Abstract
Myeloid, CD1a-sorted dendritic cells (MDC) productively replicated human immunodeficiency virus strains encoding envelope genes of either primary X4R5 or R5 strains for up to 45 days. Cell-free supernatant collected from long-term infected MDC, which had been exposed to an X4R5 virus 45 days earlier, was still infectious when placed over activated T cells. These data imply that DC can act as a persistent reservoir of infectious virus.
Dendritic cells (DC) play a crucial role in the immune defense against pathogens (4, 24). Two subsets, plasmacytoid DC and myeloid DC (MDC), were identified in humans based on their phenotypes (5). MDC are involved in uptake, processing, and presentation of foreign antigens. Immature MDC are disseminated throughout tissues, where they capture antigens, mature, and home to lymph nodes; DC then present the processed antigens to T cells (20).
A number of groups have examined the ability of MDC to interact with human immunodeficiency virus (HIV) (1, 2, 6, 17, 23, 30, 33). Recent studies demonstrated that HIV replication depends on the stage of DC differentiation (3, 19, 21). DC express CD4 and the chemokine receptors CCR5 and CXCR4 (2, 13, 29, 34); the expression of the latter molecules varies with the DC maturation stage (3, 7), thus influencing the cells' susceptibility to productive HIV infection. Also, HIV can interact with DC via the C-type lectin DC-SIGN without undergoing replication; captured virus remains infectious for days and can be transmitted to T cells (12).
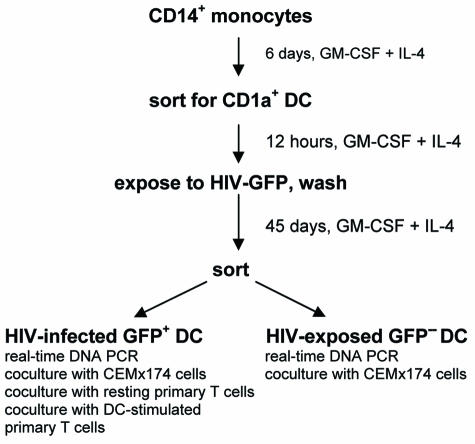
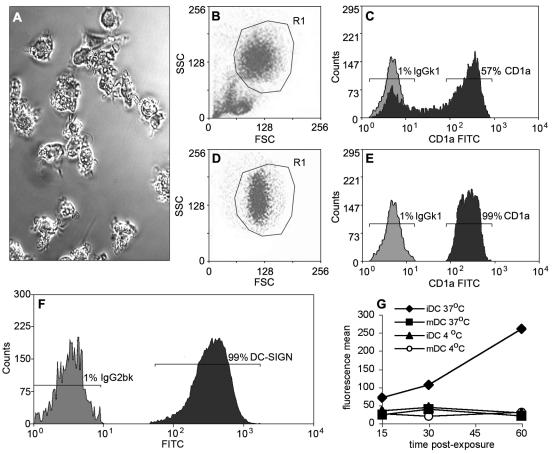
The aim of this study was to determine the period of time during which highly purified MDC can support replication of HIV to explore whether virus-exposed MDC could represent a potential long-lived reservoir of infectious HIV. For this purpose, we established long-term cultures of MDC that were differentiated from peripheral blood monocytes (Fig. 1). After 6 days of differentiation in complete RPMI 1640 medium supplemented with interleukin-4 (IL-4) and granulocyte-macrophage colony-stimulating factor (GM-CSF) (25, 26), we observed cells with a veiled appearance (Fig. 2A). These cells were purified by positive selection against CD1a (Fig. 2B to E). Staining of CD1a-sorted MDC with monoclonal antibodies (MAbs) against CD3, CD14, and CD19 did not show contamination with T cells, macrophages, and B cells (data not shown). After sorting, these MDC were DC-SIGN+ (Fig. 2F) and expressed markers typical for immature MDC; a minor fraction of these cells expressed CCR5 or CXCR4 (Table 1). CD1a-sorted MDC performed macropinocytosis efficiently; when the cells were matured with lipopolysaccharide (LPS), their ability to perform macropinocytosis decreased dramatically (Fig. 2G). Thus, our DC were highly pure and exhibited properties of immature MDC.
