Abstract
Background
While eating disorders (EDs) are thought to result from a combination of environmental and psychological stressors superimposed on genetic vulnerability, the neurobiological basis of EDs remains incompletely understood. We recently reported that a rare missense mutation in the gene for the transcriptional repressor histone deacetylase 4 (HDAC4) is associated with the risk of developing an ED in humans.
Methods
To understand the biological consequences of this missense mutation, we created transgenic mice carrying this mutation by introducing the alanine to threonine mutation at position 778 of mouse Hdac4 (corresponding to position 786 of the human protein). Bioinformatic analysis to identify Hdac4-regulated genes was performed using available databases.
Results
Male mice heterozygous for HDAC4A778T did not show any metabolic or behavioral differences. In contrast, female mice heterozygous for HDAC4A778T display several ED-related feeding and behavioral deficits depending on housing condition. Individually housed HDAC4A778T female mice exhibit reduced effortful responding for high-fat diet and compulsive grooming, whereas group-housed female mice display increased weight gain on high-fat diet, reduced behavioral despair, and increased anxiety-like behaviors. Bioinformatic analysis identifies mitochondrial biogenesis including synthesis of glutamate/gamma-aminobutyric acid as a potential transcriptional target of HDAC4A778T activity relevant to the behavioral deficits identified in this new mouse model of disordered eating.
Conclusions
The HDAC4A778T mouse line is a novel model of ED-related behaviors and identifies mitochondrial biogenesis as a potential molecular pathway contributing to behavioral deficits.
Keywords: Anorexia nervosa, Behavior, Eating disorders, Estrogen-related receptor alpha, Feeding, Histone deacetylase 4
Eating disorders (EDs), including anorexia nervosa (AN) and bulimia nervosa, are a serious class of mental illness with a 1-year prevalence in young women of 0.37% and 1.0%, respectively (1). Because EDs disproportionately affect younger individuals and often have a relapsing and remitting course, they have among the highest rates of morbidity and mortality within psychiatric illnesses (1). EDs are highly heritable, indicating that genetic variants likely contribute to the risk of developing an ED. For example, one large study utilizing the Swedish Twin Registry estimates a heritability of 0.56 for narrowly defined AN (2). However, to date, the genetic basis of ED risk is not well understood, and two recent genome-wide association studies failed to identify a variant with genome-wide significance (3,4). Additionally, an association study of 182 candidate genes also failed to identify a significant genetic variation in a sample of patients with AN (5).
As an alternative approach to the problem, we utilized familial segregation and whole exome sequencing to identify missense mutations in two large families with multiple members affected with EDs and identified missense mutations in the estrogen-related receptor alpha (ESRRAR188Q) and histone deacetylase 4 (HDAC4A786T) proteins (6). Importantly, knockout mice lacking ESRRA display behavioral deficits relevant to the study of EDs, including reduced body weight, decreased motivation to work for high-fat diet, behavioral compulsivity, and social subordination (7). Because the HDACA786T mutation increases transcriptional repression in in vitro transcriptional assays (6), conventional and conditional knockout approaches are not well suited to study the role of HDAC4 on the risk of developing an ED. Therefore, we utilized clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 nuclease (Cas9) technology to introduce the alanine to threonine mutation into the corresponding position of mouse HDAC4 protein (HDAC4A778T) to closely mimic the human genetic risk of developing an ED.
In the current study, we characterize mice heterozygous for HDAC4A778T to better understand the potential contribution of this variant to the human illness. We selected a battery of behavioral tests based on the Research Domain Criteria system to facilitate clinical translation (8) of our findings including tasks in potential threat (light/dark box), willingness to work for reward (operant responding for high-fat diet), reward learning and/or habit formation (compulsive grooming), social dominance (resident-intruder paradigm), attachment affiliation (social interaction), and executive function (Barnes maze). We then performed a bioinformatic analysis of HDAC4 and ESRRA target genes to find potential biochemical pathways that may contribute to the observed behavioral deficits.
Methods and Materials
Animal Usage
All animal procedures were performed in accordance with University of Iowa Institutional Animal Care and Use Committee guidelines. Mice were handled in accordance with the Guide for the Care and Use of Laboratory Animals as adopted by the U.S. National Institutes of Health. Specific protocols were approved by the Institutional Animal Care and Use Committee. Mice were housed in the University of Iowa vivarium in a temperature-controlled environment (lights on: 6:00 AM to 6:00 PM) with ad libitum access to water and regular chow (7913 NIH-31 modified open formula mouse sterilized diet, Harlan-Teklad, Madison, WI) or high-fat diet (HFD) (42.8% calories from fat, TD.88137; Harlan-Teklad) as noted. Tissue samples from ESRRA-null mice were collected in a previous study (7).
