Abstract
In response to the current global health emergency posed by the Zika virus (ZIKV) outbreak and its link to microcephaly and other neurological conditions, we performed a drug repurposing screen of ~6,000 compounds that included approved drugs, clinical trial drug candidates and pharmacologically active compounds, and we identified compounds that either inhibit ZIKV infection or suppress infection-induced caspase-3 activity in different neural cells. A pan-caspase inhibitor, Emricasan, inhibited ZIKV-induced increases in caspase-3 activity and protected human cortical neural progenitors in both monolayer and 3-dimensional organoid cultures. Ten structurally unrelated inhibitors of cyclin-dependent kinases inhibited ZIKV replication. Niclosamide, an FDA approved category B anthelmintic drug, also inhibited ZIKV replication. Finally, combination treatments using one compound from each category (neuroprotective and antiviral) further increased protection of human neural progenitors and astrocytes from ZIKV-induced cell death. Our results demonstrate the efficacy of this screening strategy and identify lead compounds for anti-ZIKV drug development.
Keywords: Zika virus, microcephaly, cortical neural progenitor, astrocyte, high-throughput screening, drug repurposing screen, cyclin-dependent kinase inhibitors, caspase-3/7
INTRODUCTION
ZIKV, a mosquito-borne flavivirus, has spread across the Western Hemisphere in the past year. First isolated in 1947 from a sentinel rhesus macaque in the Ziika Forest region of Uganda1, ZIKV remained in relative obscurity for many years until outbreaks in the Pacific islands and then the Americas in the past decade. A large outbreak started in Brazil in late 2014 and is a growing public health concern2. Active transmission has been reported in approximately 60 countries and territories globally. Most human infections are transmitted by mosquitos, yet ZIKV can also spread directly through sexual contact3–6 and vertically from mother to fetus7–9. About 20% of ZIKV-infected individuals develop symptoms, which mostly resemble those caused by other arboviruses, such as dengue viruses or chikungunya virus. Unlike these viruses, however, ZIKV causes congenital defects, including microcephaly10,11, and is associated with Guillain-Barré syndrome, meningoencephalitis and myelitis in infected adults8,11–14.
Since the recent declaration by the World Health Organization (WHO) that ZIKV is a global health concern, rapid progress has been made to understand its pathogenesis and to develop human in vitro models and animal in vivo models15–23. Following clinical observations of ZIKV in fetal brains obtained from infected women10,24, we reported that ZIKV efficiently target human neural progenitor cells (hNPCs) and attenuate their growth15. This finding provides a potential mechanism for ZIKV-induced microcephaly as hNPCs drive the development of human cortex. Furthermore, we and others have shown that ZIKV infection of brain organoids, 3D cellular models of early human brain development, leads to reduced thickness of hNPC and neuronal layers, and an overall reduction in organoid size16,17,20,25, again consistent with features of microcephaly. These results have also been recapitulated in mouse models20,21,23. Despite these advancements in understanding how ZIKV causes developmental abnormalities and preclinical studies that are underway to develop vaccines26,27, there is currently no drug approved to treat or prevent ZIKV infection.
Drug repurposing screens have recently emerged as an alternative approach to accelerate drug development28,29. Following a repurposing phenotypic screen, new indications for existing drugs may be rapidly identified and clinical trials can be carried out quickly, which is especially critical for rapidly spreading infectious diseases. For example, recent drug repurposing screens have led to discoveries of potential new candidate therapies for Ebola virus disease30,31, Giardiasis32, Entamoeba histolytica infection33, malaria gametocytes34, Exserohilum rostratum infection35, hepatitis C virus infection36, and, very recently, ZIKV infection37. Based on our previous finding that ZIKV infection of hNPCs results in an increase of caspase-3 activation, followed by cell death15, we designed a compound screening approach using caspase-3 activity as the primary screening assay, and a secondary cell viability assay for confirmation (Supplementary Fig. 1a). We identified two classes of effective compounds, one is antiviral and the other is neuroprotective, capable of protecting neural cells from ZIKV-induced cell death.
RESULTS
Development of high-throughput compound screening approaches
We first quantified caspase-3 activity and cell viability of hNPCs and astrocytes derived from human induced pluripotent stem cells (iPSCs), as well as glioblastoma SNB-19 cells, after ZIKV infection in a 1536-well plate format (Supplementary Tables 1 and 2). The prototypic ZIKV strain, MR766, was used in the primary screen because it produced the strongest cell death signal in cell culture experiments. The signal-to-basal (S/B) ratios and coefficient of variations (CV) obtained in the caspase-3 activity assay after 6-hour ZIKV exposure were 2.1-fold and 7.0% for hNPCs, 7.0-fold and 5.9% for SNB-19 cells, and 11.0-fold and 9.1% for astrocytes (Supplementary Fig. 1b). The Z′ factors, a measure of statistical effect size and an index for assay quality control38, for hNPCs, SNB-19, and astrocytes were 0.20, 0.68, and 0.72, respectively. Since a Z′ factor over 0.5 indicates a robust screening assay38, the caspase assay, using SNB-19 cells or astrocytes is suitable for high-throughput screening.
To measure cell viability, we performed an ATP content assay following ZIKV infection for 3 days (Supplementary Table 2). Cell viability was reduced by 39%, 82%, and 69% in hNPCs, SNB-19 cells, and human astrocytes, respectively (Supplementary Fig. 1c). The Z′ factors in these three cell types were 0.06, 0.37 and 0.32, respectively. These results indicated that measuring caspase-3 activity is a better assay for high-throughput compound screening than the cell viability assay.
High-throughput screen of compound collections
We carried out a screening campaign using the caspase-3 activity assay and SNB-19 cells with the Library of Pharmacologically Active Compounds (LOPAC, 1280 compounds), the NCATS Pharmaceutical Collection of approved drugs (2816 compounds), and a collection of clinical candidate compounds (2000 compounds). Primary hits included a total of 116 compounds that suppressed ZIKV-induced caspase-3 activity in SNB-19 cells. We also carried out an independent primary screen using hNPCs with same libraries. This second screen resulted in 173 primary hits that included all 116 compounds from the first caspase-3 screen in SNB-19 cells. All results of the primary screen of the approved drug collection and hit confirmation were deposited into the open-access PubChem database (https://pubchem.ncbi.nlm.nih.gov/).
Next, the activity of these primary hits from the caspase-3 activity assay was re-evaluated in ZIKV-infected SNB-19 cells, hNPCs, and astrocytes, and, importantly, in parallel with the compound cytotoxicity assay (Supplementary Fig. 1d–e and Supplementary Table 3). Cytotoxic compounds were then eliminated from the confirmed hit list. Consistent with the screening design, we identified compounds that reduced virally-induced caspase activation and apoptosis by either directly preventing ZIKV-induced cell death or suppressing ZIKV replication (Supplementary Table 4).
Protection from ZIKV-induced cell death by Emricasan
Emricasan, a pan-caspase inhibitor, was identified as the most potent anti-death compound with IC50 values of 0.13 – 0.9 μM in both caspase activity and cell viability assays for SNB-19 cells against three ZIKV strains: MR766 (1947 Ugandan strain), FSS13025 (2010 Cambodian strain), and PRVABC59 (2015 Puerto Rican strain) (Fig. 1a). It was also effective for all three cell types tested (Supplementary Fig. 1f and Supplementary Table 5). In addition, Emricasan reduced the number of active (cleaved) caspase-3-expressing forebrain-specific hNPCs infected by FSS13025 in both monolayer and 3D organoid cultures (Fig. 1b–c). Emricasan treatment of ZIKV-exposed brain organoids did not appear to affect hNPC proliferation compared to the mock treatment, as evaluated by phospho-Histone3 (PH3) expression (106 ± 10%; n = 8; P = 0.7; One-way ANOVA). Notably, ZIKV antigen persisted in both 2D and 3D cultures after Emricasan treatment (Fig. 1b–c). Therefore, Emricasan displays neuroprotective activity for hNPCs, but does not suppress ZIKV replication.
