Abstract
Objective
Glioblastoma (GBM) is the most malignant primary brain tumor. Collagen is present in low amounts in normal brain, but in GBMs, collagen gene expression is reportedly upregulated. However, to the authors’ knowledge, direct visualization of collagen architecture has not been reported. The authors sought to perform the first direct visualization of GBM collagen architecture, identify clinically relevant collagen signatures, and link them to differential patient survival.
Methods
Second-harmonic generation microscopy (SHG) was used to detect collagen in a GBM patient tissue microarray. Focal and invasive GBM mouse xenografts were stained with Picrosirius red. Quantitation of collagen fibers was performed using custom software. Multivariate survival analysis was done to determine if collagen is a survival marker for patients.
Results
In focal xenografts, collagen was observed at tumor brain boundaries. For invasive xenografts, collagen was intercalated with tumor cells. Quantitative analysis showed significant differences in collagen fibers for focal and invasive xenografts. The authors also found that GBM patients with more organized collagen had longer median survival than those with less organized collagen.
Conclusions
Collagen architecture can be directly visualized and is different in focal versus invasive GBMs. The authors also demonstrate that collagen signature is associated with patient survival. These findings suggest that there are collagen differences in focal versus invasive GBMs and that collagen is a survival marker for GBM.
Keywords: biomarker, cancer stem-like cells, collagen, glioblastoma, second harmonic generation
Introduction
An estimated 22,000 new primary malignant central nervous system tumors are diagnosed annually in the United States and cause 1.5% of all cancer-related deaths29. Gliomas, cancers arising from the most abundant glial cells, accounts for more than 14,000 of these cases29. The most aggressive glioma is glioblastoma (GBM). Even after the current standard of care (maximal safe resection followed by adjuvant chemo-/radiation therapy), the median survival of patients with GBM is only 14.6 months30.
Invasive glioma cells are integrated with components of the tumor microenvironment, including the extracellular matrix (ECM)18. The ECM is usually composed of collagens9. Paulus et al. reported associations of collagen with tumor vessels in GBM17. These authors later reported the association of collagen in glioblastoma cells with tumor adhesion26. While these few reports provide evidence that there may be a role of collagen in GBM, very little is still known, particularly regarding its spatial organization and possible role in the disease state. This is partly due to the low levels of collagen present in normal brain parenchyma, found mostly in perivascular regions18. Although collagen is not abundantly expressed in normal brain, traditional glioma cell lines show upregulated collagen gene expression3.
While collagen gene expression has been shown to be upregulated in traditional glioma lines3,16, these cell lines are not the optimal models for GBM11. Recent reports have shown that GBM is a genetically heterogeneous tumor with different clinically relevant molecular subtypes7. We reported classification of GBM subtypes based on GBM stem-like cells (GSC) that show highly efficient tumor initiation and therapeutic resistance34, and the utility of GSCs for discovering clinically relevant biomarkers19. An advantage of the GSC xenograft model is that it maintains the genotype and phenotype of a GBM obtained from a patient more closely than traditional glioma cell lines and is likely to yield more clinically relevant results11.
Furthermore, even though collagen is implicated in gliomagenesis, direct visualization of collagen architecture in GBMs has not been previously reported. This is key, as direct spatial studies of collagen morphology have great promise to show how collagen organization might differ in GBMs and provide cues to possible tumor heterogeneity. Direct visualization of collagen in other cancers, such as breast cancer, has been performed and found useful as a prognostic signature5,12,20. Based on this, we hypothesized that collagen might also be a prognostic marker for GBM.
In this study, two independent techniques (Picrosirius red and second harmonic generation [SHG] microscopy) were used to analyze GSC-derived xenografts. Picrosirius red is a widely accepted histological technique used to detect collagen with polarized light14,23. SHG signal is generated when two photons of incident light interact with the non-centrosymmetric structure of collagen fibers, such that the resulting photons are half the wavelength of the incident photons21. SHG microscopy has been used for direct collagen visualization in other tumors5,21 and has the distinct advantages of nondestructive, high-speed intrinsic signature analysis (no stains needed) of thicker sections (improved optical sectioning). We show GBM collagen can be directly visualized, and different collagen alignments are correlated with tumor invasiveness. This study also examines collagen’s correlation with patient survival, establishing that patients with more organized GBM collagen survive longer than patients with less organized GBM collagen.
Methods
Ethics statement
All procedures performed in studies involving human participants were in accordance with the ethical standards of the University of Wisconsin-Madison (UW) Institutional Review Board and in compliance with the 1964 Helsinki declaration and its later amendments. Informed consent was obtained from all individual participants included in the study. All procedures performed in studies involving animals were in accordance with the ethical standards of UW.
Isolation of Glioblastoma Stem-Like Cells (GSC)
GBM stem-like cells were isolated according to previously reported protocols with a few modifications4,7. Tumor tissue was collected directly from the operating room, weighed, coarsely minced using a scalpel blade, and subsequently chopped twice at 200 µm using a tissue chopper (Sorvall TC-2 Smith-Farquahar). Chopped tissue was directly plated in suspension at 10 mg/ml in growth medium (passage medium: 70% DMEM-high glucose, 30% Ham's F12, 1× B27 supplement, 5 µg/ml heparin, penicillin-streptomycin-amphotericin (PSA), and 20 ng/ml each of epidermal growth factor and basic fibroblast growth factor). Cultures were passaged approximately every 7 to 10 days by tissue chopping twice at 200 µm. For these studies, we used four different GSC cell lines (12.1 GSC, 22 GSC, 44 GSC, and 107 GSC). Each cell line was validated for self-renewal by neurosphere formation, multipotency, and tumor initiation (see below) before experiments were performed. Establishing of cell cultures came from cryopreservation of cell cultures ranging from passages 15 to 22. Cells used for experiments ranged from passage 20 to 25.