Generation of HDAC4A778T Mice
The single nucleotide polymorphism rs61754648 corresponds to chr2:240011722-240011722 of hg19, which in turn corresponds to chr1:91961470-91961470 of mm10. The CRISPR Design Tool algorithm (Zhang Lab, MIT, Cambridge, MA; http://CRISPR.mit.edu/) was used to identify potential CRISPR guide RNAs targeting Hdac4 to make a single nucleotide change of G to A, corresponding to amino acid 778 (alanine, A778T). Two guide RNAs were tested in vitro for Cas9 cleavage activity by lipofectamine transfection in mouse 3T3 cells. After transfection with guide RNA/Cas9 DNA, genomic extracts were prepared and polymerase chain reaction (PCR) products spanning the Cas9 cleavage site were analyzed using a T7 endonuclease I assay (T7E1; New England BioLabs, Ipswich, MA). Digestion by T7E1 indicates cleavage activity by Cas9. The most active guide RNA was chosen for in vivo injections. This guide RNA targets Cas9 cleavage to six nucleotides downstream from the desired mutation. T7 promoter was added to Cas9 coding region and guide RNA by PCR. T7 PCR products were gel purified and used for in vitro transcription (mMessage mMachine Ultra kit, Megashortscript kit; Life Technologies, Thermo Fisher Scientific, Waltham, MA). Messenger RNA (mRNA) was purified using MEGAclear columns (Life Technologies). Ultramer single-stranded oligo donor (Integrated DNA Technologies, Coralville, IA) was used as the donor DNA to introduce the mutation.
To produce founder animals, embryos from B6/CBA F1 hybrid mice (#100011; Jackson Laboratory, Bar Harbor, ME) were microinjected with 50 ng/μL Cas9 wild-type RNA, 25 ng/μl HDAC4 guide RNA-1 RNA, 20 ng/μL sense single-stranded oligo donor via pronuclear delivery. Injected embryos were implanted into recipient ICR pseudopregnant female mice. Mosaic founders were genotyped by pyrosequencing (Qiagen, Hilden, Germany). Mosaic founders were then bred to female C57BL/6 mice (#000664; Jackson Laboratory), and resulting pups were genotyped by Sanger sequencing (TCTACAGATCCATCACAGAATGTGAACA, GGTTACTGGTGGGTACAACATGATATTTC) to identify whole-body heterozygous HDAC4A778T mice. Male mice from these litters were subsequently bred to female C57BL/6 mice to generate heterozygous HDAC4A778T and wild-type littermate mice for study (resulting in experimental mice that were 87.5% C57BL/6 background).
Body Weight Homeostasis
Mice were weaned and genotyped by tail snip and Sanger sequencing at 3 weeks of age. At 6 weeks of age, mice were either individually housed with weekly monitoring of body weight and food intake (chow or high-fat diet), or group housed with chow or HFD and weekly monitoring of body weight only. At the end of the study, body composition was determined by the University of Iowa Metabolic Phenotyping Core using a Bruker Minispec LF50 (Bruker, Billerica, MA).
Behavioral Studies
Twelve- to 16-week-old animals were used for behavioral studies.
Operant Responding
Mice were trained to press a lever to obtain a 20-mg HFD pellet reward as previously reported (9) in standard operant conditioning chambers (model ENV307A, Med Associates, St Albans, VT). Mice were rewarded for lever presses in the middle portal only; the side portals were monitored but inactive. HFD pellets were custom prepared by Bio-serv (#F06245; Frenchtown, NJ), and provided 4.5 kcal/g of metabolizable energy of which 45.4% of energy comes from fat, 35.0% comes from carbohydrate, and 21.0% comes from protein. The main components of these pellets were casein (233 g/kg), palm oil (207 g/kg), dextrates (197 g/kg), sucrose (197 g/kg), cellulose (58 g/kg), and soybean oil (20 g/kg). During the training period, mice were kept on a restricted feeding schedule and allowed access to regular chow 4 hours per day (12:00 PM to 4:00 PM). For the training sessions, mice initially received the HFD pellet rewards under a fixed ratio (FR) schedule. In order to pass training, mice had to obtain 30 reinforcements within 1 hour for FR1 (once), FR3 (twice), and FR5 (three times) before moving on to the progressive ratio schedule. Following completion of the training period, mice were then kept on the restricted feeding schedule and advanced to a progressive ratio schedule in which they had to perform increasing numbers of lever presses to obtain the pellet according to the following series: 5, 10, 20, 30, 50, 70, 100, 130, etc. A relatively steep progressive ratio was chosen to ensure that only differences in motivation were measured and not satiation. After three days of stable responding, mice were allowed free access to chow and effortful responding was assessed for HFD pellets under ad lib conditions for 2 additional days. Total number of lever presses and pellets earned within a 2-hour session were recorded and used for statistical analysis.
Grooming and Home Cage Activity
Home cage grooming was measured using the laboratory animal behavior observation registration and analysis system (LABORAS) as previously reported (10). Briefly, LABORAS, A Comprehensive Grooming Assay: LABORAS (Metris, Hoofddorp, the Netherlands), is a system that utilizes a carbon fiber plate to detect behavior-specific vibration patterns created by animals. Various behavioral parameters are determined by LABORAS software processing of the vibration pattern. Data was collected uninterrupted over a 24-hour time period, enabling comprehensive quantification of basal grooming time, bouts, and locomotor activity in the home-cage environment throughout the light/dark cycle. Before data collection, test animals were acclimated in the test room for 1 week. Then, test animals were placed in a standard cage atop the carbon fiber platforms. Vibrations were recorded for 24 hours, and then the animals were removed. Vibration data was processed via LABORAS software.
Forced Swim Test
Mice were videotaped in a 4-L Pyrex glass beaker (Corning, Corning, NY) containing 3 L of water at 24 ± 1°C for 6 minutes. Data were analyzed using automated ANY-maze Behavioral Tracking software (Stoelting, Wood Dale, IL), using the immobility detection feature set at sensitivity 66% and minimum immobility time 500 ms.