Figure 1.
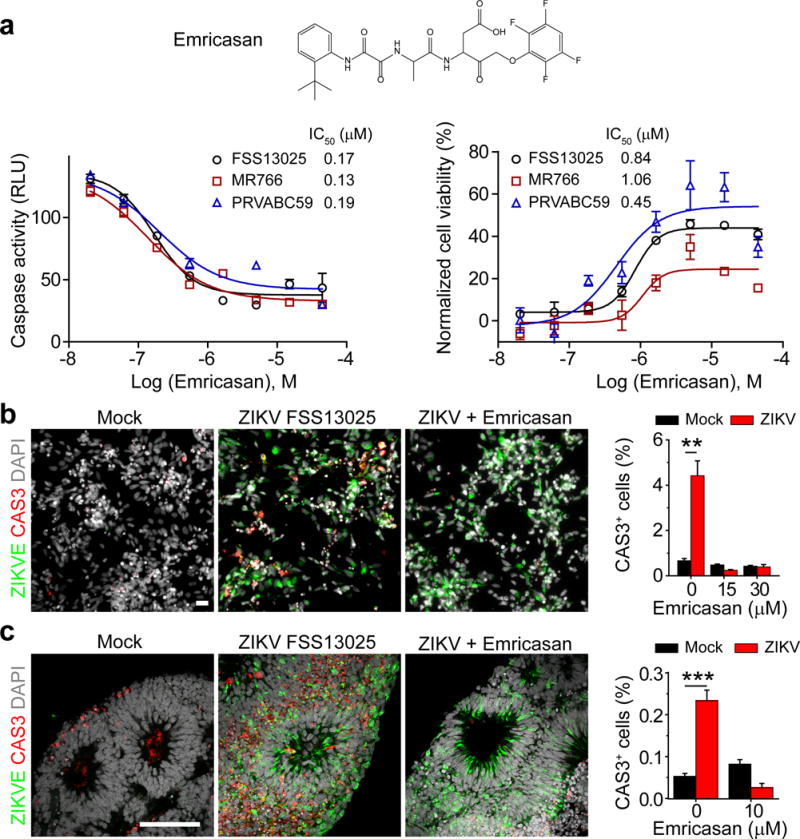
Emricasan suppresses cell death of ZIKV-infected human astrocytes and hNPCs in 2D monolayer cultures and in 3D brain organoids. (a) Top, chemical structure of Emricasan. Bottom, dose-response curves displaying the effect of Emricasan treatment on caspase activity (left) and cell viability (right) in SNB-19 glioblastoma cells exposed to 3 different ZIKV strains. Values represent mean ± s.d. (n = 3 cultures). Curves represent best fits for calculating IC50, and the insets in each panel report the calculated IC50 value against each strain. (b) Left, example images of 2D monolayer cultures of forebrain-specific hNPCs immunostained for ZIKV envelop protein (ZIKVE; green), cleaved-caspase-3 (Cas3; red) and DAPI (gray) 72 hours after ZIKV FSS-13025 exposure (with or without Emricasan treatment) compared to mock infections (multiplicity of infection/MOI = 0.04 – 0.08; scale bar: 20 μm, applies to all image panels in b). Right, quantification comparing the percentage of CAS3+ cells over DAPI in mock or ZIKV-infected hNPCs cultures treated with the indicated concentrations of Emricasan. Values represent mean ± s.e.m. (n = 3 cultures; **P < 0.01; One-way ANOVA). (c) Left, example images of 3D human forebrain-specific organoid cultures (28 days in vitro) immunostained for ZIKVE (green), Cas3 (red) and DAPI (gray), 10 days after ZIKV FSS-13025 exposure (Scale bar: 100 μm, applies to all image panels in c). Right, quantification of the percentage of CAS3+ cells over DAPI in the ventricular zone. Values represent mean ± s.e.m. (n = 6 organoids; ***P < 0.001; One-way ANOVA).
Identification of antiviral compounds against ZIKV
Using expression of ZIKV protein NS1 as a read-out to screen anti-ZIKV activity, we identified two compounds from the primary hit list that significantly inhibited ZIKV infection in SNB-19 cells (Supplementary Fig. 2a). The first was Niclosamide, an FDA-approved drug for treating worm infections in both humans and domestic livestock; the other was PHA-690509, an investigational compound that functions as a cyclin-dependent kinase inhibitor (CDKi). Both compounds inhibited all three strains of ZIKV, as measured by NS1 expression, in a dose-dependent manner (Fig. 2a–d and Supplementary Fig. 2b–c).
Figure 2.
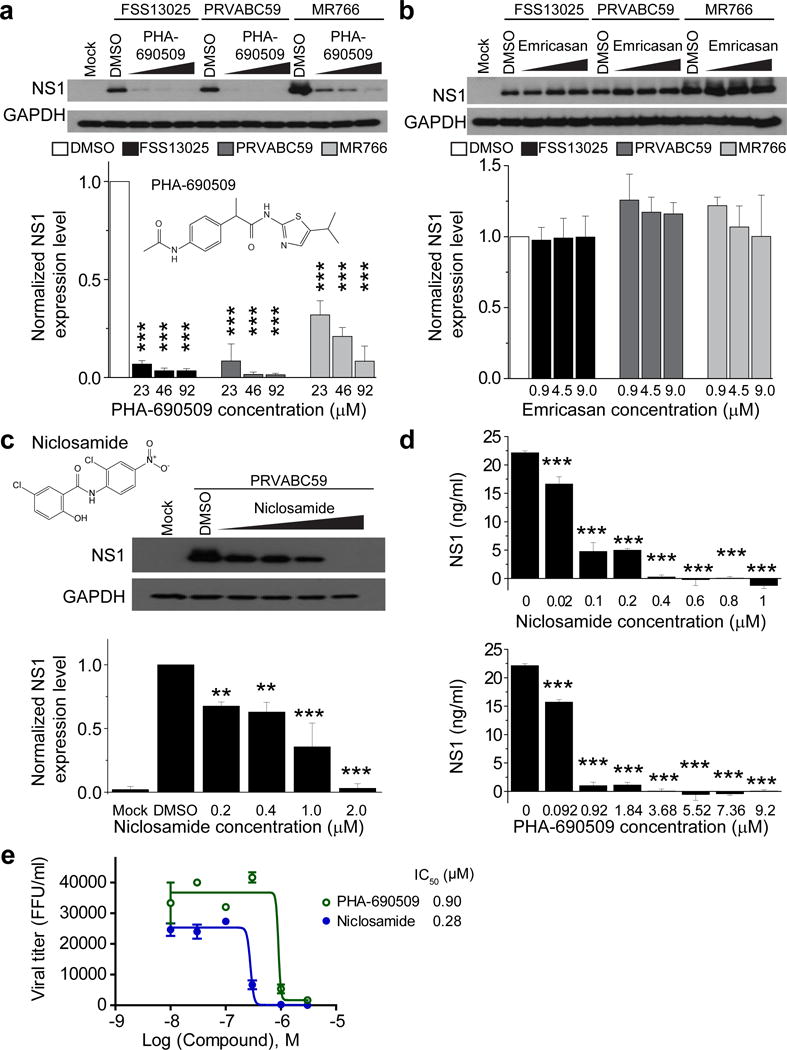
Identification of PHA-690509 and Niclosamide as antiviral compounds. (a) Top, example western blot images of titration of PHA-690509 on SNB-19 cells for anti-ZIKV activity. Glioblastoma SNB-19 cells were treated with the indicated compound for 1 hour prior to inoculation with ZIKV FSS-13025, PRVABC59, or MR766 (MOI = 1) and cells were harvested 24 hours post infection for western blot analysis. Bottom, quantification of NS1/GAPDH protein band intensities. Data were normalized to that with the DMSO treatment. Values represent mean ± s.d. (n = 3 cultures; ***P < 0.001; One-way ANOVA for comparison with the DMSO treatment). Insert shows chemical structure of PHA-690509. (b) Top, example western blot images of titration of Emricasan on SNB-19 cells for anti-ZIKV activity. Similar to (a). Values represent mean ± s.d. (n = 3 cultures; P > 0.1; One-way ANOVA for comparison with the DMSO treatment). (c) Top, chemical structure of Niclosamide and example western blot images of titration of Niclosamide on SNB-19 cells for anti-ZIKV activity using the PRVABC59 strain (MOI = 1). Bottom, quantification similar to (a). Values represent mean ±s.d. (n = 3 cultures; **P < 0.01; ***P < 0.001; One-way ANOVA for comparison with the DMSO treatment). (d) Summary of ELISA quantifications of secreted ZIKV-NS1 protein in the medium of infected cells upon treatment of different doses of Niclosamide (top) or PHA-690509 (bottom) for 24 hours. Values represent mean ± s.d. (n = 3 cultures; ***P < 0.001; One-way ANOVA for comparison with the 0 μM group). (e) Summary of titration of Niclosamide and PHA-690509 on ZIKV viral production. SNB-19 cells were treated with each indicated compound at increasing concentrations for 1 hour prior to infection with PRVABC59 at MOI = 0.5. Cell culture supernatant was collected 24 hours post infection and infectious virions were quantified for focus-forming units (FFU) using Vero cells. Data represent mean ± s.d. (n = 3 cultures). Curves represent best fits for calculating IC50, and the insets report the calculated IC50 value for each compound.
