN,40 p16INK4A,41 p53, and p2142promoters. p53 is a well-known tumor suppressor that may also mediate the aging process. Knockdown of the Caenorhabditis elegans p53 gene (Cep-1) can increase lifespan.43 Furthermore, an epidemiological study showed that a mutation in the p53 protein (R72P) increases survival and contributes to aging.44 Recent data have suggested that p53 is an important inducer of organismal aging; for example, p53-mediated skin aging involves the loss of subdermal fat and declining sebaceous gland activity.45 In our study, we found that methylation of the p53 promoter was significantly altered after alterations in miR-377 expression in HSFs. We further confirmed that miR-377 could induce p53 expression and that this effect could be reversed by upregulation of DNMT1. Thus, our study provided evidence to support that the miR-377-DNMT1-p53 axis plays an important role in HSF senescence. Identification of the roles of miR-377 and DNMT1 in skin fibroblast senescence would facilitate the identification of potential new targets or drugs to prevent or cure skin aging.
Collectively, our study demonstrates that miR-377 directly inhibit DNMT1, thereby regulating p53 methylation and promoting senescence in HSFs. These data provide a new target for the prevention of skin aging. Further study should focus on comprehensive exploration of upstream regulation of DNMT1 and how to regulate miR-377 during the natural aging of skin.
Materials and Methods
Skin tissues and cell culture
UV-unexposed normal skin tissues were collected from normal UV-unexposed areas surrounding the surgical sites from patients with benign dermatosis in the Department of Dermatology, Xiangya Hospital of Central South University. Patients were informed in advance and provided written consent for the collection and use of their tissues for research purposes. This study was conducted in accordance with the principles of the Declaration of Helsinki. Skin tissues from individuals ages 1–10 years old were used as the young group, whereas those from patients ages 65 years and older were used as the old group. Primary HSFs were isolated by digesting human skin with type II collagenase (Sigma-Aldrich, St. Louis, MO, USA), as described previously, and then cultured in Dulbecco's modified Eagle's medium (Thermo Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (Gibco/Thermo Scientific, Waltham, MA, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 °C in an atmosphere containing 5% CO2. The HSFs isolated from the young group were divided into the young HSF group (PD<10) and passage-aged group (PD>50). Similarly, HSFs (PD<10) cultured from the two groups of skin tissues were designated young HSFs and old HSFs, respectively.
HSF photoaging model
In UVA-induced senescent HSFs, young HSFs at 60% confluence were washed with phosphate-buffered saline (PBS) at 37 °C twice, covered with 3 ml PBS, and treated with 10 J/cm2 UVA/day (Philips TL20W/12 RS UVA lamps; American Ultraviolet Company, Murray Hill, NJ, USA) for 3 consecutive days.
SA-β-gal staining
Senescence was determined using a β-galactosidase activity staining kit (Cell Signaling Technology, Danvers, MA, USA) following the manufacturer's recommendations. HSFs were cultured until 80% confluence and washed with PBS twice. The cells were fixed at room temperature for 15 min. After fixation, the cells were washed with PBS three times and incubated with freshly prepared β-galactosidase staining solution at 37 °C free of CO2 overnight. The stained cells were imaged using a microscope, and the aging ratio of the cells was calculated.
RNA isolation and miRNA quantification
Total RNA from the HSFs was isolated with TRIzol (Thermo Scientific), and cDNA was prepared using a RevertAid First Strand cDNA Synthesis Kit (Gibco/Thermo Scientific). real time-quantitative PCR (RT-qPCR) was performed using the Thermo Scientific miRNA assay protocol with miR-377 or U6-specific primers (Ribobio Company, Guangzhou, China) with a Bio-Rad real-time PCR system (Bio-Rad, Hercules, CA, USA). The relative expression levels of miR-377 were calculated with the 2−ΔΔCt method using U6 as the endogenous control.46 DNMT1 and GAPDH mRNA levels were determined as described above with GAPDH as the endogenous normalization control. Primers are available on request.
Transfection
HSFs were transfected with the miR-377 mimic, miR-377 inhibitor, or scrambled controls at a final concentration of 20 μM using Lipofectamine reagent (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's instructions. The cells were subsequently incubated at 37 °C for 48 h.