Orthotopic Glioblastoma Xenograft Model
GBM xenografts were initiated as previously described4,34, under a protocol approved by the UW Institutional Animal Care and Use Committee. Briefly, GBM GSCs were enzymatically dissociated to single cells, and 2×105 cells were suspended in 5 µl of phosphate-buffered saline. Using a Hamilton syringe, cells were stereotactically implanted into the right striatum of anesthetized non-obese diabetic severe combined immunodeficient (NOD SCID) mice at 0.33 µl/min at the following coordinates (with respect to bregma point): 0 mm anteroposterior, +2.5 mm mediolateral, and −3.5 mm dorsoventral. AT least 3 mice were examined for each cell line. At either 3 months or onset of neurological symptoms, tumor formation was verified using MRI. Mice were anesthetized, underwent administration of contrast enhanced using 10 mmol/kg of intraperitoneal gadolinium (Omniscan; GE Healthcare, Piscataway, NJ), and placed onto a small-animal MRI scanner (4.7-T horizontal bore imaging/spectroscopy system; Varian, Palo Alto, CA), and T1- and T2-weighted images were obtained. As per animal protocol, once MRI showed tumor xenograft growth or when neurological symptoms were observed, NOD SCID mice were immediately euthanized by perfusion fixation with 4% paraformaldehyde, even if animals were asymptomatic. Brains were then excised, embedded in paraffin, and processed for general histology. Survival analysis was done using a log-rank test and presented as a Kaplan-Meier survival plot with the addition of hazard ratio analysis; p-values <0.05 were considered statistically significant. Plots were generated using GraphPad Prism 6.
Glioblastoma Tissue Microarray (TMA)
With institutional review board approval, we created a clinically annotated GBM tissue microarray (TMA) from 205 GBMs resected between 1999 and 2009, archived in the UW Department of Pathology and Laboratory Medicine and clinical data accessed from UW patient records. One to three representative tissue punches/cores were obtained for each tumor sample depending on morphologic heterogeneity and tissue availability. Neuropathology designated patient diagnosis and tissue punch location was determined based on the most representative section of the whole GBM specimen. Each additional tissue punch contained classic GBM features (nuclear atypia, high mitotic indices, vascular endothelial proliferation, and/or necrosis). Grades II and III astrocytoma, grade II oligodendroglioma, meningioma, hippocampus and neocortex tissue punches were used as controls. Of the patients on the TMA, 149 have a recorded value for length of survival. Based on histological processing, 111 patient samples were used in SHG collagen analysis. Patients were divided into Group A and Group B based on fribllar collagen appearance. Patients in Group A had collagen with a typical fibrillar collagen appearance; whereas those in Group B had collagen that was atypical fibrillar collagen was represented by a punctate or no collagen signal. In cases of multiple punches/cores for one tumor sample, at least one tissue punch showed fibrillar collagen to classify the patient into group A. Survival analysis was done as described above. Multivariate analysis with Cox’s regression model using SPSS was also performed, taking into account patient sex, age, Karnofsky Performance Scale (KPS) score, temozolomide treatment, radiation treatment, alcohol use, and tobacco use.
SHG Analysis
A custom multiphoton workstation at the UW Laboratory for Optical and Computational Instrumentation (LOCI) was used to image GBM tissue microarray slides with SHG microscopy using a TE300 inverted microscope (Nikon) equipped with a CFI Plan Fluor ×10 (numerical aperture= 0.5; Nikon) objective lens and a CFI Plan Apo ×20 (numerical aperture = 0.75; Nikon) objective lens and a mode-locked Ti: Sapphire laser (Mai Tai Deepsee; Spectra Physics). The excitation wavelength was tuned to 890 nm; a 445 nm ± 25 nm narrow bandpass emission filter (Semrock) was used to detect the SHG signal of collagen in the backscattered mode using a H7422P GaAsP photon counting PMT (Hamamatsu Photonics). Images of 1024 × 1024 pixels were acquired using WiscScan (LOCI, University of Wisconsin, Madison).
Masson’s Trichrome Staining, Picrosirius Red Staining, and Immunohistochemistry
According to an approved animal protocol (University of Wisconsin Institutional Animal Care and Use Committee), NOD SCID mice whose brains were implanted with 200,000 GSCs from GSC lines 12.1, 22, 44, and 107 were euthanatized for paraffin embedding, and 5-µm thick tissue sections were obtained in the UW Experimental Pathology Shared Service Translational Research Initiatives in Pathology (TRIP) laboratory. Masson’s trichrome staining was performed by UW Carbone Cancer Center experimental pathology shared services on paraffin-embedded samples. For Picrosirius red staining, sections were deparaffinized in xylene and hydrated through decreasing grades of alcohol. These sections were incubated in 0.1 % (wt/vol) Sirius red F3B (Bayer chemicals) in saturated Picric acid solution for 1 hour at room temperature and then rinsed with distilled water, and stained with Mayer’s haematoxylin, followed by differentiation in 1% Hcl, alkalinization with tap water, dehydration, and final mounting. Sections were then examined using circular polarized light. The same methods of staining were used for TMA slides. Immunohistochemistry was performed as previously described28. A human specific anti-NUMA antibody (ab36999, Abcam) was used at the concentrations of 1:200 on mouse xenograft slides.
Collagen Fiber Analysis
CT-FIRE analysis software was used as previously described to automatically extract collagen fibers and quantify fiber angle, fiber length, fiber straightness and fiber width2. Images of the entire specimen of each tumor on the tissue microarray were used in this automated analysis to avoid observation bias. Picrosirius red images collected for 12.1 GSC-, 22 GSC-, 44 GSC- and 107 GSC-derived xenografts were converted to 8-bit images using FIJI ImageJ25. Images were processed to threshold values between 0 and 255. These images were uploaded to CT-FIRE, collagen fiber extraction parameters were set to default values except the thresh_im value=30 and thresh_xlinkbox=5. Extracted values for fiber angle, fiber length, fiber straightness and fiber width were then binned based on relative frequency for GSC-derived xenografts. Mean values were calculated from the extracted values for focal versus invasive GBM xenografts and compared. The same method was used to calculate fiber angle, length, straightness and width for the SHG collagen image used to determine correlation with patient survival; p-values <0.05 were considered statistically significant.
