Abstract
Point-of-care tools are needed in sub-Saharan Africa (SSA) to improve pediatric Burkitt lymphoma (BL) diagnosis and treatment. We evaluated plasma Epstein-Barr virus (pEBV) DNA as a pediatric BL biomarker in Malawi. Prospectively enrolled children with BL were compared to classical Hodgkin lymphoma (cHL) and non-lymphoma diagnoses. Pediatric BL patients received standardized chemotherapy and supportive care. pEBV DNA was measured at baseline, mid-treatment, and treatment completion. Of 121 assessed children, pEBV DNA was detected in 76/88 (86%) with BL, 16/17 (94%) with cHL, and 2/16 (12%) with non-lymphoma, with proportions higher in BL versus non-lymphoma (p<0.001) and similar in BL versus cHL (p=0.69). If detected, median pEBV DNA was 6.1 log10copies/mL for BL, 4.8 log10copies/mL for cHL, and 3.4 log10copies/mL for non-lymphoma, with higher levels in BL versus cHL (p=0.029), and a trend toward higher levels in BL versus non-lymphoma (p=0.062). pEBV DNA declined during treatment in the cohort overall and increased in several children before clinical relapse. Twelve-month overall survival was 40% in the cohort overall, and for children with baseline pEBV detected, survival was worse if baseline pEBV DNA was ≥6 log10copies/mL versus <6 log10copies/mL (p=0.0002), and also if pEBV DNA was persistently detectable at mid-treatment versus undetectable (p=0.041). Among children with baseline pEBV DNA detected, viremia was the only significant risk factor for death by 12 months in multivariate analyses (adjusted hazard ratio 1.35 per log10copies/mL, 95% CI 1.04–1.75, p=0.023). Quantitative pEBV DNA has potential utility for diagnosis, prognosis, and response assessment for pediatric BL in SSA.
Keywords: Burkitt lymphoma, Epstein-Barr virus, sub-Saharan Africa, Hodgkin lymphoma
INTRODUCTION
Burkitt lymphoma (BL) is the most frequent childhood cancer in sub-Saharan Africa (SSA) and accounts for 50% of pediatric cancer in Malawi.1 Although it is highly curable in resource-rich settings with intensive chemotherapy, outcomes are much worse in SSA.2 Epstein-Barr virus (EBV) is causally associated with the endemic form of the disease seen most commonly throughout the region, in which more than 90% of tumor specimens are positive for EBV.3,4 Additionally, EBV can be detected in peripheral blood in endemic BL. Studies from Tanzania, Uganda, and Kenya demonstrated elevated EBV loads in whole blood among children with BL,5–7 and a Brazil study demonstrated plasma EBV load declines were associated with chemotherapy response.8 In contrast, a study of Kenyan children with endemic BL found no association between EBV load in whole blood at presentation and survival.9 For other tumor types causally associated with EBV, plasma or whole blood EBV DNA has also shown potential utility as a biomarker, including classical Hodgkin lymphoma (cHL),10 HIV-associated non-Hodgkin lymphoma (NHL),11,12 natural killer/T-cell lymphoma,13 and nasopharyngeal carcinoma.14,15
In SSA, BL diagnosis is often based on fine needle aspiration (FNA) without immunohistochemistry (IHC) or molecular confirmation, leading to diagnostic inaccuracy.16–19 In addition to diagnostic challenges, SSA centers typically lack advanced imaging like computed tomography (CT) and fluorodeoxyglucose positron emission tomography (FDG-PET), which are routinely used for staging and risk stratification, and to assess treatment response in resource-rich settings. As a result, there exists immense unmet need for implementable point-of-care tools to improve diagnosis and treatment for children with BL in SSA, for whom balancing benefits and risks of cytotoxic chemotherapy is difficult in environments with low supportive care infrastructure and significant treatment-related toxicity.
In this context, we hypothesized that plasma EBV DNA would be an implementable and valuable clinical biomarker for BL diagnosis and treatment in SSA, and undertook a study evaluating its utility for this purpose. Our evaluation was nested within a prospective longitudinal cohort of children with BL at a national teaching hospital in Malawi receiving standardized evaluation and treatment.
METHODS
Setting and population
Kamuzu Central Hospital (KCH) is located in the Malawian capital, Lilongwe, and receives cancer referrals from the northern and central regions, serving approximately 8–9 million people. Malawi has 10% HIV prevalence, 67% ART coverage, annual gross domestic product per capita of 343 US dollars, and Human Development Index rank of 173 out of 188 countries.20–22 The KCH Lymphoma Study is a prospective observational cohort initiated in June 2013. All patients with pathologically confirmed lymphoproliferative disorders are eligible to participate after informed consent. For these analyses, we focused on consecutively enrolled children <18 years with BL between June 1, 2013 and October 31, 2015. To evaluate the performance of plasma EBV DNA specifically for BL diagnosis, we also compared children with BL to enrolled children with cHL, the second most common pediatric lymphoma in Malawi which is also causally associated with EBV, as well as to enrolled children initially suspected to have lymphoma but pathologically confirmed to have non-lymphoproliferative disorders.
Pathologic diagnosis
All enrolled cases were confirmed using a novel weekly telepathology consultative platform involving 2–4 pathologists in Malawi and the United States who rendered a consensus opinion, after review of cytology slides or hematoxylin and eosin stained tissue sections. This model has been described in detail previously and demonstrated excellent concordance with subsequent United States review.23–25 Manual IHC for biopsy specimens or centrifuged cell blocks was performed locally, including CD3, CD20, CD30, CD45, CD138, Ki-67, BCL2, and terminal deoxynucleotidyl transferase (TDT). Other stains including synaptophysin and AE1/AE3 were used to distinguish lymphomas from neuroendocrine or epithelial tumors respectively when morphology was uncertain. Although we sought to obtain tissue or centrifuged cell blocks whenever possible, given frequent abdominal presentations and difficulty obtaining tissue from visceral sites with no interventional radiology, along with limited pediatric surgery and anesthesia, diagnosis was often made via cytology alone as is typical in SSA. All specimens were then shipped to the United States for secondary hematopathologist review and diagnostic confirmation, using a larger panel of automated IHC stains, including CD3, CD15, CD20, CD30, and PAX5 as required.