Light/Dark Box
Anxiety-like behavior testing was videotaped under red light using the ANY-maze Light/Dark Box (Stoelting). Time on light side, number of entries to light side, and distance traveled were calculated using the Behavioral Tracking software.
Social Dominance
Mice were trained six times over 2 days to run through a clear 1.5-inch plastic tube into a dark box at the other end. In contrast to the original description of the task (11), mice were not food restricted or provided a food reward as motivation for running through the tube. After training, mice were pair matched as knock-in/wild-type. The two mice were placed on opposite ends of the tube, and when both mice reached the center of the tube, a barrier was removed. The victor was scored as the mouse that did not retreat out of the tube. If neither mouse retreated from the tube after 2 minutes, the match was considered a draw.
Three-Chamber Social Interaction
Tests consisted of three sequential stages (12). In the first stage, test mice were placed in the center of a three-chambered box divided by acrylic glass walls and doors for a period of 5 minutes to acclimate to the apparatus. In the second stage, a novel mouse was placed inside a wire cage on one side of the apparatus. The Plexiglas door was removed to allow the test mouse to freely explore all three chambers for 10 minutes. At the start of the third stage, the test mouse was placed back in the center, and a new novel mouse was placed in a wire cage on the opposite side of the apparatus as the now-familiar mouse. The test mouse was again allowed to explore freely for a period of 10 minutes. All behavior was recorded and score by ANY-maze software.
Barnes Maze
Barnes maze was performed as recently described (13). Briefly, mice were trained to locate an escape hole using four trials per day for 4 days. On the fifth day, the escape hole was removed, and a probe trial assessed memory formation by measuring time spent around the previous location of the escape hole by video tracking (ANY-maze, Stoelting). At the beginning of the following week, the location of the escape hole was moved to an opposite quadrant, and the mice again received four trials per day for 4 days. The next day, time spent at the location of the previous escape hole (old target) and the new escape hole (new target) was measured by video tracking.
Buried Food Test
The buried food test was performed as previously reported (14).
Bioinformatics
A list of all genomic sites identified by chromatin immunoprecipitation with an anti-ESRRA antibody (#sc-66882; Santa Cruz Biotechnology, Santa Cruz, CA) was downloaded from the ENCODE Project (University of California-Santa Cruz, Santa Cruz, CA; http://genome.ucsc.edu/ENCODE/search.html). To determine which genes in the human genome were likely regulated by ESRRA, the coordinates for the binding sites were extended by 1000 base pairs (bps) on either side. The University of California-Santa Cruz (UCSC) Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables?command=start) was used to find all genes that intersected one of these 1000-bp-padded ESRRA binding sites. This list of genes was further filtered by retaining only those genes in which the 1000-bp-padded ESRRA binding site intersected with a putative promoter region as defined in the ENCODE project's chromatin state segmentation analysis (15–17). Specifically, for a gene to be retained, the associated 1000-bp-padded ESRRA binding site had to intersect with a region of the genome classified as either an “active promoter,” “weak promoter,” or “inactive/poised promoter” across all nine cell lines used in the ENCODE project (GM12878, H1-hESC, K562, HepG2, HUVEC, HMEC, HSMM, NHEK, NHLF). ENCODE chromatin state segmentation data was downloaded from UCSC Table Browser for all nine of the ENCODE cell lines using the Regulation group and Broad ChromHMM track. This set of genes was then cross-referenced with the list of Hdac4-regulated genes in mouse cortical neurons reported previously (18).
Quantitative Reverse Transcriptase PCR
Tissue punches of cingulate and medial prefrontal cortex were rapidly collected from female mice, flash frozen on liquid nitrogen, and kept at −80° until processing. Purification of mRNA (#74134; Qiagen) and preparation of complementary DNA (#11755-050; Invitrogen) was performed as per the manufacturer's instructions. Quantitative PCR was performed using an Applied Biosystems model 7900HT system (Thermo Fisher Scientific) standardized to expression of 36B4 via the ΔΔCt method. Primers used are as follows:
36b4: CACTGGTCTAGGACCCGAGAAG, GGTGCCTCTGAAGATTTTCG
Esrra: AGCAAGCCCCGATGGA, GAGAGGCCTGGGATGCTCTT
Eno1: GCCGGCACCCTGAAGT, TGAGAACCACCGTTGAT-CACA
Eno3: CCCTGTGCCTGCCTTTAAT, CCTCATTGTTCT-CCAGGATGTT
Mdh2: CTCTACGATATCGCTCACACAC, GAGCTGGATCC-AAACCCTTTA
Slc1a5: GCTACCTCATATGAACCCAAAGA, CGTACCAC-ATAATCCAGGAGAC
Ogdh: TCTGGACTCCTCCGTGCC, GGTCAGACTCGT-GTAGGCCA
Got1: GTGAGGAAGGTCGAACAGAAG, CCCTAAGAAG-TCAGCTCCAATC
Western Blot
Twelve- to 16-week-old experimentation naïve wild-type and ESRRA-null female mice were killed by cervical dislocation and the cortex, the thalamus, and the hippocampus were rapidly dissected out, immediately frozen in liquid nitrogen, and stored at −80°C until use. Frozen tissue was homogenized with ice-cold radioimmunoprecipitation assay lysis buffer (#89901; Thermo Fisher Scientific) containing cocktails of protease inhibitor (#04906845001, Roche Diagnostics, Basel, Switzerland). Protein concentration was determined by bicin-choninic acid assay method (#23228; Thermo Fisher Scientific), and equal amounts of each sample (20 μg) were separated by 4% to 20% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gradient gels (BioRad, Hercules, CA) and transferred to a polyvinylidene fluoride membrane by electroblotting. Primary antibodies were diluted as follows: malate dehydrogenase 2 (1/1000; #HPA019714; Sigma-Aldrich, St. Louis, MO), oxoglutarate dehydrogenase (1/1000; #PA5-28195, Thermo Fisher Scientific), enolase 3 (1/1000; #AV48204; Sigma-Aldrich), enolase 1 (1/1000; #AV34376, Sigma-Aldrich), valosin-containing protein (1/1000; #2648, Cell Signaling Technology, Danvers, MS). Membranes were then incubated with horseradish peroxidase–conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA), and signals were detected by chemiluminescence. Signal intensity was measured and analyzed by BioSpectrum 810 Imaging System (Upland, CA). For each experiment, signal intensity of targeted proteins was normalized to loading control valosin-containing protein signal intensity and then compared between the groups.