Results
GBM Collagen is Directly Visualized by Picrosirius Red and SHG Microscopy Better Than Masson’s Trichrome
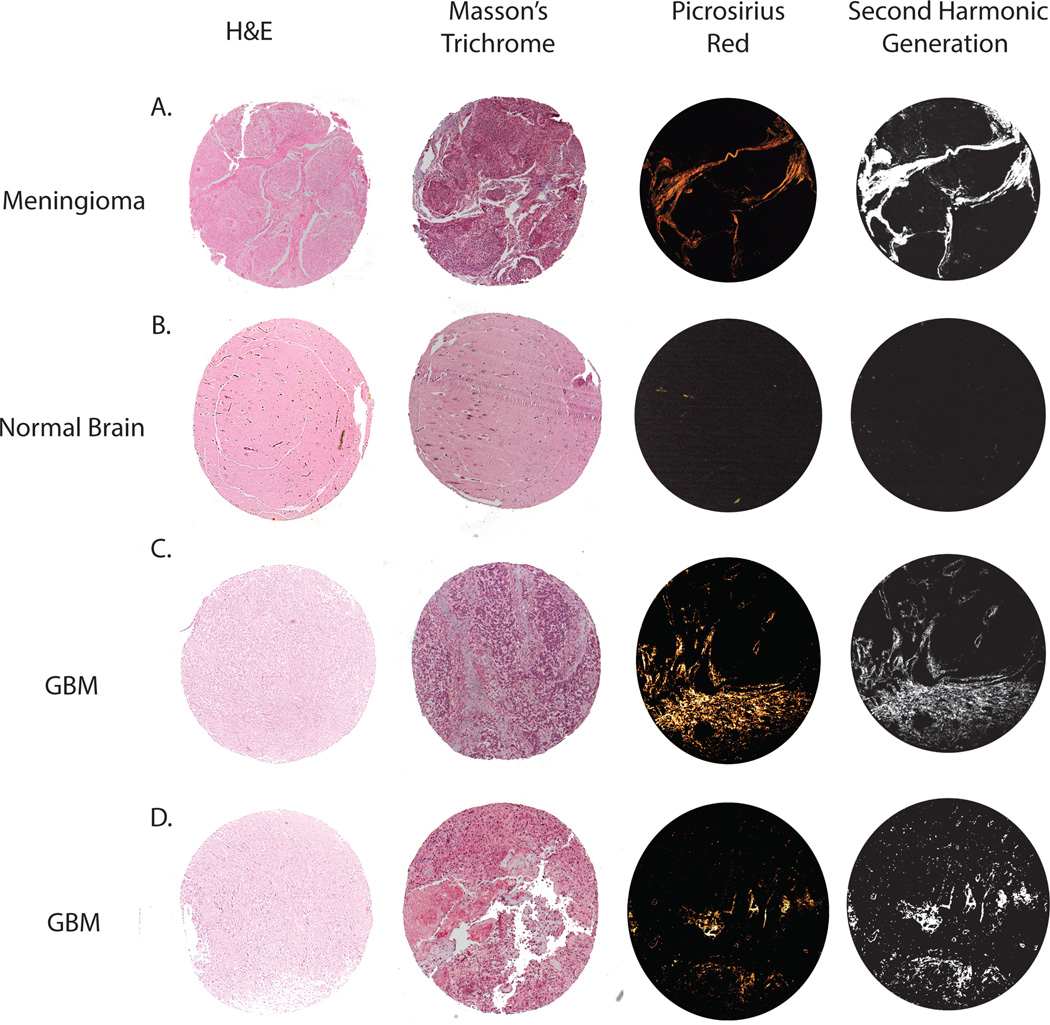
As an initial test to determine whether collagen could be directly visualized in GBM specimens, TMA sections were stained with Masson’s Trichrome and Picrosirius red. Picrosirius red is a widely accepted histological technique used to detect collagen with polarized light, whereas collagen may be missed by trichrome staining because it can be obscured by masses of other stained details14. Paraffin-embedded sections were also examined with SHG microscopy as an independent analysis method. SHG microscopy has been shown to detect collagen in other tumors5,20. Since large amounts of collagen have been shown to be present in the meninges15, meningioma specimens were chosen as a positive control. While Masson’s trichrome was able to detect collagen, very little signal was apparent. In contrast, Picrosirius red and SHG microscopy provided clear detection of collagen that was not obscured by other stained tissues. Picrosirius red and SHG microscopy appeared to detect collagen equally (Fig. 1A). Normal brain parenchyma has very low amounts of collagen, except around a few some blood vessels, and was used as a negative control. Collagen in normal brain was not detected with Masson’s trichrome. Very little collagen, only appearing around some small blood vessels, was detected with Picrosirius red and SHG microscopy (Fig. 1B). Collagen was directly visualized with Masson’s trichrome, Picrosirius red, and SHG microscopy in GBM specimens (Fig. 1C, 1D, Supplemental Fig 1). As in the control, collagen was better visualized with Picrosirius red and SHG microscopy.
Figure 1. Visualization of collagen in GBM.
Tissue specimens from 1mm core punches are used to directly visualize collagen. Masson’s trichrome, picrosirius red, and second harmonic generation microscopy detect collagen in meningioma (positive control), non-tumor brain (negative control), and GBM specimens. Corresponding H&E sections are also imaged.