FIG. 1.
Experimental design schema. MDC were differentiated from monocytes by culturing 3 × 107 CD14+ cells/flask in 75-cm3 flasks (Costar-Falcon, Franklin Lakes, N.J.) for 6 days in complete RPMI 1640 medium (Gibco BRL, Invitrogen, Gaithersburg, Md.) supplemented with 10% fetal calf serum (Sigma, St. Louis, Mo.), 100 U of penicillin/ml, 100 μg of streptomycin/ml, 2 mM l-glutamine (both from Gibco BRL), and 20-ng/ml concentrations of both IL-4 and granulocyte-macrophage colony-stimulating factor (GM-CSF) (Stem Cell Technology, Vancouver, Canada) (MDC medium). On day 6, cells were stained with anti-CD1a mouse MAbs conjugated with fluorescein isothiocyanate (BD Pharmingen, San Diego, Calif.) and purified by positive selection for CD1a. Twelve hours later, purified MDC (3 × 107 cells) were exposed to dual-tropic HIV-GFP (0.5 50% tissue culture infectious doses per cell). HIV-exposed cells were cultured in MDC medium in 75-cm3 flasks (Costar-Falcon) for 45 days. Infected (GFP+) DC were isolated by flow cytometry. Both GFP+ and GFP− fractions (5 × 103 cells) were analyzed by real-time DNA PCR and cultured in the top well of 24-well transwell units (Costar-Falcon) with 2 × 105 CEMx174 cells in the bottom well. In parallel, GFP+ MDC (2 × 103 cells) were placed into the top wells of transwell units, and autologous T cells alone or autologous T cells (2 × 105 cells) mixed with autologous, freshly differentiated MDC (2 × 104 cells) were placed into the bottom wells.
FIG. 2.
Differentiation, purification, and characterization of MDC. (A) Phase-contrast microscopy of MDC on day 6, prior to CD1a sorting (magnification, ×20). (B) forward scatter (FSC) versus side scatter (SSC) of MDC before sorting. (C) MDC were stained with fluorescein isothiocyanate (FITC)-conjugated mouse isotype control antibody (BD Pharmingen) (light plot) and an anti-CD1a mouse MAb conjugated with FITC (BD Pharmingen) (150 μg of MAb per 2 × 107 cells) (dark plot). (D) The gated population of CD1a-stained MDC (R1) was sorted using a MoFlow cytosorter (Cytomation, Fort Collins, Colo.). (E) CD1a-sorted MDC (dark plot) are shown compared to isotype control (light plot). (F) CD1a-sorted MDC (105 cells) were stained with FITC-conjugated mouse anti-human DC-SIGN MAb and FITC-conjugated mouse isotype control antibody (both from BD Pharmingen) (dark and light histogram plots, respectively) and analyzed by flow cytometry. (G) Macropinocytosis by CD1a-sorted MDC. Immature MDC (iDC) (5 × 105 cells/well in 24-well plates; Costar-Falcon) were incubated in duplicates in the presence or absence of LPS with 2.5 mg of fluorescein-isothiocyonate-dextran (Sigma)/ml for 1 h at 37°C or at 4°C. After 15, 30, and 60 min, cells were washed three times with phosphate-buffered saline and 1% fetal calf serum and analyzed by flow cytometry. One out of three representative experiments is shown. mDC, DC matured with LPS.
TABLE 1.
Surface marker expression of uninfected MDC during long-term culture
| Donor and surface marker | % Positive cells at the following no. of days in culture:
|
||
|---|---|---|---|
| 1a | 30 | 60 | |
| Donor 1 | |||
| DC-SIGN | 98.6 | 98.1 | 66.9 |
| CD1a | 94.8 | 14.3 | 0 |
| HLA-DR | 70.5 | 98.5 | 75.7 |
| CD80 | 15.7 | 4.0 | 1.4 |
| CD83 | 1.1 | 6.8 | 11.5 |
| CD4 | 56.1 | 3.8 | 2.5 |
| CCR5 | 24.8 | 4.8 | 2.1 |
| CXCR4 | 17.4 | 14.8 | 7.2 |
| Donor 2 | |||
| DC-SIGN | 97.6 | 96.8 | 75.2 |
| CD1a | 95.4 | 45.1 | 12.0 |
| HLA-DR | 75.0 | 85.8 | 78.2 |
| CD80 | 25.0 | 30.5 | 10.0 |
| CD83 | 5.8 | 12.5 | 45.3 |
| CD4 | 30.5 | 45.0 | 15.0 |
| CCR5 | 20.2 | 8.8 | 2.6 |
| CXCR4 | 2.3 | 34.2 | 18.2 |
Day 0 was designated the day of CD1a sorting.