Data Analysis and Figure Preparation
Two-way analysis of variance and Student's t test were performed, and graphs were generated in GraphPad Prism (version 6.0 for Mac; San Diego, CA). No image manipulation was applied to any figure panel other than size changes and color. A value of p < .05 was considered statistically significant.
Results
Body Weight Homeostasis
Male HDAC4A778T mice were bred to heterozygous HDAC4A778T or wild-type littermate mice for study. At 6 weeks of age, mice were individually housed and provided access to either regular chow or 42.8% HFD. No difference in body weight was noted in female or male mice on either regular chow or HFD (Figure 1A–B). It was noted, however, that female HDAC4A778T mice weighed more than their wild-type cagemates did when group housed. Therefore, we repeated body weight analysis in group-housed mice and observed that female, but not male, HDAC4A778T mice gained significantly more weight on HFD when group housed (Figure 1C–D). At present, we are unable to measure food intake of individual mice under group-housing conditions. However, when group-housed mice were individually housed temporarily, a significant increase in consumption of HFD overnight was observed (Figure 1D), suggesting that hyperphagia may contribute to the increased weight gain. No differences in body composition were noted in female or male group-housed HDAC4A778T mice (Figure 1E).
Figure 1.
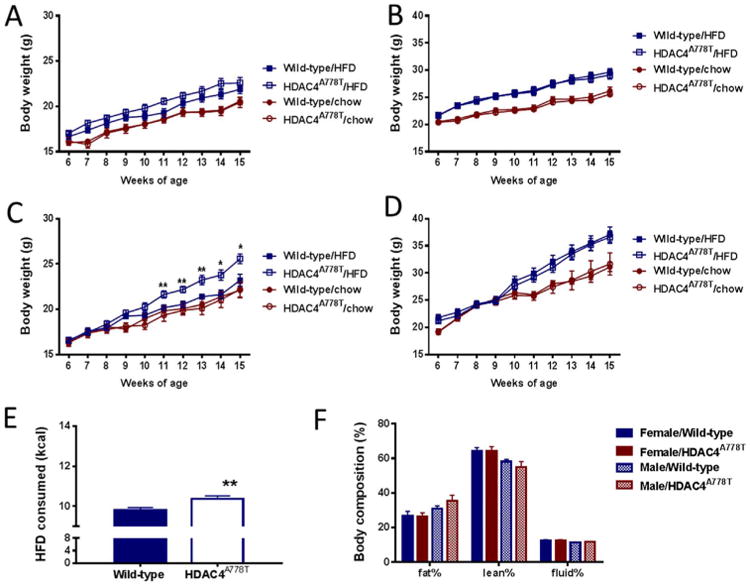
Body weight homeostasis in HDAC4A778T mice. Six-week old HDAC4A778T mice were given access to regular chow or high-fat diet (HFD) and body weight was monitored until 15 weeks of age for individually housed (A) female (n = 10–14 per group) and (B) male mice (n = 8–12 per group) and group-housed (C) female (n = 8–9 per group, significant diet × genotype interaction by two-way analysis of variance F9,160 = 2.576, p = .0085) and (D) male mice (n = 7–11 per group). (E) Food intake group-housed female mice receiving HFD (n = 8–9 per group, p = .008 by t test). (F) Body composition of group-housed mice. Data presented as mean ± SEM. *p < .05, **p < .01 indicate significant differences.
Behavioral Analysis
EDs are frequently comorbid with other psychiatric conditions including mood, anxiety, and obsessive-compulsive disorders (19). Therefore, we performed a behavioral analysis of female and male mice to determine the consequences of the HDAC4A778T mutation on behaviors relevant to eating disorders. Because we noted an effect of housing on body weight homeostasis in HDAC4A778T mice, we performed our analyses on both groups of mice. In the forced swim test, group-housed female HDAC4A778T mice, compared with individually housed female HDAC4A778T mice, showed reduced immobility (Figure 2A). No difference in immobility time was noted in wild-type female mice, male HDAC4A778T mice, or their male wild-type littermates (Figure 2A–B). Anxiety-like behavior was measured using the light/dark box and further revealed that group-housed female HDAC4A778T mice showed reduced time exploring the light side than individually housed female HDAC4A778T mice did, and no difference was noted in any other group (Figure 2C–D).
Figure 2.