We further validated the antiviral activity of these two compounds using independent approaches. First, measurement of intracellular ZIKV RNA levels showed IC50 values of 1.72 μM and 0.37 μM for PHA-690509 and Niclosamide, respectively (Supplementary Fig. 2d–e). Second, both compounds suppressed production of infectious ZIKV particles at sub-micromolar concentrations (Fig. 2e). To investigate the underlying cellular mechanism, we performed time-of-addition experiments in SNB-19 cells (Fig. 3a). Both compounds effectively inhibited ZIKV infection when added either 1 hour before or 4 hours after virus inoculation (Fig. 3b). In contrast, a monoclonal antibody against AXL, a putative ZIKV entry factor39,40, was only effective when added prior to inoculation (Fig. 3b). Furthermore, the reduction of ZIKV RNA by treatment of these compounds was only apparent after the entry phase (0–4 hours after infection), and was correlated with the replication phase (4–24 hours) of the infection cycle (Supplementary Fig. 3). Together, these results indicate that Niclosamide and PHA-690509 inhibit ZIKV infection at a post-entry stage, likely at the viral RNA replication step.
Figure 3.
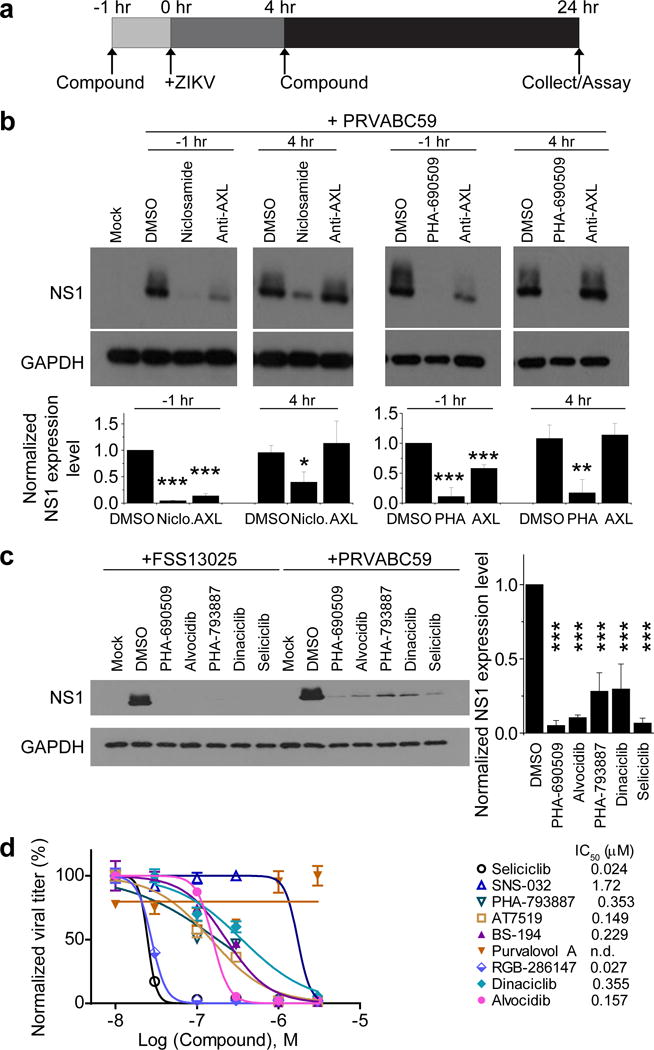
Inhibition of ZIKV infection by Niclosamide and PHA-690509 at a post-entry step and blockade of ZIKV infection by additional CDKis. (a) Schematic illustration of time-of-addition experiment for Niclosamide and PHA-690509. (b) Top, example western blot images of SNB-19 cells treated with 10 μg/ml anti-AXL antibody, 2 μM Niclosamide, or 92 μM PHA-690509 for 1 hour prior to or 4 hours post infection with PRVABC59 (MOI = 1). Bottom, quantification of NS1/GAPDH protein band intensities. Data were normalized to that with the DMSO treatment. Values represent mean ± s.d. (n = 3 cultures; *P < 0.05; **P < 0.01; ***P < 0.001; One-way ANOVA for comparison with the DMSO treatment). (c) Inhibition of ZIKV infection in CDKi-treated cells. Left, example western blot images of SNB-19 cells treated with the indicated compound at 92 μM for one hour prior to infection with FSS13025 or PRVABC59 (MOI = 1). Right, quantification similar to (b). Values represent mean ± s.d. (n = 3 cultures; ***P < 0.01; One-way ANOVA for comparison with the DMSO treatment). (d) Effect of various CDKis on ZIKV production. Similar to Figure 2e. All data were normalized to 0 μM for each compound. Data represent mean ± s.d. (n = 3 cultures). Curves represent best fits for calculating IC50 (listed on the right panel).
Pharmacological CDKis have been shown to inhibit replication of diverse viruses in culture, including herpes viruses and HIV41–46, and depletion of CDK9 impairs influenza A viral replication47. The possibility of ZIKV inhibition by a CDKi is particularly intriguing, given our recent observation that ZIKV infection and cell cycle regulation are intimately connected15. We therefore tested 27 additional structurally distinct CDKis for inhibition of ZIKV infection (Supplementary Table 6). We identified 9 active compounds in SNB-19 cells (Fig. 3c–d and Supplementary Fig. 4a), whereas 4 non-CDK kinase inhibitors exhibited minimal anti-ZIKV activity (Supplementary Fig. 4b). Analyses of ZIKV production showed IC50 values at sub-micromolar concentrations for nine of the ten CDKis tested, with Seliciclib and RGB-286147 at 24 nM and 27 nM, respectively (Fig. 3d).
Using the clinical isolate from the 2015 Puerto Rico Zika outbreak, PRVABC59, we next examined the effectiveness of identified compounds in hNPCs and astrocytes, both of which are target cells for ZIKV infection in the fetal brain9. Niclosamide and PHA-690509 inhibited ZIKV infection and production in these central nervous system cells (Fig. 4a–c). ZIKV targets astrocytes in the adult mouse brain48. Analysis of ZIKV production upon infection of human astrocytes showed IC50 values around 0.2 μM for both Niclosamide and PHA-690509 (Fig. 4c). Quantification of astrocyte viability showed minimal toxicity of Niclosamide, PHA-690509 and four CDKis at levels lower than 3 μM (Supplementary Fig. 5a). Given the critical role of CDKs in cell cycle regulation, we examined the effect of PHA-690509 and Seliciclib, the most potent CDKi for ZIKV inhibition (Fig. 3d), on hNPC proliferation. ZIKV infection led to a drastic reduction in hNPC proliferation, which was partially rescued by treatment with either compound (Supplementary Fig. 5b). Furthermore, both PHA-690509 (1 μM) and Seliciclib (5 μM) treatment alone had a minimal effect on hNPC proliferation in brain organoids (Supplementary Fig. 5c).
