Abstract
Purpose
To assess the performance of a four-kallikrein panel, with and without microseminoprotein-beta (MSP), to predict high-grade (Gleason 7+, Gleason Grade Group 2+) prostate cancer (PCa) on biopsy in a multiethnic cohort from the Prostate, Lung, Colorectal, and Ovarian (PLCO) cancer screening trial.
Materials and Methods
Levels of free, intact, total prostate-specific antigen (PSA), human kallikrein-2, and MSP were measured blinded to outcome in cryo-preserved serum from men in the intervention arm of PLCO. Marker levels from 946 men, of whom 100 were African-American, were incorporated into a prespecified statistical model to predict high-grade PCa on biopsy.
Results
The detection of high-grade PCa (n=94, 10%) was enhanced by the four-kallikrein panel with an AUC of 0.79 compared to 0.73 for the Prostate Cancer Prevention Trial (PCPT) risk calculator (increase of 0.060; 95% C.I. 0.032, 0.088; p<0.01). Additionally, the AUC increased from 0.79 to 0.81 when MSP was added to the four-kallikrein panel. In African-American men, the four-kallikrein panel model also enhanced high-grade PCa detection over PSA (AUC 0.80 versus 0.67). As an illustration of clinical implications, use of one cut-point for biopsy (6% risk of high-grade PCa) with the four-kallikrein panel model would have eliminated unnecessary biopsies in 42% (397/946) of our cohort while detecting 88% (83/94) of highgrade PCa.
Conclusions
In a multiethnic United States population, the four-kallikrein panel demonstrates improved risk discrimination for high-grade PCa over conventional clinical variables (age, PSA, digital rectal examination) as well as the PCPT risk calculator.
Keywords: prostate cancer, screening, prostate-specific antigen, biopsy
INTRODUCTION
Over the last 20 years, prostate-specific antigen (PSA) has been employed as a screening tool for prostate cancer (PCa). Although PSA provides a significant benefit over digital rectal examination (DRE) alone and is associated with clinically significant PCa,1 a moderately elevated PSA has poor specificity and positive predictive value. For instance, the rate of positive biopsy was 24% in the European Randomized Study of Prostate Cancer Screening (ERSPCS) and 35% in the Prostate, Lung, Colorectal, and Ovarian (PLCO) cancer screening trial.2,3 Furthermore, of those men with positive biopsy, less than a third have clinically aggressive disease that requires consideration for curative intervention.4
A series of retrospective cohort studies have investigated a multivariable model that combines four kallikrein marker levels in blood — total PSA, free PSA, intact PSA, and human kallikrein-related peptidase 2 (hK2)—with clinical characteristics (age, DRE and history of prior negative biopsy). The fourkallikrein (4K) panel demonstrated improved risk discrimination compared to PSA and clinical characteristics alone. Decision analyses have demonstrated that use of the panel would reduce the number of biopsies performed, while missing few cases of high-grade.5–9 The accuracy of the 4K panel to predict aggressive disease on radical prostatectomy has also been demonstrated.10
Although studied and validated in European populations,5–10 the 4K panel has not been extensively evaluated in United States men.11 More importantly, as the European population was almost exclusively Caucasian, the performance characteristics of the 4K panel need to be studied in African-Americans, who more often present with advanced and aggressive disease, and are more likely to die from PCa.12,13 We examine a prespecified prediction model using the 4K panel to diagnose high-grade PCa, using preserved serum samples of men from the PLCO screening trial, who had a biopsy for elevated PSA (≥4.0 ng/mL). As a secondary aim, we examine the benefit of another candidate blood biomarker, microseminoprotein-beta (MSP),14 in combination with the 4K panel.
MATERIALS AND METHODS
Study cohort
With institutional review board approval from all study centers, we received pre-biopsy patient serum samples from the PLCO bio-repository and assayed the levels of the four kallikrein markers (free PSA, intact PSA, total PSA, and hK2) as well as MSP. The 4K panel was combined with various clinical parameters in a prespecified statistical model. Biopsy results were released to the study statisticians only after predictions were submitted to a third party. Accordingly, assays, modeling and outcome were assessed independently and blindly.
Our study cohort included patients from the intervention arm of PLCO, who provided consent, had no prior history of cancer, had no biopsies prior to randomization, were aged <75 years, had PSA ≥4 ng/mL during the biopsy year, documented Gleason score on biopsy, and has serum from the biopsy year available in the bio-repository. In PLCO, a positive screening result (suspicious DRE or PSA ≥4 ng/mL) in the intervention arm resulted in a notification to the patients’ primary care physician, who determined the subsequent diagnostic evaluation.3 The biopsy Gleason score was obtained by PLCO trial staff and collected by medical data abstractors. No central pathologic review was performed. All samples from eligible African-American, non-Hispanic men were included (n=113), and 887 samples were selected randomly from the remaining eligible men to build our study cohort (n=1000). Patients with missing data were excluded from further analysis (n=54), leaving a final sample of 946, 100 of whom were non-Hispanic African-Americans.