We exposed the CD1a-sorted MDC to HIV-GFP, an X4R5 virus containing the envelope gene of the dual-tropic, primary HIV isolate 89.6 (9) and the gene for green fluorescent protein (GFP). Like the parental HIV89.6, HIV-GFP was able to use both CXCR4 and CCR5 for virus entry, while the reporter gene GFP placed in lieu of nef allowed detection of infection at the single-cell level. Because we wanted to show that DC could represent a reservoir of productive HIV replication during chronic infection, we chose this dual-tropic HIV strain as a representative of a primary HIV isolate that can be found during chronic infection. Throughout disease progression, some HIV strains expand their usage of coreceptors for viral entry, shifting from exclusive use of CCR5 to use of CXCR4; this shift has been described for approximately half of the HIV clade B-infected individuals with progressive disease (10, 27, 32).
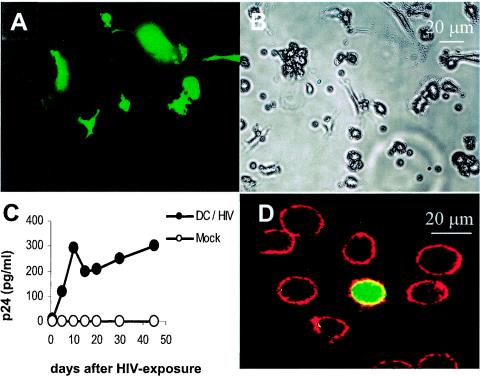
Our purified MDC were highly positive for DC-SIGN and expressed both CCR5 and CXCR4 (Fig. 2F; Table 1). As expected, these MDC were productively infected by the dual-tropic X4R5 HIV-GFP (Fig. 3). Virus-exposed MDC were cultured for 45 days, and throughout this time period, GFP-expressing and hence productively infected MDC were detected (Fig. 3A and B). In general, between 1.5 to 4.5% of the virus-exposed MDC were GFP+ and thus HIV+. Supernatants from infected DC collected at various time points contained p24 Gag that peaked on day 10 and was present in culture supernatants during the next 35 days (Fig. 3C). MDC on day 30 still expressed DC-SIGN at high levels (Table 1), and we were able to colocalize GFP and DC-SIGN, indicating productive HIV infection of DC at the single-cell level (Fig. 3D). On day 45 of culture, the viability of virus-exposed MDC and that of unexposed controls was 63 and 70%, respectively (Table 2). Both cultures were CD14 negative; no surface CD14 expression was found on GFP-expressing, HIV-infected cells (data not shown). The absolute number of virus-exposed and nonexposed control cells decreased from 3 × 107 on day one to 1.9 × 107 and 2.1 × 107 on day 45 after viral exposure, respectively. These data indicate that MDC can be maintained in culture for extended periods of time with the majority of the cells remaining viable. Furthermore, productive HIV replication by a small fraction of GFP+ cells did not appreciably influence the long-term viability of the entire culture.
FIG. 3.