Clinical care
A detailed description of baseline characteristics, treatment course and toxicities, follow-up, and survival for pediatric BL patients receiving care at our center has been published.26 Briefly, children with BL were treated uniformly with prophase COP (cyclophosphamide, vincristine, prednisone) reduction, followed by six cycles of CHOP (cyclophosphamide, doxorubicin, vincristine, prednisone) administered every 21 days. Intrathecal treatment was administered with each chemotherapy cycle. Hematopoietic growth factors were not available, and supportive care and anti-infective prophylaxis were standardized. Stage was assigned and response assessed using physical exam, chest x-ray, and abdominal ultrasound. Presence of concurrent infections such as malaria was assessed at the treatment team’s discretion based on symptoms, but not systematically, and rapid point-of-care diagnostic tests and treatment for malaria were reliably available throughout the study period.
EBV assessment
EBV was assessed in tumors and serially collected plasma specimens. EBV was evaluated in tumors by EBV-encoded RNA in situ hybridization (EBER ISH) (Leica Biosystems, Wetzlar, Germany). For quantitative plasma EBV DNA, anticoagulated plasma was collected and cryopreserved at −80°C prior to cytotoxic treatment initiation, at mid-treatment (cycle 3 day 21), at treatment completion (cycle 6 day 21), and whenever possible at clinical relapse. All samples were shipped to the University of North Carolina at Chapel Hill where plasma EBV was measured using a real-time quantitative polymerase chain reaction (qPCR) assay performed at the University of North Carolina Vironomics Core with a linear detection range of 2.0–8.0 log10copies/mL. We used a primer pair targeting a conserved region of the EBV EBNA3C gene, details of which were previously described for the detection of EBV DNA and RNA.27,28 This assay targets position 88933-89033 of the EBVI reference genome (NC_007605) and positions 89735-89835 of the EBVII reference genome (NC_009334) to yield a small 100 bp amplicon, and is able to detect total as well as fragmented DNA. The targeted region is highly conserved across EBV isolates, and because the assay uses SYBR green as the method of detection, it can accommodate single nucleotide polymorphisms without loss of sensitivity, as we have shown for other targets using the same assay design.29 Given our focus on evaluating clinical utility of a potentially implementable assay within the Malawi context, we did not conduct additional molecular investigations to distinguish encapsidated from non-encapsidated plasma EBV DNA.
Statistical analysis
Cohort characteristics were summarized using simple descriptive statistics. Plasma EBV DNA was analyzed using log10 transformed values. At baseline, we compared the proportion of children with detectable viremia and median viral loads across diagnostic groups. For children with BL, we analyzed changes in the proportion of children with detectable viremia and median viral loads during and after cytotoxic treatment. Proportions were compared using Fisher’s exact test and medians using Wilcoxon rank sum, and correlations assessed using Pearson’s correlation coefficient. Follow-up time was calculated from enrollment until progression or death, loss to follow-up, or administrative censoring on May 15, 2016. Overall survival (OS) and progression-free survival (PFS) were estimated using Kaplan-Meier methods, and the log-rank test was used to assess survival differences between groups. Cox proportional hazards were used to estimate bivariate and multivariate hazard ratios for OS and PFS. Given non-standardized criteria for response assessment for pediatric BL in SSA, and evaluation of response using relatively crude methods in Malawi where advanced imaging is not routinely available, we principally focused time-to-event analyses on OS as the clearer and more defensible clinical endpoint in our setting. Cause of death was determined by consensus review involving two study clinicians. All analyses were performed using STATA SE version 12.1 (College Station, Texas).
Ethical approval
The study was conducted in accordance with the Helsinki Declaration, after approval by the Biomedical Institutional Review Board of the University of North Carolina at Chapel Hill, the Protocol Review Committee of the Lineberger Comprehensive Cancer Center, and the Malawi National Health Sciences Research Committee.
RESULTS
Of 137 children enrolled during the study period, baseline plasma EBV DNA was assessed in 121 (88%), including 88/94 (94%) with BL, 17/18 (94%) with cHL, and 16/25 (64%) with non-lymphoma diagnoses (Table 1). Non-lymphoma diagnoses with plasma EBV DNA assessed included children who were pathologically confirmed to have tuberculosis (n=4), rhabdomyosarcoma (n=3), Kaposi sarcoma (n=2), Wilm’s tumor (n=2), other infectious/reactive lymphadenitis (n=2), acute myeloid leukemia (n=1), salivary gland tumor (n=1), and non-hematopoietic small round blue cell tumor (n=1). Among patients tested, plasma EBV DNA was detected in 76/88 (86%) children with BL, 16/17 (94%) children with cHL, and 2/16 (12%) children with non-lymphoma diagnoses, with proportions being higher in BL versus non-lymphoma (p<0.001) and similar in BL versus cHL (p=0.69). If detected, median plasma EBV DNA level was 6.1 log10copies/mL [interquartile range (IQR) 4.9–6.9] for children with BL, 4.8 log10copies/mL (IQR 4.1–5.8) for cHL, and 3.4 log10copies/mL (IQR 3.0–3.7) for non-lymphoma, with higher levels observed in BL versus cHL (p=0.029), and a trend toward higher levels in BL versus non-lymphoma (p=0.062) when detected.
Table 1.
Quantitative Epstein-Barr virus plasma DNA at baseline for pathologically confirmed Burkitt lymphoma, classical Hodgkin lymphoma, and non-lymphoma diagnoses in Lilongwe, Malawi.
| Plasma EBV DNA | Burkitt | Hodgkin | Non-lymphoma | P value, Burkitt vs Hodgkin | P value, Burkitt vs non-lymphoma |
|---|---|---|---|---|---|
| Assessed | 88/94 (94%) | 17/18 (94%) | 16/25 (64%) | ― | ― |
| Detected | 76/88 (86%) | 16/17 (94%) | 2/16 (12%) | 0.69 | <0.001 |
| Median level if detected, log10copies per mL (IQR) | 6.1 (4.9–6.9) | 4.8 (4.1–5.8) | 3.4 (3.0–3.7) | 0.029 | 0.062 |
IQR = interquartile range.