Laboratory methods
Immunoassay measurements of total and free PSA,15 intact PSA,16 hK2,17 and MSP,18 were conducted on AutoDelfia® 1235 automatic immunoassay systems (Wallenberg Research Laboratories, Department of Translational Medicine, Lund University, Skåne University Hospital, Malmö, Sweden). Free and total PSA were measured using the dual-label DELFIA Prostatus® total/free PSA-Assay (Perkin-Elmer, Turku, Finland) calibrated against the World Health Organization (WHO) 96/670 (PSA-WHO) and WHO 68/668 (free PSA-WHO) standards. The measurements of intact PSA and hK2 were performed with F(ab‘)2 fragments of the monoclonal capture antibodies as previously reported.19 The intact PSA assay measures only single-chain intact forms of free PSA as the monoclonal 4D4 IgG used in this assay does not bind multi-chain forms of free PSA cleaved at Lys145 or Lys146. Production and purification of the polyclonal rabbit anti-MSP antibody, protocols for biotinylation and Europium labeling of the anti-MSP antibody, and performance of the MSP-immunoassay were performed as previously reported.14 All assay measurements were conducted blinded to biopsy result.
Predictive Models
We assessed the accuracy of models to predict prostate biopsy outcome among men in our study cohort. Primarily we were interested in the accuracy of a model utilizing age, the 4K panel, and DRE to predict high-grade cancer on biopsy. Gleason score ≥7 or Gleason Grade Group20 (GGG) ≥2 were considered to be high-grade PCa. In order to assess the added clinical utility of the 4K panel, we compared these results to a model based on age, total PSA, and DRE, as well as the Prostate Cancer Prevention Trial Risk Calculator (PCPTRC).21 Additional predictive models that included various combinations of the clinical variables (age, PSA, DRE, 4K panel), as well as MSP, were examined. All models, other than those testing the value of incorporating MSP, were prespecified. The 4K and PSA models were derived from the Rotterdam section of the ERSPCS.6
Statistical Methods
The discrimination of the predictive models was assessed using receiver operating characteristic (ROC) curves and calculations of the area under the curve (AUC). The calibration of the models was also assessed visually by plotting the predicted risk against the observed risk of PCa. A perfectly calibrated model would have predicted risk and observed risk fall along a 45 degree line, indicating the risk obtained from the model reflects the actual observed risk of PCa on biopsy. Decision curve analysis was used to assess the clinical impact of the models.22 Due to the study sampling scheme, with intended over-sampling of African-American patients, all cohort-level statistics (e.g. AUC, calibration, decision curve analysis, and clinical consequences) have been weighted to represent the original PLCO cohort. Differences in discrimination were calculated and confidence intervals for the difference were estimated using clustered bootstrap re-sampling.
The analyses were then repeated separately for African-American men and for men of other races to assess differences in the performance of the model by race. As differences in prostate biopsy outcomes were observed between African-American men and men of other races, the models were refit to include race as a covariate. Constrained logistic regression was utilized to assess the magnitude of the adjustment to the risk prediction required for African-American men. Race and the risk prediction were entered into the regression model. Model risk was entered on the inverse-logit scale and the coefficient was constrained to be one.
Logistic regression was also utilized to assess whether MSP improves the performance of the models including the kallikrein makers. Ten-fold cross validation was utilized when assessing the performance of the model including MSP to account for the fact that models were built and evaluated on the same data set. All analyses were conducted using Stata 13.0 (Stata Corporation, College Station, Texas).
RESULTS
Characteristics of the study sample are shown in Table 1 and Supplementary Table 1; 24% (230/946) of patients had low-grade (Gleason 6, GGG 1) PCa and 10% (94/946) of patients had high-grade (Gleason ≥7, GGG ≥2) PCa. African-Americans constituted 11% (100/946) of the final cohort compared to 7% (113/1864) of the PLCO intervention arm.
Table 1.
Characteristics of selected patients with complete data. Statistics presented are median (IQR) or frequency (percent).