Long-term productive HIV infection of MDC. CD1a-sorted MDC were exposed to 0.5 50% tissue culture infectious doses per MDC of the dual-tropic strain HIV-GFP, which encodes GFP in lieu of nef. The cells were thoroughly washed and incubated in 10 ml of DC medium in 75-cm3 flasks (Costar-Falcon). Fresh DC medium (1 ml) was added every third day throughout the time of the experiment without removing medium. To visualize DC productively infected with HIV-GFP on day 45 postinoculation, MDC were subjected to fluorescent (A) and phase-contrast (B) microscopy (magnification, ×20). (C) Aliquots of supernatants (200 μl) from HIV-exposed MDC cultures were collected on days 5, 10, 15, 20, 30, and 45 and assayed for p24 Gag by enzyme-linked immunosorbent assay (Beckman Coulter, Miami, Fla.); the p24 levels were corrected for the dilution of the medium during continued long-term culture. (D) Surface expression of DC-SIGN on MDC was tested 30 days after virus exposure. Briefly, 5 × 104 cells were washed with phosphate-buffered saline supplemented with 2% bovine serum albumin (Sigma) (PBS-BSA) and incubated with 0.5 μg of mouse anti-DC-SIGN MAb conjugated with phycoerythrin (BD Pharmingen) for 30 min without cell permeabilization at 4°C in the dark. After washing with PBS-BSA, images were analyzed by fluorescent microscope (Nikon Instech Co., Kawasaki, Japan) at a magnification of ×40 and processed with the MetaMorph software (Universal Imaging Corp., Downingtown, Pa.). Results of one out of four representative experiments are shown.
TABLE 2.
Viability of HIV-infected and nonexposed MDC during long-term culture
| Myeloid DC | % Viability at the following no. of days in culture:a
|
|||||
|---|---|---|---|---|---|---|
| 1 | 10 | 20 | 30 | 45 | 60 | |
| Noninfected cells | 98 | 95 | 80 | 76 | 70 | 64 |
| HIV-exposed cells | 97 | 86 | 76 | 68 | 63 | 55 |
Viability of cells in the long-term MDC culture was determined by staining of noninfected and HIV-exposed MDC with 0.4% trypan blue.
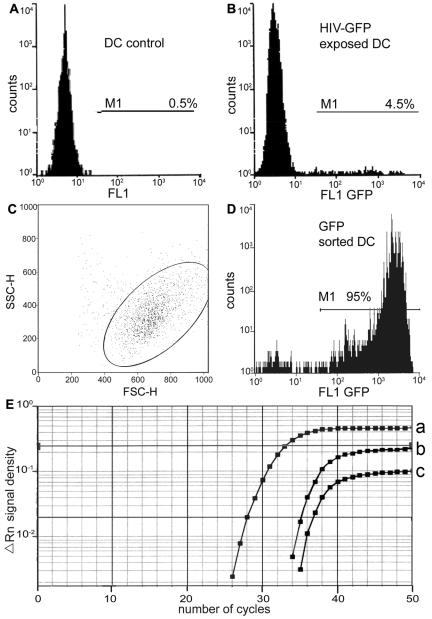
The use of dual-tropic HIV-GFP allowed us to examine whether long-term productively infected MDC could transfer viral progeny to T cells. On day 45 after virus exposure, MDC were separated by flow cytometry into GFP+ and GFP− fractions (Fig. 1 and Fig. 4B, C, and D); approximately 4.5% of the MDC were GFP+ and thus productively infected. Real-time DNA-PCR with primers specific for HIV pol was used to demonstrate the presence of proviral DNA. A strong signal was detected in GFP+ cells (Fig. 4E), indicating that HIV-GFP entered MDC, proceeded through the early steps of the viral life cycle, and completed reverse transcription.
FIG. 4.
Purification of long-term HIV-infected MDC. (A) Negative control for sorting of HIV-GFP-exposed MDC; unexposed MDC cultured for 45 days were subjected to flow cytometry. (B) On day 45 after HIV exposure, GFP+ and GFP− MDC (total of 5 × 106 cells) were separated using the MoFlow cytosorter (Cytomation); M1, marker 1 (GFP). (C) Forward scatter (FSC) versus side scatter (SSC) and (D) the histogram plot, respectively, represent patterns of HIV-GFP-infected MDC after sorting. (E) Real-time DNA PCR for HIV pol of GFP+ and GFP− MDC by TaqMan methodology. The oligonucleotides 5′-CAAGGGAAGGCCAGGGAAT-3′, 5′-CCCCAAACCTGAAGCTCTCTT-3′, and 5′-CTTCAGAGCAGACCAGAGCCAACAGCC-3′ (Integrated DNA Technologies Inc., Coralville, Iowa) represent the sequences of forward and reverse primers for PCR and the sequence of the probe, respectively. The latter was labeled on the 5′ end with the reporter dye FAM (6-carboxy-fluorescein) and at the 3′ end with the quencher dye TAMRA (6-carboxytetramethyl-rhodamine) to identify PCR products, respectively (curves a and b represent analysis of GFP+ and GFP− fractions of HIV-exposed MDC, respectively; curve c represents values of nonexposed control MDC).