Characteristics for 88 children with pathologically confirmed BL for whom baseline plasma EBV DNA was assessed are shown in Table 2, stratified by baseline plasma EBV detection. EBER ISH was available for 18 children (20%). Plasma EBV was detected in 12/12 patients with positive EBER ISH and 2/6 patients with negative EBER ISH (p=0.005), with median levels when detected 6.7 in EBER ISH positive cases versus 4.6 log10copies/mL in EBER ISH negative children (p=0.14). Additionally, children with detectable baseline plasma EBV were sicker, with worse Lansky performance status and trends toward lower albumin and higher lactate dehydrogenase (LDH). Baseline plasma EBV DNA levels were positively correlated with LDH (r=0.22, p=0.040).
Table 2.
Baseline characteristics of pathologically confirmed pediatric Burkitt lymphoma patients in Lilongwe, Malawi, stratified by plasma EBV DNA detection at diagnosis.
| pEBV DNA+ (n=76) | pEBV DNA− (n=12) | P value | |
|---|---|---|---|
| Female, n (%) | 27 (36) | 3 (25) | 0.74 |
| Age, years, median (IQR) | 9.1 (7.1–12.1) | 10.8 (7.0–12.6) | 0.51 |
| Stage III/IV, n (%) | 60 (78) | 8 (67) | 0.46 |
| HIV positive, n (%) | 2 (3) | 0 (0) | 1.00 |
| Histology diagnosisA, n (%) | 21 (28) | 4 (33) | 0.74 |
| Tumor EBER ISH positiveB, n (%) | 12/14 (86) | 0/4 (0) | 0.005 |
| UnderweightC, n (%) | 27 (37) | 5 (42) | 0.76 |
| Lansky performance status ≤70, n (%) | 57 (76) | 5 (42) | 0.034 |
| White blood cells, 103/μL, median (IQR) | 8.7 (6.6–12.9) | 9.4 (7.6–11.3) | 0.70 |
| Absolute neutrophil count, 103/μL, median (IQR) | 4.3 (2.6–7.2) | 4.4 (2.8–6.9) | 1.00 |
| Hemoglobin, g/dL, median (IQR) | 10.2 (8.8–11.7) | 10.4 (8.5–12.6) | 0.86 |
| Platelets, 103/μL, median (IQR) | 414 (270–570) | 480 (307–567) | 0.70 |
| Albumin, g/dL, median (IQR) | 3.3 (2.9–3.7) | 3.7 (3.2–4.6) | 0.078 |
| Lactate dehydrogenaseD, IU/L, median (IQR) | 668 (394–1,535) | 365 (256–983) | 0.12 |
pEBV = plasma Epstein-Barr virus. EBER ISH = Epstein-Barr virus encoded RNA in situ hybridization. IQR = interquartile range.
Diagnosis for non-histology cases was based on consensus telepathology review of cytology without tissue or cell block.
EBER ISH was only available for a subset of histology specimens evaluated in the United States.
Underweight was defined as weight-for-age z-score <−2 if <5 years or body mass index (BMI) z-score <−2 if ≥5 years.
Laboratory upper limit of normal was 250 IU/L.
During chemotherapy, plasma EBV DNA declined in most pediatric BL patients (Figure 1 and Supplemental Figure). Compared with baseline, children had less frequently detectable plasma EBV DNA at mid-treatment (86% vs 65%, p=0.003) and completion (86% vs 70%, p=0.033). When detected, plasma EBV DNA levels also declined during cytotoxic treatment (median 6.1 log10copies/mL baseline vs 4.4 mid-treatment vs 3.4 completion), with significant differences between viremia levels at mid-treatment versus baseline, and completion versus baseline (p<0.0001 for both comparisons). Significant differences were not observed between mid-treatment and treatment completion, for either the proportion of children with detectable plasma EBV DNA or median viremia levels when detected. Although few children had assessable plasma for EBV DNA at clinical relapse, the proportion with detectable viremia was similar to mid-treatment and completion timepoints, but viremia level was higher at relapse when detected (median 5.2 log10copies/mL, p=0.068 vs mid-treatment, p=0.014 vs completion). Additionally, three children had markedly elevated outlier plasma EBV DNA values at the completion timepoint as shown in Figure 1A, all of whom had BL that was clinically refractory to first-line treatment and resulted in death within three months of CHOP completion, without plasma EBV DNA being subsequently assessed.
Figure 1. Quantitative plasma Epstein-Barr virus DNA during pediatric Burkitt lymphoma treatment in Lilongwe, Malawi.
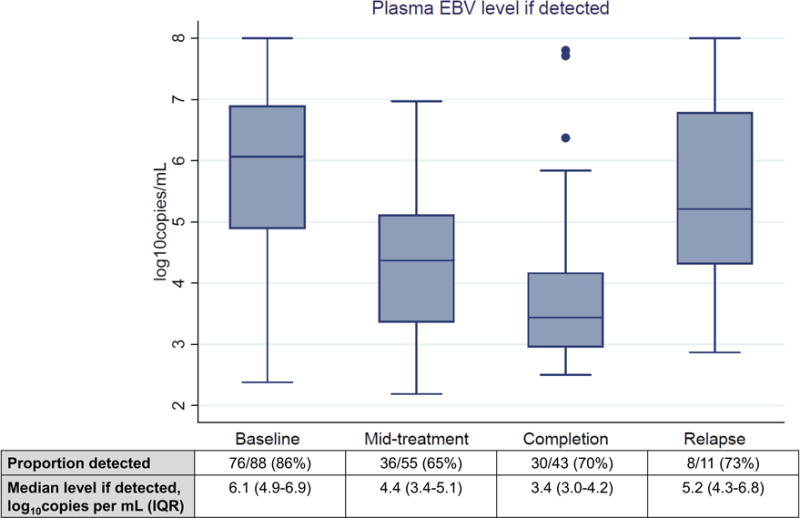
EBV = Epstein-Barr virus. IQR = interquartile range.