Figure 4.
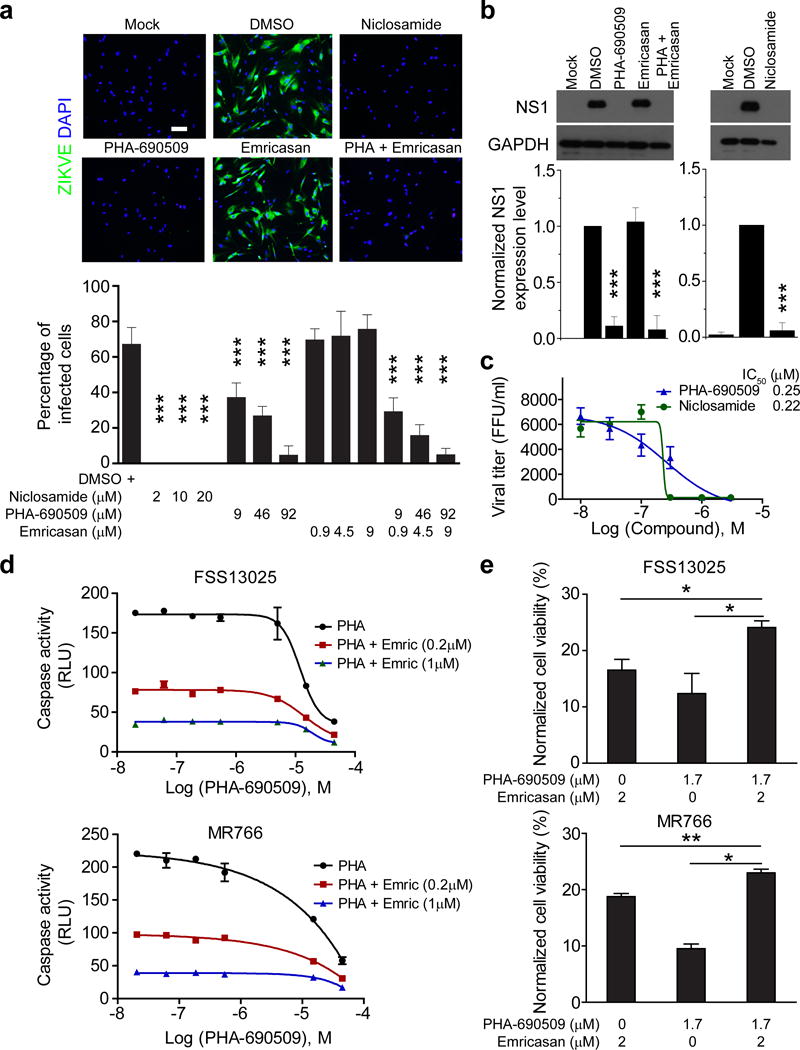
Niclosamide and PHA-690509 inhibit ZIKV infection in human astrocytes and forebrain-specific hNPCs. (a) Top, example immunostaining images of astrocytes treated with 2 μM Niclosamide, 92 μM PHA-690509, 9 μM Emricasan, or a combination of 92 μM PHA-690509 and 9 μM Emricasan for 1 hour prior to infection with PRVABC59 (MOI = 0.5). Cells were fixed 24 hours post infection and stained for ZIKVE (green) and DAPI (blue; Scale bar: 50 μm, applies to all image panels in a). Bottom, quantification of the percentage of ZIKV-infected astrocytes over DAPI. Values represent mean ± s.d. (n = 6 cultures; ***P < 0.001; One-way ANOVA for comparison with the DMSO treatment). (b) Top, example western blot images of hNPCs infected at a MOI of 0.1 and analyzed 48 hours post infection. Bottom, quantifications of NS1/GAPDH protein band intensities. Data were normalized to that with the DMSO treatment. Values represent mean ± s.d. (n = 3 cultures; ***P < 0.01; One-way ANOVA for comparison with the DMSO treatment). (c) Virus production from compound treated iPSC-derived human astrocytes. Similar to Figure 2e. Data represent mean ± s.d. (n = 3 cultures). (d–e) Summary of additive effects of a combination of Emricasan and PHA-690509 on inhibiting increased caspase-3 activity in human astrocytes infected with FSS13025-ZIKV or MR766 ZIKV (d), and on improving cell viability as measured by ATP production (e). Similar to Figure 1a. Values represent mean ± s.d. (n = 3 cultures; *P < 0.05; **P < 0.01; One-way ANOVA).
Benefit of combining neuroprotective and antiviral compounds
Finally, we explored combining these two categories of compounds (neuroprotective and antiviral). The two-drug combination of Emricasan and PHA-690509 exhibited an additive effect in inhibiting caspase-3 activity in SNB-19 cells (Fig. 4d). A similar additive effect was found to preserve astrocyte viability after ZIKV infection (Fig. 4a, e). Notably, Emricasan did not interfere with PHA-690509’s ability to inhibit ZIKV infection in the combination treatment (Fig. 4b and Supplementary Fig. 6a–c).
We also tested the effect of sequential treatment of the two categories of compounds. We found that treatment of PRVABC59-infected hNPCs with Emricassan for 72 hours followed by Niclosamide treatment for 48 hours led to the recovery of ZIKV-negative hNPCs, suggesting a beneficial effect of “buying time” through inhibition of apoptosis to allow infected cells to recover (Supplementary Fig. 6d).
DISCUSSION
Here we developed two ultra-high throughput assays using human neural cells that measure ZIKV-induced caspase-3 activity and cell viability and screened over 6,000 approved drugs and drug candidate compounds. Our efforts so far have led to identification of small molecules that either protect against cell death of multiple neural cell types or suppress ZIKV replication.
Emricasan, also named IDN-6556 or PF-03491390, is an inhibitor of activated caspases with sub-to nano-molar activity in vitro49, 50. Emricasan is currently being evaluated in Phase II clinical trials to reduce hepatic injury and liver fibrosis caused by chronic HCV infection51,52. Emricasan was well tolerated in human trials without significant adverse events52. It was reported that overall and maximum concentrations of Emricasan (oral delivery, BID, × 4 days) in human blood were 1.90 μg/ml (3.35 μM) and 2.36 μg/ml (4.15 μM), respectively. Therefore, the reported human plasma concentration of Emricasan is about 10-fold higher than the IC50 for inhibition of increased caspase-3 activity and cell death caused by ZIKV infection in vitro. Future animal studies will be critical to evaluate the efficacy of Emricasan in vivo. Whether it is safe to use Emricasan during pregnancy for ZIKV infection in humans will need to be evaluated in preclinical toxicology studies and clinical trials.
Ten CDKis with unrelated structures inhibit ZIKV replication and production, supporting a role for CDKs in the life cycle of ZIKV in human cells. As flaviviruses are not known to encode any CDKs, these results suggest that one or more cellular CDKs in the host may be important for ZIKV replication. Further studies on target identification, including targeted kinome siRNA or CRISPR screens, may reveal additional insights into the mechanism of action for these inhibitors. Although many CDKis are being evaluated in clinical trials for various cancers, Cystic Fibrosis, and Cushings Disease, these compounds may not be suitable for pregnant women because of potentially hazardous effects on the fetus53. In our in vitro analyses, treatment of PHA-690509 or Seliciclib partially rescued ZIKV-induced reduction of hNPC proliferation and the treatment itself exhibited a minimal effect on hNPC proliferation in brain organoid cultures, which model early human brain development in vitro (Supplementary Fig. 5c–d). Future studies in animal models, including both rodents and primates, are critical to test the efficacy and toxicity of target compounds in vivo. Notably, these CDKis and their derivatives may be useful for treating non-pregnant humans who face an increased risk of Guillain-Barré syndrome and other conditions following ZIKV infection. For example, viral RNA and infectious virus have been detected in the semen of men weeks after acute symptoms have resolved54. In addition, these CDKis will be valuable chemical tools for probing the role of a specific type of CDKs in ZIKV infection, which can serve as a new target for anti-ZIKV drug development. The structural diversity of effective CDKis also provides multiple scaffolds for lead compound optimization using medicinal chemistry.