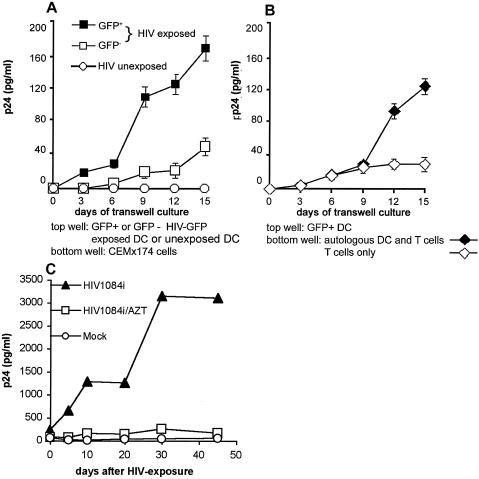
Next, the sorted GFP+ cells were plated in the upper wells of transwell tissue culture units (103 cells/well), while the lower wells contained CEMx174 cells (5 × 104 cells). As negative controls, nonexposed MDC were placed into the upper wells in parallel plates. GFP+ MDC released HIV that crossed the semipermeable membrane of the transwell unit and infected CEMx174 cells; release of p24 Gag into the culture supernatant increased as a function of time (Fig. 5A). In parallel, we also examined the MDC fraction that was GFP negative 45 days after initial exposure to HIV-GFP. Only a low level of viral replication was detected in the transwell system involving CEMx174 cells in the bottom wells (Fig. 5A), possibly due to the presence of a few MDC, in which infectious HIV-GFP had initiated the replication cycle without yet expressing GFP at sufficient levels at the time of sorting.
FIG. 5.
Transfer of cell-free HIV produced by long-term infected MDC. (A) p24 Gag was analyzed by enzyme-linked immunosorbent assay (Beckman-Coulter) in supernatants of CEMx174 cells placed in the bottom wells of a transwell unit. In the upper wells, HIV-infected GFP+, HIV-exposed GFP−, or noninfected MDC were cultured. (B) p24 Gag analysis of samples collected from the bottom wells of transwell units containing either primary resting T cells or T cells mixed with autologous, freshly differentiated MDC that had not been exposed to HIV-GFP. In the upper inserts, GFP+ HIV-exposed cells were cultured. Data are presented as averages of p24 values, which were calculated from triplicates; error bars represent standard deviations. Results of one out of two representative experiments are shown. (C) Long-term cultured MDC support replication of R5 HIV. CD1a-sorted MDC (105 cells per well) were plated in 24-well tissue culture plates (Costar-Falcon). Next, MDC were exposed to 0.1 50% tissue culture infectious doses per cell of the R5 HIV1084i. After washing, MDC were cultured for 45 days in 1.5 ml of DC medium. In parallel, mock-infected MDC and HIV-exposed MDC were cultured in the presence of AZT (10 μM). Supernatants (200 μl) were collected and assayed for p24 Gag by enzyme-linked immunosorbent assay (Beckman Coulter).
We then investigated whether sorted GFP+ MDC could transmit HIV to primary autologous T cells. GFP+ MDC were placed into the upper wells of transwell units, while the lower wells contained either autologous T cells alone or T cells cocultured with autologous, freshly differentiated, noninfected MDC at a ratio of 10:1. In the latter culture, p24 Gag increased as a function of time (Fig. 5B); in the absence of MDC, virus replicated at a substantially lower level (120 ng/ml versus 27 ng/ml of p24 on day 15 of incubation). This indicated that GFP+ MDC in the upper wells shed virus that was propagated efficiently by the T cells activated by autologous DC but not by T cells cultured alone. Together, our data clearly demonstrate that 45 days after virus exposure, MDC harbored infectious virus that could be transmitted in the absence of cell-cell contact.