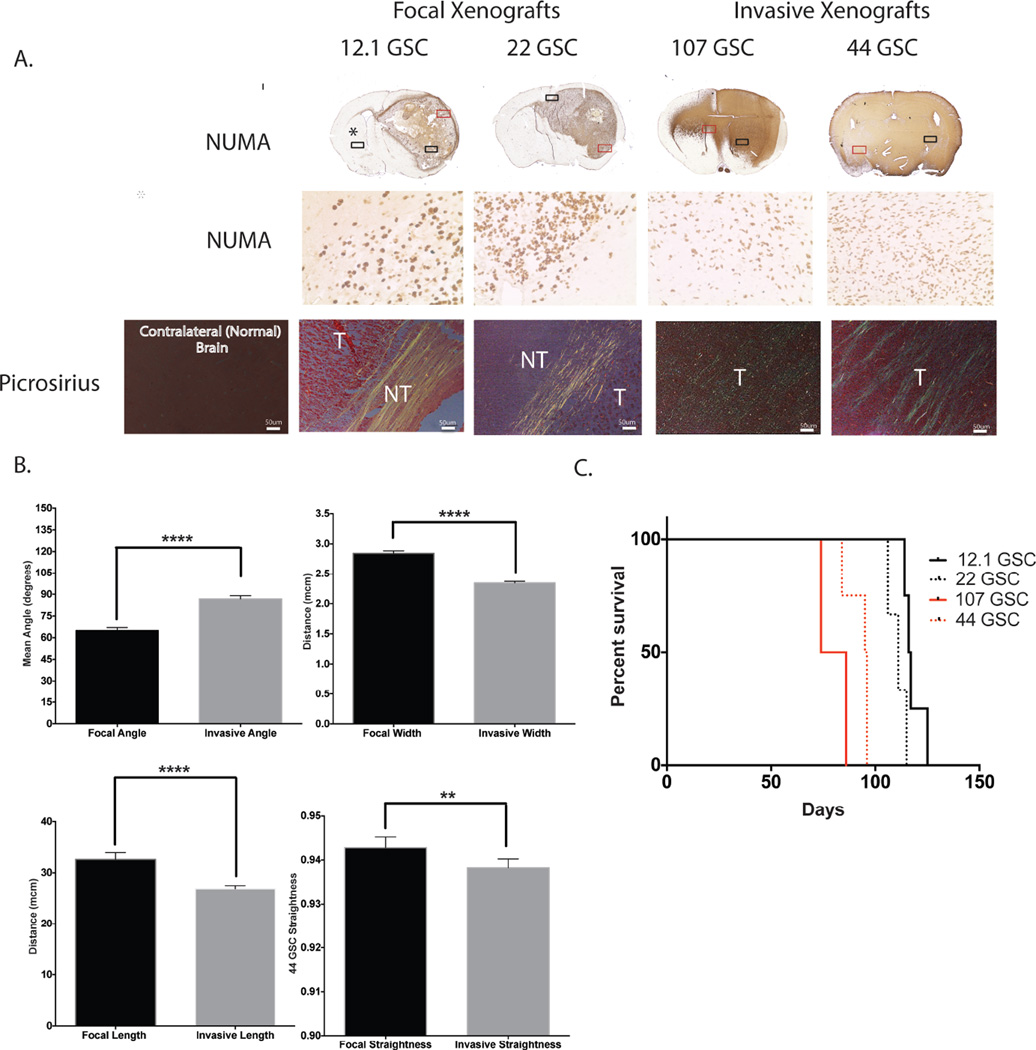
After demonstrating that collagen was present and could be directly visualized in GBM specimens, GSC lines that form more focal tumors (12.1GSC and 22 GSC lines), and highly invasive tumors (44 GSC and 107 GSC lines) were orthotopically implanted into NOD SCID mice for tumor xenograft formation. There was no difference in proliferation rates between focal and highly invasive GBM derived xenografts34. Five-micron-thick tumor xenograft sections were imaged using Picrosirius red. Collagen in focal tumors was found at the border of normal brain and tumor and appeared to encapsulate the tumor. In contrast, collagen observed in invasive tumor xenografts appeared intertwined within the tumor (Fig. 2A). Visualized collagen fibers were extracted and analyzed for fiber width, angle, length and straightness using CT-FIRE software. The mean values of collagen between focal tumors (12.1 GSC and 22 GSC lines) and invasive tumors (44 GSC and 107 GSC lines) were determined and compared. Collagen fiber width, angle between fibers, fiber length and fiber straightness were binned and the relative frequency for each bin, and mean values for each variable were determined. Comparison of focal and invasive tumors showed significant differences in fiber width, angle between fibers, and fiber length, and straightness of collagen fibers between the two groups. The mean collagen width for focal tumors was 2.85±0.02µM compared with 2.35±0.01µM for invasive tumors. The mean angle between collagen fibers for focal tumors was 64.97±1.05 degrees compared with 86.78±1.12 degrees for invasive tumors. The mean length for collagen fibers for focal tumors was 32.63±0.68µM compared with 26.89±0.31µM for invasive tumors. The mean straightness for focal collagen tumors was 0.943±0.001 compared with 0.938±0.001 for invasive tumors (a value of 1 indicates complete straightness) (Fig 2B). These numbers suggest that mice with focal tumors have more organized collagen, based on having collagen fibers being more aligned with one another, being longer, wider, and slightly straighter compared to mice with more invasive tumors (statistically significant for all measurements). Mice with focal tumors (12.1 GSC and 22 GSC lines) had a significantly better survival probability than mice with invasive tumors (44 GSC and 107 GSC lines) (p=0.0003) with HR of .2104 (95% CI 0.0086 – 0.1759) and a median survival of 115 days (N=7) for mice with focal tumors (12.1 GSC line: 116.5 days; 22GSC line: 111 days) and 90.5 days (N=6) for mice with invasive tumors (44 GSC line: 95.5 days; 107 GSC line: 80 days) (Fig 2C).
Figure 2. Visualization and analysis of collagen fibers in focal vs. invasive GBM xenografts.