As of May 15, 2016, disease and vital status were known for 80/88 (91%) children after median follow-up of 13.2 months (IQR 9.3–26.9) among those not known to have died. For children who were untraceable by cellphone, four were from neighboring Mozambique and had traveled with their families to receive care in Malawi and subsequently returned home. Despite significant efforts to standardize care with dedicated pediatric oncology support, outcomes in the cohort overall with anthracycline-based treatment were suboptimal with OS 40% [95% confidence interval (CI) 29–51%] and PFS 31% (95% CI 21–41) at 12 months, and OS 28% (95% CI 18–40%) and PFS 27% (95% CI 17–38) at 24 months. Of 55 deaths in the cohort, 41 (75%) were attributed to relapsed or refractory BL on central adjudication and 14 (25%) to treatment-related complications.
OS did not differ within the cohort based on baseline plasma EBV detection (Figure 2). However, among children with baseline plasma EBV detected, survival was significantly worse for patients with baseline level ≥6 log10copies/mL versus <6 log10copies/mL (p=0.0002). Additionally, after cytotoxic treatment initiation, survival was worse for children with persistent mid-treatment plasma EBV detection versus those without (p=0.041). Findings were similar as well in analyses using PFS as the primary outcome. During the study period, 15 confirmed episodes of clinical malaria were documented, and no differences were observed in baseline plasma EBV DNA detection or quantitative levels between children who developed malaria versus those who did not.
Figure 2. Kaplan-Meier overall survival for pediatric Burkitt lymphoma in Lilongwe, Malawi.
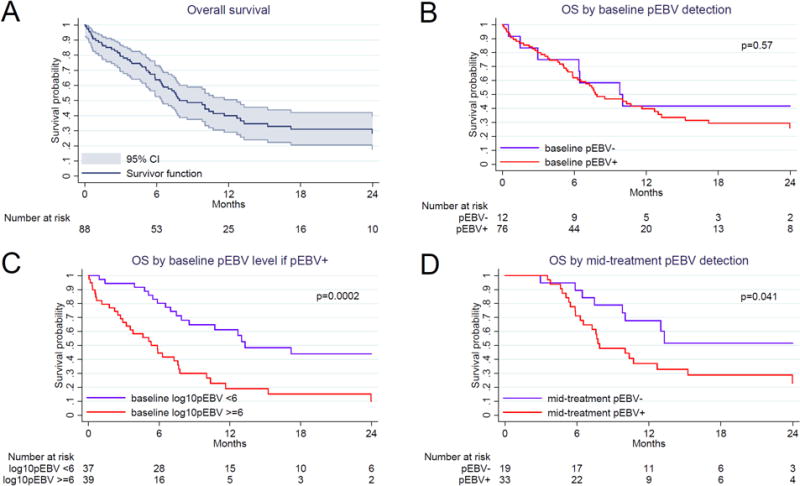
(A) Overall cohort with 95% confidence intervals. (B) Stratified by baseline plasma Epstein-Barr virus DNA detection. (C) Stratified by baseline plasma Epstein-Barr virus DNA level if detected. (D) Stratified by mid-treatment plasma Epstein-Barr virus DNA detection. OS = overall survival. CI = confidence interval. pEBV = plasma Epstein-Barr virus.
Risk factors for mortality by 12 months are shown in Table 3. In adjusted analyses in the cohort overall, mortality was associated with Lansky performance status <70 [adjusted hazard ratio (AHR) 2.38, 95% CI 1.02–5.59, p=0.046], increased LDH (AHR 1.02 per 100 IU/L, 95% CI 1.00–1.03, p=0.015), and possibly age ≥9 years (AHR 1.89, 95% CI 0.98–3.63, p=0.056). In adjusted analyses among only children with plasma EBV DNA detected at baseline, viremia level was the only significant risk factor for death by 12 months (AHR 1.35 per log10copies/mL, 95% CI 1.04–1.75, p=0.023). Again, findings were similar in analyses using PFS as the primary outcome.
Table 3.
Baseline characteristics associated with death by 12 months among children with Burkitt lymphoma in Lilongwe, Malawi.
| Variable | Unadjusted hazard ratio | 95% CI | P value | Adjusted hazard ratio | 95% CI | P value |
|---|---|---|---|---|---|---|
| Entire cohort (n=88) | ||||||
| Baseline pEBV DNA, per log10copies/mL | 1.19 | 1.03–1.37 | 0.019 | 1.05 | 0.91–1.22 | 0.50 |
| Female gender | 1.65 | 0.93–2.91 | 0.085 | 1.39 | 0.76–2.54 | 0.28 |
| Age ≥9 years | 2.24 | 1.22–4.13 | 0.009 | 1.89 | 0.98–3.63 | 0.056 |
| Stage III/IV | 2.23 | 1.00–4.97 | 0.049 | 1.32 | 0.56–3.13 | 0.52 |
| Lansky performance status <70 | 2.80 | 1.31–5.98 | 0.008 | 2.38 | 1.02–5.59 | 0.046 |
| Lactate dehydrogenase, per 100 IU/L | 1.03 | 1.01–1.04 | <0.001 | 1.02 | 1.00–1.03 | 0.015 |
| Underweight statusA | 1.74 | 0.99–3.05 | 0.056 | 1.33 | 0.72–2.44 | 0.36 |
| Baseline pEBV+ only (n=76) | ||||||
| Baseline pEBV DNA, per log10copies/mL | 1.64 | 1.27–2.12 | <0.001 | 1.35 | 1.04–1.75 | 0.023 |
| Female gender | 1.89 | 1.03–3.48 | 0.041 | 1.49 | 0.78–2.85 | 0.23 |
| Age ≥9 years | 2.47 | 1.28–4.76 | 0.007 | 1.49 | 0.73–3.05 | 0.27 |
| Stage III/IV | 2.54 | 1.00–6.46 | 0.051 | 1.37 | 0.48–3.94 | 0.56 |
| Lansky performance status <70 | 2.94 | 1.15–7.48 | 0.024 | 2.29 | 0.81–6.47 | 0.12 |
| Lactate dehydrogenase, per 100 IU/L | 1.03 | 1.01–1.04 | <0.001 | 1.01 | 1.00–1.03 | 0.13 |
| Underweight statusA | 2.24 | 1.22–4.13 | 0.010 | 1.63 | 0.84–3.15 | 0.15 |
CI = confidence interval. pEBV = plasma Epstein-Barr virus.