| Overall N=946 |
No Cancer (N=622; 66%) |
Low-Grade Cancer** (N=230; 24%) |
High-Grade Cancer*** (N=94; 10%) |
Other Races (N=846; 89%) |
Black, Non-Hispanic (N=100; 11%) |
|
|---|---|---|---|---|---|---|
| Age, years | 64 (60, 68) | 64 (60, 68) | 65 (61, 69) | 66 (61, 69) | 64 (60, 68) | 64 (60, 69) |
| Abnormal DRE | 141 (15%) | 67 (11%) | 45 (20%) | 29 (31%) | 123 (15%) | 18 (18%) |
| Race | ||||||
| White, non-Hispanic | 796 (84%) | 549 (88%) | 179 (78%) | 68 (72%) | 796 (94%) | 0 (0%) |
| Black, Non-Hispanic | 100 (11%) | 41 (6.6%) | 37 (16%) | 22 (23%) | 0 (0%) | 100 (100%) |
| Hispanic | 22 (2.3%) | 11 (1.8%) | 9 (3.9%) | 2 (2.1%) | 22 (2.6%) | 0 (0%) |
| Asian | 20 (2.1%) | 16 (2.6%) | 3 (1.3%) | 1 (1.1%) | 20 (2.4%) | 0 (0%) |
| Pacific Islander | 4 (0.4%) | 3 (0.5%) | 1 (0.4%) | 0 (0%) | 4 (0.5%) | 0 (0%) |
| American Indian | 2 (0.2%) | 1 (0.2%) | 1 (0.4%) | 0 (0%) | 2 (0.2%) | 0 (0%) |
| Unknown | 2 (0.2%) | 1 (0.2%) | 0 (0%) | 1 (1.1%) | 2 (0.2%) | 0 (0%) |
| Family History of PCa | 102 (11%) | 61 (10%) | 23 (10%) | 18 (19%) | 93 (11%) | 9 (9.0%) |
| Unknown | 8 (0.8%) | 3 (0.5%) | 3 (1.3%) | 2 (2.1%) | 7 (0.8%) | 1 (1.0%) |
| Total PSA, ng/mL | 5.8 (4.8, 7.6) | 5.6 (4.8, 7.0) | 6.0 (4.8, 8.2) | 8.0 (5.3, 17.4) | 5.8 (4.8, 7.5) | 5.9 (5.0, 8.5) |
| Free PSA, ng/mL | 1.1 (0.8, 1.5) | 1.2 (0.9, 1.6) | 1.0 (0.7, 1.4) | 1.1 (0.7, 1.8) | 1.1 (0.8, 1.6) | 1.0 (0.8, 1.4) |
| Intact PSA, ng/mL | 0.72 (0.54, 1.01) | 0.74 (0.57, 1.00) | 0.67 (0.49, 0.93) | 0.75 (0.50, 1.24) | 0.73 (0.55, 1.01) | 0.68 (0.50, 0.93) |
| hK2, ng/mL | 0.08 (0.05, 0.12) | 0.07 (0.05, 0.11) | 0.08 (0.05, 0.12) | 0.10 (0.06, 0.18) | 0.08 (0.05, 0.12) | 0.06 (0.04, 0.09) |
| MSP, ng/mL (N=938) | 21 (14, 30) | 23 (15, 32) | 18 (13, 27) | 16 (11, 25) | 22 (15, 31) | 15 (11, 22) |
| Risk of Cancer on Biopsy (%) | ||||||
| Age + PSA | 24 (22, 28) | 24 (22, 27) | 25 (22, 30) | 30 (23, 54) | 24 (22, 28) | 24 (22, 30) |
| Age + PSA + DRE | 19 (17, 26) | 19 (17, 22) | 21 (17, 39) | 31 (19, 51) | 19 (17, 25) | 20 (17, 35) |
| Age + Kallikrein Markers | 38 (30, 51) | 35 (26, 44) | 45 (37, 64) | 61 (44, 90) | 38 (30, 51) | 42 (32, 55) |
| Age + Kallikrein Markers + DRE | 35 (27, 50) | 31 (24, 41) | 42 (32, 64) | 62 (39, 89) | 34 (27, 50) | 38 (30, 55) |
| *PCPT risk calculator estimate | 30 (26, 35) | 29 (26, 33) | 32 (26, 39) | 38 (31, 49) | 29 (26, 33) | 40 (36, 48) |
| Risk of Gleason ≥7 Cancer on Biopsy (%) | ||||||
| Age + PSA | 8 (6, 10) | 7 (6, 9) | 8 (6, 12) | 10 (7, 24) | 8 (6, 10) | 8 (6, 12) |
| Age + PSA + DRE | 5 (4, 7) | 4 (3, 6) | 5 (4, 12) | 8 (4, 23) | 5 (3, 7) | 5 (4, 10) |
| Age + Kallikrein Markers | 7 (4, 16) | 6 (3, 10) | 12 (6, 28) | 25 (10, 59) | 7 (4, 16) | 10 (5, 21) |
| Age + Kallikrein Markers + DRE | 5 (3, 13) | 4 (2, 8) | 10 (4, 23) | 25 (7, 54) | 5 (3, 13) | 8 (3, 16) |
| PCPT risk calculator estimate* | 9 (7, 14) | 8 (7, 12) | 11 (7, 18) | 15 (10, 25) | 9 (7, 12) | 20 (17, 30) |
| Biopsy Gleason | ||||||
| No Cancer | 622 (66%) | 622 (100%) | 0 (0%) | 0 (0%) | 581 (69%) | 41 (41%) |
| Low-Grade Cancer** | 230 (24%) | 0 (0%) | 230 (100%) | 0 (0%) | 193 (23%) | 37 (37%) |
| Gleason Score 7 (GGG 2 or 3) | 72 (7.6%) | 0 (0%) | 0 (0%) | 72 (77%) | 54 (6.4%) | 18 (18%) |
| Gleason Score ≥8 (GGG 4 or 5) | 22 (2.3%) | 0 (0%) | 0 (0%) | 22 (23%) | 18 (2.1%) | 4 (4.0%) |
DRE = digital rectal examination, PCa = prostate cancer, PSA = prostate-specific antigen, hK2 = human kallikrein-2, MSP = microseminoprotein-beta, PCPT = Prostate Cancer Prevention Trial, GGG = Gleason Grade Group
PCPT risk calculator estimate = prespecified PCPT risk calculator version 2.0 model
Low-grade Cancer includes Gleason score 6 (Gleason Grade Group 1)
High-grade Cancer includes Gleason score ≥7 (Gleason Grade Group ≥2)
Discrimination of the prespecified statistical models are shown in Table 2. The 4K panel significantly improved the AUC for predicting high-grade PCa compared to the base model of PSA, age and DRE (increase in AUC of 0.080; 95% C.I. 0.059, 0.105; p<0.01) and compared to the PCPT risk calculator (increase of 0.060; 95% C.I. 0.032, 0.088; p<0.01). When stratifying the cohort by race, the 4K panel discrimination for high-grade PCa was higher for African-Americans (AUC 0.80) than for men of other races (AUC 0.78); however, the confidence intervals for the differences in AUC by race were not statistically significant (95% C.I. −0.10, 0.13).