(A) Focal tumors (12.1 GSC and 22 GSC) and invasive tumors (44 GSC and 107 GSC) are stained with picrosirius red. Collagen is visualized with polarized light. Human NUMA immunostaining of serial section delineate tumor cells (T=tumor, NT= non-tumor, red box= numa selection, black box= picrosirius selection). (B) Collagen fibers are extracted from focal tumors (12.1 GSC and 22 GSC) and invasive tumors (44 GSC and 107 GSC) using CT-FIRE software. Collagen fiber width, the angle between fibers, fiber length, and fiber straightness are calculated. Mean values are determined for each variable. There is a significant difference between fiber width, the angle between fibers, fiber length, and fiber straightness between focal vs. more invasive xenografts. (C) A Kaplan-Meier survival, curve for focal GBM xenografts (12.1 GSC and 22 GSC) and invasive GBM xenografts (44 GSC and 107 GSC) shows a statistically significant survival difference (p=0.0003). **=P<0.001, ****=P<0.0001
Organized Collagen Fibers Are Correlated With Better Patient Survival
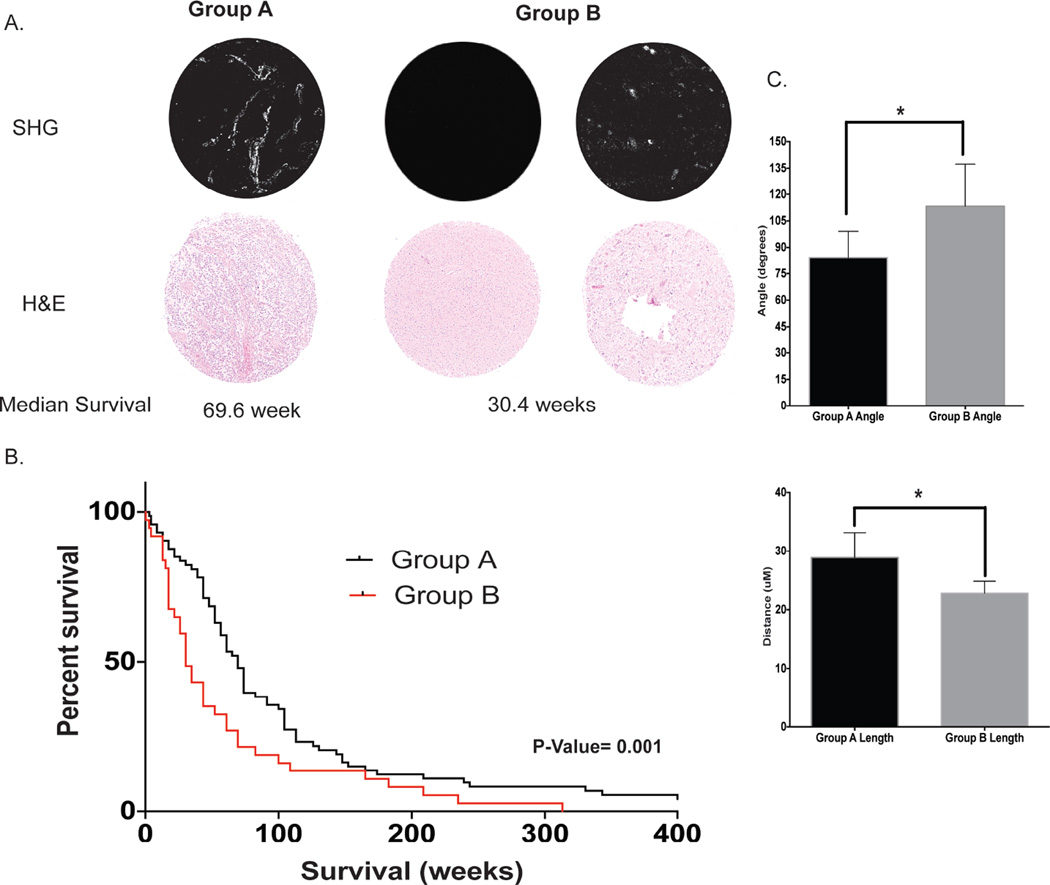
We extended the GSC-derived xenograft observations by directly visualizing collagen in patient samples in a human GBM tissue microarray (TMA) with SHG micrscopy. Two groups emerged. Patients in Group A had collagen with a typical fibrillar collagen appearance, whereas those in Group B had collagen with an atypical fibrillar collagen appearance represented by punctate or absent signal (Fig. 3A). Patient punches were blindly scored and patients in Group A demonstrated a significantly better survival probability than those in Group B (p= 0.001). The median survival of patients in Group A was 69.6 weeks (N=74) compared with 30.4 weeks (N=37) for patients in Group B (Fig 3B). Collagen fiber analysis was conducted on representative images for Group A and Group B with CT-FIRE software as previously described. There was a statistically significant difference between Groups A and B for collagen angle to other fibers, with a mean angle to other fibers of 84.37 ± 7.298 degrees and 113.2 ± 11.60 degrees respectively. There was also a statistically significant difference between Groups A and B for collagen length, with mean lengths of 28.86 ± 2.13µM and 22.79 ± 1µM respectively. There was no difference between collagen width and straightness between the two groups (Supplemental Fig. 2). Patients in Group A had more organized collagen based on having fibers that were statistically significantly more aligned with one another and that were longer. Patients with a fibrillar signal have a median survival that was better than to the overall median GBM survival of 14.6 months (63.5 weeks) for GBM patients receiving the current standard clinical care of surgery and temozolomide chemo-/radiotherapy.
Figure 3. Collagen signature correlates with patient survival.
A GBM tissue microarray with 1 mm specimen punches is used for second harmonic generation collagen imaging. (A) Groups A and B are observed. Group A is defined as typical fibrillar collagen fibers and group B is defined as atypical collagen fibers represented by punctate or no collagen. (B) Kaplan-Meier survival of the two collagen groups show a significant survival difference (p= 0.001). (C) Collagen fibers are extracted from the represented fibrillar and punctate signatures using CT-FIRE software. There is a significant difference between the angle between fibers and fiber length. *= P<0.05
Addressing Confounding variables for collagen signature with multivariate analysis
To address the potential confounding variables that might influence signature differences between Groups A and B collagen, univariate analysis and multivariate analyses were conducted. Patient sex, age, KPS score, temozolomide treatment, radiation treatment, tobacco use, and alcohol use were all assessed as potential confounding variables. Univariate analysis on the aforementioned factors was conducted to determine which factors influence patient survival rates. There was not a statistically significant difference in survival based on patient sex or alcohol use; however, there was a statistically significant difference in survival based on patient age (p=0.002), KPS score (p=0.023), temozolomide treatment (p=<0.000), radiation treatment (p=0.000) and tobacco use (p=0.008). Multivariate survival analysis using Cox’s regression model was used to determine whether collagen signature is an independent biomarker for survival taking into account patient age, KPS score, temozolomide treatment, radiation treatment and tobacco use. Based on multivariate analysis collagen signature is an independent marker for GBM patient survival with Group A having a statistically significant better survival than Group B (p=0.032) (Table 1).
Table 1.