Niclosamide is an FDA-approved drug (trade name Niclocide) used in humans to treat worm infections for nearly 50 years and is well tolerated55,56. It is known to inhibit several viruses in culture systems, including the Japanese encephalitis flavivirus57–59. Its broad antiviral activity has been attributed to its ability to neutralize endo-lysosomal pH and interfere with pH-dependent membrane fusion58, which is an essential step in the common virus entry pathway. Our time-of-addition and time-course analyses, however, suggest inhibition by Niclosamide occurs at a post-entry step, such as replication. Future molecular studies of its mechanism of action may provide additional targets for drug development. Niclosamide is a category B drug, which indicates that no risk to fetuses has been found in animal studies. It has low toxicity in mammals with an oral LD50 of 5000 mg/kg in rats60. The potency of Niclosamide on inhibition of ZIKV replication is in the submicromolar range, whereas clinically it can be delivered at micromolar levels60. Additionally, a pro-drug formulation may further increase the bio-availability of this compound currently indicated for treating intestinal parasites. WHO recommended that Niclosamide may be used during pregnancy because it has not been shown to be mutagenic, teratogenic or embryotoxic (http://apps.who.int/medicinedocs/en/d/Jh2922e/3.1.3.html). United States Centers for Disease and Prevention (CDC) concurs with the WHO recommendation and further recommends that “for individual patients in clinical settings, the risk of treatment (with Niclosamide) in pregnant women who are known to have an infection needs to be balanced with the risk of disease progression in the absence of treatment” (http://www.cdc.gov/parasites/hymenolepis/health_professionals/). Independent of evaluating potential benefits and risks for pregnant women, Niclosamide could be used to reduce viral load in infected men and nonpregnant women, which could reduce transmission and potentially prevent Guillain-Barré syndrome and other ZIKV complications in humans.
Despite rapid progress in the preclinical development of anti-ZIKV vaccines26,27, testing the safety and efficacy of vaccines in humans can take a significant amount of time. Effective countermeasures against ZIKV, including small molecule therapeutics, are also urgently needed. Our findings and tools provided here will significantly advance current ZIKV research and have an immediate impact on development of anti-ZIKV therapeutics. Furthermore, our findings could have implications for other flaviviruses, such as dengue viruses, chikungunya virus and West Nile virus, many of which can cause devastating illness.
METHODS
Methods and any associated references are available in the online version of the paper.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
ONLINE METHODS
Human forebrain-specific NPCs, astrocytes, organoids and cell line cultures
The human iPSC line (C1–2 line) was previously generated from a skin biopsy sample of a healthy male newborn and has been fully characterized and passaged on MEF feeder layers61. The BJ line was derived from healthy control fibroblasts obtained from the American Type Culture Collection (CRL-2522, male, neonatal). All studies followed institutional IRB protocols approved by Florida State University, Johns Hopkins University School of Medicine, and Icahn School of Medicine at Mount Sinai. Human iPSCs were differentiated into forebrain-specific hNPCs following the previously established protocol61. Briefly, human iPSC colonies were detached from the feeder layer with 1 mg/ml collagenase treatment for 1 hour and suspended in embryonic body (EB) medium, consisting of FGF-2-free iPSC medium supplemented with 2 μM Dorsomorphin and 2 μM A-83 in non-treated polystyrene plates for 4 days with a daily medium change. After 4 days, EB medium was replaced by neural induction medium (NPC medium) consisting of DMEM/F12, N2 supplement, NEAA, 2 μg/ml heparin, and 2 μM cyclopamine. Floating EBs were then transferred to Matrigel-coated 6-well plates at day 7 to form neural tube-like rosettes. Attached rosettes were kept for 15 d with NPC medium change every other day. On day 22, rosettes were picked mechanically and transferred to low attachment plates (Corning) to form neurospheres in NPC medium containing B27. Neurospheres were then dissociated with Accutase at 37 °C for 10 min and placed onto Matrigel-coated 6-well plates at day 24 to form monolayer hNPCs in NPC medium containing B27. These hNPCs expressed forebrain-specific progenitor markers61, including NESTIN, PAX6, EMX-1, FOXG1, and OTX2.
Human BJ iPSC line and forebrain NPCs were derived as described previously62,63. Human iPSC-derived forebrain NPCs were maintained at high density, grown on Matrigel (BD bioscience) in media consisting of DMEM/F12, 1× N2, 1× B27-RA (Invitrogen), 1 μg/ml laminin (Invitrogen) and 20 ng/ml FGF2 (Invitrogen). hNPCs were differentiated to astrocytes and cultured on Matrigel-coated plates in astrocyte medium (ScienCell). Astrocytes were split 1:3 every week with Accutase (Millipore) and cultured up to 120 days.
Forebrain-specific organoids were generated from the C1–2 human iPSC line as previously described17. Briefly, on day 1, human iPSC colonies were detached 7 days after passage with Collagenase Type IV, washed with fresh stem cell medium and transferred to an ultra-low attachment 6-well plate (Corning Costar), containing 3 ml of stem cell medium (without FGF-2), plus 2 μM Dorsomorphine (Sigma) and 2 μM A83-01 (Tocris). On days 5–6, half of the medium was replaced with induction medium consisting of DMEM:F12, 1× N2 Supplement (Invitrogen), 10 μg/ml Heparin (Sigma), 1× Penicillin/Streptomycin, 1× Non-essential Amino Acids, 1× Glutamax, 4 ng/ml WNT-3A (R&D Systems), 1 μM CHIR99021 (Cellagentech), and 1 μM SB-431542 (Cellagentech). On day 7, organoids were embedded in Matrigel (BD Biosciences) and continued to grow in induction medium for 6 more days. On day 14, embedded organoids were mechanically dissociated from Matrigel by pipetting up and down onto the plate with a 5 ml pipette tip. Typically, 10 – 20 organoids were transferred to each well of a 12-well spinning bioreactor (SpinΩ) containing differentiation medium, consisting of DMEM:F12, 1× N2 and B27 Supplements (Invitrogen), 1× Penicillin/Streptomycin, 1× 2-Mercaptoenthanol, 1× Non-essential Amino Acids, 2.5 μg/ml Insulin (Sigma). Day 18 forebrain organoids were exposed to ZIKV FSS-13025 in SpinΩ for 24 hours and then in replaced fresh medium for additional 9 days before fixation and immunostaining as previously described17. To test the potential toxic effect on hNPC proliferation, day 20 forebrain organoids were also treated with PHA-690509 (1 μM) or Seliciclib (5 μM) for three days without ZIKV exposure. On day 23 (20+3), treated forebrain organoids were pulsed with 10 μM EdU for 1 hour and immediately fixed for analysis.
The glioblastoma SNB-19 cell line (part of the National Cancer Institute 60 human tumor cell line) was a gift from Dr. David Meckes (Florida State University, Tallahassee, FL). SNB-19 cells were maintained at 37 °C in 5% CO2 in RPMI-60, 1× penicillin/streptomycin, and 10% fetal bovine serum (Invitrogen). The Aedes albopictus C6/36 cell line (ATCC) was maintained at 28 °C in 5% CO2.