To investigate whether MDC can support long-term productive infection of a primary HIV isolate, we exposed CD1a-sorted MDC to HIV1084i, an R5 primary HIV clade C strain from Africa (14). This virus, which had been isolated from a 4-month-old infant infected either intrapartum or by breast feeding, was molecularly cloned and tested for its tropism (14). We maintained HIV-exposed and nonexposed MDC cultures for 45 days in the presence or absence of azidothymidine (AZT) (Fig. 5C). Clearly, MDC supported the replication of this strain, indicating permissiveness to both X4R5 (Fig. 3C) and R5 viruses.
In summary, CD1a+ sorted MDC supported HIV replication for 45 days. Progeny virus released by GFP+ MDC on day 45 was fully infectious and readily replicated in the CEMx174 cell line and in primary T cells cocultured with uninfected autologous MDC. T cells upon contact with noninfected MDC undergo activation, which leads to the upregulation of HLA-DR, CD25, and CD69 (31). According to earlier data, activation of T cells is one of prerequisites for productive HIV replication (8, 22, 28). We found that even a low amount of HIV released by the long-term infected MDC was sufficient to rapidly spread infection in the MDC-T-cell microenvironment.
We demonstrated here for the first time that CD1a-sorted, DC-SIGN+, HIV-exposed MDC survive and release infectious virus during a protracted period of time. It would be interesting to compare the life span of our long-term in vitro HIV-infected DC with the life span of human DC in vivo. Few data are available about the life span of DC. Earlier studies reported that Langerhans cells represent a long-lived cell population (15, 16, 18). For instance, Langerhans cells were identified throughout 9 weeks in human skin grafts, which were transplanted to BALB/c nude mice (18). MDC are motile cells circulating in peripheral blood, residing in mucosa, and easily migrating across vascular endothelium (5, 11). Our data imply that MDC could form a long-lived, motile HIV reservoir with an important role in disseminating infectious virus through peripheral blood and in lymphoid and nonlymphoid tissues. Both productive infection and DC-SIGN-mediated virus capture could contribute to HIV transmission in vivo.
Acknowledgments
We thank H. Göttlinger, Worcester, Mass., for providing the dual-tropic HIV-GFP strain, P. Autissier for sorting of HIV-infected DC, S. Sharp for help in preparing the manuscript, and D. S. Popov for assistance with the figures.
This work was supported by NIH grants RO1 AI43839, RO1 DE12937, and RO1 DE016013 to R.M.R. and by Center for AIDS Research core grant IP3028691 awarded to the Dana-Farber Cancer Institute as support for AIDS research efforts by the institute.
REFERENCES
- 1.Ancuta, P., Y. Bakri, N. Chomont, H. Hocini, D. Gabuzda, and N. Haeffner-Cavaillon. 2001. Opposite effects of IL-10 on the ability of dendritic cells and macrophages to replicate primary CXCR4-dependent HIV-1 strains. J. Immunol. 166:4244-4253. [DOI] [PubMed] [Google Scholar]
- 2.Ayehunie, S., E. A. Garcia-Zepeda, J. A. Hoxie, R. Horuk, T. S. Kupper, A. D. Luster, and R. M. Ruprecht. 1997. Human immunodeficiency virus-1 entry into purified blood dendritic cells through CC and CXC chemokine coreceptors. Blood 90:1379-1386. [PubMed] [Google Scholar]
- 3.Bakri, Y., C. Schiffer, V. Zennou, P. Charneau, E. Kahn, A. Benjouad, J. C. Gluckman, and B. Canque. 2001. The maturation of dendritic cells results in postintegration inhibition of HIV-1 replication. J. Immunol. 166:3780-3788. [DOI] [PubMed] [Google Scholar]
- 4.Banchereau, J., F. Briere, C. Caux, J. Davoust, S. Lebecque, Y. J. Liu, B. Pulendran, and K. Palucka. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767-811. [DOI] [PubMed] [Google Scholar]