Underweight was defined as weight-for-age z-score <−2 if <5 years or body mass index (BMI) z-score <−2 if ≥5 years.
DISCUSSION
To our knowledge, this is the first study from SSA to systematically assess utility of plasma EBV DNA for pediatric BL diagnosis, prognosis, and response assessment. Study strengths include serial assessment of EBV DNA at pre-specified clinical timepoints, within a prospective cohort of children receiving standardized treatment with minimal loss to follow-up. Additionally, high-quality pathologic diagnoses were rendered using a novel telepathology platform which is innovative for the region,23–25 comparison was made to simultaneously enrolled children with cHL and non-lymphoproliferative disorders, and detailed evaluation facilitated adjustment for other key BL prognostic variables.
With respect to diagnosis, frequent EBV DNA detection (86%) distinguished children with BL from those referred for clinical suspicion of lymphoma who were pathologically confirmed to have non-lymphoproliferative disorders (12%). Moreover, EBV DNA levels if detected distinguished children with BL (median 6.1 log10copies/mL) from those with cHL (median 4.8 log10copies/mL), the second commonest pediatric lymphoma in Malawi which is also associated with EBV. Scarcity of diagnostic pathology in SSA has been amply documented,30 and even where pathology services exist, diagnosis of BL and other lymphoproliferative disorders often relies on cytology with absent or limited IHC and absent molecular tools.16–19 As a result, studies in the region are invariably affected by small proportions of other aggressive NHL subtypes diagnosed as endemic BL, including lymphoblastic lymphoma, diffuse large B-cell lymphoma, or sporadic EBV-negative BL, which also occurs in SSA albeit less frequently than endemic BL and typically among older children.26,31 In Lilongwe, we have made significant efforts to increase use of biopsies or centrifuged FNA cell blocks for IHC confirmation of BL diagnosis, but frequent visceral and/or abdominal presentations, limited interventional radiology, limited pediatric surgery, and limited pediatric anesthesia, often render cytologic diagnosis unavoidable despite these efforts. In such instances, plasma EBV DNA alone may not have sufficient utility to diagnose or exclude lymphoma and distinguish lymphoma subtypes, but could be included within composite diagnostic algorithms incorporating real-time pathologic, clinical, and laboratory data generated locally. We believe such an approach is more practical for SSA than tissue-based fluorescence in situ hybridization for MYC translocation to confirm all pediatric BL cases, and could substantially reduce current diagnostic uncertainties which remain a major issue for BL care and research throughout the region. IHC for MYC or Epstein-Barr nuclear antigen 1 (EBNA1) might also have applicability for improving local diagnostic accuracy. Indeed, with support from the National Cancer Institute Center for Global Health, we are now undertaking efforts to derive and validate a diagnostic score specifically for molecularly confirmed BL which incorporates plasma EBV DNA qPCR performed onsite in our Malawi laboratory, using a commercially available assay with our existing HIV RNA platform,32 along with age, anatomic site, symptom duration, LDH, and telepathology review. Moreover, analyzable DNA can be recovered from FNA specimens,33 which might also allow EBV detection directly from cytology slides.
With respect to prognosis and response assessment, we observed markedly elevated plasma EBV DNA among children with BL at baseline with subsequent declines during chemotherapy, as observed in Brazil,8 and often increasing levels prior to clinical relapse. Among children with plasma EBV detected at baseline, outcomes were significantly worse for those with higher viremia, as well as those with persistent plasma EBV DNA at the midpoint of treatment. Additionally, baseline plasma EBV DNA was the only significant predictor of outcomes in multivariate analyses for these children, with each unit increase in log10copies/mL being associated with a 35% increased hazard of death by 12 months. Patients with baseline plasma EBV detected are arguably those children most likely to have truly endemic EBV-positive BL, as opposed to other aggressive NHL subtypes, given diagnostic challenges referenced above.
Our findings suggest EBV load monitoring could improve risk stratification and response assessment in SSA, particularly in light of the critical unmet need to optimize balance between treatment efficacy and toxicity for vulnerable, often malnourished children receiving chemotherapy in poor supportive care environments. Analogously, HIV RNA assessment is well established for antiretroviral therapy monitoring and early infant diagnosis in SSA,34 and existing qPCR instruments could likely be adapted for EBV. In resource-rich settings, EBV DNA assessment has greater utility in plasma than peripheral blood mononuclear cells (PMBCs) for EBV-positive lymphoproliferative disorders.35 In Kenyan children, the strength of association between cellular and plasma EBV DNA varies between healthy children and those with BL, which may also have implications for post-treatment monitoring during remission.36 Moreover, assays utilizing plasma or whole blood can be successfully implemented even in resource-limited settings.37
Despite efforts to intensify treatment at our center including anthracycline-based chemotherapy with pediatric oncology support, given historical experience with frequent relapses after less intensive regimens, outcomes were suboptimal as previously described, with the majority of deaths due to relapsed or refractory BL on central adjudication.26 This is further supported by persistent EBV detection in 70% of children even after chemotherapy completion, suggesting that despite treatment intensification, achievable cumulative dose and dose intensity using this approach in our environment were insufficient to eradicate disease and cure children with advanced, aggressive BL. Better therapeutic strategies are therefore essential, which we are actively pursuing with regional colleagues through the National Cancer Institute Burkitt Lymphoma Trial Network, including greater incorporation of systemic methotrexate into front-line chemotherapy as has been reported by other groups to result in better outcomes.38–41 Incorporating newer non-cytotoxic agents, like the anti-CD20 monoclonal antibody rituximab, should also be an urgent priority especially in settings where a cytotoxic ‘ceiling’ is imposed by limited supportive care, rendering more intensive regimens from high-income countries impractical.
Apart from diagnostic and treatment challenges intrinsic to the Malawi setting, which we continue to address but which remain substantial, limitations of our study include referral bias at a national teaching hospital, assigning cause of death based on inference after centralized review, non-systematic assessment of concurrent malaria, and lack of distinction between encapsidated and non-encapsidated plasma EBV DNA. Additionally, plasma EBV DNA assessments were missing in some children at disease progression, and not performed after treatment completion in children without subsequent disease progression, making it difficult to clearly distinguish different plasma EBV DNA trajectories among groups of children defined by their chemotherapy response.