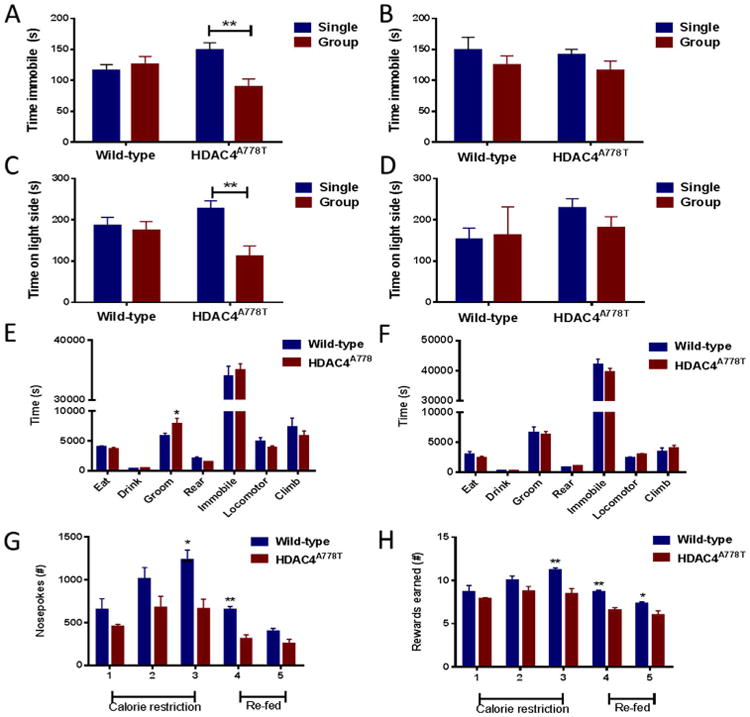
Behavioral analysis of HDAC4A778T mice. Twelve 16-week-old single- and group-housed (A) female (n = 10–16 per group), significant housing × genotype interaction by two-way analysis of variance (F1,49 = 8.187, p = .0062) and (B) male mice (n = 7–13 per group) were tested in the forced swim test. Twelve 16- week-old single- and group-housed (C) female (n = 10–14 per group), significant housing × genotype interaction by two-way analysis of variance (F1,45 = 5.394, p = .0248) and (D) male mice (n = 7–13 per group) were tested in the light/dark box. Sixteen-week-old individually housed (E) female (n = 6 per group) and (F) male (n = 6 per group) mice were monitored for home-cage activity by vibration plate. There was a significant difference in time grooming (p = .0105 by t test). Individually housed female mice were assessed by operant responding for total (G) nose pokes (n = 6–7 per group, p = .0044 at day 3 and p = .0012 at day 4 by t test) and (H) HFD pellets earned (n = 6–7 per group, p = .0042 at day 3, p = .0017 at day 4, p = .0377 at day 5 by t test). Data presented as mean ± SEM. *p < .05, **p < .01 indicate significant differences between the groups.
Quantitative assessment of several home-cage behaviors can be measured using vibration plate monitoring. Because this test requires individual housing, it was not performed in the group setting. Although no differences were noted in most behavioral measures, including activity and time spent eating, drinking, rearing, and climbing, female HDAC4A778T mice did display increased time spent grooming (Figure 2E). No differences were noted in male mice (Figure 2F).
Because patients with AN frequently exhibit reduced preference for foods high in fat content (20), we sought to assess the willingness to work for HFD. Here we used an operant-responding task under a progressive ratio to measure the number of nose pokes a mouse would perform to acquire an HFD pellet. This test was also performed only on individually housed mice, as it requires individual housing for both operant responding and timed feeding. Female HDAC4A778T mice manifested significantly fewer nose pokes and earned fewer rewards than their wild-type littermates did (Figure 2G–H). Buried food test revealed no significant impairment of olfaction in HDAC4A778T mice (Supplemental Figure S3A–B). Additional tasks in which no significant differences were noted include three-chamber social interaction, Barnes maze probe trial, and social dominance (data not shown and Supplemental Figure S1). Behavioral differences are summarized in Table 1.
Table 1. Summary of Metabolic and Behavioral Phenotypes.
| Female Group | Female Single | Male Group | Male Single | |
|---|---|---|---|---|
| Body Weight-Chow | – | – | – | – |
| Body Weight–High Fat | ↑ | – | – | – |
| Forced Swim Test (Immobility) | ↓ | – | – | – |
| Elevated Plus Maze (Open Arm) | ↓ | – | – | – |
| Operant Responding | ND | ↓ | ND | ND |
| Social Interaction | – | – | ND | ND |
| Home Cage | ND | ↑ Grooming | ND | – |
| Barnes Maze Probe | – | – | ND | ND |
ND, not determined.
Bioinformatic Analysis
We next sought to determine the transcriptional targets of HDAC4 in neurons that might mediate behavioral deficits observed in HDAC4A778T mice. Because we previously hypothesized that greater HDAC4-mediated repression of ESRRA activity increases the risk of developing an ED in humans (6), and because ESRRA-null (7) and HDAC4A778T mice share several behavioral deficits relevant to the development of EDs, we decided to focus on the subset of genes regulated by both ESRRA and HDAC4 in cortical neurons.