Preparation of ZIKV and cell infection
The MR766-ZIKV stock with the titer of 1×105 Tissue Culture Infective Dose (TCID)/ml in the form of culture fluid from an infected rhesus Macaca cell line LLC-MK2, was originally obtained from ZeptoMetrix (Buffalo, NY). The FSS13025-ZIKV strain was obtained from Drs. Robert Tesh and Pei-Yong Shi (University of Texas Medical Branch, Galveston, Texas). The PRVABC59 strain was obtained from ATCC (Manassas, VA). Original viral stocks were then amplified in Aedes albopictus clone C6/36 cells. Briefly, C6/36 cells were inoculated with viral inoculum for one hour at 28 °C in a low volume of media (3 ml per T-75 flask), with rocking every 15 minutes, before the addition of 17 ml media. Virus inoculated cells were incubated at 28 °C for 6–7 days before harvesting of supernatant. C6/36-amplified ZIKV titer was determined by infecting Vero cells for 48 hours with a methyl-cellulose overlay and analyzed for focus-forming units per ml (FFU/ml). In mock infections, an equal volume of spent uninfected C6/36 culture medium was used. For infections, cells were seeded into 12-well plates one day prior to virus addition. For all cell types, compound was added 1 hour prior to viral addition unless otherwise specified. Cells were harvested at 24–72 hours post-infection.
Compound libraries
The Library of Pharmacologically Active Compounds (LOPAC), consisting of 1280 compounds, was purchased from Sigma-Aldrich. The NCATS pharmaceutical collection64, a collection consisting of 2816 clinically approved and investigational drugs, was established in 2011. All compounds were dissolved in DMSO as 10 mM stock solutions, then diluted in DMSO at a 1:3 ratio in 384-well plates, followed by reformatting into 1536-well compound plates for use in high-throughput screening (HTS).
Caspase-3/7 assay
Caspase-Glo 3/7 assay kit (catalog number G8092; Promega, Madison, WI) was used to detect caspase-3/7 activity. Reagents were reconstituted as described in the protocol from the manufacturer. Polystyrene plates (384-well and 1536-well; regular tissue culture treated and PDL coated) were purchased from Greiner Bio-One (Monroe, NC). Cells were seeded in 384- and 1536-well assay plates and cultured at 37°C with 5% CO2 for 16 hours. ZIKV solution was added to cells, followed by incubation at 37°C with 5% CO2 for 6 hours. Caspase-Glo-3/7 was added to each well, unless otherwise specified, and incubated at room temperature for 30 minutes. The luminescence intensity of the assay plates was measured using a ViewLux plate reader (PerkinElmer). Data were normalized by using the cell-containing wells without ZIKV as a negative control (0% induction of caspase-3/7 activity) and wells containing ZIKV infected cells that induced caspase-3/7 activity were used as a positive control (100% induction of caspase 3/7 activity).
ATP content assay for cell viability and compound cytotoxicity
The ATPlite luminescence assay system assay kit (catalog number 6016731; PerkinElmer) was used to determine cell viability. The reagent was reconstituted and prepared as described by the manufacturer. In order to measure the cell death caused by ZIKV infection, cells were cultured for 16 hours at 37°C with 5% CO2 in assay plates, followed by addition of ZIKV solution and incubation at 37°C with 5% CO2 for 72 hours. ATPlite, the ATP monitoring reagent, was then added to the assay plates and they were incubated for 15 minutes. The resulting luminescence was measured using the ViewLux plate reader. Data were normalized using wells without cells as a control for 100% cell killing, and cell-containing wells without ZIKV infection as full cell viability (0% cell killing). For analysis of potential toxicity of select compounds, cells were seeded in 96-well plates. One day later, cells were treated with indicated compounds and concentrations for 24–48 hours prior to the addition of Cell Titer-Glo substrate (Promega) and measured according to manufacturer instructions.
Large-scale compound screening
A quantitative high-throughput screening (qHTS, ref65), in which each compound was assayed in four concentrations (0.37, 1.84, 9.2, and 46 μM), was performed in singlet for the primary compound screen. While a single compound concentration (in singlet) has been traditionally used for HTS of large compound collections (such as 1 to 3 million compounds), the qHTS format with multiple compound concentrations has recently been used for medium or small compound collections such as approved drug library. Specifically, SNB-19 cells and hNPCs were seeded onto PDL coated 1536-well assay plates at 250 cells per 3 μl/well and incubated at 37°C in 5% CO2 for 16 hours. Test compounds dissolved in DMSO were transferred to assay plates at a volume of 23 nl/well by an automated pintool workstation (Wako Automation, San Diego, CA). Compounds were incubated with cells for 30 minutes at 37°C in 5% CO2, immediately followed by the addition of 2 μl/well of ZIKV (2 FFU/cell). Incubation time of compound-treated cells with ZIKV varied based on assay format. Experiments measuring virus-induced caspase-3/7 activity required a 6-hour incubation of ZIKV in the presence compounds at 37°C in 5% CO2. Following this incubation, 3.5 μl/well of caspase-3/7 reagent mixture was added to assay plates. The plates were incubated for 30 minutes at room temperature, and the resultant luminescence signal was measured using a ViewLux plate reader (Perkin Elmer). Experiments measuring virus-induced cell death required a 72-hour incubation of ZIKV in the presence of compounds at 37°C in 5% CO2. Following this incubation, 3.5 μl/well of ATP content detection reagent was added to assay plates. The plates were incubated for 30 minutes at room temperature, and the resultant luminescence signal was measured in a ViewLux plate reader. Step-by-step assay protocols are listed in Supplementary Tables 1 and 2.
Confirmation of primary hits and counter-screen
The primary hits were selected with criteria of IC50 values equal or smaller than 30 μM and maximal inhibition greater than 50%. The hit compounds were diluted at 1:3 ratio in DMSO to 11 concentrations for the confirmation experiments. The same Caspase-3 activity assay was first used to confirm the activity of primary hit compounds (n = 3). In order to further confirm the activity of these compounds, the protective effect of these primary hits on the cell death caused by incubation of cells with ZIKV for three days was also determined using an ATP content cell viability assay (n = 3).
Because compound cytotoxicity could nonspecifically reduce the caspase activity induced by ZIKV, we also used the ATP content assay to measure compound cytotoxicity in the absence of ZIKV infection. Cells were seeded in the same way as described above in 1536-well assay plates. After a 6 hour incubation with compounds in the absence of ZIKV, 3.5 μl/well of ATP content reagent mixture was added to the assay plates and incubated for 30 minutes at room temperature. The luminescence signal in the assay plates was measured using a ViewLux plate reader (Supplementary Table 3). Any compounds that exhibited cytotoxicity were eliminated as false positive compounds.
Western blot
Cells were harvested by trypsinization, pelleting, and subsequent lysis in 1× Laemlli buffer and boiled, or directly lysed in 1× Laemlli buffer and boiled. Antibodies used were anti-ZIKV NS1 (1:2000; BF-1225-36, BioFront Technologies, Tallahassee, FL) or GAPDH (Santa Cruz Biotechnology, Texas). The screening of two subsets of compounds for antiviral activity (Supplemental Fig. 2a and Supplemental Fig. 4b) was blinded, while all other experiments were nonblinded.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde (Sigma) for 15 minutes at room temperature. Samples were permeabilized and blocked with 0.25% Triton X-100 (Sigma) and 10% donkey or goat serum in Phosphate Buffer Saline (PBS) for 20 minutes as previously described (Chiang et al., 2011; Wen et al., 2014; Yoon et al., 2014). Samples were then incubated with primary antibodies at 4°C overnight, followed by multiple PBST (PBS plus Tween-20) washes and incubation with secondary antibodies for 1 hour at room temperature. Slides were mounted using VECTASHIELD with 4′6-diamidino-2-phenylindole (DAPI; Vectorlabs, Burlingame, CA). The following primary antibodies were used: anti-flavivirus group antigen (clone D1-4G2-4-15; mouse; 1:500; Millipore), anti-cleaved caspase-3 (Asp15; Rabbit; 1:500; Cell Signaling Technology) and Phospho-Histone H3 (PH3; 9701S; Rabbit; 1:300; Cell Signaling Technology) as previously described17. Antibodies were prepared in PBS containing 0.25% Triton X-100 and 10% donkey serum. For EdU assay, the Click-iT EdU Alexa Fluor 488 Imaging Kit (Thermo Fisher Scientific) was used according to the manufacturer’s protocol. Images were taken by Zeiss LSM 700 and 880 confocal microscopes, Olympus BX61, or Zeiss Axiovert 200M microscope.