Univariate and multivariate cox regression analysis of GBM collagen signature and confounding variables
| Univariate Analysis |
Multivariate Analysis |
|||||||
|---|---|---|---|---|---|---|---|---|
| N | Median Survival (Weeks) |
95% CI | P-Value (Log Rank) | Hazard Ratio (HR) |
HR 95% CI | P-Value | ||
| Sex | 0.833 | NS | ||||||
| Male | 78 | 52.2 | 40.6–63.8 | |||||
| Female | 33 | 73.9 | 41.3–106.5 | |||||
| Age | 0.002 | 1.244 | 0.797–1.940 | 0.331 | ||||
| ≤ 55 | 54 | 69.6 | 53.9–85.3 | |||||
| > 55 | 56 | 43.5 | 22.2–64.8 | |||||
| KPS | 0.023 | 0.944 | 0.597–1.494 | 0.843 | ||||
| ≤ 70 | 56 | 39.1 | 27.1–51.1 | |||||
| > 70 | 55 | 73.9 | 59.9–87.9 | |||||
| Temozolomide | 0.000 | 0.471 | 0.285–0.778 | 0.004 | ||||
| Yes | 69 | 73.9 | 66.1–81.7 | |||||
| No | 42 | 26.1 | 13.8–38.3 | |||||
| Radiation | 0.000 | 0.373 | 0.200–0.696 | 0.002 | ||||
| Yes | 94 | 65.2 | 54.8–75.6 | |||||
| No | 17 | 17.4 | 13.2–21.6 | |||||
| Tobacco Use | 0.008 | 1.204 | 0.755–0.1.921 | 0.572 | ||||
| Yes | 44 | 47.8 | 37.2–58.4 | |||||
| No | 67 | 60.9 | 54.1–85.1 | |||||
| Alcohol Use | 0.313 | NS | ||||||
| Yes | 23 | 43.5 | 23.2–63.8 | |||||
| No | 88 | 60.9 | 48.6–73.2 | |||||
|
Collagen Signature |
0.001 | 0.624 | 0.405–0.963 | 0.032 | ||||
| Group A | 74 | 69.6 | 59.1–80.1 | |||||
| Group B | 37 | 30.4 | 21.8–39.0 | |||||
NS = not significant
Discussion
GBM is a devastating disease with a median survival of less than two years. While collagen gene expression has been reported as upregulated in GBM, direct visualization of collagen architecture and its role in patient survival has not been reported. We demonstrated that collagen architecture can be directly visualized in GBM specimens with both Picrosirius red and the new technique of SHG microscopy in the brain. We also demonstrate that collagen structure is different in focal tumors from that in invasive tumors. Focal tumors have more organized collagen based on collagen fibers being more aligned with each other, longer, wider, and being slightly straighter. While it is difficult to know whether collagen causes a more invasive tumor type or that more invasive tumors produce different collagen variations, our data also show collagen signature is correlated with survival. Patients harboring GBMs with the more organized collagen (Group A) had a longer survival than those with GBMs with less organized collagen (Group B). Patients in Group A had GBM collagen similar to those observed in focal GBM xenografts, while patients with in Group B had GBM collagen that more closely resembled more invasive tumors. Focal tumors and patients in group A had more organized collagen, since observed collagen fibers were more aligned with each other based on the collagen angle between fibers and their fiber length was also longer compared to invasive tumors and patients in group B. This suggests that fibrillar collagen is associated with a benefit for GBM patients. Collagen fiber width was significantly greater in focal vs. more invasive GBM xenografts, even though the greater fiber width in the fibrillar GBM was not statistically significant compared to the punctate signature fiber width. While the differences noted in collagen architecture changes are small, they are significant. In breast cancer, small but statistically significant changes in collagen alignment have been shown to promote tumor invasive and to be a prognostic marker5,20. Alteration of collagen structure too much would probably result in a non-functioning protein.
Collagen typically forms a stiff extracellular matrix. Observations that the tumor is stiffer than the surrounding brain parenchyma and that cancer cells typically invade between boundaries of stiffness, such as white matter tracts and blood vessels, are seen in gliomas24. Research has shown that collagen gels, in vitro, can influence glioma cell migration24,33. Other evidence suggests that collagen scaffolding occurs in concert with an angiogenic shift that accompanies faster tumor growth in gliomas16. These previous studies and our results, on collagen differences seen in focal and fibrillar vs. more invasive and punctate/non-fibrillar GBM, suggest that collagen may influence GBM cell invasion of surrounding brain parenchyma, and that fibrillar collagen may act as a barrier to tumor invasion. Collagen barrier disruption may result in tumor invasion. GBM cells might remodel their microenvironment through collagen degradation, collagen synthesis, or exert physical forces on collagen to alter collagen structure and cause tumor invasion. Conversely, other stromal cells in the tumor’s vicinity might influence collagen production similarly to collagen production in wound healing.
Our data can not determine exactly which collagen subtypes are involved in the GBM progress, SHG only picks up on fibrillar collagens (I, II, III, V, XI, XXIV, XXVII)1,8,13,32 and picrosirius red has been shown to detect similar collagens (I, II, III, V)6,10. While it is possible that the collagen detected in GBM is related to increased tumor angiogenesis, this is unlikely since neither SHG or picrosirius red under birefringent light detect collagen IV, the major collagen seen in blood vessel basement membranes. Furthermore, collagen is not seen in regions of blood vessels seen in H&E stained slides. This suggests that the fibrillar class of collagen plays an important role in GBM progression and should be further investigated.
Our data also suggest that collagen may be a potential survival marker for patients with GBM. TMA sections were chosen as the most representative of individual patient GBM specimens; however, these punches represent only a small piece of the tumor. Use of SHG analysis may overcome these problems in the future by providing the unique ability to image collagen directly in intact tissue20, without having to stain tissue. SHG imaging can also be performed rapidly on pathology slides already previously examined with standard immunohistochemical methods. SHG also enables live imaging of cells and animals, deep tissue imaging, and imaging of paraffin embedded slides with minimal photobleaching and phototoxicity21. While the tumor is being removed in the operating room, different tumor regional biopsies could be examined with SHG intraoperatively in order to survey the entire GBM (unlike current GBM analysis that is predicated on a small portion of excised tumor). SHG could be used to examine tumor heterogeneity and account for possible sampling bias of current clinical analysis and tissue microarrays. While SHG technology can provide insight into GBM progression, multiphoton microscopes are costly and could limit the number of facilities that could do this analysis in house. For samples that can be processed with histopathology approaches, picrosirius might provide an accurate and cost effective strategy for looking at collagen alignment.