We first interrogated the < 1800 ESRRA binding sites within the genome by downloading all ESRRA binding sites identified by chromatin immunoprecipitation-high throughput sequencing reported by the ENCODE project. Next, to identify genes that were potential transcriptional targets of ESRRA, all genes±1000 bps of these binding sites were selected. The list was further refined to eliminate all sites that did not intersect with a putative promoter site as defined by the ENCODE project's chromatin state segmentation analysis. This list of 572 genes was then cross-referenced with a list of genes repressed by HDAC4 in cortical neurons, as previously reported (18), to generate a list of genes regulated by HDAC4 in neurons with an ESRRA binding site in an active promoter (Table 2). Analysis of this set of seven genes revealed that six of the seven are involved in mitochondrial metabolism (including the production of alpha-ketoglutarate) and synthesis of vesicular glutamate (Figure 3A–B).
Table 2. Common Transcriptional Targets of HDAC4 and ESRRA.
| Chromosome | Start | Stop | Gene Name | Protein |
|---|---|---|---|---|
| chr1 | 8931005 | 8939151 | ENO1 | Enolase 1 |
| chr17 | 4854383 | 4860426 | ENO3 | Enolase 3 |
| chr10 | 101166095 | 101190530 | GOT1 | Glutamic-oxaloacetic transaminase 1 |
| chr7 | 75677392 | 75695930 | MDH2 | Malate dehydrogenase 2 |
| chr7 | 44646120 | 44748669 | OGDH | Oxoglutarate dehydrogenase |
| chr19 | 47278139 | 47291842 | SLC1A5 | Neutral amino acid transporter |
| chr20 | 47729875 | 47805288 | STAU1 | Staufen homolog 1 |
Figure 3.
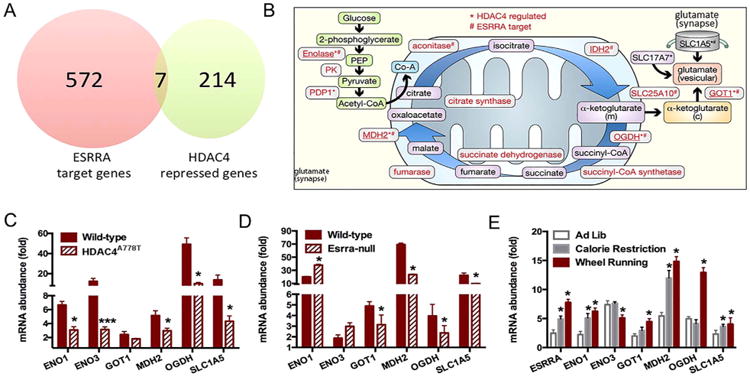
Identification of a HDAC4-ESRRA target pathway. (A) Venn diagram of ESRRA target genes and HDAC4 repressed genes. (B) Schematic representation of synaptic glutamate synthesis pathway. (C) Measure of messenger RNA (mRNA) abundance by quantitative polymerase chain reaction in cortex of female group-housed HDAC4A778T mice (n = 6 per group), (D) female ESRRA-null mice (n = 6 per group), and (E) wild-type female mice subjected to 60% calorie restriction or wheel running (n = 6 per group). Data presented as mean ± SEM. *p < .05 and ***p < .001 indicate significant differences between the groups by t test.
To determine whether expression of these genes is affected in HDAC4A778T mice, we measured relative abundance of their mRNA from the cortex of both group-housed and single-housed female HDAC4A778T mice using quantitative PCR. Compared with wild-type cagemates, expression of several genes, including Eno1, Eno3, Mdh2, Ogdh, and Slc1a5, were significantly reduced in group-housed mice (Figure 3C), whereas no differences were seen in the expression of any of these genes in single-housed mice (Supplementary Figure S3A). Additionally, we measured the relative abundance of the mRNA for this set of genes in the cortex of female ESRRA-null mice and wild-type littermates (Figure 3D). Expression of four of the six genes, including Got1, Mdh2, Ogdh, and Slc1a5, were also significantly decreased in ESRRA-null mice. These quantitative PCR results were confirmed by analyzing protein levels by Western blot using the four validated antibodies available (anti-MDH2, anti-OGDH, anti-ENO3, and anti-ENO1) (Supplemental Figure S2B–C).
After establishing that levels of these genes were altered in HDAC4A778T and ESRRA-null mice, we next determined the effect of calorie restriction and physical activity on their expression, as negative energy states have been shown to trigger episodes of AN (21). Esrra levels were increased in female mice exposed to either 60% of baseline calories for 10 days, as noted above, or to voluntary wheel running for 10 days (Figure 3E). In addition, five of the six target genes tested also displayed increased mRNA abundance in response to one or both conditions.
Discussion
Despite abundant evidence supporting the biological basis of EDs, the underlying neural substrate remains poorly understood, in part because of the lack of validated rodent models available for rigorous testing (8). In the present study, we have characterized the metabolic and behavioral phenotypes of a new mouse line carrying a single nucleotide polymorphism in the Hdac4 gene associated with the risk of developing an ED to better understand the potential contribution of this variant to the metabolic and behavioral phenotypes relevant to human illness. We find that HDAC4A778T mice display several phenotypes with face validity (feeding and behavioral abnormalities), construct validity (reduced preference for high-fat food), and predictive validity (preferentially affects female mice). Furthermore, several behavioral abnormalities reported here are similar to those previously noted in ESRRA-null mice (7), including decreased operant responding for HFD, compulsive grooming, and decreased immobility in the forced swim test. These findings support our overall hypothesis that dysregulation of the HDAC4-ESRRA pathway predisposes to development of EDs in patients (6).