- 5.Banchereau, J., S. Paczesny, P. Blanco, L. Bennett, V. Pascual, J. Fay, and A. K. Palucka. 2003. Dendritic cells: controllers of the immune system and a new promise for immunotherapy. Ann. N. Y. Acad. Sci. 987:180-187. [DOI] [PubMed] [Google Scholar]
- 6.Blauvelt, A., H. Asada, M. W. Saville, V. Klaus-Kovtun, D. J. Altman, R. Yarchoan, and S. I. Katz. 1997. Productive infection of dendritic cells by HIV-1 and their ability to capture virus are mediated through separate pathways. J. Clin. Investig. 100:2043-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canque, B., Y. Bakri, S. Camus, M. Yagello, A. Benjouad, and J. C. Gluckman. 1999. The susceptibility to X4 and R5 human immunodeficiency virus-1 strains of dendritic cells derived in vitro from CD34(+) hematopoietic progenitor cells is primarily determined by their maturation stage. Blood 93:3866-3875. [PubMed] [Google Scholar]
- 8.Cicala, C., J. Arthos, S. M. Selig, G. Dennis, Jr., D. A. Hosack, D. Van Ryk, M. L. Spangler, T. D. Steenbeke, P. Khazanie, N. Gupta, J. Yang, M. Daucher, R. A. Lempicki, and A. S. Fauci. 2002. HIV envelope induces a cascade of cell signals in non-proliferating target cells that favor virus replication. Proc. Natl. Acad. Sci. USA 99:9380-9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collman, R., J. W. Balliet, S. A. Gregory, H. Friedman, D. L. Kolson, N. Nathanson, and A. Srinivasan. 1992. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J. Virol. 66:7517-7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geijtenbeek, T. B., D. J. Krooshoop, D. A. Bleijs, S. J. van Vliet, G. C. van Duijnhoven, V. Grabovsky, R. Alon, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN-ICAM-2 interaction mediates dendritic cell trafficking. Nat. Immunol. 1:353-357. [DOI] [PubMed] [Google Scholar]
- 12.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 13.Granelli-Piperno, A., B. Moser, M. Pope, D. Chen, Y. Wei, F. Isdell, U. O'Doherty, W. Paxton, R. Koup, S. Mojsov, N. Bhardwaj, I. Clark-Lewis, M. Baggiolini, and R. M. Steinman. 1996. Efficient interaction of HIV-1 with purified dendritic cells via multiple chemokine coreceptors. J. Exp. Med. 184:2433-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grisson, R. D., A.-L. Chenine, L.-Y. Yeh, J. He, C. Wood, G. J. Bhat, W. Xu, C. Kankasa, and R. M. Ruprecht. 2004. Infectious molecular clone of a recently transmitted pediatric human immunodeficiency virus clade C isolate from Africa: evidence of intraclade recombination. J. Virol. 78:14066-14069. [DOI] [PMC free article] [PubMed]
- 15.Holt, P. G., S. Haining, D. J. Nelson, and J. D. Sedgwick. 1994. Origin and steady-state turnover of class II MHC-bearing dendritic cells in the epithelium of the conducting airways. J. Immunol. 153:256-261. [PubMed] [Google Scholar]
- 16.Katz, S. I., K. Tamaki, and D. H. Sachs. 1979. Epidermal Langerhans cells are derived from cells originating in bone marrow. Nature 282:324-326. [DOI] [PubMed] [Google Scholar]
- 17.Kawamura, T., M. Qualbani, E. K. Thomas, J. M. Orenstein, and A. Blauvelt. 2001. Low levels of productive HIV infection in Langerhans cell-like dendritic cells differentiated in the presence of TGF-beta1 and increased viral replication with CD40 ligand-induced maturation. Eur. J. Immunol. 31:360-368. [DOI] [PubMed] [Google Scholar]
- 18.Krueger, G. G., R. A. Daynes, and M. Emam. 1983. Biology of Langerhans cells: selective migration of Langerhans cells into allogeneic and xenogeneic grafts on nude mice. Proc. Natl. Acad. Sci. USA 80:1650-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald, D., L. Wu, S. M. Bohks, V. N. KewalRamani, D. Unutmaz, and T. J. Hope. 2003. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 300:1295-1297. [DOI] [PubMed] [Google Scholar]
- 20.Mellman, I., and R. M. Steinman. 2001. Dendritic cells: specialized and regulated antigen processing machines. Cell 106:255-258. [DOI] [PubMed] [Google Scholar]