To conclude, quantitative plasma EBV DNA demonstrated potential utility for diagnosis, prognosis, and response assessment in a prospective pediatric BL cohort in Malawi receiving standardized anthracycline-based treatment. These findings require validation in larger regional studies, but suggest plasma EBV DNA may be an implementable and valuable biomarker for improving outcomes, through better diagnosis and more effectively risk-stratified and response-guided therapy for children with BL in SSA.
Supplementary Material
Quantitative plasma Epstein-Barr virus DNA in individual children during pediatric Burkitt lymphoma treatment in Lilongwe, Malawi. EBV = Epstein-Barr virus. IQR = interquartile range.
Novelty and impact.
This is the first study to systematically assess plasma EBV DNA for pediatric Burkitt lymphoma diagnosis, prognosis, and response assessment in sub-Saharan Africa. Plasma EBV DNA may be an implementable biomarker to facilitate better diagnosis and risk-stratified, response-guided therapy for this challenging population in the region.
Acknowledgments
We are sincerely grateful to the children and their families for agreeing to participate in the study and entrusting their care to our team. We also thank Wiza Kumwenda for developing the study database and the University of North Carolina Translational Pathology Laboratory for performing additional stains including extensive technical contributions from Ms. Michelle Mathews. We are also grateful to leadership of Kamuzu Central Hospital, Malawi Ministry of Health, UNC Project-Malawi, Lineberger Comprehensive Cancer Center, and Baylor College of Medicine Children’s Foundation Malawi for their support. This work was supported by grants from the National Institutes of Health (K01TW009488, R21CA180815, and U54CA190152 to S.G. and P01CA019014 to D.P.D.), the Medical Education Partnership Initiative (U2GPS001965), the Lineberger Comprehensive Cancer Center (P30CA016086), and Fogarty Global Health Fellows Program (R25TW009340).
Abbreviations
- BL
Burkitt lymphoma
- cHL
classical Hodgkin lymphoma
- COP
cyclophosphamide vincristine, prednisone
- CHOP
cyclophosphamide doxorubicin, vincristine, prednisone
- CI
confidence interval
- CT
computed tomography
- DNA
deoxyribonucleic acid
- EBER ISH
Epstein-Barr virus-encoded ribonucleic acid in situ hybridization
- EBV
Epstein-Barr virus
- FDG-PET
fluorodeoxyglucose positron emission tomography
- FNA
fine needle aspirate
- IHC
immunohistochemistry
- IQR
interquartile range
- KCH
Kamuzu Central Hospital
- LANA
latency-associated nuclear antigen
- LDH
lactate dehydrogenase
- NHL
non-Hodgkin lymphoma
- OS
overall survival
- pEBV
plasma Epstein-Barr virus
- PFS
progression-free survival
- qPCR
quantitative polymerase chain reaction
- RNA
ribonucleic acid
- SSA
sub-Saharan Africa
- TDT
terminal deoxynucleotidyl transferase
References
- 1.Sinfield RL, Molyneux EM, Banda K, Borgstein E, Broadhead R, Hesseling P, Newton R, Casabonne D, Mkandawire N, Nkume H, Hodgson T, Liomba G. Spectrum and presentation of pediatric malignancies in the HIV era: experience from Blantyre, Malawi, 1998–2003. Pediatr Blood Cancer. 2007;48:515–20. doi: 10.1002/pbc.20917. [DOI] [PubMed] [Google Scholar]
- 2.Molyneux EM, Rochford R, Griffin B, Newton R, Jackson G, Menon G, Harrison CJ, Israels T, Bailey S. Burkitt’s lymphoma. Lancet. 2012;379:1234–44. doi: 10.1016/S0140-6736(11)61177-X. [DOI] [PubMed] [Google Scholar]
- 3.Mutalima N, Molyneux E, Jaffe H, Kamiza S, Borgstein E, Mkandawire N, Liomba G, Batumba M, Lagos D, Gratrix F, Boshoff C, Casabonne D, Carpenter LM, Newton R. Associations between Burkitt lymphoma among children in Malawi and infection with HIV, EBV and malaria: results from a case-control study. PLoS One. 2008;3:e2505. doi: 10.1371/journal.pone.0002505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen JI. Epstein-Barr virus infection. N Engl J Med. 2000;343:481–92. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 5.Kabyemera R, Masalu N, Rambau P, Kamugisha E, Kidenya B, De Rossi A, Petrara MR, Mwizamuholya D. Relationship between non-Hodgkin’s lymphoma and blood levels of Epstein-Barr virus in children in north-western Tanzania: a case control study. BMC Pediatr. 2013;13:4. doi: 10.1186/1471-2431-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orem J, Sandin S, Mbidde E, Mangen FW, Middeldorp J, Weiderpass E. Epstein-Barr virus viral load and serology in childhood non-Hodgkin’s lymphoma and chronic inflammatory conditions in Uganda: implications for disease risk and characteristics. J Med Virol. 2014;86:1796–803. doi: 10.1002/jmv.23988. [DOI] [PubMed] [Google Scholar]