Tumor cells are frequently left behind despite maximal safe resection, contributing to tumor recurrence in the majority of patients. While more studies need to be conducted, the correlation with focal tumors with fibrillar collagen signature and invasive tumors with punctate collagen signature, suggests that collagen signature might also be correlated with tumor invasion and used in optimizing surgeries. Furthermore, GBM classification is changing with new data revealing individual patient heterogeneity of these tumors. Collagen may be used as part of a new classification system of GBMs in the future to aid patients and physicians with better understanding of prognosis and individualize treatments. Understanding the role of collagen in GBM may lead to better therapeutic targets as well. Drugs that target the tumor microenvironment, such as COX-2 inhibitors, have therapeutic advantages in GBM27,31. Thus, drug development targeted at increasing collagen production or related mechanisms may improve patient survival.
As the collagen alignment story in GBMs is better understood, future research on detection approaches that are compatible with medical imaging need to be pursued. Optical imaging is currently necessary to detect the high-resolution changes we report in GBM collagen architecture. While there are possible future avenues to explore in optical brain imaging that could clinically detect collagen such as Optical Coherence Tomography (OCT) and endoscopy, potential use of lower resolution but deeper imaging approaches such as ultrasound to examine bulk collagen density is also being explored22.
This study provides some insight into the different collagen topologies in GBM and their correlation to GBM patient survival; however, further research is required to elucidate the mechanisms determining collagen’s role in GBM is the necessary next step. While we have considered some of the confounding variables that influence GBM survival, there are likely others that may play a role. For instance, we cannot readily discern the patient subgroups presenting with imaging showing radiologically less invasive or more invasive patterns. Analysis with current and possibly more advanced clinical imaging to determine clinically useful and relevant parameters in the context of collagen classification might further increase collagen’s validity as a clinical biomarker, and link it to known GBM biomarkers such as MGMT methylation and IDH mutation status. Additionally, a limited panel of patient-derived glioblastoma cancer stem cell-generated xenografts were used as an initial screen for possible collagen differences in different patient GBMs. The observed collagen difference in more focal vs. more invasive GBM xenografts suggests that more mechanistic studies with a large number of patient-derived lines will be useful to robustly determine the signaling or biomechanical properties that underlie those differences. Further investigations of collagen changes over temporal and spatial tumor heterogeneity are also interesting and planned. Planned future studies using chemical or genetic strategies to alter collagen expression and structure are important to determine how collagen architecture could affect GBM tumorigenesis and clinical outcomes. Our study is the first to correlate fibrillar collagen with patient survival and is an important initial step to understand how collagen is associated with clinically relevant GBM biology.
Conclusions
More organized collagen is associated with less invasive GBM xenografts and longer patient survival. These findings suggest that collagen is important for GBM progression and may be a useful survival marker for patients.
Supplementary Material
Acknowledgments
We also wish to acknowledge Drs. Corinne Vokoun, Yuming Liu, Anna Huttenlocher, Gail Robertson, Ruth Sullivan, and Amy Schendel for project advice.
Funding:
Kelli Pointer: NIH Grants R01NS75995 and T32GM008692, Laboratory for Optical and Computational Instrumentation (LOCI)
Paul Clark: UW Carbone Cancer Center UL1RR025011 and P30CA014520
Kevin Eliceiri: Laboratory for Optical and Computational Instrumentation (LOCI)
John Kuo: NIH Grant R01NS75995, UW Carbone Cancer Center UL1RR025011 and P30CA014520, Headrush Brain Tumor Research Professorship, Roger Loff Memorial Fund “Farming against Brain Cancer”
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Portions of this work were presented in abstract form at the 2014 Cancer Stem Cell Conference in Cleveland, Ohio and the 2014 Society for Neuro-Oncology Conference in Miami, Florida.
References
- 1.Ajeti V, Nadiarnykh O, Ponik SM, Keely PJ, Eliceiri KW, Campagnola PJ. Structural changes in mixed Col I/Col V collagen gels probed by SHG microscopy: implications for probing stromal alterations in human breast cancer. Biomed Opt Express. 2011;2:2307–2316. doi: 10.1364/BOE.2.002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bredfeldt JS, Liu Y, Pehlke CA, Conklin MW, Szulczewski JM, Inman DR, et al. Computational segmentation of collagen fibers from second-harmonic generation images of breast cancer. J Biomed Opt. 2014;19:16007. doi: 10.1117/1.JBO.19.1.016007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chernov AV, Baranovskaya S, Golubkov VS, Wakeman DR, Snyder EY, Williams R, et al. Microarray-based transcriptional and epigenetic profiling of matrix metalloproteinases, collagens, and related genes in cancer. J Biol Chem. 2010;285:19647–19659. doi: 10.1074/jbc.M109.088153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark PA, Iida M, Treisman DM, Kalluri H, Ezhilan S, Zorniak M, et al. Activation of multiple ERBB family receptors mediates glioblastoma cancer stem-like cell resistance to EGFR-targeted inhibition. Neoplasia. 2012;14:420–428. doi: 10.1596/neo.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conklin MW, Eickhoff JC, Riching KM, Pehlke CA, Eliceiri KW, Provenzano PP, et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol. 2011;178:1221–1232. doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dayan D, Hiss Y, Hirshberg A, Bubis JJ, Wolman M. Are the polarization colors of picrosirius red-stained collagen determined only by the diameter of the fibers? Histochemistry. 1989;93:27–29. doi: 10.1007/BF00266843. [DOI] [PubMed] [Google Scholar]