Table 2.
Model discrimination for predicting high-grade cancer
| a) Overall findings | ||
|---|---|---|
| AUC | 95% CI | |
| Age + PSA | 0.691 | 0.641, 0.735 |
| Age + Four-Kallikrein Panel | 0.786 | 0.748, 0.816 |
| Age + PSA + DRE | 0.706 | 0.660, 0.746 |
| PCPT risk calculator* | 0.726 | 0.684, 0.771 |
| Age + Four-Kallikrein Panel + DRE | 0.786 | 0.748, 0.815 |
| Age + Four-Kallikrein Panel + MSP | 0.809 | 0.774, 0.838 |
| Age + Four-Kallikrein Panel + MSP + DRE | 0.810 | 0.775, 0.840 |
| b) Discrimination by race | ||||
|---|---|---|---|---|
| African- American |
Other Races |
Difference | 95% CI | |
| Age + PSA | 0.671 | 0.694 | −0.023 | −0.19, 0.14 |
| Age + Four-Kallikrein Panel | 0.803 | 0.781 | 0.022 | −0.10, 0.13 |
| Age + PSA + DRE | 0.691 | 0.710 | −0.019 | −0.18, 0.14 |
| Age + Four-Kallikrein Panel + DRE | 0.790 | 0.783 | 0.007 | −0.12, 0.12 |
PCPT risk calculator estimate = prespecified PCPT risk calculator version 2.0 model
AUC = area under the curve
CI = confidence interval
PSA = prostate-specific antigen
DRE = digital rectal examination
MSP = beta-microseminoprotein
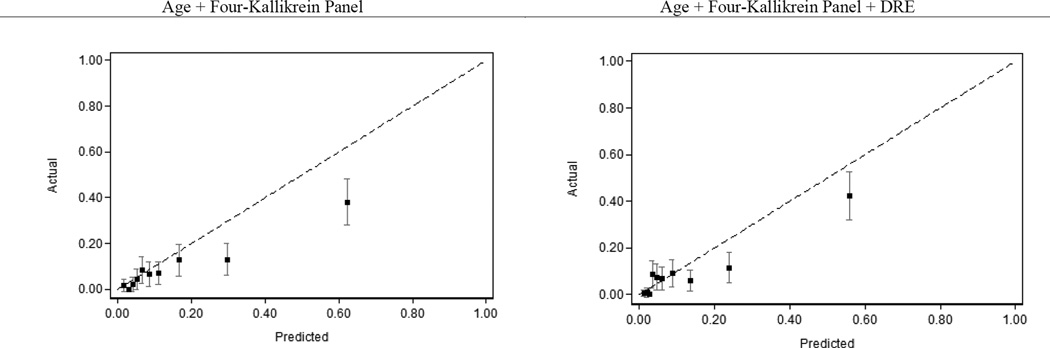
The calibration plot for the 4K model with and without DRE is provided in Figure 1. The calibration plot demonstrated good agreement for patients with estimated risk of high grade PCa of 20% of less.
Figure 1.
Calibration plot of the four-kallikrein model predicting high-grade prostate cancer.