One interesting observation here is that both ESRRA-null and group-housed HDAC4A778T female mice have reduced immobility in the forced swim test, a phenotype traditionally associated with an antidepressant-like effect. This observation would stand in contrast to the well-known association of EDs with depressed mood (19). It should be noted, however, that the forced swim test was originally designed as a rodent test for screening potential antidepressant compounds and not as a measure of endogenous mood per se (22). Acutely, administration of antidepressant compounds is often associated with a “jitteriness/anxiety syndrome” characterized by agitation, irritability, and akathisia (23). Indeed, irritability is frequently reported in patients with EDs (24) and is consistent with our observations of these mice.
One surprising issue that arose from this work is the effect of housing on behavior. We observed greater HFD-induced weight gain, anxiety-like behavior, and reduced immobility in the forced swim test in group-housed mice. Likewise, the changes in the expression of HDAC4 target genes Eno1, Eno3, Mdh2, Ogdh, and Slc1a5 were only observed in the group-housed mice suggesting that a genotype × environment interaction is required for transcriptional alterations. Little is known about the effects of housing status on body weight homeostasis in rodents, in which individual housing is required for the gold-standard method of assessing energy metabolism (25). Further studies utilizing more sophisticated measures of social functioning in female mice may be warranted to better understand the consequences of the HDAC4A778T variant on the observed behavioral and metabolic phenotypes.
Finally, we utilized a bioinformatic approach to identify multiple mitochondrial and synaptic genes as a common target of HDAC4 and ESRRA transcriptional regulation. Consistent with our hypothesis that disruption of HDAC4-ESRRA activity could lead to neuronal dysfunction by affecting mitochondrial function, ESRRA is known to increase mitochondrial biogenesis in the brain (26). Notably, an important role for mitochondria in the support of synaptic function has recently emerged, including bioenergetics (27), calcium buffering and synaptic vesicle endocytosis (28), and neurotransmitter synthesis (29,30). Consistent with this possibility, female ESRRA-null mice were recently found to have alterations in glutamatergic synapse functioning including increased frequency and amplitude of miniature excitatory postsynaptic currents, decreased paired-pulse facilitation, and reduced number of presynaptic glutamatergic vesicles (31). Although the initial expression data are consistent with a potential role for disrupted mitochondrial metabolism contributing to the behavioral phenotypes of ESRRA-null mice, more work will need to be done to validate that mitochondrial biogenesis is a key mechanism by which the HDAC4A778T variant exerts its metabolic and behavioral effects. Additionally, future studies, such as RNA sequencing and chromatin immunoprecipitation sequencing, will be required to determine whether HDAC4-ESRRA targets additional pathways that might contribute to the observed metabolic and behavioral alterations.
Supplementary Material
Acknowledgments
This material is based on work supported by the Dylan Tauber Researcher Award from the Brain and Behavior Foundation (to ML), the Klarman Family Foundation Grants Program in Eating Disorder Research (to ML), Grant No. R21 MH109920-01 (to HC), American Heart Association Scientist Developmental Grant No. 14SDG20140054 (to HC), and a pilot grant from the Fraternal Order of Eagles Diabetes Research Center at the University of Iowa.
Footnotes
Disclosures: The authors report no biomedical financial interests or potential conflicts of interest.
Supplementary material cited in this article is available online at http://dx.doi.org/10.1016/j.biopsych.2016.09.024.
References
- 1.Smink FR, van Hoeken D, Hoek HW. Epidemiology of eating disorders: Incidence, prevalence and mortality rates. Curr Psychiatry Rep. 2012;14:406–414. doi: 10.1007/s11920-012-0282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bulik CM, Sullivan PF, Tozzi F, Furberg H, Lichtenstein P, Pedersen NL. Prevalence, heritability, and prospective risk factors for anorexia nervosa. Arch Gen Psychiatry. 2006;63:305–312. doi: 10.1001/archpsyc.63.3.305. [DOI] [PubMed] [Google Scholar]
- 3.Boraska V, Franklin CS, Floyd JA, Thornton LM, Huckins LM, Southam L, et al. A genome-wide association study of anorexia nervosa. Mol Psychiatry. 2014;19:1085–1094. doi: 10.1038/mp.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang K, Zhang H, Bloss CS, Duvvuri V, Kaye W, Schork NJ, et al. for the Price Foundation Collaborative Group. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Molecular Psychiatry. 2011;16:949–959. doi: 10.1038/mp.2010.107. [DOI] [PubMed] [Google Scholar]