MTS assay
HSFs were seeded in 96-well culture dishes at 5000 cells/well. Next, 20 μl CellTiter 96 AQueous MTS Reagent (Promega, Madison, WI, USA) was added to each well. The culture dishes were further incubated for 4 h at 37 °C in an atmosphere containing 5% CO2 in the dark. The absorbance of the samples was measured at 490 nm. All measurements were repeated three times.
Western blotting
Western blotting was performed as previously described.47 Briefly, the cells were rinsed twice with ice-cold PBS and collected with radioimmunoprecipitation assay buffer. Total protein was quantified, separated on 10% SDS/polyacrylamide gels, and transferred to PVDF membranes. The membranes were blocked in 5% nonfat milk for 1 h at room temperature and probed with specific antibodies or anti-β-actin antibodies at 4 °C overnight, followed by incubation with a secondary antibody conjugated to horseradish peroxidase. Immunoreactivity was detected with Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific).
Luciferase reporter assays
The 3′-UTRs of the DNMT1 gene were amplified by PCR from human genomic DNA and cloned into the pMIR reporter vector using the SacІ and HindIII sites immediately downstream from the stop codon of luciferase. The primers were as follows: forward 5′-TCGGAGCTCGGAGGAGGAAGCTGCTAAG-3′ and reverse 5′-GCGAAGCTTTTGGTTTATAGGAGAGAT-3′. We also constructed a mutant report gene using a QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Stratagene Company, La Jolla, San Diego, CA, USA). Each vector, along with 50 nM miR-377, was transfected into HEK293 cells with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Firefly and renilla luciferase activities were measured consecutively using dual luciferase assays (Promega, Madison, WI, USA) at 48 h after transfection.
Detection of promoter methylation of senescent genes
Genomic DNA extraction was accomplished using a TIANamp Genomic DNA Kit (TianGen Biotech, Beijing, China) according to the manufacturer's instructions. Bisulfite conversion was carried out using an EZ DNA Methylation-Lightning Kit (ZymoResearch, Irvine, CA, USA), and bisulfite sequencing primer design for each gene was achieved using the online design tool Methprimer (http://www.urogene.org/methprimer/). High-throughput microfluidic PCR for target enrichment, next-generation bisulfite sequencing, sequencing data alignment, and methylation analysis were performed as previously described.48
Statistical analysis
All data were expressed as the mean±standard error of the mean. For comparisons, paired-samples t test, a one-way analysis of variance with least significant difference post-hoc tests or analysis of variance for multiple comparisons were performed as appropriate(SPSS Software 19.0). Linear regression analysis (R2) was used for the correlation analysis. Differences with P-values of <0.05 were considered significant.
Acknowledgments
We greatly thank the Department of Dermatology in Xiangya Hospital of Central South University, especially the individuals participated in this study. This research was supported by the National Natural Science Foundation of China (Grant Nos. 81171520, 81371756, 81271775 and 81472904) and the Hunan Provincial Natural Science Foundation of China (Grant No. 2015JJ1023). The authors have no conflicts of interest to declare.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Raschella'
The authors declare no conflict of interest.