- 21.Messmer, D., J. Bromberg, G. Devgan, J. M. Jacque, A. Granelli-Piperno, and M. Pope. 2002. Human immunodeficiency virus type 1 Nef mediates activation of STAT3 in immature dendritic cells. AIDS Res. Hum. Retrovir. 18:1043-1050. [DOI] [PubMed] [Google Scholar]
- 22.Ostrowski, M. A., D. C. Krakauer, Y. Li, S. J. Justement, G. Learn, L. A. Ehler, S. K. Stanley, M. Nowak, and A. S. Fauci. 1998. Effect of immune activation on the dynamics of human immunodeficiency virus replication and on the distribution of viral quasispecies. J. Virol. 72:7772-7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pope, M., S. Gezelter, N. Gallo, L. Hoffman, and R. M. Steinman. 1995. Low levels of HIV-1 infection in cutaneous dendritic cells promote extensive viral replication upon binding to memory CD4+ T cells. J. Exp. Med. 182:2045-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rescigno, M., and P. Borrow. 2001. The host-pathogen interaction: new themes from dendritic cell biology. Cell 106:267-270. [DOI] [PubMed] [Google Scholar]
- 25.Romani, N., S. Gruner, D. Brang, E. Kampgen, A. Lenz, B. Trockenbacher, G. Konwalinka, P. O. Fritsch, R. M. Steinman, and G. Schuler. 1994. Proliferating dendritic cell progenitors in human blood. J. Exp. Med. 180:83-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sallusto, F., and A. Lanzavecchia. 1994. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 179:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scarlatti, G., E. Tresoldi, A. Bjorndal, R. Fredriksson, C. Colognesi, H. K. Deng, M. S. Malnati, A. Plebani, A. G. Siccardi, D. R. Littman, E. M. Fenyo, and P. Lusso. 1997. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 3:1259-1265. [DOI] [PubMed] [Google Scholar]
- 28.Shapira-Nahor, O., S. Maayan, K. W. Peden, R. Rabinowitz, M. Schlesinger, A. Alian, and A. Panet. 2002. Replication of HIV-1 deleted Nef mutants in chronically immune activated human T cells. Virology 303:138-145. [DOI] [PubMed] [Google Scholar]
- 29.Sozzani, S., W. Luini, A. Borsatti, N. Polentarutti, D. Zhou, L. Piemonti, G. D'Amico, C. A. Power, T. N. Wells, M. Gobbi, P. Allavena, and A. Mantovani. 1997. Receptor expression and responsiveness of human dendritic cells to a defined set of CC and CXC chemokines. J. Immunol. 159:1993-2000. [PubMed] [Google Scholar]
- 30.Steinman, R. M., A. Granelli-Piperno, M. Pope, C. Trumpfheller, R. Ignatius, G. Arrode, P. Racz, and K. Tenner-Racz. 2003. The interaction of immunodeficiency viruses with dendritic cells. Curr. Top. Microbiol. Immunol. 276:1-30. [DOI] [PubMed] [Google Scholar]
- 31.Vanham, G., L. Penne, H. Allemeersch, L. Kestens, B. Willems, G. van der Groen, K. T. Jeang, Z. Toossi, and E. Rich. 2000. Modeling HIV transfer between dendritic cells and T cells: importance of HIV phenotype, dendritic cell-T cell contact and T-cell activation. AIDS 14:2299-2311. [DOI] [PubMed] [Google Scholar]
- 32.van Rij, R. P., M. Worobey, J. A. Visser, and H. Schuitemaker. 2003. Evolution of R5 and X4 human immunodeficiency virus type 1 gag sequences in vivo: evidence for recombination. Virology 314:451-459. [DOI] [PubMed] [Google Scholar]
- 33.Weissman, D., Y. Li, J. Ananworanich, L. J. Zhou, J. Adelsberger, T. F. Tedder, M. Baseler, and A. S. Fauci. 1995. Three populations of cells with dendritic morphology exist in peripheral blood, only one of which is infectable with human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 92:826-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaitseva, M., A. Blauvelt, S. Lee, C. K. Lapham, V. Klaus-Kovtun, H. Mostowski, J. Manischewitz, and H. Golding. 1997. Expression and function of CCR5 and CXCR4 on human Langerhans cells and macrophages: implications for HIV primary infection. Nat. Med. 3:1369-1375. [DOI] [PubMed] [Google Scholar]