For organoid studies, quantitative analyses were conducted on randomly selected cortical structures captured by confocal microscope in a blind fashion (example as shown in Figure 1c). Cell death and cell proliferation were quantified by counting activated Caspase-3+ and PH3+ nuclei, respectively, over total nuclei stained by DAPI in the ventricular structures in at least 6 samples as previously described17. We determined our sample sizes based on previous studies conducted in our lab17.
NS1 ELISA
The anti-ZIKV NS1 ELISA kit (ZKV-NS1-EK) was obtained from BioFront Technologies (Tallahassee, FL) and used according to the manufacturer’s protocol.
Viral titer by focus forming unit (FFU) assay
ZIKV titer from cell supernatant was determined by infecting Vero cells for 48 hours with a methyl-cellulose overlay and analyzed for focus-forming units per mL (FFU/ml). Briefly, cell supernatant was titrated in triplicates onto a monolayer culture of Vero cells in 96-well plates and incubated at 37 °C for 2 hours. Viral inoculum was removed and replaced with a methyl-cellulose overlay. Vero cells were incubated for an additional 48–72 hours prior to fixation and incubated with anti-flavivirus group antigen overnight at 4 °C. The next day, fixed cells were washed three times with PBS and incubated with anti-mouse-HRP secondary antibody for 1 hour at room temperature, washed again three times with PBS, and incubated with DAB peroxidase substrate for ten minutes (Vector Labs).
Antiviral compound analysis
SNB-19 cells, human astrocytes, or hNPCs were seeded in 12-well plates at approximately 3×105 cells/well. The next day, cells were treated with compound at 1–10× IC50 or indicated concentration for 1 hour prior to inoculation with ZIKV (MOI = 0.5–1). Cells and supernatant were harvested 24–48 hours post infection and analyzed by western blot, immunostaining, or quantitative RT-PCR analyses. Western blot bands were quantified and immunostaining images counted using ImageJ (NIH, Bethesda, MD). For qRT-PCR analysis, total cellular RNA was purified using the RNeasy Mini Plus kit (Qiagen). About 500–1000 ng total RNA was reverse transcribed into cDNA using random hexamers according to manufacturer’s instructions. (Superscript III First Strand Synthesis System, Invitrogen). Quantitative PCR was performed using SYBR green PCR master mix (Invitrogen), gene specific primers (ZIKV-NS1-Forward: TGGAGTTCAACTGACGGTCG; ZIKV-NS1-Reverse: TACCCCGAACCCATGATCCT; GAPDH-Forward: TCACTGCCACCCAGAAGACTG; and GAPDH-Reverse: GGATGACCTTGCCCACAGC), and an Applied Biosystems 7500 Fast real-time PCR system. A 60°C to 95°C melt curve analysis following PCR was performed using default settings. Relative quantitation was performed using the ΔΔCT method with GAPDH as the endogenous control and the relative fold change was calculated by normalizing to control cells.
Data analysis and statistics
The primary screen data and concentration response curves were analyzed using software developed internally66. The IC50 values of compound confirmation data were calculated using the Prism software (GraphPad Software, Inc. San Diego, CA). All values are expressed as the mean ± s.d. unless specified otherwise. Western blots were quantified using ImageJ (NIH, Bethesda, MD). We did not exclude any samples. Experiments were not blinded, except for those were described. The sample sizes were estimated according to previous studies and the known variability of the assays. Statistical analyses were carried out with the one-way ANOVA using the Prism software. The Z′ factor, a measure of statistical effect size and an index for assay quality control, was calculated by: Z′= 1 − (3*s.d.signal + 3*s.d.basal)/(Meansignal − Meanbasal).
Supplementary Material
Acknowledgments
We thank Dr. Robert Tesh at UTMB and the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) for FSS-13025 isolate of ZIKV. This work was supported by the Intramural Research Program of the National Center for Advancing Translational Sciences, National Institutes of Health (NIH) (W.Z.); by ZIKV seed funding from Florida State University (H.T.), Start-up fund from Emory University (Z.W.), Brain and Behavior Research Foundation and the New York Stem Cell Foundation (K.J.B.), Maryland Stem Cell Research Fund (G-l.M. and H.S.), and partially by NIH grants NS048271 and NS095348 (G-l.M.), NS047344, NS097206 and MH106434 (H.S.), MH101454, MH106056, P50AG005138, and AG046170 (K.J.B.), and AI119530 (H.T.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
M.X., E.M.L., Z.W., A.S., K.M.C., M-H.X., G-L.M., W.Z., H.S., and H.T. conceived of the research, designed the study, interpreted data, and wrote the manuscript. M.X., E.M.L., Z.W., Y.C., W-K.H., X.Q., J.K., S.C.Q., C.H., F.J., H.N.N., M.I., C.H., P.S., C.A., S.G.M., and W.H. performed experiments. J.TCW, A.G. and K.B. provided astrocytes. R.H. analyzed data. G-L.M., W.Z., H.S. and H.T. are co-senior authors who directed the research.
References
- 1.Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Heymann DL, et al. Zika virus and microcephaly: why is this situation a PHEIC? Lancet. 2016;387:719–721. doi: 10.1016/S0140-6736(16)00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Ortenzio E, et al. Evidence of Sexual Transmission of Zika Virus. N Engl J Med. 2016;374:2195–2198. doi: 10.1056/NEJMc1604449. [DOI] [PubMed] [Google Scholar]
- 4.Foy BD, et al. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis. 2011;17:880–882. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarthy M. Zika virus was transmitted by sexual contact in Texas, health officials report. Bmj. 2016;352:i720. doi: 10.1136/bmj.i720. [DOI] [PubMed] [Google Scholar]
- 6.Musso D, et al. Potential sexual transmission of Zika virus. Emerg Infect Dis. 2015;21:359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brasil P, et al. Zika Virus Infection in Pregnant Women in Rio de Janeiro – Preliminary Report. N Engl J Med. 2016 doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen E, et al. Rapid Spread of Zika Virus in The Americas–Implications for Public Health Preparedness for Mass Gatherings at the 2016 Brazil Olympic Games. International journal of infectious diseases: IJID: official publication of the International Society for Infectious Diseases. 2016;44:11–15. doi: 10.1016/j.ijid.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Driggers RW, et al. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N Engl J Med. 2016;374:2142–2151. doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- 10.Mlakar J, et al. Zika Virus Associated with Microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 11.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika Virus and Birth Defects–Reviewing the Evidence for Causality. N Engl J Med. 2016;374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 12.Cao-Lormeau VM, et al. Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araujo LM, Ferreira ML, Nascimento OJ. Guillain-Barre syndrome associated with the Zika virus outbreak in Brazil. Arq Neuropsiquiatr. 2016;74:253–255. doi: 10.1590/0004-282X20160035. [DOI] [PubMed] [Google Scholar]
- 14.Fontes CA, Dos Santos AA, Marchiori E. Magnetic resonance imaging findings in Guillain-Barre syndrome caused by Zika virus infection. Neuroradiology. 2016 doi: 10.1007/s00234-016-1687-9. [DOI] [PubMed] [Google Scholar]