- 7.Moormann AM, Chelimo K, Sumba OP, Lutzke ML, Ploutz-Snyder R, Newton D, Kazura J, Rochford R. Exposure to holoendemic malaria results in elevated Epstein-Barr virus loads in children. J Infect Dis. 2005;191:1233–8. doi: 10.1086/428910. [DOI] [PubMed] [Google Scholar]
- 8.Machado AS, Da Silva Robaina MC, Magalhaes De Rezende LM, Apa AG, Amoedo ND, Bacchi CE, Klumb CE. Circulating cell-free and Epstein-Barr virus DNA in pediatric B-non-Hodgkin lymphomas. Leuk Lymphoma. 2010;51:1020–7. doi: 10.3109/10428191003746331. [DOI] [PubMed] [Google Scholar]
- 9.Buckle G, Maranda L, Skiles J, Ong’echa JM, Foley J, Epstein M, Vik TA, Schroeder A, Lemberger J, Rosmarin A, Remick SC, Bailey JA, Vulule J, Otieno JA, Moormann AM. Factors influencing survival among Kenyan children diagnosed with endemic Burkitt lymphoma between 2003 and 2011: A historical cohort study. Int J Cancer. 2016;139:1231–40. doi: 10.1002/ijc.30170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandhi MK, Lambley E, Burrows J, Dua U, Elliott S, Shaw PJ, Prince HM, Wolf M, Clarke K, Underhill C, Mills T, Mollee P, Gill D, Marlton P, Seymour JF, Khanna R. Plasma Epstein-Barr virus (EBV) DNA is a biomarker for EBV-positive Hodgkin’s lymphoma. Clin Cancer Res. 2006;12:460–4. doi: 10.1158/1078-0432.CCR-05-2008. [DOI] [PubMed] [Google Scholar]
- 11.Fan H, Kim SC, Chima CO, Israel BF, Lawless KM, Eagan PA, Elmore S, Moore DT, Schichman SA, Swinnen LJ, Gulley ML. Epstein-Barr viral load as a marker of lymphoma in AIDS patients. J Med Virol. 2005;75:59–69. doi: 10.1002/jmv.20238. [DOI] [PubMed] [Google Scholar]
- 12.Bonnet F, Jouvencel AC, Parrens M, Leon MJ, Cotto E, Garrigue I, Morlat P, Beylot J, Fleury H, Lafon ME. A longitudinal and prospective study of Epstein-Barr virus load in AIDS-related non-Hodgkin lymphoma. J Clin Virol. 2006;36:258–63. doi: 10.1016/j.jcv.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Kim HS, Kim KH, Kim KH, Chang MH, Ji SH, Lim DH, Kim K, Kim SJ, Ko Y, Ki CS, Jo SJ, Lee JW, Kim WS. Whole blood Epstein-Barr virus DNA load as a diagnostic and prognostic surrogate: extranodal natural killer/T-cell lymphoma. Leuk Lymphoma. 2009;50:757–63. doi: 10.1080/10428190902803669. [DOI] [PubMed] [Google Scholar]
- 14.Chan KC, Zhang J, Chan AT, Lei KI, Leung SF, Chan LY, Chow KC, Lo YM. Molecular characterization of circulating EBV DNA in the plasma of nasopharyngeal carcinoma and lymphoma patients. Cancer Res. 2003;63:2028–32. [PubMed] [Google Scholar]
- 15.Ryan JL, Fan H, Swinnen LJ, Schichman SA, Raab-Traub N, Covington M, Elmore S, Gulley ML. Epstein-Barr Virus (EBV) DNA in plasma is not encapsidated in patients with EBV-related malignancies. Diagn Mol Pathol. 2004;13:61–8. doi: 10.1097/00019606-200406000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Orem J, Sandin S, Weibull CE, Odida M, Wabinga H, Mbidde E, Wabwire-Mangen F, Meijer CJ, Middeldorp JM, Weiderpass E. Agreement between diagnoses of childhood lymphoma assigned in Uganda and by an international reference laboratory. Clin Epidemiol. 2012;4:339–47. doi: 10.2147/CLEP.S35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naresh KN, Raphael M, Ayers L, Hurwitz N, Calbi V, Rogena E, Sayed S, Sherman O, Ibrahim HA, Lazzi S, Mourmouras V, Rince P, Githanga J, Byakika B, Moshi E, Durosinmi M, Olasode BJ, Oluwasola OA, Akang EE, Akenova Y, Adde M, Magrath I, Leoncini L. Lymphomas in sub-Saharan Africa–what can we learn and how can we help in improving diagnosis, managing patients and fostering translational research? Br J Haematol. 2011;154:696–703. doi: 10.1111/j.1365-2141.2011.08772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naresh KN, Ibrahim HA, Lazzi S, Rince P, Onorati M, Ambrosio MR, Bilhou-Nabera C, Amen F, Reid A, Mawanda M, Calbi V, Ogwang M, Rogena E, Byakika B, Sayed S, Moshi E, Mwakigonja A, Raphael M, Magrath I, Leoncini L. Diagnosis of Burkitt lymphoma using an algorithmic approach–applicable in both resource-poor and resource-rich countries. Br J Haematol. 2011;154:770–6. doi: 10.1111/j.1365-2141.2011.08771.x. [DOI] [PubMed] [Google Scholar]
- 19.Ogwang MD, Zhao W, Ayers LW, Mbulaiteye SM. Accuracy of Burkitt lymphoma diagnosis in constrained pathology settings: importance to epidemiology. Arch Pathol Lab Med. 2011;135:445–50. doi: 10.1043/2009-0443-EP.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.UNAIDS. Malawi Progress Report for 2015. http://www.unaids.org/en/regionscountries/countries/malawi. Accessed October 9, 2016.
- 21.United Nations Statistics Division. Malawi country profile. http://data.un.org/CountryProfile.aspx?crName=malawi. Accessed October 9, 2016.
- 22.United Nations Development Program. Human Development Report 2015. http://hdr.undp.org/en/2015-report. Accessed October 9, 2016.