Supplementary Material
References
- Farage MA, Miller KW, Elsner P, Maibach HI. Characteristics of the aging Skin. Adv Wound Care 2013; 2: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammeyer A Luiten. Oxidation events and skin aging. Ageing Res Rev 2015; 21: 16–29. [DOI] [PubMed] [Google Scholar]
- Thangapazham RL, Darling TN, Meyerle J. Alteration of skin properties with autologous dermal fibroblasts. Int J Mol Sci 2014; 15: 8407–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinnerthaler M, Bischof J, Streubel MK, Trost A, Richter K. Oxidative stress in aging human skin. Biomolecules 2015; 5: 545–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon GK, Dal Farra C, Botto JM, Domloge N. Mitochondria: a new focus as an anti-aging target in skin care. J Cosmet Dermatol 2010; 9: 122–131. [DOI] [PubMed] [Google Scholar]
- Vukmanovic-Stejic M, Rustin MH, Nikolich-Zugich J, Akbar AN. Immune responses in the skin in old age. Curr Opin Immunol 2011; 23: 525–531. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell 2013; 153: 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol 1988; 203: 971–983. [DOI] [PubMed] [Google Scholar]
- Bormann F, Rodríguez-Paredes M, Hagemann S, Manchanda H, Kristof B, Lyko F et al. Reduced DNA methylation patterning and transcriptional connectivity define human skin aging. Aging Cell 2016; 15: 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandiver AR, Irizarry RA, Hansen KD, Garza LA, Runarsson A, Feinberg AP et al. Age and sun exposure-related widespread genomic blocks of hypomethylation in nonmalignant skin. Genome Biol 2015; 16: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Suschek CV, Bork S, Goergens M, Joussen S, Wagner W et al. Specific age-associated DNA methylation changes in human dermal fibroblasts. PLoS ONE 2011; 6: e16679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz G, Hagemann S, Söhle J, Kulkarni PP, Kaderali L, Lyko F et al. Aging is associated with highly defined epigenetic changes in the human epidermis. Epigenetics Chromatin 2013; 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Xu X. Reduction in DNA methyltransferases and alteration of DNA methylation pattern associate with mouse skin ageing. Exp Dermatol 2014; 23: 357–359. [DOI] [PubMed] [Google Scholar]
- Li J, Jiang TX, Hughes MW, Wu P, Yu J, Widelitz RB et al. Progressive alopecia reveals decreasing stem cell activation probability during aging of mice with epidermal deletion of DNA methyltransferase 1. J Invest Dermatol 2012; 132: 2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar S, Deb M, Sengupta D, Shilpi A, Parbin S, Torrisani J et al. An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function. Epigenetics 2012; 7: 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang HY, Woetmann A, Raghunath PN, Odum N, Wasik MA. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood 2006; 108: 1058–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JF, Li XJ, Si XX, Li AD, Ding HJ, Han X et al. ERalpha positively regulated DNMT1 expression by binding to the gene promoter region in human breast cancer MCF-7 cells. Biochem Biophys Res Commun 2012; 427: 47–53. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Su PF, Huang YF, Yew TL, Hung SC. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol Cell 2012; 47: 169–182. [DOI] [PubMed] [Google Scholar]
- Pan K, Chen Y, Roth M, Wang W, Wang S, Yee AS et al. HBP1-mediated transcriptional regulation of DNA methyltransferase 1 and its impact on cell senescence. Mol Cell Biol 2013; 33: 887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell 2014; 25: 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Song J, Ewing R, Wang Z. Regulation of protein stability of DNA methyltransferase 1 by post-translational modifications. Acta Biochim Biophys Sin 2014; 46: 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RK, Wang YC. Dysregulated transcriptional and post-translational control of DNA methyltransferases in cancer. Cell Biosci 2014; 4: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. Faseb J 2008; 22: 4126–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Huang W, Feng Y, Cai W, Wang Y, Wang X et al. MicroRNA-377 regulates mesenchymal stem cell-induced angiogenesis in ischemic hearts by targeting VEGF. PLoS ONE 2014; 9: e104666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Lu L, Liu C, Shan L, Yuan D. MicroRNA-377 suppresses cell proliferation and invasion by inhibiting TIAM1 expression in hepatocellular carcinoma. PLoS ONE 2015; 10: e117714. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang L, Shao J, Zhang X, Xu M, Zhao J. microRNA-377 suppresses the proliferation of human osteosarcoma MG-63 cells by targeting CDK6. Tumour Biol 2015; 36: 3911–3917. [DOI] [PubMed] [Google Scholar]