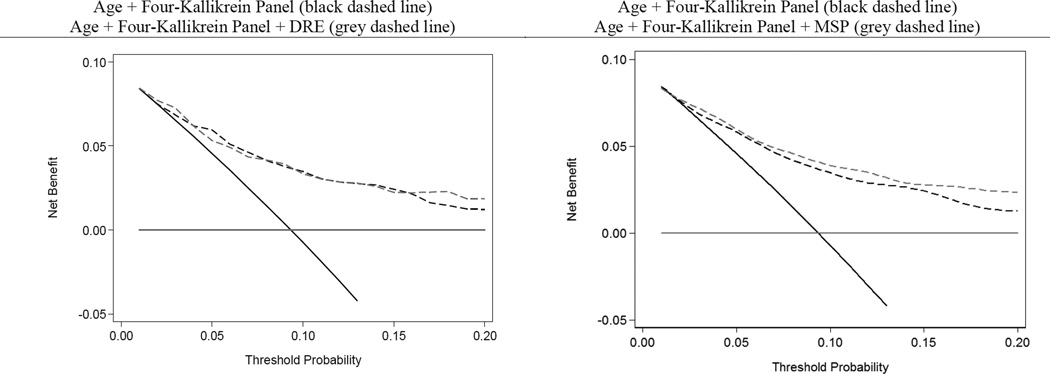
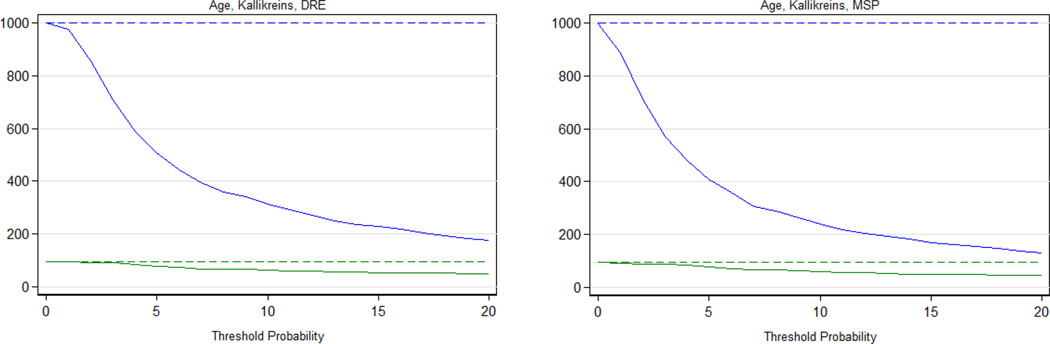
The decision curve analysis for the 4K model with and without MSP is provided in Figure 2. The decision curve analysis demonstrates improved clinical outcomes over biopsy all or no biopsy across the range of reasonable threshold probabilities for high-grade PCa. The clinical performance of using these models at various risk thresholds to perform biopsy as compared to PSA alone is provided in Table 3. At one illustrative threshold, 6% risk of high-grade disease, use of the panel in 1000 men would reduce the number of biopsies by 420 and the number of low-grade PCa diagnoses by 63. Of the 420 men avoiding biopsy, 11 (2.6%) would harbor high-grade disease. The clinical outcomes of using the 4K model to perform biopsy across a range of thresholds is illustrated separately in Figure 3.
Figure 2.
Decision curve analysis of the four-kallikrein model predicting high-grade prostate cancer.
Table 3.
Clinical implications of using models to determine biopsy.
| Biopsy Threshold (risk of high-grade* cancer) |
No. Biopsied |
Biopsies Avoided (%) |
Cancers Found |
Cancers Delayed |
High-grade* Cancers Found |
High-grade* Cancers Delayed |
|---|---|---|---|---|---|---|
| Biopsy All Men | 1000 | 0 (0%) | 330 | 0 | 93 | 0 |
| Age and PSA | ||||||
| 4% | 946 | 54 (5.4%) | 310 | 20 | 88 | 5 |
| 6% | 734 | 266 (27%) | 255 | 75 | 79 | 14 |
| 8% | 460 | 540 (54%) | 192 | 139 | 66 | 28 |
| 10% | 285 | 715 (72%) | 144 | 186 | 52 | 42 |
| 4K model | ||||||
| 4% | 749 | 251 (25%) | 302 | 28 | 89 | 4 |
| 6% | 580 | 420 (42%) | 256 | 74 | 83 | 11 |
| 8% | 466 | 534 (53%) | 227 | 104 | 75 | 18 |
| 10% | 380 | 620 (62%) | 197 | 133 | 69 | 24 |
PSA = prostate-specific antigen, 4K = four-kallikrein panel
High-grade cancer includes Gleason score ≥7 (Gleason Grade Group ≥2)
Figure 3.
Clinical consequences of using age, the four-kallikrein panel and DRE (left panel) and the four-kallikrein panel where DRE was replaced with MSP (right panel) to identify men for biopsy at varying biopsy thresholds. Results presented per 1000 men. The blue solid lines represent the number of men requiring prostate biopsy utilizing the respective models, dashed blue line marks the biopsy all men alternative. The solid green lines represent the number of high-grade cancers found, dashed green line marks the number of high-grade cancers found if all men are biopsied.
African-American race was significantly associated with high-grade PCa (OR 2.83; 95% C.I. 1.38, 5.81) after adjusting for the 4K model score. Inclusion of race with the 4K model improved discrimination for high-grade PCa (AUC 0.80 versus 0.79).