- 23.Gopal S, Krysiak R, Liomba G. Building a pathology laboratory in Malawi. Lancet Oncol. 2013;14:291–2. doi: 10.1016/S1470-2045(13)70109-8. [DOI] [PubMed] [Google Scholar]
- 24.Gopal S, Krysiak R, Liomba NG, Horner MJ, Shores CG, Alide N, Kamiza S, Kampani C, Chimzimu F, Fedoriw Y, Dittmer DP, Hosseinipour MC, Hoffman IF. Early experience after developing a pathology laboratory in Malawi, with emphasis on cancer diagnoses. PLoS One. 2013;8:e70361. doi: 10.1371/journal.pone.0070361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montgomery ND, Liomba NG, Kampani C, Krysiak R, Stanley CC, Tomoka T, Kamiza S, Dhungel BM, Gopal S, Fedoriw Y. Accurate Real-Time Diagnosis of Lymphoproliferative Disorders in Malawi Through Clinicopathologic Teleconferences: A Model for Pathology Services in Sub-Saharan Africa. Am J Clin Pathol. 2016;146:423–30. doi: 10.1093/ajcp/aqw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanley CC, Westmoreland KD, Heimlich BJ, El-Mallawany NK, Wasswa P, Mtete I, Butia M, Itimu S, Chasela M, Mtunda M, Chikasema M, Makwakwa V, Kaimila B, Kasonkanji E, Chimzimu F, Kampani C, Dhungel BM, Krysiak R, Montgomery ND, Fedoriw Y, Rosenberg NE, Liomba NG, Gopal S. Outcomes for paediatric Burkitt lymphoma treated with anthracycline-based therapy in Malawi. Br J Haematol. 2016;173:705–12. doi: 10.1111/bjh.13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilscher C, Vahrson W, Dittmer DP. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic Acids Res. 2005;33:e182. doi: 10.1093/nar/gni181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobson MA, Ditmer DP, Sinclair E, Martin JN, Deeks SG, Hunt P, Mocarski ES, Shiboski C. Human herpesvirus replication and abnormal CD8+ T cell activation and low CD4+ T cell counts in antiretroviral-suppressed HIV-infected patients. PLoS One. 2009;4:e5277. doi: 10.1371/journal.pone.0005277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papin JF, Vahrson W, Dittmer DP. SYBR green-based real-time quantitative PCR assay for detection of West Nile Virus circumvents false-negative results due to strain variability. J Clin Microbiol. 2004;42:1511–8. doi: 10.1128/JCM.42.4.1511-1518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adesina A, Chumba D, Nelson AM, Orem J, Roberts DJ, Wabinga H, Wilson M, Rebbeck TR. Improvement of pathology in sub-Saharan Africa. Lancet Oncol. 2013;14:e152–7. doi: 10.1016/S1470-2045(12)70598-3. [DOI] [PubMed] [Google Scholar]
- 31.Stefan DC, Lutchman R. Burkitt lymphoma: epidemiological features and survival in a South African centre. Infect Agent Cancer. 2014;9:19. doi: 10.1186/1750-9378-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abbott. RealTime EBV Assay. https://www.molecular.abbott/int/en/products/infectious-disease/realtime-ebv. Accessed January 19, 2017.
- 33.Treece AL, Montgomery ND, Patel NM, Civalier CJ, Dodd LG, Gulley ML, Booker JK, Weck KE. FNA smears as a potential source of DNA for targeted next-generation sequencing of lung adenocarcinomas. Cancer Cytopathol. 2016;124:406–14. doi: 10.1002/cncy.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lecher S, Ellenberger D, Kim AA, Fonjungo PN, Agolory S, Borget MY, Broyles L, Carmona S, Chipungu G, De Cock KM, Deyde V, Downer M, Gupta S, Kaplan JE, Kiyaga C, Knight N, MacLeod W, Makumbi B, Muttai H, Mwangi C, Mwangi JW, Mwasekaga M, Ng’Ang AL, Pillay Y, Sarr A, Sawadogo S, Singer D, Stevens W, Toure CA, Nkengasong J. Scale-up of HIV Viral Load Monitoring–Seven Sub-Saharan African Countries. MMWR Morb Mortal Wkly Rep. 2015;64:1287–90. doi: 10.15585/mmwr.mm6446a3. [DOI] [PubMed] [Google Scholar]
- 35.Kanakry JA, Hegde AM, Durand CM, Massie AB, Greer AE, Ambinder RF, Valsamakis A. The clinical significance of EBV DNA in the plasma and peripheral blood mononuclear cells of patients with or without EBV diseases. Blood. 2016;127:2007–17. doi: 10.1182/blood-2015-09-672030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulama DH, Bailey JA, Foley J, Chelimo K, Ouma C, Jura WG, Otieno J, Vulule J, Moormann AM. Sickle cell trait is not associated with endemic Burkitt lymphoma: an ethnicity and malaria endemicity-matched case-control study suggests factors controlling EBV may serve as a predictive biomarker for this pediatric cancer. Int J Cancer. 2014;134:645–53. doi: 10.1002/ijc.28378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens SJ, Pronk I, Middeldorp JM. Toward standardization of Epstein-Barr virus DNA load monitoring: unfractionated whole blood as preferred clinical specimen. J Clin Microbiol. 2001;39:1211–6. doi: 10.1128/JCM.39.4.1211-1216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouda C, Traore F, Atteby JJ, et al. A multicenter study of the Groupe Franco Africain d’Oncologie Pediatrique for the treatment of Burkitt lymphoma in sub-Saharan African countries. In Fifth International Symposium on Childhood, Adolescent and Young Adult Non-Hodgkin Lymphoma Abstracts. Br J Haematol. 2015;171:1–88. [Google Scholar]
- 39.Ngoma T, Adde M, Durosinmi M, Githang’a J, Aken’Ova Y, Kaijage J, Adeodou O, Rajab J, Brown BJ, Leoncini L, Naresh K, Raphael M, Hurwitz N, Scanlan P, Rohatiner A, Venzon D, Magrath I. Treatment of Burkitt lymphoma in equatorial Africa using a simple three-drug combination followed by a salvage regimen for patients with persistent or recurrent disease. Br J Haematol. 2012;158:749–62. doi: 10.1111/j.1365-2141.2012.09236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hesseling P, Broadhead R, Mansvelt E, Louw M, Wessels G, Borgstein E, Schneider J, Molyneux E. The 2000 Burkitt lymphoma trial in Malawi. Pediatr Blood Cancer. 2005;44:245–50. doi: 10.1002/pbc.20254. [DOI] [PubMed] [Google Scholar]
- 41.Harif M, Barsaoui S, Benchekroun S, Bouhas R, Doumbe P, Khattab M, Ladjaj Y, Moreira C, Msefer-Alaoui F, Patte C, Rakotonirina G, Raphael M, Raquin MA, Lemerle J. Treatment of B-cell lymphoma with LMB modified protocols in Africa–report of the French-African Pediatric Oncology Group (GFAOP) Pediatr Blood Cancer. 2008;50:1138–42. doi: 10.1002/pbc.21452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative plasma Epstein-Barr virus DNA in individual children during pediatric Burkitt lymphoma treatment in Lilongwe, Malawi. EBV = Epstein-Barr virus. IQR = interquartile range.