- 5.Pinheiro AP, Bulik CM, Thornton LM, Sullivan PF, Root TL, Bloss CS, et al. Association study of 182 candidate genes in anorexia nervosa. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1070–1080. doi: 10.1002/ajmg.b.31082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui H, Moore J, Ashimi SS, Mason BL, Drawbridge JN, Han S, et al. Eating disorder predisposition is associated with ESRRA and HDAC4 mutations. J Clin Invest. 2013;123:4706–4713. doi: 10.1172/JCI71400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui H, Lu Y, Khan MZ, Anderson RM, McDaniel L, Wilson HE, et al. Behavioral disturbances in estrogen-related receptor alpha-null mice. Cell Rep. 2015;11:344–350. doi: 10.1016/j.celrep.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutter M, Croghan AE, Cui H. Escaping the golden cage: Animal models of eating disorders in the post-diagnostic and statistical manual era. Biol Psychiatry. 2016;79:17–24. doi: 10.1016/j.biopsych.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Perello M, Sakata I, Birnbaum S, Chuang JC, Osborne-Lawrence S, Rovinsky SA, et al. Ghrelin increases the rewarding value of high-fat diet in an orexin-dependent manner. Biol Psychiatry. 2010;67:880–886. doi: 10.1016/j.biopsych.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu P, Grueter BA, Britt JK, McDaniel L, Huntington PJ, Hodge R, et al. Double deletion of melanocortin 4 receptors and SAPAP3 corrects compulsive behavior and obesity in mice. Proc Natl Acad Sci U S A. 2013;110:10759–10764. doi: 10.1073/pnas.1308195110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindzey G, Winston H, Manosevitz M. Social dominance in inbred mouse strains. Nature. 1961;191:474–476. doi: 10.1038/191474a0. [DOI] [PubMed] [Google Scholar]
- 12.Kaidanovich-Beilin O, Lipina T, Vukobradovic I, Roder J, Woodgett JR. Assessment of social interaction behaviors. J Vis Exp. 2011;25 doi: 10.3791/2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin TC, Britt JK, De Jesus-Cortes H, Lu Y, Genova RM, Khan MZ, et al. P7C3 neuroprotective chemicals block axonal degeneration and preserve function after traumatic brain injury. Cell Rep. 2014;8:1731–1740. doi: 10.1016/j.celrep.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, Crawley JN. Simple behavioral assessment of mouse olfaction. Curr Protoc Neurosci. 2009;Chapter 8 doi: 10.1002/0471142301.ns0824s48. Unit 8.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sando R, 3rd, Gounko N, Pieraut S, Liao L, Yates J, 3rd, Maximov A. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell. 2012;151:821–834. doi: 10.1016/j.cell.2012.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Brien KM, Vincent NK. Psychiatric comorbidity in anorexia and bulimia nervosa: Nature, prevalence, and causal relationships. Clin Psychol Rev. 2003;23:57–74. doi: 10.1016/s0272-7358(02)00201-5. [DOI] [PubMed] [Google Scholar]
- 20.Steinglass J, Foerde K, Kostro K, Shohamy D, Walsh BT. Restrictive food intake as a choice—A paradigm for study. Int J Eat Disord. 2014;48:59–66. doi: 10.1002/eat.22345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbarich-Marsteller NC, Foltin RW, Walsh BT. Does anorexia nervosa resemble an addiction? Curr Drug Abuse Rev. 2011;4:197–200. doi: 10.2174/1874473711104030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castagne V, Moser P, Roux S, Porsolt RD. Rodent models of depression: Forced swim and tail suspension behavioral despair tests in rats and mice. Curr Protoc Pharmacol. 2010;Chapter 5 doi: 10.1002/0471141755.ph0508s49. Unit 5.8. [DOI] [PubMed] [Google Scholar]
- 23.Harada T, Inada K, Yamada K, Sakamoto K, Ishigooka J. A prospective naturalistic study of antidepressant-induced jitteriness/anxiety syndrome. Neuropsychiatr Dis Treat. 2014;10:2115–2121. doi: 10.2147/NDT.S70637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giovanni AD, Carla G, Enrica M, Federico A, Maria Z, Secondo F. Eating disorders and major depression: Role of anger and personality. Depress Res Treat. 2011;2011:194732. doi: 10.1155/2011/194732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tschop MH, Speakman JR, Arch JR, Auwerx J, Bruning JC, Chan L, et al. A guide to analysis of mouse energy metabolism. Nat Methods. 2012;9:57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strum JC, Shehee R, Virley D, Richardson J, Mattie M, Selley P, et al. Rosiglitazone induces mitochondrial biogenesis in mouse brain. J Alzheimers Dis. 2007;11:45–51. doi: 10.3233/jad-2007-11108. [DOI] [PubMed] [Google Scholar]
- 27.Faits MC, Zhang C, Soto F, Kerschensteiner D. Dendritic mitochondria reach stable positions during circuit development. Elife. 2016;5:e11583. doi: 10.7554/eLife.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marland JR, Hasel P, Bonnycastle K, Cousin MA. Mitochondrial calcium uptake modulates synaptic vesicle endocytosis in central nerve terminals. J Biol Chem. 2016;291:2080–2086. doi: 10.1074/jbc.M115.686956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamberger AC, Chiang GH, Nylen ES, Scheff SW, Cotman CW. Glutamate as a CNS transmitter: I. Evaluation of glucose and glutamine as precursors for the synthesis of preferentially released glutamate. Brain Res. 1979;168:513–530. doi: 10.1016/0006-8993(79)90306-8. [DOI] [PubMed] [Google Scholar]
- 30.Waagepetersen HS, Sonnewald U, Larsson OM, Schousboe A. Synthesis of vesicular GABA from glutamine involves TCA cycle metabolism in neocortical neurons. J Neurosci Res. 1999;57:342–349. [PubMed] [Google Scholar]
- 31.De Jesus-Cortes H, Lu Y, Anderson RM, Khan MZ, Nath V, McDaniel L, et al. Loss of estrogen-related receptor alpha disrupts ventral-striatal synaptic function in female mice. Neuroscience. 2016;329:66–73. doi: 10.1016/j.neuroscience.2016.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