Adjusting for the 4K panel result, MSP was a significant predictor of high-grade PCa (OR 0.96, 95% C.I. 0.94, 0.98; p<0.01). The addition of MSP to the 4K panel provided improvement in risk discrimination (0.81, 95% C.I. 0.77, 0.84 versus 0.79, 95% C.I. 0.75, 0.82). The 4K panel with MSP model demonstrated an increased net benefit over the 4K panel alone at all threshold probabilities in the decision curve analysis (Figure 2).
DISCUSSION
In our study of the 4K panel in participants from the intervention arm of PLCO, we found that a prespecified 4K panel model improved predictive discrimination for high-grade PCa compared to other clinical risk determinants (age, PSA, DRE, and the PCPTRC). Calibration was generally good, and decision curve analysis demonstrated that the 4K panel would improve clinical outcomes as compared to biopsy in all men. As such, use of the 4K panel to determine biopsy indication would importantly reduce the number of biopsies performed, while missing few high-grade PCa.
As our study cohort was generated from the PLCO trial, the study results are constrained to the inclusion and exclusion criteria of the original trial. Specifically, the improved risk discrimination demonstrated with the use of the 4K panel model cannot necessarily be applied to men aged greater than 74 years, with PSA less than 4 ng/mL, or who have undergone a prior biopsy. Given the questionable benefit of PCa screening, diagnosis, or treatment in older men,23 the most clinically relevant limitation listed above is the inability to apply our study results to men with prior biopsy. In these men, particularly those with prior negative biopsies, adjuvant diagnostic tests (such as magnetic resonance imaging) may be necessary to improve risk stratification and reduce unnecessary subsequent biopsies.
The 4K panel provided a slightly greater improvement in risk discrimination for African-Americans (AUC 0.803) as compared to other races (AUC 0.781). In common with other studies, African-American race was found to be a significant predictor for high-grade PCa, independent of the 4K panel. Although we were unable to demonstrate that the 4K panel is significantly superior in African-Americans compared to other races, an analysis for which we had limited power, our finding suggests that improvements afforded by the 4K panel are highly unlikely to be worse in this racial group.
MSP was found to improve discrimination when added to the 4K panel. Decision analysis illustrates a small improvement in clinical outcome by using MSP with the 4K panel compared to the 4K panel alone. An extension of this concept has been demonstrated in the Stockholm 3 study; in a prospective fashion, Gronberg et al. used a combination of serum biomarkers (of which 5/6 are the same as the ones used in the current study but measured on a different instrument platform), single nucleotide polymorphisms, and traditional clinical characteristics to refine high grade PCa detection.24
The risk discrimination found in our study was similar to that of a prospective study of a commercialized 4K based model (“4Kscore”) applied to United States men accrued in routine urologic practice (AUC 0.82, 95%CI 0.79 to 0.85).25 The improvement in risk discrimination for high-grade PCa over commonly used clinical tools (e.g. PSA, PCPTRC) was also similar in both studies. There is also similarity in findings with respect to race, with both studies finding that the discrimination of the 4K panel was higher in African-American men compared to other races, although the limited number of African-Americans in both studies prevent a definitive conclusion. The key result in each case is that the 4K model significantly improves high-grade PCa risk discrimination in a multi-ethnic United States population, confirming the initial European results, despite differences in racial composition.
Given the current controversy with PCa screening and associated overdiagnosis and overtreatment,23 the need for clinical tools to provide improved risk stratification prior to prostate biopsy, particularly for high-grade PCa,26 is critical to improving PCa care in the United States. The results of this study confirm that the 4K panel can be utilized as such a tool to reduce the number of biopsies performed while minimizing the high-grade PCa that would be delayed or missed. Reduction in prostate biopsies performed, and thus reduction in the diagnosis of low-grade PCa, with minimal sacrifices in the detection of high-grade PCa, would likely provide a net positive impact on both total healthcare costs as well as patient morbidity.27,28 Future studies must consider the costs associated with the use of each additional biomarker (e.g. MSP), the comparative effectiveness of magnetic resonance imaging as a risk stratification tool over biomarkers,29 and the optimal cost-effective strategy for timing these tests prior to biopsy.
This study is not without limitations. As discussed above, the study results may not clearly be applied to men aged greater than 74 years, with PSA less than 4 ng/mL, or who have undergone prior biopsy. Although our study cohort was built to oversample African-American participants in PLCO and approached the most recent United States racial demographics (12.6% the U.S. population on the most recent census identified as African-American),30 the study was likely underpowered to detect a significant difference in the risk discrimination with the 4K panel in African-Americans over other races. The overall incidence of high-grade PCa in our study was lower than in some contemporary studies, and as a result the 4K panel overestimated high-grade PCa risk at higher clinical risk thresholds (greater than 20%, Figure 1). This is likely less clinically meaningful as those with substantially high clinical risk do not represent the majority of the screened population, and furthermore, these patients would be recommended for biopsy whether their risk of high-grade PCa was 40% based on PSA or 60% with the 4K panel model.