- 15.Tang H, et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell. 2016;18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcez PP, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 17.Qian X, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossi SL, et al. Characterization of a Novel Murine Model to Study Zika Virus. Am J Trop Med Hyg. 2016 doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazear HM, et al. A Mouse Model of Zika Virus Pathogenesis. Cell Host Microbe. 2016 doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cugola FR, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, et al. Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 22.Miner JJ, et al. Zika Virus Infection during Pregnancy in Mice Causes Placental Damage and Fetal Demise. Cell. 2016;165:1081–1091. doi: 10.1016/j.cell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu KY, et al. Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell research. 2016;26:645–654. doi: 10.1038/cr.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Driggers RW, et al. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N Engl J Med. 2016 doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- 25.Dang J, et al. Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larocca RA, et al. Vaccine protection against Zika virus from Brazil. Nature. 2016 doi: 10.1038/nature18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbink P, et al. Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science. 2016 doi: 10.1126/science.aah6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang YM, et al. A small molecule screen in stem-cell-derived motor neurons identifies a kinase inhibitor as a candidate therapeutic for ALS. Cell Stem Cell. 2013;12:713–726. doi: 10.1016/j.stem.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun W, Sanderson PE, Zheng W. Drug combination therapy increases successful drug repositioning. Drug Discov Today. 2016;21:1189–1195. doi: 10.1016/j.drudis.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouznetsova J, et al. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg Microbes Infect. 2014;3:e84. doi: 10.1038/emi.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansen LM, et al. A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Sci Transl Med. 2015;7:290ra289. doi: 10.1126/scitranslmed.aaa5597. [DOI] [PubMed] [Google Scholar]
- 32.Chen CZ, et al. High-throughput Giardia lamblia viability assay using bioluminescent ATP content measurements. Antimicrob Agents Chemother. 2011;55:667–675. doi: 10.1128/AAC.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Debnath A, et al. A high-throughput drug screen for Entamoeba histolytica identifies a new lead and target. Nat Med. 2012;18:956–960. doi: 10.1038/nm.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun W, et al. Chemical signatures and new drug targets for gametocytocidal drug development. Sci Rep. 2014;4:3743. doi: 10.1038/srep03743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun W, et al. Rapid identification of antifungal compounds against Exserohilum rostratum using high throughput drug repurposing screens. PLoS One. 2013;8:e70506. doi: 10.1371/journal.pone.0070506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He S, et al. Repurposing of the antihistamine chlorcyclizine and related compounds for treatment of hepatitis C virus infection. Sci Transl Med. 2015;7:282ra249. doi: 10.1126/scitranslmed.3010286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrows NJ, et al. A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe. 2016 doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 39.Hamel R, et al. Biology of Zika Virus Infection in Human Skin Cells. J Virol. 2015;89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nowakowski TJ, et al. Expression Analysis Highlights AXL as a Candidate Zika Virus Entry Receptor in Neural Stem Cells. Cell Stem Cell. 2016;18:591–596. doi: 10.1016/j.stem.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schang LM, St Vincent MR, Lacasse JJ. Five years of progress on cyclin-dependent kinases and other cellular proteins as potential targets for antiviral drugs. Antivir Chem Chemother. 2006;17:293–320. doi: 10.1177/095632020601700601. [DOI] [PubMed] [Google Scholar]
- 42.Badia R, et al. Inhibition of herpes simplex virus type 1 by the CDK6 inhibitor PD-0332991 (palbociclib) through the control of SAMHD1. The Journal of antimicrobial chemotherapy. 2016;71:387–394. doi: 10.1093/jac/dkv363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nemeth G, et al. Novel, selective CDK9 inhibitors for the treatment of HIV infection. Current medicinal chemistry. 2011;18:342–358. doi: 10.2174/092986711794839188. [DOI] [PubMed] [Google Scholar]
- 44.Okamoto M, et al. Selective inhibition of HIV-1 replication by the CDK9 inhibitor FIT-039. Antiviral research. 2015;123:1–4. doi: 10.1016/j.antiviral.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 45.Van Duyne R, et al. Effect of mimetic CDK9 inhibitors on HIV-1-activated transcription. Journal of molecular biology. 2013;425:812–829. doi: 10.1016/j.jmb.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto M, et al. CDK9 inhibitor FIT-039 prevents replication of multiple DNA viruses. The Journal of clinical investigation. 2014;124:3479–3488. doi: 10.1172/JCI73805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Li G, Ye X. Cyclin T1/CDK9 interacts with influenza A virus polymerase and facilitates its association with cellular RNA polymerase II. J Virol. 2010;84:12619–12627. doi: 10.1128/JVI.01696-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bell TM, Field EJ, Narang HK. Zika virus infection of the central nervous system of mice. Arch Gesamte Virusforsch. 1971;35:183–193. doi: 10.1007/BF01249709. [DOI] [PubMed] [Google Scholar]
- 49.Haddad JJ. Current opinion on 3-[2-[(2-tert-butyl-phenylaminooxalyl)-amino]-propionylamino]-4-oxo-5-(2,3,5,6-tetrafluoro-phenoxy)-pentanoic acid, an investigational drug targeting caspases and caspase-like proteases: the clinical trials in sight and recent anti-inflammatory advances. Recent Patents on Inflammation & Allergy Drug Discovery. 2013;7:229–258. doi: 10.2174/1872213x113079990017. [DOI] [PubMed] [Google Scholar]
- 50.Hoglen NC, et al. Characterization of IDN-6556 (3-[2-(2-tert-butyl-phenylaminooxalyl)-amino]-propionylamino]-4-oxo-5-(2,3,5,6-te trafluoro-phenoxy)-pentanoic acid): a liver-targeted caspase inhibitor. J Pharmacol Exp Ther. 2004;309:634–640. doi: 10.1124/jpet.103.062034. [DOI] [PubMed] [Google Scholar]
- 51.Barreyro FJ, et al. The pan-caspase inhibitor Emricasan (IDN-6556) decreases liver injury and fibrosis in a murine model of non-alcoholic steatohepatitis. Liver International. 2015;35:953–966. doi: 10.1111/liv.12570. [DOI] [PubMed] [Google Scholar]
- 52.Shiffman ML, et al. Clinical trial: the efficacy and safety of oral PF-03491390, a pancaspase inhibitor – a randomized placebo-controlled study in patients with chronic hepatitis C. Alimentary Pharmacology and Therapeutics. 2010;31:969–978. doi: 10.1111/j.1365-2036.2010.04264.x. [DOI] [PubMed] [Google Scholar]
- 53.Ibrance (palbociclib) capsules [prescribing information] Pfizer, Inc; New York, NY: 2015. [Google Scholar]
- 54.Mansuy JM, et al. Zika virus: high infectious viral load in semen, a new sexually transmitted pathogen? The Lancet Infectious diseases. 2016;16:405. doi: 10.1016/S1473-3099(16)00138-9. [DOI] [PubMed] [Google Scholar]
- 55.Andrews P, Thyssen J, Lorke D. The biology and toxicology of molluscicides, Bayluscide. Pharmacol Ther. 1982;19:245–295. doi: 10.1016/0163-7258(82)90064-x. [DOI] [PubMed] [Google Scholar]
- 56.Al-Hadiya BMH. Niclosamide: Comprehensive profile. Profiles of Drug Substances, Excipients, and Related Methodology. 2005;32:67–96. doi: 10.1016/S0099-5428(05)32002-8. [DOI] [PubMed] [Google Scholar]
- 57.Wu CJ, et al. Inhibition of severe acute respiratory syndrome coronavirus replication by niclosamide. Antimicrob Agents Chemother. 2004;48:2693–2696. doi: 10.1128/AAC.48.7.2693-2696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jurgeit A, et al. Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects. PLoS Pathog. 2012;8:e1002976. doi: 10.1371/journal.ppat.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang J, et al. Identification of three antiviral inhibitors against Japanese encephalitis virus from library of pharmacologically active compounds 1280. PLoS One. 2013;8:e78425. doi: 10.1371/journal.pone.0078425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khanim FL, et al. Redeployment-based drug screening identifies the anti-helminthic niclosamide as anti-myeloma therapy that also reduces free light chain production. Blood Cancer J. 2011;1:e39. doi: 10.1038/bcj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wen Z, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Topol A, et al. Increased abundance of translation machinery in stem cell-derived neural progenitor cells from four schizophrenia patients. Transl Psychiatry. 2015;5:e662. doi: 10.1038/tp.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang R, et al. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci Transl Med. 2011;3:80ps16. doi: 10.1126/scitranslmed.3001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inglese J, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Jadhav A, Southal N, Huang R, Nguyen DT. A grid algorithm for high throughput fitting of dose-response curve data. Curr Chem Genomics. 2010;4:57–66. doi: 10.2174/1875397301004010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