- 7.Denysenko T, Gennero L, Roos MA, Melcarne A, Juenemann C, Faccani G, et al. Glioblastoma cancer stem cells: heterogeneity, microenvironment and related therapeutic strategies. Cell Biochem Funct. 2010;28:343–351. doi: 10.1002/cbf.1666. [DOI] [PubMed] [Google Scholar]
- 8.Gong B, Sun J, Vargas G, Chang Q, Xu Y, Srivastava D, et al. Nonlinear imaging study of extracellular matrix in chemical-induced, developmental dissecting aortic aneurysm: evidence for defective collagen type III. Birth Defects Res A Clin Mol Teratol. 2008;82:16–24. doi: 10.1002/bdra.20408. [DOI] [PubMed] [Google Scholar]
- 9.Halfter W, Candiello J, Hu H, Zhang P, Schreiber E, Balasubramani M. Protein composition and biomechanical properties of in vivo-derived basement membranes. Cell Adh Migr. 2013;7:64–71. doi: 10.4161/cam.22479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Junqueira LC, Bignolas G, Brentani RR. Picrosirius staining plus polarization microscopy, a specific method for collagen detection in tissue sections. Histochem J. 1979;11:447–455. doi: 10.1007/BF01002772. [DOI] [PubMed] [Google Scholar]
- 11.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Leeming DJ, Koizumi M, Qvist P, Barkholt V, Zhang C, Henriksen K, et al. Serum N-Terminal Propeptide of Collagen Type I is Associated with the Number of Bone Metastases in Breast and Prostate Cancer and Correlates to Other Bone Related Markers. Biomark Cancer. 2011;3:15–23. doi: 10.4137/BIC.S6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutz V, Sattler M, Gallinat S, Wenck H, Poertner R, Fischer F. Characterization of fibrillar collagen types using multi-dimensional multiphoton laser scanning microscopy. Int J Cosmet Sci. 2012;34:209–215. doi: 10.1111/j.1468-2494.2012.00705.x. [DOI] [PubMed] [Google Scholar]
- 14.Montes GS, Junqueira LC. The use of the Picrosirius-polarization method for the study of the biopathology of collagen. Mem Inst Oswaldo Cruz. 1991;86(Suppl 3):1–11. doi: 10.1590/s0074-02761991000700002. [DOI] [PubMed] [Google Scholar]
- 15.Nam MH, Baek M, Lim J, Lee S, Yoon J, Kim J, et al. Discovery of a novel fibrous tissue in the spinal pia mater by polarized light microscopy. Connect Tissue Res. 2014;55:147–155. doi: 10.3109/03008207.2013.879864. [DOI] [PubMed] [Google Scholar]
- 16.Noreen R, Chien CC, Chen HH, Bobroff V, Moenner M, Javerzat S, et al. FTIR spectro-imaging of collagen scaffold formation during glioma tumor development. Anal Bioanal Chem. 2013;405:8729–8736. doi: 10.1007/s00216-013-7337-8. [DOI] [PubMed] [Google Scholar]
- 17.Paulus W, Roggendorf W, Schuppan D. Immunohistochemical investigation of collagen subtypes in human glioblastomas. Virchows Arch A Pathol Anat Histopathol. 1988;413:325–332. doi: 10.1007/BF00783025. [DOI] [PubMed] [Google Scholar]
- 18.Payne LS, Huang PH. The pathobiology of collagens in glioma. Mol Cancer Res. 2013;11:1129–1140. doi: 10.1158/1541-7786.MCR-13-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pointer KB, Clark PA, Zorniak M, Alrfaei BM, Kuo JS. Glioblastoma cancer stem cells: Biomarker and therapeutic advances. Neurochem Int. 2014;71:1–7. doi: 10.1016/j.neuint.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Provenzano PP, Eliceiri KW, Campbell JM, Inman DR, White JG, Keely PJ. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Provenzano PP, Eliceiri KW, Keely PJ. Multiphoton microscopy and fluorescence lifetime imaging microscopy (FLIM) to monitor metastasis and the tumor microenvironment. Clin Exp Metastasis. 2009;26:357–370. doi: 10.1007/s10585-008-9204-0. [DOI] [PubMed] [Google Scholar]
- 22.Reusch LM, Feltovich H, Carlson LC, Hall G, Campagnola PJ, Eliceiri KW, et al. Nonlinear optical microscopy and ultrasound imaging of human cervical structure. J Biomed Opt. 2013;18:031110. doi: 10.1117/1.JBO.18.3.031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rich La WP. Collagen and Picrosirius red staining: A polarized light assessment of fibrillar hue and spatial distribution. Braz J morphol Sci. 2005;22:97–104. [Google Scholar]
- 24.Rubenstein BM, Kaufman LJ. The role of extracellular matrix in glioma invasion: a cellular Potts model approach. Biophys J. 2008;95:5661–5680. doi: 10.1529/biophysj.108.140624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senner V, Ratzinger S, Mertsch S, Grassel S, Paulus W. Collagen XVI expression is upregulated in glioblastomas and promotes tumor cell adhesion. FEBS Lett. 2008;582:3293–3300. doi: 10.1016/j.febslet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Sharma V, Dixit D, Ghosh S, Sen E. COX-2 regulates the proliferation of glioma stem like cells. Neurochem Int. 2011;59:567–571. doi: 10.1016/j.neuint.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 28.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 29.Society AC. Cancer Facts and Figures 2009. American Cancer Society. 2009 [Google Scholar]
- 30.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki K, Gerelchuluun A, Hong Z, Sun L, Zenkoh J, Moritake T, et al. Celecoxib enhances radiosensitivity of hypoxic glioblastoma cells through endoplasmic reticulum stress. Neuro Oncol. 2013;15:1186–1199. doi: 10.1093/neuonc/not062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tiaho F, Recher G, Rouede D. Estimation of helical angles of myosin and collagen by second harmonic generation imaging microscopy. Opt Express. 2007;15:12286–12295. doi: 10.1364/oe.15.012286. [DOI] [PubMed] [Google Scholar]
- 33.Yang YL, Motte S, Kaufman LJ. Pore size variable type I collagen gels and their interaction with glioma cells. Biomaterials. 2010;31:5678–5688. doi: 10.1016/j.biomaterials.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 34.Zorniak M, Clark PA, Leeper HE, Tipping MD, Francis DM, Kozak KR, et al. Differential expression of 2',3'-cyclic-nucleotide 3'-phosphodiesterase and neural lineage markers correlate with glioblastoma xenograft infiltration and patient survival. Clin Cancer Res. 2012;18:3628–3636. doi: 10.1158/1078-0432.CCR-12-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.