CONCLUSIONS
The use of the 4K panel in a selected population of PLCO participants confirms the results of prior studies, that the 4K panel can improve detection of high-grade PCa in a multi-ethnic United States cohort. Furthermore, the improvement in risk discrimination is similar among African-American men. Finally, the addition of a fifth serum biomarker, MSP, provides an incremental improvement in the ability to identify patients with high-grade PCa.
Supplementary Material
Acknowledgments
Source of Funding: R01-CA160816 from NCI to Dr. Lilja and Dr. Vickers, funds from David H. Koch provided through the Prostate Cancer Foundation, the Sidney Kimmel Center for Prostate and Urologic Cancers, P50-CA92629 SPORE grant from the National Cancer Institute to Dr. H Scher, the P30-CA008748 NIH/NCI Cancer Center Support Grant to MSKCC, Swedish Cancer Society project number 14-0722 to Dr. Lilja, the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre Program in UK, and contracts from the NCI to PLCO investigators.
Dr. Lilja holds patents for free PSA, hK2, and intact PSA assays, and is named, along with Dr. Vickers, on a patent application for a statistical method to detect prostate cancer, which is commercialized by OPKO Health. Drs. Vickers and Lilja receive royalties from any sales of the test. Dr. Lilja owns stock in OPKO and Dr. Vickers is on the advisory board of OPKO Health and has OPKO stock options. Dr. Vickers has a consulting or advisory role in Genome DX and Genomic Health. Dr. Lilja served on an advisory panel for Roche Diagnostics during 2014, and an immediate family member of Dr. Lilja is an employee at Ferring Pharmaceuticals. Dr. Andriole is a consultant for Augmenix and Tolmar Pharmaceuticals; clinical investigator for Blue Earth Diagnostics, Medivation, Progenics, and Traxxsson.
ABBREVIATIONS
- PSA
prostate-specific antigen
- PCa
prostate cancer
- DRE
digital rectal examination
- ERSPC
European Randomized Study of Prostate Cancer Screening
- PLCO
Prostate, Lung, Colorectal, Ovarian
- hK2
human kallikrein-related peptidase 2
- 4K
four-kallikrein
- MSP
microseminoprotein-beta
- WHO
World Health Organization
- GGG
Gleason Grade Group
- PCPTRC
Prostate Cancer Prevention Trial Risk Calculator
- ROC
receiver operating characteristic
- AUC
area under the curve
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
Role of the for-profit health care companies in the writing of the manuscript: None.
REFERENCES
- 1.Catalona WJ, Richie JP, Ahmann FR, et al. Comparison of digital rectal examination and serum prostate specific antigen in the early detection of prostate cancer: results of a multicenter clinical trial of 6,630 men. J Urol. 1994;151:1283–1290. doi: 10.1016/s0022-5347(17)35233-3. [DOI] [PubMed] [Google Scholar]
- 2.Schrӧder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. NEJM. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 3.Andriole GL, Crawford ED, Grubb RG, et al. Mortality results from a randomized prostate-cancer screening trial. NEJM. 2009;360:1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albertsen PC, Hanley JA, Fine J. 20-year outcomes following conservative management of clinically localized prostate cancer. JAMA. 2005;293:2095–2101. doi: 10.1001/jama.293.17.2095. [DOI] [PubMed] [Google Scholar]
- 5.Vickers AJ, Cronin AM, Aus G, et al. A panel of kallikreins markers can reduce unnecessary biopsy for prostate cancer: data from the European Randomized Study of Prostate Cancer Screening in Goteborg, Sweden. BMC Med. 2008;6:19. doi: 10.1186/1741-7015-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vickers A, Cronin A, Roobol M, et al. Reducing unnecessary biopsy during prostate cancer screening using a four-kallikrein panel: an independent replication. J Clin Oncol. 2010;28:2493–2498. doi: 10.1200/JCO.2009.24.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vickers AJ, Cronin AM, Aus G, et al. Impact of recent screening on predicting the outcome of prostate cancer biopsy in men with elevated PSA: data from the European Randomized Study of Prostate Cancer screening in Gothenburg, Sweden. Cancer. 2010;116:2612–2620. doi: 10.1002/cncr.25010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta A, Roobol MJ, Savage CJ, et al. A four-kallikrein panel for the prediction of repeat prostate biopsy: data from the European Randomized Study of Prostate Cancer screening in Rotterdam, Netherlands. Br J Cancer. 2010;103:708–714. doi: 10.1038/sj.bjc.6605815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant RJ, Sjoberg DD, Vickers AJ, et al. Predicting high-grade cancer at ten-core prostate biopsy using four kallikrein markers measured in blood in the ProtecT study. J Natl Cancer Inst. 2015;107(7):djv095. doi: 10.1093/jnci/djv095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlsson S, Maschino A, Schrӧder F, et al. Predictive value of four kallikreins markers for pathologically insignificant compared with aggressive prostate cancer in radical prostatectomy specimens: results from the European Randomized Study of Screening for Prostate Cancer section Rotterdam. Eur Urol. 2013;64:693–699. doi: 10.1016/j.eururo.