- Wen X, Wu JQ, Peng W, Feng JF, Tang JH. MicroRNA-377 predicts poor clinical outcome of gastric cancer and induces tumorigenesis by targeting multiple tumor-suppressor genes. Oncol Rep 2015; 34: 203–210. [DOI] [PubMed] [Google Scholar]
- Wang W, Ding XQ, Gu TT, Song L, Li JM, Xue QC et al. Pterostilbene and allopurinol reduce fructose-induced podocyte oxidative stress and inflammation via microRNA-377. Free Radic Biol Med 2015; 83: 214–226. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 2005; 102: 10604–10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AY, Jung JW, Lee S, Kim HS, Kang KS. DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs. PLoS ONE 2011; 6: e19503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet 2011; 43: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Wu A, Wilkinson JE, Murphy HS, Lu Q, Kluve-Beckerman B et al. Aging in heterozygous Dnmt1-deficient mice: effects on survival, the DNA methylation genes, and the development of amyloidosis. J Gerontol A Biol Sci Med Sci 2006; 61: 115–124. [DOI] [PubMed] [Google Scholar]
- Qadir XV, Han C, Lu D, Zhang J, Wu T. miR-185 inhibits hepatocellular carcinoma growth by targeting the DNMT1/PTEN/Akt pathway. Am J Pathol 2014; 184: 2355–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard AP, Mooso BA, Libertini SJ, Lim RM, Nakagawa RM, Vidallo KD et al. miR-148a dependent apoptosis of bladder cancer cells is mediated in part by the epigenetic modifier DNMT1. Mol Carcinog 2015; 55: 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Ma Y, Yu D, Zhao J, Ma P. miR-377 functions as a tumor suppressor in human clear cell renal cell carcinoma by targeting ETS1. Biomed Pharmacother 2015; 70: 64–71. [DOI] [PubMed] [Google Scholar]
- Zehavi L, Schayek H, Jacob-Hirsch J, Sidi Y, Leibowitz-Amit R, Avni D. MiR-377 targets E2F3 and alters the NF-kB signaling pathway through MAP3K7 in malignant melanoma. Mol Cancer 2015; 14: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Luo H, Wang S, Chen W, Chen Z, Wang HW et al. MicroRNA-377 inhibited proliferation and invasion of human glioblastoma cells by directly targeting specificity protein 1. Neuro Oncol 2014; 16: 1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa A, Markert EK, Lena AM, Italiano D, Finazzi-Agro' E, Levine AJ et al. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene 2014; 33: 5173–5182. [DOI] [PubMed] [Google Scholar]
- Johnson AA, Akman K, Calimport SR, Wuttke D, Stolzing A, de Magalhaes JP. The role of DNA methylation in aging, rejuvenation, and age-related disease. Rejuvenation Res 2012; 15: 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Che Q, Jiang FZ, Wang HH, Wang FY, Liao Y et al. Piwil1 causes epigenetic alteration of PTEN gene via upregulation of DNA methyltransferase in type I endometrial cancer. Biochem Biophys Res Commun 2015; 463: 876–880. [DOI] [PubMed] [Google Scholar]
- Jung JK, Arora P, Pagano JS, Jang KL. Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res 2007; 67: 5771–5778. [DOI] [PubMed] [Google Scholar]
- Liu CC, Lin JH, Hsu TW, Su K, Li AF, Hsu HS et al. IL-6 enriched lung cancer stem-like cell population by inhibition of cell cycle regulators via DNMT1 upregulation. Int J Cancer 2015; 136: 547–559. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy 2008; 4: 870–873. [DOI] [PubMed] [Google Scholar]
- Bojesen SE, Nordestgaard BG. The common germline Arg72Pro polymorphism of p53 and increased longevity in humans. Cell Cycle 2008; 7: 158–163. [DOI] [PubMed] [Google Scholar]
- Kim J, Nakasaki M, Todorova D, Lake B, Yuan CY, Jamora C et al. p53 Induces skin aging by depleting Blimp1+ sebaceous gland cells. Cell Death Dis 2014; 5: e1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Xie H, Liu F, Liu L, Dan J, Luo Y, Yi Y et al. Protective role of AQP3 in UVA-induced NHSFs apoptosis via Bcl2 up-regulation. Arch Dermatol Res 2013; 305: 397–406. [DOI] [PubMed] [Google Scholar]
- Li Z, Guo X, Wu Y, Li S, Yan J, Peng L et al. Methylation profiling of 48 candidate genes in tumor and matched normal tissues from breast cancer patients. Breast Cancer Res Treat 2015; 149: 767–779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.