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Center MM, Jemal A, Lortet-Tieulent J, et al. International variation in prostate cancer incidence and mortality rates. Eur Urol. 2012;61:1079–1092. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman RM, Gilliland FD, Eley JW, et al. Racial and ethnic differences in advanced-stage prostate cancer: the prostate cancer outcomes study. J Natl Cancer Inst. 2001;93:388–395. doi: 10.1093/jnci/93.5.388. [DOI] [PubMed] [Google Scholar]
- 13.Taksler GB, Keating NL, Cutler DM. Explaining racial differences in prostate cancer mortality. Cancer. 2012;118:4280–4289. doi: 10.1002/cncr.27379. [DOI] [PubMed] [Google Scholar]
- 14.Haiman CA, Stram DO, Vickers AJ, et al. Levels of beta-microseminoprotein in blood and risk of prostate cancer in multiple populations. J Natl Cancer Inst. 2013;105:237–243. doi: 10.1093/jnci/djs486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitrunen K, Pettersson K, Piironen T, et al. Dual-label one-step immunoassay for simultaneous measurement of free and total prostate-specific antigen concentrations and ratios in serum. Clin Chem. 1995;41:1115–1120. [PubMed] [Google Scholar]
- 16.Peltola MT, Niemela P, Alanen K, et al. Immunoassay for the discrimination of free prostate-specific antigen (fPSA) forms with internal cleavages at Lys(145) or Lys(146) from fPSA without internal cleavages at Lys(145) or Lys(146) J Immunol Methods. 2011;369:74–80. doi: 10.1016/j.jim.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vӓisӓnen V, Eriksson S, Ivaska KK, et al. Development of sensitive immunoassays for free and total human glandular kallikrein 2. Clin Chem. 2004;50:1607–1617. doi: 10.1373/clinchem.2004.035253. [DOI] [PubMed] [Google Scholar]
- 18.Valtonen-Andre C, Sӓvblom C, Fernlund P, et al. Beta-microseminoprotein in serum correlates with the levels in seminal plasma of young, healthy males. J Androl. 2008;29:330–337. doi: 10.2164/jandrol.107.003616. [DOI] [PubMed] [Google Scholar]
- 19.Vӓisӓnen V, Peltola MT, Lilja H, et al. Intact free prostate-specific antigen and free and total human glandular kallikrein 2. Elimination of assay interference by enzymatic digestion of antibodies to F(ab’)2 fragments. Anal Chem. 2006;15:7809–7815. doi: 10.1021/ac061201+. [DOI] [PubMed] [Google Scholar]
- 20.Epstein JI, Egevad L, Amin MB, et al. The 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading for prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am J Surg Pathol. 2016;40:244–252. doi: 10.1097/PAS.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 21.Ankerst DP, Hoefler J, Bock S, et al. Prostate cancer prevention trial risk calculator 2.0 for the prediction of low- vs high-grade prostate cancer. Urology. 2014;83:1362–1368. doi: 10.1016/j.urology.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim EH, Andriole GL. Prostate-specific antigen-based screening: controversy and guidelines. BMC Med. 2015;13:61. doi: 10.1186/s12916-015-0296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grӧnberg H, Adolfsson J, Aly M, et al. Prostate cancer screening in men aged 50–69 years (STHLM3): a prospective population-based diagnostic study. Lancet Oncol. 2015;16:1667–1676. doi: 10.1016/S1470-2045(15)00361-7. [DOI] [PubMed] [Google Scholar]
- 25.Parekh DJ, Punnen S, Sjoberg DD, et al. A multi-institutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer. Eur Urol. 2015;68:464–470. doi: 10.1016/j.eururo.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X, Albertsen PC, Andriole GL, et al. Risk-based prostate cancer screening. Eur Urol. 2012;61:652–661. doi: 10.1016/j.eururo.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heijnsdijk EAM, der Kinderen A, Wever EM, et al. Overdetection, overtreatment and costs in prostate-specific antigen screening for prostate cancer. Br J Cancer. 2009;101:1833–1838. doi: 10.1038/sj.bjc.6605422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heijnsdijk EAM, Wever EM, Auvinen A, et al. Quality-of-life effects of prostate-specific antigen screening. N Engl J Med. 2012;367:595–605. doi: 10.1056/NEJMoa1201637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salami SS, Vira MA, Turkbey B, et al. Multiparametric magnetic resonance imaging outperforms the prostate cancer prevention trial risk calculator in predicting clinically significant prostate cancer. Cancer. 2014;120:2876–2882. doi: 10.1002/cncr.28790. [DOI] [PubMed] [Google Scholar]
- 30.Humes K, Jones NA, Ramirez RR. Overview of race and Hispanic origin, 2010. US Department of Commerce, Economics and Statistics Administration, US Census Bureau; 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.